Abstract
Oocyte and preimplantation embryo development entail dynamic changes in chromatin structure and gene expression, which are regulated by a number of maternal and zygotic epigenetic factors. Histone deacetylases (HDACs), which tighten chromatin structure, repress transcription and gene expression by removing acetyl groups from histone or non-histone proteins. HDAC1 and HDAC2 are two highly homologous Class I HDACs and display compensatory or specific roles in different cell types or in response to different stimuli and signaling pathways. We summarize here the current knowledge about the functions of HDAC1 and HDAC2 in regulating histone modifications, transcription, DNA methylation, chromosome segregation, and cell cycle during oocyte and preimplantation embryo development. What emerges from these studies is that although HDAC1 and HDAC2 are highly homologous, HDAC2 is more critical than HDAC1 for oocyte development and reciprocally, HDAC1 is more critical than HDAC2 for preimplantation development.
Facts
HDAC1 and HDAC2 show different expression profiles during oocyte and preimplantation development.
HDAC1 and HDAC2 regulate oocyte development through transcription in a dosage-dependent manner.
HDAC2 is the major HDAC in mouse oocytes and regulates global DNA methylation and imprinting marks by interacting with DNMT3A2.
HDAC2 regulates chromosome segregation and kinetochore function via H4K16 deacetylation during oocyte maturation.
HDAC1 is the responsible HDAC involved in cell cycle regulation and zygotic genomic activation during preimplantation development.
Open Questions
How do HDAC1 and HDAC2 regulate cross-talk between histone acetylation and other epigenetic modifications during oocyte and preimplantation embryo development?
What is the catalog of chromatin remodeling complexes that contain HDAC1 and/or HDAC2 in oocytes and preimplantation embryos, and what is the relative contribution of these HDAC1/2-containing complexes in defining steady-state levels of acetylated histones?
What non-histone proteins are deacetylated by HDAC1 and/or HDAC2 in oocytes and preimplantation embryos, and what are the functional consequences of their deacetylation?
In eukaryotes, DNA is organized into a highly ordered nucleoprotein assembly called chromatin, whose fundamental unit is the nucleosome. The nucleosome consists of 146 bp of DNA wrapped around a histone core comprised of two molecules each of histones H2A, H2B, H3 and H4. Histone H1 is bound to linker DNA between nucleosomes.1, 2 Histones are subject to multiple post-translational modifications (PTMs), including acetylation, methylation, ubiquitylation, phosphorylation, and sumoylation. These PTMs determine open and closed chromatin conformations, which, in turn, regulate the differential access and recruitment of transcription factors and other regulatory chromatin-binding proteins to DNA.3, 4, 5 Among these histone modifications, histone acetylation is the most well-studied modification, which occurs at the ɛ-amino groups of evolutionarily conserved lysine residues located at the N termini. Although all core histones are acetylated in vivo, modifications of histones H3 and H4 are more extensively characterized than those of H2A and H2B.6
Histone Deacetylases
Lysine acetylation of histones is controlled by histone acetyl transferases (HATs) and histone deacetylases (HDACs).7 The balance between the actions of these enzymes serves as a key regulatory mechanism for gene expression and governs numerous developmental processes and disease states. HATs catalyze the transfer of an acetyl group to lysine residues of histone tails, thereby neutralizing the positive charge of histones. The decrease in net positive histone charge decreases the affinity between histones and DNA, which relaxes chromatin structure to make it more accessible to transcription factors. Therefore, HATs are considered as transcription co-activators. In contrast, HDACs remove acetyl groups from histone tails and are therefore considered as transcriptional co-repressors.8, 9 In addition to histones, HDACs can also deacetylate non-histone proteins, for example, transcription factors and a growing list of other proteins.8, 10 As a result, HDACs are now also called KDACs, or lysine deacetylases.11
In mammals, 18 HDACs have been identified and are grouped into four classes based on their homology with yeast proteins.12 Class I, which are homologous to the yeast protein RPD3 and ubiquitously expressed in human cell lines and tissues, include HDAC 1, 2, 3, and 8 that have a nuclear localization.6, 13 Class II is homologous to yeast Hda1 and can be subdivided into two subclasses: IIa (HDAC 4, 7, and 9) and IIb (HDAC 6 and 10). Class II exhibits tissue-specific expression and can shuttle between the nucleus and cytoplasm, which suggests that this class of HDACs is involved in acetylation of non-histone proteins.6 Class I and Class II HDACs are inhibited by trichostatin A (TSA). Class III HDACs, or sirtuins (SIRT1-7), are homologous with the yeast SIRT2 family of proteins and require NAD+ as a cofactor.14 The subcellular distribution and pattern of tissue-specific expression of this class are unknown.9, 15, 16 HDAC11, the only member of class IV, is homologous to both class I and class II HDACs.17 The multiplicity of histone deacetylases reflects diversification of functions in different tissues and biological processes.18
Structure and Complexes of Mammalian HDAC1 and HDAC2
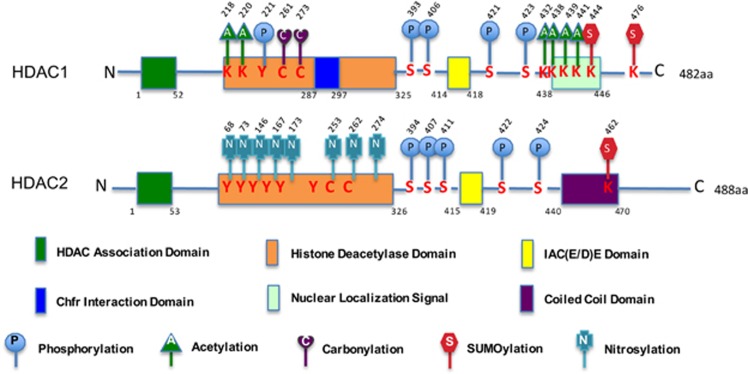
Two highly homologous Class I enzymes, HDAC1 and HDAC2, are expressed ubiquitously, localized predominantly to the nucleus, and display high enzymatic activity toward histone substrates.8, 19 The genes for HDAC1 and HDAC2 originated by gene duplication8, 20 and the two proteins exhibit ~86% amino-acid sequence identity in mice and human, suggesting a high functional redundancy between HDAC1 and HDAC2.21 Both HDAC1 and HDAC2 contain several domains with defined function (Figure 1). Some domains are common to both, whereas other domains are specific for each HDAC.8 The histone deacetylase domain common to all class I HDACs is formed by a stretch of more than 300 amino acids that constitute a large portion of the protein.6 An N-terminal HDAC association domain (HAD; residues 1 to ~50) is essential for homo- and hetero-dimerization.21 The C-terminal portion contains an IAC(E/D)E motif (IACEE in HDAC1 and IACDE in HDAC2) involved in the interaction with the pocket proteins PRb, P107, and P130.8 HDAC1 has a 2-residue Chfr interaction domain that is essential for the interaction with Chfr, an ubiquitin ligase regulating protein degradation,22 and a nuclear localization signal at the C terminus.21 A coiled-coil domain is only found at the C terminus of HDAC2.8 In addition, HDAC1 and HDAC2 are not only protein-modifiers but also undergo numerous post-translational modification. These modifications can be either chemical moieties (acetylation, phosphorylation, methylation, nitrosylation, ADP-ribosylation, glycosylation, and carbonylation) or proteins (ubiquitin, SUMO, NEDD8, FAT10, ISG15, and ATG8/ATG12).18 Many of the sites modified in either HDAC1 or HDAC2 are conserved between both proteins (Figure 1).
Figure 1.
A schematic diagram of mammalian HDAC1 and HDAC2 structures with functional domains and post-translational modifications. HDAC1 and HDAC2 share a highly conserved N-terminal HDAC association domain (HAD) that is essential for homo- and hetero-dimerization. The C-terminal part contains an IAC(E/D)E motif (IACEE in HDAC1 and IACDE in HDAC2) involved in the interaction with the pocket proteins. HDAC1 has a 2-residue Chfr interaction domain and a nuclear localization signal (NLS) at the C terminus. HDAC2 contains a coiled-coil domain at the C terminus. HDAC1 and HDAC2 are regulated by different post-translational modifications, such as phosphorylation, acetylation, nitrosylation, carbonylation, and sumoylation. K, lysine; C, cysteine; S, Serine; Y, tyrosine. Numbers indicate the corresponding amino-acidic (aa) position
HDAC1 and HDAC2 lack a DNA-binding domain, as do all deacetylases, and execute their function by interacting with transcription factors as either homo- or heterodimers, or being part of multi-component repressor complexes.8, 9 The best characterized HDAC1/2-containing complexes in mammals are the SIN3 co-repressor complex,23 nucleosome remodeling and deacetylase (NuRD) complex,24 CoREST complex,25 Nanog and Oct4 (POU5F1)-associated deacetylase complex that is specifically found in embryonic stem (ES) cells,26 and PRC2 complex.27
The ubiquitous expression, high deacetylase activity toward common substrates and high homology between HDAC1 and HDAC2 suggest that each could compensate for loss of function of the other. Indeed, loss-of-function studies in mice provide important insights regarding the compensatory functions of HDAC1 and HDAC2 in regulating cell proliferation, apoptosis, and differentiation in different cell types and tissues.8, 28 Tissue-specific conditional knockout of Hdac1 or Hdac2 alone does not evoke an obvious phenotype in cardiomyocytes,29 neuron precursors,30 oligodendrocyte,31 B cells,32 embryonic epidermis,33 and T cells,34 whereas deletion of both genes results in severe phenotypes in all tissues examined. In contrast, results of other studies support the notion that HDAC1 and HDAC2 have distinct functions. For example, germ-line deletion of HDAC1 causes mouse embryo lethality before embryonic day 10.5, even though HDAC2 is upregulated,35 and HDAC2 specifically regulates synaptic plasticity and memory formation, a function that is not compensated by overexpressing HDAC1.36 A unique role of HDAC1 is also observed in embryonic stem cells.37 In addition, HDAC2 has a unique role by controlling the fate of neural progenitors during normal brain development.38 Taken together, these results indicate that the functions of HDAC1 and HDAC2 are diverse in different tissues or cell types. The reader is referred to several excellent reviews discussing the physiological role of HDAC1 and HDAC2.8, 9, 16, 28, 39 Here we summarize and discuss the results of experiments using conditional mutant mice in combination with RNA interference (RNAi) to dissect the roles of HDAC1 and HDAC2 in oocytes and preimplantation embryos.
Expression of HDAC1 and HDAC2 During Mouse Oocyte and Preimplantation Development
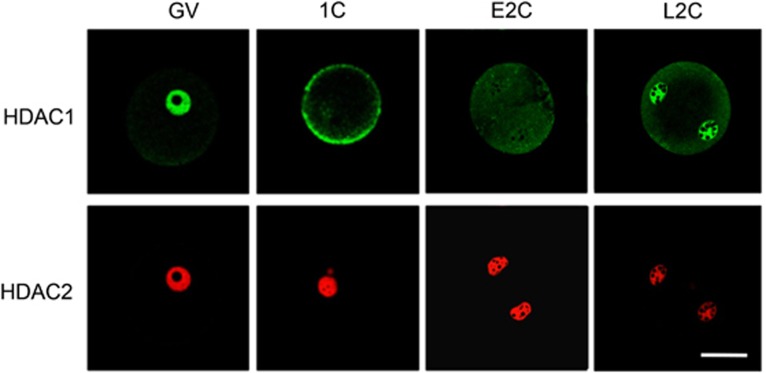
Oocyte and preimplantation embryo development, which are critical processes for reproduction and have important implications for both maternal and fetal health, comprise a series of developmental stages that occur over the course of some 4 weeks in mice, that is, oocyte growth during folliculogenesis, oocyte maturation, fertilization, embryo cleavage, and morula and blastocyst formation.40, 41 Detailed temporal and spatial patterns of HDAC1 and HDAC2 expression have been established by immunocytochemical analysis.35, 42, 43 HDAC1 and HDAC2 are concentrated in the nucleus throughout oocyte growth.43 The intensity of HDAC1 nuclear staining displays a progressive decrease during the course of oocyte growth and following resumption of meiosis (i.e., germinal vesicle breakdown (GVBD)) HDAC1 co-localizes with chromosomes. In contrast, nuclear HDAC2 staining increases between days 5 and 12 post-partum and then remains relatively constant for the duration of the growth phase. After GVBD, HDAC2 is uniformly dispersed throughout the cytoplasm.42, 43 Following fertilization, HDAC2 again appears in the pronucleus/nucleus, and the intensity of HDAC2 nuclear staining progressively decreases during preimplatation development.42 HDAC1 staining, on the other hand, is largely confined to the cytoplasm in fertilized eggs and early 2-cell embryos with enrichment in the nucleus becoming visible in the late 2-cell embryo, concomitant with zygotic genome activation (Figure 2). Thereafter, the intensity of HDAC1 nuclear staining increases until the morula stage but the morula–blastocyst transition is accompanied by a decrease in staining intensity.42 The different expression profiles of HDAC1 and HDAC2 during oocyte and preimplantation development suggest distinct functions of the two proteins in oocyte and preimplantation development.
Figure 2.
Expression of HDAC1 and HDAC2 in full-grown oocytes, 1- and 2-cell embryos. HDAC2 is concentrated in the nucleus in oocytes, 1-cell and 2-cell embryos. HDAC1 appears in the nucleus in full-grown oocytes but then is essentially located in the cytoplasm in fertilized eggs and early 2-cell embryos. HDAC1 nuclear staining becomes clearly enriched at late 2-cell embryo stage. All samples for a given HDAC were processed for immunocytochemistry together, and all images were taken at the same laser power, thereby enabling direct comparison of signal intensities. GV, fully grown oocyte; 1C, one-cell embryo; E2C, early two-cell embryo; L2C, late two-cell embryo. Scale Bar, 25 μm
HDAC1 and HDAC2 Regulate Oocyte Development Through Transcription
Because global germ-line ablation of HDAC1 and HDAC2 cause early lethality, conditional Cre recombinase-mediated deletions were required to reveal the functions of these deacetylases in specific tissues. Deletion of either Hdac1 or Hdac2 in a variety of tissues/cell types, including the heart, brain, endothelial cells, smooth muscle, and neural crest cells, ES cells, fibroblasts, B cells, thymocytes, keratinocytes, and various cell lines causes no or only mild effects.9, 16, 28 Likewise, deletion of either Hdac1 or Hdac2 does not evoke an obvious effect on oocyte development to the full-grown oocyte stage.43 Deletion of both Hdac1 and Hdac2, however, results in infertility due to oocyte development arresting at the secondary follicle stage, which is accompanied by mis-regulation of histone modifications, a significant reduction of global transcription, and an increased incidence of apoptosis. These results suggest an apparent compensatory function of HDAC1 and HDAC2 in oocytes.43 Nevertheless, deletion of three of the four Hdac1/Hdac2 alleles shows that HDAC1 and HDAC2 function unequally during oocyte development. Haplo-insufficency of Hdac1 in the absence of HDAC2 (Hdac1−/+/Hdac2−/−) reveals a more important function of HDAC2 in oocytes, the mice are infertile,43 whereas in the opposite allelic combination, a single Hdac2 allele in the absence of HDAC1 does not produce any apparent phenotypes. These results suggest that HDAC2 function is more critical for oocyte development than HDAC1. A similar situation is also observed recently in the developing nervous system, where a single allele of Hdac2, but not Hdac1, is sufficient for normal mouse brain development in the absence of its paralog.38
Deletion of both Hdac1 and Hdac2 leads to DNA damage that ultimately results in apoptosis in a variety of cell types and tissues including B cells,32 T cells,34 thymocyte,44 and brain.38 A significant increased incidence of apoptosis accompanied by pronounced DNA damage is also observed in growing oocytes following loss of both Hdac1 and Hdac2 and likely attributed to hyperacetylation of TRP53.43, 45 Interestingly, no apparent apoptosis is detected in Hdac1−/+/Hdac2−/− oocytes although oocyte development is mainly blocked at the secondary follicle stage, suggesting that increased apoptosis is not the major cause for the oocyte developmental defects following loss of Hdac1 and Hdac2.46
Histone acetylation is typically linked with transcriptional activation, whereas histone deacetylation is associated with gene repression.47 Thus, HDACs have long been regarded as transcriptional repressors. Recent studies, however, challenge this conventional notion and show that HDACs can also function in transcriptional activation.48, 49 A positive correlation between HDAC1/HDAC2 binding and transcription was discovered by a genome-wide ChIP-seq analysis in human CD4+ cells.49 Indeed, a combined loss of HDAC1 and HDAC2 in oocytes results in a significant reduction of transcription that is accompanied by global histone hyperacetylation and decreased histone H3K4 methylation, a transcription active mark.43 Thus, HDAC1/2 may act as transcription activators in female germ cells. Consistent with such a role, microarray analysis revealed that the top four categories of downregulated transcripts in Hdac1:2−/− oocytes are related to transcription or positive regulation of transcription.43
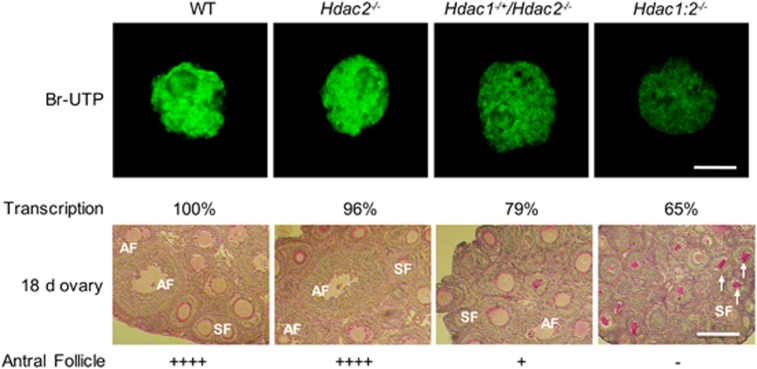
Oocyte development is characterized by progressive changes in gene expression with dramatic changes occurring during the primordial-to-primary follicle transition and then again during the secondary follicle-to-antral follicle transition;50 deleting transcription factors always leads to impaired oocyte development during these two transitions.51, 52 A similar phenotype with failure to undergo the secondary follicle-to-antral follicle transition during folliculogenesis is observed in females that lack Taf4b (a germ cell-specific subunit of TFIID),53 Yy1,54 or Tbp2 (TATA-binding protein 2).55 In particular, female mice deficient in the oocyte-specific TBP2 display phenotypes very similar to that observed for Hdac1:2−/− mice, including infertility, small ovaries, arrest of follicle growth before antrum formation, a decrease in Pol II CTD S2 phosphorylation and histone H3K4 methylation.55 The similarity in phenotypes suggests that Hdac1 and Hdac2 double mutants and these transcriptional factor mutants share a common mechanism that is linked to a decrease in transcription. Indeed, a detailed analysis using mice lacking different combinations of Hdac1 and Hdac2 suggests a threshold level of transcription is required for oocyte development (Figure 3).45 In Hdac1:2−/− oocytes, transcription is reduced ~40%, which results in a complete block of oocyte development at the secondary follicle stage with no antral follicles observed. An ~20% reduction in transcription in oocytes allows a small fraction of growing oocytes develop to the antral follicle stage in Hdac1−/+/Hdac2−/− oocytes. Deletion of only Hdac2 shows little effect on transcription in oocytes and normal oocyte development to the full-grown stage is observed (Figure 3). These results strongly suggest that HDAC1/2 regulate oocyte development through transcription in a dosage-dependent manner and that there is a threshold of transcription activity required for normal oocyte development. Should transcription be reduced below this threshold, oocyte development will be impaired and apoptosis ensues. From this perspective transcription regulates oocyte development in a manner similar to that of cell cycle checkpoints.
Figure 3.
HDAC1 and HDAC2 regulate oocyte development through transcription. Transcription in oocytes obtained from mice 12-days-of-age and lacking different combinations of Hdac1 and Hdac2 was detected by BrUTP incorporation (upper panel), Quantification of immunofluorescence data shown below the panel, the nuclear staining intensity of Br-UTP in the WT oocytes was set to 100%. The bar corresponds to 10 μm. Histological analysis of ovaries obtained from mice 18-days-of-age and lacking different combinations of Hdac1 and Hdac2 is shown in lower panel. Only secondary follicles and antral follicles are indicated in the figures. Numbers of antral follicles in WT and Hdac2−/− ovaries are similar, significantly fewer antral follicles are observed in Hdac1−/+Hdac2−/− ovaries. In ovaries from Hdac1:2−/− mice, no antral follicles are found, and there is an increase in the number of degenerating follicles (white arrows). Scale bar, 200 μm. AF, antral follicle; SF, secondary follicle
HDAC2 Regulates Global DNA Methylation and Genomic Imprinting Marks in Oocytes Through Targeting DNMT3A2
Deleting both Hdac1 and Hdac2 in oocytes not only results in histone H3K4 demethylation,43 but also leads to a global decrease of DNA methylation,45 that is, HDACs can regulate maternal-specific de novo DNA methylation. Maternal-specific de novo DNA methylation occurs in postnatal growing oocytes,56, 57, 58 and methylation acquisition is related to oocyte size.57, 58 DNA methyltransferases A (DNMT3A) and its cofactor DNA methyltransferases L (DNMT3L) are required to establish DNA methylation in growing oocytes.59 Maternal-specific de novo DNA methylation likely contributes to gene regulation in the oocyte and marks specific genes for activity in the embryo, as in the case of imprinted genes.60 Indeed, Hdac1/2 mutant oocytes fail to establish normal imprinting DNA methylation marks.45
Surprisingly, a significant decrease of nuclear-associated DNMT3A2, the predominant isoform of DNMT3A in oocytes, is observed in Hdac1:2−/− oocytes, and could in principle be responsible for the decreased global DNA methylation and failure to establish DNA methylation of imprinted genes.45 A moderate decrease in DNMT3A2 nuclear staining is also observed in Hdac2−/− oocytes, demonstrating that loss of HDAC2 is not compensated by upregulation of HDAC1 that occurs in Hdac2−/− oocytes. These results also implicate HDAC2 as the major HDAC in regulating DNA methylation during oogenesis. HDAC1/2 likely affects DNA methylation in oocytes through DNMT3A2 independent of its enzymatic deacetylase function because inhibiting HDAC activity with TSA has no effect on localization of DNMT3A2.45
DNMT3A2 can interact with HDAC2 as detected by co-immunoprecipitation, which suggests that HDAC2 and DNMT3A2 exist in a complex in oocytes that is essential for DNMT3A2 to target germline differentially methylated regions (gDMRs) and methylate DNA. In agreement with this proposal, disruption of the SIN3A HDAC1/2-containing complex by conditional deletion of Sin3A leads to a significant reduction in both nuclear HDAC2 and DNMT3A2, and hypomethylation of imprinted genes, a phenotype similar to that observed in Hdac1:2−/− oocytes.45 These results suggest that HDAC2 provides a structural role, rather than its enzymatic function, in regulating de novo DNA methylation by interacting with DNMT3A2 in growing oocytes.45 In a similar vein, G9a, a histone methyltransferase (HMTase) primarily responsible for histone H3 dimethylation at K9 (H3K9me2), promotes DNA methylation in ES cells.61 In these cells G9a mediates DNA methylation independent of its HMTases activity but depends on its interactions with DNMT3B; G9a directly recruits DNMT3B to the Pou5f1 promoter by forming a specific complex with DNMT3B.62 It should be noted, however, that a recent study reveals that HDACs can also affect DNA methylation; inhibiting HDAC activity reversed DNA methylation by repressing DNMT1 expression in LNCaP cells.63
HDAC2 Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation During Oocyte Maturation
The process of oocyte maturation traditionally has been described by changes in chromosome morphology during meiosis64 and is accompanied by dynamic changes of chromatin structure and histone modifications. HDACs, as chromatin remodeling proteins, regulate mammalian oocyte maturation as assessed by using HDAC inhibitors. Histone deacetylation accompanies oocyte maturation and inhibiting HDAC activity with TSA results in global histone hyperacetylation and chromosome segregation defects.65, 66, 67, 68 The responsible HDACs, however, have not been fully identified.
Conditional null alleles for HDACs have been used to investigate the functions of individual HDACs in oocyte maturation.46 Conditional deletion of Hdac2 in oocytes results in hyperacetylation of H4K16 without affecting normal deacetylation of other lysine residues of histone H3 or H4, and defective chromosome condensation and segregation and compromised kinetochore function during oocyte maturation occur in a sub-population of oocytes.46 Significant upregulation of HDAC1 in Hdac2−/− oocytes could not compensate loss of Hdac2, suggesting that HDAC2 is the HDAC specifically responsible for deacetylation of H4K16 that is essential for normal chromosome condensation and segregation during oocyte maturation.46 No changes of histone acetylation are observed in the Hdac1 mutant eggs,46 demonstrating that HDAC1 is not required for histone deacetylation during oocyte maturation although HDAC1 is associated with chromosome during the process.42, 43, 65
Deletion of Hdac2 leads to global histone hyperacetylation in growing oocytes and hyperacetyation of histone H4K16 in MII eggs, which is linked with disruption of a normal chromatin configuration.46 Acetylation of H4K16 inhibits formation of the higher order 30 nm chromatin structure, and loss of H4K16 shows defects equivalent to the loss of the H4 tails.69 The increased acetylation of H4K16 observed in Hdac2-deficient oocytes likely contributes to the failure of chromosomes to condense fully during oocyte maturation and compromised kinetochore function in mutant eggs, because reducing H4K16 acetylation by overexpressing HDAC2 in Hdac2−/− oocytes restores normal chromosome condensation and segregation.46 A recent report also provides further evidence that H4K16 deacetylation has an important role in maintaining normal chromatin structure in oocyte maturation; H4K16 hyperacetylation induced by knockdown of SIRT2, a member of Class III HDACs, leads to spindle defects, missegregation of chromosome and impaired kinetochore–microtubule interaction.70 It is not surprising that SIRT2 is also involved in regulating deacetylation of H4K16 during oocyte maturation because class III HDACs (sirtuins) specifically deacetylate histone H4K16 in other systems.71 SIRT2, however, is localized on the meiotic spindle,70 so how SIRT2 leads to H4K16 deacetylation is not apparent.
Although HDAC2 is responsible for H4K16 deacetylation during oocyte maturation, nuclear HDAC2 in full-grown oocytes appears unable to deacetylate H4K16 because similar amounts of acetylated H4K16 are observed in full-grown WT and Hdac1−/+/Hdac2−/− oocytes.46 These observations suggest that HDAC2 in full-grown oocytes is inactive or inaccessible to histone substrates and then becomes functional or has access to histone substrates during oocyte maturation. In agreement with this proposal, a previous study using porcine oocytes showed that global histone deacetylation induced by HDACs is correlated with mixing of nuclear contents and cytoplasmic factors following GVBD.72, 73 Thus, HDAC2 activity during oocyte maturation may be regulated by cytoplasmic factors or factors only expressed/degraded after GVBD, which activate HDAC2 or form a complex with HDAC2 and mediate HDAC2's ability to bind chromatin; HDACs lack a DNA-binding domain and execute their function by being a component of repressor complexes.74, 75
A recent study provided strong evidence to support this notion. Knockdown of retinoblastoma binding protein P46 (RBAP46, also called RBBP7), an integral subunit in HDAC1/2-containing complexes, results in similar phenotypes including H4K16 hyperacetylation and missegregation of chromosomes to that observed in Hdac2 mutant eggs.76 Immunoblot analysis revealed that RBBP7 protein level increases significantly from GV to MII eggs;76 this maturation-associated increase in RBBP7 protein is well correlated with activation of HDAC2. In addition, RBBP7 co-localizes with chromosomes during meiosis division,76 suggesting that it may promote HDAC2 association with chromatin and thereby facilitate deacetylation of H4K16.
It should also be noted that deacetylation of other histone H3 and H4 lysine residues except H4K16 are observed in both Hdac2 and Sirt2 mutant eggs, suggesting that other HDACs are responsible for deacetylation of these lysine residues. Further experiments are necessary to identify the responsible HDACs.
HDAC1 is the Responsible HDAC Involved in Cell Cycle Regulation and Zygotic Genomic Activation During Preimplantation Development
As described above, the difference in expression profiles for HDAC1 and HDAC2 suggest distinct functions during preimplantation development. Consistent with this proposal is that ablation of both maternal and zygotic Hdac1 by RNA interference (RNAi) leads to hyperacetylation of histone H4 and a developmental delay even though expression of HDAC2 and HDAC3 is significantly induced in Hdac1-suppresssed embryos. RNAi-mediated reduction of HDAC2 has no noticeable effect on histone acetylation and preimplantation development, suggesting that HDAC1 function is more important than HDAC2 for preimplantation development.42 Consistent with this notion is that germline deletion of Hdac1 results in an early embryonic lethal phenotype due to severe developmental problems, with loss of HDAC1 during embryogenesis not being compensated by HDAC2.29, 35 In contrast, depletion of HDAC2 does not display a phenotype during early development.29, 77, 78 These results demonstrate a unique function of HDAC1 in early embryos.
HDAC1-deficient embryos and embryonic stem cells are characterized by an overall reduced cellular proliferation rate in response to increased levels of the cyclin-dependent kinase (CDK) inhibitor P21 and P27.29 Similarly, increased expression of P21Cip1/Waf may contribute to the observed developmental delay from morula to blastocyst after depleting Hdac1 in preimplantation embryos,42 suggesting a general function of HDAC1 in repressing the cyclin-dependent kinase inhibitor P21 and thus positively regulating cell cycle proliferation.16 Indeed, the proliferation defect in embryonic stem cells is rescued by deleting P21.79
Deleting both Hdac1 and Hdac2 in dividing cells results in a similar cell cycle block in G1 phase.39 Fibroblasts lacking both HDAC1 and HDAC2 fail to proliferate in culture and exhibit a strong cell cycle block in the G1 phase that is associated with upregulation of the CDK inhibitors p21Cip1/Waf1 and p57Kip2.32 Combined deletion of Hdac1 and Hdac2, or inactivation of their deacetylase activity in primary or oncogenic-transformed fibroblasts, results in a senescence-like G1 cell cycle arrest, accompanied by upregulation of the cyclin-dependent kinase inhibitor P21 and P53.80 A similar situation is also observed in one-cell embryos; reduction in maternal Hdac1 combined with Hdac2 deficiency (Hdac1−/+/Hdac2−/−) in fertilized eggs results in developmental arrest from 1-cell to 2-cell transition due to a cell cycle block in G1.46 However, the cell cycle block is not caused by upregulation of any CDK inhibitors, but rather due to failing to replicate DNA,46 pointing to different mechanisms of HDAC1 and HDAC2 in cell cycle regulation in different cell types or tissues.
A fundamental event in early mouse development is transforming the highly differentiated oocyte into totipotent blastomeres by the 2-cell stage. This transition, which is called the maternal-to-zygotic transition, first entails degradation of maternal mRNAs and then activation of the embryonic genome.40, 81, 82 Superimposed on genome activation, which results in a dramatic reprogramming of gene expression, is the development of a chromatin-mediated transcriptionally repressive state that is likely critical for generating the correct pattern of gene expression required for continued development.40 HDACs likely underlie development of the transcriptionally repressive state because inducing histone hyperacetylation using HDAC inhibitors not only relieves the enhancer requirement for efficient expression of plasmid-borne reporter genes in 2-cell embryos,83, 84 but also results in a further increase in global transcription.85
Several lines of evidence support a role for HDAC1 in development of a transcriptionally repressive state that initiates in 2-cell embryos.42 First, microarray data and qRT-PCR show that Hdac1 is zygotically expressed beginning from 2-cell stage,42, 86, 87 accompanying genome activation. Second, HDAC1 protein enters into nucleus in late 2-cell embryos that is concomitant with zygotic genome activation at this stage (Figure 2).42 Third, microarray data reveal that only HDAC1 is sensitive to α-amanitin among the HDACs expressed in 2-cell embryos and Ingenuity Pathway Analysis placed HDAC1 at the hub of numerous interactions in a gene network that could contribute to the development of the transcriptionally repressive state.87 Finally, partial knockdown of HDAC1 results in histone H4 hyperacetylation and relieves the repression of a subset of genes at the late 2-cell embryos.42 These results indicate that HDAC1 is a major contributor to the development of the transcriptionally repressive state.
Conclusions and Future Perspectives
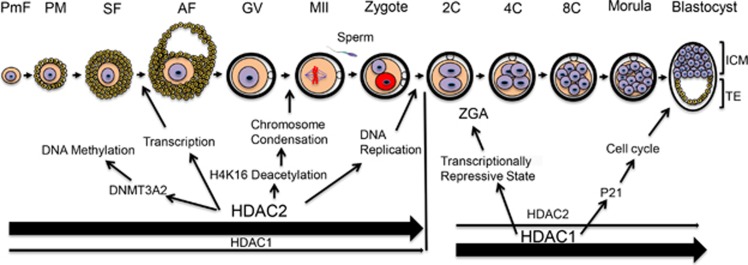
In summary, HDAC1 and HDAC2 have an essential role during oogenesis and preimplantation embryogenesis (Figure 4). HDAC1 and HDAC2 exert their function in regulating dynamic changes in chromatin structure and gene expression that occur during oocyte and preimplantation development through two distinct epigenetic activities, their catalytic activity and a structural role. HDAC2 is the major HDAC for oocyte development by regulating histone acetylation, transcription, and DNA methylation. HDAC2 is also critical for oocyte maturation by fostering histone H4K16 deacetylation, which is required for normal chromosome segregation and kinetochore function. HDAC1 appears to have little role in these processes but is critical for preimplantation development by regulating histone acetylation, cell cycle progression and the development of a transcriptionally repressive state that initiates in 2-cell embryos (Figure 4).
Figure 4.
Model for the role of HDAC1 and HDAC2 during oocyte and preimplantation embryo development. HDAC1 and HDAC2 are essential for oocyte and preimplantation embryo development. HDAC2 is the major HDAC during oogenesis and regulates histone acetylation, transcription, DNA methylation through forming a complex with DNMT3A2 and is essential for oocyte development. HDAC2 is responsible for histone H4K16 deacetylation during oocyte maturation, which is essential for normal chromosome segregation and kinetochore function. In contrast, HDAC1 is critical for preimplantation embryogenesis and regulates histone acetylation, cell cycle progression, development of a transcriptionally repressive state that initiates in 2-cell embryos. AF, antral follicle; GV, full-grown oocyte; ICM, inner cell mass; MII, metaphase II egg; PF, primary follicle; PmF, primordial follicle; SF secondary follicle; 1C, one-cell embryo; 2C, two-cell embryo; 4C, four-cell embryo; 8C, eight-cell embryo; TE, trophoectoderm; ZGA, Zygotic genome activation. Bold arrows indicate major contribution
Despite this recent progress, much remains to be learned about how HDAC1/HDAC2 regulates chromatin structure and gene expression in oocytes and preimplantation embryos. Future genetic analyses of components in HDAC1/HDAC2-containing complexes will provide important information on the roles of HDAC1/HDAC2 in oocytes and preimplantation embryos. Given the limited amounts of readily obtainable biomaterial, new emerging technologies, such as high-throughput sequencing and microChIP techniques, will be of great value to define how HDAC1/HDAC2 regulate gene expression in oocytes and early embryos. Use of novel, more specific HDAC inhibitors will also be instrumental to identify target genes of HDAC1/HDAC2 in oocytes and early embryos. Collectively, a more detailed analysis of HDAC1/HDAC2 function will provide new insights into how epigenetic-mediated dynamic changes in chromatin structure and gene expression result in formation of a developmentally competent oocyte that in turn supports appropriate genome activation, cell fate and lineage commitment during preimplantation development. This knowledge can be also used to develop methods of cellular reprogramming for the production of cloned animals for agricultural and biomedical uses, and to create autologous cells for transplantation therapies in animals and humans. Furthermore, such an understanding may help elucidate how mis-regulation of epigenetic factors contributes to infertility, miscarriages, birth defects, ovarian cancer, and other reproductive and developmental disorders.
Acknowledgments
We apologize to all colleagues whose work could not be cited due to space limitations. We thank Eric N. Olson and Rusty L. Montgomery for providing mice containing floxed alleles of Hdac1 and Hdac2. The work referenced from the authors' laboratory was supported by a grant from NIH (HD022681) to RMS.
Glossary
- HDAC
histone deacetylase
- HAT
histone acetyl transferase
- PTM
post-translational modification
- KDAC
lysine deacetylase
- TSA
trichostatin A
- NAD+
nicotinamide adenine dinucleotide
- HAD
HDAC association domain
- GVBD
germinal vesicle breakdown
- ChIP-seq
chromatin immunoprecipitation sequencing
- TFIID
transcription factor II D
- YY1
yin yang 1
- Pol II CTD S2
serine 2 within the RNApolymerase II C-terminal domain
- H3K4
lysine 4 of histone 3
- H3K9
lysine 9 of histone 3
- H4K16
lysine 16 of histone 4
- SIRT
NAD-dependent deacetylase sirtuin
- TBP2
TATA-binding protein 2
- DNMT
DNA methyltransferases
- RBAP46
retinoblastoma binding protein P46
- RNAi
RNA interference
- aa
amino acidic
- BrUTP
5-Bromouridine 5′-triphosphate
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- CDK
cyclin-dependent kinase
- gDMRs
germline differentially methylated regions
- HMTase
histone methyltransferase
- SCNT
somatic cell nuclear transfer
- ZGA
zygotic genome activation
The authors declare no conflict of interest.
Footnotes
Edited by M Piacentini
References
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol 2003; 15: 172–183. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Iizuka M, Smith MM. Functional consequences of histone modifications. Curr Opin Genet Dev 2003; 13: 154–160. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 2013; 20: 259–266. [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 2003; 15 370 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkühler C. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res 2007; 17: 195–211. [DOI] [PubMed] [Google Scholar]
- Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol 2009; 53: 275–289. [DOI] [PubMed] [Google Scholar]
- Reichert N, Choukrallah MA, Matthias P. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell Mol Life Sci 2012; 69: 2173–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res 2007; 5: 981–989. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 2008; 9: 206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 2006; 5: 769–784. [DOI] [PubMed] [Google Scholar]
- Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol 2007; 1: 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem 2004; 93: 57–67. [DOI] [PubMed] [Google Scholar]
- Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: what are the cancer relevant targets? Cancer Lett 2009; 277: 8–21. [DOI] [PubMed] [Google Scholar]
- Moser MA, Hagelkruys A, Seiser C. Transcription and beyond: the role of mammalian class I lysine deacetylases. Chromosoma 2014; 123: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem 2002; 277: 25748–25755. [DOI] [PubMed] [Google Scholar]
- Segré CV, Chiocca S. Regulating the regulators: the post-translational code of class I HDAC1 and HDAC2. J Biomed Biotechnol 2011; 2011: 690848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 2009; 10: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khier H, Bartl S, Schuettengruber B, Seiser C. Molecular cloning and characterization of the mouse histone deacetylase 1 gene: integration of a retro- virus in 129SV mice. Biochim Biophys Acta 1999; 1489: 365–373. [DOI] [PubMed] [Google Scholar]
- Taplick J, Kurtev V, Kroboth K, Posch M, Lechner T, Seiser C. Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J Mol Biol 2001; 308: 27–38. [DOI] [PubMed] [Google Scholar]
- Oh YM, Kwon YE, Kim JM, Bae SJ, Lee BK, Yoo SJ et al. Chfr is linked to tumour metastasis through the downregulation of HDAC1. Nat Cell Biol 2009; 11: 295–302. [DOI] [PubMed] [Google Scholar]
- Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet 2005; 47: 1–17. [DOI] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene 2007; 26: 5433–5438. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005; 437: 432–435. [DOI] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 2008; 10: 731–739. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev 2003; 13: 143–153. [DOI] [PubMed] [Google Scholar]
- Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans 2013; 41: 741–749. [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 2007; 21: 1790–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci USA 2009; 106: 7876–7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci 2009; 12: 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Cubizolles F, Zhang Y, Reichert N, Kohler H, Seiser C et al. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev 2010; 24: 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBoeuf M, Terrell A, Trivedi S, Sinha S, Epstein JA, Olson EN et al. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell 2010; 19: 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey OM, Foster CT, Conte N, Edwards SA, Edwards JM, Singh R et al. Histone deacetylase (HDAC) 1 and 2 are essential for normal T cell development and genomic stability in mice. Blood 2013; 121: 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J 2002; 21: 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 2009; 459: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA 2010; 107: 8242–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelkruys A, Lagger S, Krahmer J, Leopoldi A, Artaker M, Pusch O et al. A single allele of Hdac2 but not Hdac1 is sufficient for normal mouse brain development in the absence of its paralog. Development 2014; 141: 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkin J, Zupkovitz G, Lagger S, Grausenburger R, Hagelkruys A, Kenner L et al. Distinct and redundant functions of histone deacetylases HDAC1 and HDAC2 in proliferation and tumorigenesis. Cell Cycle 2011; 10: 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update 2002; 8: 323–331. [DOI] [PubMed] [Google Scholar]
- Rodrigues P, Limback D, McGinnis LK, Plancha CE, Albertini DF. Oogenesis: prospects and challenges for the future. J Cell Physiol 2008; 216: 355–365. [DOI] [PubMed] [Google Scholar]
- Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol 2008; 319: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Pan H, Montgomery RL, Olson EN, Schultz RM. Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc Natl Acad Sci USA 2012; 109: E481–E489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideman MR, Wilting RH, Yanover E, Velds A, de Jong J, Kerkhoven RM et al. Dosage-dependent tumor suppression by histone deacetylases 1 and 2 through regulation of c-Myc collaborating genes and p53 function. Blood 2013; 121: 2038–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, De Waal E, Weaver JR, Bartolomei MS, Schultz RM. A DNMT3A2-HDAC2 complex is essential for genomic imprinting and genome integrity in mouse oocytes. Cell Rep 2015; 13: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Schultz RM. Histone deacetylase 2 (HDAC2) regulates chromosome segregation and kinetochore function via H4K16 deacetylation during oocyte maturation in mouse. PLoS Genet 2013; 9: e1003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature 1997; 389: 349–352. [DOI] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 2002; 298: 1412–1414. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009; 138: 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, O'Brien M J, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol 2005; 286: 493–506. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Rajkovic A. Transcriptional regulation of early oogenesis: in search of masters. Hum Reprod Update 2006; 12: 65–76. [DOI] [PubMed] [Google Scholar]
- Jagarlamudi K, Rajkovic A. Oogenesis: transcriptional regulators and mouse models. Mol Cell Endocrinol 2012; 356: 31–39. [DOI] [PubMed] [Google Scholar]
- Falender AE, Shimada M, Lo YK, Richards JS. TAF4ba TBP associated factor, is required for oocyte development and function. Dev Biol 2005; 288: 405–419. [DOI] [PubMed] [Google Scholar]
- Griffith GJ, Trask MC, Hiller J, Walentuk M, Pawlak JB, Tremblay KD et al. Yin-yang1 is required in the mammalian oocyte for follicle expansion. Biol Reprod 2011; 84: 654–663. [DOI] [PubMed] [Google Scholar]
- Gazdag E, Santenard A, Ziegler-Birling C, Altobelli G, Poch O, Tora L et al. TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes Dev 2009; 23: 2210–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Yamazaki K, Suzuki R, Fujimoto H, Sasaki H, Sakaki Y et al. Parental methylation patterns of a transgenic locus in adult somatic tissues are imprinted during gametogenesis. Development 1992; 116: 831–839. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet 2004; 13: 839–849. [DOI] [PubMed] [Google Scholar]
- Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells 2006; 11: 353–361. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Hirasawa R, Chiba H, Okano M, Li E, Sasaki H. Genetic evidence for Dnmt3a-dependent imprinting during oocyte growth obtained by conditional knockout with Zp3-Cre and complete exclusion of Dnmt3b by chimera formation. Genes Cells 2010; 15: 169–179. [DOI] [PubMed] [Google Scholar]
- Tomizawa S, Nowacka-Woszuk J, Kelsey G. DNA methylation establishment during oocyte growth: mechanisms and significance. Int J Dev Biol 2012; 56: 867–875. [DOI] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 2008; 27: 2691–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J et al. De novo DNA methylation promoted by G9a prevents repro- gramming of embryonically silenced genes. Nat Struct Mol Biol 2008; 15: 1176–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Abujamra AL, Loew JE, Forman LW, Perrine SP, Faller DV. Histone deacetylase inhibitors reverse CpG methylation by regulating DNMT1 through ERK signaling. Anticancer Res 2011; 31: 2723–2732. [PubMed] [Google Scholar]
- Voronina E, Wessel GM. The regulation of oocyte maturation. Curr Top Dev Biol 2003; 58: 53–110. [DOI] [PubMed] [Google Scholar]
- Kim JM, Liu H, Tazaki M, Nagata M, Aoki F. Changes in histone acetylation during mouse oocyte meiosis. J Cell Biol 2003; 162: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R, Viveiros MM, Wigglesworth K, Eppig JJ. ATRXa member of the SNF2 family of helicase/ATPases, is required for chromosome alignment and meiotic spindle organization in metaphase II stage mouse oocytes. Dev Biol 2004; 272: 1–14. [DOI] [PubMed] [Google Scholar]
- Sarmento OF, Digilio LC, Wang Y, Perlin J, Herr JC, Alice CD et al. Dynamic alterations of specific histone modifications during early murine development. J Cell Sci 2004; 117: 4449–4459. [DOI] [PubMed] [Google Scholar]
- Endo T, Naito K, Aoki F, Kume S, Tojo H. Changes in histone modifications during in vitro maturation of porcine oocytes. Mol Reprod Dev 2005; 71: 123–128. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006; 311: 844–847. [DOI] [PubMed] [Google Scholar]
- Zhang L, Hou X, Ma R, Moley K, Schedl T, Wang Q. Sirt2 functions in spindle organization and chromosome alignment in mouse oocyte meiosis. FASEB J 2014; 28: 1435–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene 2007; 26: 5505–5520. [DOI] [PubMed] [Google Scholar]
- Endo T, Naito K, Kume S, Nishimura Y, Kashima K, Tojo H. Activities of maturation-promoting factor (MPF) and mitogen-activated protein kinase (MAPK) are not required for the global histone deacetylation observed after germinal vesicle breakdown (GVBD) in porcine oocytes. Reproduction 2006; 131: 447. [DOI] [PubMed] [Google Scholar]
- Endo T, Kano K, Naito K. Nuclear histone deacetylases are not required for global histone deacetylation during meiotic maturation in porcine oocytes. Biol Reprod 2008; 78: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 1998; 391: 601. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998; 393: 389. [DOI] [PubMed] [Google Scholar]
- Balboula AZ, Stein P, Schultz RM, Schindler K. Knockdown of RBBP7 unveils a requirement of histone deacetylation for CPC function in mouse oocytes. Cell Cycle 2014; 13: 600–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med 2007; 13: 324–331. [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Kiefer F, Prudenziati M, Spiller C, Hansen J, Floss T et al. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res 2007; 67: 9047–9054. [DOI] [PubMed] [Google Scholar]
- Zupkovitz G, Grausenburger R, Brunmeir R, Senese S, Tischler J, Jurkin J et al. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol Cell Biol 2010; 30: 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting RH, Yanover E, Heideman MR, Jacobs H, Horner J, van der Torre J et al. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and ha- ematopoiesis. EMBO J 2010; 29: 2586–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Wu J. Epigenetic regulation in mammalian preimplantation embryo development. Reprod Biol Endocrinol 2009; 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry GN, Tanasijevic B, Barry ER, Krueger W, Rasmussen TP. Epigenetic regulatory mechanisms during preimplantation development. Birth Defects Res C Embryo Today 2009; 87: 297–313. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol 1993; 159: 366–378. [DOI] [PubMed] [Google Scholar]
- Henery CC, Miranda M, Wiekowski M, Wilmut I, DePamphilis ML. Repression of gene expression at the beginning of mouse development. Dev Biol 1995; 169: 448–460. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181: 296–307. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol 2004; 272: 483–496. [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol 2005; 283: 40–57. [DOI] [PubMed] [Google Scholar]