Abstract
During apoptosis, proapoptotic BAX and BAK trigger mitochondrial outer membrane (MOM) permeabilization by a mechanism that is not yet fully understood. BH3-only proteins such as tBID, together with lipids of the MOM, are thought to play a key role in BAX and BAK activation. In particular, cardiolipin (CL) has been shown to stimulate tBID-induced BAX activation in vitro. However, it is still unclear whether this process also relies on CL in the cell, or whether it is more dependent on MTCH2, a proposed receptor for tBID present in the MOM. To address this issue, we deleted both alleles of cardiolipin synthase in human HCT116 cells by homologous recombination, which resulted in a complete absence of CL. The CL-deficient cells were fully viable in glucose but displayed impaired oxidative phosphorylation and an inability to grow in galactose. Using these cells, we found that CL was not required for either tBID-induced BAX activation, or for apoptosis in response to treatment with TRAIL. Downregulation of MTCH2 in HCT116 cells also failed to prevent recruitment of tBID to mitochondria in apoptotic conditions. However, when both CL and MTCH2 were depleted, a significant reduction in tBID recruitment was observed, suggesting that in HCT116 cells, CL and MTCH2 can have redundant functions in this process.
Mitochondrial outer membrane (MOM) permeabilization (MOMP) is a crucial event in the so-called intrinsic pathway of apoptosis, which is triggered by a large number of death stimuli. This process requires activation of the proapoptotic members of the BCL-2 family, BAX and BAK, which are directly responsible for MOMP.1, 2 Activation of BAX and BAK results from complex conformational changes that induce oligomerization of these proteins and the formation of pores in the MOM.3 Permeabilization of the MOM then leads to the efflux into the cytosol of cytochrome c (cyt c) and other apoptogenic factors normally confined within the mitochondrial intermembrane space (IMS), triggering the activation of the caspase cascade and cell death.4, 5
Activation of BAX and BAK requires a subclass of the BCL-2 family members known as the BH3-only proteins.6 Among them, BID has been particularly well studied and found to activate BAX and BAK directly following cleavage by caspase 8 or other proteases.7, 8 Caspase 8 is activated in the so-called extrinsic pathway of apoptosis, which is initiated upon engagement of TNF superfamily receptors, such as FAS and TRAIL receptors, by their respective ligands.9 Once activated, caspase 8 can cleave and activate executioner caspases leading to the ordered dismantling of the cell. This direct activation of downstream caspases by caspase 8 is characteristic of ‘type I cells'. On the other hand, in ‘type II cells' in which caspase activation is inhibited by high expression of inhibitors of apoptosis (IAPs), engagement of the intrinsic apoptotic pathway through caspase 8-mediated cleavage of BID is essential for apoptosis to proceed.10 In type II cells, the cleaved form of BID (tBID) translocates to the MOM where it recruits and activates BAX and/or BAK, leading to MOMP and cell death.7, 8 Thus, formation of tBID is a crucial link between the extrinsic and intrinsic apoptotic pathways, and therefore of central importance to the onset of apoptosis.
The mitochondrial-specific phospholipid cardiolipin (CL) is thought to be required for many aspects of mitochondrial physiology, including oxidative phosphorylation and mitochondrial dynamics.11 It has also been proposed to have an important role in the regulation of MOMP,12 and particularly in the recruitment of tBID and BAX to the mitochondria.13, 14, 15, 16, 17, 18, 19 Moreover, in type II cells exposed to FASL, it has been suggested that complete activation of caspase 8 is facilitated by the presence of CL in the MOM.20 In apoptotic conditions, CL becomes peroxidized, and this event is thought to weaken its interaction with cyt c, thus facilitating cyt c release after MOMP.21 In addition, monolyso-CL (a CL metabolite whose level increases during apoptosis) may also interact with tBID and thus have a role in its recruitment to mitochondria.22 Redistribution of CL from the mitochondrial inner membrane (MIM) to the MOM, which was shown to occur under apoptotic conditions, is also thought to favor these protein–lipid interactions.23, 24 Despite these observations, other studies have questioned the requirement for CL in apoptosis and MOMP, and have suggested that CL is much less important for tBID-induced BAX activation than previously anticipated.25, 26, 27, 28 In addition, it has been recently shown that MTCH2, a transmembrane protein of the MOM, can also act as a receptor for tBID in the mitochondria during apoptosis.29
To investigate the function of CL during apoptosis, we have generated a derivative of the human colorectal cancer cell line HCT116, in which we have deleted the cardiolipin synthase (CLS1) gene. The cells obtained were completely devoid of CL. Even though mitochondrial respiration was impaired in CL-depleted cells, they were viable. Looking at MOMP and apoptosis, we could observe that these processes function normally in the absence of CL. To investigate this further, we used these cells to generate other cell lines stably expressing a shRNA directed against MTCH2. Our results show that depletion of either CL or MTCH2 alone had no effect on tBID recruitment to mitochondria following the induction of apoptosis. In contrast, we observed a strong reduction in tBID recruitment in cells depleted for both CL and MTCH2, indicating that in HCT116 cells, they appear to have a redundant function. However, this impairment in tBID recruitment did not correlate with reduced cell death, suggesting that apoptosis can still proceed normally in the absence of CL and MTCH2. Interestingly, when MTCH2 downregulation alone was performed in HeLa cells, tBID recruitment to mitochondria after apoptotic stimulation was impaired, indicating that the dependency for CL and/or MTCH2 in this process is affected by the cellular context.
Results
Generation of HCT116 cells lacking CLS1
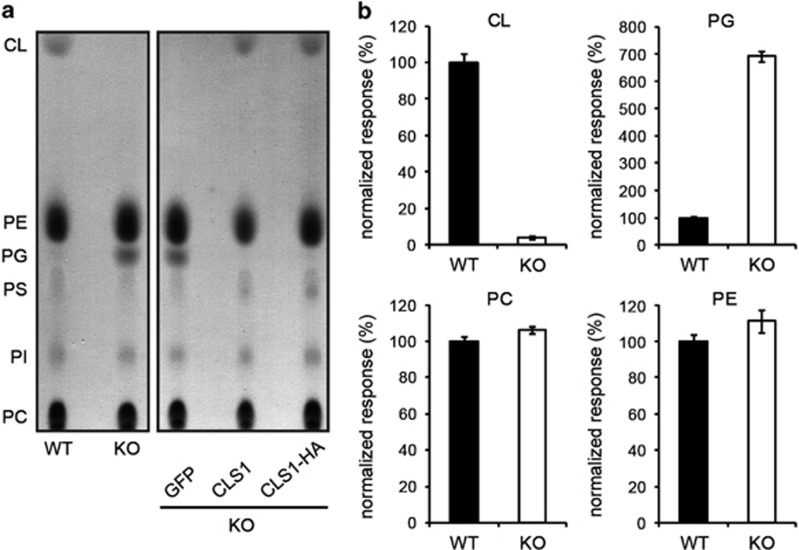
To investigate the function of CL in apoptosis, we deleted both alleles of the CLS1 gene in the human colorectal cancer cell line HCT116 by homologous recombination (Supplementary Figure 1). The resulting cells were completely devoid of CL as evidenced by thin-layer chromatography (TLC) of mitochondrial lipid extracts (Figure 1a). In accordance with previous observations in yeast, we also observed a concomitant increase in the level of phosphatidylglycerol (PG), the precursor of CL.30, 31 Stable expression of the CLS1 cDNA in knock-out (KO) cells using lentiviral vectors restored CL to the normal level, and both wild-type (WT) and C-terminally HA-tagged proteins were equally functional. As a negative control, KO cells stably expressing GFP were also produced using the same procedure, and as expected, no change in CL was observed in these cells. These data were confirmed by mass spectrometry analysis of total cell lipid extracts (Figure 1b).
Figure 1.
Analysis of mitochondrial lipids in WT and CLS1 KO cells using TLC and mass spectrometry. (a) Mitochondrial lipid extracts from the cell lines indicated were separated by TLC. (b) Mass spectrometric analysis of total lipids extracted from WT and CLS1 KO cells. WT values were set at 100%. Mean±S.E.M. of at least eight experiments. CL, cardiolipin; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PS, phosphatidylserine; PI, phosphatidylinositol; PC, phosphatidylcholine
Effects of CLS1 KO on mitochondrial respiration and morphology
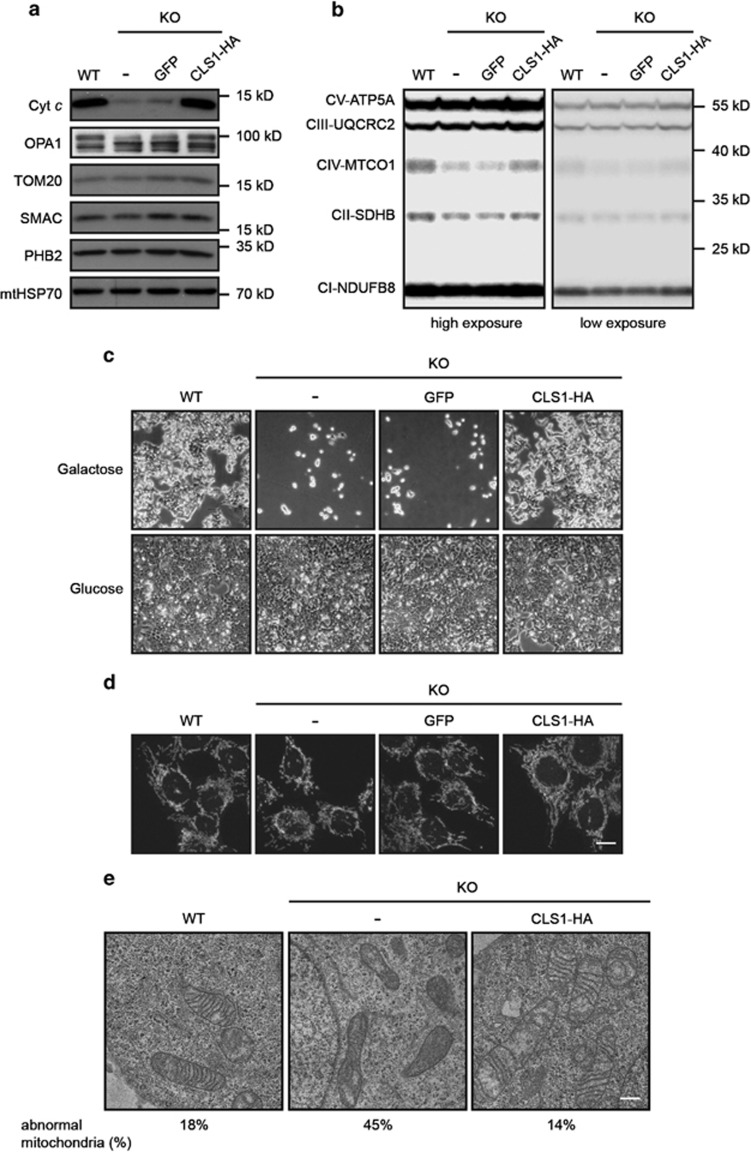
We then used western blotting to investigate the consequences of CL deficiency on different mitochondrial proteins. Interestingly, we found a dramatic decrease in the amount of cyt c in KO cells, suggesting that CL has a role in its biogenesis and/or stability (Figure 2a). Depletion of CL also led to an altered profile of OPA1 forms (Figure 2a). This is in agreement with the work of others showing that perturbation of mitochondrial lipids results in cleavage of OPA1 by the protease OMA1.32 OPA1 has been shown to have a role in cristae morphogenesis and mitochondrial fusion.33 Strikingly, despite the lack of CL and alterations in OPA1 processing profile, the morphology of the mitochondrial network was not affected (Figure 2d). We do not exclude the possibility that compensatory mechanisms in KO cells may be responsible for this absence of phenotype. However, mitochondrial ultrastructure was altered in CLS1 KO cells, as expected. Indeed, these mitochondria exhibited increased electron density with less and disorganized cristae (Figure 2e), which is in agreement with previously published results.33, 34, 35, 36, 37
Figure 2.
Effect of CLS1 KO on mitochondrial respiration and morphology. (a) Western blot analysis of mitochondrial proteins from the cell lines indicated. Controls for each mitochondrial compartment were: TOM20 (MOM), SMAC (IMS), PHB2 (MIM) and mtHSP70 (matrix). (b) Presence of respiratory chain protein components as monitored by western blot. An antibody mix which recognizes members of all five complexes of the oxidative phosphorylation machinery was used. (c) The indicated cell lines were seeded at 5% confluency and grown for 5 days in medium containing galactose or glucose as carbon source. Final confluency was monitored using phase contrast microscopy. (d) Visualization of mitochondrial morphology by confocal microscopy after staining with MitoTracker Red. Scale bar, 10 μm. (e) Mitochondrial ultrastructural analysis of the lines indicated using transmission electron microscopy. Scale bar, 0.5 μm. The percentage of abnormal mitochondria is indicated for each condition
In addition to reduction in cyt c, we also observed a decrease in the protein level of MTCO1 (Figure 2b). A reduction in the amount of this labile protein suggests a defect in respiratory chain complex IV assembly. On the basis of these observations and on the fact that other studies have shown a requirement for CL in proper functioning of the respiratory chain,38, 39 we decided to investigate whether mitochondrial respiration was affected in the CLS1 KO line. To test this, cells were incubated in a medium containing galactose in which cell growth depends on functional oxidative phosphorylation.40 As a control, cells were grown in normal glucose-containing medium in which energy production occurs mainly via glycolysis. Strikingly, in the absence of CL, cells were unable to grow in galactose medium (Figure 2c), which points to a defective mitochondrial respiratory chain. However, no increase in ROS production was observed (data not shown).
Absence of CL does not affect apoptosis
To characterize the role of CL in the regulation of apoptosis and, in particular, in the activation of BCL-2 family proteins, we took advantage of the TRAIL cytokine. TRAIL is a potent inducer of the mitochondrial apoptotic pathway in HCT116 cells, which induces caspase 8 activation at the plasma membrane, cleavage of BID, BAX activation at the MOM and cyt c release.7
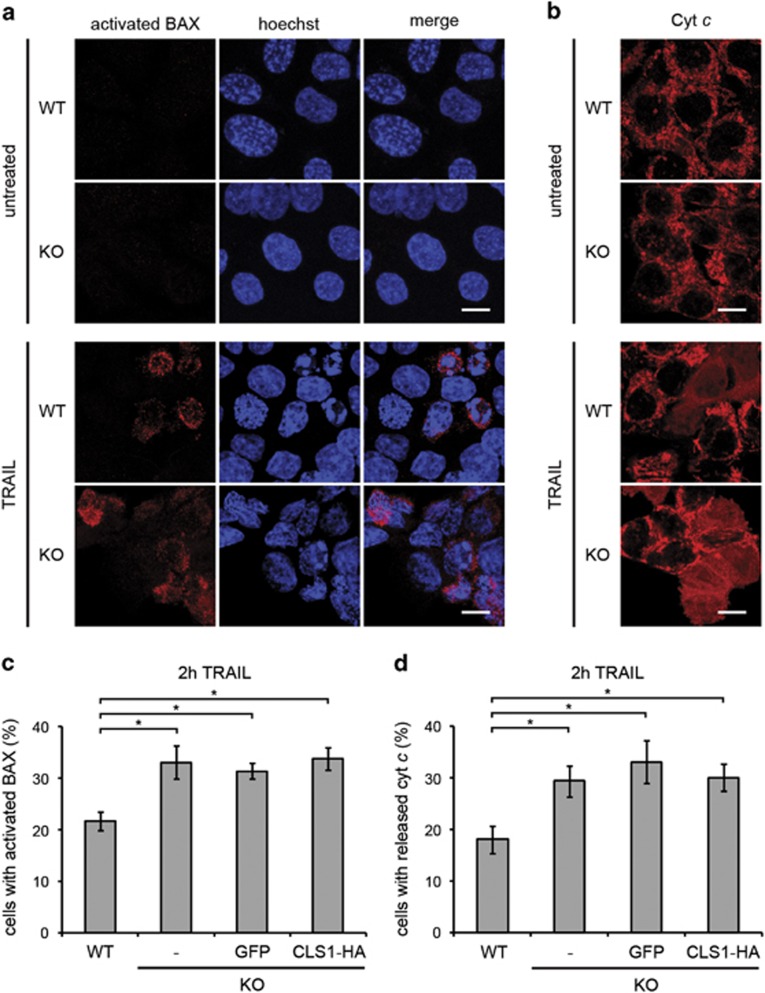
We found that the lack of CL did not reduce the apoptotic response of HCT116 cells following the treatment with TRAIL. Indeed, activation of BAX as assessed by exposure of its N terminus (Figure 3a and c), and release of cyt c (Figures 3b and d) were both found to be slightly increased in CL-deficient cells compared with WT. However, this was also the case in cells rescued by expression of recombinant CLS1 in which CL was restored to normal levels, suggesting that this effect was CL-independent.
Figure 3.
CL depletion does not affect BAX activation and cyt c release. (a and b) Immunostaining of BAX N terminus and cyt c in WT and KO cells, without or after 2 h exposure to TRAIL (50 ng/ml). Cells were imaged by confocal microscopy. Scale bars, 10 μm. (a) The BAX 2D2 antibody used recognizes residues 3–16 of human BAX. (b) Laser power was increased to image cyt c in CLS1 KO cells to obtain a comparable signal intensity to that obtained in WT cells. (c and d) Quantification of the data obtained in a and b for cells treated with TRAIL. Data from KO cells expressing GFP and CLS1-HA were also included. For untreated cells, quantification was not performed owing to an extremely low count of positive cells in all the conditions. Graphs represent mean±S.E.M. of three independent experiments. *P<0.05 based on a Student's unpaired t-test
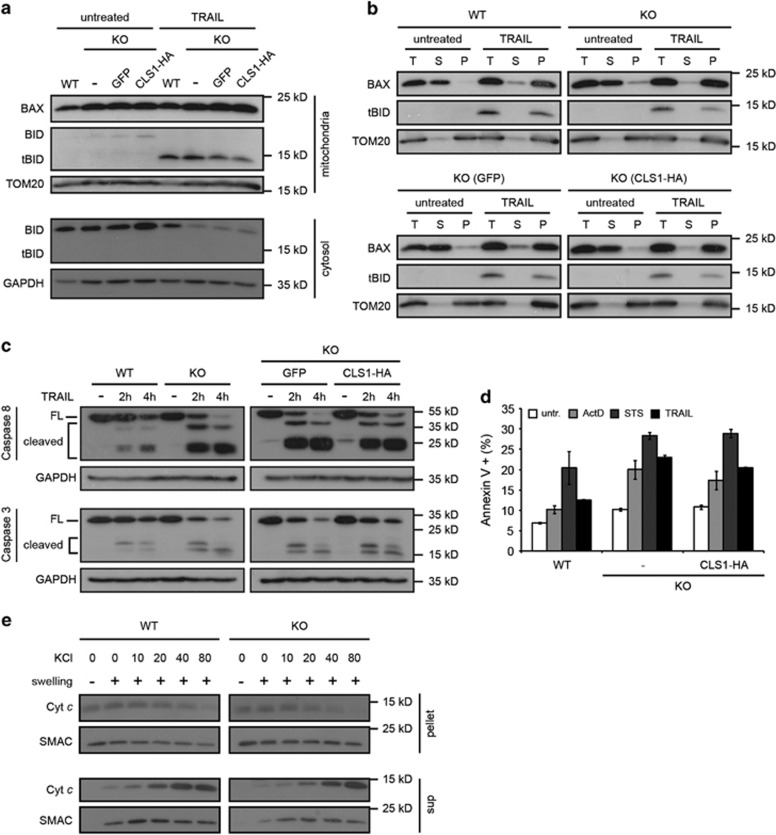
Consistent with these data, we found that cleavage of BID and translocation of tBID to mitochondria after apoptotic stimulation was not significantly affected in the absence of CL (Figure 4a), and insertion of BAX and tBID into the MOM as monitored by resistance to sodium carbonate extraction was also normal (Figure 4b). We then looked at caspase activation after induction of apoptosis and found equivalent amounts of cleaved fragments of caspase 8 and caspase 3 in KO and rescued lines (Figure 4c). In addition, when FITC-coupled annexin V was used to assess induction of apoptosis after treatment with TRAIL or other death stimuli, such as staurosporine or actinomycin D, KO and rescued cells showed comparable staining (Figure 4d), suggesting that CL is also not required in apoptotic pathways that do not use tBID as BAX activator. Finally, we also assessed the availability of cyt c in the IMS of WT and KO mitochondria. This was achieved by incubating isolated swelled mitochondria in solutions of different ionic strengths. In both the cases, we observed that cyt c dissociated similarly from the MIM, and this in a salt concentration-dependent manner (Figure 4e), suggesting that CL does not affect cyt c affinity for the MIM. Altogether, these experiments show that CL is not required for tBID-induced BAX activation, caspase activation and cell death in HCT116 cells.
Figure 4.
CL is neither required for insertion of tBID and BAX into the MOM, nor for caspase activation and apoptosis. (a and b) Cells were left untreated or exposed to 50 ng/ml TRAIL for 2 h. (a) Mitochondrial and cytosolic fractions were subjected to immunoblot analysis using the antibodies indicated. (b) Sodium carbonate extraction of proteins from purified mitochondria followed by ultracentrifugation to separate solubilized proteins from the insoluble, membrane-integrated proteins. T, total extract; S, supernatant; P, pellet. (c) Analysis of caspase 3 and caspase 8 cleavage by immunoblot analysis of total cell extracts untreated or exposed to 50 ng/ml TRAIL as indicated. FL, full-length protein. (d) Indicated cell lines were left untreated or stimulated as follows: 24 h with actinomycin D (10 μM), 6 h with staurosporine (2 μM) or 3 h with TRAIL (50 ng/ml). The proportion of apoptotic cells was quantified by FACS analysis after exposure of cells to FITC-coupled annexin V. Graphs represent mean±S.E.M. of three independent experiments. (e) Isolated mitochondria from WT and CLS1 KO cells were left intact or swelled in hypotonic buffer before addition of an increasing concentration of KCl to dissociate IMS proteins from the MIM. Final concentrations of KCl in μM are indicated. After centrifugation, soluble and membrane-bound proteins were subjected to immunoblot analysis using the antibodies indicated. Exposure time was adjusted to obtain comparable signals for cyt c in WT and CLS1 KO cells. SMAC was used as a control as it is not affected by salt concentration
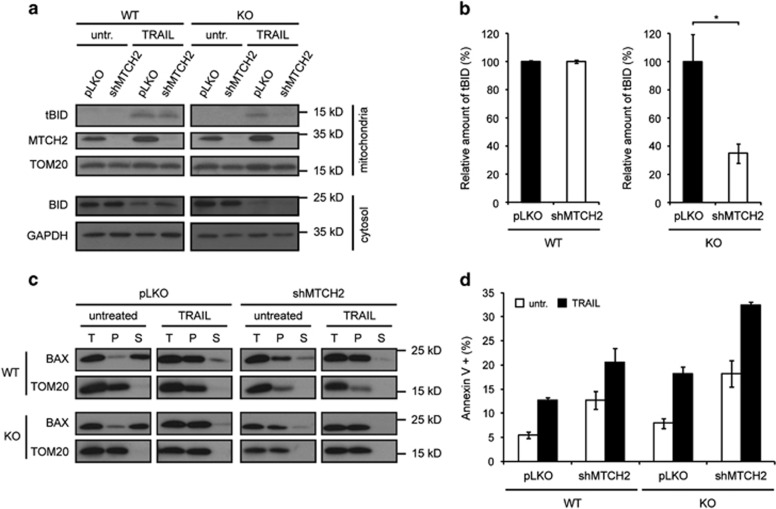
MTCH2 downregulation in CLS1 KO cells reduces mitochondrial recruitment of tBID
In the light of recent studies indicating that MTCH2 may be a receptor for tBID in the MOM,29, 41 we wished to study the effect of MTCH2 downregulation on tBID recruitment to mitochondria, following induction of cell death in WT and CLS1 KO cells. We found that in cells depleted of MTCH2, tBID was normally recruited to mitochondria upon treatment with TRAIL. However, downregulation of MTCH2 in the CLS1 KO cell line resulted in impaired tBID recruitment (Figure 5a and b), indicating that even though CL may not be required for MOMP in normal HCT116 cells, it is nevertheless able to recruit tBID to mitochondria in the absence of MTCH2. Therefore, CL and MTCH2 appear to have a redundant function during apoptosis in HCT116 cells. However, whether it is CL or MTCH2 that is most important in this process seems to be dependent on the cellular context. Indeed, MTCH2 downregulation alone in HeLa cells was sufficient to impair tBID recruitment to mitochondria after TRAIL treatment (Supplementary Figure 2).
Figure 5.
Effects of MTCH2 depletion on tBID recruitment, BAX activation and apoptosis in WT and CLS1 KO HCT116 cells. (a–d) Lentiviral infections with a shRNA was performed to downregulate MTCH2 in WT and CLS1 KO cells. Apoptosis was induced by exposing cells to 50 ng/ml TRAIL. (a) Mitochondrial and cytosolic fractions of untreated and 2 h TRAIL-treated cells were subjected to immunoblot analysis with the antibodies indicated. (b) Quantification of the data obtained in a for cells treated with TRAIL as described in the Materals and Methods. Percentages indicate the amount of tBID relative to the loading control. Graphs represent mean±S.E.M. of three independent experiments. *P<0.05 based on Student's unpaired t-test. (c) Sodium carbonate extraction of mitochondrial proteins from untreated and 2 h TRAIL-treated cells, followed by ultracentrifugation to separate soluble from insoluble membrane-integrated proteins. T, total; S, supernatant; P, pellet. (d) PS exposure of untreated and 3 h TRAIL-treated cells was quantified by FACS analysis after exposure of cells to FITC-coupled annexin V. Graphs represent mean±S.E.M. of three independent experiments
Reduced tBID recruitment to the MOM does not prevent BAX insertion and induction of apoptosis
We then checked whether the reduced tBID recruitment seen previously resulted in impaired BAX activation and apoptosis. Surprisingly, sodium carbonate extraction of mitochondrial proteins revealed a pre-insertion of BAX into the MOM of MTCH2-depleted WT and KO cells cultured under normal conditions (Figure 5c). This was accompanied by increased spontaneous apoptosis (Figure 5d), suggesting that depletion of MTCH2 in HCT116 cells may produce some form of cellular stress, which in turn leads to a tBID-independent pre-activation of BAX. After treatment with TRAIL, BAX insertion into the MOM (Figure 5c) and the percentage of apoptotic cells (Figure 5d) increased even further, regardless of CL presence. This suggested that apoptosis could still proceed normally, independent of both MTCH2 and CL.
Discussion
In this study, we have used homologous recombination to disrupt the CLS1 gene in HCT116 cells (Supplementary Figure 1a). This resulted in total absence of CL as judged by TLC and mass spectrometric analysis (Figure 1a and b). The KO cells were perfectly viable when grown under standard conditions (Figure 2c), making them a suitable model for the study of apoptotic cell death. To our knowledge, this is the first report of a mammalian cell completely devoid of CL that is capable of proliferating normally. As a control, we also generated rescued cells by stably reintroducing the CLS1 cDNA into KO cells.
The main goal of our work was to characterize the importance of CL in apoptosis, and more specifically its role in tBID-induced activation of BAX and MOMP. According to the ‘embedded together' model, a subgroup of BH3-only proteins, including BID, BIM and PUMA, described as ‘activators' due to their ability to bind directly to BAX or BAK, recruit BAX into the MOM and promote its activation.42 These events can be simulated in vitro using synthetic membranes, and in this system, it has been shown that the presence of CL in the membrane was required for tBID recruitment and BAX insertion.13, 15, 16, 17, 18, 19 These results raised the possibility that CL could have a similar role in mitochondria during apoptosis. However, involvement of CL in BAX activation has been questioned, in particular, in yeast. Indeed, it has been shown that CL is not essential for BAX-mediated MOMP in this organism.25, 27 Additional studies in mammalian cells and liposomes have also questioned a specific requirement for CL in MOMP.26, 28 Some authors even showed increased kinetics of apoptosis in cells with less CL.35 Importantly, MTCH2, a protein of the mitochondrial carrier family inserted in the MOM, has been recently proposed to act as a receptor for tBID under apoptotic conditions.29, 41
To test the involvement of CL in apoptosis, we chose to focus on TRAIL-induced apoptosis in HCT116 cells. These cells show a type II response43 and apoptosis is dependent on tBID and BAX, but not on BAK that is tightly inhibited by MCL-1.44 To our surprise, we observed that caspase 8 was normally cleaved in CL-depleted cells (Figure 4c). This result is in contrast with previous observations showing that CL is required for caspase 8 activation.20 However the latter findings were made using tafazzin-deficient cells in which the structure of CL is abnormal, and thus the cellular context is different from the CL-deficient HCT116 cells used here. In addition, we found that the subsequent steps of the apoptotic pathway were also not affected. Indeed, BID was cleaved normally (Figure 4a), and tBID and BAX were efficiently recruited to mitochondria (Figure 4a and b), resulting in cyt c release (Figures 3b and d). Finally, caspase 3, the major executioner of apoptosis, was normally processed (Figure 4c). These observations suggest that CL appears to be much less important than initially anticipated from experiments using synthetic membranes. Alternatively, it is possible that cells display strategies to compensate CL deficiency, although we do not think that accumulation of PG, the precursor of CL, could be part of such compensatory mechanisms, as previous experiments showed that PG cannot substitute for CL in the regulation of MOMP.13, 27 In contrast, MTCH2, which has previously been reported to act as a tBID receptor,29, 41 could compensate for the CL deficit. In favor of this hypothesis, we observed an effect of MTCH2 depletion on tBID recruitment in cells depleted of CL (Figures 5a and b), suggesting that CL and MTCH2 may have compensatory roles in directly binding and recruiting tBID to the MOM, an idea already proposed by others.19 It should also be mentioned that the relative importance of CL and/or MTCH2 for tBID recruitment seems to depend on the cellular environment, as tBID recruitment is impaired in CL-containing MTCH2-depleted HeLa cells (Supplementary Figure 2).
Despite a significant decrease in tBID recruitment at the mitochondria, we found that cells lacking both CL and MTCH2 underwent apoptosis normally (Figure 5d). This is in contradiction with previous results from the Gross laboratory showing that MTCH2 KO cells displayed less tBID to the MOM in apoptotic conditions and resisted apoptosis induced by various stimuli.29 Slight variations in the apoptotic response of the different cell types used may possibly explain this discrepancy. The fact that we used cells only partially depleted for MTCH2 could also explain this observation. In addition, they also reported in the same study that in the absence of MTCH2, BAX activation was delayed. In contrast to these results, our experiments revealed that depletion of MTCH2 was sufficient to prime HCT116 cells to apoptosis through pre-insertion of BAX into the MOM of untreated cells (Figure 5c), indicating that depletion of MTCH2 is somehow stressful for these cells. It is thus possible that these stressful conditions lower the required amount of tBID at the MOM for apoptosis induction, therefore, allowing the death of MTCH2-deficient CLS1 KO cells after TRAIL treatment. MTCH2 belongs to the family of mitochondrial carriers, but its function remains unclear.45 A reduction in its carrier activity or in another putative function may be responsible for the priming observed in HCT116 cells. To overcome this issue and thus assess the role of MTCH2 during cell death in the absence of CL, it would be necessary to express a mutant of MTCH2 that fails to bind tBID but displays a normal activity. This experiment will become possible when the exact function of MTCH2 will be identified.
In conclusion, we have generated mammalian cells that are devoid of CL and grow normally under standard growth conditions. These cells have been instrumental in assessing the role of CL in tBID-induced BAX activation and apoptosis since they allowed us to understand that MTCH2 and CL participate in the recruitment and integration of tBID into the MOM. This recruitment possibly occurs in membrane microdomains, such as contact sites between the MOM and the MIM,14 where CL is expected to be enriched.46 Even though we do not know whether MTCH2 is also enriched in these microdomains, this raises the possibility that CL and MTCH2 might be involved in their formation, an intriguing question to address in the future.
Materials and Methods
Cell culture
HCT116 cells were cultured in McCoy's 5a medium supplemented with 10% fetal bovine serum, 2 mM glutamine and 100 U/ml penicillin+0.1 mg/ml streptomycin. Other cell lines were maintained in DMEM containing the same supplements. For growth studies of HCT116 cells in galactose and glucose, media compositions were as follows: DMEM without glucose (Thermo Fisher Scientific, Ecublens, Switzerland) supplemented as described above, and containing 1 mM sodium pyruvate and 10 mM galactose or glucose as specified.
Gene targeting
Disruption of the CLS1 locus in HCT116 cells was based on previously published procedures47, 48 with some modifications. Vectors were a gift of R. Youle. The CLS1 gene comprises seven exons. We decided to target the third exon for the following reasons. First, the putative catalytic site (CDP-OH-P motif) of CLS1 is located mainly in this exon,49 and second, the disruption of the third exon leads to a frameshift and the presence of a stop codon at the sixth position of the fourth exon. Thus, targeting this exon results in the formation of an inactive truncated protein. The targeting construct used was made of several elements: two ~1 kb homology arms corresponding to the end of the second intron and the beginning of the third intron. These sequences were analyzed online with RepeatMasker to verify that they did not contain repeated sequences found elsewhere in the genome. Between these homology arms were located a Neomycin cassette flanked by two FRT sites and the third exon flanked by two LoxP sites. The complete insert was synthesized by GenScript. It was then cloned into the NotI site of pAAV-MCS, resulting in the formation of the pAAV-CLS1 KO plasmid. This vector was co-transfected with pAAV-RC and pHelper in AAV-293 cells to produce adeno-associated virus particles. HCT116 cells were then infected with these viral particles, diluted and plated in ten 96-well plates at an estimated confluency that would result in the formation of less than 15 resistant clones per plate. Selection with G418 was performed until resistant colonies of a manageable size were obtained. PCR screening of genomic DNA was used to verify correct integration of the targeting construct. Positive clones were expanded and frozen. Transient transfection of pPGKFLPobpA allowed the expression of flippase. After that, cells were diluted to obtain single colonies. Cassette excision was confirmed by G418 sensitivity and PCR screening. Positive clones were again expanded and frozen. Targeting of the second allele was performed by repeating the procedure described above except that in this case we did not remove the resistance cassette. PCR screening was again used to confirm correct integration of the construct. Finally, the Cre recombinase was expressed using Ad-Cre-GFP adenovirus (Vector Biolabs, Malvern, PA, USA). Cells were diluted to obtain single colonies, which were then tested for double excision of the third exon by PCR. Positive clones were considered as CLS1 KO, expanded and frozen. To generate rescued cell lines, we cloned the CLS1 cDNA with or without the HA tag into pWPT (gift from P. Salmon). Lentiviral particles were produced by co-transfecting this vector with psPAX2 and pMD2.G in HEK 293 T cells. HCT116 CLS1 KO cells were subsequently infected with these viral particles. Infection efficiency was assessed by immunofluorescence using an anti-HA antibody (Covance, Geneva, Switzerland). More than 90% of the cells showed a strong mitochondrial staining. A control cell line stably expressing GFP was also produced using the same procedure.
Lipid extraction and TLC
Lipids from crude mitochondrial fractions (500 μg proteins quantified by Bradford assay) were extracted as described below. Mitochondria were resuspended in a solution of 0.9% w/v NaCl, 10 mM HCl in a final volume of 260 μl. One milliliter of MeOH/CHCl3 at a 2/1 ratio was added. After vortexing, the mixture was incubated for 10 min on ice. A total 666 μl CHCl3/(0.9% w/v NaCl, 10 mM HCl) at a ratio of 1/1 was added. The mix was again vortexed and incubated on ice for a further 10 min. After centrifugation at 200 g for 5 min at 4 °C, the lower organic phase was recovered and dried under a N2 flow. Dried lipids were then resuspended in 50 μl CHCl3. A silica gel TLC plate was predeveloped in MeOH/CHCl3 at a 1/1 ratio. After air-drying for 5 min at room temperature (RT), the plate was incubated in a solution of 1.8% w/v boric acid in 100% EtOH for a few seconds and again air-dried for 5 min at RT. The plate was then heated at 100 °C for 15 min in an oven. Samples and lipid standards were subsequently spotted before separation in a closed chamber. The composition of the mobile phase was CHCl3/EtOH/H2O/Et3N (30/35/7/35). After separation, the plate was air-dried for 5 min at RT, then for 4 h at 100 °C in the oven, and finally overnight at RT. Staining was performed using iodine vapor.
Lipid measurements by mass spectrometry
Cell pellets were resuspended in water and sonicated for 30 s (8 W) using a tip sonicator. Phospholipids were extracted using a single-phase extraction. A defined amount of internal standards (0.1 nmol of CL(14:0)4, 0.2 nmol of BMP(14:0)2, 2.0 nmol of PC(14:0)2, 0.1 nmol of PG(14:0)2, 5.0 nmol of PS(14:0)2, 0.5 nmol of PE(14:0)2, 1.0 nmol of PA(14:0)2, 2.0 nmol of SM(14:0)2, 0.02 nmol of LPG(14:0), 0.1 nmol of LPE(14:0), 0.5 nmol of LPC(14:0), 0.1 nmol of LPA(14:0) (Avanti Polar Lipids, Alabaster, AL, USA), dissolved in 120 μl of CHCl3/MeOH (1:1, v/v)) and 1.5 ml of CHCl3/MeOH (1:1, v/v) were added to cell homogenates that contained 1 mg of protein (quantified by Bradford assay). Subsequently, the mixture was sonicated in a water bath for 5 min, followed by centrifugation at 15 000 × g for 5 min. The organic layer was transferred to a glass vial and taken to dryness under a nitrogen steam at 60 °C. Subsequently, the residue was dissolved in 200 μl of CHCl3/MeOH/H2O (50:45:5, v/v/v) containing 0.01% of NH4OH, and 10 μl of the solution was injected into the liquid chromatography/mass spectrometry (LC/MS) system. LC/MS analysis was performed as described previously.50 In the negative mode, mass spectra of phospholipid molecular species were obtained by continuous scanning from m/z 380 to m/z 1500 with a scan time of 2 s. In positive mode, mass spectra of PC, LPC and SM were obtained by continuous scanning for product ions with m/z 184 from m/z 400 to 1000. Mass spectra for PE, LPE and PE plasmalogens were obtained by scanning on neutral losses of 141 from m/z 400 to 1000. The raw LC/MS data were converted to mzXML format using msConvert51 for the negative scan data and ReAdW for the positive specific scan data. Details on the metabolic data pre-processing pipeline will be described elsewhere (manuscript in preparation). Briefly, the data set was processed using a semi-automated metabolomics pipeline written in the R programming language that includes pre-processing (peak-finding and quantification), identification of metabolites, isotope correction, normalization/scaling based on internal added standards and statistical analysis of the results.
Western blotting
For total cell extracts, cells were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl pH 8, 0.5% w/v deoxycholate, 1% v/v NP-40) supplemented with a protease inhibitor mix (Roche, Switzerland). For all the other samples, they were resuspended directly in sample buffer. Proteins were separated by SDS-PAGE in 15% gels, transferred to PVDF membranes and probed with antibodies against cyt c (homemade), OPA1 (BD Biosciences, Allschwil, Switzerland), TOM20 (Santa Cruz, Dallas, TX, USA), SMAC (Enzo Life Sciences, Lausen, Switzerland), PHB2 (Upstate, Lake Placid, NY, USA), mtHSP70 (Thermo Fisher Scientific), Total OXPHOS (Mitosciences, Eugene, OR, USA), BAX (2D2, Thermo Fisher Scientific), BID (R&D Systems, Minneapolis, MN, USA), caspase 8 (Merck Millipore, Schaffhausen, Switzerland), caspase 3 (Cell Signalling, Danver, MA, USA), GAPDH (gift form D Picard) or MTCH2 (Sigma-Aldrich).
Fluorescence microscopy
Mitochondrial morphology was assessed by staining with MitoTracker Red (Thermo Fisher Scientific) as described by the manufacturer. For immunofluorescence, cells were fixed in 4% w/v paraformaldehyde for 5 min and washed in PBS. Incubation with antibodies was performed in PBS containing 3% w/v BSA and 0.1% v/v Triton X-100. BAX (2D2, Thermo Fisher Scientific) and cyt c (BD Biosciences) antibodies were used. Imaging was performed with a ZEISS LSM700 (Feldbach, Switzerland) confocal microscope.
Transmission electron microscopy
For electron microscopy, sample preparations and imaging was performed as described previously.52
Isolation of mitochondria
Crude mitochondrial fractions were prepared by differential centrifugation. Briefly, cells were resuspended in MB buffer (210 mM mannitol, 70 mM sucrose, 10 mM Hepes-KOH pH 7.4, 1 mM EDTA) supplemented with a protease inhibitor mix (Roche), and mechanically disrupted by 20 passages through a 25G needle. The extract was centrifuged at 2000 × g for 5 min at 4 °C to pellet nuclei and unbroken cells, and then the supernatants were centrifuged at 13 000 × g for 10 min at 4 °C. Pellets were designated as crude mitochondria, and supernatants as cytosolic fractions.
Sodium carbonate extraction of mitochondrial proteins
For each condition, 90 μl of 20 mM Hepes-KOH pH 7.4 was added to 10 μl of mitochondria (50 μg of proteins quantified by Bradford assay). After incubation on ice for 20 min, 100 μl of 200 mM Na2CO3 was added and samples were kept on ice for 30 min. Ultracentrifugation at 100 000 × g for 30 min at 4 °C was subsequently performed to separate membrane-integrated from soluble proteins. Supernatant proteins were concentrated by TCA precipitation.
Assessment of cyt c availability
For each condition, 90 μl of 20 mM Hepes-KOH pH 7.4, or MB buffer for the control, were added to 10 μl of mitochondria (50 μg of proteins quantified by Bradford assay). After incubation for 15 min at RT, 10 μl of KCl solutions were added to the sample to obtain a final concentration of KCl ranging from 0 to 80 μM. Following an incubation for 10 min at RT, samples were centrifuged at 10 000 × g for 5 min at RT to separate soluble from membrane-bound proteins.
MTCH2 downregulation
MTCH2 was downregulated using shRNA. A set of five shRNAs (Sigma-Aldrich) cloned in pLKO.1 was used and Lentiviral particles were generated from these five vectors. HCT116 WT and CLS1 KO, as well as HeLa cells, were infected and selected with puromycin (Merck Millipore). Out of the five, the shRNA giving the best downregulation efficiency was kept for further experiments. Its sequence is: 5′-CCGGCCCTTTGTGCTTGTCTCCAATCTCGAGATTGGAGACAAGCACAAAGGGTTTTTG-3′ (ref, TRCN0000059395).
FACS analysis
Cells were stained with FITC-coupled annexin V (BD Biosciences) following the instructions of the manufacturer. Analysis was performed with a Gallios apparatus (Beckman Coulter, Nyon, Switzerland).
Quantification of tBID
Band intensities following western blotting were quantified from three independent experiments, using the ImageJ software. Values were normalized to the loading control TOM20, and set at 100% for WT or CLS1 KO pLKO-infected cells. Values for WT and CLS1 KO MTCH2-downregulated cells were determined relative to these respective controls.
Statistical analysis
Data are represented as mean±S.E.M. of at least three independent experiments. *P<0.05 based on a Student's unpaired two-tailed t-test.
Acknowledgments
This work was supported by the Swiss National Science Foundation (31003A_141068/1), IGE3, and the State of Geneva. We thank all members of the Martinou laboratory, and especially Dr. Kinsey Maundrell, for their input and comments on the manuscript, Dr. Christoph Bauer for electron microscopy imaging, Professor Atan Gross for providing us with MTCH2 antibody, Professor Andera Ladislav for the TRAIL cytokine, Dr. Chunxin Wang and Professor Richard Youle who kindly provided materials for the generation of the KO cell lines, and Martin Vervaart, Mia Pras-Raves and Angela Luyf for technical assistance.
Glossary
- MOM
mitochondrial outer membrane
- MOMP
mitochondrial outer membrane permeabilization
- cyt c
cytochrome c
- IMS
intermembrane space
- IAPs
inhibitors of apoptosis
- CL
cardiolipin
- MIM
mitochondrial inner membrane
- CLS1
cardiolipin synthase
- TLC
thin-layer chromatography
- KO
knock-out
- WT
wild type
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PS
phosphatidylserine
- PI
phosphatidylinositol
- PC
phosphatidylcholine
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by R Youle
Supplementary Material
References
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59. [DOI] [PubMed] [Google Scholar]
- Jourdain A, Martinou JC. Mitochondrial outer-membrane permeabilization and remodelling in apoptosis. Int J Biochem Cell Biol 2009; 41: 1884–1889. [DOI] [PubMed] [Google Scholar]
- Westphal D, Dewson G, Czabotar PE, Kluck RM. Molecular biology of Bax and Bak activation and action. Biochim Biophys Acta 2011; 1813: 521–531. [DOI] [PubMed] [Google Scholar]
- Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem 2011; 80: 1055–1087. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997; 91: 479–489. [DOI] [PubMed] [Google Scholar]
- Happo L, Strasser A, Cory S. BH3-only proteins in apoptosis at a glance. J Cell Sci 2012; 125(Pt 5): 1081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantari C, Walczak H. Caspase-8 and bid: caught in the act between death receptors and mitochondria. Biochim Biophys Acta 2011; 1813: 558–563. [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998; 94: 491–501. [DOI] [PubMed] [Google Scholar]
- Walczak H. Death receptor-ligand systems in cancer, cell death, and inflammation. Cold Spring Harb Perspect Biol 2013; 5: a008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost PJ, Grabow S, Gray D, McKenzie MD, Nachbur U, Huang DC et al. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature 2009; 460: 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol 2011; 192: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raemy E, Martinou JC. Involvement of cardiolipin in tBID-induced activation of BAX during apoptosis. Chem Phys Lipids 2014; 179: 70–74. [DOI] [PubMed] [Google Scholar]
- Lutter M, Fang M, Luo X, Nishijima M, Xie X, Wang X. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat Cell Biol 2000; 2: 754–761. [DOI] [PubMed] [Google Scholar]
- Lutter M, Perkins GA, Wang X. The pro-apoptotic Bcl-2 family member tBid localizes to mitochondrial contact sites. BMC Cell Biol 2001; 2: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 2002; 111: 331–342. [DOI] [PubMed] [Google Scholar]
- Lucken-Ardjomande S, Montessuit S, Martinou JC. Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ 2008; 15: 929–937. [DOI] [PubMed] [Google Scholar]
- Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R et al. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 2010; 142: 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrones O, Antonsson B, Yamaguchi H, Wang HG, Liu J, Lee RM et al. Lipidic pore formation by the concerted action of proapoptotic BAX and tBID. J Biol Chem 2004; 279: 30081–30091. [DOI] [PubMed] [Google Scholar]
- Shamas-Din A, Bindner S, Chi X, Leber B, Andrews DW, Fradin C. Distinct lipid effects on tBid and Bim activation of membrane permeabilization by pro-apoptotic Bax. Biochem J 2015; 467: 495–505. [DOI] [PubMed] [Google Scholar]
- Gonzalvez F, Schug ZT, Houtkooper RH, MacKenzie ED, Brooks DG, Wanders RJ et al. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J Cell Biol 2008; 183: 681–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol 2005; 1: 223–232. [DOI] [PubMed] [Google Scholar]
- Esposti MD, Cristea IM, Gaskell SJ, Nakao Y, Dive C. Proapoptotic Bid binds to monolysocardiolipin, a new molecular connection between mitochondrial membranes and cell death. Cell Death Differ 2003; 10: 1300–1309. [DOI] [PubMed] [Google Scholar]
- Liu J, Epand RF, Durrant D, Grossman D, Chi NW, Epand RM et al. Role of phospholipid scramblase 3 in the regulation of tumor necrosis factor-alpha-induced apoptosis. Biochemistry 2008; 47: 4518–4529. [DOI] [PubMed] [Google Scholar]
- Schlattner U, Tokarska-Schlattner M, Ramirez S, Bruckner A, Kay L, Polge C et al. Mitochondrial kinases and their molecular interaction with cardiolipin. Biochim Biophys Acta 2009; 1788: 2032–2047. [DOI] [PubMed] [Google Scholar]
- Iverson SL, Enoksson M, Gogvadze V, Ott M, Orrenius S. Cardiolipin is not required for Bax-mediated cytochrome c release from yeast mitochondria. J Biol Chem 2004; 279: 1100–1107. [DOI] [PubMed] [Google Scholar]
- Manara A, Lindsay J, Marchioretto M, Astegno A, Gilmore AP, Esposti MD et al. Bid binding to negatively charged phospholipids may not be required for its pro-apoptotic activity in vivo. Biochim Biophys Acta 2009; 1791: 997–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polcic P, Su X, Fowlkes J, Blachly-Dyson E, Dowhan W, Forte M. Cardiolipin and phosphatidylglycerol are not required for the in vivo action of Bcl-2 family proteins. Cell Death Differ 2005; 12: 310–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer B, Quispe J, Choudhary V, Chipuk JE, Ajero TG, Du H et al. Mitochondrial outer membrane proteins assist Bid in Bax-mediated lipidic pore formation. Mol Biol Cell 2009; 20: 2276–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman Y, Shachnai L, Yivgi-Ohana N, Schwarz M, Maryanovich M, Houtkooper RH et al. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat Cell Biol 2010; 12: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Heacock PN, Mileykovskaya E, Voelker DR, Dowhan W. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J Biol Chem 1998; 273: 14933–14941. [DOI] [PubMed] [Google Scholar]
- Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F, Daum G. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett 1998; 421: 15–18. [DOI] [PubMed] [Google Scholar]
- Richter-Dennerlein R, Korwitz A, Haag M, Tatsuta T, Dargazanli S, Baker M et al. DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metab 2014; 20: 158–171. [DOI] [PubMed] [Google Scholar]
- Landes T, Leroy I, Bertholet A, Diot A, Khosrobakhsh F, Daloyau M et al. OPA1 (dys)functions. Semin Cell Dev Biol 2010; 21: 593–598. [DOI] [PubMed] [Google Scholar]
- Acehan D, Malhotra A, Xu Y, Ren M, Stokes DL, Schlame M. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys J 2011; 100: 2184–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Gonzalvez F, Jenkins GM, Slomianny C, Chretien D, Arnoult D et al. Cardiolipin deficiency releases cytochrome c from the inner mitochondrial membrane and accelerates stimuli-elicited apoptosis. Cell Death Differ 2007; 14: 597–606. [DOI] [PubMed] [Google Scholar]
- Pineau B, Bourge M, Marion J, Mauve C, Gilard F, Maneta-Peyret L et al. The importance of cardiolipin synthase for mitochondrial ultrastructure, respiratory function, plant development, and stress responses in Arabidopsis. Plant Cell 2013; 25: 4195–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Inoue T, Otomo Y, Yokomori N, Ohno M, Arai H et al. Deficiency of cardiolipin synthase causes abnormal mitochondrial function and morphology in germ cells of Caenorhabditis elegans. J Biol Chem 2012; 287: 4590–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Vaz FM. Cardiolipin, the heart of mitochondrial metabolism. Cell Mol Life Sci 2008; 65: 2493–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem Phys Lipids 2014; 179: 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 2004; 64: 985–993. [DOI] [PubMed] [Google Scholar]
- Katz C, Zaltsman-Amir Y, Mostizky Y, Kollet N, Gross A, Friedler A. Molecular basis of the interaction between proapoptotic truncated BID (tBID) protein and mitochondrial carrier homologue 2 (MTCH2) protein: key players in mitochondrial death pathway. J Biol Chem 2012; 287: 15016–15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B, Lin J, Andrews DW. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 2007; 12: 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekharan SP, Koc M, Morizot A, Micheau O, Sorensen PH, Gaide O et al. TRAIL promotes membrane blebbing, detachment and migration of cells displaying a dysfunctional intrinsic pathway of apoptosis. Apoptosis 2013; 18: 324–336. [DOI] [PubMed] [Google Scholar]
- Wang C, Youle RJ. Predominant requirement of Bax for apoptosis in HCT116 cells is determined by Mcl-1's inhibitory effect on Bak. Oncogene 2012; 31: 3177–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AJ, Kunji ER, Gross A. Mitochondrial carrier homolog 2 (MTCH2): the recruitment and evolution of a mitochondrial carrier protein to a critical player in apoptosis. Exp Cell Res 2012; 318: 1316–1323. [DOI] [PubMed] [Google Scholar]
- Ardail D, Privat JP, Egret-Charlier M, Levrat C, Lerme F, Louisot P. Mitochondrial contact sites. Lipid composition and dynamics. J Biol Chem 1990; 265: 18797–18802. [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 2010; 191: 1141–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rago C, Vogelstein B, Bunz F. Genetic knockouts and knockins in human somatic cells. Nat Protoc 2007; 2: 2734–2746. [DOI] [PubMed] [Google Scholar]
- Lu B, Xu FY, Jiang YJ, Choy PC, Hatch GM, Grunfeld C et al. Cloning and characterization of a cDNA encoding human cardiolipin synthase (hCLS1). J Lipid Res 2006; 47: 1140–1145. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Rodenburg RJ, Thiels C, van Lenthe H, Stet F, Poll-The BT et al. Cardiolipin and monolysocardiolipin analysis in fibroblasts, lymphocytes, and tissues using high-performance liquid chromatography-mass spectrometry as a diagnostic test for Barth syndrome. Anal Biochem 2009; 387: 230–237. [DOI] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 2012; 30: 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 2009; 28: 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.