Abstract
Aim:
Many currently available pharmacogenomic assays and algorithms interrogate a set of ‘tag’ polymorphisms for inferring haplotypes. We wanted to test the accuracy of such haplotype inferences across different populations.
Materials & methods:
We simulated haplotype inferences made by existing pharmacogenomic assays for seven important pharmacogenes based on full genome data of 2504 persons in the 1000 Genomes dataset.
Results:
A sizable fraction of samples did not match any of the haplotypes in the star allele nomenclature systems. We found no clear population bias in the accuracy of results of simulated assays.
Conclusion:
Haplotype nomenclatures and inference algorithms need to be improved to adequately capture pharmacogenomic diversity in human populations.
Keywords: : errors, genetic diversity, genetic testing, medical nomenclature, pharmacogenomics
Pharmacogenomics can potentially improve the safety and effectiveness of medications in individual patients [1–3]. While data from functional and association studies can be compelling, large-scale trials on representative patient populations have not been published for all pharmacogenomic tests, and in some cases studies have found conflicting results. For example, the results of a randomized controlled trial on utilizing pharmacogenomic data for optimizing warfarin dosing recently published by Kimmel et al. [4] indicated that the incorporation of pharmacogenomic data worsened, rather than improved, treatment outcomes for the subgroup of patients self-identifying as ‘black’. These findings are contrasted by another recent study reporting positive effects of genotype-based warfarin dosing in a cohort of European patients [1]. The authors of this study reported that the vast majority of participants were of ‘white European ethnic origin’.
Many currently available genetic tests, as well as algorithms for inferring haplotypes and subsequent treatment recommendations, interrogate genetic variants that ‘tag’ haplotypes, in other words, the data they generate only offer a constrained view on a defined set of polymorphisms that are tested. Due to population-specific allele frequency, it is possible that some tests might perform very well for some populations but poorly in populations that have significant genetic variability outside these constrained views.
In this study we analyzed how the design of different pharmacogenomic assays could affect the haplotypes called by those assays, and whether some populations were more prone to be affected by potential errors in haplotype calls than other populations. To this end, we analyzed the 1000 Genomes Phase 3 dataset, which contains full genome data of 2504 persons from diverse populations [5]. We simulated the inferences that would be made on these data using different constrained views that were derived from existing pharmacogenomic assays, and compared the results of the haplotype calls among different constrained views and different populations. At the outset of our study, we hypothesized that assays that test a larger number of polymorphisms would yield more accurate results than assays that test a smaller number of polymorphisms. We also hypothesized that we may observe a difference in the accuracy of haplotype calls among populations due to differences in allele frequency among populations and representation of different populations in the definition of the haplotype tables.
Materials & methods
Gene selection & haplotype definitions
We included genes for which clinical pharmacogenomic guidelines backed by good evidence were available [3,6], and for which haplotype definitions had been published. The following genes were included in our final analysis: CYP2C19, CYP2C9, CYP3A5, DPYD, SLCO1B1, TPMT and VKORC1. We excluded CYP2D6 from our final analysis because of uncertainties about the effects of copy-number variations on the data reported in the 1000 Genomes datasets and concerns about the accuracy of haplotype assignment based on data from that gene.
Haplotype definitions were downloaded from the Pharmacogenomics Knowledge Base (PharmGKB) [7] in February 2014 and subjected to additional manual curation and automated mapping to achieve a consistent representation across all genes. The resulting haplotype definitions contained all 163 polymorphisms in the PharmGKB tables of the selected genes that could be mapped to dbSNP reference SNP identifiers and that were validated by the 1000 Genomes project, as well as an unfiltered list of 295 haplotypes of the selected genes described in the PharmGKB tables. We refer to these definitions as the FULL view, as they were not constrained by the limitations of any particular assay and acted as a gold standard for judging the performance of the other constrained views.
Assay selection & constrained haplotype definitions
We selected four pharmacogenomic assays which test for a broad panel of important pharmacogenes and which were previously used in pilot projects geared toward the implementation of clinical pharmacogenomic testing:
Affymetrix DMET™ Plus assay [8];
Illumina VeraCode® ADME Core Panel [9];
TaqMan® OpenArray® PGx Panel [10];
University of Florida and Stanford University Personalized Medicine Program Custom Array [11].
Since each platform interrogates a different subset of variant sites, we created several different constrained views for each gene, based on the polymorphisms tested by these assays and the haplotypes that are claimed to be covered by the assay in its documentation. The concept of constrained views and its implications are illustrated in Figure 1 and described in more detail below. We created four constrained views for our analysis: DMET (derived from the Affymetrix DMET™ Plus assay), VERA (Illumina VeraCode® ADME Core Panel), TAQM (TaqMan® OpenArray® PGx Panel) and FLOR (University of Florida and Stanford University Personalized Medicine Program Custom Array).
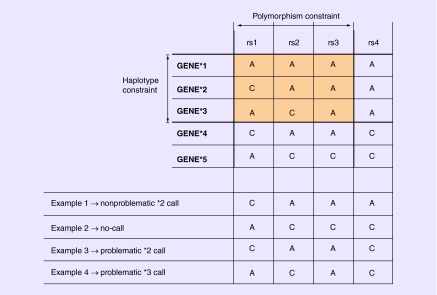
Figure 1. . Illustration of the concept of ‘constrained views’ based on restricted sets of polymorphisms and haplotypes (top); examples of haplotype calls produced by the constrained view (bottom).
The constrained view only takes the highlighted variants into account for haplotype calling, excluding other polymorphisms and haplotypes in the full haplotype definition table. In this example, the assay tests three (rs1, rs2 and rs3) of the four variant sites and reports three (GENE*1, GENE*2 and GENE*3) of the five defined haplotypes. Therefore, while the hypothetical dataset contains results for all four variant sites (examples 1–4 at bottom), only three of them are used in haplotype assignment. In example 1, the assigned haplotype (GENE*2) is correct. A haplotype cannot be assigned for example 2 because the genotype patterns at rs1, rs2 and rs3 do not match any of the reported haplotype alleles (*1–*3). The assigned haplotypes for examples 3 and 4 are incorrect due to the assay's constrained view regarding variant sites and/or defined haplotypes, and they are therefore categorized as ‘problematic calls’. Problematic calls are those in which a constrained view leads to calling a haplotype that is not called with the FULL view.
Genetic dataset
We retrieved 1000 Genomes data by downloading VCF files covering polymorphisms for the selected pharmacogenes from all patients in the dataset through the Genome Browser web interface [12].
Haplotype analysis
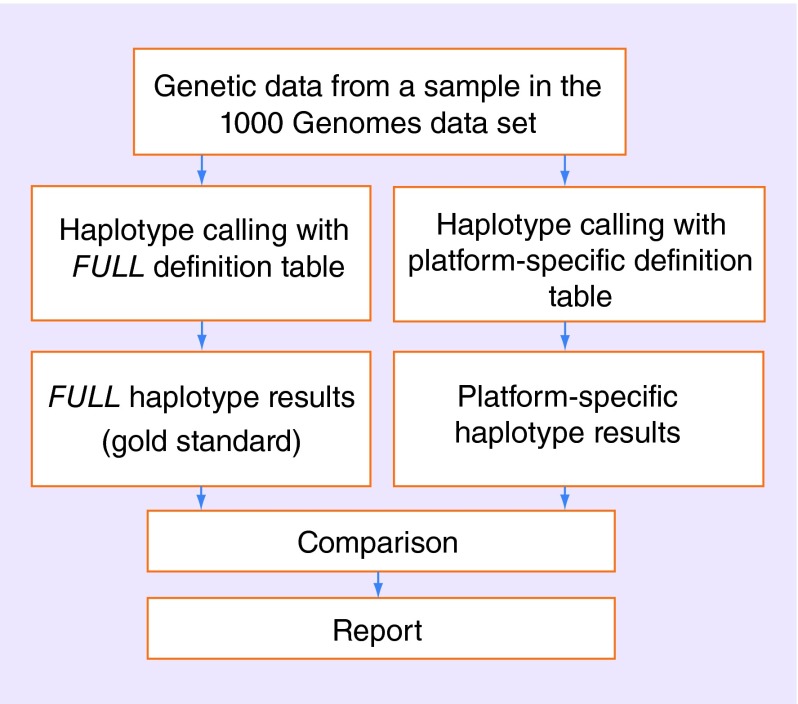
We automatically processed the 1000 Genomes dataset by inferring matching haplotypes based on the TAQM, VERA, DMET, FLOR and FULL views. A haplotype from one of these views was assigned if all the SNPs available in a view matched all the SNPs in the 1000 Genomes sample (as illustrated in Figure 1). If using the TAQM, VERA, DMET or FLOR view would lead to calling a haplotype that was not called when using the gold standard (i.e., the FULL view), this was an indicator that the haplotype might be inferred in error because additional constraints imposed by the assay led to loss of relevant genetic data. In this paper, we refer to such cases as ‘problematic’ haplotype calls. Figure 2 provides an outline of the processing steps for individual samples.
Figure 2. . Flow diagram outlining data processing of individual samples from the 1000 Genomes dataset.
Examples of problematic and nonproblematic haplotype calls based on a constrained view are provided at the bottom of Figure 1. Example 1 in Figure 1 leads to the inference of a nonproblematic *2 call, because both the constrained view and the full haplotype definition table lead to a *2 call. In example 2, the constrained view leads to a no-call (i.e., no haplotype in the assay's constrained view matches the available data), while the FULL view would lead to a *5 call. Example 3 leads to a problematic *2 call, because the FULL view would lead to a *4 call instead. Finally, example 4 leads to a problematic *3 call, because the FULL view would lead to a no-call instead. The possible outcomes of the analysis, comparing gold-standard results (FULL view) and the results of other constrained views of assays, are further outlined in Table 1. Based on the inferred haplotypes, we calculated statistics on nonproblematic calls, problematic calls and no-calls for all possible combinations of constrained views, genes and 1000 Genomes populations.
Table 1. . Overview of possible discrepancies between gold-standard results and the results of other constrained views of assays.
|
Gold-standard haplotype result (based on FULL view) |
Assay-specific haplotype result |
||
|---|---|---|---|
| Call | No-call | ||
| |
Call |
If haplotype called by assay is also called by gold standard: ‘nonproblematic call’. If haplotype called by assay is not also called by gold standard: ‘problematic call’ |
No-call due to lack of coverage of assay |
| No-call | ‘Problematic call’ because sample does not match any defined haplotype, but platform calls haplotype because it interrogates only some genetic loci | No-call due to lack of coverage of both assay and haplotype definition table | |
Code availability
The curated resources, the IPython notebook for conducting the processing described above and detailed results were made publicly available on the web at [13].
Results
Our analysis included data from all 2504 samples in the 1000 Genomes final release, so 2504 * 2 = 5008 gene copies were included. The data included samples from persons of African ancestry (AFR, 1322 gene copies), American ancestry (694 gene copies), south Asian ancestry (978 gene copies), European ancestry (EUR, 1006 gene copies) and east Asian ancestry (ASN, 1008 gene copies).
A sizable fraction of polymorphisms used in the definition of rare haplotypes were not observed in the 1000 Genomes samples (Table 2). On the other hand, the total number of polymorphisms observed in the 1000 Genome samples far exceeded the number of polymorphisms in haplotype definitions.
Table 2. . Statistics on differences between numbers of polymorphisms in haplotype definitions versus number of polymorphisms in the 1000 Genomes Phase 3 dataset.
| Gene | SNPs in haplotype definition | SNPs from haplotype definition not found in 1000 Genomes dataset | Total SNPs in 1000 Genomes dataset (in exons/in entire gene) |
|---|---|---|---|
|
CYP2C9 |
24 |
9 |
50/1561 |
|
CYP2C19 |
42 |
8 |
71/9188 |
|
CYP3A5 |
14 |
7 |
321/6515 |
|
DPYD |
14 |
7 |
83/21007 |
|
SLCO1B1 |
27 |
13 |
27/1661 |
|
TPMT |
32 |
17 |
30/422 |
| VKORC1 | 5 | 0 | 63/397 |
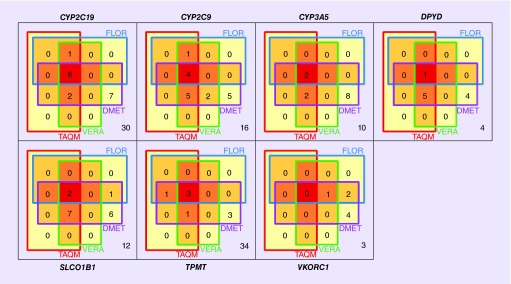
Basic statistics on the polymorphisms and haplotypes included in the constrained views derived from assays are listed in Table 3; overlaps in the polymorphisms covered by different constrained views are displayed in Figure 3. It is noteworthy that for each constrained view the number of polymorphisms considered is smaller than the number of haplotypes considered. There are two reasons for this: first, some of the haplotypes are defined not only through a single tagging polymorphism, but through distinct combinations of several polymorphisms that differ from the reference haplotype, allowing for a greater number of haplotypes being defined through a smaller number of polymorphisms. Second, some of the haplotypes formed sets that were mutually indistinguishable in some of the views because one or more variant sites that distinguish between the haplotypes were not interrogated by the assay. For example, a specific view might be able to infer that *3A or *3B are present, without being able to distinguish between these haplotypes.
Table 3. . Basic statistics on the ‘constrained views’ used in the analysis.
| Constrained view | Polymorphisms considered | Haplotypes considered |
|---|---|---|
|
FULL |
163 |
295 |
|
DMET |
85 |
123 |
|
VERA |
46 |
60 |
|
TAQM |
44 |
57 |
| FLOR | 25 | 34 |
Only polymorphisms and haplotypes for the genes selected for this study were included in the analysis. Detailed data per gene are contained in the associated web repository.
Figure 3. . Venn diagram displaying the numbers and overlaps of polymorphisms covered by constrained views derived from four pharmacogenomic assays.
Numbers of polymorphisms not covered by any assay-derived constrained view but covered by haplotype definition tables are shown on the lower right of each panel. See text for details.
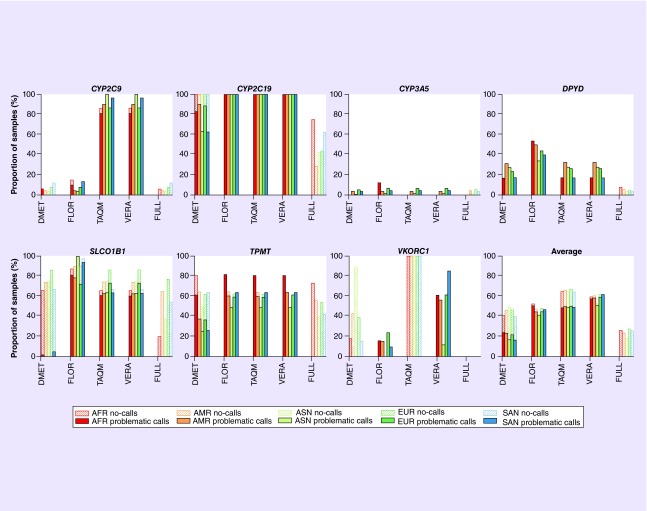
Figure 4 shows the fractions of samples with problematic calls and no-calls for all possible combinations of constrained views, genes and populations, as well as averaged data across all genes (detailed numerical results are available in Supplementary Data). Our analysis led to several unanticipated results.
Figure 4. . Fractions of gene copies in the 1000 Genomes sample with problematic calls and no-calls for all possible combinations of constrained views, genes and populations.
VKORC1 is not covered by the TAQM view, so all samples resulted in no-calls. nAFR = 1322, nAMR = 694, nSAN = 978, nEUR = 1006, nASN = 1008.
AFR: African ancestry; AMR: American ancestry; ASN: East Asian ancestry; EUR: European ancestry; SAN: South Asian ancestry.
We observed that the FULL view, which is based on the maximum of information in the haplotype definition tables, resulted in a large fraction of no-calls for some of the genes, in other words, none of the known haplotypes in the allele definition tables matched the patient data. This observation was especially striking for CYP2C19, SLCO1B1 and TPMT. For CYP2C19 and TPMT, the fraction of no-calls was highest for the AFR populations (74.6 and 72.6%, respectively). Surprisingly, however, the AFR population had the lowest fraction of no-calls with the FULL view for SLCO1B1 (19.9%), while the EUR population had the largest fraction of no-calls for this gene (76.7%), suggesting that the haplotype tables for these genes do not contain alleles that are common in those respective populations. We observed that the reason for many no-calls was the combination of nonreference sequence nucleotides at three or more variant sites that did not match any haplotype in the definition table. Examples are the combination of rs17885098:T and rs3758581:G with one or more additional variant(s) in CYP2C19, and the combination of rs12529220:A and rs2518463:G with one or more additional variant(s) in TPMT.
For those genes that resulted in large fractions of no-calls for the FULL view, we observed a large fraction of problematic calls in the constrained views derived from pharmacogenomic assays, in other words, these views did not utilize some of the genetic data and inferred haplotypes that are likely to be incorrect (i.e., problematic calls). The fractions of the samples with problematic calls were >49% for CYP2C19, TPMT and SLCO1B1 across all populations with the FLOR, TAQM and VERA views, reaching up to 100% for CYP2C19. In general, the DMET view interrogated more sites than the other assays, which tended to decrease the fractions of problematic calls compared with the other assay-derived views and increased the fractions of no-calls.
For the DPYD gene, the constrained views derived from assays made erroneous *1 calls with high frequency, while the FULL view resulted in other haplotype calls, but few no-calls.
The views derived from pharmacogenomic assays differed significantly in the number of polymorphisms they considered, ranging from 25 polymorphisms (FLOR) to 85 polymorphisms (DMET). Overall, assays with more highly constrained views (e.g., smaller numbers of polymorphisms) generated fewer nonproblematic calls than assays that interrogated more polymorphic sites. This difference in result quality was especially marked for DPYD and SLCO1B1, with the lowest number of nonproblematic calls observed for SLCO1B1 and the ASN population in the assay with the fewest interrogated sites (0% of samples resulted in nonproblematic calls with the FLOR view).
The fractions of problematic calls and no-calls for different populations differed considerably for each gene, but there were no clear patterns in direction or magnitude for a given population. For example, in terms of nonproblematic calls of assay-derived constrained views, while the AFR population had the worst results among all populations for TPMT, the EUR population had the worst results for CYP3A5 and SLCO1B1, and the AMR population had the worst results for DPYD. Averaged overall genes considered in our analysis, the EUR population had the lowest proportion of correct haplotype calls (72.7% of samples resulted in nonproblematic calls), while the ASN population had the highest proportion of correct calls (82.2% of samples resulting in nonproblematic calls).
We conducted an informal comparison of haplotype frequencies reported with our methodology for the 1000 Genomes samples with several published haplotype frequency datasets that are not based on the 1000 Genomes data (e.g., the Clinical Pharmacogenetics Implementation Consortium [6] provides averaged haplotype frequency data derived from multiple different studies for all of the genes included in our analysis). Since most of these studies are quite old, a large number of recently discovered haplotypes included in our script are not yet represented in the published frequency tables. In cases where comparisons were possible, we did not find any unexpected discrepancies between the allele frequencies we observed in the 1000 Genomes data and those that were published previously. Tables summarizing these comparisons are included in the Supplementary Data.
The Supplementary Data contains statistics on inferred calls by gene and population. Further result statistics as well as detailed information on inferences made for each individual sample are publicly available on the web at [13].
Discussion
We anticipated that different platforms interrogated different subsets of known variant sites. The purpose of this work was to determine the effect of site selection in clinical pharmacogenomics platforms on the resulting (simulated) haplotype calls. The main objective of our study was not to prove the existence of this expected effect, but to analyze the magnitude of the effect.
We found that a sizable fraction of the genomes in the 1000 Genomes dataset could not be assigned a haplotype using the existing haplotype tables. This was not unexpected, as whole genome sequence data will identify more variants than those that are present on array-based genotyping platforms, and the additional data will complicate haplotype assignment. However, the magnitude of this problem was not expected at the outset of this study. These results resonate with recent, preliminary results of another study based on next-generation sequencing that indicates the presence of significant numbers of previously unknown polymorphisms in pharmacogenes [14].
In genes where a large fraction of samples did not fit any of the defined haplotypes, using a larger number of polymorphisms for haplotype calling resulted in a smaller fraction of erroneous haplotype assignments, in other words, interrogating more sites increased the number of no-calls and decreased the number of problematic calls. This confirmed our hypothesis that assays interrogating a larger number of polymorphisms might result in fewer errant haplotype calls. It follows that haplotype assignments based on complete genotype data and haplotype definitions will be more accurate than those that rely on tag SNPs and an assumption of high linkage disequilibrium between interrogated and noninterrogated (or imputed) sites. In this regard, the results of our study add to current knowledge by quantifying the magnitude of this effect and comparing it between different simulated assays.
We hypothesized that we may observe a difference in the accuracy of haplotype calls among populations due to differences in allele frequencies among populations and the relative representation of different populations in the definition of the haplotype tables. While the accuracy of haplotype assignment varied widely within and between populations among the seven genes in this study, there were no obvious patterns to these deviations.
Our findings demonstrate that different platforms can produce the same haplotype calls while interrogating different sites. Furthermore, haplotype naming schemes, such as the star allele system widely used in pharmacogenetics, do not specify which site(s) were interrogated (which vary by platform), do not include all observed haplotypes (which vary by population), assert genotype results for sites that were specified in the definition table but were not interrogated, and are ‘lossy’ because genotype information is lost for sites that were interrogated but were not specified in the definition table. Therefore, we suggest that genetic test results should not be reported as named haplotypes (star alleles) without a clear accompanying statement that includes a list of interrogated sites, the genotype at each interrogated site and an explanation of the haplotype calling algorithm.
The potential impact of the shortcomings of reporting genetic variation as named haplotypes, which include star allele nomenclatures, requires further scrutiny both in academic research and in clinical scenarios. In particular, there is widespread ambiguity as to whether a named haplotype is a statement about the presence of one or more defining SNP(s) with functional consequences independent of variability at other sites, or a whether it is a statement about the entire genetic sequence of the gene. The former interpretation seems to be used – at least implicitly – by manufacturers of pharmacogenomic assays that advertise assays as testing for a specific set of star alleles, while in fact only a one or two SNPs per gene are being tested (e.g., [15]). It is also noteworthy that the current inclusion criteria of the Human Cytochrome P450 (CYP) Allele Nomenclature database states that ‘So-called sub-alleles containing additional nonfunctional variations in addition to the functional ones described (e.g., CYP2D6*10B) will no longer be designated’ [16]. On the other hand, comprehensive haplotype translation tables (e.g., as found in PharmGKB) seem to rest on the assumption that nondefining SNPs in haplotypes default to a reference sequence. This ambiguity can impact the interpretation of genetic test results and complicate retrospective analyses.
Limitations
This study has several limitations. Most importantly, the results generated by constrained views derived from genetic assays are rough approximations of the actual results that these assays would report based on the sites they interrogate and the named haplotypes they return, since we do not have access to the precise haplotype calling algorithms used by each platform. The aim of this study is to provide insight into how different constrained views on genetic data can affect haplotype calls across populations, not to evaluate the performance of the actual genetic assays. Therefore, we urge readers not to draw conclusions about the accuracy of actual assays based on the results of these simulations without considering these limitations.
This study focused solely on polymorphisms that could be mapped to dbSNP identifiers and which were confirmed by the 1000 Genomes project. The full 1000 Genomes data for the pharmacogenes we analyzed contained hundreds of polymorphisms not covered by existing haplotype definitions (haplotype constraint), and which are not interrogated by most genotyping-based assays (polymorphism constraint). For some genes with high rates of no-calls in constrained views derived from assays, the results might actually point to shortcomings in haplotype definitions, rather than errors caused by constrained views. The current system of star nomenclatures, which focuses on defining haplotypes through tag SNPs rather than full sequences, might increase the occurrence of such errors. A modernized system for identifying, defining and sharing haplotype definitions that makes better use of next generation sequencing and information technologies is needed to reduce the likelihood of errors and ambiguities due to incomplete allele definition tables.
Finally, an analysis of the phenotypic effects of haplotype calls was not in the scope of this research. It is likely that a portion of problematic haplotype calls identified in this study might be assigned functional phenotypes that are similar to the phenotypes of the correct haplotypes, but the rate and clinical consequence of this occurrence remains to be determined.
Conclusion
Current haplotype definitions, including star allele nomenclature systems, are incomplete relative to the actual genetic data in the 1000 Genomes dataset for a large fraction of samples. In addition, constraints on the number of polymorphisms available for haplotype calling by pharmacogenomic assays led to erroneous assignment of haplotypes across all populations studied. Taken together, these constraints can significantly impact the accuracy of haplotype calls in clinical sequencing data.
The potential significance of these findings for drug-response phenotypes and clinical outcomes needs to be evaluated in follow-up studies.
Executive summary.
A sizable fraction of samples in the 1000 Genomes database did not match any of the haplotypes defined in the star allele nomenclature systems of important pharmacogenes.
Simulated assays interrogating smaller numbers of polymorphisms produced larger numbers of potentially incorrect results when calling pharmacogenomic haplotypes.
While the number of potentially incorrect results for different populations varied drastically for each gene, we found no clear population bias across all investigated genes.
Our findings indicate that haplotype definitions, nomenclatures and inference algorithms need to be improved to adequately capture pharmacogenomic diversity in human populations.
The significance of these findings for clinical research and practice needs to be evaluated in follow-up studies.
Supplementary Material
Footnotes
Author note
The study was based on publicly available and de-identified human data (1000 Genomes dataset).
Financial & competing interests disclosure
This work was supported in part by the Austrian Science Fund (FWF; P 25608-N15) and the NIH/NIGMS (U19 GM61388; the Pharmacogenomics Research Network; RR Freimuth). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Open Access
This work is licensed under the Creative Commons Attribution-NonCommercial 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
References
- 1.Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N. Engl. J. Med. 2013;369(24):2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 2.Mallal S, Phillips E, Carosi G, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N. Engl. J. Med. 2008;358(6):568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 3.Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte – an update of guidelines. Clin. Pharmacol. Ther. 2011;89(5):662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 4.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med. 2013;369(24):2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium T1000 GP. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr. Drug Metab. 2014;15(2):209–217. doi: 10.2174/1389200215666140130124910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonagh EM, Whirl-Carrillo M, Garten Y, Altman RB, Klein TE. From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomark. Med. 2011;5(6):795–806. doi: 10.2217/bmm.11.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.http://media.affymetrix.com/support/technical/brochures/dmet_drug_metabolism_brochure.pdf DMET™ Plus Solution (product sheet)
- 9.http://res.illumina.com/documents/products/datasheets/datasheet_veracode_adme_core_panel.pdf VeraCode ADME Core Panel (product sheet)
- 10.http://tools.lifetechnologies.com/content/sfs/brochures TaqMan OpenArray Pharmacogenomics (PGx) Panel (product sheet)
- 11.Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin. Pharmacol. Ther. 2012;92(4):437–439. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.www.1000genomes.org/ 1000 Genomes – a deep catalog of human genetic variation.
- 13.https://gitlab.com/medication-safety/ms-ipython/tree/master/pharmacogenomic_test_marker_coverage Medication-safety/ms-ipython @ GitLab.
- 14.Freimuth RR, Gordon AS, Zhu Q, Nickerson DA, Chute CG. Pharmacogenomics Research Network (PGRN) Scientific Meeting. FL, USA: 2–3 April 2014. Evaluating the application of star allele nomenclature for pharmacogenomics in the era of high-throughput sequencing. Presented at. [Google Scholar]
- 15.http://pharmgenomics.com/index.php/products?id=83 GenoChip Toxo.
- 16.www.cypalleles.ki.se/criteria.htm Human Cytochrome P450 (CYP) Allele Nomenclature inclusion criteria.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.