Abstract
The PIM1 protein is an important regulator of cell proliferation, the cell cycle, apoptosis, and metabolism in various human cancers. MicroRNAs (miRNAs) are powerful post‐transcriptional gene regulators that function through translational repression or transcript destabilization. Therefore, we aimed to identify whether a close relationship exists between PIM1 and miRNAs. PIM1 protein levels and mRNA levels were significantly upregulated in astrocytoma tissues, indicating the oncogenic role of PIM1 in astrocytoma. Further bioinformatics analysis indicated that miR‐124‐3p targeted the 3′‐UTR of PIM1. We also observed an inverse correlation between the miR‐124‐3p levels and PIM1 protein or mRNA levels in astrocytoma samples. Next, we experimentally confirmed that miR‐124‐3p directly recognizes the 3′‐UTR of the PIM1 transcript and regulates PIM1 expression at both the protein and mRNA levels. Furthermore, we examined the biological consequences of miR‐124‐3p targeting PIM1 in vitro. We showed that the repression of PIM1 in astrocytoma cancer cells by miR‐124‐3p suppressed proliferation, invasion, and aerobic glycolysis and promoted apoptosis. We observed that the restoration or inhibition of PIM1 activity resulted in effects that were similar to those induced by miR‐124‐3p inhibitors or mimics in cancer cells. Finally, overexpression of PIM1 rescued the inhibitory effects of miR‐124‐3p. In summary, these findings aid in understanding the tumor‐suppressive role of miR‐124‐3p in astrocytoma pathogenesis through the inhibition of PIM1 translation.
Keywords: Apoptosis, bioenergetics, invasion, miR‐124‐3p, PIM1
Human astrocytomas are the most common and lethal primary tumors of the central nervous system (CNS). The WHO classification divides astrocytomas into four grades (I–IV), with increasing degrees of malignancy.1, 2 Currently, surgical resection and adjuvant chemotherapy with temozolomide combined with radiotherapy are standard treatment strategies for this disease.3 Unfortunately, anaplastic astrocytomas (WHO III) and glioblastomas (GBM; WHO IV) constitute the majority of astrocytomas and are essentially incurable. The median survival time of GBM is only 15 months under standard treatment.4, 5 Hence, with an increasing understanding of the molecular pathology of malignant astrocytoma, novel signaling pathways driving tumorigenesis and progression may become candidates for therapeutic targets and novel agents because effective treatments are urgently needed.
The PIM kinases are a family of serine/threonine kinases that consist of three members, PIM1, PIM2, and PIM3.6 The PIM oncogenes are frequently overexpressed in solid tumors and hematological malignancies.7 Among them, PIM1 is the most interesting because it has been reported to play important roles in regulating cellular proliferation, the cell cycle, apoptosis, and metabolism in various human cancers.7 Despite advances in understanding the important roles of PIM1 in cancer, the precise molecular mechanism through which PIM1 contributes to astrocytoma progression remains largely unknown. MicroRNAs (miRNAs) are a group of small non‐coding regulatory RNA molecules that regulate protein expression by binding to the 3′‐UTR of mRNA, resulting in mRNA degradation or translation inhibition.8, 9 In the past decade, the emergence of miRNAs is one of the most striking developments in cancer biology. Numerous studies have shown that miRNAs play important roles in various physiological and pathological processes during cancer development by controlling the expression of their target genes.10 There is evidence that the deregulation or dysfunction of miRNAs contributes to aberrant cell proliferation, apoptosis, and metabolism in different types of cancer, causing human carcinogenesis and cancer progression.11, 12, 13
In this study, we addressed the key questions on the expression pattern of PIM1 in astrocytoma, the regulatory mechanism between PIM1 and miRNA, and the functional characterization of PIM1 and miRNA in astrocytoma. We found that the PIM1 protein levels and mRNA levels were both upregulated in primary astrocytomas. We detected an inverse correlation between miR‐124‐3p expression and PIM1 at both the protein and mRNA levels in human astrocytomas. Additionally, PIM1 was identified as a direct functional target of miR‐124‐3p in astrocytoma. Furthermore, miR‐124‐3p inhibited cell proliferation, invasion, and bioenergetics and promoted apoptosis in astrocytoma. Finally, PIM1 was responsible for the antitumor effects induced by miR‐124‐3p.
Materials and Methods
Human tissues and cell lines
Fresh tumor specimens and paired normal adjacent tissues (NATs) were obtained from patients with astrocytomas who underwent surgical treatment at the Department of Neurosurgery at the Third Affiliated Hospital of Soochow University (Changzhou, China). All of the cases were histologically confirmed primary astrocytomas. Each participant signed an informed consent form, and the Research Ethics Board of the Third Affiliated Hospital of Soochow University approved this study. The collected tissues were immediately snap‐frozen in liquid nitrogen at the time of surgery and stored at −80°C. The human astrocytoma cell line U251 was purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in DMEM (Thermo Fisher Scientific, Waltham, Massachusetts, USA) supplemented with 100 U penicillin/mL, 100 mg streptomycin/mL, and 10% FBS (Thermo Fisher Scientific, Waltham, Massachusetts, USA) at 37°C with 5% CO2.
RNA isolation and quantitative real‐time PCR
Total RNA was extracted from cultured cells or human tissues using TRIzol Reagent (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer's instructions. The levels of mature miR‐124‐3p were quantified with TaqMan miRNA probes using the ABI 7500 System (Thermo Fisher Scientific, Waltham, Massachusetts, USA) as previously reported.14, 15 Briefly, 2 μg total RNA was reverse transcribed to cDNA using the AMV reverse transcriptase (TaKaRa, Otsu, Shiga, Japan) and a stem‐loop primer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The mixture was incubated at 16°C for 15 min, 42°C for 60 min, and 85°C for 5 min. Quantitative real‐time PCR (qRT‐PCR) was performed using a TaqMan PCR kit and the ABI 7500 System (Thermo Fisher Scientific, Waltham, Massachusetts, USA) under the conditions of 95°C for 5 min followed by 40 cycles consisting of 95°C for 15 s and 60°C for 1 min. The relative levels of miR‐124‐3p in cells and tissues were normalized to that of U6. To quantify PIM1 mRNA, 2 μg total RNA was reverse transcribed to cDNA using oligo(dT) (Takara, Otsu, Shiga, Japan) with the conditions of 42°C for 60 min and 70°C for 10 min. Quantitative RT‐PCR was carried out using the SYBR Green PCR Master Mix (Takara, Otsu, Shiga, Japan) and ABI 7500 System (Thermo Fisher Scientific, Waltham, Massachusetts, USA). β‐Actin served as an endogenous control. The sequences of the primers were as follows: PIM1 (sense), 5′‐CGGCAAGTTGTCGGAGACG‐3′; PIM1 (antisense), 5′‐CCTGGAGGTTGGGATGCTCT‐3′; β‐actin (sense), 5′‐AGGGAAATCGTGCGTGAC‐3′; and β‐actin (antisense), 5′‐CGCTCATTGCCGATAGTG‐3′. The reactions were incubated at 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The expression level of PIM1 mRNA was normalized to that of β‐actin. All real‐time PCRs were carried out in triplicate.
Protein isolation and Western blot analysis
The cells were washed twice with ice‐cold PBS (pH 7.4) and lysed on ice in RIPA buffer (Beyotime, Shanghai, China) supplemented with a protease and phosphatase inhibitor cocktail tablet (Roche, Basel, Switzerland). The tissues were frozen solid with liquid nitrogen, ground into a powder, and lysed on ice in RIPA buffer. The protein concentration was calculated using the Pierce BCA protein assay kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Western blot analyses were carried out according to the standard protocol, as previously described.15, 16 The following antibodies were used: PIM1 (Abcam, Cambridge, UK), β‐actin (Cell Signaling Technology, Danvers, Massachusetts, USA), and IgG‐HRP (Sigma, St. Louis, Missouri, USA). Protein levels were detected using the ECL detection solution (Thermo Fisher Scientific, Waltham, Massachusetts, USA), visualized on the Bio‐Rad ChemiDocXRS (Bio‐Rad, Hercules, California, USA) and quantified by Quantity One software (Bio‐Rad, Hercules, California, USA).
Cell transfection
The miR‐124‐3p mimic (pre‐miR‐124‐3p), mimic negative control (pre‐ncRNA), miR‐124‐3p inhibitor (anti‐miR‐124‐3p), and inhibitor negative control (anti‐ncRNA) were purchased from GenePharma (Shanghai, China). Three siRNAs targeting PIM1 and an siRNA negative control were designed by and purchased from GenePharma. The PIM1‐targeting sequence of siRNA‐1 was 5′‐GGUGUGUGGAGAUAUUCCUTT‐3′ and 5′‐AGGAAUAUCUCCACACACCTT‐3′. The PIM1‐targeting sequence of siRNA‐2 was 5′‐GGUUUCUCCGGCGUCAUUATT‐3′ and 5′‐ UAAUGACGCCGGAGAAACCTT‐3′. The PIM1‐targeting sequence of siRNA‐3 was 5′‐GGCCAACCUUCGAAGAAAUTT‐3′ and 5′‐AUUUCUUCGAAGGUUGGCCTT‐3′. The siRNA negative control sequence was 5′‐UUCUCCGAACGUGUCACGUTT‐3′ and 5′‐ACGUGACACGUUCGGAGAATT‐3′. Cells were transfected with oligonucleotides at indicated concentrations with Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer's protocol.
Luciferase assay
The 3′‐UTR of PIM1 was amplified and cloned into the p‐MIR‐reporter plasmid (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and confirmed by sequencing. The PIM1 3′‐UTR was mutated and inserted into the firefly luciferase reporter to test for binding specificity. The luciferase assay was carried out using a similar strategy to that previously reported, according to the manufacturer's instructions.15 The data were acquired from three independent experiments.
Cell proliferation assay
U251 cells were seeded at 5000 cells/well in 96‐well plates and transfected with different oligonucleotides. At various time points, 20 μL MTT (Sigma, St. Louis, Missouri, USA) was added to each well and incubated for another 4 h at 37°C. The plates were centrifuged, and the supernatant was discarded. Next, 150 μL DMSO was added to each well at room temperature. The absorbance was measured at a wavelength of 570 nm using the BioTek Elx800 (BioTek, Winooski, Vermont, USA). Experiments were carried out in triplicate.
Cell invasion assay
In vitro cell invasion was quantified using Transwell plates coated with Matrigel (BD, Franklin Lakes, New Jersey, USA), as previously reported.15 Briefly, serum‐starved cells were trypsinized and added to the upper Transwell chambers coated with Matrigel (BD, Franklin Lakes, New Jersey, USA). The lower chambers were filled with fresh medium containing 10% FBS as a chemoattractant. After a 48‐h incubation, non‐invading cells were removed from the upper surface with a cotton swab, and the lower surface was fixed in 4% formaldehyde (Sigma, St. Louis, Missouri, USA) and stained with 0.1% crystal violet (Sigma, St. Louis, Missouri, USA). The cells were photographed in six randomly selected fields using an IX71 microscope (Olympus, Shinjuku‐ku, Tokyo, Japan) with Image‐Pro Insight software (Olympus, Shinjuku‐ku, Tokyo, Japan). The mean number of invading cells was expressed as a percentage relative to the control. The data represent the mean ± SD from at least three independent experiments.
Apoptosis assay
The transfected cells were collected, washed twice with cold PBS, and resuspended in binding buffer. The apoptosis assay was carried out using an annexin V‐FITC apoptosis detection kit (Beyotime, Shanghai, China) on the Guava EasyCyte 6HT‐2L flow cytometer (Merck Millipore, Darmstadt, Germany) according to the manufacturer's protocol, as previously reported.15
Seahorse XF‐96 metabolic flux analysis
The extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were measured using a Seahorse XF‐96 extracellular flux analyzer (Seahorse, Boston, Massachusetts, USA). The cells were cultured on Seahorse XF‐96 plates at a density of 8 × 104 cells per well until 85% confluence. Before analysis, the cell culture medium was replaced with unbuffered DMEM (DMEM supplemented with 25 mM glucose, 1 mM sodium pyruvate, 31 mM NaCl, 2 mM GlutaMax, and phenol red, pH 7.4) and incubated at 37°C for 1 h in a non‐CO2 incubator. Baseline rates were measured four times at 37°C. The mitochondrial inhibitors oligomycin, carbonylcyanide p‐(trifluoromethoxy) phenylhydrazone, and rotenone were sequentially injected. All of the injection reagents were adjusted to pH 7.4. Four readings were taken after the addition of each inhibitor. The ECAR and OCR were automatically calculated by Seahorse XF‐96 software (Seahorse, Boston, Massachusetts, USA). Each data point represents an average of six different wells.
Statistical analysis
Data were analyzed with GraphPad Prism 5 (GraphPad Software, La Jolla, California, USA). The data shown are presented as the mean ± SD of at least three independent experiments. Two‐tailed Student's t‐test was used for the comparison of two independent groups, and a P‐value < 0.05 was considered statistically significant.
Results
PIM1 is upregulated in astrocytomas
First, we determined the expression patterns of PIM1 in astrocytoma tissues by Western blotting and qRT‐PCR. The clinical features of astrocytoma patients are shown in Table 1. By measuring the protein expression levels of PIM1 in 12 paired astrocytomas and their corresponding NATs, we found that the PIM1 protein levels were commonly overexpressed in astrocytomas (Fig. 1a,b). Subsequently, we used qRT‐PCR to measure the mRNA levels of PIM1 in the same 12 paired samples. We found that the PIM1 mRNA levels were also significantly upregulated in astrocytomas (Fig. 1c). Next, we analyzed the relationship between the WHO grade and PIM1 expression levels based on the 12 samples. As shown in Figure 1(d), there was an obvious trend that the mean fold‐change of PIM1 progressively increased from WHO grade II to WHO grade IV astrocytomas.
Table 1.
Clinical features of patients with astrocytoma
| Case no. | Sex | Age, years | WHO grade |
|---|---|---|---|
| 1 | Male | 69 | III |
| 2 | Male | 52 | III |
| 3 | Male | 49 | III |
| 4 | Male | 47 | IV |
| 5 | Female | 49 | IV |
| 6 | Male | 37 | IV |
| 7 | Female | 43 | II |
| 8 | Female | 60 | IV |
| 9 | Female | 48 | IV |
| 10 | Male | 49 | IV |
| 11 | Female | 53 | III |
| 12 | Male | 48 | II |
Figure 1.
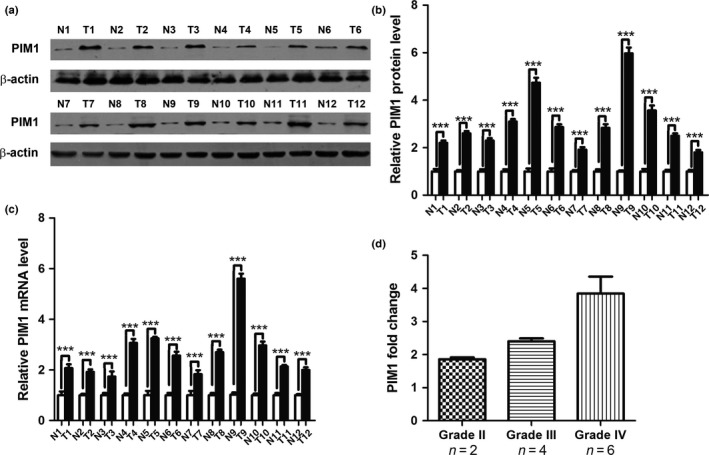
PIM1 protein and mRNA expression levels in astrocytoma tissues. (a) Western blot analysis of PIM1 protein expression levels in 12 pairs of astrocytoma (T) and normal adjacent tissue (N) samples. (b) Quantitative analysis of the PIM1 protein expression levels in 12 pairs of N and T samples (***P < 0.001). (c) Quantitative RT‐PCR analysis of relative PIM1 mRNA expression levels in 12 pairs of N and T samples (***P < 0.001). (d) Fold changes of PIM1 in WHO grade II, III, and IV samples. The fold change of PIM1 protein expression in each sample was calculated by comparing the PIM1 expression in astrocytoma and its normal adjacent tissue.
PIM1 3′‐UTR is targeted by miR‐124‐3p
Because miRNAs play key roles in the regulation of gene expression through post‐transcriptional mechanisms, we asked whether the oncogenic kinase PIM1 is a target of miRNAs in astrocytoma. Two computational algorithms, TargetScan17 and microRNA.org,18 were used in combination to search for potential miRNAs that target PIM1. In our previous study, miR‐124‐3p was confirmed to be downregulated in astrocytomas and was therefore selected for further analysis.19 The predicted interactions between miR‐124‐3p and the target sites in the PIM1 3′‐UTR are illustrated in Figure 2(a). Two potential miR‐124‐3p target sites were found in the PIM1 3′‐UTR mRNA sequence. The minimum free energy values of the two hybrids were −14.6 and −19.8 kcal/mol, respectively, which were well within the range of genuine miRNA target pairs. Moreover, there is perfect base‐pairing between the cognate targets and seed region. Furthermore, the miR‐124‐3p binding sequences in the PIM1 3′‐UTR are highly conserved across species (Fig. 2a). Based on the notion that miRNAs have an opposite expression pattern compared with their target genes, we examined the expression of miR‐124‐3p in the same 12 pairs of astrocytoma tissues and corresponding NATs. Using qRT‐PCR analysis, we found that miR‐124‐3p levels were significantly lower in astrocytoma tissues than in NATs (Fig. 2b). The correlation between the miR‐124‐3p expression levels and PIM1 protein levels (Fig. 2c), and the correlation between the miR‐124‐3p expression levels and PIM1 mRNA levels (Fig. 2d) in the 12 pairs of astrocytoma tissues and corresponding NATs were further illustrated using Pearson's correlation scatter plots. The results showed that there was an inverse correlation between the protein levels of PIM1 or mRNA levels of PIM1 and miR‐124‐3p (r = −0.8955, P < 0.001; r = −0.8120, P < 0.001) in human astrocytomas. These results suggest that the mechanism for the miR‐124‐3p regulation of PIM1 expression is not only through a post‐transcriptional mechanism but also through mRNA stability.
Figure 2.
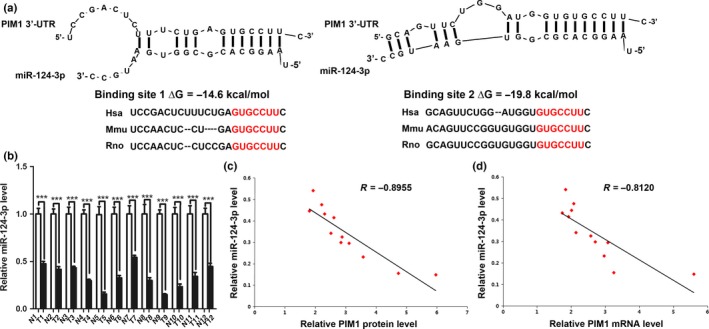
Inverse correlation between MicroRNA (miR)‐124‐3p and PIM1 levels in astrocytomas. (a) Schematic illustration of the hypothesized duplexes formed from interactions between the PIM1 3′‐UTR binding sites and miR‐124‐3p, with the predicted free energy of each hybrid indicated. The seed matching sequences are highly conserved across species. (b) Quantitative RT‐PCR analysis of miR‐124‐3p expression levels in 12 pairs of astrocytoma (T) and normal adjacent tissue (N) samples (***P < 0.001). (c) Pearson's correlation scatter plot for the fold change of miR‐124‐3p and PIM1 protein levels in human astrocytomas. (d) Pearson's correlation scatter plot of the fold change of miR‐124‐3p and PIM1 mRNA levels in astrocytomas.
To determine whether the regulatory mechanism of miR‐124‐3p on PIM1 expression occurred through miR‐124‐3p binding to the presumed sites in the PIM1 3′‐UTR, U251 cells were infected with the miR‐124‐3p mimic and inhibitor, along with their respective negative controls. The efficient overexpression and knockdown of miR‐124‐3p in U251 cells is shown in Figure 3(a). The miR‐124‐3p expression level was significantly increased after transfection with pre‐miR‐124‐3p, whereas its expression was decreased after transfection with anti‐miR‐124‐3p, compared with their corresponding negative controls. We next investigated whether miR‐124‐3p influenced PIM1 expression at the protein or mRNA level. The miR‐124‐3p mimic and inhibitor, along with their respective negative controls, were transfected into U251 cells, and the levels of PIM1 protein and mRNA were examined. At 48 h post‐transfection, the expression levels of PIM1 protein were assessed by Western blot analysis. MicroRNA‐124‐3p overexpression significantly reduced PIM1 expression by approximately 50%, while miR‐124‐3p inhibition slightly enhanced PIM1 expression by approximately 20%, relative to their respective controls (Fig. 3b,c). Because one miRNA might decrease the levels of its specific target gene by destabilizing the mRNA, PIM1 transcript levels were also evaluated at 24 h post‐transfection. The alterations in PIM1 mRNA levels induced by pre‐miR‐124‐3p or anti‐miR‐124‐3p were similar to the alterations at the protein level (Fig. 3d). However, the upregulation of PIM1 mRNA induced by miR‐124‐3p downregulation was higher than the variation in PIM1 protein level. To determine whether the negative regulatory effects of miR‐124‐3p on PIM1 expression were mediated by the binding of miR‐124‐3p to the presumed binding sites in the PIM1 mRNA 3′‐UTR, we fused the full‐length PIM1 3′‐UTR, containing the two presumed miR‐124‐3p binding sites, downstream of the firefly luciferase reporter in a plasmid. The plasmid was transfected into U251 cells along with β‐gal as the control plasmid and pre‐ncRNA, pre‐miR‐124‐3p, anti‐ncRNA, or anti‐miR‐124‐3p. As shown in Figure 3(e), pre‐miR‐124‐3p significantly decreased the luciferase reporter activity compared with pre‐ncRNA treatment, while anti‐miR‐124‐3p significantly increased the reporter activity. Furthermore, we introduced point mutations into the corresponding complementary sites in the 3′‐UTR of PIM1 to eliminate the predicted miR‐124‐3p binding sites (both binding positions were mutated). This mutated luciferase reporter was unaffected by either the overexpression or knockdown of miR‐124‐3p (Fig. 3e). These results strongly indicated that PIM1 was directly targeted by miR‐124‐3p, which downregulated PIM1 expression by directly recognizing its 3′‐UTR. Our data suggest that miR‐124‐3p regulates the expression of PIM1 at both the transcript and protein levels.
Figure 3.
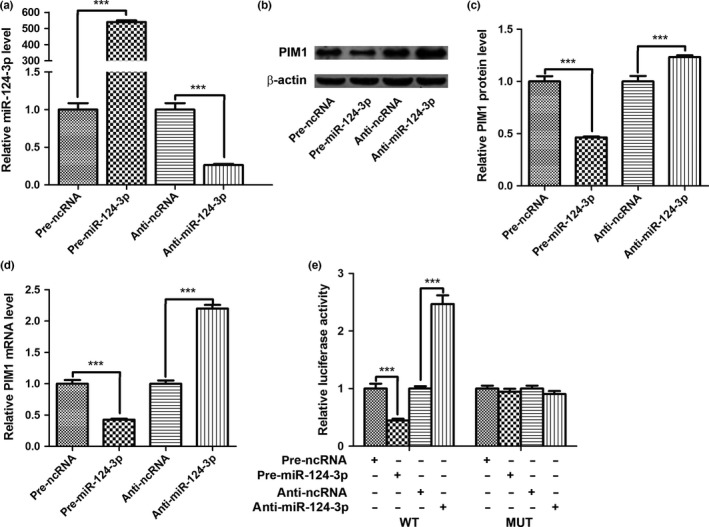
MicroRNA (miR)‐124‐3p directly targets the PIM1 gene. (a) Quantitative RT‐PCR analysis of miR‐124‐3p expression in U251 cells after transfection with miR‐124‐3p mimic negative control (pre‐ncRNA), miR‐124‐3p mimic (pre‐miR‐124‐3p), inhibitor negative control (anti‐ncRNA), or miR‐124‐3p inhibitor (anti‐miR‐124‐3p). (b) Representative Western blots showing the PIM1 protein levels in U251 cells after transfection with pre‐ncRNA, pre‐miR‐124‐3p, anti‐ncRNA, or anti‐miR‐124‐3p. (c) Quantitative analysis of PIM1 protein expression levels after different treatments. Statistical analysis was carried out on three independent experiments. (d) Quantitative RT‐PCR analysis of PIM1 mRNA expression levels in U251 cells treated with pre‐ncRNA, pre‐miR‐124‐3p, anti‐ncRNA, or anti‐miR‐124‐3p. The results represent data from three independent experiments. (e) Firefly luciferase reporters containing wild‐type (WT) or mutant (MUT) miR‐124‐3p binding sites in the PIM1 3′‐UTR were cotransfected into U251 cells along with pre‐ncRNA, pre‐miR‐124‐3p, anti‐ncRNA, or anti‐miR‐124‐3p. Twenty‐four hours after transfection, the cells were assayed using a luciferase assay kit. ***P < 0.001.
MicroRNA‐124‐3p inhibits cell proliferation, invasion, and bioenergetics, while promoting apoptosis
Because PIM1 is known to be involved in the regulation of cell proliferation, invasion, and apoptosis, we investigated the biological functions of miR‐124‐3p in astrocytoma cells. We transfected equal concentrations of pre‐ncRNA, pre‐miR‐124‐3p, anti‐ncRNA, or anti‐miR‐124‐3p into U251 cells. The MTT assay was used to analyze cell proliferation, and the results showed that the ectopic expression of miR‐124‐3p caused proliferation to be reduced, whereas the reduced expression of miR‐124‐3p caused the enhancement of cell proliferation. The relative cell survival rate of the pre‐miR‐124‐3p‐transfected cells at 72 h was 74.8%, and the relative cell survival rate of the anti‐miR‐124‐3p transfected cells at 72 h was 115.8% (Fig. 4a). Next, we assessed the role of miR‐124‐3p in cell invasion using the Transwell assay. As shown in Figure 4(b), the invasive ability of cancer cells transfected with pre‐miR‐124‐3p was reduced to only 19% of that of the controls, whereas the invasive ability of cancer cells transfected with anti‐miR‐124‐3p was slightly enhanced to 120% of that of the controls; the results of the statistical analysis are shown in Figure 4(c). Next, we investigated the effects of miR‐124‐3p on cell apoptosis using flow cytometry analysis. As shown in Figure 4(d), the percentage of apoptotic cells in the pre‐miR‐124‐3p‐transfected group was significantly increased; in contrast, the percentage of apoptotic cells in the anti‐miR‐124‐3p‐transfected group was slightly decreased. Furthermore, we determined the effect of miR‐124‐3p on cellular energetics by measuring ECAR and OCR, which are indicators of lactic acid production through aerobic glycolysis and mitochondrial respiration in cells, respectively. A substantial block in glycolysis was also observed in the pre‐miR‐124‐3p group, as shown by a significant decrease in ECAR relative to the pre‐ncRNA group, whereas a slight increase in glycolysis, compared with the anti‐ncRNA group, was observed in the anti‐miR‐124‐3p group (Fig. 4e). However, the OCR was not significantly changed between these groups (Fig. 4f). In summary, these results suggested that the ectopic expression of miR‐124‐3p in cancer cells in vitro inhibited cell growth, invasion, and aerobic glycolysis while promoting apoptosis.
Figure 4.
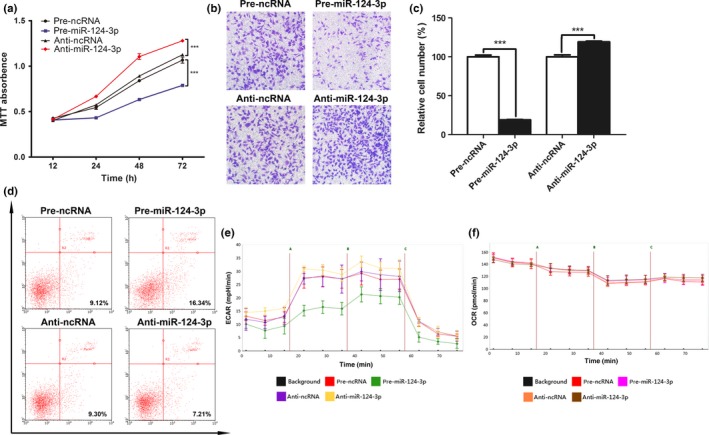
Role of microRNA (miR)‐124‐3p in cell proliferation, invasion, apoptosis, and bioenergetics in vitro. (a) Role of miR‐124‐3p in cell proliferation. The MTT assay was carried out at 0, 12, 24, 48, and 72 h. ***P < 0.001. (b) Role of miR‐124‐3p in cell invasion. The representative images are shown from at least three independent experiments. (c) Statistical analysis of the cell invasion rate. ***P < 0.001. (d) Role of miR‐124‐3p in cell apoptosis. The experiment was repeated three times, and representative data are shown. (e) Role of miR‐124‐3p in aerobic glycolysis. (f) Role of miR‐124‐3p in mitochondrial respiration. (a, c, e, f) Data represent the mean ± SD from three separate experiments. Anti‐miR‐124‐3p, miR‐124‐3p inhibitor; anti‐ncRNA, inhibitor negative control; ECAR, extracellular acidification rate; OCR, oxygen consumption rate; pre‐miR‐124‐3p, miR‐124‐3p mimic; pre‐ncRNA, miR‐124‐3p mimic negative control.
MicroRNA‐124‐3p inhibits proliferation and invasion by targeting PIM1
To confirm that PIM1 was involved in the antitumor effects of miR‐124‐3p, we first carried out targeted knockdown of PIM1 expression by RNAi using si‐PIM1 in U251 cells. Three siRNAs were designed to target PIM1, and Western blot analysis was carried out to evaluate their inhibition efficiencies. The most efficient inhibition was shown by siRNA‐1, followed by siRNA‐2 and siRNA‐3 (Fig. 5a). In the following experiments, siRNA‐1 was chosen to knockdown PIM1. A vector containing the full‐length ORF of PIM1 without its 3′‐UTR was constructed and transfected into U251 cells, and PIM1 expression was significantly enhanced compared with the negative control (Fig. 5b). The cell proliferation assay indicated that PIM1 overexpression significantly promoted cell growth, whereas knocking down PIM1 had the reverse effect in U251 cells (Fig. 5c). Consequently, the inhibitory effect induced by si‐PIM1 was stronger than that induced by pre‐miR‐124‐3p. PIM1 overexpression or downregulation caused similar results in the cell invasion assay (Fig. 5d). These results indicated that PIM1 regulation could phenocopy the effects of miR‐124‐3p. Next, we examined whether the introduction of PIM1 without the 3′‐UTR could rescue the antitumor effects of miR‐124‐3p. Vectors encoding PIM1 without the miR‐124‐3p‐responsive 3′‐UTR and miR‐124‐3p were cotransfected into U251 cells. The ectopic expression of PIM1 significantly rescued the suppression of proliferation (Fig. 5e) and invasion (Fig. 5f) induced by miR‐124‐3p in vitro in U251 cells, indicating that miR‐124‐3p exerts its tumor suppression function by suppressing PIM1 expression through direct binding to the 3′‐UTR of PIM1 in astrocytomas.
Figure 5.
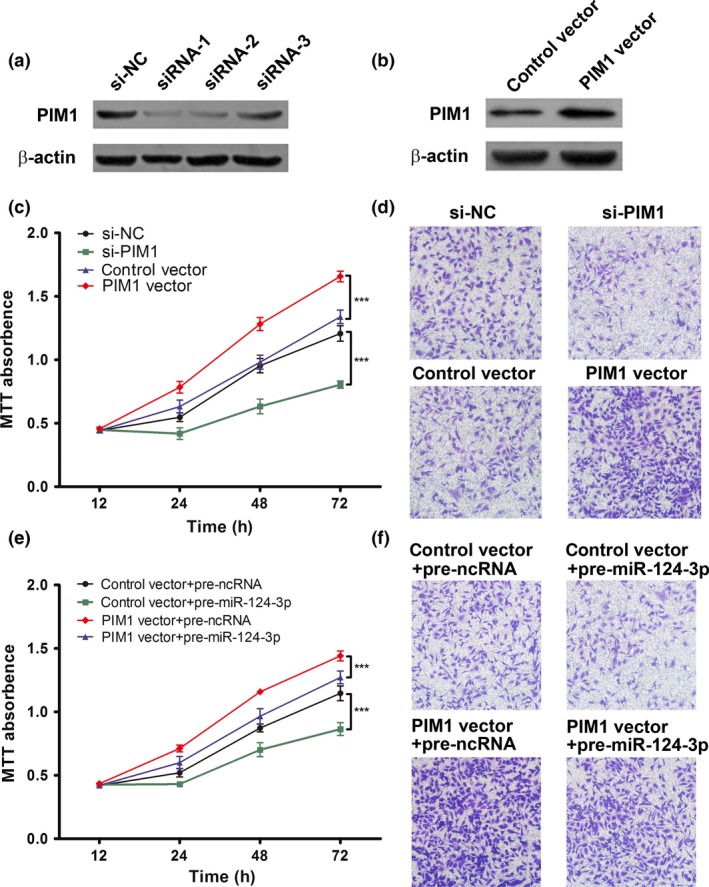
PIM1 is involved in the antitumor effects of microRNA (miR)‐124‐3p. (a) Three siRNAs targeting different PIM1 sites were transfected into U251 cells. The PIM1 protein levels were determined by Western blot analysis. (b) Representative Western blot image of PIM1 protein levels in U251 cells treated with control vector or PIM1 vector. (c) Role of PIM1 in cell proliferation. MTT cell viability assay was carried out at 0, 12, 24, 48, and 72 h. Results are presented as the mean ± SD of three independent experiments. ***P < 0.001. (d) Role of PIM1 in cell invasion. The images shown represent at least three independent experiments. (e) Ectopic expression of PIM1 significantly reversed the proliferation suppression induced by miR‐124‐3p. (f) Ectopic expression of PIM1 significantly reversed the invasion suppression induced by miR‐124‐3p. The experiment was repeated three times, and representative data are shown. Anti‐miR‐124‐3p, miR‐124‐3p inhibitor; anti‐ncRNA, inhibitor negative control; pre‐miR‐124‐3p, miR‐124‐3p mimic; pre‐ncRNA, miR‐124‐3p mimic negative control; si‐NC, siRNA negative control.
Discussion
In this study, we first used a set of clinical samples to identify the overexpression of PIM1 protein and mRNA in human astrocytomas. Next, PIM1 was experimentally confirmed as a direct target gene of miR‐124‐3p. Furthermore, miR‐124‐3p inhibited cell proliferation, invasion, and bioenergetics, while promoting apoptosis, in vitro. Finally, miR‐124‐3p exerted its antitumor effects in astrocytomas by regulating PIM1. Our results suggest the potential value of miR‐124‐3p and PIM1 as novel therapeutic targets of astrocytoma.
PIM1 belongs to the PIM family, a group of highly evolutionarily conserved serine/threonine kinases. The PIM family proteins aid in the regulation of apoptosis, metabolism, the cell cycle, homing, and migration, explaining why these proteins are interesting targets for anticancer drug discovery.7 PIM1 is frequently overexpressed or aberrantly activated in a range of cancers. In hepatocellular carcinoma, PIM1 enhances glycolysis and promotes tumor progression.20 In acute myeloid leukemia, PIM1 regulates cell migration, invasion, and cell scattering by regulating MET.21 In prostate cancer, PIM1 overexpression is associated with increased proliferation by stabilizing the prostate‐specific tumor suppressor NKX3.1.22 In nasopharyngeal carcinoma, PIM1 promotes cancer cell proliferation and migration.23 In diffuse large B‐cell lymphoma, nuclear PIM1 expression is associated with disease stage and increased proliferative activity through the activation of the JAK/STAT signaling pathway.24 In non‐small‐cell lung cancer, PIM1 promotes radioresistance by regulating irradiation‐induced signaling pathways.25 In breast cancer, PIM1 overexpression is responsible for tumor progression and carcinogenesis.26 In this study, we showed that silencing PIM1 expression using siRNA inhibits cell proliferation and invasion in astrocytomas, whereas overexpressing PIM1 had opposite effects, thus validating its role as an essential oncogene during tumorigenesis. MicroRNAs are important regulators of gene expression, and we experimentally validated miR‐124‐3p as a direct regulator of PIM1. In summary, our results identified an miRNA as a link between the PIM1 regulatory pathway and pathogenesis of astrocytoma. It is well known that multiple miRNAs can target a single gene. PIM1 is also suppressed by miR‐486‐5p in breast cancer27 and by miR‐33a in colon carcinoma.28 However, this is the first report that PIM1 is directly regulated by miR‐124‐3p in astrocytoma.
MicroRNA‐124‐3p is the most abundant miRNA in the brain and is involved in stem cell regulation and neurodevelopment.29, 30 Currently, it is widely accepted that miR‐124‐3p affects a broad spectrum of biological processes in the CNS, such as CNS development, neurodegeneration, CNS stress, neuroimmunity, stroke, and brain tumor.31 MicroRNA‐124‐3p could inhibit tumor progression from many biological aspects by targeting different genes in glioma and specifically inhibits cell proliferation by targeting IQ motif containing GTPase activating protein 1 (IQGAP1),32 son of sevenless homolog 1 (SOS1),33 protein phosphatase 1 regulatory subunit 13 like (PPP1R13L),34 and cell differentiation agent‐2 (CDA‐2).35 MicroRNA‐124‐3p inhibits cell migration and invasion by targeting IQGAP1,32 rho‐associated coiled‐coil containing protein kinase 1,36 PPP1R13L,34 and calpain small subunit37 in glioma. MicroRNA‐124‐3p also induces cell differentiation in glioma by targeting CDA‐235 or with decreased CDK6 expression38 and inhibits glioma stem‐like traits through Snail family zinc finger 2.39 MicroRNA‐124‐3p inhibits glioma angiogenesis and enhances chemosensitivity by targeting R‐Ras and N‐Ras.40 It also inhibits tumor progression by counteracting pro‐survival stress responses in GBM.41 The antitumor effect of miR‐124‐3p in GBM cells is enhanced through gap junctions by partly decreasing CDK6 expression.42 Furthermore, miR‐124‐3p is associated with malignant tumor progression and poor prognosis in glioma patients43 and is related to recurrent high‐grade gliomas in Mexican children,44 indicating that miR‐124‐3p is a potential biomarker for glioma. Therefore, one miRNA can target multiple genes and affect various cellular pathways, and this study clearly showed that PIM1 is also a direct target gene of miR‐124‐3p. MicroRNA‐124‐3p greatly suppresses the malignancy of astrocytoma through the inhibition of cell proliferation, invasion, and bioenergetics, while inducing apoptosis through PIM1 suppression. In summary, our results show that miR‐124‐3p acts as a tumor suppressor by targeting PIM1 in astrocytoma and may provide new therapeutic strategies for astrocytoma prevention and treatment.
Altered energy metabolism is widely observed in cancer cells and has been accepted as a core hallmark of cancer.45 MicroRNA‐7‐5p inhibits cellular growth and glucose metabolism by directly targeting insulin‐like growth factor 1 receptor in glioma.46 MicroRNA‐106a‐5p inhibits glioma cell glucose uptake by targeting solute carrier family 2 (facilitated glucose transporter), member 3 in glioblastoma.47 MicroRNA‐143‐3p inhibits glycolysis and promotes differentiation by targeting hexokinase 2 in glioblastoma stem‐like cells.48 MicroRNA‐451a regulates cellular adaptation to metabolic stress in glioma by regulating the serine/threonine kinase 11/AMP‐activated protein kinase pathway.49 MicroRNA‐495‐3p increases glucose uptake and lactate production by targeting Glut1 in glioma.50 PIM1 has been reported to promote the activity of the rapamycin‐sensitive mammalian target of rapamycin, which regulates cell growth and metabolism,51 thereby increasing hypoxia‐inducible factor 1α abundance and the expression of hypoxia‐inducible factor 1α‐associated glycolytic enzyme and Glc transporter.52 It is very possible that miR‐124‐3p overexpression inhibits aerobic glycolysis through its target gene PIM1 downregulation. Aberrant miRNA expression enables cancer cells to meet the elevated anabolic and energetic demands during tumor growth that may be exploited as new drug targets. Angiogenesis is an important aspect in malignant glioma. Shi et al.40 reported that miR‐124‐3p suppresses glioma angiogenesis in nude mice by targeting R‐Ras and N‐Ras. However, there are no other reports concerning the role of miR‐124‐3p or PIM1 in glioma angiogenesis. In bladder cancer, Wang et al.53 reported that miR‐124‐3p inhibits cell proliferation, motility, and angiogenesis by targeting ubiquitin‐like, containing PHD and RING finger domains, 1. Chen et al.54 reported that PIM1 kinase promotes angiogenesis in vascular endothelial cells by regulating the activation of endothelial NO synthase at Ser‐633. It is believed that further studies are needed to determine the role of miR‐124‐3p or PIM1 in glioma angiogenesis.
In summary, our data indicated that miR‐124‐3p is a tumor suppressor gene in astrocytomas. Furthermore, we showed that PIM1 is a direct target of miR‐124‐3p. Our results suggest the potential value of miR‐124‐3p and PIM1 as new therapeutic targets for astrocytoma.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81302197, 31071046), Jiangsu Provincial Special Program of Medical Science (Grant No. BL2014035), Changzhou Social Development Project (Grant No. CS20102010), and Changzhou Health Bureau Project (Grant No. ZD201007).
Cancer Sci 107 (2016) 899–907
Funding Information
National Natural Science Foundation of China (31071046, 81302197); Jiangsu Provincial Special Program of Medical Science (BL2014035); Changzhou Social Development Project (CS20102010); Changzhou Health Bureau Project (ZD201007).
References
- 1. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008; 359: 492–507. [DOI] [PubMed] [Google Scholar]
- 2. Rousseau A, Mokhtari K, Duyckaerts C. The 2007 WHO classification of tumors of the central nervous system – what has changed? Curr Opin Neurol 2008; 21: 720–7. [DOI] [PubMed] [Google Scholar]
- 3. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro‐oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin 2010; 60: 166–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gabayan AJ, Green SB, Sanan A et al GliaSite brachytherapy for treatment of recurrent malignant gliomas: a retrospective multi‐institutional analysis. Neurosurgery 2006; 58: 701–9; discussion ‐9. [DOI] [PubMed] [Google Scholar]
- 5. Luo JW, Wang X, Yang Y, Mao Q. Role of micro‐RNA (miRNA) in pathogenesis of glioblastoma. Eur Rev Med Pharmacol Sci 2015; 19: 1630–9. [PubMed] [Google Scholar]
- 6. Xu J, Zhang T, Wang T, You L, Zhao Y. PIM kinases: an overview in tumors and recent advances in pancreatic cancer. Future Oncol 2014; 10: 865–76. [DOI] [PubMed] [Google Scholar]
- 7. Narlik‐Grassow M, Blanco‐Aparicio C, Carnero A. The PIM family of serine/threonine kinases in cancer. Med Res Rev 2014; 34: 136–59. [DOI] [PubMed] [Google Scholar]
- 8. Esteller M. Non‐coding RNAs in human disease. Nat Rev Genet 2011; 12: 861–74. [DOI] [PubMed] [Google Scholar]
- 9. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 10. Dvinge H, Git A, Graf S et al The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013; 497: 378–82. [DOI] [PubMed] [Google Scholar]
- 11. Esquela‐Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–69. [DOI] [PubMed] [Google Scholar]
- 12. Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010; 10: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu J, Getz G, Miska EA et al MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–8. [DOI] [PubMed] [Google Scholar]
- 14. Zhi F, Zhou G, Shao N et al miR‐106a‐5p inhibits the proliferation and migration of astrocytoma cells and promotes apoptosis by targeting FASTK. PLoS ONE 2013; 8: e72390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhi F, Wang Q, Deng D et al MiR‐181b‐5p downregulates NOVA1 to suppress proliferation, migration and invasion and promote apoptosis in astrocytoma. PLoS ONE 2014; 9: e109124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhi F, Gong G, Xu Y et al Activated beta‐catenin forces N2A cell‐derived neurons back to tumor‐like neuroblasts and positively correlates with a risk for human neuroblastoma. Int J Biol Sci 2012; 8: 289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lewis BP, Shih IH, Jones‐Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003; 115: 787–98. [DOI] [PubMed] [Google Scholar]
- 18. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol 2004; 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhi F, Chen X, Wang S et al The use of hsa‐miR‐21, hsa‐miR‐181b and hsa‐miR‐106a as prognostic indicators of astrocytoma. Eur J Cancer 2010; 46: 1640–9. [DOI] [PubMed] [Google Scholar]
- 20. Leung CO, Wong CC, Fan DN et al PIM1 regulates glycolysis and promotes tumor progression in hepatocellular carcinoma. Oncotarget 2015; 6: 10880–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cen B, Xiong Y, Song JH et al The Pim‐1 protein kinase is an important regulator of MET receptor tyrosine kinase levels and signaling. Mol Cell Biol 2014; 34: 2517–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Padmanabhan A, Gosc EB, Bieberich CJ. Stabilization of the prostate‐specific tumor suppressor NKX3.1 by the oncogenic protein kinase Pim‐1 in prostate cancer cells. J Cell Biochem 2013; 114: 1050–7. [DOI] [PubMed] [Google Scholar]
- 23. Jie W, He QY, Luo BT et al Inhibition of Pim‐1 attenuates the proliferation and migration in nasopharyngeal carcinoma cells. Asian Pac J Trop Med 2012; 5: 645–50. [DOI] [PubMed] [Google Scholar]
- 24. Brault L, Menter T, Obermann EC et al PIM kinases are progression markers and emerging therapeutic targets in diffuse large B‐cell lymphoma. Br J Cancer 2012; 107: 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim W, Youn H, Kwon T et al PIM1 kinase inhibitors induce radiosensitization in non‐small cell lung cancer cells. Pharmacol Res 2013; 70: 90–101. [DOI] [PubMed] [Google Scholar]
- 26. Malinen M, Jaaskelainen T, Pelkonen M et al Proto‐oncogene PIM‐1 is a novel estrogen receptor target associating with high grade breast tumors. Mol Cell Endocrinol 2013; 365: 270–6. [DOI] [PubMed] [Google Scholar]
- 27. Zhang G, Liu Z, Cui G, Wang X, Yang Z. MicroRNA‐486‐5p targeting PIM‐1 suppresses cell proliferation in breast cancer cells. Tumour Biol 2014; 35: 11137–45. [DOI] [PubMed] [Google Scholar]
- 28. Thomas M, Lange‐Grunweller K, Weirauch U et al The proto‐oncogene Pim‐1 is a target of miR‐33a. Oncogene 2012; 31: 918–28. [DOI] [PubMed] [Google Scholar]
- 29. Lee MR, Kim JS, Kim KS. miR‐124a is important for migratory cell fate transition during gastrulation of human embryonic stem cells. Stem Cells 2010; 28: 1550–9. [DOI] [PubMed] [Google Scholar]
- 30. Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR‐124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 2009; 12: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun Y, Luo ZM, Guo XM, Su DF, Liu X. An updated role of microRNA‐124 in central nervous system disorders: a review. Front Cell Neurosci 2015; 9: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu SH, Jiang XJ, Xiao GL, Liu DY, Yuan XR. miR‐124a restoration inhibits glioma cell proliferation and invasion by suppressing IQGAP1 and beta‐catenin. Oncol Rep 2014; 32: 2104–10. [DOI] [PubMed] [Google Scholar]
- 33. Lv Z, Yang L. MiR‐124 inhibits the growth of glioblastoma through the downregulation of SOS1. Mol Med Rep 2013; 8: 345–9. [DOI] [PubMed] [Google Scholar]
- 34. Zhao WH, Wu SQ, Zhang YD. Downregulation of miR‐124 promotes the growth and invasiveness of glioblastoma cells involving upregulation of PPP1R13L. Int J Mol Med 2013; 32: 101–7. [DOI] [PubMed] [Google Scholar]
- 35. Xie YK, Huo SF, Zhang G et al CDA‐2 induces cell differentiation through suppressing Twist/SLUG signaling via miR‐124 in glioma. J Neurooncol 2012; 110: 179–86. [DOI] [PubMed] [Google Scholar]
- 36. An L, Liu Y, Wu A, Guan Y. microRNA‐124 inhibits migration and invasion by down‐regulating ROCK1 in glioma. PLoS ONE 2013; 8: e69478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cai JJ, Qi ZX, Chen LC, Yao Y, Gong Y, Mao Y. miR‐124 suppresses the migration and invasion of glioma cells in vitro via Capn4. Oncol Rep 2016; 35: 284–90. [DOI] [PubMed] [Google Scholar]
- 38. Silber J, Lim DA, Petritsch C et al miR‐124 and miR‐137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 2008; 6: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xia H, Cheung WK, Ng SS et al Loss of brain‐enriched miR‐124 microRNA enhances stem‐like traits and invasiveness of glioma cells. J Biol Chem 2012; 287: 9962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shi Z, Chen Q, Li C et al MiR‐124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R‐Ras and N‐Ras. Neuro Oncol 2014; 16: 1341–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mucaj V, Lee SS, Skuli N et al MicroRNA‐124 expression counteracts pro‐survival stress responses in glioblastoma. Oncogene 2015; 34: 2204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suzhi Z, Liang T, Yuexia P et al Gap junctions enhance the antiproliferative effect of microRNA‐124‐3p in glioblastoma cells. J Cell Physiol 2015; 230: 2476–88. [DOI] [PubMed] [Google Scholar]
- 43. Chen T, Wang XY, Li C, Xu SJ. Downregulation of microRNA‐124 predicts poor prognosis in glioma patients. Neurol Sci 2015; 36: 131–5. [DOI] [PubMed] [Google Scholar]
- 44. Eguia‐Aguilar P, Perezpena‐Diazconti M, Benadon‐Darszon E et al Reductions in the expression of miR‐124‐3p, miR‐128‐1, and miR‐221‐3p in pediatric astrocytomas are related to high‐grade supratentorial, and recurrent tumors in Mexican children. Childs Nerv Syst 2014; 30: 1173–81. [DOI] [PubMed] [Google Scholar]
- 45. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012; 21: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang B, Sun F, Dong N et al MicroRNA‐7 directly targets insulin‐like growth factor 1 receptor to inhibit cellular growth and glucose metabolism in gliomas. Diagn Pathol 2014; 9: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dai DW, Lu Q, Wang LX et al Decreased miR‐106a inhibits glioma cell glucose uptake and proliferation by targeting SLC2A3 in GBM. BMC Cancer 2013; 13: 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao S, Liu H, Liu Y et al miR‐143 inhibits glycolysis and depletes stemness of glioblastoma stem‐like cells. Cancer Lett 2013; 333: 253–60. [DOI] [PubMed] [Google Scholar]
- 49. Godlewski J, Nowicki MO, Bronisz A et al MicroRNA‐451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell 2010; 37: 620–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nie S, Li K, Huang Y, Hu Q, Gao X, Jie S. miR‐495 mediates metabolic shift in glioma cells via targeting Glut1. J Craniofac Surg 2015; 26: e155–8. [DOI] [PubMed] [Google Scholar]
- 51. Zhang F, Beharry ZM, Harris TE et al PIM1 protein kinase regulates PRAS40 phosphorylation and mTOR activity in FDCP1 cells. Cancer Biol Ther 2009; 8: 846–53. [DOI] [PubMed] [Google Scholar]
- 52. Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG Jr. TSC2 regulates VEGF through mTOR‐dependent and ‐independent pathways. Cancer Cell 2003; 4: 147–58. [DOI] [PubMed] [Google Scholar]
- 53. Wang X, Wu Q, Xu B et al miR‐124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine‐tuning UHRF1. FEBS J 2015; 282: 4376–88. [DOI] [PubMed] [Google Scholar]
- 54. Chen M, Yi B, Zhu N et al Pim1 kinase promotes angiogenesis through phosphorylation of endothelial nitric oxide synthase at Ser‐633. Cardiovasc Res 2016; 109: 141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]


