Abstract
DNA-encoded synthesis can generate vastly diverse screening libraries of arbitrarily complex molecules as long as chemical reaction conditions do not compromise DNA’s informational integrity, a fundamental constraint that “DNA-compatible” reaction development does not presently address. We devised DNA-encoded reaction rehearsal, an integrated analysis of reaction yield and impact on DNA, to acquire these key missing data. Magnetic DNA-functionalized sensor beads quantitatively report the % DNA template molecules remaining viable for PCR amplification after exposure to test reaction conditions. Analysis of solid-phase bond forming (e.g., Suzuki-Miyaura cross-coupling, reductive amination) and deprotection reactions (e.g., allyl esters, silyl ethers) guided the definition and optimization of DNA-compatible reaction conditions (> 90% yield, > 30% viable DNA molecules), most notably in cases that involved known (H+, Pd) and more obscure (Δ, DMF) hazards to DNA integrity. The data provide an empirical yet mechanistically consistent and predictive framework for designing successful DNA-encoded reaction sequences for combinatorial library synthesis.
Graphical Abstract
At its inception, combinatorial synthesis represented a revolutionary departure from the throughput limitations of serial compound library synthesis. Split-and-pool diversification1,2 readily yields one bead one compound (OBOC) collections3 that contain in excess of one million members, though library chemotypes that facilitate post-screening mass spectrometric (MS) hit structure elucidation (e.g. α-peptides, peptoids) dominate the landscape. Encoded synthesis circumvents this MS structure elucidation problem by storing structural information in co-synthesized tags,4,5 such as nucleic acids,4,6–13 electrophores,14 peptides,5,15–17 and peptoids.18
DNA-encoded synthesis in particular has recently garnered intense interest. The widespread availability of advanced high-throughput DNA sequencing technology for decoding,19 molecular libraries that can eclipse OBOC library diversity by 2 orders of magnitude, and selection-mode hit identification collectively provide a small molecule discovery platform that is highly complementary to conventional high-throughput screening.11,20–25 Further DNA-encoded reaction development has yielded increasingly “drug-like” libraries20–23,26–28 and adaptation to DNA-encoded solid-phase synthesis (DESPS) promises a broader scope of chemistry for library preparation by liberating synthesis from the constraints of DNA’s limited solubility profile.9,29
The pursuit of expanded DNA-encoded chemical reaction scope has renewed interest in the concept itself: what constitutes “DNA-compatible” chemistry? Unlike other encoding strategies that utilize inert encoding chemotypes (e.g., PNAs, hydrocarbons),14,30 most DNA-encoded library synthesis protocols involve chemical synthesis on an unprotected DNA substrate,11,20–28,31–33 thus compatible reactions generally must proceed under conditions that solubilize the DNA substrate and do not modify or destroy the information encoded therein. HPLC-based analysis of reaction product yield on a DNA substrate is now a routine method for evaluating a reaction’s synthesis compatibility with unprotected DNA,9 and as a result, recent reaction development using this approach focuses solely on solubility and synthesis yield. The impact of synthesis conditions on DNA’s informational integrity, and consequently its viability in PCR to generate templates for sequencing-based decoding, is currently not a consideration in the development of new DNA-encoded reactions.32–35
The issue of DNA’s informational integrity became a pressing issue during DNA-encoded solid-phase reaction development.9,29 Most solid-phase synthesis conditions, which tend to be highly aggressive, resulted in significant loss of PCR-amplifiable encoding DNA, often below the limit of robust PCR detection (~100 molecules/resin particle).29 However, further investigation revealed trends in reactivity, guiding the development of optimal acylation conditions that generated product in high yield while preserving sufficient PCR-amplifiable encoding DNA for post-synthesis structure elucidation. DNA-encoded syntheses routinely employ up to 3 such transformations. Therefore, a DESPS resin particle harboring ~10,000 DNA-encoding tags could undergo a sequence of 3 chemical transformations, each leaving at least 30% of PCR amplifiable DNA-encoding tags, and maintain a sufficient quantity of DNA for robust PCR amplification and decoding. These considerations highlighted the need for a general method to evaluate candidate reaction conditions for both high product yield and compatibility with the DNA-encoding tag to guide the design of successful DNA-encoded libraries.
Results and Discussion
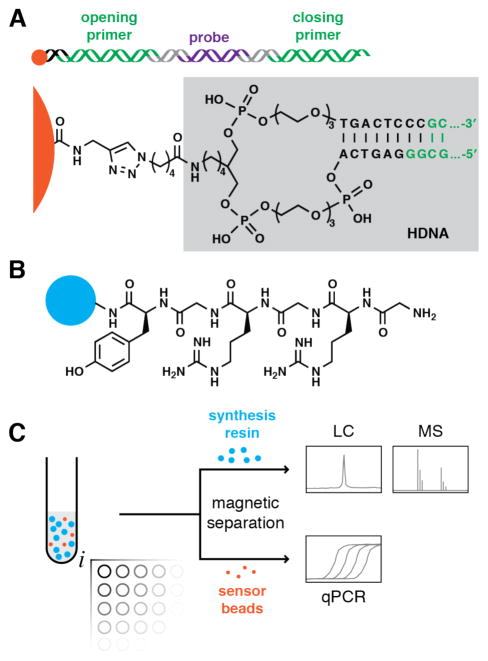
Early work in combinatorial synthesis that highlighted the importance of testing each monomer and reaction condition of a library synthesis, dubbed “rehearsal,”36 inspired us to explore an analogous strategy for encoded synthesis. DNA-encoded reaction rehearsal (Figure 1), a quantitative measurement of reaction product yield and the impact of synthesis conditions on the amplifiability of DNA, uses a mixture of magnetic beads functionalized with a dsDNA encoding tag (“sensor beads”) and synthesis resin functionalized with a short peptide linker. Aliquots of mixed sensor beads and synthesis resin then undergo test reaction conditions, followed by magnetic separation of sensor beads from synthesis resin and analysis. Cleavage and HPLC/MALDI-TOF MS analysis of resin-bound material provides synthesis product yield (henceforth “yield”). Quantitative PCR (qPCR) analysis of sensor beads measures the amplifiable DNA remaining after exposure to the test reaction conditions (normalized to the unexposed sensor bead stock, henceforth “% DNA remaining”), which corresponds to the amplifiable DNA tags remaining for post-synthesis PCR detection and subsequent sequencing. Using this strategy, we investigated and optimized (where needed) a variety of solid-phase reactions.
Figure 1. DNA-encoded reaction rehearsal.
(A) Each reaction condition test includes dsDNA-functionalized magnetic “sensor beads” (orange). The 131-base pair dsDNA contains a 5′-exonuclease probe binding site (purple) with flanking PCR primer binding sites (green), and is attached via DNA headpiece (HDNA, gray box).20 (B) Synthesis resin (blue) for assessing solid-phase reactions displays a linker containing tyrosine for UV quantitation and arginine to facilitate MS analysis. (C) Aliquots of mixed synthesis resin and sensor beads are subjected to test reaction conditions (e.g., varying reactant, solvent, concentration) in filtration microplates. Sensor beads are magnetically separated from synthesis resin, and analyzed in parallel by qPCR to determine % DNA remaining. Yield of cleaved synthesis resin bound products is analyzed by HPLC and MALDI-TOF MS.
Protecting group manipulations figure prominently in solid-phase synthesis, however, numerous deprotection strategies involve conditions known to compromise DNA structure. We examined a suite of deprotection reactions (Table 1), optimizing them for DESPS compatibility by identifying conditions that minimally impact DNA amplifiability while generating high product yield. Exposure to Pd(PPh3)4 during O-Allyl deprotection results in 0.1% DNA remaining, however, changing the order of addition (phenylsilane addition prior to Pd(PPh3)4) significantly preserves DNA (40% DNA remaining) while still affording > 95% yield. Pd likely coordinates the N7 position of adenine, causing depurination and concomitant phosphodiester bond cleavage.37 Deprotection of silyl ethers to reveal alcohols using the oligonucleotide synthesis reagent TBAF is poor (3% DNA remaining), but TEA • 3HF treatment38 gives satisfactory yield of alcohol and 70% DNA remaining. In addition to the previously examined DESPS Fmoc deprotection,29 Mtt-deprotection in the presence of 1% TFA proceeds with 50% amplifiable DNA remaining. An alternate route to primary amines, the Staudinger reduction of an azide with TCEP, proceeded with high yield and negligible impact on DNA after overnight incubation in buffered solution. Amine product yield is equally high after TCEP treatment in DMF, but TCEP • HCl acidifies the DMF, degrading the DNA (< 0.2% DNA remaining).
Table 1.
Solid-phase DNA-encoded reaction rehearsal.
| Solid-phase reaction | % Yield | % Amplifiable DNA remaining |
|---|---|---|
|
|
>95 | 60 |
|
|
>95 | 50 |
|
|
>95 | 90 |
|
|
>95 | 40 |
|
|
64–88 | 70 |
|
|
>95 | 80 |
|
|
>95 | 51 |
|
|
>95 | 17 |
|
|
>95 | 57 |
|
|
>95 | 40 |
|
|
>95 | 70 |
|
|
>95 | 80 |
|
|
>95 | 61 |
|
|
90 | ~100 |
|
|
93 | 47 |
See Supporting Information for full structures. a) Piperidine (20%, DMF, RT), b) TFA (1%, DCM, 15 min, RT), c) TCEP (pH 7.6, 16 h, 50 °C), d) Pd(PPh3)4/phenylsilane (4.3/610 mM, DCM, 30 min, RT), e) TEA•3HF (16 h, RT), f) benzylamine (1 M, DMF, 3 h, 37 °C), g) Fmoc-Pro-OH/HOAt/DIC (40/40/57 mM, DMF, 1 h, 37 °C), h) Fmoc-Ser(tBu)-OH/Oxyma/DIC/TMP (80/80/100/80 mM, DMF, 3 h, 37 °C), i) chloroacetic acid/HOAt/DIC (40/40/57 mM, DMF, 1 h, 37 °C), j) (R/E)-5-chloro-2,4-dimethylpent-3-enoic acid/HOAt/DIC (40/40/57 mM, DMF, 1 h, 37 °C), k) acetic anhydride (20%, DMF, 15 min, RT), l) PyAOP/HOAt/DIEA (45/93/134 mM, DMF, 3 h, 37 °C), m) 4-(chloromethyl)phenyl isocyanate (40 mM, DMF, 15 min, RT), n) 4-iodobenzaldehyde (0.5 M, 1% AcOH in DMF, 10 min, RT), o) NaCNBH3 (0.5 M, 1% AcOH in MeOH, 1 h, RT), p) Pd(PPh3)4/4-isopropylphenylboronic acid/DIEA (0.13/344/690 mM, NMP, 7 h, 70 °C).
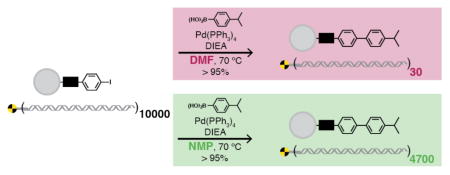
Bond forming reactions are a second important class of transformations in solid-phase synthesis, and like deprotection, can compromise DNA’s viability as a template for PCR amplification. We evaluated and optimized common bond forming reaction conditions for DESPS compatibility. Amide bond formation is the quintessential high-yielding solid-phase reaction. Conditions for acylation of either primary or secondary amines at > 95% yield are different due to the reduced reactivity of secondary amines.29 Harsher secondary amine acylation conditions leave only 17% DNA remaining compared to 51% DNA remaining after exposure to milder conditions of primary amine acylation. Formation of urea linkages following nucleophilic addition to an isocyanate proceeds with high yield and 61% DNA remaining. Reductive amination39 of an aldehyde with a resin-bound amine proved to be an ideal reaction, proceeding with 90% yield and no detectable loss in DNA amplifiability. Suzuki-Miyaura cross-coupling,40 a carbon-carbon bond forming reaction of a boronic acid and resin-bound aryl iodide, was optimized to 93% yield of biphenyl product and 47% DNA remaining.
DNA-encoded reaction rehearsal is a quantitative and predictive tool for selecting library synthesis reaction conditions. Optimal reaction conditions balance high single-step yield and preserve sufficient DNA remaining for robust post-synthesis detection by PCR amplification. High yield (ideally > 90%) ensures that the DNA-encoded synthesis history predicts the major product. Meanwhile, the synthesis resin DNA-encoding tag loading capacity and PCR detection limit jointly define the sustainable severity and frequency of DNA-damaging reaction conditions. For example, a 10-μm resin particle with an encoding tag loading capacity of 10,000 molecules can undergo a sequence of up to 2 cycles of secondary amine acylation with a haloacid followed by nucleophilic displacement with an amine, and maintain at least ~100 DNA molecules necessary for detection using quantitative PCR and subsequent sequencing.29 The same resin could support ~10 iterative reductive amination and deprotection cycles.
While DNA-encoded rehearsal data directly inform reaction selection for solid-phase library synthesis, solution-phase libraries are much more prevalent in the literature and introduce potentially new considerations for defining chemistry compatibility. Whereas DESPS resin particle products exhibit quantifiable encoding tag redundancy, in that each resin particle displays multiple copies of the same encoding tag, solution-phase DNA-encoded synthesis products are single compounds each displaying a single encoding tag. A handful of studies now exist describing the preparation and screening of solution-phase libraries that incorporate diverse reactions—including those rehearsed above—though only with conversion yield data.20–23,32–34 We sought to determine whether our assay could be extensible to the investigation of solution-phase DNA-encoded reaction development, and whether and how those data could apply to solution-phase library planning and synthesis.
DNA-encoded reaction rehearsal can simulate solution-phase DNA-encoded library synthesis by combining magnetic sensor beads with a 5′-amino-modified oligonucleotide to evaluate synthesis product yield on a soluble DNA substrate.9,20,33–35 Following a test reaction, sensor beads are analyzed as before, and the solution is analyzed by HPLC and MALDI-TOF MS to determine the conversion of 5′-amino oligonucleotide starting material to DNA-conjugated product (Table 2). Condensation of 5′-amino oligonucleotide substrate with Fmoc-Lys(Alloc)-OH in the presence of the activator DMT-MM, a staple amide bond forming reaction in DNA-encoded library synthesis,20,41 proceeds with 65% yield and 69% DNA remaining. Subsequent Alloc deprotection34 yields a mix of Alloc-deprotected (70%) and fully deprotected product (30%), and 50% DNA remaining. Published Suzuki-Miyaura cross-coupling conditions33 leave 30% of amplifiable DNA remaining. Conventional solution-phase library preparation includes HPLC purification, likely eliminating synthesis products attached to Pd-catalyzed DNA cleavage products (see Supporting Information).37 The particularly aggressive pH and temperature of published DNA-compatible quinazolinone formation34 unsurprisingly proved to be quite detrimental to encoding tag integrity (< 0.2% DNA remaining).
Table 2.
| Solution-phase reaction | % Yield | % Amplifiable DNA remaining |
|---|---|---|
|
|
95 | 70 |
|
|
70* | 50 |
|
|
N.D. | 90/80 |
|
|
65 | 69 |
|
|
N.D. | 0.2 |
|
|
>95 | 30 |
See Supporting Information for full structures. a) Piperidine (10%, H2O, 4 h, RT), b) Pd(PPh3)4/NaBH4 (1.4/29 mM, 1:1:7 ACN/DMA/250 mM borate pH 9.4, 2 h, RT), c) NaOAc/MgCl2 (30/200 mM, H2O, 16 h, 70 ºC), d) 250 mM borate, pH 9.4 (16 h, 90 ºC), e) Fmoc-Lys(Alloc)-OH/DMT-MM (27/27 mM, 18% DMF/H2O, 18 h 4 ºC), f) NaOH (0.5 M, 125 mM borate pH 9.4, 16 h, 90 ºC), g) 3,4-(methylenedioxy)phenylboronic acid/Pd(PPh3)4/Na2CO3 (18/0.89/36 mM, 2:4:9:226 DCM/Toluene/ACN/H2O, 1.5 h, 80 ºC).
70% yield of expected product, 30% yield of expected product-Fmoc.
Employing reactions that significantly degrade DNA-encoding tag PCR amplifiability during library synthesis may have serious consequences for later sequencing and decoding. Consider, for example, a 1-pmol aliquot of a 25-million member solution-phase library, which would contain an average of 24,000 copies of each library member if all library reaction conditions cause an equal loss of amplifiable DNA. However, if a single reaction condition results in ~1,000-fold greater loss of DNA (e.g., quinazolinone formation) than average conditions, only 24 detectable copies of any library member product of that reaction will remain. Whether such inadvertent bias in encoding tag amplifiability will negatively impact solution-phase library selections is unclear since published libraries20–23,26–28 do not yet incorporate such aggressive chemistry. Rehearsal data suggest, though, that chemistry incompatibility can silence single library members or whole structural families within the library if the loss in DNA due to any single reaction or sequence of reactions decreases the number of amplifiable DNA copies below the PCR limit of detection. These observations with respect to encoding fidelity draw parallels to recent theoretical studies,42 which predict that variable synthetic yield during DNA-encoded library synthesis may bias next-generation sequencing analysis of hits.
DNA-encoded reaction rehearsal provides a practical window into reaction compatibility with DNA, but is by no means comprehensive. First, % DNA remaining measured with sensor beads may not correlate exactly with the actual % DNA remaining after solution-phase library synthesis. The kinetic barrier associated with reactants diffusing to the sensor bead surface is not present in solution-phase encoded synthesis, thus the sensor beads provide a conservative basis for establishing solution-phase chemistry compatibility. Second, the sensor beads are quantitative but phenotypic sensors in that they only report DNA viability in PCR. Some reagents (e.g., hydroxylamine) induce mutation, which would destroy information stored in DNA, yet minimally impact amplifiability in PCR and thus escape notice. Furthermore, the assay yields neither mechanistic insight into how the reaction compromises DNA amplifiability, nor routes to mitigate the damage.
Assay imperfections aside, DNA-encoded reaction rehearsal provides a critical layer of detail for identification of problematic reaction components and potential strategies for reaction optimization. Chemistry incompatibility can be due to known reactivities of DNA,43 such as acid-promoted depurination and concomitant phosphodiester bond cleavage.44 However, some issues are apparent only after rehearsal, and dissection of the suspect reaction into components can illuminate problematic species and hint at remedies. Rehearsal pinpointed Pd as problematic in allyl ester deprotection, consistent with previous data.37 Adding phenylsilane prior to Pd(PPh3)4 mitigates the effect. In solid-phase Suzuki-Miyaura40 reactions conditions,45 DMF decomposition at elevated temperature was principally responsible for the loss in amplifiable DNA, likely the product of radical-mediated DNA cleavages due to DMF decomposition.46 The optimized reaction conditions accommodate the key change of solvent to NMP, yielding a truly DNA-compatible reaction.
These studies comprise an expanded suite of reactions for DNA-encoded synthesis, but more importantly, DNA-encoded reaction rehearsal provides a general and quantitative tool for defining DNA-compatible reactions and reaction sequences for library synthesis. Starting material solubility and yield of DNA-conjugated product9,32–35 provide important preliminary constraints on reaction selection, but DNA-compatible reaction conditions ultimately must preserve sufficient detectable encoding DNA following synthesis and screening while maintaining sufficiently high synthesis product yield. Coupling rehearsal data with knowledge of specific mechanisms of DNA degradation provides a further layer of insight into selecting and designing ideal reactions for DNA-encoded library synthesis that proceed with high yield and negligible loss of amplifiable DNA (e.g., reductive amination). The resultant expanded latitude in DNA-encoded synthesis will yield libraries encompassing more diverse connectivities and molecular content, translating broadly to the discovery of more “drug-like” leads or larger oligomers exhibiting self-assembly or catalytic properties.
Supplementary Material
Acknowledgments
We thank Mr. Vuong Dang for experimental assistance.
Funding
A Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC) to M.L.M., and awards from the NIH Director’s New Innovator (OD008535) and DARPA Fold F(X) programs to B.M.P. (N66001-14-2-4057) supported this research.
Abbreviations
- OBOC
one bead one compound
- DESPS
DNA-encoded solid-phase synthesis
- HDNA
headpiece DNA
- TCEP
tris(2-carboxyethyl)phosphine
- DMT-MM
4-(4,6-eimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride
- qPCR
quantitative polymerase chain reaction
- DMF
N,N-dimethylformamide
- NMP
N-methyl-2-pyrrolidone
Footnotes
Notes
The authors declare no competing financial interests.
The Supporting Information is available free of charge on the ACS Publications website at http://pubs.acs.org. Detailed experimental procedure, linker structures, compound characterization data, qPCR sample data.
References
- 1.Furka A, Sebestyén F, Asgedom M, Dibo G. General-Method for Rapid Synthesis of Multicomponent Peptide Mixtures. Int J Pept Protein Res. 1991;37(6):487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 2.Houghten RA, Pinilla C, Blondelle SE, Appel JR. Generation and Use of Synthetic Peptide Combinatorial Libraries for Basic Research and Drug Discovery. Nature. 1991;354(6348):84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 3.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A New Type of Synthetic Peptide Library for Identifying Ligand-Binding Activity. Nature. 1991;354(6348):82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 4.Brenner S, Lerner RA. Encoded Combinatorial Chemistry. Proc Natl Acad Sci USA. 1992;89(12):5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu R, Marik J, Lam KS. A Novel Peptide-Based Encoding System for “One-Bead One-Compound” Peptidomimetic and Small Molecule Combinatorial Libraries. J Am Chem Soc. 2002;124(26):7678–7680. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- 6.Needels MC, Jones DG, Tate EH, Heinkel GL, Kochersperger LM, Dower WJ, Barrett RW, Gallop MA. Generation and Screening of an Oligonucleotide-Encoded Synthetic Peptide Library. Proc Natl Acad Sci USA. 1993;90(22):10700–10704. doi: 10.1073/pnas.90.22.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen J, Brenner S, Janda KD. Synthetic Methods for the Implementation of Encoded Combinatorial Chemistry. J Am Chem Soc. 1993;115(21):9812–9813. [Google Scholar]
- 8.Winssinger N, Ficarro S, Schultz PG, Harris JL. Profiling Protein Function with Small Molecule Microarrays. Proc Natl Acad Sci USA. 2002;99(17):11139–11144. doi: 10.1073/pnas.172286899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halpin DR, Lee JA, Wrenn SJ, Harbury PB. DNA Display III. Solid-Phase Organic Synthesis on Unprotected DNA. PLoS Biol. 2004;2(7):1031–1038. doi: 10.1371/journal.pbio.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. DNA-Templated Organic Synthesis and Selection of a Library of Macrocycles. Science. 2004;305(5690):1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melkko S, Scheuermann J, Dumelin CE, Neri D. Encoded Self-Assembling Chemical Libraries. Nat Biotechnol. 2004;22(5):568–574. doi: 10.1038/nbt961. [DOI] [PubMed] [Google Scholar]
- 12.Svensen N, Díaz-Mochón JJ, Bradley M. Decoding a PNA Encoded Peptide Library by PCR: the Discovery of New Cell Surface Receptor Ligands. Chem Biol. 2011;18(10):1284–1289. doi: 10.1016/j.chembiol.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Zambaldo C, Barluenga S, Winssinger N. PNA-Encoded Chemical Libraries. Curr Opin Chem Biol. 2015;26:8–15. doi: 10.1016/j.cbpa.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Ohlmeyer M, Swanson RN, Dillard LW, Reader JC, Asouline G, Kobayashi R, Wigler M, Still WC. Complex Synthetic Chemical Libraries Indexed with Molecular Tags. Proc Natl Acad Sci USA. 1993;90(23):10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerr JM, Banville SC. Encoded Combinatorial Peptide Libraries Containing Non-Natural Amino Acids. J Am Chem Soc. 1993;115(6):2529–2531. [Google Scholar]
- 16.Geysen HM, Wagner CD, Bodnar WM, Markworth CJ, Parke GJ, Schoenen FJ, Wagner DS, Kinder DS. Isotope or Mass Encoding of Combinatorial Libraries. Chem Biol. 1996;3(8):679–688. doi: 10.1016/s1074-5521(96)90136-2. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Zhang J, Song A, Lebrilla CB, Lam KS. Encoding Method for OBOC Small Molecule Libraries Using a Biphasic Approach for Ladder-Synthesis of Coding Tags. J Am Chem Soc. 2004;126(18):5740–5749. doi: 10.1021/ja049322j. [DOI] [PubMed] [Google Scholar]
- 18.Song A, Zhang J, Lebrilla CB, Lam KS. A Novel and Rapid Encoding Method Based on Mass Spectrometry for “One-Bead-One-Compound” Small Molecule Combinatorial Libraries. J Am Chem Soc. 2003;125(20):6180–6188. doi: 10.1021/ja034539j. [DOI] [PubMed] [Google Scholar]
- 19.Mannocci L, Zhang Y, Scheuermann J, Leimbacher M, De Bellis G, Rizzi E, Dumelin C, Melkko S, Neri D. High-Throughput Sequencing Allows the Identification of Binding Molecules Isolated From DNA-Encoded Chemical Libraries. Proc Natl Acad Sci USA. 2008;105(46):17670–17675. doi: 10.1073/pnas.0805130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, Ding Y, Franklin GJ, Franzen KD, Gefter ML, Hale SP, Hansen NJV, Israel DI, Jiang J, Kavarana MJ, Kelley MS, Kollmann CS, Li F, Lind K, Mataruse S, Medeiros PF, Messer JA, Myers P, O’Keefe H, Oliff MC, Rise CE, Satz AL, Skinner SR, Svendsen JL, Tang L, van Vloten K, Wagner RW, Yao G, Zhao B, Morgan BA. Design, Synthesis and Selection of DNA-Encoded Small-Molecule Libraries. Nat Chem Biol. 2009;5(9):647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 21.Franzini RM, Ekblad T, Zhong N, Wichert M, Decurtins W, Nauer A, Zimmermann M, Samain F, Scheuermann J, Brown PJ, Hall J, Graeslund S, Schueler H, Neri D. Identification of Structure-Activity Relationships From Screening a Structurally Compact DNA-Encoded Chemical Library. Angew Chem Int Ed. 2015;54(13):3927–3931. doi: 10.1002/anie.201410736. [DOI] [PubMed] [Google Scholar]
- 22.Deng H, Zhou J, Sundersingh FS, Summerfield J, Somers D, Messer JA, Satz AL, Ancellin N, Arico-Muendel CC, Sargent Bedard KL, Beljean A, Belyanskaya SL, Bingham R, Smith SE, Boursier E, Carter P, Centrella PA, Clark MA, Chung C-W, Davie CP, DeLorey JL, Ding Y, Franklin GJ, Grady LC, Herry K, Hobbs C, Kollmann CS, Morgan BA, Pothier Kaushansky LJ, Zhou Q. Discovery, SAR, and X-Ray Binding Mode Study of BCATm Inhibitors From a Novel DNA-Encoded Library. ACS Med Chem Lett. 2015;6(8):919–924. doi: 10.1021/acsmedchemlett.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Y, O’Keefe H, DeLorey JL, Israel DI, Messer JA, Chiu CH, Skinner SR, Matico RE, Murray-Thompson MF, Li F, Clark MA, Cuozzo JW, Arico-Muendel C, Morgan BA. Discovery of Potent and Selective Inhibitors for ADAMTS-4 Through DNA-Encoded Library Technology (ELT) ACS Med Chem Lett. 2015;6(8):888–893. doi: 10.1021/acsmedchemlett.5b00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddavide FV, Lin W, Lehnert S, Zhang Y. DNA-Encoded Dynamic Combinatorial Chemical Libraries. Angew Chem Int Ed. 2015;54(27):7924–7928. doi: 10.1002/anie.201501775. [DOI] [PubMed] [Google Scholar]
- 25.Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, Neri D, Scheuermann J. Dual-Display of Small Molecules Enables the Discovery of Ligand Pairs and Facilitates Affinity Maturation. Nat Chem. 2015;7(3):241–249. doi: 10.1038/nchem.2158. [DOI] [PubMed] [Google Scholar]
- 26.Wu Z, Graybill TL, Zeng X, Platchek M, Zhang J, Bodmer VQ, Wisnoski DD, Deng J, Coppo FT, Yao G, Tamburino A, Scavello G, Franklin GJ, Mataruse S, Bedard KL, Ding Y, Chai J, Summerfield J, Centrella PA, Messer JA, Pope AJ, Israel DI. Cell-Based Selection Expands the Utility of DNA-Encoded Small-Molecule Library Technology to Cell Surface Drug Targets: Identification of Novel Antagonists of the NK3 Tachykinin Receptor. ACS Comb Sci. 2015;17(12):722–731. doi: 10.1021/acscombsci.5b00124. [DOI] [PubMed] [Google Scholar]
- 27.Scheuermann J, Dumelin CE, Melkko S, Zhang Y, Mannocci L, Jaggi M, Sobek J, Neri D. DNA-Encoded Chemical Libraries for the Discovery of MMP-3 Inhibitors. Bioconjug Chem. 2008;19(3):778–785. doi: 10.1021/bc7004347. [DOI] [PubMed] [Google Scholar]
- 28.Encinas L, O’Keefe H, Neu M, Remuiñán MJ, Patel AM, Guardia A, Davie CP, Pérez-Macías N, Yang H, Convery MA, Messer JA, Pérez-Herrán E, Centrella PA, Álvarez-Gómez D, Clark MA, Huss S, O’Donovan GK, Ortega-Muro F, McDowell W, Castañeda P, Arico-Muendel CC, Pajk S, Rullás J, Angulo-Barturen I, Álvarez-Ruíz E, Mendoza-Losana A, Ballell Pages L, Castro-Pichel J, Evindar G. Encoded Library Technology as a Source of Hits for the Discovery and Lead Optimization of a Potent and Selective Class of Bactericidal Direct Inhibitors of Mycobacterium Tuberculosis InhA. J Med Chem. 2014;57(4):1276–1288. doi: 10.1021/jm401326j. [DOI] [PubMed] [Google Scholar]
- 29.MacConnell AB, McEnaney PJ, Cavett VJ, Paegel BM. DNA-Encoded Solid-Phase Synthesis: Encoding Language Design and Complex Oligomer Library Synthesis. ACS Comb Sci. 2015;17(9):518–534. doi: 10.1021/acscombsci.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chouikhi D, Ciobanu M, Zambaldo C, Duplan V, Barluenga S, Winssinger N. Expanding the Scope of PNA-Encoded Synthesis (PES): Mtt-Protected PNA Fully Orthogonal to Fmoc Chemistry and a Broad Array of Robust Diversity-Generating Reactions. Chemistry. 2012;18(40):12698–12704. doi: 10.1002/chem.201201337. [DOI] [PubMed] [Google Scholar]
- 31.Halpin DR. Harbury PBDNA Display I. Sequence-Encoded Routing of DNA Populations. PLoS Biol. 2004;2(7):1015–1021. doi: 10.1371/journal.pbio.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franzini RM, Samain F, Abd Elrahman M, Mikutis G, Nauer A, Zimmermann M, Scheuermann J, Hall J, Neri D. Systematic Evaluation and Optimization of Modification Reactions of Oligonucleotides with Amines and Carboxylic Acids for the Synthesis of DNA-Encoded Chemical Libraries. Bioconjug Chem. 2014;25(8):1453–1461. doi: 10.1021/bc500212n. [DOI] [PubMed] [Google Scholar]
- 33.Ding Y, Clark MA. Robust Suzuki-Miyaura Cross-Coupling on DNA-Linked Substrates. ACS Comb Sci. 2015;17(1):1–4. doi: 10.1021/co5001037. [DOI] [PubMed] [Google Scholar]
- 34.Satz AL, Cai J, Chen Y, Goodnow R, Gruber F, Kowalczyk A, Petersen A, Naderi-Oboodi G, Orzechowski L, Strebel Q. DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjug Chem. 2015;26(8):1623–1632. doi: 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- 35.Goodnow RA., Jr A Handbook for DNA-Encoded Chemistry: Theory and Applications for Exploring Chemical Space and Drug Discovery. 2014 [Google Scholar]
- 36.Gordon EM, Gallop MA, Patel DV. Strategy and Tactics in Combinatorial Organic Synthesis. Applications to Drug Discovery. Acc Chem Res. 1996;29:144–154. [Google Scholar]
- 37.Iverson BL, Dervan PB. Adenine Specific DNA Chemical Sequencing Reaction. Nucleic Acids Res. 1987;15(19):7823–7830. doi: 10.1093/nar/15.19.7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westman E, Strömberg R. Removal of T-Butyldimethylsilyl Protection in RNA-Synthesis, Triethylamine Trihydrofluoride (TEA, 3HF) Is a More Reliable Alternative to Tetrabutylammonium Fluoride (TBAF) Nucleic Acids Res. 1994;22(12):2430–2431. doi: 10.1093/nar/22.12.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pels K, Kodadek T. Solid-Phase Synthesis of Diverse Peptide Tertiary Amides by Reductive Amination. ACS Comb Sci. 2015;17(3):152–155. doi: 10.1021/acscombsci.5b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyaura N, Suzuki A. Palladium-Catalyzed Cross-Coupling Reactions of Organoboron Compounds. Chem Rev. 1995;95:2457–2483. [Google Scholar]
- 41.Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB. Synthetic Ligands Discovered by in Vitro Selection. J Am Chem Soc. 2007;129(43):13137–13143. doi: 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satz AL. DNA Encoded Library Selections and Insights Provided by Computational Simulations. ACS Chem Biol. 2015;10(10):2237–2245. doi: 10.1021/acschembio.5b00378. [DOI] [PubMed] [Google Scholar]
- 43.Gates KS. The Chemical Reactions of DNA Damage and Degradation. In: Platz MS, Moss RA, MJ, editors. Reviews of Reactive Intermediate Chemistry. John Wiley & Sons, Inc; 2006. pp. 333–378. [Google Scholar]
- 44.Zoltewicz JA, Clark DF, Sharpless TW. Kinetics and Mechanism of the Acid-Catalyzed Hydrolysis of Some Purine Nucleosides. J Am Chem Soc. 1970;92(6):1741–1750. doi: 10.1021/ja00709a055. [DOI] [PubMed] [Google Scholar]
- 45.Guiles JW, Johnson SG, Murray WV. Solid-Phase Suzuki Coupling for C-C Bond Formation. J Org Chem. 1996;61(15):5169–5171. [Google Scholar]
- 46.Midorikawa K, Murata M, Oikawa S, Tada-Oikawa S, Kawanishi S. DNA Damage by Dimethylformamide: Role of Hydrogen Peroxide Generated During Degradation. Chem Res Toxicol. 2000;13(4):309–315. doi: 10.1021/tx990139r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.