Abstract
Vorinostat is an FDA-approved histone deacetylase inhibitor (HDACi) that has proven clinical success in some patients; however, it remains unclear why certain patients remain unresponsive to this agent and other HDACis. Constitutive STAT (signal transducer and activator of transcription) activation, overexpression of prosurvival Bcl-2 proteins and loss of HR23B have been identified as potential biomarkers of HDACi resistance; however, none have yet been used to aid the clinical utility of HDACi. Herein, we aimed to further elucidate vorinostat-resistance mechanisms through a functional genomics screen to identify novel genes that when knocked down by RNA interference (RNAi) sensitized cells to vorinostat-induced apoptosis. A synthetic lethal functional screen using a whole-genome protein-coding RNAi library was used to identify genes that when knocked down cooperated with vorinostat to induce tumor cell apoptosis in otherwise resistant cells. Through iterative screening, we identified 10 vorinostat-resistance candidate genes that sensitized specifically to vorinostat. One of these vorinostat-resistance genes was GLI1, an oncogene not previously known to regulate the activity of HDACi. Treatment of vorinostat-resistant cells with the GLI1 small-molecule inhibitor, GANT61, phenocopied the effect of GLI1 knockdown. The mechanism by which GLI1 loss of function sensitized tumor cells to vorinostat-induced apoptosis is at least in part through interactions with vorinostat to alter gene expression in a manner that favored apoptosis. Upon GLI1 knockdown and vorinostat treatment, BCL2L1 expression was repressed and overexpression of BCL2L1 inhibited GLI1-knockdown-mediated vorinostat sensitization. Taken together, we present the identification and characterization of GLI1 as a new HDACi resistance gene, providing a strong rationale for development of GLI1 inhibitors for clinical use in combination with HDACi therapy.
Epigenetic-based drugs show great promise for treatment of a wide range of cancers. Targeting HDACs has proven effective as a cancer therapy strategy and many new histone deacetylase inhibitors (HDACis) are being developed with varying target specificities. HDACis exert their anticancer effects through a variety of tumor-cell- and non-tumor-cell-autonomous effects, and induction of apoptosis is an important mechanism underpinning the anticancer activity of these compounds. Although HDACis have been clinically approved for the treatment of cutaneous T-cell lymphoma and are in advanced clinical trials for other malignancies, many patients remain unresponsive to single-agent HDACi treatment. There exists a need to identify novel genes and pathways that can be targeted in combination with HDACi to improve clinical utility of these compounds. Furthermore, clinical biomarkers are required to predict patient response to inform which patients should receive HDACi therapy.
There have been a number of cellular factors implicated in resistance to HDACi treatment such as drug efflux, alterations in the molecular targets (i.e. HDAC overexpression or mutation), changes in the epigenetic landscape of HDACi target genes, responses to oxidative stress and antiapoptotic and/or prosurvival mechanisms (reviewed in Fantin and Richon1). Hyperactive JAK/STAT (Janus kinase/signal transducer and activator of transcription) signaling can mediate resistance of cutaneous T-cell lymphoma cells to vorinostat.2 In addition, we and others have demonstrated that overexpression of prosurvival Bcl-2-family proteins protects cells against HDACi-induced apoptosis.3, 4, 5, 6 HR23B has also been implicated in vorinostat resistance as a protein through which vorinostat induces apoptosis via impaired proteasome function.7, 8, 9 As the identification of these mechanisms of resistance has not yet impacted the clinical use of HDACi through the generation of combination therapy approaches or clinical biomarkers, a genome-wide study was conducted to identify further mechanisms of resistance to HDACis that may improve the clinical application of these agents.
To elucidate vorinostat-resistance mechanisms, we used human tumor cells with acquired resistance to vorinostat and conducted a protein-coding genome-wide functional genomics screen to identify novel genes that resensitize cells to vorinostat-induced apoptosis. High-throughput RNA interference (RNAi) screening has demonstrated utility for the identification of novel drug targets for cancer therapy and biomarkers for drug response.10, 11, 12, 13 In addition, screening for synthetic lethality has successfully identified targetable cancer-specific vulnerabilities alone and in combination with existing therapies.14, 15, 16, 17, 18, 19, 20, 21 The novel identification of GLI1 as a gene that regulates vorinostat-mediated apoptosis and the demonstration that a small-molecule GLI1 inhibitor functions cooperatively with vorinostat to induce tumor cell death provides a new avenue for combination therapies using vorinostat and other HDACis.
Results
Development of tumor cells with acquired resistance to vorinostat and other HDACi
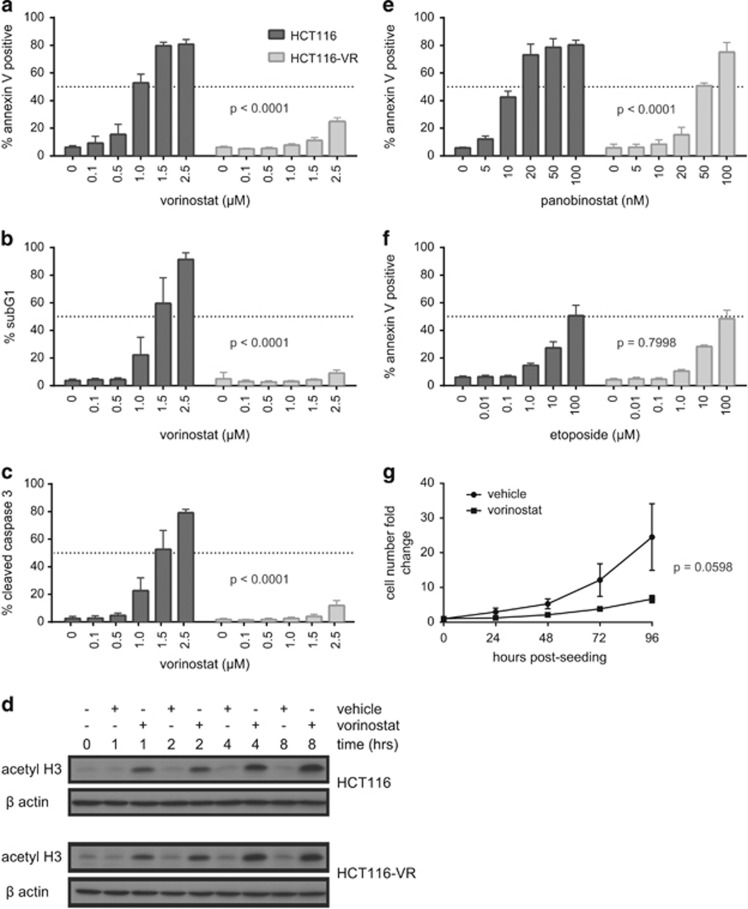
To identify genes that confer resistance to vorinostat, the vorinostat-sensitive HCT116 colon cancer cell line was serially passaged in the presence of vorinostat at apoptotic concentrations resulting in the development of stable vorinostat-resistant cells (HCT116-VR). Compared with HCT116 cells, the HCT116-VR cells were resistant to vorinostat-induced apoptosis as assessed by annexin V staining and assays for DNA fragmentation and caspase-3 activation (Figures 1a–c and Supplementary Figure 1). The resistance to vorinostat-induced apoptosis was not because of a lack of on-target drug activity within the HCT116-VR cells as equivalent histone H3 hyperacetylation was observed in vorinostat-treated HCT116 and HCT116-VR cells (Figure 1d). HCT116-VR cells demonstrated relative cross-resistance to the HDACi panobinostat (Figure 1e); however, the cells remained sensitive to apoptosis mediated by the chemotherapeutic drug etoposide (Figure 1f). Using a cell counting assay over 96 h, HCT116-VR cells cultured in the presence of vorinostat continued to proliferate albeit at a much slower rate compared with vehicle-treated cells (Figure 1g). Taken together, these data indicate that HCT116-VR cells show acquired and selective apoptotic resistance to the HDACi vorinsostat and panobinostat and therefore represent an appropriate model to screen for genes that mediate the resistance phenotype.
Figure 1.
Characterization of a vorinostat-resistant HCT116 (HCT116-VR) cell line. The induction of apoptosis by vorinostat was assessed in HCT116 and HCT116-VR cells by (a) annexin V positivity (as a marker of phosphatidylserine externalization), (b) sub-G1 DNA content as a marker of DNA fragmentation) and (c) cleaved caspase-3 (as a marker of caspase activation) 48 h after vorinostat treatment at the doses indicated. (d) Western blot for acetylated histone H3 (acetyl H3) as a biomarker for intracellular vorinostat activity. HCT116 and HCT116-VR cells were treated with 2.5 μM vorinostat or equivalent amounts of the vehicle, dimethyl sulfoxide (DMSO), for 1, 2, 4 or 8 h and harvested for protein extraction. β-Actin serves as a loading control. Annexin V staining of HCT116 and HCT116-VR cells after treatment with (e) panobinostat and (f) etoposide at the doses indicated for 48 h. Mean and S.D. are plotted for at least three independent biological replicates. P-values indicate statistical significance of cell type and dose–response interactions as measured by two-way analysis of variance (ANOVA) comparing back with HCT116 cells. (g) Cell counting assay using HCT116-VR cells treated with 2.5 μM vorinostat or DMSO at cell seeding (0 h)
Genome-wide RNAi screens for genes that confer resistance to HDACi-induced apoptosis
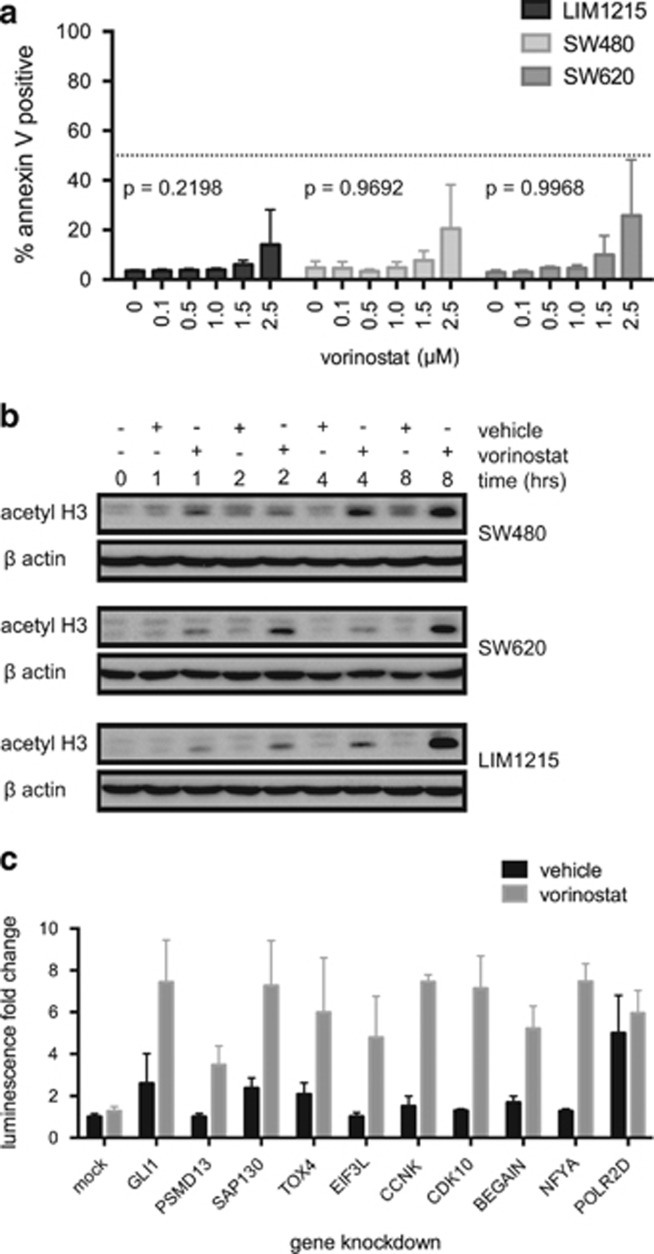
Multiple tiers of RNAi screening were used to identify genes that can regulate vorinostat-induced apoptosis.22 Genes were considered candidate vorinostat-resistance genes if gene knockdown in combination with vorinostat induced cell death of HCT116-VR cells, but gene knockdown alone did not. HCT116-VR cells were robotically transfected in arrayed 384-well format with annotated SMARTpool siRNAs to putatively knockdown all known human protein-coding genes. The cells were subsequently incubated for a further 24 h in the presence and absence of vorinostat.22 Cell death in the control, minus drug, arm of the screen was assessed with DAPI nuclear staining using high content microscopy. Cell death readouts for the vorinostat-treated plus drug arm of the screen were caspase-3 and -7 activity (Caspase-Glo) and cell viability using CellTitre-Fluor (CTF). The primary SMARTpool siRNA screen yielded 450 gene hits comprising 317 Caspase-Glo 3/7 hits and 150 CTF hits, with 17 genes identified in both assays (Supplementary Figure 2A). A secondary validation screen was conducted by deconvolution of the SMARTpools into the four individual constituent siRNAs. Of the 450 primary targets, 106 genes validated with high confidence, scoring 2 or more individual siRNAs recapitulating the SMARTpool phenotype (Supplementary Figure 2B). As a final screen, we identified three human colon cancer cell lines, SW480, SW620 and LIM1215, that demonstrated intrinsic resistance to vorinostat similar to that observed in HCT116-VR cells (Figures 2a and b). A tertiary knockdown screen of the 106 candidate genes in SW480, SW620 and LIM1215 cells resulted in the identification of 10 genes that specifically conferred resistance to vorinostat in two or more of the four cell lines tested (Supplementary Table 1). Nine of the ten vorinostat-resistance candidate genes were hits in the Caspase-Glo assay (GLI1, PSMD13, SAP130, CCNK, CDK10, EIF3L, NFYA, BEGAIN and TOX4) and the tenth hit was identified through the CTF assay (POLR2D). For the nine Caspase-Glo hits, gene knockdown alone induced low or no caspase activation above mock transfection levels; however, upon vorinostat treatment, caspase activity was greatly increased (Figure 2c).
Figure 2.
Sensitivity human tumor cell lines to vorinostat-induced apoptosis. (a) Annexin V staining of LIM1215, SW480 and SW620 after treatment with vorinostat at the doses indicated for 48 h. Mean and S.D. are plotted for three independent biological replicates. P-values indicate statistical significance of cell type and dose–response interactions as measured by two-way analysis of variance (ANOVA) comparing back with HCT116-VR cells (Figure 1a). (b) Western blot for acetylated histone H3 (acetyl H3) as a biomarker for intracellular vorinostat activity. LIM1215, SW480 and SW620 cells were treated with 2.5 μM vorinostat or equivalent amounts of the vehicle, dimethyl sulfoxide (DMSO), for 1, 2, 4 or 8 h and harvested for protein extraction. β-Actin serves as a loading control. (c) Caspase-Glo 3/7 luminescence upon gene knockdown of each of the 10 vorinostat-resistance candidates from the tertiary screen in HCT116-VR cells, treated with vehicle or 2.5 μM vorinostat for 24 h
Complete data sets for each of the primary, secondary and tertiary screens can be found online (PubChem IDs: AID743454, AID743458, AID743448). Specific depletion of target gene mRNA of >90% for all 10 candidate vorinostat-resistance genes was confirmed using quantitative real-time PCR (qRT-PCR) (data not shown).
Using the tiered screening approach outlined above, the 10 candidate vorinostat-resistance genes were: GLI1, PSMD13, SAP130, CCNK, CDK10, POLR2D, EIF3L, NFYA, BEGAIN and TOX4. With the exception of two genes, PSMD1323 and SAP130,24 these genes had not previously been linked to HDACi activities and represent potentially novel mechanisms of resistance to HDACi. Analysis of RNAseq gene expression profiles upon knockdown of each of these 10 candidates did not identify common signatures or pathway modulation between vorinostat-resistance candidates.22 As GLI1 has known oncogenic function and pharmacological inhibitors already exist for GLI1 and pathway members, we chose to focus on this candidate for follow-up mechanistic studies.
GLI1 regulates sensitivity to HDACis and this occurs independently of SMO
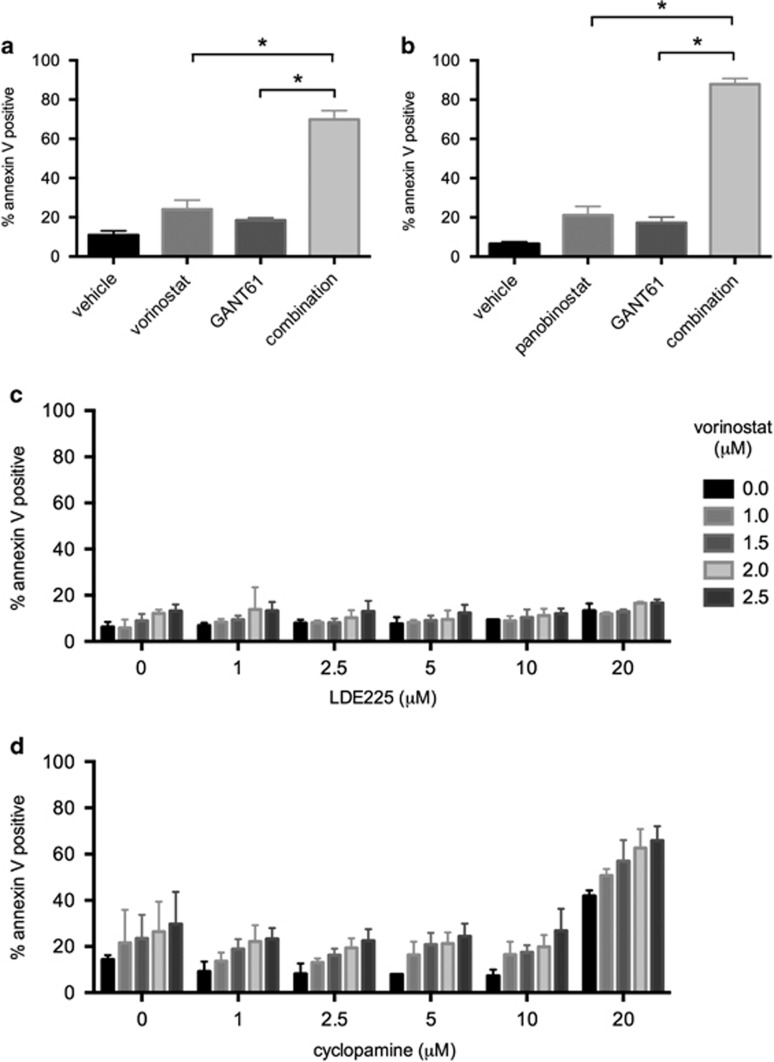
GLI1 functions as an oncogene in certain contexts through amplification in glioblastoma and alternative splicing in a range of tumors including glioblastoma, breast, prostate, pancreatic and gastric cancers.25 GLI1, however, has not previously been linked to HDACi response or HDACi mechanism of action and was therefore selected for further investigation. Transfection of siGLI1 and treatment with vorinostat showed a significant induction of cell death, as measured by ATP levels using CellTiter-Glo (Promega, Madison, WI, USA) in SW480, SW620 and LIM1215 cell lines consistent with the primary screen phenotype in the HCT116-VR cells (Supplementary Figure 3). To confirm that GLI1 conferred resistance to HDACi-mediated apoptosis, the GLI1 small-molecule inhibitor, GANT61,26, 27 was used in combination with two broad-spectrum HDACi, vorinostat and panobinostat. HCT116-VR cells were treated simultaneously with GANT61 and either vorinostat or panobinostat. Pharmacological inhibition of GLI1 successfully phenocopied genetic knockdown of GLI1 resulting in even greater sensitization of HCT116-VR cells to HDACi-induced apoptosis (Figures 3a and b) and a similar outcome was observed for SW480, SW620 and LIM1215 cell lines (Supplementary Figure 3). The canonical role of GLI1 is as a transcription factor involved in Hedgehog signaling,25, 28 where upon binding of the Hedgehog ligand to the cell surface receptor Patched (PTCH) releases its inhibitory effect on Smoothened (SMO), which in turn activates GLI1 and a prosurvival and proproliferation transcriptional response. To test for dependence on the Hedgehog pathway in this context, two SMO inhibitors, LDE22529, 30 and cyclopamine,31 were evaluated in combination with vorinostat. No cooperation between SMO and HDAC inhibition was observed (Figures 3c and d), suggesting that in the context of vorinostat resistance, GLI1 functions independently of SMO and Hedgehog signaling. Furthermore, other members of the pathway such as GLI2, SMO, PTCH1 and SUFU did not read out in the RNAi screen (data not shown).
Figure 3.
Pharmacological inhibition of GLI1 but not SMO sensitizes to HDACi-induced apoptosis. The effect of combining GLI1 inhibition using GANT61 with HDACi treatment was assessed for apoptosis by annexin V staining in HCT116-VR cells. Cells were treated with 15 μM GANT61 and (a) 2.5 μM vorinostat or (b) 15 nM panobinostat alone and in combination. Mean and S.D. are plotted for at least three independent biological replicates. The effect of each single treatment was compared with that of the combination using a Student's t-test: *P<0.05. The effect of combining vorinostat treatment with SMO inhibition using (c) LDE225 or (d) cyclopamine was assessed for apoptosis by annexin V staining in HCT116-VR cells. Cells were treated for 48 h with vorinostat (1–2.5 μM) and/or (c) LDE225 (1–20 μM) or (d) cyclopamine (1–20 μM). No combination effect was seen between vorinostat and either of the SMO inhibitors. Mean and S.D. are plotted for at least three independent biological replicates
GLI1 expression is induced by vorinostat treatment
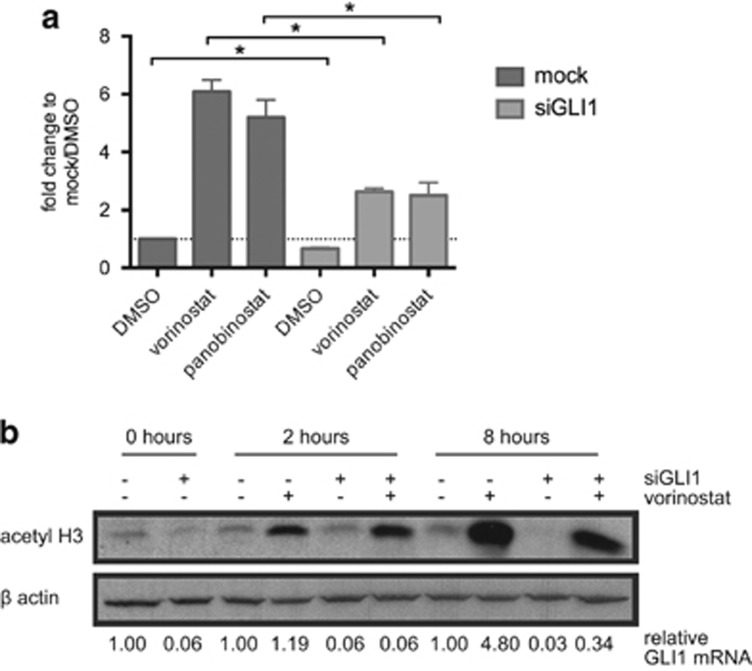
As vorinostat mediates its effects at least in part through transcriptional mechanisms, the effect of vorinostat treatment on GLI1 expression levels was assessed. GLI1 mRNA was induced by vorinostat (6.1-fold) and panobinostat (5.2-fold) in HCT116-VR cells after 8 h (Figure 4a). We also observed increased expression of GLI1 mRNA following treatment of parental HCT116 cells with vorinostat (data not shown). Transcriptome analysis (RNAseq) following treatment of HCT116-VR cells with vorinostat indicated that of the 10 candidate vorinostat-resistance genes, GLI1 was the most upregulated candidate upon vorinostat treatment (8.6-fold after 8 h). Furthermore, GLI1 was among the top 10% of all genes upregulated after 4 and 8 h of vorinostat treatment. SMARTpool siRNA-mediated GLI1 knockdown was reduced by 33% in untreated cells and this was sufficient to significantly attenuate the HDACi-induced induction of GLI1 expression (Figure 4a).
Figure 4.
GLI1 expression is induced by HDACi treatment and knockdown does not affect vorinostat-induced histone H3 hyperacetylation. (a) HCT116-VR cells were reverse transfected with lipid only (mock) or siGLI1. After 48 h transfection, cells were treated with vorinostat (2.5 μM), panobinostat (15 nM) or dimethyl sulfoxide (DMSO) and expression of GLI1 mRNA was assessed by qRT-PCR after 8 h. Data were normalized to the housekeeping gene L32 as L32 expression is not affected by HDACi treatment. Vorinostat and panobinostat induced expression of GLI1. Even with minimal GLI1 knockdown (33% in DMSO control), GLI1 induction by vorinostat and panobinostat was significantly suppressed. Data are expressed as the mean and S.D. of three independent biological experiments. Student's t-test analysis: *P<0.05. The dotted line indicates a fold change of 1. (b) Western blot analysis for acetylated histone H3 (acetyl H3) as a biomarker for intracellular vorinostat activity. HCT116-VR cells were reverse transfected with lipid only (mock) or siGLI1 for 48 h and then treated with 2.5 μM vorinostat or equivalent amounts of the vehicle, DMSO, for 2 or 8 h and harvested for protein extraction. β-Actin serves as a loading control. Histone H3 hyperacetylation is seen in cells with and without GLI1 knockdown upon vorinostat treatment in a time-dependent manner. Levels of GLI1 mRNA normalized to untreated cells are shown below the image. Representative image from one of three independent biological replicates
To confirm that GLI1 knockdown did not interfere with the on-target activity of vorinostat, acetylated histone H3 was used as a biomarker of vorinostat activity in HCT116-VR cells transfected with siGL1. HCT116-VR cells with and without GLI1 knockdown were treated with vorinostat or DMSO. Vorinostat treatment induced histone H3 hyperacetylation regardless of GLI1 expression level (Figure 4b).
GLI1 knockdown and vorinostat treatment shift the balance of prosurvival and proapoptosis genes in favor of apoptosis
As vorinostat induces changes in phenotype at least in part through modulation of transcription3 and GLI1 is a transcriptional factor,25 whole-genome transcriptome analysis (RNAseq) was used to investigate potential mechanism(s) of action. HCT116-VR cells were subjected to GLI1 knockdown and then treated with vorinostat or DMSO for 4 h (no cell death detectable) and 8 h (low level of cell death detectable). All conditions were analyzed for differential gene expression compared with mock-transfected, DMSO-treated cells. Complete RNAseq data sets can be found online (Gene Expression Omnibus ID: GSE57871). GLI1 knockdown resulted in reduced expression of validated GLI1 target genes including Cyclin D1, Bcl2, Jagged 2, IL7, FoxF1 and FoxL1 and resulted in fewer gene expression changes than vorinostat treatment. The combination of GLI1 knockdown and vorinostat resulted in more differentially expressed genes than either treatment alone. Pathway enrichment analysis using MetaCore (ThomsonReuters, New York, NY, USA) was conducted on differentially expressed genes in each of the treatment groups for identification of differentially expressed pathways. Both timepoints were analyzed simultaneously, thereby giving greater weight to genes differentially expressed at both timepoints. Apoptosis pathways were enriched in the combination of vorinostat and GLI1 knockdown compared with single treatments (Table 1).
Table 1. MetaCore pathway enrichment for apoptosis pathways.
| Pathway enrichment: GLI1 knockdown and vorinostat |
| Apoptosis and survival_Endoplasmic reticulum stress response pathway |
| Apoptosis and survival_Lymphotoxin-β-receptor signaling |
| Apoptosis and survival_p53-dependent apoptosis |
| Apoptosis and survival_Regulation of Apoptosis by Mitochondrial Proteins |
| Pathway enrichment: vorinostat only |
| Apoptosis and survival_Anti-apoptotic TNFs/NF-κB/Bcl-2 pathway |
| Apoptosis and survival_Lymphotoxin-beta receptor signaling |
| Pathway enrichment: GLI1 knockdown only |
| No apoptosis pathways |
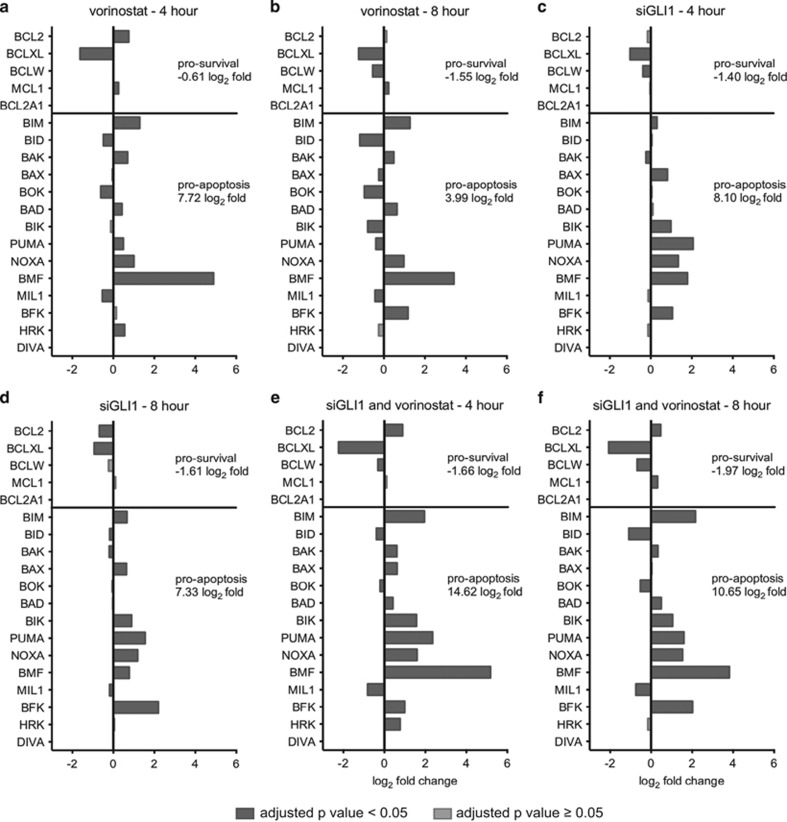
To investigate the enrichment of apoptosis pathways further, the differential expression values of Bcl-2-family apoptosis genes were extracted from the complete RNAseq data set under each experimental condition and time point (Figure 5). Expression of five prosurvival genes (BCL2, BCL2L1, BCL2L2, MCL1 and BCL2A1) and 14 proapoptotic genes (BCL2L11, BID, BAK1, BAX, BOK, BAD, BIK, BBC3, PMAIP1, BMF, BCL2L13, BCL2L15, HRK and BCL2L10) was assessed. No expression of BCL2A1 or BCL2L10 (DIVA) was detected in this experiment. Vorinostat treatment shifted the balance of pro- and anti-apoptosis genes towards apoptosis and this was due, in part, to a large increase in BMF expression (Figures 5a and b). GLI1 knockdown also shifted the balance towards apoptosis because of induction of a range of pro-apoptosis genes including BMF, BAX, BIK, BBC3 (PUMA), PMAIP1 (NOXA) and BCL2L15 (BFK) and downregulation of BCL2L1 (BCLXL) (Figures 5c and d). Taken together, the combination of GLI1 knockdown and vorinostat treatment resulted in greater induction of pro-apoptosis genes such as upregulation of BCL2L11 (BIM) and greater suppression of pro-survival genes such as BCL2L1 (Figures 5e and f). This supports the identification of apoptosis pathways in the enrichment analysis and identified a shift in expression of pro- and anti-apoptosis genes in favor of apoptosis as a result of vorinostat treatment and GLI1 knockdown.
Figure 5.
Changes in expression of apoptosis genes as a result of GLI1 knockdown and vorinostat treatment alone and in combination. Differential expression of Bcl-2-family apoptosis genes as determined by RNAseq. Differential expression is expressed as log2 fold change in expression compared with mock-transfected dimethyl sulfoxide (DMSO)-treated cells. Prosurvival genes (BCL2, BCL2L1, BCL2L2, MCL1 and BCL2A1) are represented above the solid line and proapoptotic genes (BCL2L11, BID, BAK1, BAX, BOK, BAD, BIK, BBC3, PMAIP1, BMF, BCL2L13, BCL2L15, HRK and BCL2L10) are represented below the solid line. Dark bars represent an adjusted P-value <0.05 and light bars represent an adjusted P-value ⩾0.05. Cumulative log2 fold change for prosurvival genes and proapoptosis genes is listed to the right of each graph (does not include genes with adjusted P-value ⩾0.05). Cells were reverse transfected with lipid only (mock) or siGLI1 and treated with vorinostat (2.5 μM) or DMSO 48 h after transfection. Cells were harvested for RNA after (a, c and e) 4 h or (b, d and f) 8 h drug treatment. Differential expression is presented upon (a and b) vorinostat treatment alone, (c and d) GLI1 knockdown alone and (e and f) the combination of GLI1 knockdown followed by vorinostat treatment. Both vorinostat treatment (a and b) and GLI1 knockdown (c and d) resulted in a net increase in proapoptotic gene expression and net decrease in prosurvival gene expression. The combination of GLI1 knockdown and vorinostat treatment (e and f) resulted in a greater net increase in proapoptotic gene expression and net decrease in prosurvival gene expression compared with either treatment alone
BCL2L1 transcriptional regulation is functionally important for GLI1 knockdown-mediated sensitization to vorinostat-induced apoptosis
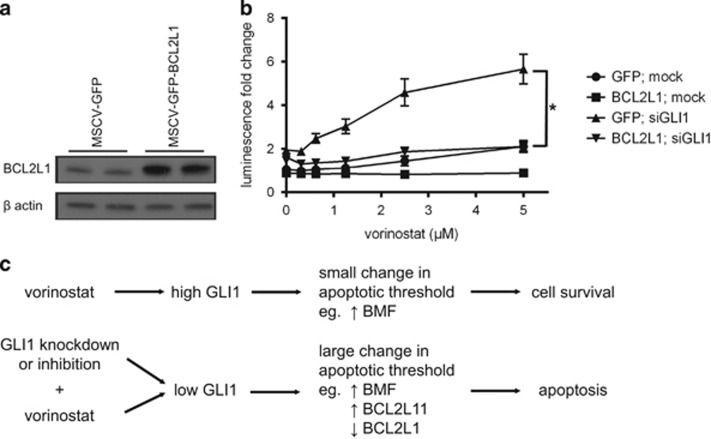
The functional effect of the antiapoptotic Bcl-2-family gene, BCL2L1, in mediating GLI1-regulated sensitization to vorinostat was assessed by stably overexpressing BCL2L1 in HCT116-VR cells concurrently with GLI1 knockdown and vorinostat treatment. HCT116-VR cells were stably transduced with a GFP or BCL2L1 overexpression construct (Figure 6a). These cells were then transfected with siGLI1 or mock transfection control and assessed for sensitivity to increasing concentrations of vorinostat. As shown in Figure 6b, BCL2L1 overexpression protected cells from vorinostat-induced apoptosis in cells with concomitant knockdown of GLI1.
Figure 6.
BCL2L1 overexpression abrogates GLI1 knockdown-mediated vorinostat sensitization. (a) Western blot showing BCL2L1 overexpression in technical replicate in HCT116-VR cells with β-actin as the loading control. (b) Caspase activation was assessed through Caspase-3/7 Glo luminescence upon GLI1 knockdown and vorinostat treatment in GFP- and BCL2L1-overexpressing HCT116-VR cells. Vorinostat treatment and GLI1 knockdown in control GFP-expressing cells but not BCL2L1-overexpressing cells resulted in a dose-dependent increase in luminescence indicating induction of cell death. BCL2L1 overexpression protected cells from cell death induced by vorinostat treatment and GLI1 knockdown. Two-way analysis of variance (ANOVA): *P<0.05. (c) Working model: Schematic representation of the proposed mechanistic pathways that regulate resistance to vorinostat-induced apoptosis by GLI1
Discussion
The biological effects and mechanisms of action of HDACis are extremely broad and there may be many mechanisms by which tumor cells exhibit or acquire resistance to these agents. Through whole-genome functional screening, GLI1 was identified as a novel vorinostat-resistance gene. The importance of GLI1 in regulating vorinostat-mediated apoptosis was originally discovered through a screen in the setting of acquired resistance to vorinostat using HCT116-VR cells. However, using other solid tumor cells that were intrinsically resistant to vorinostat (LIM1215, SW480, SW620), we showed that the importance of GLI1 as a regulator of vorinostat sensitivity was more widespread. A synthetic lethal interaction between GLI1 knockdown and vorinostat treatment resulted in changes in expression of genes that directly regulate apoptosis.3, 32 Many studies have demonstrated the ability of HDACi to regulate apoptosis gene expression in favor of apoptosis and this is important in HDACi mechanism of action (reviewed in Bolden et al.3 and Falkenberg and Johnstone33). Although vorinostat treatment alone induced gene expression changes in HCT116-VR cells in favor of apoptosis, these were clearly not sufficient to induce cell death. However, following GLI1 knockdown, this balance between pro-survival and pro-apoptosis gene expression was shifted further in favor of apoptosis, resulting in death of previously vorinostat-resistant cells. A working model is presented in Figure 6c.
Baseline expression levels of GLI1 in HCT116-VR cells were low and these were markedly increased upon vorinostat treatment. A similar induction of GLI1 was observed in MEFs treated with the pan-HDACi trichostatin A.34 The canonical role of GLI1 is as a key transcription factor and effector molecule in the Hedgehog signaling pathway.25 As SMO inhibition did not sensitize to vorinostat-induced apoptosis, GLI1 appears to function through a noncanonical pathway in this system. Other studies have also identified SMO-independent GLI1 dependence in cancer (reviewed in Shevde and Samant35), such as the SMO-independent role of GLI1 in neuroblastoma demonstrated using pharmacological and genetic approaches.36 GLI1 function is regulated through acetylation and ubiquitination.37 Unlike its family members, GLI1 is a constitutive transcriptional activator and proteasomal cleavage results in degradation of the protein.38, 39 GLI1 transcriptional activity is enhanced by HDAC1-mediated deacetylation leading to enhanced cellular proliferation and transformation.40 An E3 ubiquitin ligase complex containing Cullin3, REN, KCASH2 and KCASK3 leads to HDAC1 degradation and reverses this phenotype.40, 41 As vorinostat inhibits HDAC1, the effect of vorinostat and GLI1 knockdown on GLI1 function may be a combination of indirect GLI1 inhibition through inhibition of HDAC1 and prevention of GLI1 induction through GLI1 knockdown. Epigenetic repression of GLI1 expression occurs in part through histone methylation. Menin recruits PRMT5 to the GLI1 promoter to lay down the H4R3me2 repressive mark in a Hedgehog signaling-independent manner42 and a recently identified noncoding RNA induces H3K27me3 at the GLI1 promoter, thereby reducing RNA polymerase II recruitment.43
GLI1 has frequently been implicated in cancer cell therapy resistance through decreased chemotherapy and radiotherapy responsiveness44, 45, 46, 47, 48 and increased expression of cell surface drug transporters.49, 50, 51 Recently, GLI1 was shown to drive resistance of AML cells to ribavirin and cytarabine through UGT1A-dependent glucuronidation of the compounds and inhibition of GLI1 with GANT61 restored sensitivity.52 Additionally, SMO-independent GLI1 activity is important for osteopontin-mediated resistance of breast cancer cells to the cytotoxic effects of doxorubicin, paclitaxel and cisplatin.49 These studies provide strong evidence for the role of GLI1 in tumor cell resistance to a range of anticancer therapies; however, this is the first study to identify a role for GLI1 in response to HDACi. The noncanonical regulation of GLI1 in this system highlights the need for the generation of GLI1 inhibitors, where SMO inhibitors that are receiving much clinical attention would be ineffective.
GLI1 is known to positively regulate expression of BCL2 through direct promoter binding53, 54 and herein we show the downregulation of BCL2 upon GLI1 knockdown. GLI2 is a more potent activator of BCL2 expression than GLI1,55 which may explain the subtle transcription repression observed (1.6-fold). Bolden et al.5 previously described a tumor-cell-selective pro-apoptosis gene expression signature upon vorinostat treatment of paired normal and tumorigenic human fibroblasts. Vorinostat treatment of BJ LTSTERas fibroblasts resulted in repression of BCL2L1 (BCLXL) and BCL2L2 (BCLW), which was also observed in the vorinostat-treated HCT116-VR cells in this study, indicating a potentially conserved transcriptional response to vorinostat treatment in transformed cells. Similarly, a similar panel of proapoptotic genes was induced in both studies including BCL2L11 (BIM), BAD, PMAIP1 (NOXA) and BAK1. Induction of BMF expression by vorinostat in lymphoma cells has also been previously observed.56
Based on our studies, we posit that combination therapy approaches involving HDACi and GLI1 inhibitors merit further investigation. At present, there is no small-molecule inhibitor of GLI1 appropriate for use in humans and this is a major hurdle before combination therapies consisting of GLI1 and HDAC inhibition can be trailed. Given the noncanonical activation of GLI1 observed in this study and in many others, there is a strong rationale for development of GLI1 inhibitors to complement significant efforts currently focusing on SMO inhibitors.57
Materials and Methods
Reagents
HDACis, vorinostat and panobinostat, were obtained directly from the manufacturers Merck (Boston, MA, USA) and Novartis (Cambridge, MA, USA), respectively. The GLI1 inhibitor, GANT61, was obtained from Sigma (St Louis, MO, USA) (cat. no. G9048). SMO inhibitors, LDE225 and cyclopamine (cat. no. C4116), were obtained from Novartis and Sigma, respectively.
siRNA screening
A detailed description of the methods used in the siRNA screen has been described previously.22, 58 Briefly, HCT116-VR cells were reverse transfected on day 1, media changed on day 2, treated with vorinostat or vehicle (DMSO) on day 3 and assessed for cell death on day 4. The two parallel arms of the screen (plus and minus drug) evaluated cell death using different measurements. The plus drug arm was evaluated for general viability using CTF (Promega) and apoptosis using Caspase-Glo 3/7 (Promega). The minus drug arm was subjected to nuclear staining by DAPI (Invitrogen, Carlsbad, CA, USA) followed by high content cell counting as a surrogate readout of cell death.
Cell culture and transfection
Vorinostat-resistant HCT116 colon carcinoma cells (HCT116-VR) were a kind gift from Merck Research Laboratories (Boston, MA, USA) and were generated by culturing the parental line in increasing concentrations of vorinostat until a vorinostat-resistant line was established. Cells were cultured in RMPI containing GlutaMAX (Life Technologies, Carlsbad, CA, USA) supplemented with 10% FBS, 2.5 g/l glucose and 1% (v/v) sodium pyruvate in humidified incubators at 37 °C and 5% CO2. Cells were reverse transfected with DharmaFECT2 (GE, Pittsburgh, PA, USA) as follows: 6-well plate – 1.5 μl, 1.8x105 cells (harvested after 72–96 h); 12-well plate – 1.0 μl, 3.5x105 cells (harvested after 24 h); 24-well plate – 0.5 μl, 3.0x104 cells (harvested after 96 h); and 384-well plate – 0.06 μl, 1.4x103 cell (harvested after 72 h). siRNA concentrations used were between 2.5 and 40 nM. The GLI1 siRNA SMARTpool (GE; cat. no. M-003896-00) consisted of the following duplex sequences: D-003896-01_GGAAAUGACUGGCAAUGCA, D-003896-02_GCACUGGUCUGUCCACUCU, D-003896-03_GUCCUCGACUUGAACAUUA and D-003896-04_AGGCUCAGCUUGUGUGUAA. SW480 (1.8x103 cells per well), SW620 (2.5x103 cells) and LIM1215 (1.3x103 cells) cell lines were cultured in RPMI-1640 containing 10% FBS and were transfected with the siGLI1 SMARTpool (40 nM) using DharmaFECT 2 (0.1 μl per well for each line) in parallel using high-throughput liquid handling in 384-well format. Plates were poly-l-lysine treated before addition of cells, as per Falkenberg et al.22 At 48 h after transfection, cells were treated with vorinostat (2.5 μM) or DMSO and cell viability measured at 24, 48 and 72 h after drug treatment using CellTiter-Glo, an end-point luminescence reagent that quantitates cellular ATP using a plate reader (Promega).
Drug treatments and cell viability assay
Capase-Glo 3/7 and CTF assays were conducted in 384-well format.22, 58 CTF buffer and substrates were combined and 5 μl was added to wells containing 20 μl cells and media. Plates were incubated at 37 °C for 1.5 h and fluorescence quantified (excitation: 380–400 nm and emission: 505 nm). Caspase-Glo 3/7 reagent and substrate were combined and 12 μl added to each well before incubation at room temperature for 30 min followed by luminescence quantification. Readings were taken on the Synergy H4 Hybrid Multi-mode Microplate Reader (BioTek, Winooski, VT, USA).
For drug combination assays, cells were treated with both drugs concurrently for 48 h before harvesting for annexin V staining. Doses of drugs used were: 1–2.5 μM vorinostat, 5–50 nM panobinostat, 1–20 μM LDE225 and 1–20 μM cyclopamine. For vorinostat treatment of BCL2L1-overexpressing cells, 0.1–5 μM was used. For flow cytometry analysis of apoptosis, cells were resuspended in 100 μl annexin-V-binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 5 mM CaCl2) containing 1/100 dilution of annexin-V-APC (BD Biosciences, North Ryde, NSW, Australia) and 1/100 dilution propidium iodide (PI) citrate buffer (69 mM PI (Sigma), 38.8 mM sodium citrate).
For the cell counting assay, HCT116 and HCT116-VR cells were seeded in 24-well plates and treated with 2.5 μM vorinostat or DMSO. Every 24 h, triplicate wells were trypsinised and counted using trypan blue and a haemocytometer.
For the drug combination studies performed in the SW480, SW620 and LIM1215 cell lines, cells were plated using liquid handling automation at the same density reported for siRNA transfections above. At 48 h after seeding, cells were treated with DMSO, vorinostat alone (2.5 μM), GANT61 alone (15 and 20 μM doses) and in combination. There were six replicate wells per treatment condition. Cell viability was measured using CellTiter-Glo as described above.
BCL2L1 overexpression
MSCV-GFP and MSCV-GFP-BCL2L1 constructs were previously generated in the laboratory. Retrovirus was packaged by calcium phosphate-transfected Phoenix E cells and used to transduce HCT116-VR cells. Cells were sorted for medium-high GFP expression and western blot was used to confirm overexpression.
Western blot analysis
Cells were lysed with 25 mM HEPES, 250 mM NaCl, 2.5 mM EDTA, 0.1% Triton-X 100, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin and 0.5 mM phenylmethylsulfonyl fluoride, and protein concentration was determined with a BCA assay (Pierce, Rockford, IL, USA). Equal amounts of protein per sample were separated by SDS-PAGE and transferred to an Immobilon-P PVDF membrane (Millipore, Bedford, MA, USA). Membranes were blocked with 5% skim milk in TBS-T, followed by incubation with antibodies. Primary antibodies were diluted in blocking buffer: β-actin (Sigma; cat. no. A2228), acetylated histone H3 (EMD Millipore, Billerica, MA, USA; cat. no. 06-599), BCL2L1 (Santa Cruz, Castle Hill, NSW, Australia; cat. no. 8392) and incubated at 4 °C overnight. Secondary antibodies (Dako, Campbellfield, VIC, Australia) were incubated in blocking buffer for 1 h at RT before detection of protein with enhanced chemoluminescence reagents (Amersham ECL, Amersham ECL Plus) and Fuji film (Super RX).
qRT-PCR
Total RNA was isolated from cells using the RNeasy Plus Mini Kit (Qiagen, Doncaster, VIC, Australia). cDNA was prepared using random primers and MMLV reverse transcriptase (Promega). qRT-PCR was performed on the 7900 HT Fast Real Time PCR system with SDS v.2.3 software (Applied Biosystems, Forster City, CA, USA) using the SYBR green detection method. A standard curve was generated in each run for each primer and results were analyzed using the adjusted Ct method followed by normalization to the housekeeping gene, L32. qRT-PCR primers: GLI1 forward – 5′-AAGCGTGAGCCTGAATCTGT-3′ and GLI1 reverse – 5′-CAGCATGTACTGGGCTTTGA-3′ L32 forward – 5′-TTCCTGGTCCACAACGTCAAG-3′ and L32 reverse – 5′-TGTGAGCGATCTCGGCAC-3′.
Next-generation sequencing
RNA was prepared for sequencing on the Illumina HighSeq 2000 using the proprietary TruSeqTM RNA Sample Preparation v2 Guide with all kit reagents purchased from Illumina (San Diego, CA, USA). RNAseq reads were aligned to the human reference genome (Homo_sapiens.GRCh37.67.gtf (from ENSEMBL release 67)) using TopHat/Bowtie.59 Sample quality control was performed simultaneously with the FastQC program (Babraham Bioinformatics, Babraham Institute, Cambridge, UK). Read counting was performed with HTSeq.60 Differential gene expression analysis was performed using EdgeR61 and limma-voom.62 All treatment samples were compared with mock transfection and differential gene expression was defined as log2 fold change ⩾1 or ⩽−1 and adjusted P-value <0.05.
Acknowledgments
We thank Richard Tothill, Maria Doyle and Jason Ellul for constructive discussions about NGS methodologies and analysis. For technical support, we also thank Yanny Handoko and Daniel Thomas (Victorian Centre for Functional Genomics), Andrea Newbold and Timothy Semple, Timothy Holloway and Aga Borcz (Molecular Genomics Facility). This work was supported by Merck Research Laboratories, and an Australian National Health and Medical Research Council (NHMRC) project grant APP1028871 (to RWJ). KJF was a recipient of an Australian Postgraduate Award and later a Cancer Council Victoria Postdoctoral Research Fellowship. RWJ is a Senior Principal Research Fellow of the National Health and Medical Research Council (NHMRC) of Australia and supported by NHMRC Program and Project Grants, Cancer Council Victoria, the Leukemia Foundation of Australia and the Victorian Cancer Agency. The VCFG (KJS) is funded by the Australian Cancer Research Foundation (ACRF), the Victorian Department of Industry, Innovation and Regional Development (DIIRD), the Australian Phenomics Network (APN) supported by funding from the Australian Government's Education Investment Fund through the Super Science Initiative, the Australasian Genomics Technologies Association (AMATA), the Brockhoff Foundation and the Peter MacCallum Cancer Centre Foundation.
Glossary
- DMSO
dimethyl sulfoxide
- HDAC
histone deacetylase
- HDACi
histone deacetylase inhibitor
- JAK
Janus kinase
- PTCH
patched
- qRT-PCR
quantitative real-time PCR
- SMO
smoothened
- STAT
signal transducer and activator of transcription
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by E Gottlieb
The Johnstone laboratory received research grant support from Merck for studies involving vorinostat.
Supplementary Material
References
- Fantin VR, Richon VM. Mechanisms of resistance to histone deacetylase inhibitors and their therapeutic implications. Clin Cancer Res 2007; 13: 7237–7242. [DOI] [PubMed] [Google Scholar]
- Fantin VR, Loboda A, Paweletz CP, Hendrickson RC, Pierce JW, Roth JA et al. Constitutive activation of signal transducers and activators of transcription predicts vorinostat resistance in cutaneous T-cell lymphoma. Cancer Res 2008; 68: 3785–3794. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 2006; 5: 769–784. [DOI] [PubMed] [Google Scholar]
- Lindemann RK, Newbold A, Whitecross KF, Cluse LA, Frew AJ, Ellis L et al. Analysis of the apoptotic and therapeutic activities of histone deacetylase inhibitors by using a mouse model of B cell lymphoma. Proc Natl Acad Sci USA 2007; 104: 8071–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden JE, Shi W, Jankowski K, Kan CY, Cluse L, Martin BP et al. HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis 2013; 4: e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart MJ, Tainton KM, Ruefli AA, Dear AE, Sedelies KA, O'Reilly LA et al. Novel mechanisms of apoptosis induced by histone deacetylase inhibitors. Cancer Res 2003; 63: 4460–4471. [PubMed] [Google Scholar]
- Fotheringham S, Epping MT, Stimson L, Khan O, Wood V, Pezzella F et al. Genome-wide loss-of-function screen reveals an important role for the proteasome in HDAC inhibitor-induced apoptosis. Cancer Cell 2009; 15: 57–66. [DOI] [PubMed] [Google Scholar]
- Khan O, Fotheringham S, Wood V, Stimson L, Zhang C, Pezzella F et al. HR23B is a biomarker for tumor sensitivity to HDAC inhibitor-based therapy. Proc Natl Acad Sci USA 2010; 107: 6532–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New M, Olzscha H, Liu G, Khan O, Stimson L, McGouran J et al. A regulatory circuit that involves HR23B and HDAC6 governs the biological response to HDAC inhibitors. Cell Death Differ 2013; 20: 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijwening JH, Beijersbergen RL. Using large-scale RNAi screens to identify novel drug targets for cancer. IDrugs 2010; 13: 772–777. [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 2004; 428: 431–437. [DOI] [PubMed] [Google Scholar]
- Tandle AT, Kramp T, Kil WJ, Halthore A, Gehlhaus K, Shankavaram U et al. Inhibition of polo-like kinase 1 in glioblastoma multiforme induces mitotic catastrophe and enhances radiosensitisation. Eur J Cancer 2013; 49: 3020–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J, von der Saal W, van Dongen MM, Reiff U, van Willigen R, Beijersbergen RL et al. Candidate biomarkers of response to an experimental cancer drug identified through a large-scale RNA interference genetic screen. Clin Cancer Res 2009; 15: 5811–5819. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ et al. A Genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell 2013; 24: 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel-Vinay S, Bajrami I, Friboulet L, Elliott R, Fontebasso Y, Dorvault N et al. A high-throughput screen identifies PARP1/2 inhibitors as a potential therapy for ERCC1-deficient non-small cell lung cancer. Oncogene 2013; 32: 5377–5387. [DOI] [PubMed] [Google Scholar]
- Fredebohm J, Wolf J, Hoheisel JD, Boettcher M. Depletion of RAD17 sensitizes pancreatic cancer cells to gemcitabine. J Cell Sci 2013; 126: 3380–3389. [DOI] [PubMed] [Google Scholar]
- Zhang S, Liu X, Zhang Y, Cheng Y, Li Y. RNAi screening identifies KAT8 as a key molecule important for cancer cell survival. Int J Clin Exp Pathol 2013; 6: 870–877. [PMC free article] [PubMed] [Google Scholar]
- Singel SM, Cornelius C, Batten K, Fasciani G, Wright WE, Lum L et al. A targeted RNAi screen of the breast cancer genome identifies KIF14 and TLN1 as genes that modulate docetaxel chemosensitivity in triple-negative breast cancer. Clin Cancer Res 2013; 19: 2061–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Gazin C, Park SM, Zhu LJ, Debily MA, Kittler EL et al. A synthetic interaction screen identifies factors selectively required for proliferation and TERT transcription in p53-deficient human cancer cells. PLoS Genet 2012; 8: e1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui M, Xie T, Orth JD, Carpenter AE, Rudnicki S, Kim S et al. An intermittent live cell imaging screen for siRNA enhancers and suppressors of a kinesin-5 inhibitor. PLoS One 2009; 4: e7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol 2005; 7: 591–600. [DOI] [PubMed] [Google Scholar]
- Falkenberg KJ, Gould CM, Johnstone RW, Simpson KJ. Genome-wide functional genomic and transcriptomic analyses for genes regulating sensitivity to vorinostat. Scientific Data 2014; 1: 140017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland-Searby S, Bhaumik SR. The 26 S proteasome complex: an attractive target for cancer therapy. Biochim Biophys Acta 2012; 1825: 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol Cell Biol 2003; 23: 3456–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter RL, Lo HW. Hedgehog pathway and GLI1 isoforms in human cancer. Discov Med 2012; 13: 105–113. [PMC free article] [PubMed] [Google Scholar]
- Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci USA 2007; 104: 8455–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Rodova M, Roy SK, Sharma J, Singh KP, Srivastava RK et al. GANT-61 inhibits pancreatic cancer stem cell growth in vitro and in NOD/SCID/IL2R gamma null mice xenograft. Cancer Lett 2013; 330: 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rubin MA, Chinnaiyan AM. Integrative biology of prostate cancer progression. Annu Rev Pathol 2006; 1: 243–271. [DOI] [PubMed] [Google Scholar]
- ClinicalTrials.gov: a service of the US National Institutes of Health (cited 29 August 2013); available at: clinicaltrials.gov; last accessed on 17 January 2016.
- Fendrich V, Wiese D, Waldmann J, Lauth M, Heverhagen AE, Rehm J et al. Hedgehog inhibition with the orally bioavailable Smo antagonist LDE225 represses tumor growth and prolongs survival in a transgenic mouse model of islet cell neoplasms. Ann Surg 2011; 254: 818–823; discussion 823. [DOI] [PubMed] [Google Scholar]
- Ma H, Li HQ, Zhang X. Cyclopamine, a naturally occurring alkaloid, and its analogues may find wide applications in cancer therapy. Curr Top Med Chem 2013; 13: 2208–2215. [DOI] [PubMed] [Google Scholar]
- Matthews GM, Newbold A, Johnstone RW. Intrinsic and extrinsic apoptotic pathway signaling as determinants of histone deacetylase inhibitor antitumor activity. Adv Cancer Res 2012; 116: 165–197. [DOI] [PubMed] [Google Scholar]
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 2014; 13: 673–691. [DOI] [PubMed] [Google Scholar]
- Dhanyamraju PK, Holz PS, Finkernagel F, Fendrich V, Lauth M. Histone deacetylase 6 represents a novel drug target in the oncogenic Hedgehog signaling pathway. Mol Cancer Ther 2015; 14: 727–739. [DOI] [PubMed] [Google Scholar]
- Shevde LA, Samant RS. Nonclassical hedgehog-GLI signaling and its clinical implications. Int J Cancer 2014; 135: 1–6. [DOI] [PubMed] [Google Scholar]
- Wickstrom M, Dyberg C, Shimokawa T, Milosevic J, Baryawno N, Fuskevag OM et al. Targeting the hedgehog signal transduction pathway at the level of GLI inhibits neuroblastoma cell growth in vitro and in vivo. Int J Cancer 2013; 132: 1516–1524. [DOI] [PubMed] [Google Scholar]
- Gulino A, Di Marcotullio L, Canettieri G, De Smaele E, Screpanti I. Hedgehog/Gli control by ubiquitination/acetylation interplay. Vitam Horm 2012; 88: 211–227. [DOI] [PubMed] [Google Scholar]
- Huntzicker EG, Estay IS, Zhen H, Lokteva LA, Jackson PK, Oro AE. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev 2006; 20: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol 2006; 8: 1415–1423. [DOI] [PubMed] [Google Scholar]
- Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol 2010; 12: 132–142. [DOI] [PubMed] [Google Scholar]
- De Smaele E, Di Marcotullio L, Moretti M, Pelloni M, Occhione MA, Infante P et al. Identification and characterization of KCASH2 and KCASH3, 2 novel Cullin3 adaptors suppressing histone deacetylase and Hedgehog activity in medulloblastoma. Neoplasia 2011; 13: 374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung B, Feng Z, Hua X. Menin directly represses Gli1 expression independent of canonical Hedgehog signaling. Mol Cancer Res 2013; 11: 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas VE, Rahman MF, Fernandez-Barrena MG, Diao Y, Liapi E, Sonkoly E et al. Identification of novel non-coding RNA-based negative feedback regulating the expression of the oncogenic transcription factor GLI1. Mol Oncol 2014; 8: 912–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, You Z, Li T, Yu C, Tao G, Hu M et al. Correlation of hedgehog signal activation with chemoradiotherapy sensitivity and survival in esophageal squamous cell carcinomas. Jpn J Clin Oncol 2011; 41: 386–393. [DOI] [PubMed] [Google Scholar]
- Steg AD, Katre AA, Bevis KS, Ziebarth A, Dobbin ZC, Shah MM et al. Smoothened antagonists reverse taxane resistance in ovarian cancer. Mol Cancer Ther 2012; 11: 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy B, Lu Y, Teng KY, Nuovo G, Li X, Shapiro CL et al. Hedgehog signaling is a novel therapeutic target in tamoxifen-resistant breast cancer aberrantly activated by PI3K/AKT pathway. Cancer Res 2012; 72: 5048–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Lin CP, Hsu ML, Shieh HR, Chao NK, Chao KS. Sonic hedgehog signaling protects human hepatocellular carcinoma cells against ionizing radiation in an autocrine manner. Int J Radiat Oncol Biol Phys 2011; 80: 851–859. [DOI] [PubMed] [Google Scholar]
- Mozzetti S, Martinelli E, Raspaglio G, Prislei S, De Donato M, Filippetti F et al. Gli family transcription factors are drivers of patupilone resistance in ovarian cancer. Biochem Pharmacol 2012; 84: 1409–1418. [DOI] [PubMed] [Google Scholar]
- Das S, Samant RS, Shevde LA. Nonclassical activation of Hedgehog signaling enhances multidrug resistance and makes cancer cells refractory to Smoothened-targeting Hedgehog inhibition. J Biol Chem 2013; 288: 11824–11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RR, Kunkalla K, Qu C, Schlette E, Neelapu SS, Samaniego F et al. ABCG2 is a direct transcriptional target of hedgehog signaling and involved in stroma-induced drug tolerance in diffuse large B-cell lymphoma. Oncogene 2011; 30: 4874–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D, Xu Q, Wang K, Che X. Gli1 is a potential target for alleviating multidrug resistance of gliomas. J Neurol Sci 2010; 288: 156–166. [DOI] [PubMed] [Google Scholar]
- Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature 2014; 511: 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow RL, Chari NS, Unden AB, Spurgers KB, Lee S, Roop DR et al. Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J Biol Chem 2004; 279: 1197–1205. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med 2009; 9: 873–886. [DOI] [PubMed] [Google Scholar]
- Regl G, Kasper M, Schnidar H, Eichberger T, Neill GW, Philpott MP et al. Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res 2004; 64: 7724–7731. [DOI] [PubMed] [Google Scholar]
- Wiegmans AP, Alsop AE, Bots M, Cluse LA, Williams SP, Banks KM et al. Deciphering the molecular events necessary for synergistic tumor cell apoptosis mediated by the histone deacetylase inhibitor vorinostat and the BH3 mimetic ABT-737. Cancer Res 2011; 71: 3603–3615. [DOI] [PubMed] [Google Scholar]
- Lin TL, Matsui W. Hedgehog pathway as a drug target: smoothened inhibitors in development. OncoTargets Ther 2012; 5: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg K, Saunders D, Simpson K. A high-throughput, multiplex cell death assay using an RNAi screening approach. Cold Spring Harbour Protocols 2014; 6: 663–676. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl P, Huber W. HTSeq – A Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: 1–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.