Abstract
Small molecule therapeutic drugs must reach their intended cellular targets (pharmacokinetics) and engage them to modulate therapeutic effects (pharmacodynamics). These processes are often difficult to measure in vivo due to their complexities and occurrence within single cells. It has been particularly difficult to directly measure cellular drug target binding.
Fluorescence polarization is commonly used in pharmacological screening assays to measure drug-protein or protein-protein interactions. We hypothesized that fluorescence polarization imaging could be adapted and used with fluorescently labeled drugs to measure drug target engagement in vivo.
Here we summarize recent results using two photon fluorescence anisotropy microscopy. Our imaging technique offers quantitative pharmacological binding information of diverse molecular interactions at the microscopic level, differentiating between bound and unbound states. Used in combination with other recent advances in the development of novel fluorescently labeled drugs, we expect that the described imaging modality will provide a window into the distribution and efficacy of drugs in real time and in vivo at the cellular and subcellular level.
Index Terms: Intravital microscopy, fluorescence polarization, fluorescence anisotropy, in vivo imaging, pharmacology, drug imaging
I. Introduction
In the typical drug development process, diverse compound libraries are commonly screened against purified targets and “hits” are then assayed and validated in cells. Companies and research institutions have mastered this approach to produce numerous promising drug candidates. But while it is generally assumed that such hits from the screen will behave similarly in vivo, the opposite is often the case.
To explore in vivo drug phenomena, accurate target identification and validation measurements in conjunction with optimized lead selection are of paramount importance in the drug development process [1]. In particular it is difficult to attribute pharmacological effects due to the perturbation of the protein of interest versus other mechanisms. Only by verifying that chemical probes directly engage in vivo with their intented target can provide sufficient pharmacological validation [2]. This determination requires the use and development of new technologies capable of measuring target engagement in cells and model organisms.
Historically, methods to directly measure molecule engagement with their cognate targets in intact cells and at the cellular level both in vitro and in a biological setting [2] have been elusive. This absence of quantitative knowledge on drug action at the molecular level, prevents therefore direct correlation of molecular pharmacology with efficacy.
Drug efficacy i.e. the ability of a drug to interact with a receptor, has been so far measured indirectly by monitoring cellular response following activation of the drug downstream signal, but recently new approaches have been developed capable of directly measuring drug binding in cells and in vivo. Drug affinity responsive target stability (DARTS)[3], positron emission tomography [4, 5], and mass spectroscopy [6] help determining the degree of drug interaction with a protein, the degree of drug accumulation in a tumor through drug radiolabeling, or to determine drug target engagement in situ through desorption of the sample and analysis. Also cellular thermal shift assay (CETSA) [7], has extended target engagement measurements to cells and soluble protein fractions extracted from cell lysates.
Unfortunately, these techniques lack the cellular spatial resolution as well as the potential to make temporal measurements in vivo. Mass spectrometry and PET provide spatial imaging but destroy the sample limiting temporal measurements or lacking cellular resolution, respectively. The other methods have high specificity with the capability of measuring off target binding, but unfortunately rely on sample disruption and quantification through western blot analysis.
Optical cellular imaging is among other recently introduced detection technologies used to determine cellular pharmacology by exploiting the capability to make measurements in complex biological environments [8–10].. Also, the development of novel labeling techniques and reporters has enabled both quantitative analysis and phenotype visualization of single cellular events.
Among the different fluorescent imaging modalities, intravital fluorescence microscopy has greatly contributed to our understanding of biological processes at the single cell level and in vivo due to its extended imaging depth, spatial and temporal resolution, and multi-reporter visualization capability. In particular it has become an indispensable tool for studying the cellular micro-dynamics in animal cancer models, enabling studies on tumor angiogenesis, cellular proliferation and tumor survival. When combined with the use fluorescently labeled drugs, prodrugs and activity based probes which can act as drug imaging companion [11–13], intravital confocal microscopy provides a window into the distribution and efficacy of agents in real time and in vivo. This is in contrast to other well established imaging technologies which highlight the macroscopic distribution of agents, but lack both the spatial and temporal resolution offered by fluorescence imaging techniques.
While the number of naturally fluorescent drugs is quite restricted, recent advances in chemical techniques have been successfully implemented to create fluorescently labeled drugs retaining targeting affinity therefore establishing a basis for the biomedical and clinical translation [14–15]. Specifically, the use of fluorescent drugs in some of our recent works has provided direct evidence of the presence of drug heterogeneity in vivo at the tissue [14], cellular [16], and subcellular levels [17].
However, intravital fluorescent drug imaging methods are based on simple fluorescence signal quantification of labeled drugs [14] (Fig. 1), and they do not differentiate between the specific, non-specific and unbound states of the labeled tracer, information which is critical to better understand the ligand-receptor interactions [18].
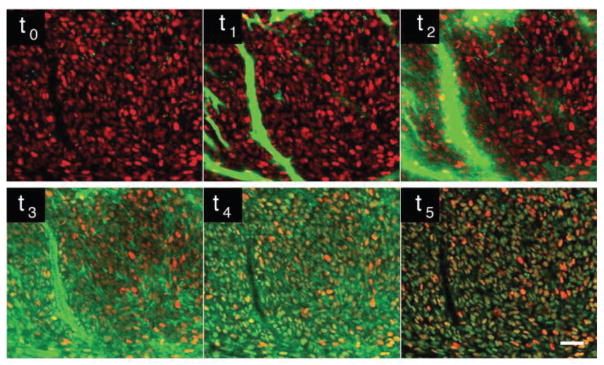
Fig. 1.
Time lapse imaging of in vivo drug distribution of a PARP inhibitor in a H2B-RFP implanted tumor, over the course of three hours. Within a few minutes after injection the drug (green) accumulates within the H2B-RFP cells (RFP, red). After a few hours the non-specifically bound drug clears out from the cells and only drug within the nuclei of the H2B-RFP cells is maintained, suggesting the drug has bound to its target. t0 before drug injection, t1 20 seconds after injection, t2–t5 at respectively 2, 14, 40 and 150 minutes. Adapted and reprinted with permission from [14].
The concept of fluorescence polarization/fluorescence anisotropy (FP/FA), which is based on the determination of the emission polarization orientation with respect to that of the excitation light, is commonly used in screening assays for drug development and pharmacological studies to characterize protein-protein interactions.
Measuring the degree of anisotropy of a fluorophore can determine, among others parameters, the rotational diffusion rate of molecules, which, in a controlled environment, is dependent on the molecular size. Therefore, the degree of interaction of a fluorescently labeled drug with a larger compound (such as a protein target) can be quantified. While most FP/FA applications are based on simple in vitro assays with fixed end points [19], the possibility to develop microscopy imaging platform based on fluorescence polarization detection could enable non-destructive mapping of fluorescence polarization phenomena in living cells with great spatiotemporal resolution for longitudinal measurements.
Based on these premises we have recently developed two photon fluorescence anisotropy microscopy [20], to measure drug distribution in vivo. This new imaging modality offers high spatial and temporal high resolution while quantitatively differentiating between bound and unbound states. The potential of the technique has been demonstrated on a phase III drug candidates both in vitro and for real-time quantification of drug target engagement in vivo in a mouse tumor, with submicron resolution and at great depth.
II. Basic Principles of Fluorescence Polarization
While we here briefly review the concept of fluorescence polarization/fluorescence anisotropy, for a more comprehensive treatment we refer the readers to more extensive reviews which are widely available in the literature [21–23].
The underlying principles of FP/FA is based on the fact that the probability of interaction of a fluorescent molecule with an exciting photon is proportional to the square of the scalar product of the molecule’s absorption transition dipole moment and the electric field vector.
If polarized light is incident on an ensemble of randomly oriented fluorescent molecules, a so-called photoselection process is induced. Emission will occur preferentially along the molecules’ emission transition dipole moment. The angular relation present between the absorption and emission determines a degree of anisotropy that can be calculated measuring the fluorescence emission in the planes parallel and perpendicular to the excitation polarization.
To characterize the extent of anisotropy a dimensionless physical parameter P, fluorescence polarization, is typically introduced and defined as the ratio of the polarized components to the total intensity, i.e. the fraction of the fluorescence light which is linearly polarized:
Here Ip and Is represent the parallel and perpendicular components of the fluorescence intensity, defined with respect to the excitation polarization. The denominator corresponds to the fluorescence intensity along the direction of observation.
If the emission is completely polarized along the parallel direction, P=1. If instead the emission is polarized along the perpendicular direction, P=−1. These values represent theoretical limits and occur only in the presence of oriented crystals or stretched polymers, when the fluorescence light is completely polarized along the perpendicular or parallel direction respectively. In the more general case of a solution, the transition dipoles are generally isotropically distributed, with each molecule oriented randomly in space. These values are therefore not reached, but instead P is included in a restricted interval between −1/3 and 1/2. Another dimensionless parameter of great use, particularly in biophysics and biochemistry, is fluorescence anisotropy r, which is defined as the ratio of the polarized components to the total intensity
Fluorescence polarization and fluorescence anisotropy are interrelated through equation p=3r/(2+r) and can be used interchangeably, even though FP is measured with respect to the fluorescence intensity in the direction of observation, while FA is determined in relation to the total fluorescence emission intensity [24]. Also the fluorescence anisotropy r is a very convenient quantity because it is additive with the different component concentration.
While for single photon excitation the limiting polarization value is equal to 0.5, for two-photon excitation the limiting value is higher [25–27]. Extensive treatments of the two photon fluorescence anisotropy theory can be found in several review papers [25,26] but essentially it can be ascribed to the increased two-photon angular photoselection. This property has been exploited in several works [28–30] offering the advantage among the others of a higher SNR and increased sensitivity due to a reduced Rayleigh scattering component and higher limiting value.
It is important to emphasize that the limiting polarization values given above are valid in the absence of molecular rotation. This is far from true for the case of solution or for example the intracellular environment. After photoselection, rotational diffusion of the excited molecules tends to scramble the orientation of the emission dipoles removing any preferential direction present in the emission fluorescence. The degree of rotation and scrambling depends ultimately on how fast the molecules can rotate before emission occurs, creating a time dependence in the anisotropy values. If the rotation of the molecule occurs on a time scale that is shorter than its fluorescence lifetime (τr≪τf), the fluorescence emission will be isotropic and the measured anisotropy will tend to zero. On the opposite side instead if the rotation occurs slowly in relation to the fluorescence lifetime, a strong anisotropy will be present.
Emission depolarization following molecular rotation can be quantitatively described using Perrins’ equation [31], which gives the relation between the anisotropy r, the fluorescence lifetime τf, and the rotational correlation time τr:
where r0 corresponds to the molecular fundamental anisotropy as measured in the absence of rotational diffusion. In principle cellular environmental factors such as pH and temperature may alter τf, and τr respectively. But in general, the anisotropy of a fluorescent molecule is largely defined by its intrinsic properties, i.e. its fundamental anisotropy, its size and its fluorescence lifetime. The fact that dyes with different values of lifetimes present distinct values of fluorescence anisotropy has been recently exploited to simultaneously resolve spectrally similar fluorophores in two photon microscopy, where the number of fluorescent labels that can be imaged simultaneously based on their spectral properties is often limited to three (Fig. 2) [32].
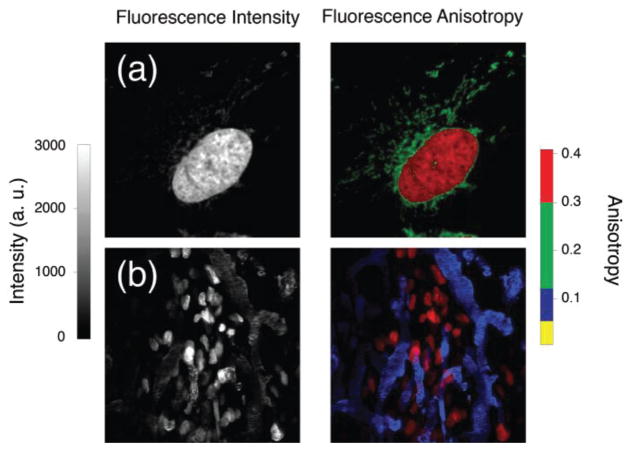
Fig. 2.
Two photon in vivo and in vitro anisotropy imaging resolves simultaneously spectrally similar fluorophores. On the left column B&W images of total fluorescence intensity are shown. On the right column color encoded images of fluorescence anisotropy (color scale on the right side) are presented. (a) HT1080 GFP cells (red, GFP) loaded with MitoTracker Green. (b) HT1080 GFP tumor implanted in a skinfold dorsal window chamber (red, GFP), perfused with a FITC-dextran vascular dye (blue). Adapted and reprinted with permission from [32]
Now as a fluorescent molecule gets larger, its rotational speed will decrease and its anisotropy value increase. This property is particularly useful for well plate based high throughput studies of binding affinity, immunoassays and assays related to drug discovery [33]. Following binding to a much slower rotating molecule such as a protein, a small fluorescent molecule undergoes an increase in fluorescence anisotropy with respect to its value in free solution. Through readings of FA, binding isotherms can then be easily obtained without any need of separation of free and bound components due to the additive property of the FA.
III. Receptor Binding Assays
Fluorescence polarization/fluorescence anisotropy has been successfully used as a screening or detection tool in non-imaging assays, for several decades. Thanks to the opportunity it offers to measure the displacement of a fluorescence molecule from a target in the presence of competitive molecules, the technique has made it possible for it to be widely adopted by the drug discovery field in the clinical and biomedical settings. Its main advantage over alternative techniques such as ELISA or enzyme activity, consists in the fact that it is an inherent separation-free homogeneous assay offering the ability to make quantitative measurements without washing one, or more, of the components of the measurements.
FP/FA does not require particularly complicated measurements setups and it readily translates in the high throughput screening formats, which greatly accelerate data acquisition. Subsequently, commercially available plate readers often have FP/FA configuration options. The intrinsic simplicity of this format and its use has made it possible for it to be quickly adopted by companies and academic labs alike with large library compounds that need to measure millions of compounds at multiple doses, facilitating drug discovery process. Its applications can be classified depending on the specific target class or molecular interaction.
Fluorescence polarization immunoassays were first developed in the 1960s to determine the binding of antigen to antibody [34]. The original experiments measured the concentration of antigen in a sample by first saturated antibody with fluorescently labeled antigen, then measuring the displacement of the fluorescent antigen by sample antigen through a decrease in FP. Using a calibration curve the sample concentration could then be determined. Contrary to others, FP immunoassays do not require tethering antibodies or washing the assay chamber. Also Because FA measurements typically probe solutions over a large volume they are representative of an average of all the fluorescent molecules present within the probed system. The degree of polarization which is correlated to the fraction bound of fluorescence molecules is therefore not limited by the total concentration of fluorescent molecules, assuming the concentration is the same across a binding measurement.
Enzymatic activity can be also readily determined through FP. In these measurements, a fluorescently labeled target protein will have a decreased FP signal if cleaved by an enzyme, due to the decrease in a mass and thus increased Brownian rotation. Therefore, using a known peptide substrate, the concentration, activity, or inhibition of enzymes can be rapidly assessed.
Despite its success and different implementations, the exploitation of FP in the imaging field has been limited. Within this context, most applications so far have been restricted to the determination of structure, orientation, or cellular membrane dynamics. Regarding binding activity measurements, FP imaging has been used to determine where calmodulin binds to myosin in fibroblasts [35]. Bigelow, et al, [36] used FP imaging to measure enzyme activity with confocal microscopy. Through this approach spatial resolution of trypsin and proteinase K enzyme activity could be obtained.
Apart from these applications, the translation of common in vitro FP/FA assay measurements to cellular, or in vivo, imaging with high spatial and temporal resolution has been quite limited.
IV. Instrumentation
FP/FA has been widely exploited due to its intrinsic procedural simplicity. Measurements are typically made exciting the probed solution with a well defined state of polarization (p), and by resolving the emission fluorescence into parallel (p) and perpendicular (s) components. Different strategies are possible and the state of polarization of the excitation light can be rotated or alternatively the fluorescence emission light is decomposed in two orthogonal components. Typically this is achieved using a combination of waveplates, linear polarizers, or polarization beam splitters. Once the two orthogonal components of the fluorescence are measured, values of FP or FA are easily determined.
While the use of polarization fluorimetry is extensively implemented across a range of disciplines in the life sciences, the possibility to measure FA intracellularly has been the object of different studies since many years. The most straightforward solution consists in implementing widefield fluorescence microscopy. Here two images are captured by a CCD camera (or two), each one representative of the two orthogonal states of polarization. While easily implemented, this method suffers of poor axial resolution which is particularly troublesome when imaging in solution under the presence of free unbound fluorescent molecules. Also variation across the field of view of the light polarization makes it difficult to perform accurate image analysis.
Another possibility consists in exploiting laser scanning microscopy in order to obtain in plane- and axial high-resolution FA images. While several authors have utilized confocal microscopy and demonstrated its use for FA imaging, the possibility to combine two photon microscopy with fluorescence anisotropy detection is particularly attractive thanks to all advantages that nonlinear imaging offers over other microscopy imaging modalities [37]. Specifically its extended imaging penetration depth enables to image deep within tissue and tumors in more physiologically relevant context. Also its low scattering component in the near infrared substantially reduces tissue scattering properties, which could bias calculated anisotropy values. In combination with its low phototoxicity and high axial resolution, all this feature contribute in making multiphoton microscopy ideally suited for high resolution FA imaging within single cells and in vivo.
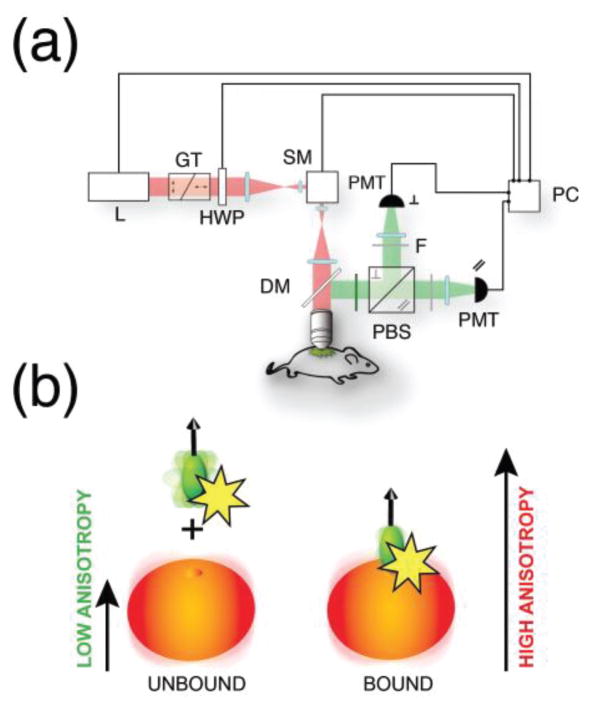
We have therefore custom adapted a commercial multiphoton microscope [20] (Fig. 3). Light from a Ti:Sapphire laser is linearly polarized along a fixed predetermined axis (p), using a Glan-Thompson polarizer and a half-wave plate. Polarized light is then focused through a water immersion objective, onto the imaging sample. Fluorescence is epi-collected in non-descanned mode, spectrally filtered and separated through a polarizing beam splitter in two orthogonal state of polarization (s,p) parallel and perpendicular with respect to the incoming exciting one. Both light components are then detected by two separate photomultiplier tubes.
Fig. 3.
(a) Schematic representation of the two-photon fluorescence anisotropy microscopy system. L laser, GT Glan-Thompson polarizer, HWP half waveplate, SM scanning mirror, PMT photomultiplier tube, DM dichroic mirror, PBS polarization beamsplitter, F polarizing filters. (b) Upon binding to their target, fluorescently labeled small molecule drugs present an increase in fluorescence anisotropy due to the larger molecular weigth of the target. Adapted and reprinted with permission from [20].
In LSM image acquisition is based on a sequential point-by-point excitation, with the laser point scanning along a preset path to cover the imaging field of view. The measured pixel value of FA is calculated in accordance to the definition of r, and corresponds to the fraction-weighted sum of the possible anisotropy values within the voxel, which for two photon microscopy corresponds roughly to a volume of a femtoliter. Whole images can then be obtained in a pixel-by-pixel basis representing FA distribution maps across the imaging field of view.
Different factors can contribute to errors in the calculation of the FA. The PMT for example can respond differently to the fluorescence signal intensity and the optical components can be polarization sensitive. It is therefore imperative to perform a calibration using dyes with known anisotropy in order to measure the correction G-factor. The numerical aperture of the objective (which is typically in the order of 1 for two photon microscopy) can also impact the values of the calculated anisotropy [38, 39], with light depolarization increasing with increasing objective’s aperture angle. Calibration measurements and corrections can compensate for objective induced artifacts, but simply restricting the field of view can easily ensure that the polarization is uniform across the entire field of view.
Also scattering can be a major source of artifacts [40], inducing a decrease of the degree of polarization. Experiments in phantoms and on fluorescent beads with known FA values injected into superficial tissue within a nude mouse dorsal window chamber indicated a slight dependend loss of anisotropy with a 10% loss at 100 microns [20].
Errors in the recorded fluorescence intensity induced by noise can also heavily affect FA values [41]. Particularly at low counts per pixel, the signal-to-noise ratio of the measurements will be increasingly higher leading to strong artifacts on the FA images. In order to account for these aleatory variations, one viable route consists in statistically weighting every anisotropy value pixel-wise by the corresponding total intensity [20].
Following these consideration and through the use of several fluorescent dyes at progressively increasing viscosities, we have demonstrated that our system is capable of providing accurate maps of FA [20]. Also using fluorescent microspheres we have shown high planar and axial FA resolution both in vitro and in vivo, demonstrating that our imaging system is particularly suitable for accurate in vivo high resolution FA imaging.
V. Quantification and imaging of drug binding
The possibility to optically detect fluorescent small molecule drugs offers new insights into drug accumulation and distribution within cells. However, fluorescence imaging alone does not provide any functional information about drug activity and binding inside the cell. The combination of FP imaging and traditional, non-imaging binding assays provides a unique opportunity to image drug binding activity in cells.
We have therefore utilized our imaging approach in a relevant drug-target system and here we illustrate results obtained in one specific instance. Poly (ADP ribose) polymerase (PARP) is a family of enzymes that, due to their role in DNA repair, are the target of numerous chemotherapeutic drug candidates [42]. Their inhibition is especially potent in cells that are BRCA1 or BRCA2 deficient [43–44].
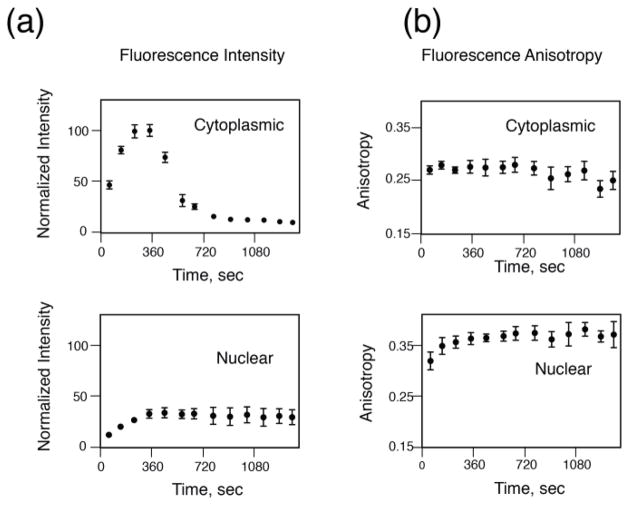
We therefore synthetized a fluorescent derivative of AZD2281 (Olaparib), a small molecule PARP inhibitor which leads to an increase in unrepaired DNA damage and, ultimately, cell death. AZD2281 was fluorescently labeled with BODIPY FL (AZD2281-BODIPY FL), which can be excited in two photon, while maintaining a high affinity for PARP1 [14]. Two photon fluorescence microscopy time lapse imaging of HT1080 cells, a well understood fibrosarcoma cell line expressing PARP, shows that throughout the course of the loading and washing the drug enters the cell through the plasma membrane (Fig. 4). The drug accumulates in the cytosolic area, likely interacting with organelle lipids, but also readily enters the nucleus. Here, where the target PARP1 mainly resides, the drug accumulates while over time during the washing phase, the drug present in the cytoplasm is cleared (Fig. 5). Because PARP1 is a large molecule (~120 kDa) there is a significant change in anisotropy between free, and target-bound AZD2281-BODIPY FL.
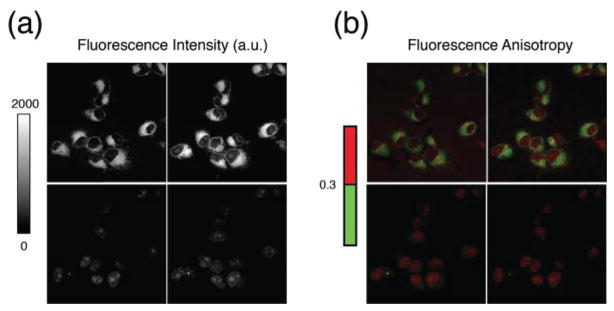
Fig. 4.
(a) Fluorescence intensity and (b) corresponding fluorescence anisotropy time lapse imaging of the fluorescently labeled drug AZD2281-BODIPY FL in live HT1080 cells after drug loading and washing. The top left and right images are collected during the drug loading phase, at respectively 173, and 375 seconds. Cells are then washed, and images (bottom left and right) are collected at 691, and 1382 sec respectively. Fluorescence anisotropy images are color encoded (right color bar). Pixels with values of anisotropy lower than 0.3 are displayed in green. Higher valeus of anisotropy are displayed in red. Adapted and reprinted with permission from [20]
Fig. 5.
Graphs representing the temporal evolution of the total fluorescence intensity (a) and the corresponding fluorescence anisotropy (b) of AZD2281-BODIPY FL in the cytoplasms (top) and nuclei (bottom) cells of Fig. 4. Time data points are obtained by fluorescence intensity and fluorescence anisotropy images acquired at different time points for the same cells. Adapted and reprinted with permission from [20]
Two photon fluorescence anisotropy microscopy can therefore quantify exactly what is the fraction of the amount of drug bound to its target in the cell.
An anisotropy threshold can be assigned to distinguish among the different states (bound/unbound) and FA intracellular time lapse imaging of drug-target engagement can be obtained. When the cells are loaded with the fluorescently labeled drug we observe non-specifically bound drug in the cytoplasmic region, while bound drug is present in the nucleoli where PARP accumulates. Following washing of the drug the cytoplasmic anisotropy signal is cleared. It is important to emphasize that because measurements occur in cells, different causes could contribute to increased values of anisotropy such for example changes in the probe lifetime or increased cellular or nuclear viscosity. Time resolved fluorescence microscopy [45] measurements have demonstrated that changes in lifetime are not responsible for FA increase [20]. Moreover competition experiments produced anisotropy values in the cytoplasm and nucleus that were similar, demonstrating that subcellular viscosity was not responsible also.
Because it is two photon based, our imaging modality offers high planar and axial resolution. Moreover is also a viable tool for an in vivo imaging setting where we have demonstrated its use measuring and mapping drug-target engagement in dorsal window chamber implanted HT1080 tumors (Fig. 6).
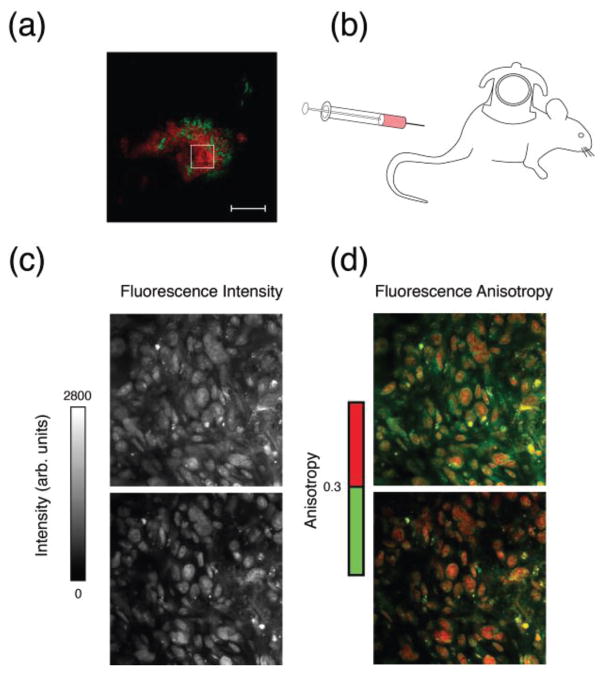
Fig. 6.
In vivo imaging of a HT1080 BTK-mCherry tumor implanted in a skinfold dorsal window chamber (a) (red, mCherry; green, vasculature). After injection of the fluorescently labeled drug AZD2281-BODIPY FL (b), drug accumulates in the cytosolic area of the cells and in the nuclei where its anisotropy value is higher due to target binding. Over time fluorescence signal from the cytoplasm decreases, and fluorescence anisotropy values in the nuclei increases. (c) B&W images of the total fluorescence intensity. (d) Color encoded images of the corresponding fluorescence anisotropy images (color scale on the right side). Top images acquired following drug intravenous infusion. Bottom images acquired 34 minutes post infusion. Adapted and reprinted with permission from [20]
This is the first time that drug engagement is measured in real time at the cellular level, both in vitro and in vivo, providing invaluable insight into the activity of drug in vivo and potential indication of inefficacy.
VI. Conclusions
Several elaborate target engagement measurement techniques have recently been developed to better understand the fate of drugs in cells and in vivo. However, to achieve subcellular resolution with sufficient temporal capabilities novel microscopy approaches are necessary. Building upon intravital microscopy modalities developed in our group to image drug pharmacokinetics and cellular heterogeneity, we have recently exploited fluorescence polarization to determine target engagement within these systems. While traditional polarization assays are limited to artificial system such as biochemical assays or cell lysates, which at best can serve as qualitative interference models and rarely accurately reflect actual drug dynamics, microscopy based imaging modalities such as two photon fluorescence anisotropy microscopy can extend drug target engagement measurements and pharmacological validation of protein function in vivo. As such, they can tremendously accelerate drug development as they allow to directly correlate treatment efficacy with target activity and open up a new way to directly quantify target binding in live intact cells and in vivo.
Acknowledgments
This project was funded in part by Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (under Contract No. HHSN268201000044C), National Cancer Institute (T32CA079443 and P50CA086355), and from the Institute of Biomedical Engineering (under R01EB006432), and in part by 1R01CA164448-01. The research leading to these results has also received funding from the EC Seventh Framework Programme under the Grant Agreement nr. 622182.
Biographies
 Claudio Vinegoni (PhD 2002) is currently Asst. Professor at Harvard Medical School, and works at the Center for Systems Biology at Massachusetts General Hospital (MGH). He has published over 80 original publications in peer reviewed journals. His research activity involves the development of novel optical imaging instruments and techniques with applications in the clinical and biomedical arena.
Claudio Vinegoni (PhD 2002) is currently Asst. Professor at Harvard Medical School, and works at the Center for Systems Biology at Massachusetts General Hospital (MGH). He has published over 80 original publications in peer reviewed journals. His research activity involves the development of novel optical imaging instruments and techniques with applications in the clinical and biomedical arena.
 J. Matthew Dubach (PhD 2012) is currently an instructor at Harvard Medical School within the Center for Systems Biology at Massachusetts General Hospital. His research focuses on imaging technique and sensor development to enable quantitative measurements in complex biological environments.
J. Matthew Dubach (PhD 2012) is currently an instructor at Harvard Medical School within the Center for Systems Biology at Massachusetts General Hospital. His research focuses on imaging technique and sensor development to enable quantitative measurements in complex biological environments.
 Paolo Fumene Feruglio (PhD 2010) is Visiting Research Fellow at the Center for Systems Biology, Harvard Medical School and Res. Assist. at the University of Verona. He graduated in Electrical Engineering at the University of Padova and received his Ph.D in Multimodal Imaging in Biomedicine at the University of Verona. His research involves the developing of models and algorithms to process and analyze data with application to in vivo and ex vivo molecular imaging techniques at macro and microscopic level.
Paolo Fumene Feruglio (PhD 2010) is Visiting Research Fellow at the Center for Systems Biology, Harvard Medical School and Res. Assist. at the University of Verona. He graduated in Electrical Engineering at the University of Padova and received his Ph.D in Multimodal Imaging in Biomedicine at the University of Verona. His research involves the developing of models and algorithms to process and analyze data with application to in vivo and ex vivo molecular imaging techniques at macro and microscopic level.
 Ralph Weissleder (PhD 1985) is a Professor at Harvard Medical School, Director of the Center for Systems Biology at Massachusetts General Hospital (MGH), and Attending Clinician (Interventional Radiology) at MGH. Dr. Weissleder is also a member of the Dana Farber Harvard Cancer Center and member of the National Academies of Medicine. He has published ~800 publications in peer-reviewed journals and has authored several textbooks.
Ralph Weissleder (PhD 1985) is a Professor at Harvard Medical School, Director of the Center for Systems Biology at Massachusetts General Hospital (MGH), and Attending Clinician (Interventional Radiology) at MGH. Dr. Weissleder is also a member of the Dana Farber Harvard Cancer Center and member of the National Academies of Medicine. He has published ~800 publications in peer-reviewed journals and has authored several textbooks.
Contributor Information
Claudio Vinegoni, Email: cvinegoni@mgh.harvard.edu, Center for System Biology, Massachusetts General Hospital and Harvard Medical School, Richard B. Simches Research Center, 185 Cambridge Street, Boston 02114, USA.
John M. Dubach, Center for System Biology, Massachusetts General Hospital and Harvard Medical School, Richard B. Simches Research Center, 185 Cambridge Street, Boston 02114, USA
Paolo Fumene Feruglio, Center for System Biology, Massachusetts General Hospital and Harvard Medical School, Richard B. Simches Research Center, 185 Cambridge Street, Boston 02114, USA and with the Department of Neurological and Movement Sciences, University of Verona, Strada Le Grazie 8, 37134 Verona, Italy.
Ralph Weissleder, Center for System Biology, Massachusetts General Hospital and Harvard Medical School, Richard B. Simches Research Center, 185 Cambridge Street, Boston 02114, USA.
References
- 1.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nature Reviews Drug Discovery. 2010 Mar;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 2.Simon GlM, Niphakis MJ, Cravatt BF. Determining target engagement in living systems. Nature Chemical Biology. 2013 Apr;9:200–205. doi: 10.1038/nchembio.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, Wohlschlegel JA, Vondriska TM, Pelletier J, Herschman HR, Clardy J, Clarke CF, Huang J. Target identification using drug affinity responsive target stability (DARTS) Proc Natl Acad Sc U S A. 2009 Dec;102(4):21984–9. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimwood S, Hartig PR. Target site occupancy: emerging generalizations from clinical and preclinical studies. Pharmacol Ther. 2009 Jun;122:281–301. doi: 10.1016/j.pharmthera.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Matthews PM, Rabiner EA, Passchier J, Gunn RN. Positron emission tomography molecular imaging for drug development. Br J Clin Pharmacol. 2012 Feb;73(2):175–186. doi: 10.1111/j.1365-2125.2011.04085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munteanu B, Meyer B, Reitzenstein C, Burgermeister E, Bog S, Pahl A, Ebert MP, Hopf C. Label-free in situ monitoring of histone deacetylase drug target engagement by matrix-assisted laser desorption ionization-mass spectrometry biotyping and imaging. Anal Chem. 2014;86(10):4642–4647. doi: 10.1021/ac500038j. [DOI] [PubMed] [Google Scholar]
- 7.Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular Thermal Shift Assay. Science. 2013 Jul;341(6141):84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 8.Pittet MJ, Weissleder R. Intravital Imaging. Cell. 2011 Nov;147(5):983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguirre AD, Vinegoni C, Sebas MM, Weissleder R. Intravital imaging of cardiac function at the single-cell level. Proc Natl Acad Sci U S A. 2014 Aug;111(31):11257–11262. doi: 10.1073/pnas.1401316111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuste R. Fluorescence microscopy today. Nature Methods. 2005;2:902–904. doi: 10.1038/nmeth1205-902. [DOI] [PubMed] [Google Scholar]
- 11.Edgington LE, Verdoes M, Ortega A, Withana NP, Lee J, Syed S, Bachmann MH, Blum G, Bogyo M. Functional imaging of legumain in cancer using a new quenched activity-based probe. J Am Chem Soc. 2013 Dec;135:174–182. doi: 10.1021/ja307083b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang KS, Budin G, Reiner T, Vinegoni C, Weissleder R. Bioorthogonal imaging of aurora kinase A in live cells. Angew Chem Int Ed Engl. 2012;51:6598–6603. doi: 10.1002/anie.201200994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Hong Lee J, Mi Jeon H, Hye Han J, Park N, He Y, Lee H, Soo Hong K, Kang C, Kim JS. Folate-based near-infrared fluorescent theranostic gemcitabine delivery. J Am Chem Soc. 2013 Jul;135:11657–11662. doi: 10.1021/ja405372k. [DOI] [PubMed] [Google Scholar]
- 14.Thurber GM, Yang KS, Reiner T, Kohler RH, Sorger P, Mitchison T, Weissleder R, et al. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo. Nat Commun. 2013;4:1504. doi: 10.1038/ncomms2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dam GM, Themelis G, Crane LMA, Harlaar NJ, Pleijhuis RG, Kelder W, Sarantopoulos A, de Jong JS, Arts HJG, van der Zee AGJ, Bart J, Low PS, Ntziachristos V. Intraoperative tumor-specific fluorescence imaging in ovarian cancer by folate receptor-alpha targeting: first in-human results. Nat Med. 2011;17:1315–1319. doi: 10.1038/nm.2472. [DOI] [PubMed] [Google Scholar]
- 16.Laughney AM, Kim E, Sprachman MM, Miller MA, Kohler RH, Yang KS, Orth JD, Mitchison TJ, Weissleder R. Single-cell pharmacokinetic imaging reveals a therapeutic strategy to overcome drug resistance to the microtubule inhibitor eribulin. Sci Transl Med. 2014 Nov;6(261):261ra152. doi: 10.1126/scitranslmed.3009318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thurber GM. Effect of Small-Molecule Modification on Single-Cell Pharmacokinetics of PARP Inhibitors. Mol Cancer Ther. 2014 Apr;13(4):986–995. doi: 10.1158/1535-7163.MCT-13-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi AM, Taylor CW. Analysis of protein-ligand interactions by fluorescence polarization. Nat Protoc. 2011;6:365–387. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bullen A. Microscopic imaging techniques for drug discovery. Nature Reviews Drug Discovery. 2008 Jan;7:54–67. doi: 10.1038/nrd2446. [DOI] [PubMed] [Google Scholar]
- 20.Dubach JM, Vinegoni C, Mazitschek R, Fumene Feruglio P, Cameron LA, Weissleder In vivo imaging of specific drug-target binding at subcellular resolution. Nat Commun. 2014;5:3946. doi: 10.1038/ncomms4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; 2006. [Google Scholar]
- 22.Axelrod D. Fluorescence polarization microscopy. Methods Cell Biol. 1989;30:333–352. [PubMed] [Google Scholar]
- 23.Weber G. Polarization of the fluorescence of macromolecules. 1. Theory and experimental method. Biochem J. 1952;51:145–155. doi: 10.1042/bj0510145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mocz G. Information Content of Fluorescence Polarization and Anisotropy. J Fluoresc. 2006 Jul;16:511–524. doi: 10.1007/s10895-006-0095-7. [DOI] [PubMed] [Google Scholar]
- 25.Kierdaszuk B, Gryczynski I, Lakowicz JR. Two-photon induced fluorescence of proteins. Topics in Fluorescence Spectroscopy. 2002;5:187–210. [Google Scholar]
- 26.Callis PR. The theory of two-photon-induced fluorescence anisotropy. Topics in Fluorescence Spectroscopy. 2002;5:1–42. [Google Scholar]
- 27.Jameson DM, Croney JC. Fluorescence polarization: past, present and future. Comb Chem High Throughput Screen. 2003;6:167–173. doi: 10.2174/138620703106298347. [DOI] [PubMed] [Google Scholar]
- 28.Brasselet S, Ferrand P, Kress A, Wang X, Ranchon H, Gasecka A. Imaging molecular order in cell membranes by polarization resolved fluorescence microscopy. Fluorescent Methods to Study Biological Membranes. 2012;13:311–337. [Google Scholar]
- 29.Ferrand P, Gasecka P, Kress A, Wang X, Bioud FZ, Duboisset J, Breasselet S. Ultimate use of two-photon fluorescence microscopy to map orientational behaviour fo fluorophores. Biophys J. 2014;106:2330–2339. doi: 10.1016/j.bpj.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vishwasrao HD, Trifilieff P, Kandel ER. In vivo imaging of the actin polymerization state with two-photon fluorescence anisotropy. Biophys J. 2012;102:1204–1214. doi: 10.1016/j.bpj.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrin F. The polarisation of fluorescence light. Average life of molecules in their excited state. Journal De Physique Et Le Radium. 1926;7:390–401. [Google Scholar]
- 32.Dubach JM, Vinegoni C, Weissleder R. Steady state anisotropy two-photon microscopy resolves multiple, spectrally similar fluorophores, enabling in vivo multilabel imaging. Optics Letters. 2014 Aug;39(15):4482–4495. doi: 10.1364/OL.39.004482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jameson DM, Ross JA. Fluorescence Polarization/Anisotropy in Diagnostics and Imaging. Chem Rev. 2010 May;110(5):2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dandliker WB, Feigen GA. Quantification of the antigen-antibody reaction by the polarization of fluorescence. Biochem Biophys Res Commun. 1961 Jul;5:299–304. doi: 10.1016/0006-291x(61)90167-x. [DOI] [PubMed] [Google Scholar]
- 35.Gough AH, Taylor DL. Fluorescence anisotropy imaging microscopy maps calmodulin binding during cellular contraction and locomotion. J Cell Biol. 1993 Jun;121(5):1095–1107. doi: 10.1083/jcb.121.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bigelow CE, Conover DL, Foster TH. Confocal fluorescence spectroscopy and anisotropy imaging system. Opt Lett. 2003;28:695–697. doi: 10.1364/ol.28.000695. [DOI] [PubMed] [Google Scholar]
- 37.Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006 Jun;50(6) doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 38.Bahlmann K, Hell SW. Electric field depolarization in high aperture focusing with emphasis on annular apertures. J Microsc. 2000;200:59–67. doi: 10.1046/j.1365-2818.2000.00739.x. [DOI] [PubMed] [Google Scholar]
- 39.Schon P, Munhoz F, Gasecka A, Brustlein S, Brasselet S. Polarization distortion effects in polarimetric two-photon microscopy. Opt Express. 2008;16:20891–20901. doi: 10.1364/oe.16.020891. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh N, Majumder SK, Gupta PK. Fluorescence depolarization in a scattering medium: effect of size parameter of a scatterer. Phys Rev E. 2002 Jan;65:026608. doi: 10.1103/PhysRevE.65.026608. [DOI] [PubMed] [Google Scholar]
- 41.Lidke KA, Rieger B, Lidke DS, Jovin TM. The role of photon statistics in fluorescence anisotropy. IEEE Trans Image Process. 2005 Sep;14(9):1237–1245. doi: 10.1109/tip.2005.852458. [DOI] [PubMed] [Google Scholar]
- 42.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell Å, Ekblad T, Öncü D, Kull B, Robertson GM, Pellicciari R, Schüler H, Weigelt J. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nature Biotechnology. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 43.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005 Apr;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 44.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 45.Suhling K, French PM, Phillips D. Time resolved fluorescence microscopy. Photochem Photobiol Sci. 2005;1:13–22. doi: 10.1039/b412924p. [DOI] [PubMed] [Google Scholar]