Abstract
HIV infection is controlled but not eradicated by combination antiretroviral therapy (cART), and persistence during therapy represents a substantial barrier to strategies to eradicate infection. The nature of persistence is uncertain, and a number of mechanisms have been proposed to explain HIV persistence in vivo, including low-level HIV replication, sanctuary sites for HIV-infected cells, and latent HIV residing within long-lived cells. Analysis of residual viraemia and of cell-associated HIV revealed evidence of multiple copies of identical HIV sequences suggesting infected cells can undergo cellular expansion. Recently, analysis of integration sites in HIV-infected cells derived from peripheral blood lymphocytes of patients undergoing long-term cART revealed direct evidence that HIV-infected cells undergo clonal expansion. These studies demonstrated that clonally expanded populations are common in HIV-infected individuals, persist for prolonged periods and increase in frequency during prolonged therapy. Several analyses reported that site of integration may affect persistence, clonal expansion, or both. As such, expanded populations may represent an important source of infectious HIV during cART. Many HIV integrants are defective for replication, however, and additional research is essential to determine to what degree clonally expanded populations represent a reservoir of replication-competent HIV.
Keywords: HIV integration, clonal expansion, viral persistence
Introduction
Infection with human immunodeficiency virus (HIV) results in a chronic, progressive immunodeficiency which, left untreated, results in death from opportunistic infections or neoplastic diseases [1]. A number of pathogenic mechanisms have been reported to contribute to progressive disease, but the control of viral replication after initiating combination antiretroviral therapy (cART) arrests these processes, and results in a degree of immune restoration. Although cART controls HIV infection, it does not eradicate infection, and lifelong therapy is essential to continue viral suppression and preserve immune recovery [2,3]. As prolonged therapy is associated with adverse effects, metabolic and cardiovascular complications, new strategies to eliminate or control HIV without antiretroviral therapy are essential. Substantial gaps in our understanding of HIV persistence limit our ability to identify specific and focused approaches to viral eradication. Several mechanisms have been reported to explain HIV persistence during therapy, including low-level cycles of replication, infection of long-lived cells and residence in sanctuary sites, but the relative contribution of each of these processes to maintain a viral ‘reservoir’, operationally defined as the population of replication-competent HIV capable of spread in the absence of therapy, remains uncertain. Over the last few years, a number of lines of evidence have suggested that HIV-infected cells may undergo cellular division, and recently several groups reported direct evidence of cellular clonal expansion of HIV-infected cells. The role of clonal expansion in HIV persistence remains uncertain. Here we review evidence of HIV clonal expansion and its potential role as a mechanism of HIV persistence.
HIV replication in vivo establishes a population of HIV-infected cells
In vivo, HIV infection spreads via a single or limited number of infecting variants [4,5]; as a consequence, viral populations emerging early in infection are relatively genetically uniform. HIV spreads to a number of different CD4 T cell subsets throughout primary and secondary lymphoid tissues, including lymph nodes and mucosa-associated lymphoid tissues, as well as to additional sites such as the central nervous system. As HIV replication is rapid and error prone, HIV populations gradually accumulate new variants, and within several years of infection, HIV populations are diverse, such that each individual genome is genetically distinct. Virus populations are also large; estimates (probably underestimates) of replicating population sizes exceed 104–105 cells [6–9]. Although substantial, the numbers of infected cells are still small compared with numbers of cells in the total lymphocyte pool; the numbers of cells infected by HIV, as measured by total cell-associated HIV-DNA in peripheral blood lymphocytes, is in the range of 1 infected cell per 100–1,000 cells. When untreated, HIV infection is accompanied by progressive immunodeficiency, reflected in a gradual loss of peripheral CD4 cells, but this loss seems not to be due to direct HIV cytopathic effect, but largely due to immune-based mechanisms. By the time antiretroviral therapy is typically introduced in chronically infected individuals, the immune system has undergone a varying degree of CD4 lymphopenia as the HIV-infected population has become large, genetically diverse, and widely distributed in lymphoid organs.
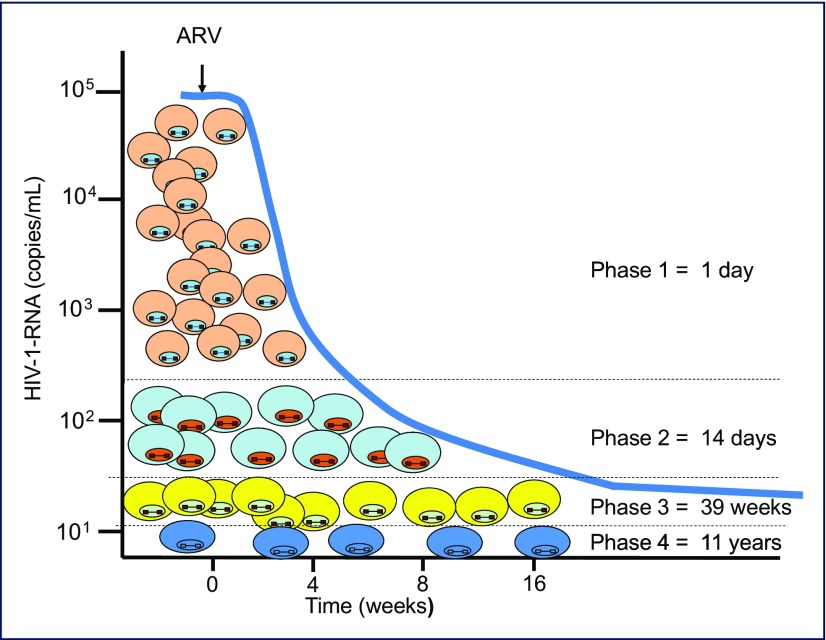
Initiation of combination antiretroviral therapy is accompanied by a dramatic decline in viraemia, as cART blocks infection to new cells and infected cells are eliminated (Figure 1). As the half-life of HIV in plasma is relatively short (15 minutes–3 hours), the decrease in viraemia reflects elimination of infected cells. Viral decay kinetics after cART is introduced are multiphasic as infected cells persist for variable periods prior to elimination; half-lives of approximately 1 day, 14 days, 39 weeks and 11 years have been measured as viraemia declines to a low residual level (c. 1 copy/mL using sensitive single-copy assays). The greatest decline in viraemia takes place in the first few weeks of therapy, indicating that the majority of circulating virus (>99.9%) is produced by cells with a relatively short half-life, typically activated T cells [10]; subsequent decay rates correspond to cells with longer half-lives [11–14]. Detailed analysis identified that resting memory CD4 cells were latently infected with HIV, demonstrating the presence of a relatively stable, long-lived reservoir harbouring replication-competent virus [15–17]. This latent reservoir declined over time in individuals undergoing cART, but the decay kinetics were slow (half-life approximately 44 months), demonstrating that a strategy of HIV eradication by attrition of these cells was essentially unachievable [18,19].
Figure 1.
Establishing a reservoir of HIV-infected cells during combination antiretroviral therapy. Diverse cell types are infected with HIV and are eliminated at varying rates following infection. After prolonged cART, long-lived cells are present, some of which may have undergone clonal expansion
In contrast to the typical >10,000-fold decline in HIV-RNA, declines in numbers of HIV-infected cells are comparatively modest. Numbers of cells with cell-associated HIV-DNA decrease by 10–50-fold [10]. Genetic analysis of HIV populations revealed that cART reduces HIV viraemia, but not viral diversity; during the first several years of therapy, HIV genetic diversity is indistinguishable from pre-therapy populations and remains both highly diverse and non-divergent from pre-therapy virus [20]. Thus, early in treatment, the large and genetically diverse populations that are present prior to therapy give way to equally diverse populations of infected cells. Characterising HIV from these infected cells has been useful in shedding new light on the nature of HIV persistence. Several sensitive quantitative assays have been developed [21] to characterise HIV populations during cART, and are broadly divided into assays that: (1) quantify HIV nucleic acid species: specifically HIV plasma viraemia, cell-associated RNA (unspliced and spliced), cell-associated total or integrated DNA; (2) characterise HIV sequences in plasma or cell-derived material; (3) quantitatively determine numbers of HIV-infected cells capable of producing infectious HIV via HIV outgrowth assays; and (4) characterise HIV nucleic acid sequences from in vitro recovery. No assay is perfect or will describe the HIV reservoir completely; quantitative nucleic acid assays determine the numbers of cells that have become HIV infected, but do not provide information regarding the replication competence of the infected cells. Quantitative recovery of infectious HIV from peripheral blood lymphocytes or other locations provides critical information, but only for a limited number of viruses that were recovered under specific in vitro conditions. The subset of cells capable of producing replication competent HIV from these cells represents the relevant HIV reservoir and is likely to be only a small fraction of the total HIV-infected population [22]. A fundamental goal is utilising an appropriate suite of assays to sufficiently characterise the HIV reservoir during therapy.
Emergence of HIV variants with identical sequences during long -term cART
Nucleic acid sequence analysis of HIV directly from patient-derived material or after in vitro reactivation has provided important additional clues in identifying mechanisms of HIV persistence during long-term cART. Early studies of residual viraemia from patients undergoing cART by Persaud and co-workers [23] demonstrated that sequences could be recovered from low-level viraemia and that these sequences did not contain mutations conferring resistance to the suppressive antiretroviral therapy regimen. These data suggested that viruses present in plasma did not represent the product of drug resistance to the concurrent regimen, but were likely to have been produced from cells infected prior to initiating effective therapy. The development of single genome sequencing techniques enabled new studies of HIV populations, and the ability to obtain numerous HIV sequences from individual time points, even in the setting of low-level viraemia (<50 copies/mL plasma) permitted detailed phylogenetic analysis. These studies identified the presence of multiple copies of identical HIV sequences in plasma after prolonged cART, which is highly unlikely in patients with genetically diverse populations of HIV. It suggests that only a limited subset of cells was producing HIV, and that these cells were all infected with identical virus; these populations comprised a substantial proportion of the virus population and were termed ‘predominant plasma clones’ (PPC) [24]. Longitudinal analyses identified that identical PPCs could be detected in samples obtained over time, suggesting cells infected with these viruses were persistently producing HIV. In individuals with genetically diverse populations, emergence of populations with identical sequences strongly suggests the viruses are being produced from cells with identical proviruses. Although it is possible that virus production is derived from original or early-infected cells that were all genetically uniform, it was more likely that cells with integrated proviruses had undergone cellular expansion. Comparison of HIV populations pre- and post-therapy demonstrated that as expected, HIV populations were highly diverse after initiation of antiretroviral therapy, and PPC sequences emerged after several years of cART; the PPCs were not divergent from pre-therapy virus and had not acquired new CTL escape mutations [20]. Thus PPCs represented a strong argument for the presence of a clonal population of infected cells chronically producing low levels of HIV.
The presence of identical sequences in patients with chronic infection and a diverse genetic background could also be detected in DNA from peripheral blood mononuclear cells (PBMC) and in cells in a number of anatomical compartments and lymphocyte subsets [25–27]. Notably, Imamichi et al. and Palmer and co-workers demonstrated long-term persistence of clonal HIV variants [28,29]. In both of these instances, the HIV provirus was defective for replication, and therefore was incapable of viral spread. Instead, the multiple copies of HIV present were likely to be the product of cells that had undergone clonal expansions. Taken together, these data suggested that the predominant plasma clones detected after prolonged cART were derived from HIV-infected cells that had undergone a degree of clonal expansion.
Identification of HIV integration sites
Direct evidence for clonal expansion of HIV-infected cells from individuals undergoing long-term antiretroviral therapy was obtained by identifying and quantifying HIV integration sites. During retroviral replication, integration of the newly reverse transcribed HIV-DNA into the host genome is an essential event in virus replication and enzymology of integration has been investigated [30]; solving the crystal structure of foamy virus retrovirus integrase, and analysis of integrase inhibitors provided critical information regarding mechanisms of integration [31,32]. Initial approaches to detect retroviral integration used a strategy of generating cellular DNA fragments digested by restriction enzymes with frequent sites in the eukaryotic genome, such as AflII, PstI or AluI; the product of this digestion is a large population of small DNA fragments with ‘sticky’ ends, which then undergo a ligation reaction, under conditions that promote self-ligation. The resulting circular DNA fragments are then subjected to an ‘inverse PCR’ amplification using primers in the HIV sequence, typically including primers amplifying HIV-LTR and gag that will amplify outwards around the circles [33,34]. This approach is ideally suited for studies of in vitro infections where the infection frequency is quite high with many integrants present, or in the analysis of retroviral-associated tumours, which are frequently clonal or oligoclonal. The presence of many identical integrants will facilitate detection. These early studies of various retroviral infections identified a number of common characteristics as well as obvious distinctions in integration profiles. It was evident from many early studies that HIV integration occurred in diverse locations and that integration preferences were present but were generally mild [33–37]. The most consistent observation was that HIV integrated in diverse sites, but was largely within transcriptional units. Co-factors in integration, notably LEDGF/p75, were identified that participated in integration-site preferences [38–42]. Integration events occurred with the HIV provirus present either in the same or opposite orientation of the host gene transcriptional unit. In vitro, some differences between resting and activated CD4 cells were evident [43]. Introduction of massively parallel sequencing platforms expanded the ability for fine structure analysis of integration sites in vitro. HIV was integrated largely in transcriptional units, but extensive comparison of experimental HIV infections in a number of cell-based latency models did not identify specific latency-associated features with integration location [44], highlighting difficulties in identifying determinants of latency.
Although extensive studies of HIV integration have been conducted in vitro, identifying HIV integration sites in vivo presents additional challenges as the frequency of infected cells is relatively low and integration sites have to be identified from within a vast background of human genomic DNA. As a result, the overall numbers of integrants, when they could be detected in these experiments, were low relative to in vitro infections. In addition, inverse PCR sequencing methods do not identify whether any of the integration sites that are identified are clonally expanded. Nevertheless, a number of useful observations were made, which drove additional research in detecting HIV integration in vivo.
Initial studies by Han and co-workers, McGrath et al. and Ikeda et al., characterised HIV integrants in PBMC, resting CD4+ memory cells or tissues from late-stage patients [45–47]. In these in vitro studies, HIV integrates largely (>80% in these studies) into transcriptional units; this is a particularly striking finding, as only a minority of the human genome (30%) contains transcriptional units. Three patients had multiple integrations in a single intron (intron 5) of the BACH2 gene [45,46]. One hypothesis raised at the time was that these multiple integrations represented a preferred site of integration, but large datasets of integration sites from other infections, including in vitro infections were not available to investigate the issue further. Subsequent in vitro studies with large yields of integration sites demonstrated HIV was capable of widespread integration without highly selective or specific hotspots, and no orientation specificity. Several integration sites were identified multiple times, suggesting the cell with this integrated provirus had undergone cell division, but artefactual PCR amplification could not be excluded as an explanation. The discrepancy between widespread integration in vitro and identification of multiple integrations in individual genes in cells from individuals undergoing antiretroviral therapy for prolonged periods suggested that cells with particular integrants were present because they had acquired an advantage in persistence or clonal expansion, or both.
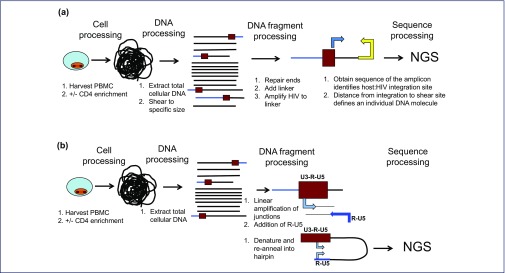
New approaches permitted more efficient identification of HIV integration sites, and unequivocally determined whether such integrants were clonally expanded. Two approaches proved useful to obtain numerous integrants from clinical material (Figure 2). Bangham and co-workers studying HTLV integration [48–50] developed techniques to detect integrants that utilised Illumina next-generation sequencing (Illumina, Inc, San Diego, USA) approaches and were capable of detecting low levels of HIV integrants in the presence of many uninfected cells, which were adapted for use with HIV infections [51]. In brief, cellular DNA is isolated and randomly sheared to a relatively defined size (usually c. 500 nucleotides), DNA ends are repaired and modified with the addition of a single A residue to the 3′ ends, facilitating ligation to linkers, and primers recognising the linker and HIV sequences are used to selectively amplify HIV–host gene sequences using an Illumina-based platform. Extensive sequence processing is essential to identify host–viral junction sequences, and the result yields a large database of junction sequences from infected patients undergoing antiretroviral therapy [52].
Figure 2.
Assays to detect HIV integration. The goal is to detect proviral integration in an unbiased fashion within a large background of DNA from uninfected cells. Assays require sensitivity and ability to unequivocally identify junction sequences, and eliminate possible artefacts from mispriming. The finding of multiple copies of individual integrants indicates clonal expansion, and has to be distinguished from PCR amplification artefacts. (a) Detecting HIV integration by linker mediated PCR amplification. (b) Detecting HIV integrants by integration site loop amplification (ISLA). NGS: next-generation sequencing
In parallel, Wagner and co-workers [53] developed a novel method, denoted integration site loop amplification (ISLA), to improve the probability of detecting integrants by: (1) increasing integrant abundance prior to amplification; and (2) removing the requirement for a specific restriction site to be present near the integration site. In this approach, specific HIV forward primers in env and nef are used in a linear amplification strategy to copy HIV and adjacent host DNA, and increase the abundance of proviral integration sites relative to the remainder of the host genome. Next, a second round of linear amplification is performed using primers consisting of: (1) a 5′ end identical to a well-conserved portion of the U5 region, followed by (2) a 3′ random decamer. The random decamers hybridise throughout the host genome, including the linearly amplified junction sequences. After a second round of linear amplification, the product molecules have two identical copies of a portion of the HIV U5 region, from the U5 in the provirus and the U5 in the primer. As a result, when the products of the synthesis are denatured and re-annealed under conditions that promote intramolecular annealing, stem loop structures consisting of U5 stems are generated; the adjacent host sequence is captured in the loop. The stem is extended into the LTR and the final stem loop structure is subjected to amplification using nested primers in the R region. The sequences of the captured host DNA adjacent to the host–viral junction are identified by Illumina platform sequencing. Integration sites are unequivocally identified by this approach. Once host gene sequences are identified, additional host primers can be synthesised for use with a portion of the linear extension products to amplify a longer sequence of HIV, yielding a sequence from env through the host gene. Comparative analysis of the env–nef sequences can identify multiple sequences with an identical env–nef-LTR sequence and a common junction sequence with the host genome.
Both approaches provide four characteristics essential for understanding HIV persistence.
(1) Substantial numbers of unequivocally identified HIV–host junction sequences
By comparing the host sequence with the sequence of the human genome (UCSC), it is possible to identify the gene or intergenic region into which the provirus is integrated. In some cases, proviruses are integrated into pseudogenes, or repetitive DNA sequences that are distributed throughout the genome, and an unequivocal chromosomal assignment is not directly possible; such integrants are classified as ‘ambiguous’. Although not nearly as extensive as libraries that can be identified from in vitro experiments, datasets are sufficiently large to investigate certain genes or pathways after prolonged cART.
(2) Identification of clonal expanded populations with proviruses
A key advance over prior approaches, these methods clearly identify lineages with a common proviral integration site and exclude the appearance of amplification due to post-processing PCR artefacts. In the Hughes and co-workers approach [52], the genomic DNA is randomly sheared, and thus the distance from an integration site to the shear site (where linkers are ligated) is random. The amplification, sequencing and processing steps yield large numbers of host viral sequences and the same host–virus junction sequence may be identified more than once. Those sequences that have identical host–viral junction sequences but multiple distinct distances to the shear site/linker were each derived from distinct DNA molecules and represent a clonally expanded provirus. In contrast, identical junction sequences with identical distances to a common shear site are obviously the product of PCR amplification of a single site, and are not counted as expanded clones. Strategies to amplify both 3′ and 5′ integration sites are used, and with sufficient efficiency that the 5′ ends and corresponding 3′ ends of proviruses can be identified. Frenkel and colleagues [53] identify clonal expansion after linear amplification to a random decamer annealing site; the primer also incorporates a second U5 domain, and only those molecules with an added U5 domain subsequently undergo PCR amplification.
(3) Orientation of the provirus relative to the host gene
As the junction sequence is clearly identified, it is possible to determine whether the integration occurred with the provirus in the same or opposite orientation as the transcription of the host gene.
(4) Integration precision
Strategies to amplify both 3′ and 5′ integration sites are used, and with sufficient efficiency, both the 5′ ends and corresponding 3′ ends of proviruses can be identified; the precise integration site and the five base-pair duplication in the host gene sequence expected from a correct integration event can be unequivocally identified. In addition, the Wagner et al. approach yields significant internal HIV sequences that can be readily used to compare to variants expressed in plasma or tissues.
Using these techniques, a total of >3,100 HIV integration sites from eight patients were unequivocally identified [52,53]. Despite the differences in approach, the conclusions of these studies were remarkably similar. As prior in vitro studies had reported, the majority of integration sites (79%) were identified in genic regions, proviruses were more commonly present in introns, but introns comprise the majority of the sequence space in genic regions. Overall, HIV clonal expansion was common in both genic and intergenic regions and over 40% of all integrants identified were present in expanded populations. Some clones were identified with just a few members, while other expansions could dominate the infected cell sample. In one patient, approximately 50% of all the integrants identified were mapped to a single proviral integration [52]. The degree of clonal expansion was not uniform across patients; several patients had extensive clonal expansions, while in others, multiple lineages were less common. Patient data remains relatively limited, and it is important to point out that the yield of integration sites in these studies, although substantial, is in no way exhaustive. Neither technique captures the entirety of the infected cell population. As a result, the finding of overall clonal expansion in 40% of all integrants in this limited sample should be interpreted as an underestimate, and suggests that many more proviruses, perhaps all, have undergone expansion, at least to some degree. Clonal lineages persisted for prolonged periods and could be detected in samples obtained 11–13 years apart. Patient numbers and sampling were not sufficiently robust to detect or characterise rates of expansion over time, and no obvious correlations between CD4 cell recovery or other immunophenotypic characteristics and the degree of clonal expansion were evident [52,53] although Wagner et al. reported that frequency of identical sequences did increase with time [54]. Recently Cohn et al. [55] studying HIV integration sites in individuals undergoing long-term cART using an amplification procedure that localises integration sites, also identified integrations in MKL2 and in BACH2.
In these datasets obtained from individuals undergoing long-term cART, genes with integrations could be analysed for broad patterns of classes of genes into which HIV was found to be integrated. While no preference for functional classes of genes was detected in analyses of large datasets of proviral integration in acute in vitro infections, analysis of clinical material from patients undergoing long-term cART revealed a striking shift in integration patterns compared to the total complement of human genes. A large proportion of genes with integrations were involved in cell growth, replication or associated with cancer. These findings raised the provocative possibility that the cells present after prolonged periods had selective advantage for persistence or expansion because they had a provirus integrated in a growth-promoting gene altering a pathway that favoured persistence, expansion, or both. The diverse nature of integration events throughout many genes in these pathways suggests these growth pathways can be disrupted at numerous points. One potential implication of these findings is that, with sufficiently extensive datasets, it may be possible to clearly identify pathways commonly associated with persistence, which represent useful targets to eliminate persistent lineages.
The vast majority of proviruses were integrated in an orientation-independent fashion with respect to the transcriptional unit of the host cell, indicating that the mechanism of persistence or expansion did not require HIV provirus to be present in a specific orientation. Similarly, no detectable correlation between position of integration within the gene and the position of the start of host-gene transcription, as has been reported for HTLV-1 [49].
In contrast to the majority of genes with integrants that were present independent of transcription profile of the host gene, integrants in several genes had a striking and distinct pattern of integration. As described above, Matsushita and co-workers had previously identified multiple integrations in the BACH2 intron gene, all of which were integrated in the same orientation as the transcriptional unit of BACH2, and all in an intron upstream of the translational start site. Subsequently Imamichi and co-workers identified an additional BACH2 integrant in one patient undergoing long-term cART with HIV suppression [29], also in the same orientation as BACH2 (Imamichi, personal communication). In more extensive datasets [52,53], BACH2 and a second gene MKL2 had multiple integrations in introns. Proviruses integrated into BACH2 genes were invariably present in intron 4 and 5, upstream of the translational start site. Similarly, proviruses integrated into MKL2 genes were all located in intron 6 or intron 4. In addition, all of the proviruses in BACH2 or MKL2 were integrated in the same orientation as the transcriptional units of the corresponding genes. Both BACH2 and MKL2 are associated with cell growth and cancer. This arrangement was clearly not the result of preferential integration into a specific region of BACH2 or MKL2 to the exclusion of the remainder of the corresponding genes. In vitro studies identified both BACH2 and MKL2 as common targets for HIV integration, but in vitro, integration was widely distributed throughout BACH2 and MKL2 genes and was not restricted to individual introns as occurs in vivo. In addition, orientation of proviruses was independent of transcription profile. These findings suggest that the entire BACH2 and MKL2 genes represent common sites for HIV integration and which occurs without orientation preference. With time on therapy, those cells with integrations in specific introns (listed above), with proviruses in the same orientation as the corresponding host gene, have a selection advantage that favours cellular persistence, or expansion, or both. The mechanisms by which proviral integration into BACH2 or MKL2 drives persistence and expansion are not known. Although the integration sites and proviral orientations are well documented, the characteristics of the cells with these integrants are not known. In addition, it is not known whether proviruses integrated in these genes are infectious or even intact, as only a portion of the HIV sequence in the region near the integration sites has been sequenced. In addition, the effect of proviral integration in introns of these genes on gene expression remains uncertain, and it is not known whether gene expression is increased or silenced. It is possible that any disruption of the host gene by proviral integration modulates gene activity to promote persistence or expansion.
BACH2, located on chromosome 6 in humans, is a transcription factor in the basic region-leucine zipper family that heterodimerises with MAF oncoprotein elements. These elements bind to Maf recognition elements (MAREs) and regulate transcription through enhancer-binding sequences and exert broad effects in lymphocyte development and function [56]. In lymphocyte differentiation, BACH2 can facilitate B cell development by suppressing myeloid differentiation, and has a prominent role in plasma cell differentiation class switch recombination, hypermutation of immunoglobulin genes and influences glycosylation of immunoglobulins [56–58]. In T cells, functions of BACH2 remain under study, but analysis of transgenic mouse model systems revealed loss of BACH2 was associated with progressive wasting, loss of regulatory T lymphocytes (Treg) function and increases T effector function [59]. BACH2 function remains under study and has been implicated, in various systems, in T cell homeostasis and survival [60,61]. BACH2 has been associated with neoplastic processes; fusion transcripts including BACH2 have been reported with BCL2L1 and IGHCdelta in lymphoma cell lines and in aggressive B cell lymphoma [62,63]. In mantle cell lymphoma, therapy with bortezomib induced apoptotic responses that were associated with BACH2 expression and transport to nucleus, and that cellular distribution of BACH2 determined whether apoptosis occurred [64].
MKL2 (myocardin-related transcription factor-B/megakaryoblastic leukemia 2) gene is also a transcription factor member of the MKL family broadly expressed in many tissues, but has been less well studied compared with BACH2. However, like BACH2, it has been reported to have a number of functions in lymphoid and non-lymphoid cells [65]. Notably, it is reported to function as a potent co-activator of serum response factor (SRF) and participates in inducing expression of a number of SRF target immediate early genes involved in induction of cell cycle, cell growth and differentiation [65–67]. MKL2 has also been implicated in the neoplastic process, and fusion oncogenes have been identified, most consistently in chondroid lipomas, in which a C11orf95-MKL2 fusion has been frequently identified [68,69]. Although useful for diagnosis, the precise role in the neoplastic process remains to be elucidated.
The presence of BACH2 and MKL2 integrants during long-term cART raise intriguing questions about the characterisation of the integrants and their effect on cell function. Although the integrants in these genes were easily detectable, and found in a number of patients across several studies, these specific integrants represent a very small fraction of all infected cells, and it has not been easy to study the expression of BACH2, MKL2 or any gene in a single lineage-specific manner. In addition, integrants in these genes have been readily detected in some patients, but not at all in others; integrations in BACH2 or MKL2 do not appear to be universal in all patients. Nevertheless, determining the mechanisms of how integrations in these genes favour persistence and expansion are likely to yield useful information regarding HIV infection.
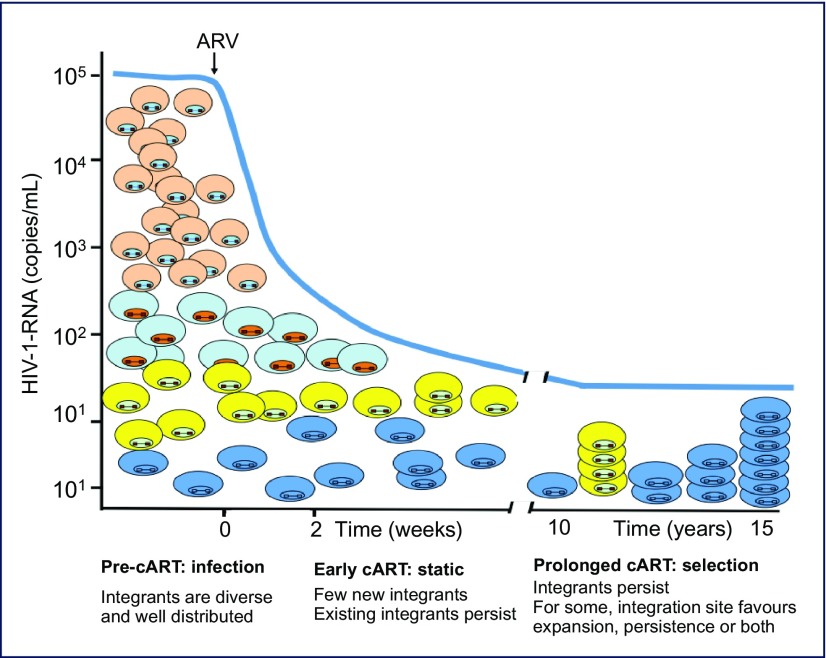
These data demonstrated that HIV integration occurs widely distributed throughout the human genome and is frequently associated with clonal expansion. A working model of this process (Figure 3) suggests that integration is relatively widespread throughout the genome with a few preferences. Clonal expansion may occur during the course of HIV infection, including prior to therapy. With time on therapy, cells with proviruses that persist or expand are frequently those where the provirus is integrated into genes associated with growth promotion. Thus proliferation and persistence are a consequence of a selective benefit resulting from an advantageous integration.
Figure 3.
Model of HIV persistence through clonal amplification. HIV proviruses integrate diversely in the human genome, largely in transcriptionally active genes. After prolonged periods, however, the persistently infected cells that remain have integrants in genes associated with cell growth, suggesting that proviral integration may be involved in selection for persistence. HIV expansion can occur at any time; thus far, integrants have been detected following prolonged cART.
Analysis of HIV integration sites and identification of expanded populations in infected individuals undergoing cART raise a number of questions that are readily and directly testable. The most critical issue is whether clonally expanded cells are capable of producing infectious HIV. As the techniques used to identify integration sites provide only limited sequence information flanking the junction site, it has not been possible to readily identify the individual HIV provirus at specific integration sites. CD4 cells commonly undergo expansion and cells from HIV-infected individuals are capable of cellular expansion to the same degree as cells from uninfected individuals [70,71]. Therefore, in one sense it is not surprising that HIV-infected cells can clonally expand. HIV is often cytopathic, however, and encodes genes (vpr), which have been shown to block cell division in in vitro experiments and so the mechanism of clonal expansion of HIV-infected cells is uncertain. In addition, most integrants, including those present after prolonged cART, are defective, and it is estimated that only 1–4% of all integrated proviruses are replication competent. As a result, it is possible that clonally expanded integrants are those with defective, or deleted proviruses, and do not contribute to the relevant HIV reservoir of replication-competent virus during long-term cART. Cohn et al. [55] reported characterisation of proviruses from a number of clonally expanded HIV-infected cells, and none (0/75) was replication competent.
Simonetti et al. [72] reported an example of a replication-competent HIV variant arising from an expanded clone. In this example, identical HIV sequences were present in plasma over a period of several years, and characterisation of expanded proviruses detected by integration-site analysis identified the provirus responsible for the predominant clone in plasma. Transfection of proviral DNA amplified from this site yielded intact HIV that gave rise to infectious virus when used to infect PBMC in vitro. In addition, the clone could be recovered directly from patient PBMC by in vitro cultivation.
These preliminary studies indicate it is possible for a clonally expanded lineage to give rise to infectious HIV, but do not determine the frequency of clonally expanded populations harbouring infectious virus. As described above, estimates of frequency of clonally expanded populations (c. 40%) are likely to be underestimates, and it is possible that most populations of infected cells persisting on therapy are the products of clonal expansion. Predominant plasma clones are found in low-level viraemia in suppressed patients, but they are not universally present, and many individual sequences are also present. Relative HIV production from these sources remains entirely unexplored, and the degree to which such lineages contribute to the HIV reservoir remains to be determined.
Additional questions raised by these observations have a bearing on HIV pathogenesis. Clonally expanded populations were detected in HIV-infected individuals undergoing cART, but the kinetics of expansion and the establishment of expanded lineages has not been extensively investigated. Wagner et al. reported expansion over time on therapy, and additional studies will further characterise the kinetics of expansion. It is not known when cellular lineages are established, whether they are present during acute HIV infection, to what degree cellular expansion is present prior to initiating antiretroviral therapy and whether the presence of expanded clones has pathogenic consequences. Introduction of cART is generally associated with recovery of CD4 cells, both memory and naïve cell subsets; it is not known whether cell expansions occur as a part of CD4 cell recovery and are correlated with the extent of increases. Cumulative analysis of HIV integration sites revealed a number of interesting features, including examples of expanded integrants with proviruses concentrated in specific introns of genes, such as MKL2 and BACH2. It is not known whether there are more such examples, and what pathways are responsible for persistence and expansion of these lineages. Infected individuals often undergo therapies for comorbid illnesses, which include cytotoxic chemotherapy for neoplastic diseases. It is not known whether such interventions affect HIV lineages or the HIV reservoir. From a pathogenesis standpoint, it is unclear whether expansion of HIV-infected cells plays a role in inhibiting CD4 cell recovery, especially in immune non-responder patients who do not experience substantial CD4 cell recovery. Whether such expansions occur in individuals who have natural control of HIV infection is also unclear. Previous analyses of HIV populations from long-term non-progressors including elite controllers, demonstrated the presence of identical sequences in low-level viraemia, but additional study is necessary to determine whether these clonal populations in plasma are the products of clonal expansion.
Although the effects of clonal expansion on the composition of the reservoir of replication-competent HIV is unknown, measuring clonal expansion will be a useful modality in analysing the effects of the eradication strategies. For instance, in current shock and kill strategies, the effects of latency-reversing agents on clonal expansion remains unknown. Although it is possible that cells activated during the ‘shock’ stage are all eliminated in the ‘kill’ phase, it is likely that some activated cells may undergo expansion. Integration sites are distributed throughout the genome, largely in transcriptional units, and it is not known whether location and epigenetic characteristics [73] of individual integrants will be more readily reactivated than others; comparative analysis of models of integration did not identify clear common chromosomal characteristics favouring reactivation. As reviewed [74,75], there are likely to be multiple mechanisms of latency that will require a multifaceted approach. In future cure research, correlating the extent of clonal expansion with other measures of HIV populations and their replication competence and chromosomal-integration site will prove useful in identifying effective eradication strategies.
Summary
HIV infection can be controlled but not eradicated by antiretroviral therapy. Viral pathogenesis is arrested by continuous antiretroviral therapy, but the mechanisms of persistence are still under study. Initial studies of patients undergoing long-term cART have identified clonally expanded HIV-infected populations that persist for prolonged periods. The observation that integrants themselves may drive persistence and expansion of infected cells raises a number of new questions regarding their role in HIV persistence during therapy. These hypotheses remain readily testable and will provide useful information regarding the source of HIV persistence during therapy.
References
- 1. Moir S, Conners M, Fauci AS.. The immunology of human immunodeficiency virus infection In: Bennett JE, Dolin R, Blaser MJ, eds. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Philadelphia PA USA, Elsevier Saunders, 2014: 1526– 1540. [Google Scholar]
- 2. Siliciano JD, Siliciano RF.. Recent developments in the search for a cure for HIV-1 infection: targeting the latent reservoir for HIV-1. J Allergy Clin Immunol 2014; 134: 12– 19. [DOI] [PubMed] [Google Scholar]
- 3. Siliciano RF. What do we need to do to cure HIV infection. Top HIV Med 2010; 18: 104– 108. [PubMed] [Google Scholar]
- 4. Kearney M, Maldarelli F, Shao W et al. . Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol 2009; 83: 2715– 2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keele BF, Giorgi EE, Salazar-Gonzalez JF et al. . Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008; 105: 7552– 7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maldarelli F, Kearney M, Palmer S et al. . HIV populations are large and accumulate high genetic diversity in a nonlinear fashion. J Virol 2013; 87: 10313– 10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rouzine IM, Coffin JM.. Linkage disequilibrium test implies a large effective population number for HIV in vivo. Proc Natl Acad Sci U S A 1999; 96: 10758– 10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rouzine IM, Coffin JM, Weinberger LS.. Fifteen years later: Hard and soft selection sweeps confirm a large population number for HIV in vivo. PLoS Genet 2014; 10: e1004179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rouzine IM, Rodrigo A, Coffin JM.. Transition between stochastic evolution and deterministic evolution in the presence of selection: general theory and application to virology. Microbiol Mol Biol Rev 2001; 65: 151– 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Besson GJ, Lalama CM, Bosch RJ et al. . HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis 2014; 59: 1312– 1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perelson AS, Neumann AU, Markowitz M et al. . HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 1996; 271: 1582– 1586. [DOI] [PubMed] [Google Scholar]
- 12. Perelson AS, Essunger P, Cao Y et al. . Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 1997; 387: 188– 191. [DOI] [PubMed] [Google Scholar]
- 13. Palmer S, Maldarelli F, Wiegand A et al. . Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 2008; 105: 3879– 3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riddler SA, Aga E, Bosch RJ et al. . Continued slow decay of residual plasma viremia in HIV-1-infected adults on long term antiretroviral therapy. J Infect Dis 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chun TW, Finzi D, Margolick J et al. . In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1995; 1: 1284– 1290. [DOI] [PubMed] [Google Scholar]
- 16. Chun TW, Carruth L, Finzi D et al. . Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387: 183– 188. [DOI] [PubMed] [Google Scholar]
- 17. Finzi D, Hermankova M, Pierson T et al. . Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278: 1295– 1300. [DOI] [PubMed] [Google Scholar]
- 18. Finzi D, Blankson J, Siliciano JD et al. . Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999; 5: 512– 517. [DOI] [PubMed] [Google Scholar]
- 19. Siliciano JD, Kajdas J, Finzi D et al. . Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9: 727– 728. [DOI] [PubMed] [Google Scholar]
- 20. Kearney MF, Spindler J, Shao W et al. . Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog 2014; 10: e1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eriksson S, Graf EH, Dahl V et al. . Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 2013; 9: e1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho YC, Shan L, Hosmane NN et al. . Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155: 540– 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hermankova M, Ray SC, Ruff C et al. . HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 2001; 286: 196– 207. [DOI] [PubMed] [Google Scholar]
- 24. Bailey JR, Sedaghat AR, Kieffer T et al. . Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol 2006; 80: 6441– 6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tobin NH, Learn GH, Holte SE et al. . Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J Virol 2005; 79: 9625– 9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chun TW. Tracking replication-competent HIV reservoirs in infected individuals. Curr Opin HIV AIDS 2013; 8: 111– 116. [DOI] [PubMed] [Google Scholar]
- 27. Chomont N, El-Far M, Ancuta P et al. . HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15: 893– 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Josefsson L, Stockenstrom S, Faria NR et al. . The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 2013; 110: E4987– 4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Imamichi H, Natarajan V, Adelsberger JW et al. . Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS 2014; 28: 1091– 1099. [DOI] [PubMed] [Google Scholar]
- 30. Craigie R, Bushman FD.. HIV DNA integration. Cold Spring Harb Perspect Med 2012; 2: a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hazuda DJ. HIV integrase as a target for antiretroviral therapy. Curr Opin HIV AIDS 2012; 7: 383– 389. [DOI] [PubMed] [Google Scholar]
- 32. Engelman A, Cherepanov P.. Retroviral integrase structure and DNA recombination mechanism. Microbiol Spectr 2014; 2: 1– 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Butler SL, Hansen MS, Bushman FD.. A quantitative assay for HIV DNA integration in vivo. Nat Med 2001; 7: 631– 634. [DOI] [PubMed] [Google Scholar]
- 34. Bushman F, Lewinski M, Ciuffi A et al. . Genome-wide analysis of retroviral DNA integration. Nature Rev Microbiol 2005; 3: 848– 858. [DOI] [PubMed] [Google Scholar]
- 35. Schroder AR, Shinn P, Chen H et al. . HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002; 110: 521– 529. [DOI] [PubMed] [Google Scholar]
- 36. Mitchell RS, Beitzel BF, Schroder AR et al. . Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2004; 2: E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewinski MK, Yamashita M, Emerman M et al. . Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog 2006; 2: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marshall HM, Ronen K, Berry C et al. . Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2007; 2: e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kvaratskhelia M, Sharma A, Larue RC et al. . Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res 2014; 42: 10209– 10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferris AL, Wu X, Hughes CM et al. . Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci U S A 2010; 107: 3135– 3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Engelman A, Cherepanov P.. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog 2008; 4: e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Craigie R, Bushman FD.. Host factors in retroviral integration and the selection of integration target sites. Microbiol Spectr 2014; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brady T, Agosto LM, Malani N et al. . HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS 2009; 23: 1461– 1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sherrill-Mix S, Lewinski MK, Famiglietti M et al. . HIV latency and integration site placement in five cell-based models. Retrovirology 2013; 10: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mack KD, Jin X, Yu S et al. . HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J Acquir Immune Defic Syndr 2003; 33: 308– 320. [DOI] [PubMed] [Google Scholar]
- 46. Ikeda T, Shibata J, Yoshimura K et al. . Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J Infect Dis 2007; 195: 716– 725. [DOI] [PubMed] [Google Scholar]
- 47. Han Y, Wind-Rotolo M, Yang HC et al. . Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol 2007; 5: 95– 106. [DOI] [PubMed] [Google Scholar]
- 48. Niederer HA, Bangham CR.. Integration site and clonal expansion in human chronic retroviral infection and gene therapy. Viruses 2014; 6: 4140– 4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gillet NA, Malani N, Melamed A et al. . The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood 2011; 117: 3113– 3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cook LB, Rowan AG, Melamed A et al. . HTLV-1-infected T cells contain a single integrated provirus in natural infection. Blood 2012; 120: 3488– 3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koh Y, Wu X, Ferris AL et al. . Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J Virol 2013; 87: 648– 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maldarelli F, Wu X, Su L et al. . HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014; 345: 179– 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wagner TA, McLaughlin S, Garg K et al. . HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014; 345: 570– 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wagner TA, McKernan JL, Tobin NH et al. . An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol 2013; 87: 1770– 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cohn LB, Silva IT, Oliveira TY et al. . HIV-1 integration landscape during latent and active infection. Cell 2015; 160: 420– 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Igarashi K, Ochiai K, Itoh-Nakadai A, Muto A.. Orchestration of plasma cell differentiation by Bach2 and its gene regulatory network. Immunol Rev 2014; 261: 116– 125. [DOI] [PubMed] [Google Scholar]
- 57. Nutt SL, Taubenheim N, Hasbold J et al. . The genetic network controlling plasma cell differentiation. Semin Immunol 2011; 23: 341– 349. [DOI] [PubMed] [Google Scholar]
- 58. Vuckovic F, Kristic J, Gudelj I et al. . Systemic lupus erythematosus associates with the decreased immunosuppressive potential of the IgG glycome. Arthritis Rheumatol 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roychoudhuri R, Hirahara K, Mousavi K et al. . BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 2013; 498: 506– 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Uittenboogaard LM, Payan-Gomez C, Pothof J et al. . BACH2: a marker of DNA damage and ageing. DNA Repair 2013; 12: 982– 992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kuwahara M, Suzuki J, Tofukuji S et al. . The Menin-Bach2 axis is critical for regulating CD4 T-cell senescence and cytokine homeostasis. Nat Commun 2014; 5: 3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Turkmen S, Riehn M, Klopocki E et al. . A BACH2-BCL2L1 fusion gene resulting from a t(6;20)(q15;q11.2) chromosomal translocation in the lymphoma cell line BLUE-1. Genes Chromosomes Cancer 2011; 50: 389– 396. [DOI] [PubMed] [Google Scholar]
- 63. Kobayashi S, Taki T, Chinen Y et al. . Identification of IGHCdelta-BACH2 fusion transcripts resulting from cryptic chromosomal rearrangements of 14q32 with 6q15 in aggressive B-cell lymphoma/leukemia. Genes Chromosomes Cancer 2011; 50: 207– 216. [DOI] [PubMed] [Google Scholar]
- 64. Chen Z, Pittman EF, Romaguera J et al. . Nuclear translocation of B-cell-specific transcription factor, BACH2, modulates ROS mediated cytotoxic responses in mantle cell lymphoma. PLoS One 2013; 8: e69126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cen B, Selvaraj A, Prywes R.. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J Cell Biochem 2004; 93: 74– 82. [DOI] [PubMed] [Google Scholar]
- 66. Lee SM, Vasishtha M, Prywes R.. Activation and repression of cellular immediate early genes by serum response factor cofactors. J Biol Chem 2010; 285: 22036– 22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cen B, Selvaraj A, Burgess RC et al. . Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol Cell Biol 2003; 23: 6597– 6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang D, Sumegi J, Dal Cin P et al. . C11orf95-MKL2 is the resulting fusion oncogene of t(11;16)(q13;p13) in chondroid lipoma. Genes Chromosomes Cancer 2010; 49: 810– 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Flucke U, Tops BB, Saint Aubain Somerhausen N et al. . Presence of C11orf95-MKL2 fusion is a consistent finding in chondroid lipomas: a study of eight cases. Histopathology 2013; 62: 925– 930. [DOI] [PubMed] [Google Scholar]
- 70. Langhoff E, McElrath J, Bos HJ et al. . Most CD4+ T cells from human immunodeficiency virus-1 infected patients can undergo prolonged clonal expansion. J Clin Invest 1989; 84: 1637– 1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cameron PU, Forsum U, Teppler H et al. . During HIV-1 infection most blood dendritic cells are not productively infected and can induce allogeneic CD4+ T cells clonal expansion. Clin Exp Immunol 1992; 88: 226– 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Simonetti F, Sobolowski M, Hill S et al. . Residual viremia caused by clonally expanded tumor-infiltrating CD4+ cells. Conference on Retroviruses and Opportunistic Infections. February 2015. Seattle, WA, USA. Abstract 105.
- 73. Hakre S, Chavez L, Shirakawa K, Verdin E.. Epigenetic regulation of HIV latency. Curr Opin HIV AIDS 2011; 6: 19– 24. [DOI] [PubMed] [Google Scholar]
- 74. Ott M, Verdin E.. Three rules for HIV latency: location, location, and location. Cell Host Microbe 2013; 13: 625– 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Katlama C, Deeks SG, Autran B et al. . Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet 2013; 381: 2109– 2117. [DOI] [PMC free article] [PubMed] [Google Scholar]