Abstract
Mitophagy is critical for cell homeostasis. Externalization of the inner mitochondrial membrane phospholipid, cardiolipin (CL), to the surface of the outer mitochondrial membrane (OMM) was identified as a mitophageal signal recognized by the microtubule-associated protein 1 light chain 3. However, the CL-translocating machinery remains unknown. Here we demonstrate that a hexameric intermembrane space protein, NDPK-D (or NM23-H4), binds CL and facilitates its redistribution to the OMM. We found that mitophagy induced by a protonophoric uncoupler, carbonyl cyanide m-chlorophenylhydrazone (CCCP), caused externalization of CL to the surface of mitochondria in murine lung epithelial MLE-12 cells and human cervical adenocarcinoma HeLa cells. RNAi knockdown of endogenous NDPK-D decreased CCCP-induced CL externalization and mitochondrial degradation. A R90D NDPK-D mutant that does not bind CL was inactive in promoting mitophagy. Similarly, rotenone and 6-hydroxydopamine triggered mitophagy in SH-SY5Y cells was also suppressed by knocking down of NDPK-D. In situ proximity ligation assay (PLA) showed that mitophagy-inducing CL-transfer activity of NDPK-D is closely associated with the dynamin-like GTPase OPA1, implicating fission–fusion dynamics in mitophagy regulation.
Appreciation of the central role of the mitochondrion as a hub controlling multiple intra- and extra-cellular metabolic responses, as well as cellular life or death decisions, has raised great interest in the processes involved in their maintenance and quality control.1, 2 The homeostatic health of mitochondria is preserved owing to coordinated functions of several control mechanisms, including proteolysis of misfolded proteins, proteasomal degradation of aberrant proteins on their surface, dynamic repair processes of fission and fusion, formation and budding off of specialized mitochondrial particles, and autophagic degradation of the entire organelle in lysosomes (mitophagy).3 In the latter process, identification of the signals distinguishing ‘fit' from dysfunctional mitochondria is particularly important, and several essential molecular interactions are beginning to emerge.3, 4 Pink1-Parkin-mediated ubiquitination of the outer mitochondrial membrane (OMM) proteins and their indirect interactions with the autophagy protein MAP1-light chain 3/Atg8 (LC3) via LC3-interacting region (LIR) domain adaptor proteins have been proposed as one of the potentially essential mitophageal signaling pathways.5
Recently, we reported that a mitochondria-unique anionic phospholipid, cardiolipin (CL), undergoes redistribution and externalization to the surface of mitsochondria injured by a variety of insults in several types of cells.6 The externalized CL is recognized by LC3, thus triggering the elimination of damaged mitochondria. However, the protein machinery responsible for CL externalization remained enigmatic. As the majority of CL, synthesized by cardiolipin synthase 1 (CLS-1), are normally confined to the inner leaflet of the inner mitochondrial membrane (IMM), three translocations are required for CL exposure on the mitochondrial outer surface. Our previous work demonstrated that an intermembrane space hexameric protein, NDPK-D (also called NM23-H4 and encoded by the NME4 gene), can bind CL and facilitate its redistribution to the OMM.7, 8 Here, we tested the hypothesis that NDPK-D regulates the elimination of mitochondria via autophagy, thus acting as a mechanism of mitochondrial quality control.
Results
To explore the role of NDPK-D in mitophagy-associated CL redistribution we assessed the amounts of CLs externalized to the surface of mitochondria in response to a standard model of carbonyl cyanide m-chlorophenylhydrazone (CCCP) – induced mitophagy in two different cell lines – MLE (mouse lung epithelial), and HeLa – in which we manipulated the content of the wild-type (w/t) NDPK-D or mutated protein.9
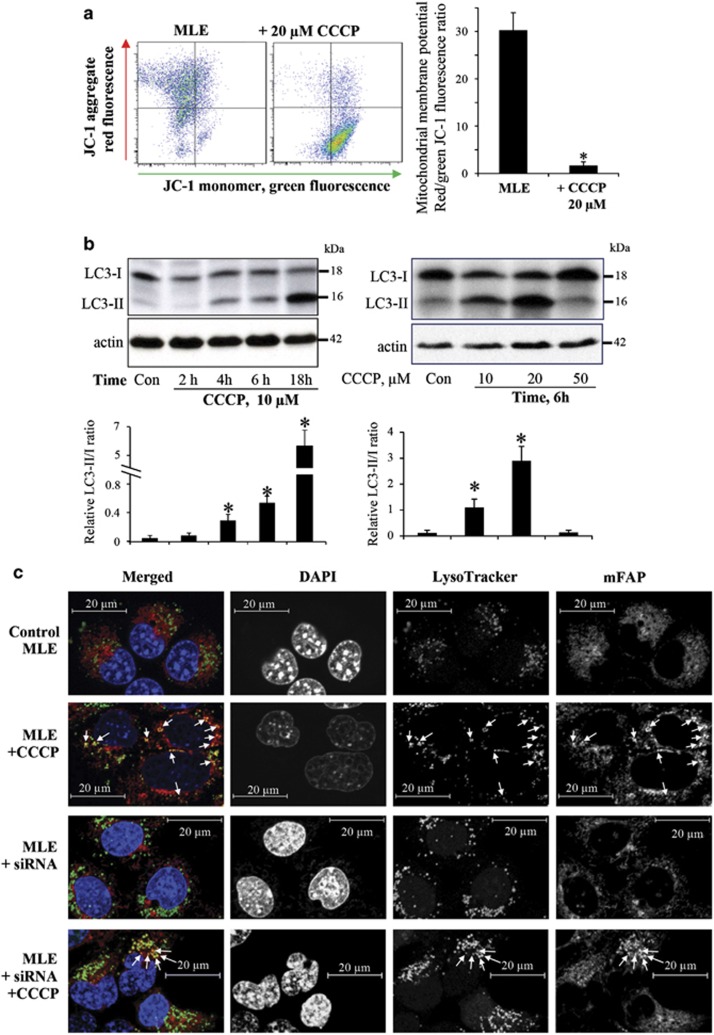
Depolarization of mitochondria by the protonophoric uncoupler, CCCP (Figure 1a), activated autophagy in MLE cells, resulting in dose- and time-dependent conversion of LC3-I into LC-3-II (Figure 1b). The activation of the autophagic machinery was accompanied by the appearance of mitochondria in the lysosomal compartment, as detected by the co-localization of mitochondrial-targeted fluorogen-activating protein (mFAP) with LysoTracker Red-labeled lysosomes (Figure 1c). Further evidence for mitophagic clearance of depolarized mitochondria was obtained by western blotting of three mitochondrial marker proteins (Supplementary Figure 1a). The OMM protein TOM40, the IMM protein TIM23, and the matrix protein MnSOD showed decreased levels after treatment with CCCP (Supplementary Figure 1a).
Figure 1.
Characterization of CCCP-induced damage of mitochondria and mitophagy in mouse lung epithelial cells. (a) Membrane potential in CCCP-treated MLE cells (20 μM/30 min) determined by JC-1 using flow cytometry (n=3). (b) Dose- and time-dependent CCCP-induced conversion of LC3-I to LC3-II using western blot analysis. Cells were treated with 10 μM CCCP for indicated time (2, 4, 6, and 18 h, left panel), or cells were treated with different concentrations (10, 20, and 50 μM, right panel) of CCCP for 6 h. The histograms under the blots show the LC3-I/II ratio calculated based on densitometry (n=3). (c) Representative immunofluorescence imaging of mitochondria (mFAP) and lysosomes (LysoTracker Red) showing co-localization (arrows), scale bar is 20 μm. *P<0.05 versus control cells without CCCP treatment
To evaluate the role of CL in CCCP-induced clearance of damaged mitochondria, we knocked down CLS-1, the enzyme catalyzing the final stage of CL biosynthesis from phosphatidylglycerol precursors.10 RNAi against CLS-1 caused a >2.5-fold decrease in the content of the CLS-1 in MLE cells (Supplementary Figure 1b) associated with a previously demonstrated robust decrease of CL levels.11 This led to marked inhibition of CCCP-induced mitophagy as assessed by the loss of mitochondrial proteins (Supplementary Figure 1).
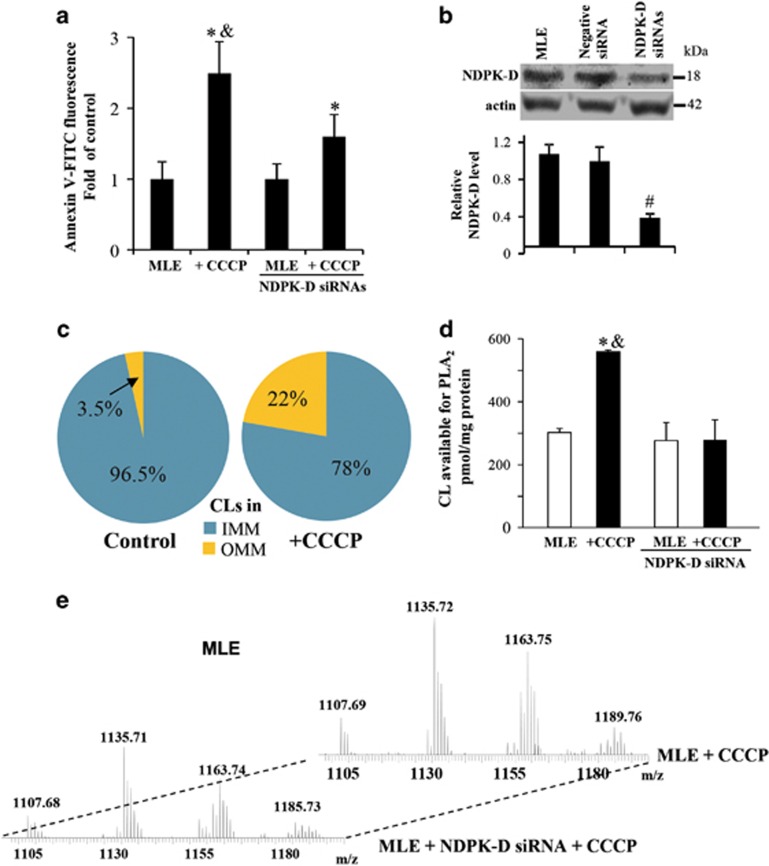
Most importantly, CCCP caused NDPK-D-dependent externalization of CL to the mitochondrial surface. We used two independent methods to detect and quantify the appearance of CL on the mitochondrial surface. In the first approach, we took advantage of the well-known lack of phosphatidylserine in mitochondria12 and used Annexin V-FITC to monitor the content of accessible CL on the surface.6 Mitochondria from MLE cells treated with CCCP responded with a ~2.5-fold increase in the fluorescence of Annexin V-FITC (Figure 2a). Notably, this response was dependent upon NDPK-D, as it was much reduced in MLE cells treated with NDPK-D RNAi (Figure 2a and b).
Figure 2.
CCCP induces externalization of cardiolipin in MLE cells. (a) Evaluation of CL in the outer leaflet of OMM using Annexin V-binding assay in w/t and NDPK-D RNAi MLE cells. Cells were treated with 20 μM CCCP for 1 h. Isolated mitochondria were incubated with FITC-labeled Annexin V to stain surface-exposed CL (anionic phospholipids) and then subjected to flow cytometric analysis (FACSCanto, Becton-Dickinson). (b) Knockdown of NDPK-D using siRNA interference in MLE cells. As controls, cells were mock transfected or transfected with non-targeted negative siRNAs. Expression of NDPK-D was evaluated by western blotting. Lower panel shows the relative NDPK-D expression calculated based on densitometry (n=3). (c) LC-MS analysis-based relative contents of CL in OMM and IMM in control and CCCP-treated MLE cells are presented. (d) Evaluation of CL in the outer leaflet of OMM in w/t and NDPK-D knocked down MLE cells using PLA2 treatment with subsequent assay of mono-lyso-CL by LC-MS analysis. (e) Representative MS spectra of mono-lyso CL from MLE cells with normal expression of endogenous NDPK-D or after treatment with NDPK-D siRNAs. *P<0.05 versus control cells without CCCP treatment. #P<0.05 versus cells transfected with non-targeting negative siRNAs. &P<0.05 versus MLE cells transfected with NDPK-D siRNAs under the same condition (20 μM CCCP/1 h)
To more directly characterize CL externalization, we performed liquid chromatography–mass spectrometry (LC-MS) characterization of CL in the OMM and IMM isolated from mitochondria of control cells and cells exposed to CCCP (Figure 2c). The content of CL in the OMM was increased almost sixfold in response to CCCP without any significant changes in the CL molecular speciation (Table 1). To estimate the amounts of CL exposed on the outer leaflet of OMM, we performed LC-MS analysis of mono-lyso-CLs generated by phospholipase A2 (PLA2) treatment of control and CCCP-depolarized mitochondria (Figure 2d and e). CCCP exposure increased the content of mono-lyso-CLs after PLA2 treatment by almost twofold (~3 mol% versus ~1.5 mol%) (300 versus 575 pmol/mg protein, P<0.05, Figure 2d). Analysis of LC-MS spectra did not reveal the appearance of new molecular species of mono-lyso-CL after the exposure to CCCP. This is consistent with redistribution of existing CL species caused by CCCP (Table 1). Of note, no oxidized CL species were detectable in CCCP-treated MLE cells. The quantity of detectable mono-lyso-CL at the mitochondrial surface was dependent on NDPK-D expression. Knockdown of NDPK-D in MLE cells prevented changes in the level of mono-lyso-CLs after the CCCP exposure (Figure 2d).
Table 1. Molecular species of CL in the outer mitochondrial membrane.
| (M-H)− m/z | Identified acyl chains | Molecular species |
|---|---|---|
| 1344 | C16:1/C16:1/C16:1/C16:1 | 64:4 |
| 1346 | C16:1/C16:1/C16:1/C16:0 | 64:3 |
| 1346 | C16:1/C16:1/C14:0/C18:1 | 64:3 |
| 1370 | C16:1/C16:1/C16:1/C18:2 | 66:5 |
| 1372 | C16:1/C16:1/C16:0/C18:2 | 66:4 |
| 1372 | C16:1/C16:1/C16:1/C18:1 | 66:4 |
| 1374 | C16:1/C16:1/C16:0/C18:1 | 66:3 |
| 1376 | C16:1/C16:0/C16:0/C18:1 | 66:2 |
| 1386 | C16:1/C17:1/C16:1/C18:1 | 67:4 |
| 1396 | C16:1/C18:2/C16:1/C18:2 | 68:6 |
| 1398 | C16:1/C16:1/C18:1/C18:2 | 68:5 |
| 1400 | C16:1/C18:1/C16:1/C18:1 | 68:4 |
| 1402 | C16:1/C18:1/C16:0/C18:1 | 68:3 |
| 1404 | C16:0/C18:1/C16:0/C18:1 | 68:2 |
| 1404 | C16:0/C16:0/C18:1/C18:1 | 68:2 |
| 1414 | C16:1/C18:1/C17:1/C18:1 | 69:4 |
| 1424 | C16:1/C18:1/C18:2/C18:2 | 70:6 |
| 1426 | C16:1/C18:1/C18:1/C18:2 | 70:5 |
| 1428 | C16:1/C18:1/C18:1/C18:1 | 70:4 |
| 1430 | C16:0/C18:1/C18:1/C18:1 | 70:3 |
| 1432 | C16:0/C18:1/C18:1/C18:0 | 70:2 |
| 1448 | C18:2/C18:2/C18:2/C18:2 | 72:8 |
| 1450 | C18:2/C18:2/C18:1/C18:2 | 72:7 |
| 1452 | C18:1/C18:2/C18:1/C18:2 | 72:6 |
| 1454 | C18:1/C18:2/C18:1/C18:1 | 72:5 |
| 1456 | C18:1/C18:1/C18:1/C18:1 | 72:4 |
Transmission electron microscopy revealed the appearance in CCCP-treated cells of autophagic structures with condensed, partially degraded mitochondria (Supplementary Figure 2). In contrast, in cells with knocked down NDPK-D, the autophagic structures induced by CCCP were devoid of mitochondria.
The delivery of mitochondria into the lysosomal compartment (Figure 1c), combined with the TEM appearance of autophagic structures at different stages of degradation (Supplementary Figure 2), indicated that autophagic maturation was intact. Notably, knocking down of NDPK-D blocked CCCP-induced co-localization of mitochondria with lysosomes (Figure 1c) and degradation of mitochondrial marker proteins (Supplementary Figure 3). To further directly assess for completion of mitophagy, we studied the loss of mitochondria content. By using mFAP, we observed the CCCP-induced loss of mitochondrial volume and increased mitochondrial fragmentation (as assessed by increased sphericity of small mitochondrial fragments) in MLE cells (Supplementary Figure 4). Both of these effects were attenuated by knocking down of NDPK-D.
Combined, these data obtained with MLE cells provide strong evidence that NDPK-D was essential for the translocation of CL to the surface of depolarized mitochondria and for their mitophagic clearance.
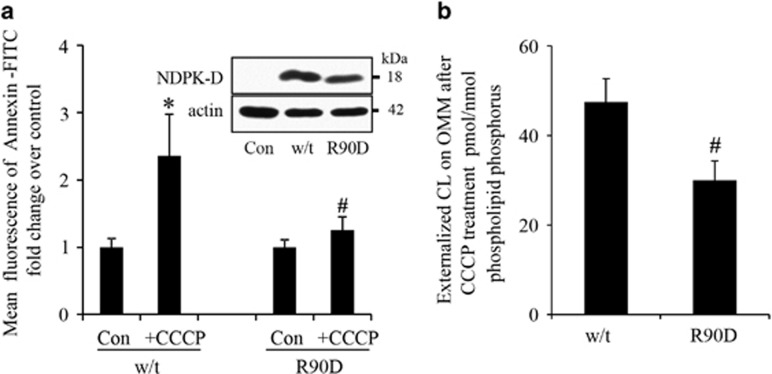
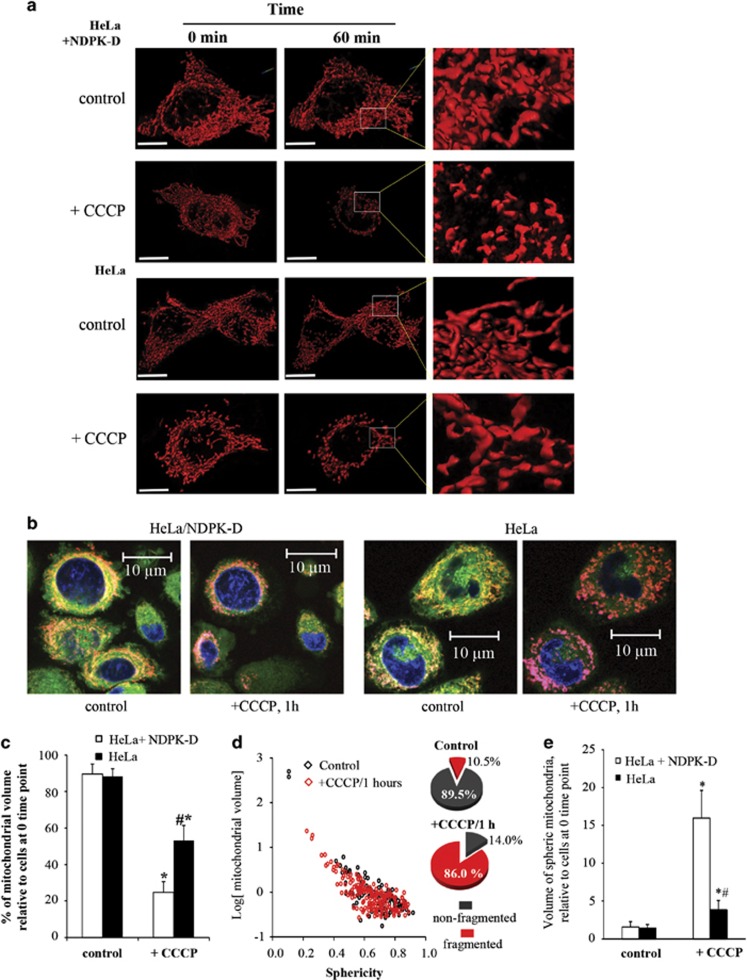
To examine whether this pathway of NDPK-D-driven CL externalization and mitophagy of damaged mitochondria could be observed in other cell types, we chose to study Parkin-expressing HeLa cells, one of the most commonly used cell type for the study of depolarization-induced mitophagy.4, 13 HeLa cells constitutively express negligible levels of NDPK-D,14 which allowed us to introduce either w/t or mutant protein. The R90D mutant is unable to translocate lipids between donor and acceptor bilayer vesicles in general,15 and it is unable to bind CL and therefore to mediate the translocation of CL from IMM to OMM.7 We found that w/t but not R90D NDPK-D facilitated CCCP-induced translocation of CL from the IMM to the mitochondrial surface in Parkin-HeLa cells. This was estimated by using either the Annexin V-FITC externalization assay (Figure 3a) or LC-MS-based quantitation of mono-lyso-CLs generated by hydrolysis of molecular species of CL accessible to externally added PLA2 (Figure 3b). In both cases, robust externalization of CL on depolarized mitochondria was detected in cells transfected with w/t NDPK-D, but not with the R90D mutant. In agreement with the important role of NDPK-D in translocating and externalizing CL as a mitophageal signal, the effectiveness of mitochondrial elimination – as documented by the loss of the mitochondrial marker proteins by western blotting (Figure 4) – the loss of mitochondrial volume, and increased mitochondrial fragmentation assessed using mFAP-infected cells (Figure 5 and Supplementary Figure 6) required w/t NDPK-D. The mitochondrial network is filamentous in HeLa cells expressing w/t NDPK-D. After treatment with CCCP, the mitochondrial filamentous network was disrupted. In addition, the LC3 labeling, mainly as puncta, was significantly increased as compared with control cells without CCCP treatment. Several yellow spots – indicative of LC3 and mitochondria co-localization – were also noticeable. By sharp contrast, only a slight increase in LC3 labeling was observed in HeLa cells expressing R90D mutant (Supplementary Figure 7).
Figure 3.
Characterization of CCCP-induced cardiolipin externalization in HeLa cells transfected with wild-type or R90D mutant of NDPK-D. (a) HeLa cells were transiently transfected with vectors expressing w/t or R90D NDPK-D for 48 h followed by treatment with 20 μM CCCP for 4 h. Crude mitochondria were isolated for determination of CL externalization using Annexin V-binding (n=3). Insert: representative western blots of NDPK-D expression in HeLa cells. (b) LC-MS-based relative amounts of CL in OMM and IMM in HeLa cells expressing w/t NDPK-D and R90D mutant. Data are means±S.D., n=3. *P<0.05 compared with cells without CCCP treatment, #P<0.05 compared with cells expressing w/t NDPK-D under the same conditions (20 μM CCCP/4 h)
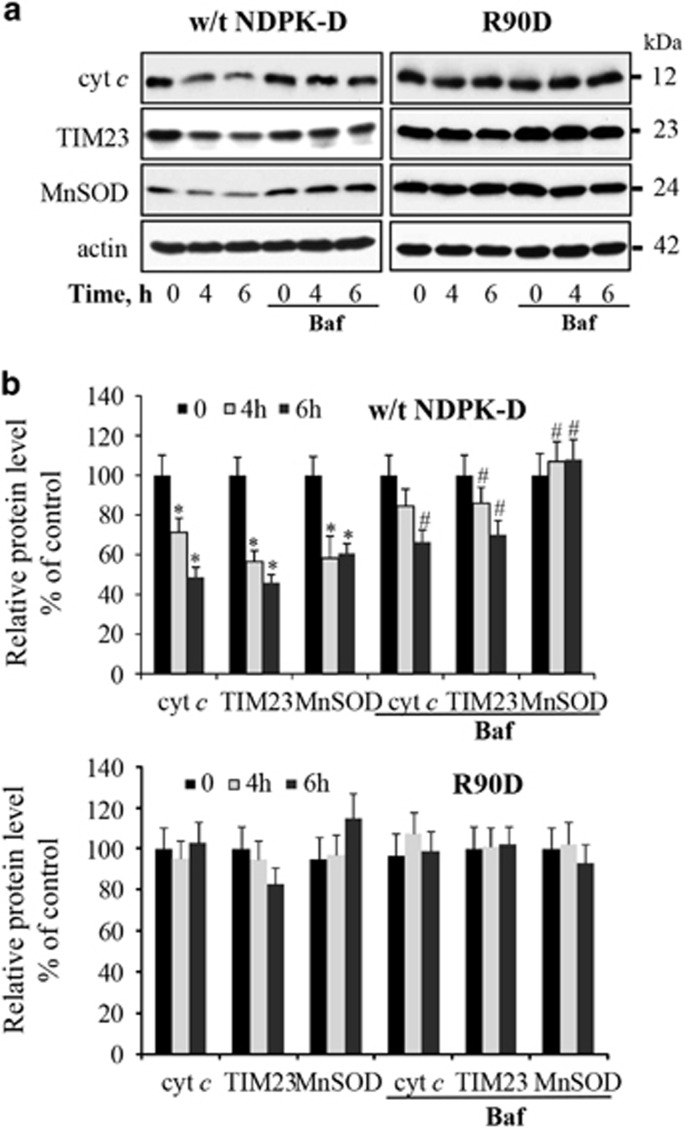
Figure 4.
Characterization of CCCP-induced mitophagy in HeLa cells. After treatment of cells with 20 μM CCCP for the indicated times, CCCP-induced degradation of mitochondrial marker proteins (cyt c, Tim23, and MnSOD) were determined by western blotting (a). Autophagy inhibitor, bafilomycin A1 (baf), was added to cells 2 h prior to harvesting. The panel (b) shows the densitometry analysis. Data are means±S.D. (n=3). *P<0.05 compared with 0 h, #P<0.05 compared with 4 h and 6 h CCCP without bafilomycin A1 treatment
Figure 5.
CCCP-induced mitochondrial volume change and fragmentation in NDPK-D-manipulated HeLa cells. (a) Representative images of 3D surface reconstructions using Imaris 8 software. Control HeLa cells or cells transfected with NPDK-D were infected with adenovirus expressing mCerulean/FAP. After addition of CCCP, control or NDPK-D-expressing cells were subjected to multi-position time-lapse imaging for 60 min with 5-min intervals, with a collection of z-stacks for each position, using a Nikon T1 microscope. Images were deconvolved using the 3D Landwebser deconvolution capabilities of Nikon Elements and imported into Imaris software. Mitochondrial volume, surface area, and sphericity were calculated using Imaris software. Scale bar is 20 μm. (b) Representative high-resolution confocal imaging of mitochondria in CCCP-treated control HeLa cells or cells transfected with w/t NDPK-D. (c) Wild-type NDPK-D facilitated CCCP-induced mitochondrial volume loss in Parkin-HeLa cells. The relative mitochondrial volume in HeLa cells after 1 h incubation with CCCP was calculated as the percentage of initial mitochondrial volume at the first time point (n=6–8 images). (d) Mitochondrial fragmentation in HeLa cells was assessed using sphericity parameter for small mitochondrial fragments (with an arbitrary threshold of sphericity <0.4) to differentiate the fragmented mitochondria and plotted against log (mitochondrial volume). Dot plots shown are representative of NDPK-D-expressing HeLa cells before and after (1 h) addition of CCCP. The inserted pie plots show the percentage of fragmented and non-fragmented mitochondrial volume. (e) The ratio of the volume of spheric (fragmented) mitochondria versus non-fragmented mitochondria in HeLa cells was calculated and compared with the ratio at the initial time point (n=6–8). *,#P<0.05 versus HeLa cells without CCCP treatment, or HeLa cells with NDPK-D under the same condition (20 μM CCCP/1 h), respectively (n=6–8)
To evaluate if NDPK-D could regulate the Parkin translocation to mitochondrial compartment after mitochondrial damage, we performed experiments in which we examined the effects of NDPK-D and its R90D mutant on Parkin translocation in CCCP-treated HeLa cells. Parkin-expressing cells were transiently transfected with vectors expressing w/t NDPK-D or R90D mutant. Studies of the time course showed that Parkin translocation was much faster in w/t NDPK-D transfected cells than in R90D transfected cells (Supplementary Figure 8a). In a different cell line, SH-SY5Y cells, assessments of translocated Parkin using fluorescence imaging of the protein demonstrated that the percentage of w/t SH-SY5Y cells with translocated Parkin after CCCP treatment was significantly decreased in cells transfected with siRNA against NDPK-D (Supplementary Figure 8b).
Although most studies of mitophagy mechanisms have used CCCP, it has become clear that mitophagy can also be induced by stimuli causing lesser degrees of depolarization (such as rotenone, ROT) or even in the absence of depolarization (PMI, P62-mediated mitophagy inducer).16 We thus examined other pharmacological inducers (ROT and 6-hydroxy-dopamine, 6-OHDA) of mitophagy that do not cause severe depolarization. We found that knocking down of NDPK-D using siRNAs significantly attenuated ROT and 6-OHDA-induced mitophagy in SH-SY5Y cells (Supplementary Figure 9). A robust mitophagy triggered by ROT treatment of w/t SH-SY5Y cells was less apparent in the NDPK-D-deficient cells (compare merged images in rows 3 and 4, Supplementary Figure 9a). Similarly, a greater level of mitophagy was observed in w/t SH-SY5Y cells versus NDPK-D-deficient cells upon treatment with 6-OHDA (compare merged images in rows 5 and 6, Supplementary Figure 8a). Quantitative analysis of the images clearly demonstrated weakening of the mitophageal responses to either ROT or 6-OHDA in NDPK-depleted cells as compared with w/t SH-SY5Y cells (Supplementary Figure 9b). In line with these imaging results, western blot analysis revealed less robust degradation of IMM marker proteins (ATP5a, UQCRC2, and COX-IV) in w/t SH-SY5Y cells depleted for NDPK-D (Supplementary Figure 9c).
To further diversify the spectrum of stimulants and type of cells undergoing mitophageal responses, we employed Saccharomyces cerevisae yeast cells in which mitophagy can be triggered by switching the cells from synthetic complete (SC) medium to synthetic medium containing a non-fermentable carbon source and depleted of essential nutrients.17 To detect mitophagy, we used an isocitrate dehydrogenase (Idh1)-GFP processing assay.18 Wild-type cells and cells deleted for yeast nucleoside diphosphate kinase Ndk1/Ynk1 (ndk1Δ) also localizing to mitochondria were pre-cultured in SC medium and then transferred to synthetic lactate medium to induce mitophagy.19 During mitophagy, mitochondria, along with Idh1-GFP, are delivered into the vacuole for degradation. Idh1 is proteolytically removed or degraded, whereas the GFP moiety is relatively stable and accumulates in the vacuole. After 6–7 h, cells were harvested and analyzed for release of free GFP, indicative of mitophagy.18, 19, 20, 21, 22, 23, 24 The free GFP band was apparent beginning at 6 h after induction of mitophagy in w/t cells, and was markedly decreased in ndk1Δ cells, indicating the impaired mitophagy in the latter (Supplementary Figure 10). These findings suggest that Ndk1/Ynk1 is an important contributor to the elimination of mitochondria induced by starvation of yeast cells.
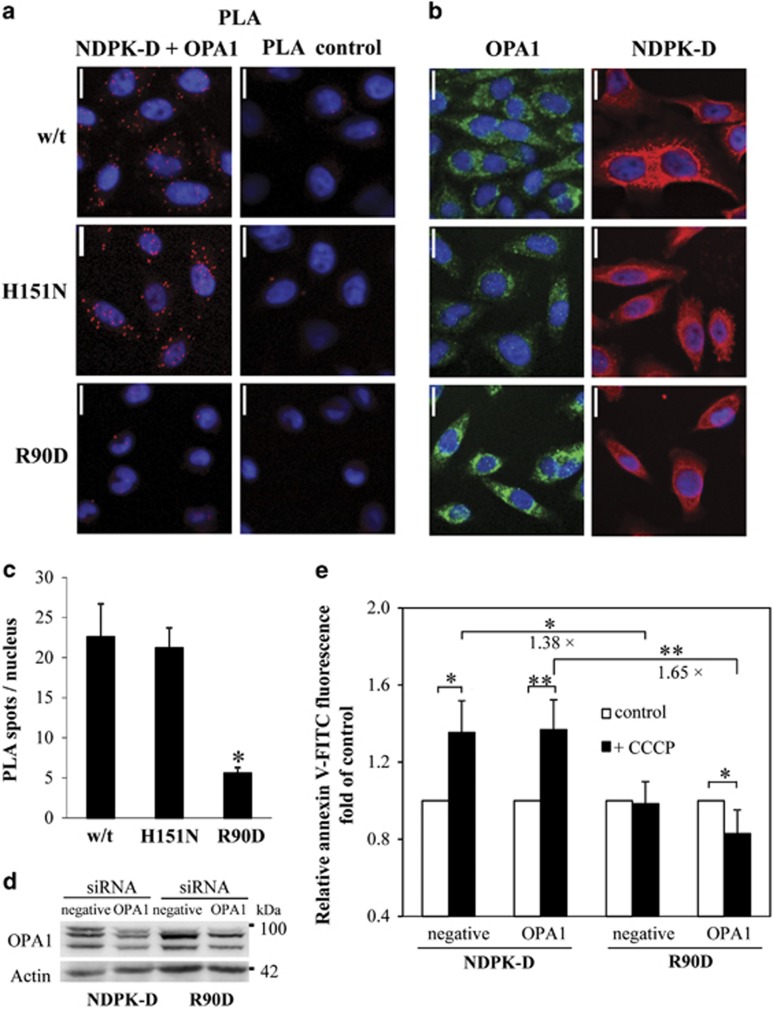
It is not known so far how NDPK-D adapts to its different functions. We recently showed that the dynamin-like GTPase OPA1 forms complexes with NDPK-D, both proteins bound to the CL-rich IMM, and that in this topology OPA1 is directly fueled with GTP by the kinase.16 Earlier, we observed that NDPK-D that cross-links IMM and OMM, a topology that is able to support CL transfer, is kinase inactive,7 suggesting that this is an uncomplexed form of the kinase. Thus, formation of OPA1/NDPK-D complexes may interfere with CL translocation. In a PLA (Figure 6a and c) only HeLa cells expressing CL-binding and principally CL-transfer competent NDPK-D proteins, i.e., w/t or the kinase-inactive H151N mutant, were found associated with OPA1, but not the CL-binding and CL-transfer-deficient R90D mutant. We then partially silenced OPA1 by siRNA to study its consequences on CCCP-induced CL transfer in NDPK-D w/t and R90D mutant HeLa cell by the Annexin V-FITC externalization assay (Figure 6d and e). OPA1 was silenced only partially (to ~60%) to observe mild phenotypes, as stronger OPA1 knockdown would itself induce mitophagy and apoptosis, independent of CCCP.25, 26, 27 The data confirm that expression of w/t NDPK-D increases CCCP-induced CL externalization. This increase occurred under both conditions, control and OPA1 silenced, but was more statistically significant (P<0.01 versus 0.05) in OPA1-silenced cells. Such an increase was not observed in cells expressing the R90D mutant; rather, OPA1 silencing led to a decrease in externalized CL. A significant effect of partial OPA1 silencing was observed when comparing between the CCCP-treated cells expressing NDPK-D w/t or R90D mutant. In this comparison, CL externalization is more increased in the OPA1-silenced background (1.65±0.24 fold) as compared with the control background (1.38±0.01 fold), and this was statistically significant (P<0.01).
Figure 6.
Association of OPA1 with NDPK-D in HeLa cells. (a–c) OPA1/NDPK-D interaction evidence by proximity ligation assay (PLA) in HeLa cells stably expressing w/t NDPK-D, kinase-inactive H151N mutant (H151N), or CL-transfer-deficient R90D mutant (R90D). Representative magnifications of confocal pictures are given, scale bar is 16 μm. (a) Left panel: PLA with NDPK-D and OPA1 primary antibodies; right panel: control PLA without primary antibodies. (b) Control immunofluorescence studies showing expression of NDPK-D (right panel) and OPA1 (left panel) as detected by the same NDPK-D and OPA1 primary antibodies. (c) Quantification of PLA as shown in a from three experiments, two quantifications each (n=6), *P<0.01 versus HeLa cells expressing w/t NDPK-D. (d, e) Effect of partial OPA1 silencing on CL externalization. Representative immunoblot of partial OPA1 silencing in HeLa cells expressing w/t NDPK-D and R90D mutant using siRNA. HeLa cells permanently transfected with w/t NDPK-D or R90D mutant were transfected with negative or siRNA against OPA1. (e) Surface assessable CL was quantified by an Annexin V-binding assay. MitoTracker-positive particles of appropriate size were analyzed for FITC fluorescence by FACS. Data are means±S.D. (n=3–4); *P<0.05; **P<0.01. Numbers indicate the fold-difference between CCCP-treated R90D- and w/t NDPK-D-expressing cells, either on a control (1.38-fold) or partial OPA1-silenced background (1.65-fold).
Discussion
This work demonstrates, for the first time, that NDPK-D (NM23-H4) is essential for the externalization of CL, having a key mechanistic role in regulating the appearance of this ‘eat-me' signal on the mitochondrial surface thus labeling depolarized organelles destined for mitophagy.
The highly asymmetric distribution of CL between the IMM and OMM in normal, uninjured mitochondria whereby 96.5 mol% of it is confined to the IMM, creates a gradient poised to respond to changes in organelle homeostasis. Based on LC-MS assessments, this asymmetry is disturbed during depolarization, resulting in a robust CL enrichment of the OMM that is dependent upon the ability of NDPK-D to mediate its redistribution from IMM to OMM. Although the OMM content of CL is approximately doubled, this still represents only a small fraction of total cellular CL. These quantitative assessments point to the high sensitivity of the autophagic machinery to externalized CLs. The surprisingly small percentage of CL translocated between the IMM and mitochondrial surface should be considered in the context of the available pool of ‘free' CLs as compared with its total abundance in the IMM. Indeed, over the last decade, extensive studies have identified multiple proteins engaged in functionally meaningful interactions with CLs. Many IMM proteins have CL-binding domains, thus limiting free exchange and redistribution of the phospholipid to the OMM.28, 29, 30, 31, 32, 33, 34, 35, 36 In some of these proteins, CL molecules are buried ‘deeply' within multimeric protein complexes (like in electron-transporting respirasomes35, 36), whereas in the others the affinity and accessibility of CL for translocations seem to be less restricted (e.g., TIM4037). Although the exact amounts of CL in the two pools – free versus bound – have not been established, it is likely that in normally functioning mitochondria, the majority of the IMM CLs is not readily mobilized. This is compatible with the high sensitivity of mitophagy to decreased CLS-1 expression and the lowered total CL levels in mitochondria. The observed redistribution of CLs in depolarized mitochondria suggests that this balance is changed favoring interactions of CL with NDPK-D, resulting in translocation to the OMM. This NDPK-D-driven mechanism is engaged in CL externalization and execution of mitophagy in neuronal cells mitochondria damaged by ROT or 6-OHDA.6 NDPK-D-dependent CL externalization is also accountable for the pro-mitophageal stimulation by protonophoric uncouplers such as CCCP, leading to the complete depolarization of mitochondria in pulmonary epithelial cells and cervical carcinoma cells. Interestingly, we found that yeast Ndk1/Ynk1 acts as an important contributor to the elimination of mitochondria induced by starvation of yeast cells. Notably, S. cerevisiae Ndk1/Ynk1 is highly homologous to other eukaryotic NDPKs, including Homo sapiens. It also forms a homo-hexamer and a fraction of it is localized to the mitochondrial intermembrane space.38, 39 Genetic studies demonstrated the suppressed mitophagy in CLS null mutant deficient in CL synthesis (Shen and Greenberg, unpublished). Combined with our results on impaired mitophagy in ndk1Δ mutant, these data are compatible with the CL-dependent mitophagy in S. cerevisiae.
The mechanisms of the NDPK-D-driven CL translocation process are not fully deciphered. According to one of the proposed models, the hexameric NDPK-D complex acts as a rotary nano-device, whereby the binding of negatively charged phosphate groups of CL with the positively charged R90-contaning domain triggers structural re-arrangements that places the nano-machine in motion.7 The very low levels of CL in the OMM are conducive for the dissociation of the complex and release of CL. Indeed, CL-induced conformational changes of NDPK-D, which result in the suppression of its kinase activity and facilitation of its lipid transfer activity, have been reported.7 Although this hypothetical mechanism may serve as a plausible explanation for the transfer of CL between the outer monolayer of the IMM and the inner leaflet of the OMM, two additional steps – translocation within the IMM and OMM – are required for the appearance of CL on the mitochondrial surface. Although we previously reported that knockdown of phospholipid scramblase 3 inhibits mitophagy, it is currently unclear whether this enzyme may act at the IMM, the OMM, or both.6
Within the time frame of the current experiments, we did not detect CL oxidation during exposure of MLE or HeLa cells to CCCP. As CL translocation in some contexts may be associated with the interactions of CL with the intermembrane hemoprotein cytochrome c – known to yield a complex with peroxidase activity40 – one may wonder why the oxidation of CL does not take place. It should be noted that the formation of the peroxidase complex is a pre-requisite for CL oxidation, which is dependent on the presence and availability of oxidizing equivalents such as H2O2. Given that oxidized CL can be released from mitochondria to participate in apoptosome formation,40 we speculate that CL oxidation status may modulate the decision between cytoprotective mitophagy and stimulation of mitochondrial death pathways.
Given that NDPK-D has two major functions, phosphorylating nucleoside diphosphates and facilitating externalization of CL, its regulation, particularly in the context of switching from the kinase to the translocase activity of the protein complex, deserves further studies. In vitro, the kinase complex shows intermembrane lipid transfer activity as soon as it can simultaneously bind to two membranes containing some CL.15 However, NDPK-D interacts not only with CL,14 but also with OPA1,7 a dynamin-like GTPase. OPA1, beyond its classical role in inner membrane fusion,41, 42 is also directly implicated in mitochondrial quality control via its stress-regulated proteolytic cleavage via OMA1.25, 26, 43 In functional mitochondria, OPA1/NDPK-D interaction helps to fuel OPA1 with GTP,44 but this topology has to be different from the one where NDPK-D is cross-linking the two mitochondrial membranes, which is kinase inactive.7 Thus, OPA1/NDPK-D complex formation may have a role in determining NDPK-D function, and two pieces of evidence in HeLa cells support this idea. First, only CL-transfer competent NDPK-D proteins also show OPA1 association, not the R90D mutant. Second, more CL is transferred in an OPA1-reduced background as compared with a control background when comparing NDPK-D w/t with R90D after CCCP treatment. This suggests a negative effect of OPA1 on NDPK-D-facilitated CL transfer. Lower OPA1 levels, by reducing OPA1/NDPK-D complexes, could liberate non-complexed NDPK-D that would engage into the well-documented cross-linking of IMM and OMM as required for CL transfer. However, exact delineation of the role of OPA1 for NDPK-D functions will require further molecular analysis of the OPA1/NDPK-D interaction.
Among several quality-control mechanisms in yeast and mammalian cells, mitophagy has recently attracted much attention.45 Most mitophagy studies in mammalian cells have focused on the use of depolarizing agents in triggering the Parkin/Pink1-mediated pathway.46 Increasing evidence, however, supports the notion that mitophagy can also occur independently of Parkin/Pink1.47, 48, 49, 50, 51 The initial report showing that selective mitophagy occurs in PINK1-deficient neuroblastoma cells51 was followed by studies showing that Parkin may be recruited through PINK1-independent mechanisms both in vitro and in vivo.52, 53 Pathways independent of both PINK1 and Parkin have also emerged, often involving mitophagy stimuli that cause lesser degrees of or no mitochondrial depolarization.6, 16 Several mitophagy adaptor proteins (NIX, BNIP3, and FUNDC1) are upregulated or post-translationally modified during developmental or hypoxia-induced mitophagy to directly interact with LC3 via a LIR to recruit autophagosomes.54 AMBRA1, an autophagy-related gene-related protein, has been also shown to bind directly with LC3 via its LIR and to induce LC3-dependent, but Parkin and p62-independent mitophagy.55 Likewise, externalized CL is capable of directly binding LC3 during mitophagy.6 In addition, Parkin-independent mitophagy can be triggered by PMI, which upregulates p62 transcriptionally to facilitate p62-LC3 interactions. As p62 is one of the ubiquitin-binding LIR domain adaptors, this suggests the possibility of additional ubiquitin ligases that can label mitochondria for degradation.16 Indeed, mitochondrial ubiquitination and mitophagy are still observed in Parkin knockout cells.56 Overall, it is clear that multiple specific molecular pathways exist for targeting damaged or unneeded mitochondria for selective clearance by autophagy. A given stimulus, such as CCCP, can trigger more than one pathway to include the canonical Pink1/Parkin pathway, a vesicular lysosomal-targeting pathway,57 and the NDPK-D-dependent CL externalization pathway described here. Based on the comparison of the data on the effects of NDPK-D on CL translocation with those illustrating the role of NDPK-D in Parkin translocation, we suggest that these processes may be related to each other. Our results are compatible with the interpretation that CL may facilitate the accumulation of Parkin in mitochondria during mitophagy. Moreover, LC3 itself interacts with multiple mitochondrial surface molecules, either directly as in the case of CL, FUNDC1 or AMBRA, or with various ubiquitinated substrates through LIR adaptors, to affect mitophagy.
In conclusion, we have found that NDPK-D has an important role in the early stages of mitophagy as induced by CCCP as well as by PINK1-Parkin-independent stimuli such as rotenone and 6-OHDA. Given its dual function of producing nucleoside triphosphates and facilitating CL transfer, as well as its close association with Opa1, NDPK-D is capable of functioning as a key integration site to determine appropriate fission–fusion or mitophagy responses. Overall, the current data – along with the previously established role of CL as a recognition signal for mitophagy of damaged mitochondria6 – identify NDPK-D as an important contributor to the quality control of mitochondrial health.
Materials and Methods
Reagents
CCCP, ROT, 6-OHDA, protease inhibitor cocktail, fetal bovine serum (FBS), and mouse or rabbit actin antibodies were purchased from Sigma (St. Louis, MO, USA). Lipofectamine 2000, Lipofectamine reagent RNAiMax, CellTracker Green CMFDA, LysoTracker Red DND-99, and MitroTracker Red CMXRos were from Life Technologies (Grand Island, NY, USA). Annexin V-FITC was purchased from Biovision (Milpitas, CA, USA). Mouse NDPK-D siRNAs were from Origene (SR404588, Rockville, MD, USA). Human OPA1 siRNAs were from Eurogentec S.A. (Seraing, Belgium). Mouse anti-cyt c, anti-Tim23, and anti-MnSOD antibodies were from BD sciences (San Jose, CA, USA). Mouse-anti-LC3 (5F10) was from Nanotools (Teningen, Germany), and mouse anti-OPA1 antibody from BD Biosciences (Le Pont De Claix, France). The rabbit polyclonal antibody against NDPK-D was obtained as previously described.14 All other reagents were from Sigma unless stated otherwise.
Cell culture
The murine lung epithelial cell line MLE-12 and human cervical adenocarcinoma cell line HeLa were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). MLE-12 cells were cultured at 37 °C and 5% CO2 in DMEM/F-12 medium (ATCC) supplemented with 0.005 mg/ml insulin, 0.01 mg/ml transferrin, 30 nM sodium selenite, 10 nM hydrocortisone, 10 nM beta-estradiol, 2 mM L-glutamine, 10 mM HEPES, and 10% FBS. HeLa cells were maintained in DMEM (Life Technologies) supplemented with 10% FBS.
Transfection
The pcDNA4/TO vectors expressing w/t NDPK-D and R90D mutant were prepared as previously described.14 HeLa cells were transiently transfected using Lipofectamine 2000 according to the manufacturer's instructions. At 48 h after transfection, cells were collected for further experiments. For knocking down the expression of NDPK-D in MLE cells, cells were transfected with 50 nM (final) siRNAs using RNAiMax. For partial silencing OPA, HeLa cells permanently transfected with w/t NDPK-D or R90D mutant were transfected with 50 nM siRNA against OPA1 using lipofectamine, and experiments were performed 72 h after transfection. As controls, cells were mock transfected or transfected with scrambled negative siRNAs. For measurements of mitochondria volume and fragmentation, MLE cells or HeLa were infected with adenovirus expressing mCerulean/FAP with COX-IV and COX-VIII mitochondrial targeting sequences, respectively.
Fluorescence microscopy
For co-localization assay of mitochondria and lysosomes, cells were grown on Matteck Dishes and transiently transfected with Mito-GFP (Clontech, Mountain View, CA, USA). Forty-eight hours after transfection, cells were treated with 20 μM CCCP for the indicated times. Cells were stained with 200 nM LysoTracker Red 30 min prior to the end point according to the manufacturer's instructions. Images were visualized and captured using a Nikon TI inverted microscope in either widefield or with and A1 scan head (Nikon Inc., Melville, NY, USA).
For determination of mitochondrial volume/fragmentation changes, cells were first infected with adenoviral vector expressing mitochondria directed mCerulean/mFAP prior to treatment. Cells were then incubated with 5 nM malachite green to reveal the transgene prior to imaging (thereby defining the mitochondria58) and mounted in a Tokai Hit environmental stage (Tokyo, Japan) in a Nikon TI inverted microscope equipped with a sweptfield confocal head. Three dimensional stacks were collected using a 1.49 NA oil objective with excitation/emission wavelength of 640/700 nm with a Z-space separation of 0.2 micron (20 Z-positions); each stack takes 1 s to collect. Image stacks are collected every 5 min for 60 min. Subsequent to collection images were deconvolved using the 3D Landweber deconvolution capabilities of Nikon Elements. Deconvolved images were imported into Imaris (Bitplane Company, Zurich, Switzerland) and the mitochondrial volume, surface area, and sphericity (defined as the ratio of the surface area of the given object to the surface area of a sphere with the same volume as the given object) were calculated. Imaging conditions included control cells and cells after addition of 20 μM CCCP.
Quantitation of CL levels in mitochondria OMM and IMM
The IMM and OMM fractions were obtained by incubating mitochondria with digitonin.59 After centrifugation, the pellet and supernatant were collected as IMM and OMM fractions, respectively. The purities of the membrane fractions were examined by western blot analysis of COX-IV and TOM40, and by assaying of marker enzymes. Lipids from OMM and IMM were extracted using the Folch procedure.60 Lipid phosphorus was determined by a micro-method.61 CL was analyzed by LC/MS with a Dionex Ultimate 3000 HPLC (using a normal phase column, Luna Silica(2) 100A, 3 μM, 150 × 1 mm, (Phenomenex, Torrance CA, USA) coupled on-line to a linear ion trap LXQ mass spectrometer (ThermoFisher Scientific, San Jose, CA, USA) as previously described.6 Separation of CL was performed usng gradient solvent A (hexane:propanol:water, 47:57:1, v/v) and solvent B (hexane:propanol:water, 47:57:10, v/v) each containing 5 mM ammonium acetate and 0.01M formic acid. The column was eluted at a flow rate of 0.05 mlmin as follows: 0–3 min, linear gradient, 14.5–37% solvent B; 3–12.5 min, isocratic at 37% solvent B; 12.5–20 min, linear gradient, 37–100% solvent B; 20–45 min, isocratic at 100% solvent B; 45–60 min, isocratic at 14.5% solvent B. The ESI probe was operated at a voltage of 3–5 kV in the negative ion mode. Capillary temperature was maintained at 150°C. MS/MS analysis was employed to determine the fatty-acid composition of CL species. Tetra-myristoyl-CL (Avanti polar lipids, Alabaster, AL, USA) was used as internal standard.
Assessment of CL externalized to the mitochondrial surface
To assess CL externalized on the outer leaflet of the OMM, mitochondria were treated with PLA2 from porcine pancreas (0.7 U/mg protein) (Sigma-Aldrich) in mitochondria isolation buffer (210 mM mannitol, 70 mM sucrose, and 5 mM HEPES, pH 7.4) containing 2 mM CaCl2 and 100 μM DTPA for 45 min at 4 °C. To prevent mitochondria damage by CL hydrolysis products, essential-fatty-acid-free human serum albumin (20 mg/ml) was added to the incubation medium. At the end of incubation, lipids were extracted using the Folch procedure.60 Mono-lyso-CL formed in PLA2-driven reaction was analyzed by LC/MS using a Dionex Ultimate 3000 HPLC coupled on-line to a Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (ThermoFisher Scientific) as previously described.62 Mono-lyso-CLs were separated on a normal phase column (Silica Luna 3 μm, 100 A, 150 × 2 mm, Phenomenex) with flow rate 0.2 ml/min using gradient solvents containing 5 mM CH3COONH4 (A – n-hexane:2-propanol:water, 43:57:1 (v/v/v) and B – n-hexane:2-propanol:water, 43:57:10 (v/v/v). Mono-lyso-CL (tri-myristoyl-lyso-CL) was prepared from tetra-myristoyl-CL (Avanti polar lipids) by PLA2 hydrolysis and used as internal standard.
Annexin V-binding assay using flow cytometry
The relative CL amount on the surface of the outer leaflet of the OMM was also evaluated using the newly developed Annexin V-binding assay.6 Cells were stained with 250 nM Mitotracker Red CMXRos (Life Technologies) for 30 min at 37 °C to label mitochondria prior to harvesting. Isolated crude mitochondria were incubated with FITC-labeled Annexin V (Biovision) to stain for anionic phospholipids and then subjected to flow cytometric analysis (FACSCanto, BD, Rutherford, NJ or FACS Fortessa SORP1P, BD, Le Pont de Claix, France) of the green FITC fluorescence. Mitochondrial analysis was performed after appropriate settings of the forward light scatter and side light scatter detectors. The FITC fluorescence from gated red fluorescent mitochondria events was determined to evaluate the binding of Annexin V to mitochondria. Data presented are the relative FITC fluorescence intensity compared with that of mitochondria isolated from non-treated cells.
Western blotting
The harvested cells were lysed on ice for 10 min using RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1% sodium deoxycholate) supplemented with protease inhibitors (Sigma), the lysates were then centrifuged at 5000 × g for 5 min. The resulting supernatants (30 μg protein) were resolved on 12% SDS-PAGE, and transferred to a nitrocellulose membrane. Immuno-blotting was performed as per the antibody manufacturer's details.
In situ proximity ligation assay
In situ PLA was performed using a Duolink kit (Olink Bioscience, Uppsala, Sweden) as previously described.63 Cells grown on chamber microscopy slides were fixed with 3% cold paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS, blocked with a Duolink blocking agent and then incubated with primary antibodies (mouse anti-OPA1, BD Biosciences and rabbit anti NDPK-D produced and characterized as described earlier9). PLA probes (secondary antibodies tagged with DNA oligonucleotides) were then added. Hybridization, ligation, amplification, and detection using Duolink Detection Reagents Red (excitation 594 nm, emission 624 nm) were realized according to the manufacturer's protocol. In brief, secondary antibodies were incubated in preheated humidity chamber for 1 h at 37 °C. Ligation was performed with a ligase-containing solution for 30 min at 37 °C. Finally, amplification step was performed with a polymerase-containing solution for 1 h 40 min at 37 °C. Nuclei were stained with Hoechst 33258. In parallel, expression of OPA1 and NDPK-D was analyzed using the same NDPK-D and OPA1 primary antibodies and standard immunofluorescence procedure (with Alexa Fluor 488- and Cy5-conjugated secondary IgGs). Images were collected with a Leica TCS SP2 AOBS inverted laser scanning confocal microscope using a × 63 oil immersion objective. Laser excitation was 351–364 nm for Hoechst 33258, 488 nm for Alexa 488, 543 nm for PLA, and 633 nm for Cy5. Fluorescence emissions adjusted with AOBS were 480 nm for Hoechst 33258, 500–535 nm for Alexa488, and 565–600 nm for PLA and 645–685 for Cy5. Experiments were performed on a randomly chosen field containing 15–25 cells. The background noise autofluorescence was removed by fine filter (Kernel 3 × 3) using Volocity software (Perkin Elmer, Waltham, MA, USA). Image quantification was performed using ImageJ (NIH images) and Volocity software. The PLA signal was quantified and normalized to cell number.
Statistical analysis
All data presented in the study are means±S.D. Data were analyzed using student's t-test and analysis of variance. For all analyses, a two-sided P-value <0.05 was considered to be statistically significant. Statistical analysis was performed using STATA version 13 (STATA Corp., College Station, TX, USA).
Acknowledgments
This study was supported in part by the National Institutes of Health P01HL114453, U19AI068021, NS076511, NS061817, ES020693, NS065789, AG026389, HL117880, National Institute of Occupational Safety and Health OH008282, Human Frontier Science Program HFSP-RGP0013/2014, and the Barth Syndrome Foundation of Canada and United States. This work was also supported by the Fondation pour la Recherche Médicale (FRM; DPM20121125557), a CMIRA Explo'ra doc fellowship of the Region Rhone Alpes, GEFLUC and the Natural Sciences and Engineering Council of Canada, grant 9848.
Glossary
- ATG
autophagy-related gene-related
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- CL
cardiolipin
- CLS
cardiolipin synthase
- cyt c
cytochrome c
- IMM
inner mitochondrial membrane
- LC-MS
liquid chromatography–mass spectrometry
- LC3
microtubule-associated protein 1A/1B-light chain 3
- LIR
LC3-interacting region
- mFAP
mitochondrial-targeted fluorogen-activating protein
- MLE
mouse lung epithelial
- NDPK-D
nucleoside diphosphate kinase D
- 6-OHDA
6-hydroxydopamine
- OMM
outer mitochondrial membrane
- OPA1
optic atrophy 1
- ROT
rotenone
- PG
phosphatidylglycerol
- PLA
proximity ligation assay
- PLA2
phospholipase A2
- PMI
p62-mediated mitophagy inducer
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Piacentini
Supplementary Material
References
- Kroemer G, Reed JC. Mitochondrial control of cell death. Nat Med 2000; 6: 513–519. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondria and cell signalling. J Cell Sci 2012; 125(Pt 4): 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni HM, Williams JA, Ding WX. Mitochondrial dynamics and mitochondrial quality control. Redox Biol 2014; 4C: 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011; 12: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem 2010; 285: 27879–27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol 2013; 15: 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlattner U, Tokarska-Schlattner M, Ramirez S, Tyurina YY, Amoscato AA, Mohammadyani D et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. J Biol Chem 2013; 288: 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlattner U, Tokarska-Schlattner M, Epand RM, Boissan M, Lacombe ML, Klein-Seetharaman J et al. Mitochondrial NM23-H4/NDPK-D: a bifunctional nanoswitch for bioenergetics and lipid signaling. Naunyn Schmiedebergs Arch Pharmacol 2015; 388: 271–278. [DOI] [PubMed] [Google Scholar]
- Milon L, Meyer P, Chiadmi M, Munier A, Johansson M, Karlsson A et al. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J Biol Chem 2000; 275: 14264–14272. [DOI] [PubMed] [Google Scholar]
- Schlame M, Rua D, Greenberg ML. The biosynthesis and functional role of cardiolipin. Prog Lipid Res 2000; 39: 257–288. [DOI] [PubMed] [Google Scholar]
- Huang Z, Jiang J, Tyurin VA, Zhao Q, Mnuskin A, Ren J et al. Cardiolipin deficiency leads to decreased cardiolipin peroxidation and increased resistance of cells to apoptosis. Free Radic Biol Med 2008; 44: 1935–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sire O, Mangeney M, Montagne J, Bereziat G, Nordmann J. Changes of fatty acid composition of phospholipids and lipid structural order in rat liver mitochondrial membrane subsequent to galactosamine intoxication. Effect of clofibrate. Biochim Biophys Acta 1986; 860: 75–83. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 2010; 189: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarska-Schlattner M, Boissan M, Munier A, Borot C, Mailleau C, Speer O et al. The nucleoside diphosphate kinase D (NM23-H4) binds the inner mitochondrial membrane with high affinity to cardiolipin and couples nucleotide transfer with respiration. J Biol Chem 2008; 283: 26198–26207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RF, Schlattner U, Wallimann T, Lacombe ML, Epand RM. Novel lipid transfer property of two mitochondrial proteins that bridge the inner and outer membranes. Biophys J 2007; 92: 126–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East DA, Fagiani F, Crosby J, Georgakopoulos ND, Bertrand H, Schaap M et al. PMI: a DeltaPsim independent pharmacological regulator of mitophagy. Chem Biol 2014; 21: 1585–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ, Okamoto K. Mitochondria autophagy in yeast. Antioxid Redox Signal 2011; 14: 1989–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem 2008; 283: 32386–32393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tu BP. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol Biol Cell 2011; 22: 4124–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Kang D, Klionsky DJ. Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy 2009; 5: 1186–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 2009; 17: 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Klionsky DJ. A genomic screen for yeast mutants defective in mitophagy. Autophagy 2010; 6: 278–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol 2011; 193: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 2009; 17: 87–97. [DOI] [PubMed] [Google Scholar]
- Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol 2009; 187: 959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVicar TD, Lane JD. Impaired OMA1-dependent cleavage of OPA1 and reduced DRP1 fission activity combine to prevent mitophagy in cells that are dependent on oxidative phosphorylation. J Cell Sci 2014; 127(Pt 10): 2313–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem 2003; 278: 7743–7746. [DOI] [PubMed] [Google Scholar]
- Fry M, Green DE. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. J Biol Chem 1981; 256: 1874–1880. [PubMed] [Google Scholar]
- Vik SB, Georgevich G, Capaldi RA. Diphosphatidylglycerol is required for optimal activity of beef heart cytochrome c oxidase. Proc Natl Acad Sci USA 1981; 78: 1456–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pember SO, Powell GL, Lambeth JD. Cytochrome P-450scc-phospholipid interactions. Evidence for a cardiolipin binding site and thermodynamics of enzyme interactions with cardiolipin, cholesterol, and adrenodoxin. J Biol Chem 1983; 258: 3198–3206. [PubMed] [Google Scholar]
- Beyer K, Klingenberg M. ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by 31P nuclear magnetic resonance. Biochemistry 1985; 24: 3821–3826. [DOI] [PubMed] [Google Scholar]
- Kadenbach B, Mende P, Kolbe HV, Stipani I, Palmieri F. The mitochondrial phosphate carrier has an essential requirement for cardiolipin. FEBS Lett 1982; 139: 109–112. [DOI] [PubMed] [Google Scholar]
- Noel H, Pande SV. An essential requirement of cardiolipin for mitochondrial carnitine acylcarnitine translocase activity. Lipid requirement of carnitine acylcarnitine translocase. Eur J Biochem 1986; 155: 99–102. [DOI] [PubMed] [Google Scholar]
- Paradies G, Ruggiero FM. Effect of hyperthyroidism on the transport of pyruvate in rat-heart mitochondria. Biochim Biophys Acta 1988; 935: 79–86. [DOI] [PubMed] [Google Scholar]
- Bazan S, Mileykovskaya E, Mallampalli VK, Heacock P, Sparagna GC, Dowhan W. Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J Biol Chem 2013; 288: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnarez C, Mazat JP, Elezgaray J, Marrink SJ, Periole X. Evidence for cardiolipin binding sites on the membrane-exposed surface of the cytochrome bc1. J Am Chem Soc 2013; 135: 3112–3120. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuan Szklarz LK, Schulze-Specking A et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J 2004; 23: 3735–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amutha B, Pain D. Nucleoside diphosphate kinase of Saccharomyces cerevisiae, Ynk1p: localization to the mitochondrial intermembrane space. Biochem J 2003; 370(Pt 3): 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bao R, Jiang C, Yang Z, Zhou CZ, Chen Y. Structure of Ynk1 from the yeast Saccharomyces cerevisiae. Acta Crystallogr Sect F Struct Biol Crystal Commun 2008; 64(Pt 7): 572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol 2005; 1: 223–232. [DOI] [PubMed] [Google Scholar]
- Alavi MV, Fuhrmann N. Dominant optic atrophy, OPA1, and mitochondrial quality control: understanding mitochondrial network dynamics. Mol Neurodegener 2013; 8: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer P, Pellegrini L. The dynamin GTPase OPA1: more than mitochondria? Biochim Biophys Acta 2013; 1833: 176–183. [DOI] [PubMed] [Google Scholar]
- Baker MJ, Lampe PA, Stojanovski D, Korwitz A, Anand R, Tatsuta T et al. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J 2014; 33: 578–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissan M, Montagnac G, Shen Q, Griparic L, Guitton J, Romao M et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science 2014; 344: 1510–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ 2013; 20: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008; 183: 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature 2008; 454: 232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 2009; 29: 2570–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 2012; 14: 177–185. [DOI] [PubMed] [Google Scholar]
- Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep 2013; 14: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem 2009; 284: 13843–13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JY, Kang MJ, Kim KH, Han PL, Kim HS, Ha JY et al. Nitric oxide induction of Parkin translocation in PTEN-induced putative kinase 1 (PINK1) deficiency: functional role of neuronal nitric oxide synthase during mitophagy. J Biol Chem 2015; 290: 10325–10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli DA, Cortez MQ, Moyzis AG, Najor RH, Lee Y, Gustafsson AB. PINK1 is dispensable for mitochondrial recruitment of parkin and activation of mitophagy in cardiac myocytes. PloS One 2015; 10: e0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 2014; 157: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ 2015; 22: 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J 2014; 33: 2798–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol 2012; 22: 135–141. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C, Schmidt BF, Creeger Y, Fisher GW, Zakel KL, Adler S et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol 2008; 26: 235–240. [DOI] [PubMed] [Google Scholar]
- Montamat EE, Blanco A. Subcellular distribution of the lactate dehydrogenase isozyme specific for testis and sperm. Exp Cell Res 1976; 103: 241–245. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957; 226: 497–509. [PubMed] [Google Scholar]
- Chalvardjian A, Rudnicki E. Determination of lipid phosphorus in the nanomolar range. Anal Biochem 1970; 36: 225–226. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Poloyac SM, Tyurin VA, Kapralov AA, Jiang J, Anthonymuthu TS et al. A mitochondrial pathway for biosynthesis of lipid mediators. Nat Chem 2014; 6: 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 2006; 3: 995–1000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.