Abstract
Mutations of HSPB5 (also known as CRYAB or αB-crystallin), a bona fide heat shock protein and molecular chaperone encoded by the HSPB5 (crystallin, alpha B) gene, are linked to multisystem disorders featuring variable combinations of cataracts, cardiomyopathy, and skeletal myopathy. This study aimed to investigate the pathological mechanisms involved in an early-onset myofibrillar myopathy manifesting in a child harboring a homozygous recessive mutation in HSPB5, 343delT. To study HSPB5 343delT protein dynamics, we utilize model cell culture systems including induced pluripotent stem cells derived from the 343delT patient (343delT/343delT) along with isogenic, heterozygous, gene-corrected control cells (WT KI/343delT) and BHK21 cells, a cell line lacking endogenous HSPB5 expression. 343delT/343delT and WT KI/343delT-induced pluripotent stem cell-derived skeletal myotubes and cardiomyocytes did not express detectable levels of 343delT protein, contributable to the extreme insolubility of the mutant protein. Overexpression of HSPB5 343delT resulted in insoluble mutant protein aggregates and induction of a cellular stress response. Co-expression of 343delT with WT prevented visible aggregation of 343delT and improved its solubility. Additionally, in vitro refolding of 343delT in the presence of WT rescued its solubility. We demonstrate an interaction between WT and 343delT both in vitro and within cells. These data support a loss-of-function model for the myopathy observed in the patient because the insoluble mutant would be unavailable to perform normal functions of HSPB5, although additional gain-of-function effects of the mutant protein cannot be excluded. Additionally, our data highlight the solubilization of 343delT by WT, concordant with the recessive inheritance of the disease and absence of symptoms in carrier individuals.
Keywords: aggregation; chaperone; induced pluripotent stem cell (iPSC); protein misfolding; small heat shock protein (sHsp); HSPB5 (CRYAB, αB-crystallin); myofibrillar myopathy
Introduction
HSPB5 (also known as CRYAB or αB-crystallin) is a small molecular weight heat shock protein encoded by the HSPB5 (crystallin, alpha B) gene and functions as a molecular chaperone. Its promoter contains a heat shock element, a stress-responsive binding site of heat shock transcription factor 1 (HSF1), that functionally up-regulates the expression of HSPB5. Increased levels of HSPB5 can then go on to provide distinct cytoprotective effects engaged in restoring cellular homeostasis (reviewed in Refs. 1, 2). Additionally, HSPB5 contains tissue-specific enhancer elements in its promoter that allow constitutive high expression in the lens, heart, and skeletal muscle (3, 4).
As a member of the small molecular weight heat shock protein family, HSPB5 contains a well conserved central “α-crystallin” domain (ACD),4 flanked by N- and C-terminal regions (5). The ACDs dimerize and assemble to form a polydisperse ensemble of oligomers, largely through dynamic interactions mediated by the terminal regions (6). The isolated ACD has been shown to have potent chaperone activity in vitro (7), and it is hypothesized that it might become exposed within the context of the wild-type protein in a manner regulated by phosphorylation (8) or cellular stress directly (9). The HSPB5 ACD contains two hydrophobic grooves, one between the β4 and β8 strands and the other at the dimer interface, both of which serve as putative binding sites for chaperone action (10). In addition, evidence also points at the involvement of the N terminus in target binding (11). Current data suggest that HSPB5 is a dynamic protein that is flexible in how it interacts with its diverse range of clients.
In striated muscle, HSPB5 acts as a chaperone for important structural client proteins, including desmin (12–14), titin (15–18), and actin (14, 19), a property that becomes particularly important under conditions of pathology or stress. HSPB5/HSPB2 double knockout (DKO) mice exhibit progressive skeletal myopathy throughout life (20), with an impact on cardiac muscle only observed under conditions of exogenous stress (21, 22). More recently, a requirement for HSPB5 in muscle homeostasis has been demonstrated via modulation of argonaute 2 activity (23). DKO mice show reduced basal levels of skeletal muscle progenitor cells or satellite cells and defective muscle regeneration with cellular injury (23).
Multiple mutations in HSPB5 are linked to human pathologies affecting the lens, heart, skeletal muscle, or some combination thereof, with the underlying disease mechanism(s) only partially understood (reviewed in Refs. 24, 25). These mutations are either dominant or recessive and have variable penetrance and expressivity. Of the known mutations in HSPB5 linked with (cardio)myopathy, all but one result in aberrant protein aggregation of the mutant protein (reviewed in Ref. 25). For the well studied R120G mutation in HSPB5, the aggregates are thought to sequester other metastable proteins, such as desmin (26–28). The relative contribution of loss- versus gain-of-toxic function effects remain unclear for specific mutations.
An enhanced understanding of HSPB5 as a chaperone is needed to inform therapy development. Here we have investigated a recessive mutation in HSPB5, 343delT, found in a patient who presented at age 4 months with profound muscle stiffness, persistent creatine kinase elevation, and histopathological features of a myofibrillar myopathy (29). This homozygous recessive, single base deletion results in a frameshift and formation of a premature stop codon following addition of 14 missense residues (S115fs129X). The truncated protein is predicted to contain residues 1–115, having lost the C-terminal extension, as well as a significant portion of the ACD. The skeletal muscle biopsy of the patient shows dense, irregular staining of HSPB5, detectable only by an antibody recognizing the N terminus of HSPB5 (NT-HSPB5). This band runs at the predicted molecular weight of 15 kDa on a Western blot, smaller than the 22-kDa WT HSPB5 protein (29). Additionally, the biopsy shows intense, irregular staining of desmin and myotilin. At presentation, the patient exhibited neither signs of an associated cardiomyopathy nor cataracts, although with age, these symptoms may develop. Of note, the non-consanguineous parents, each heterozygous for 343delT, have no known symptoms attributed to this mutation; however, potential development of late-onset symptoms cannot be excluded.
To understand 343delT protein dynamics and their contribution to disease, we have incorporated the use of induced pluripotent stem cells (iPSCs) (30) derived from the patient as a unique system to permit the study of endogenous 343delT protein within cell types of interest. Additionally, we used systems of overexpression in a cell line that lacks endogenous HSPB5 expression (reviewed in Ref. 31) because of the recessive nature of the mutation as well as in vitro analysis of 343delT. Here we describe our findings, which provide insights into the molecular defects of 343delT in disease pathogenesis.
Results
Generation of iPSCs for the Investigation of 343delT
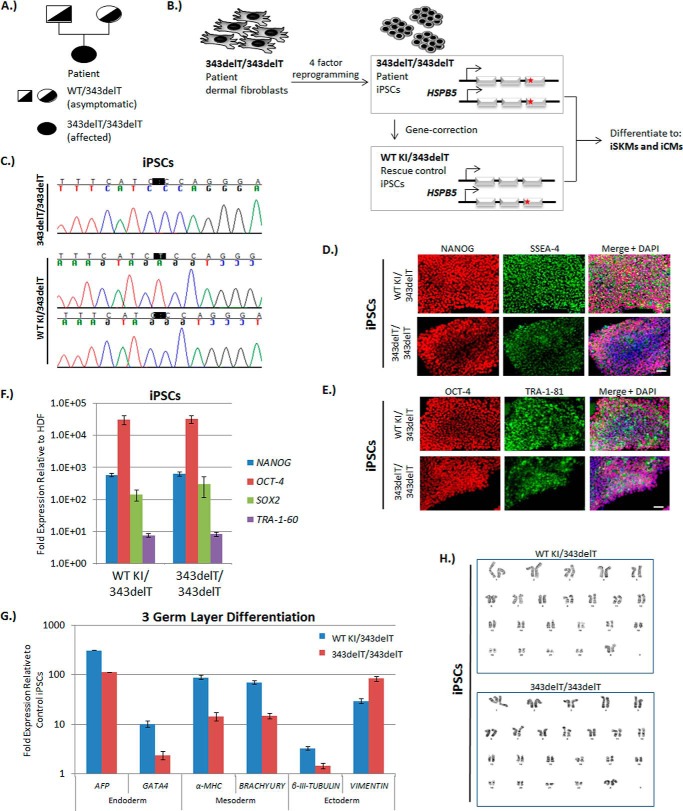
Fig. 1A shows the family pedigree of the patient harboring the homozygous, recessive 343delT mutation. The non-consanguineous parents are each heterozygous for the mutation and are asymptomatic, whereas the homozygous 343delT patient exhibits a severe, infantile-onset myopathy (29). To examine the effects of 343delT (strategy outlined in Fig. 1B), we generated iPSCs from dermal fibroblasts of the patient through four-factor reprogramming. Isogenic, heterozygous, gene-corrected control cells were then engineered from the patient iPSCs with a strategy adapted from Yusa et al. (32) through zinc finger nuclease-stimulated homologous recombination (see “Experimental Procedures”). The genotypes of both the patient (343delT/343delT) and heterozygous wild-type knockin control (WT KI/343delT) iPSCs are outlined in Fig. 1B and were confirmed through direct Sanger sequencing (Fig. 1C). Both iPSC lines expressed relevant pluripotency markers, including NANOG, stage-specific embryonic antigen 4 (SSEA-4), octamer-binding protein 4 (OCT-4), and TRA-1–81 at the protein level (Fig. 1, D and E) and NANOG, OCT-4 (POU5F1), SOX2, and TRA-1-60 at the RNA level (Fig. 1F). In vitro EZ sphere differentiations of both cell lines resulted in up-regulation of markers from all three germ layers (Fig. 1G). Karyotypes were normal (Fig. 1H). These data validate the use of generated iPSC lines for further investigations.
FIGURE 1.
343delT iPSC generation and pluripotency characterization. A, representative pedigree for the family of the homozygous recessive 343delT patient. Females are represented by circles, males by squares. Filled shapes represent homozygous 343delT. Half-filled shapes represent heterozygous 343delT. B, schematic of the experimental strategy. Dermal fibroblasts derived from the homozygous 343delT patient were reprogrammed into iPSCs (343delT/343delT), with the 343delT mutation indicated by red stars. Heterozygous gene-corrected control iPSCs were generated from the 343delT/343delT iPSCs through gene editing and have one copy of WT knocked into the HSPB5 locus (WT KI/343delT). WT KI/343delT and 343delT/343delT iPSCs were differentiated to iCMs and iSKMs, respectively. C, direct Sanger sequencing of a PCR product covering the region of interest reveals deletion of the T at position 343 (highlighted in black) in the 343delT/343delT iPSCs. The PCR product from the WT KI/343delT iPSC line was TOPO-cloned into a vector, and multiple clones were sequenced. Reads showing WT (top panel) and 343delT (bottom panel) were observed, indicating that the WT KI/343delT iPSC line is heterozygous for this mutation. D and E, WT KI/343delT and 343delT/343delT iPSCs were stained with immunocytochemistry for the pluripotency markers nanog (red) and SSEA-4 (green) (D) and OCT-4 (red) and TRA-1–81 (green) (E). Nuclei were counterstained and merged with DAPI (blue). Scale bars = 50 μm. F, qRT-PCR was performed on WT KI/343delT and 343delT/343delT iPSCs, with the graph depicting mean ± S.E. relative expression levels of the pluripotency markers NANOG, OCT-4, SOX2, and TRA-1–60 normalized to 18S rRNA and compared with human dermal fibroblasts (HDF) derived from the patient. G, WT KI/343delT and 343delT/343delT iPSCs differentiated to EZ spheres were taken at week 5 of differentiation for qRT-PCR analysis with markers of the three germ layers: endoderm (AFP and GATA4), mesoderm (α-MHC and BRACHYURY), and ectoderm (β-III-TUBULIN and VIMENTIN). The graph shows mean ± S.E. expression values normalized to 18s rRNA and compared with WT KI/343delT iPSCs. H, 20 proliferating cells were counted and fully analyzed using G-banding for each cell line, with representative karyotype images showing no consistent abnormalities.
343delT Protein Is Not Detectable in iSKMs or iCMs although RNA Is Present
Validated iPSC lines were differentiated into skeletal myotubes (iSKMs) and cardiomyocytes (iCMs) using established protocols (see “Experimental Procedures”). iCMs contracted rhythmically in the dish, stained positive for cardiomyocyte markers, including Troponin T (TnT) (Fig. 2A), and exhibited cardiomyocyte-like action potential profiles (Fig. 2C). iSKMs elongate as they mature in culture, many appear multinucleated, some cells can be observed to twitch spontaneously, and they stain positively for muscle markers, including TnT (Figs. 2B and 4A) and desmin (Fig. 4B). No obvious phenotypic differences were observed between WT KI/343delT and 343delT/343delT iCMs or iSKMs when characterizing cell differentiation in this way.
FIGURE 2.
343delT protein is not detectable in iSKMs or iCMs although RNA is present. A and B, WT KI/343delT and 343delT/343delT iPSCs were differentiated to iCMs (A) or iSKMs (B)and stained with antibodies that recognized NT HSPB5 (red) and TnT (green) and merged with DAPI (blue). Scale bars = 50 μm. C, representative action potentials recorded from spontaneously beating WT KI/343delT (top panel) and 343delT/343delT iPSCs (bottom panel) cardiomyocytes (n = 6). Western blotting and qRT-PCR were performed on paired samples derived from WT KI/343delT and 343delT/343delT cells. D and E, Western blotting of WCL from iCMs (D) and iSKMs (E) probed with antibodies that recognize NT HSPB5 and GAPDH as a loading control. F and G, qRT-PCR of iCMs (F) and iSKMs (G) showing mean ± S.E. gene expression for HSPB5 and TROPONIN T (TNNT2) normalized to 18s rRNA. Expression is shown as -fold change compared with undifferentiated iPSCs.
FIGURE 4.
343delT forms visible aggregates when overexpressed and induces a cellular stress response. A and B, 343delT/343delT iSKMs were transfected with pCMV-MYC-WT or pCMV-MYC-343delT constructs. Cells were stained 48 h post-transfection with antibodies recognizing NT HSPB5 (red) and TnT (green) (A) and MYC tag (green) and desmin (red) (B). Scale bars = 25 μm. C–E, BHK21 cells were transfected with pCMV-MYC-WT or pCMV-MYC-343delT constructs for 24 h and co-stained with antibodies against NT HSPB5 (red) and MYC tag (green) (C) MYC tag (green) and desmin (red) (D), and MYC-tag (green) and HSP70 (red) (E). Scale bars = 50 μm. In all merged immunocytochemistry images, nuclei were counterstained and merged with DAPI (blue). Enlargements of the area within the white dotted box are shown at the right. Arrows indicate 343delT aggregates. F, WCL from BHK21 cells transfected as above or similarly with pCMV-MYC-empty vector (EV) were analyzed by Western blotting with antibodies recognizing HSP70, MYC (9E10), and α-tubulin as a loading control. Samples were run in duplicate.
The major difference observed between the muscle-differentiated cell lines was a complete lack of detectable 343delT protein both through immunocytochemistry (Fig. 2, A and B) and Western blotting (Fig. 2, D and E) in the 343delT/343delT line when probed with an NT HSPB5 antibody. In the WT KI/343delT control, predicted to express both a 22-kDa-sized WT protein and a 15-kDa-sized 343delT protein, only the WT form of the protein is detectable (Fig. 2, D and E), indicating that lack of detectable 343delT is not due to immaturity of the cells. This result was surprising to us because the muscle biopsy from the patient showed dense, irregular staining of HSPB5 through immunohistochemistry and a 15-kDa-sized protein detectable by Western blotting with the NT HSPB5 antibody (29). Lack of 343delT protein was confirmed in a second iPSC line derived from the patient as well as the isogenic, heterozygous, gene-corrected control line derived from that iPSC line (supplemental Fig. S1). Additionally, homozygous knockin of the 343delT mutation into the endogenous HSPB5 locus of an iPSC line derived from an unrelated, healthy individual also exhibited a complete lack of detectable 343delT protein (data not shown).
Because 343delT protein was not detectable, we next investigated whether this was due to defects in RNA levels. The 343delT transcript is not predicted to be a target for nonsense-mediated mRNA decay because the premature stop codon is downstream of the final exon-exon junction complex (33). Therefore, we hypothesized that 343delT transcript levels would be comparable with the WT. Quantitative real-time PCR (qRT-PCR) of iCMs and iSKMs revealed expression of HSPB5 mRNA in both WT KI/343delT and 343delT/343delT (Fig. 2, F and G). HSPB5 mRNA levels may differ depending on the efficiency of differentiation, which varies between iPSC lines and differentiation attempts, as noted by differences in expression of the muscle-specific gene Troponin T (TNNT2) (Fig. 2, F and G). Therefore, although the level of HSPB5 mRNA cannot be directly compared between samples, we confirm that 343delT mRNA is present. Additionally, in contrast to an unstable mRNA control, MYC, levels of HSPB5 mRNA did not decline in WT KI/343delT or 343delT/343delT iCMs with treatment of actinomycin D to inhibit transcription over 24 h (supplemental Fig. S2), indicating similar transcript stability in WT and 343delT. These results show that the defect in 343delT protein expression is not due to deficient transcript levels.
Lack of 343delT Protein Is Not Due to Degradation, MicroRNA Targeting, or Defective Translation
To further investigate the reason for lack of 343delT protein detection, we next examined protein degradation as a possible mechanism. iCMs were treated with the proteasomal inhibitor MG132 (34) (Fig. 3A). In contrast to the increase in the short-lived cell cycle inhibitor p21 with MG132 treatment (35), 343delT protein (15 kDa) remained undetectable in both WT KI/343delT and 343delT/343delT with inhibition of proteasomal degradation. Inhibition of autophagosome maturation with bafilomycin A1 for 12 h (36) was also insufficient to evoke 343delT protein detection in iCMs, although LC3-II (lower band) increased as expected with treatment (supplemental Fig. S3A). Additionally, treatment of 343delT/343delT iSKMs with the proteasomal inhibitor bortezomib for up to 48 h (37) (supplemental Fig. S3B) or MG132 for 12 h (supplemental Fig. S3C) and inhibition of autophagosome maturation with bafilomycin A1 for 12 h (supplemental Fig. S3C) were also insufficient to evoke 343delT detection. These data suggest that the lack of detectable 343delT protein is not due to degradation by the proteasome or autophagy. Complementary experiments of overexpression of 343delT in a human breast cancer cell line (MCF7 cells) showed that both inhibition of the proteasome (MG132) and autophagy (bafilomycin A1) resulted in slight increases in 343delT protein (supplemental Fig. S3, D and E). This result suggests that 343delT protein is likely degraded to some extent by the proteasome and autophagy, but this is not the major cause for our inability to detect 343delT protein in our iPSC model.
FIGURE 3.
Lack of 343delT protein is not due to degradation, microRNA targeting, or disrupted translation. A, WT KI/343delT and 343delT/343delT iCMs were treated with DMSO or 10 μm MG132 to inhibit the proteasome for 12 h. Western blots of WCL were probed with antibodies recognizing NT HSPB5, p21, and GAPDH. Results are representative of two independent experiments. B, WT KI/343delT and 343delT/343delT iCMs were transfected with scrambled siRNA control (−) or either 40 pmol (+) or 80 pmol (++) of siRNA targeting dicer. Samples were harvested 72 h post-transfection, and WCL was run on a Western blot and probed with antibodies against NT HSPB5, dicer, c-MYC (N-262), and GAPDH. C, equal amounts of pCSC-MYC-WT and pCS2-MYC-343delT plasmids were entered into an in vitro transcription/translation (TnT) assay along with 35S-labeled methionine/cysteine. Aliquots were taken from reactions at 30 and 90 min as indicated and separated by SDS-PAGE, transferred to nitrocellulose membranes, and exposed to film (top panel). The image is representative of three independent experiments, with the quantification (bottom panel) expressing synthesis over time as arbitrary units, indicating band intensity, showing mean ± S.D. There is no significant difference in the rate of synthesis over time for WT (21.8 ± 3.4) and 343delT (19.1 ± 6.9) as calculated by Student's t test (p > 0.5).
Another possibility we considered is that 343delT mRNA is differentially targeted compared with WT by a microRNA that inhibits translation. To examine this possibility, we transfected iCMs with siRNA to knock down dicer, the protein responsible for processing pre-microRNAs into mature microRNAs (38). Although knockdown of dicer was confirmed through decreased levels of dicer protein and elevated levels of c-MYC, a known target of dicer-dependent microRNAs (39), we were unable to detect 343delT protein in either WT KI/343delT or 343delT/343delT (Fig. 3B). This result suggests that 343delT is not differentially targeted by a dicer-dependent microRNA.
To confirm that there is no inherent defect in the translation efficiency of 343delT compared with WT, we performed an in vitro transcription/translation assay using rabbit reticulocyte lysate labeling-translated proteins with [35S]l-methionine and [35S]l-cysteine. We utilized a plasmid containing N-terminally MYC-tagged versions of WT (pCS2-MYC-WT) and 343delT (pCS2-MYC-343delT) and an SP6 promoter as the template for this reaction. Both WT and 343delT are visible after 30 min of incubation and increase after 90 min (Fig. 3C). No significant difference is observed between WT and 343delT in the rate of synthesis over time (21.8 ± 3.4 and 19.1 ± 6.9, respectively) of the recombinant protein. This result shows that 343delT is translated efficiently, at least in this reticulocyte lysate. In toto, these results demonstrate that lack of detectable 343delT protein in iCMs and iSKMs generated from 343delT/343delT and WT KI/343delT is not attributable to proteasomal degradation, autophagic degradation, microRNA targeting, or defective translation.
343delT Forms Insoluble Protein Aggregates and Induces a Cellular Stress Response When Overexpressed
The results presented above utilizing iCMs and iSKMs show an absence of detectable 343delT protein. The presence of mutant protein in the biopsy specimen of the protein suggests that accumulation of 343delT in the affected tissue may contribute to disease. For this reason, we attempted to simulate these conditions with our model to determine whether high levels of 343delT expression would result in aggregation in muscle cells.
We transiently transfected N-terminally MYC-tagged WT or 343delT constructs (pCMV-MYC-WT and pCMV-MYC-343delT) into 343delT/343delT iSKMs. Overexpression of 343delT resulted in aggregation in iSKMs marked by TnT (Fig. 4A) or desmin (Fig. 4B), whereas overexpression of WT resulted in diffuse cytoplasmic staining (Fig. 4, A and B). 343delT aggregates exclude TnT and desmin. Similar aggregates of 343delT were observed with overexpression in 343delT/343delT iCMs (data not shown). These results indicate that, at high levels, 343delT forms aggregates in muscle cells.
Because overexpression of 343delT in iSKMs recapitulates aggregation observed in the biopsy of the patient, we examined whether overexpression in non-muscle cell lines also results in 343delT aggregation. Transfection is difficult in iCMs and iSKMs, and variances in differentiation efficiency may impact results, making overexpression studies in commercially available cells lines that lack endogenous HSPB5 expression, such as BHK21 cells (baby hamster kidney fibroblasts), more appealing. Overexpression of pCMV-MYC-343delT forms cytoplasmic aggregates in BHK21 cells (Fig. 4C), which are detectable with both NT HSPB5 and MYC tag antibodies. The absence of additional cell staining with NT HSPB5 relative to the MYC tag exemplifies the lack of endogenous HSPB5 expression. Aggregates in BHK21 cells typically appear in the perinuclear region and look similar to aggregates observed with overexpression in iSKMs (Fig. 4, A and B). Overexpression of pCMV-MYC-WT in BHK21 cells results in diffuse cytoplasmic staining (Fig. 4C). These experiments provide a proof of concept for the use of overexpression of 343delT in BHK21 cells as a platform to investigate 343delT aggregation and protein dynamics.
We noted that levels of 343delT protein expression were consistently lower than WT levels by immunocytochemistry and Western blotting when overexpressed in all cell lines tested. We confirmed equal transfection efficiency through co-transfection with a pCMV-dsRed plasmid (data not shown). Additionally, the levels of WT and 343delT mRNA were assessed between 2 and 24 h after transfection and determined to be similar (supplemental Fig. S4A). RNA levels correlated with transfection efficiency for the WT construct (supplemental Fig. S4B). Transfection with either WT or 343delT impacted neither cell proliferation nor viability (supplemental Fig. S5, A and B) when compared with empty vector transfection over a similar period. These results indicate that transfection efficiencies and RNA levels were equal and that cell proliferation and viability were unaffected by transfection.
The metastable desmin protein is a known client for HSPB5 (12–14) and is present in aggregates in patients with desmin-related (cardio)myopathy (25, 26, 40). Although 343delT aggregates did not colocalize with desmin in iSKMs, we wanted to examine this in BHK21 cells, which express endogenous desmin as well (26, 31). 343delT aggregates in BHK21 cells colocalize with desmin in some (Fig. 4D, center panels) but not all cells (Fig. 4D, bottom panels), potentially indicating stochastic incorporation of desmin into aggregates.
Misfolded proteins and protein aggregates are triggers for the induction of a cellular stress response driven by the master regulatory transcription factor HSF1 (41). This response results in the up-regulation of genes whose promoters contain heat shock response elements, including HSP70 (as reviewed in Ref. 2). Indeed, overexpression of 343delT in BHK21 cells resulted in induction of HSP70 in some but not all transfected cells (Fig. 4E), whereas, in WT, it does not result in HSP70 induction (Fig. 4E). We further confirmed this induction by Western blotting (Fig. 4F). These results indicate that 343delT can induce a cellular stress response when overexpressed. Consistent up-regulation of HSP70 in 343delT/343delT iCMs under basal conditions was not observed (data not shown), indicating that perhaps the detectable, aggregated form of 343delT and/or the presence of other proteins in the aggregates is required for up-regulation of HSP70.
Together, these results show the ability of 343delT to form visible, cytoplasmic aggregates when overexpressed in muscle and non-muscle cells, with potentially stochastic incorporation of desmin into these aggregates, at least in non-muscle cells. Additionally, HSP70 induction demonstrates disruption in overall protein homeostasis with overexpression of 343delT.
Co-Expression with WT Solubilizes 343delT
Because 343delT is a recessively inherited mutation, to mimic the unaffected heterozygous parents, we co-transfected HA-tagged WT and MYC-tagged 343delT constructs (pCMV-HA-WT HSPB5 and pCMV-MYC-343delT HSPB5) into cell lines in equimolar ratios. In the presence of WT, 343delT no longer formed aggregates in BHK21 cells (Fig. 5A, bottom panels). Additionally, when expressed alone, 343delT was present almost entirely in the insoluble fraction (99 ± 1%, mean ± S.D., n = 3), whereas, when co-expressed with WT, the percentage of insoluble 343delT was reduced 6.6-fold (15% ± 6%, mean ± S.D., n = 3, p < 0.01 as calculated by Student's t test) (Fig. 5, B and C). The percentage of insoluble 343delT was quantified using the 15-kDa-sized band ([insoluble] density × 100/([insoluble] density +[soluble] density)). Larger molecular weight bands may be posttranslationally modified forms of the protein. Inclusion of these bands in the analysis does not impact the relative percentage of insoluble 343delT. These results indicate that the presence of WT shifts 343delT from the insoluble to the soluble fraction.
FIGURE 5.
WT co-expression reduces 343delT aggregates and increases solubility of the mutant protein. A, BHK21 cells were transfected with pCMV-HA-WT, pCMV-MYC-343delT, or both (co-transfection) and stained with antibodies against HA tag and MYC tag after 24 h. Nuclei were counterstained and merged with DAPI (blue). Enlargements of the area within the white dotted box are shown at the right. Arrows indicate 343delT aggregates. Scale bar = 50 μm. B, BHK21 cells were transfected with pCMV-HA-WT, pCMV-HA-WT, and pCMV-MYC-343delT (co-transfection), pCMV-MYC-343delT, or pCMV-MYC-WT, harvested 24 h post-transfection, and fractionated into soluble and insoluble fractions or WCL. Equal amounts of protein were loaded for Western blotting using an anti-MYC (9E10) antibody and an HA tag antibody with β-actin as a loading control. The image depicts representative blots from three independent experiments. C, the percentage of insoluble 343delT, quantified using the 15-kDa-sized band ([insoluble] density × 100/([insoluble] density + [soluble] density)). Insoluble 343delT is reduced 6.6-fold by co-expression with WT (343delT alone = 99% ± 1% insoluble 343delT, 343delT + WT = 15% ± 6% insoluble 343delT, mean ± S.D., n = 3, p < 0.01 as calculated by Student's t test). D, BHK21 cells transfected as in B were harvested after 24 h for co-IP using RIPA buffer and immunoblotted with anti-HA tag or anti-MYC (9E10) antibodies. Western blotting was performed, showing IP fractions as well as WCL and the insoluble fraction (the remaining pellet from the sample solubilized in RIPA buffer) and stained with antibodies against MYC (9E10), HA tag, and α-tubulin as a loading control.
To determine whether 343delT and WT are interacting, we co-transfected HA-tagged WT and MYC-tagged 343delT and performed co-immunoprecipitation (Co-IP). IP of HA resulted in the pulldown of MYC-tagged 343delT only in the co-transfected sample (Fig. 5D). Reciprocally, IP of MYC resulted in the pulldown of HA-tagged WT only in the co-transfected sample (Fig. 5D). The insoluble pellet fraction demonstrated that much of the 343delT protein, when transfected alone, was insoluble (Fig. 5D), consistent with the idea that WT solubilizes 343delT. These results demonstrate an interaction between WT and 343delT proteins within the cell.
343delT Solubility: in Vitro and in Vivo
In line with overexpression in mammalian cell lines, the overexpression of recombinant 343delT in Escherichia coli resulted in an insoluble product exclusively found in inclusion bodies (Fig. 6A). Refolding of 343delT in the absence of WT predominantly led to the formation of insoluble aggregates (Fig. 6B). Conversely, the presence of an excess of WT during the entire refolding process resulted in the formation of a soluble product (Fig. 6B). This mixture eluted from gel filtration in a single peak with a slight shoulder at an elution volume of ∼10 ml, slightly earlier than refolded WT, which eluted at 10.5 ml (Fig. 6C). SDS-PAGE of the resulting fractions showed the co-elution of 343delT (14.7 kDa) with WT (20.2 kDa) across the entire peak, whereas only a single band at the WT mass was visible in the control (Fig. 6C, inset). To obtain a higher-resolution view of the oligomers formed, we performed native mass spectrometry (42). A spectrum of WT reveals a broad region of signal centered around 10,000 m/z, resulting from the overlap of a large number of charge states (Fig. 6D), and consistent with a polydisperse ensemble of oligomers (43). A spectrum obtained for the 343delT:WT mixture at identical mass spectrometry conditions reveals an additional charge series at ∼2000 m/z, corresponding to a mass of 14,695 Da, which can be assigned to the monomeric form of 343delT (Fig. 6D). Interestingly, the charge states populated are relatively low, indicative of a largely folded conformation of the 343delT monomers free in solution. Examination of the high m/z region of the spectrum for WT allows the assignment of overlapping charge state envelopes to the oligomers comprising the ensemble. Strikingly, many additional features are observed in the spectra for the 343delT:WT mixture (Fig. 6D, inset). The complexity of the spectrum is such that individual oligomers cannot be directly identified; however, by comparison with the WT data it is clear that they cannot be explained by integer stoichiometries of WT HSPB5 but, rather, that an ensemble of hetero-oligomers is present, resulting from a direct interaction between the two proteins. These in vitro data further support the notion that WT solubilizes 343delT through a direct interaction.
FIGURE 6.
In vitro refolding of 343delT in the presence of WT and SUMOylation in cells rescue 343delT insolubility. A, recombinant 343delT expressed in E. coli was found to accumulate within insoluble inclusion bodies (I) and not within the soluble lysate (S). B, refolding WT or a 343delT:WT mixture resulted in a soluble product that was not removed upon passing through a 0.22-μm filtration device. Refolding of 343delT in the absence of WT resulted in aggregates that were removed from solution upon filtration. U, unfiltered; F, filtered. C, 300 μl of refolded WT and refolded 343delT:WT mixture from B was loaded in subsequent runs onto a Superdex 200 10/300 column (pre-equilibrated into 200 mm ammonium acetate (pH 6.9). Separation was carried out at a flow rate of 0.4 ml/min, and absorbance was monitored at 280 nm. The chromatogram for the 343delT:WT mixture (orange) had a slight shoulder and was shifted to earlier elution volumes compared with WT alone (purple). 9 μl of fractions 9–17 were analyzed by SDS-PAGE for each run, alongside molecular weight markers. These revealed the co-elution of 343delT with WT. D, corresponding fractions from each run in C were analyzed by means of native mass spectrometry. The control containing WT displayed a complicated overlapping signal centered near 10,000 m/z, relating to multiple charge state series of WT oligomers. For the mixture, two areas of interest were identified, a region around 10,000 m/z akin to the WT spectra and a charge state series at 2000 m/z. This was identified to correspond to monomeric 343delT (molecular mass = 14,695 Da). Close inspection of the region at high m/z (inset) reveals peaks in the 343delT:WT mixture that cannot arise from WT homo-oligomers but, rather, correspond to an ensemble of hetero-oligomers comprising both HSPB5 components. E, BHK21 cells were transfected with pCMV-MYC-WT, pCMV-MYC-SUMO3 Q89P-WT, pCMV-MYC-343delT, or pCMV-MYC-SUMO3 Q89P-343delT and harvested 24 h post-transfection, and WCL was isolated. Equal amounts of protein were loaded for Western blotting using an anti-MYC (9E10) antibody with β-actin as a loading control. The image depicts a representative blot from three independent experiments, with longer exposure shown at the right. The relative increase in WT and 343delT proteins with the addition of SUMO is indicated below the respective lanes. WT increased 6 ± 1-fold, whereas 343delT increased 130 ± 60-fold (mean ± S.D. of three independent experiments) with p < 0.05% as calculated by Student's t test. F, 343delT/343delT iCMs were transfected with pCMV-MYC-343delT or pCMV-MYC-SUMO3 Q89P-343delT and harvested 24 h post-transfection. Analysis of WCL by Western blotting was performed using an anti-MYC (9E10) antibody with β-actin as a loading control, with longer exposure shown at the right. The relative increase in 343delT protein with the addition of SUMO is indicated (811 ± 218-fold) as mean ± S.D. of three independent experiments.
We hypothesized that extreme insolubility of 343delT was responsible for our inability to detect the mutant protein in iSKMs and iCMs and for lower levels of 343delT compared with WT upon overexpression. We have excluded many reasons for the absence of detectable 343delT protein noted above, including mRNA instability, degradation by the proteasome or autophagy, targeting by a dicer-dependent microRNA, and defective translation. Decreased detectable 343delT protein compared with WT was consistently observed both through analyses by Western blotting and immunocytochemistry. We tested additional lysis buffers to determine whether we could detect 343delT by Western blotting, including RIPA buffer, 100% formic acid, and 8 m urea plus 2% SDS. We observed a consistent lack of 343delT protein in iCMs with all tested buffers (data not shown).
Addition of a small ubiquitin-related modifier (SUMO) to proteins has been shown to enhance their solubility (44, 45). To directly test our hypothesis that the insolubility of 343delT is the reason for low levels of detection with overexpression, we generated SUMO-HSPB5 fusion constructs. These plasmids contain a version of WT and 343delT HSPB5 that are N-terminally MYC-tagged followed by SUMO, preceding the HSPB5 variant. We used a non-cleavable version of SUMO3 Q89P (46) in this construct to prevent removal of SUMO by endogenous deSUMOylases. MYC tag-only versions of the constructs were used as controls. Expression of SUMOylated constructs in BHK21 cells results in a 130 ± 60 and 5 ± 1 (mean ± S.D.) -fold increase in 343delT and WT, respectively (Fig. 6E). The larger increase of 343delT compared with WT through the addition of SUMO (p < 0.05, calculated by Student's t test) suggests that the increase in mutant protein is likely not due to inherent stabilization and/or enhanced transcription/translation of proteins by SUMO addition. If this were the case, similar increases in WT and 343delT would be expected. Elevated detection of 343delT was also observed with overexpression of 343delT and SUMOylated 343delT in 343delT/343delT iCMs, resulting in 811 ± 218-fold (mean ± S.D.) enhanced detection of the SUMOylated protein by Western blotting (Fig. 6F). These results suggest that our suboptimal ability to detect 343delT is due to extreme insolubility of the mutant protein in both non-muscle and muscle cells.
Discussion
Together, the data presented here describe molecular defects observed with disease-causing 343delT. Although mRNA is unaffected, basal levels of 343delT protein in iCMs or iSKMs were undetectable in both WT KI/343delT and 343delT/343delT cell lines. Detection of the WT-sized protein, but not the truncated 343delT-sized protein in the heterozygous WT KI/343delT iPSC-derived cells, indicates that the absence of 343delT is not due to lack of maturation. Several possibilities, such as proteasomal degradation, autophagic degradation, Dicer-dependent microRNA targeting, and inherent translational defects, were examined and proved to be unlikely mechanisms responsible for the lack of detectable 343delT protein. Overexpression of 343delT consistently resulted in much lower levels of protein compared with WT as well. Greatly enhanced detection of 343delT when fused to SUMO3 Q89P, a non-cleavable form of SUMO known to enhance protein solubility, is consistent with extreme insolubility of 343delT as the reason for lack of protein detection.
It is conceivable that insoluble 343delT would be unavailable to perform essential, normal functions of HSPB5 in muscle cells, suggesting that loss of HSPB5 function contributes to myopathy in the patient harboring homozygous 343delT. HSPB5 specifically plays key chaperone roles for proteostasis and protein quality control, differentiation, and maintenance in muscle cells (12–23). Loss of these or other functions of HSPB5 may leave myotubes vulnerable to stress, prone to muscle breakdown with increased usage, and/or have an impact on regeneration of skeletal muscle from satellite cells. Although absent at presentation, the early onset of the skeletal myopathy in the patient does not preclude later development of cardiac manifestations.
Although 343delT protein is observed in the patient skeletal muscle biopsy, it does appear at lower levels when compared with a control individual (29). Nonetheless, endogenous detection of the mutant protein was not observed in our iPSC-derived cells. Potentially, our model does not recapitulate 343delT expression observed in the patient for reasons related to the culture environment (pH differences, two-dimensional cell culture versus three-dimensional tissue structure, oxygen levels, etc.), stress (contraction, muscle turnover), variability in experimental techniques, age- and tissue-dependent, and other factors. This may illustrate the inherent limitations of iPSCs in modeling diseases because of an accumulation of stressors over time, also corresponding to the observation that the patient was entirely normal until disease onset at 4 months of age. Therefore, we hypothesize that the iPSCs recapitulate what may have occurred early in the patient, i.e. prior to disease manifestation. 343delT may be packaged into a completely insoluble state when expressed at basal levels, and its accumulation over time could result in 343delT surpassing a threshold, thereby allowing detection. The potential change in protein status may or may not contribute to disease. It is of interest that the severity and early-onset nature of the autosomal recessive disease in the 343delT patient is not consistent with the DKO mouse (20), which develops progressive myopathy into adulthood. We recognize that these differences could be due to species variation and/or the DKO mouse also lacking HSPB2. Alternatively, the presence of 343delT protein could exacerbate the phenotypes observed in the complete loss of function scenario (DKO mouse).
Co-aggregation of 343delT with desmin was observed upon overexpression of 343delT in some but not all BHK21 cells but was not observed with overexpression in iSKMs. Other mutations in HSPB5, such as R120G (26, 40), result in co-aggregation of the mutant protein with desmin, and patients are termed to have desmin-related (cardio)myopathy. The stochastic accumulation of desmin in 343delT aggregates is likely due to loss of 343delT chaperone function toward aggregation-prone desmin, resulting in the two proteins clustered together, among other proteins, in aggregates. The extent of desmin accumulation in aggregates likely depends on overall protein homeostasis within the cell. The finding that overexpression of 343delT induced elevated levels of HSP70 in BHK21 cells is consistent with cells suffering from an imbalance in protein homeostasis. We did not observe consistently elevated HSP70 levels in 343delT/343delT iCMs under basal conditions, which indicates that the presence of detectable 343delT (i.e. the visible aggregated form) and potentially other proteins present in the aggregates are required for disruption of protein homeostasis.
The recessive nature of 343delT led us to investigate the interaction between 343delT and WT HSPB5. It is unknown whether the heterozygous parents, who are non-consanguineous and healthy, express detectable levels of the 343delT protein. Potentially, expression of 343delT in the heterozygous state may not be detrimental, as we have shown increased solubility of 343delT and loss of aggregate formation upon co-expression with equimolar WT. Our co-IP experiments demonstrate an interaction between 343delT and WT within the cell. Together, these data suggest that WT solubilizes 343delT. Additionally, the presence of WT during refolding of 343delT rescues the protein from forming insoluble aggregates in vitro. The shift in elution volume during size exclusion and co-elution of the proteins, as observed by SDS-PAGE, suggest the formation of a complex between WT and 343delT, which is confirmed directly by native mass spectrometry experiments. The observation of a large proportion of what appears, from the absence of highly charged 343delT species, to be solution-phase monomeric 343delT in the mass spectrum suggests a labile interaction, especially under the conditions used, consistent with a chaperone-like interaction. The ability of WT to solubilize 343delT and the presence of unaffected WT protein to perform normal cellular functions likely result in the parents being asymptomatic.
Overall, our data support loss of HSPB5 function as a likely mechanism contributing to the myopathy. Given the substantial number of protein aggregation diseases, the findings reported here have direct implications for understanding the mechanisms of variable expressivity of both disease susceptibility and resistance. Although we cannot exclude the presence of the mutant protein directly contributing to disease, any detrimental effects of the 343delT protein would likely be ameliorated by the presence of WT through solubilization. Exogenous expression of WT HSPB5 by a gene therapy approach may therefore improve symptoms in mutant HSPB5-induced myopathy. Design of effective delivery methods for exogenous chaperones to the tissue or gene/cell therapy would be necessary for this treatment.
Experimental Procedures
Reprogramming Patient Dermal Fibroblasts to iPSCs
Dermal fibroblasts were isolated from the patient containing the 343delT mutation (29) with written consent of the mother of the patient and with research ethics committee approval at Guy's and St. Thomas' NHS Foundation Trust (London, UK) for biobank storage and transfer of samples for use in an ethically approved research project. Fibroblast culture and further research in the laboratory were performed with appropriate institutional review board approval at the University of Utah (Salt Lake City, UT) and the Medical College of Wisconsin (Milwaukee, WI) for use of human cells. Dermal fibroblasts expanded in DMEM supplemented with 10% FBS and 100 units of penicillin/100 μg of streptomycin/ml of medium (P/S) (all from Life Technologies) were reprogrammed into iPSCs through transduction with four retroviruses carrying the genes OCT-4, SOX2, c-MYC, and KLF4 as described previously (30). Following transduction, transduced fibroblasts were seeded onto SNL feeder cells (Cell Biolabs) that were mitotically inactivated through treatment with Mitomycin C (ATCC) in ESC medium composed of knockout DMEM supplemented with 20% knockout serum replacement, minimum essential medium-non-essential amino acids, 2 mm l-glutamine (all from Life Technologies), P/S, 0.1 mm β-mercaptoethanol (Sigma), 10 ng/ml human basic FGF (Cell Signaling Technology), and 50 ng/ml l-ascorbic acid (Sigma). The resulting iPSC clones were manually picked and maintained on feeder cells in ESC medium for four passages before transitioning to feeder-free culture as described below.
iPSC Culture
iPSCs were routinely cultured under feeder-free conditions on Matrigel (Corning)-coated dishes with mTeSR1 (Stem Cell Technologies) or StemMACS iPS-Brew XF (Miltenyi Biotec). Cells were passaged every 3–4 days using Accutase (Life Technologies) and seeded in medium containing 10 μm Rho-associated kinase inhibitor (Y-27632, Selleck) for 24 h following passaging.
Gene Editing in iPSCs, Genotyping, and Sequencing
iPSCs derived from the homozygous recessive 343delT patient (343delT/343delT) were gene-corrected to generate heterozygous 343delT iPSCs (WT KI/343delT) using a protocol modified from a previously defined strategy that involves knockin/excision of a selection cassette (32). Briefly, zinc finger nucleases were designed and generated by Sigma (CompoZr custom zinc finger nucleases). The targeting vector was designed and synthesized using Gene Art (Life Technologies) containing a portion of the WT HSPB5 sequence with a silent mutation (435 G>T) to generate a TTAA site for piggyBac excision. The puromycin/thymidine kinase selection cassette driven by the phosphoglycerate kinase promoter (PGK-puroΔtk) and flanked by piggyBac repeats was cloned from the pPB-CAG-OSKM-puDtk vector (Sanger Institute) into the targeting vector through In-Fusion Cloning (CloneTech). Components were transfected into iPSCs using a 4D-Nucleofector and P4 nucleofector solution with program CB-150 (Lonza), followed by seeding onto SNL feeder cells and selection with 0.5–1 μg/ml puromycin (InvivoGen) from days 5–12 post-transfection. The resulting puromycin-resistant clones were manually picked and genotyped. Heterozygous knockin clones were selected for excision of the selection cassette and transfected with an excision-only piggyBac transposase (PBx, Transposagen), followed by negative selection with 0.5 μm ganciclovir (Cayman Chemicals). The resulting colonies were picked and genotyped. PCR products from WT KI/343delT iPSCs were cloned into a TOPO vector (Life Technologies), and plasmids were isolated from bacterial colonies and Sanger-sequenced by Retrogen (San Diego, CA) to confirm heterozygosity for the 343delT mutation and incorporation of the engineered silent mutation. WT KI/343delT iPSCs were expanded and transitioned to feeder-free culture as described above.
Immunocytochemistry
Cells on 4% paraformaldehyde (Sigma)-fixed glass coverslips were permeabilized with 0.1% Triton X-100 (Sigma), blocked with 3% bovine serum albumin (Ultrapure BSA, Cell Signaling Technology), and stained appropriately before finally mounting with Ultracruz Hard Set mounting medium plus DAPI (Santa Cruz Biotechnology). Primary antibodies were as follows: NANOG (Cell Signaling Technology, 4903p, 1:200), SSEA-4 (Stem Cell Technologies, 60062AD, 1:40), OCT-4 (Cell Signaling Technology, 2840, 1:400), TRA-1–81 (Abcam, ab16289, 1:100), NT HSPB5 (NovaCastra ABCrys513, Leica, 1:100), TnT (Abcam, 45932, 1:500), MYC tag (Cell Signaling Technology, 2278, 1:100), desmin (Dako, M0760, 1:100), HSP70 (Stress Marq, SMC-100A/B, 1:100), and HA tag (Invivogen, ab-hatag, 1:500). Secondary antibodies were as follows: Alexa Fluor 488 donkey anti-rabbit IgG and Alexa Fluor 555 donkey anti-mouse IgG (Life Technologies, 1:500). Images were taken on a Nikon A1 confocal microscope.
qRT-PCR
RNA was isolated using the PureLink RNA MiniPrep kit (Life Technologies), followed by treatment with the DNA-free DNA removal kit (Life Technologies). cDNA was generated using the high-capacity cDNA reverse transcription kit (Life Technologies). qRT-PCR was performed using a Bio-Rad CFX 96 thermocycler with Qiagen QuantiTect SYBR Green PCR mixture and QuantiTect primer assays (Qiagen). Bio-Rad CFX Manager software was used for data analysis.
Karyotyping
Karyotyping was performed by Wisconsin Diagnostic Laboratories (formerly Dynacare Laboratories, Milwaukee, WI).
iCM and iSKM Differentiation
iCM differentiation was performed using chemical modulation of the Wnt pathway modified from a previously described protocol (47). Briefly, iPSCs at 100% confluency were overlaid with Matrigel in mTeSR1 or StemMACS iPS-Brew XF medium on day 1. On day 0, cells were treated with 12 μmol/liter Chir-99021 (Selleck) and 10 ng/ml human Activin-A (R&D Systems) in RPMI minus insulin medium (RPMI supplemented with 1× B27 supplement minus insulin (both from Life Technologies). On day 2, the medium was changed to RPMI minus insulin. On day 3, cells were treated with 5 μmol/liter IWP-2 (Stem Cell Technologies) in RPMI minus insulin, with medium subsequently changed on day 5 to RPMI minus insulin. The medium was changed to Complete medium, RPMI supplemented with 1× B27 supplement (Life Technologies) and P/S, on day 7 and changed every 2–3 days thereafter until cells were utilized for experiments around day 30 of differentiation. iCMs were passaged prior to experiments using TrypLE Express (Life Technologies) and seeded in Complete medium containing 10% FBS and 10 μm Rho-associated kinase inhibitor on 0.1% gelatin-coated surfaces.
iSKMs were differentiated using the EZ sphere protocol as described previously (48). Week 5 EZ spheres where treated with Accutase and pelleted for qRT-PCR of markers for all three germ layers. Week 6 EZ spheres were terminally differentiated as described previously (48) for 3 weeks prior to the experiments. Immunocytochemistry was performed directly on cells differentiated on coverslips.
Electrophysiology of iCMs
Action potentials were recorded using intracellular sharp electrodes. Coverslips containing beating clusters of iCMs were placed in Tyrode solution and held at physiological temperature with a heated recording chamber (TC-344B, Warner Instruments). Sharp electrodes were pulled from borosilicate glass (FHC, Inc.) using a Sutter model P-97 to resistances of 50–70 MΩ when backfilled with 3 m KCl. Recordings were acquired using a Multiclamp 700B amplifier (Molecular Devices) interfaced with a Digidata 1449A (Molecular Devices). The pClamp software suite (version 10, Molecular Devices) was used for data acquisition and analysis. Graphing of representative action potentials was performed using Origin software (OriginLab).
Cell Fractionation Assay and Western Blotting
Detergent-soluble, detergent-insoluble, and whole cell lysate (WCL) fractions were isolated, and Western blotting was performed as described previously (49), except equal amounts of protein were separated on Mini-Protean TGX gels, 4–20% (Bio-Rad). Primary antibodies were as follows: NT HSPB5 (NovaCastra ABCrys513, Leica, 1:100), GAPDH (Cell Signaling Technology, 2118, 1:10,000), p21 (Santa Cruz Biotechnology, sc-sc397, 1:250), LC3 (Novus, NB100–2220, 1:10,000), dicer (Santa Cruz Biotechnology, sc-393328, 1:100), c-MYC N-262 (Santa Cruz Biotechnology, sc-764, 1:500), MYC 9E10 (Santa Cruz Biotechnology, sc-40, 1:200), HSP70 (Stress Marq, SMC-100A/B, 1:1,000), α-tubulin (Santa Cruz Biotechnology, sc-5546, 1:500), β-actin (Abcam, ab6276, 1:5000), and HA tag (Invivogen, ab-hatag, 1:1000). Quantification of band intensity was performed using ImageJ or Image Lab 5.2.1 (Bio-Rad) software.
Inhibition of the Proteasome and Autophagosome Maturation
iCMs and iSKMs were treated with DMSO, 10 μm MG132, or 10 or 100 nm bortezomib to inhibit the proteasome or 80 nm bafilomycin A1 to inhibit autophagosome maturation for 12 h.
Dicer Knockdown
iCMs were transfected with scrambled siRNA control (−) or either 40 pmol (+) or 80 pmol (++) of siRNA targeting dicer (Santa Cruz Biotechnology) using Lipofectamine RNAi Max (Life Technologies). Samples were harvested 72 h post-transfection.
HSPB5 Vector Constructs
pCMV N-terminal MYC-tagged WT and HA-tagged WT (pCMV-MYC-WT and pCMV-HA-WT, respectively) were generated previously (49). The pCMV-MYC-343delT construct was produced by in vitro site-directed mutagenesis as described previously (49). MYC-WT and MYC-343delT were PCR-amplified from pCMV-MYC vectors and cloned into a pCS2 vector that contained an SP6 promoter required for the TnT assay (see below). For SUMOylated constructs, a gBlock gene fragment (Integrated DNA Technologies) was purchased, containing the non-cleavable SUMO3 Q89P sequence obtained from the pcDNA3 MYC-SUMO3Q89P vector (Addgene plasmid 48963) (46), and cloned into the pCMV-MYC-WT and 343delT constructs. The resulting constructs, pCMV-MYC-SUMO3 Q89P-WT and pCMV-MYC-SUMO3 Q89P-343delT, were a fusion in the N to C-terminal direction of MYC-SUMO3Q89P-WT and 343delT HSPB5, respectively.
In Vitro Transcription/Translation Assay
The TnT SP6 quick-coupled transcription/translation system (Promega) was used with EXPRE35S35S protein labeling mix (PerkinElmer Life Sciences) containing both [35S]l-methionine and [35S]l-cysteine. The pCS2-MYC-WT and 343delT plasmids were used as template DNA in the reaction according to the instructions of the manufacturer. Samples were subsequently run on a 12% SDS-PAGE gel, transferred to nitrocellulose membranes, and exposed to film at −80 °C. Quantification of band intensity was performed using ImageJ software.
Transfection
iSKMs were transfected using Lipofectamine 3000 (Life Technologies) according to the instructions of the manufacturer.
BHK21 cells (ATCC) were cultured in Glasgow medium supplemented with 5% tryptose phosphate broth (both from Life Technologies), 15% FBS, and P/S. BHK21 cells were transfected with Lipofectamine 3000 according to the instructions of the manufacturer. For co-transfection of two plasmids, equal amounts of each plasmid were used, with empty vector plasmid added to single-plasmid transfections to control for the total DNA amount.
iCMs were passaged and resuspended in P3 nucleofection solution (Lonza) and transected using the CA-137 program on the 4D nucleofector system (Lonza). Following transfection, cells were seeded onto fibronectin-coated surfaces (Corning) in Complete medium supplemented with 10% FBS and Rho-associated kinase inhibitor.
Co-immunoprecipitation
Cell pellets were resuspended in RIPA buffer (Thermo Scientific) supplemented with Halt protease and phosphatase inhibitor (Thermo Scientific), sonicated on ice, and centrifuged at 4 °C for 10 min at 12,000 × g. A portion was removed here for WCL, diluted with 4× XT sample buffer (Bio-Rad), and boiled for 5 min. The remaining WCL was used for IP performed with protein G-Sepharose 4 Fast Flow beads (Genesee). Samples were precleared through incubation with beads for 45 min on a rotisserie shaker at 4 °C, followed by preincubation with 200 μg/ml antibody, either MYC 9E10 (Santa Cruz Biotechnology, sc-40) or HA (Invivogen, ab-hatag) for 30 min at 4 °C with intermittent vortexing. Beads were added to the sample/antibody mixture and incubated on a rotisserie shaker for 1 h at 4 °C. Bead-bound samples were washed six times with RIPA buffer and finally resuspended in RIPA buffer with 4× XT sample buffer, boiled for 5 min, and centrifuged for 30 s at 12,000 × g. The insoluble pellet, prior to IP, was washed twice with RIPA buffer, resuspended in 4× XT sample buffer, boiled for 5 min, and centrifuged for 30 s at 12,000 × g. All fractions were loaded onto Mini-Protean TGX gels, 4–20% (Bio-Rad). Western blotting was performed as described above.
In vitro 343delT Protein Expression, Refolding, and Purification
The 343delT plasmid, purchased from GenScript, was transformed into BL21 (DE3) Stratagene Gold-competent cells (Agilent Technologies) and plated on kanamycin-selective plates. A single colony was picked and used to inoculate 5 ml of Luria-Bertani broth (Fisher). This culture was incubated at 37 °C, 220 rpm overnight until a high optical density was achieved. 1 liter of Luria-Bertani broth was then inoculated with 10 ml of the overnight culture and grown at 37 °C, 200 rpm, until an optical density of between 0.6–0.8 was obtained. To induce 343delT expression, a final concentration of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside (Merck) was added, and cultures were grown for a further 3 h before harvesting by centrifugation. Cell pellets were then stored at −80 °C for future use. Cells were thawed, resuspended, and lysed in 50 mm Tris (Sigma), 0.1 mm EDTA (Sigma), 0.1 mm DTT (Sigma), 0.1 m NaCl (Sigma), 5% glycerol (Fisher) (pH 7.9) (lysis buffer) containing a cOmplete EDTA-free protease inhibitor tablet (Roche). Post-clarification, overexpressed 343delT was shown to reside within insoluble inclusion bodies and not within the soluble lysate. The insoluble pellet was resuspended in lysis buffer, and any remaining cells were lysed by an additional sonication step. Membrane elements were solubilized by the addition of a final concentration of 1% Triton X-100 (Promega). This preparation was left to incubate for 15 min on ice before centrifugal clarification. Two further washes of the inclusion body pellet were carried out in lysis buffer to remove the remaining Triton X-100. Clean inclusion body pellets were then solubilized in 6 m GuHCl (Sigma), 50 mm Tris, 0.1 mm EDTA, 0.1 mm DTT, 5% glycerol (pH 7.9) and agitated at room temperature for 1 h, with any remaining insoluble material being removed by centrifugation. The protein concentration of the sample was estimated from UV absorbance measurements, and 2 molar equivalents of WT HSPB5 were added. Protein refolding was achieved by gradually reducing the concentration of GuHCl by stepwise dialysis, initially to 2 m, 1 m, and then 0 m to ensure that refolding was complete. Samples were passed through a 0.22 μm Spin X centrifuge tube (Fisher Scientific) and loaded onto a Superdex 200 10/300 column (GE Healthcare) to further purify and exchange buffer into 200 mm ammonium acetate (Sigma, pH 6.9). Fractions could then be analyzed by SDS-PAGE before being selected for mass spectrometry analysis.
Mass Spectrometry
Complex-containing samples were analyzed by nanoelectrospray mass spectrometry without the need for further sample preparation or concentration. Transmission pressures and energies on a Synapt G1 HDMS (Waters Corp.) were optimized to preserve non-covalent interactions but to maintain the transmission of large assemblies. Spectra were obtained using a previously defined protocol (42) under the following conditions: capillary, 1.5 kV; cone, 50 V; extractor, 3.0 V; trap, 8.0 V; transfer, 8.0 V; analyzer pressure, 1.58 e−3 millibar; backing pressure, 5.8 e0 millibar. All spectra were processed and analyzed in MassLynx V4.1.
Author Contributions
K. A. M., P. L., F. D. L. K., H. J., E. W., A. M. G., J. L. P. B., M. R., E. S. C., A. C. M., and I. J. B. conceived and designed the experiments. K. A. M., P. L., M. J. C., F. D. L. K, S. L., K. D. K., Q. D., M. N. G., H. Z., G. M. T., Q. L., J. A. H., R. B., and J. L. P. B. performed the experiments. K. A. M., P. L., F. D. L. K., W. M. K., M. T. T., K. B., A. M. G., J. L. P. B., M. R., E. S. C., A. C. M., and I. J. B interpreted the data. K. A. M. wrote the manuscript. F. D. L. K., K. D. K., H. J., J. L. P. B., E. S. C., A. C. M., and I. J. B. edited the article.
Supplementary Material
Acknowledgments
We are grateful to the clinicians involved in the care of our patient, including Dr. Andrew Durward and Dr. Jane Heraghty at the Evelina Children's Hospital, Guy's and St. Thomas NHS Foundation Trust (London, UK). We also thank Stefan Buk and Dr. Safa Al-Saraj (both King's College, London, UK) and Dr. Caroline Sewry (Dubowitz Neuromuscular Centre, Great Ormond Street Children's Hospital) for expert neuropathological input. We are grateful to Dr. Allison Ebert and Dr. Jered McGivern (both Medical College of Wisconsin, Milwaukee, WI) for instruction in the EZ sphere protocol for skeletal muscle differentiation.
This work was supported by National Institute of Health Director's Pioneer Award Grant 8DP1HL17650-04 (to I. J. B.) and a Royal Society University Research Fellowship (to J. L. P. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We dedicate this work to the patient and her family.

This article contains supplemental Figures S1–S5.
- ACD
- α-crystallin domain
- DKO
- double knockout
- NT
- N-terminal
- iPSC
- induced pluripotent stem cell
- iSKM
- iPSC-derived skeletal myotube
- iCM
- iPSC-derived cardiomyocyte
- qRT-PCR
- quantitative real-time PCR
- TnT
- Troponin T
- IP
- immunoprecipitation
- RIPA
- radioimmune precipitation assay
- SUMO
- small ubiquitin-related modifier
- WCL
- whole cell lysate.
References
- 1. Morimoto R. I. (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12, 3788–3796 [DOI] [PubMed] [Google Scholar]
- 2. Christians E. S., Yan L. J., and Benjamin I. J. (2002) Heat shock factor 1 and heat shock proteins: critical partners in protection against acute cell injury. Crit. Care Med. 30, S43–S50 [PubMed] [Google Scholar]
- 3. Gopal-Srivastava R., and Piatigorsky J. (1993) The murine α B-crystallin/small heat shock protein enhancer: identification of α BE-1, α BE-2, α BE-3 and MRF control elements. Mol. Cell. Biol. 13, 7144–7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gopal-Srivastava R., Haynes J. I. 2nd, and Piatigorsky J. (1995) Regulation of the murine aB-crystallin/small heath shock protein gene in cardiac muscle. Mol. Cell. Biol. 15, 7081–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poulain P., Gelly J. C., and Flatters D. (2010) Detection and architecture of small heat shock protein monomers. PLoS ONE 5, e9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hochberg G. K., and Benesch J. L. (2014) Dynamical structure of αB-crystallin. Prog. Biophys. Mol. Biol. 115, 11–20 [DOI] [PubMed] [Google Scholar]
- 7. Hochberg G. K., Ecroyd H., Liu C., Cox D., Cascio D., Sawaya M. R., Collier M. P., Stroud J., Carver J. A., Baldwin A. J., Robinson C. V., Eisenberg D. S., Benesch J. L., and Laganowsky A. (2014) The structured core domain of αB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc. Natl. Acad. Sci. U.S.A. 111, E1562–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peschek J., Braun N., Rohrberg J., Back K. C., Kriehuber T., Kastenmüller A., Weinkauf S., and Buchner J. (2013) Regulated structural transitions unleash the chaperone activity of αB-crystallin. Proc. Natl. Acad. Sci. U.S.A. 110, E3780–E3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rajagopal P., Tse E., Borst A. J., Delbecq S. P., Shi L., Southworth D. R., and Klevit R. E. (2015) A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis. eLife 4, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clark A. R., Naylor C. E., Bagnéris C., Keep N. H., and Slingsby C. (2011) Crystal structure of R120G disease mutant of human αB-crystallin domain dimer shows closure of a groove. J. Mol. Biol. 408, 118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mainz A., Peschek J., Stavropoulou M., Back K. C., Bardiaux B., Asami S., Prade E., Peters C., Weinkauf S., Buchner J., and Reif B. (2015) The chaperone αB-crystallin uses different interfaces to capture an amorphous and an amyloid client. Nat. Struct. Mol. Biol. 22, 898–905 [DOI] [PubMed] [Google Scholar]
- 12. Houck S. A., Landsbury A., Clark J. I., and Quinlan R. A. (2011) Multiple sites in αB-crystallin modulate its interaction with desmin filaments assembled in vitro. PLoS ONE 6, e25859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perng M. D., Cairns L., van den Ijssel P., Prescott A., Hutcheson A. M., and Quinlan R. A. (1999) Intermediate filament interactions can be altered by HSP27 and αB-crystallin. J. Cell Sci. 112, 2099–2112 [DOI] [PubMed] [Google Scholar]
- 14. Bennardini F., Wrzosek A., and Chiesi M. (1992) α B-crystallin in cardiac tissue: association with actin and desmin filaments. Circ. Res. 71, 288–294 [DOI] [PubMed] [Google Scholar]
- 15. Bullard B., Ferguson C., Minajeva A., Leake M. C., Gautel M., Labeit D., Ding L., Labeit S., Horwitz J., Leonard K. R., and Linke W. A. (2004) Association of the chaperone αB-crystallin with titin in heart muscle. J. Biol. Chem. 279, 7917–7924 [DOI] [PubMed] [Google Scholar]
- 16. Golenhofen N., Arbeiter A., Koob R., and Drenckhahn D. (2002) Ischemia-induced association of the stress protein α B-crystallin with I-band portion of cardiac titin. J. Mol. Cell. Cardiol. 34, 309–319 [DOI] [PubMed] [Google Scholar]
- 17. Golenhofen N., Perng M. D., Quinlan R. A., and Drenckhahn D. (2004) Comparison of the small heat shock proteins αB-crystallin, MKBP, HSP25, HSP20, and cvHSP in heart and skeletal muscle. Histochem. Cell Biol. 122, 415–425 [DOI] [PubMed] [Google Scholar]
- 18. Kötter S., Unger A., Hamdani N., Lang P., Vorgerd M., Nagel-Steger L., and Linke W. A. (2014) Human myocytes are protected from titin aggregation-induced stiffening by small heat shock proteins. J. Cell Biol. 204, 187–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singh B. N., Rao K. S., Ramakrishna T., Rangaraj N., and Rao C. M. (2007) Association of αB-crystallin, a small heat shock protein, with actin: role in modulating actin filament dynamics in vivo. J. Mol. Biol. 366, 756–767 [DOI] [PubMed] [Google Scholar]
- 20. Brady J. P., Garland D. L., Green D. E., Tamm E. R., Giblin F. J., and Wawrousek E. F. (2001) aB-Crystallin in Lens development and muscle integrity: a gene knockout approach. Invest. Ophthalmol. Vis. Sci. 42, 2924–2934 [PubMed] [Google Scholar]
- 21. Morrison L. E., Whittaker R. J., Klepper R. E., Wawrousek E. F., and Glembotski C. C. (2004) Roles for αB-crystallin and HSPB2 in protecting the myocardium from ischemia-reperfusion-induced damage in a KO mouse model. Am. J. Physiol. Heart Circ. Physiol. 286, H847–H855 [DOI] [PubMed] [Google Scholar]
- 22. Kumarapeli A. R., Su H., Huang W., Tang M., Zheng H., Horak K. M., Li M., and Wang X. (2008) α B-crystallin suppresses pressure overload cardiac hypertrophy. Circ. Res. 103, 1473–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neppl R. L., Kataoka M., and Wang D. Z. (2014) Crystallin-αB regulates skeletal muscle homeostasis via modulation of Argonaute2 activity. J. Biol. Chem. 289, 17240–17248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Christians E. S., Ishiwata T., and Benjamin I. J. (2012) Small heat shock proteins in redox metabolism: implications for cardiovascular diseases. Int. J. Biochem. Cell Biol. 44, 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mitzelfelt K. A., and Benjamin I. J. (2015) in The Big Book on Small Heat Shock Proteins (Tanguay R. M., and Hightower L. E., eds.), 1st ed., pp. 269–299, Springer International Publishing, Switzerland [Google Scholar]
- 26. Vicart P., Caron A., Guicheney P., Li Z., Prévost M. C., Faure A., Chateau D., Chapon F., Tomé F., Dupret J. M., Paulin D., and Fardeau M. (1998) A missense mutation in the αB-crystallin chaperone gene causes a desmin-related myopathy. Nat. Genet. 20, 92–95 [DOI] [PubMed] [Google Scholar]
- 27. Elliott J. L., Der Perng M., Prescott A. R., Jansen K. A., Koenderink G. H., and Quinlan R. A. (2013) The specificity of the interaction between αB-crystallin and desmin filaments and its impact on filament aggregation and cell viability. Philos. Trans. R Soc. Lond.B Biol. Sci. 368, 20120375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X., Osinska H., Klevitsky R., Gerdes A. M., Nieman M., Lorenz J., Hewett T., and Robbins J. (2001) Expression of R120G- B-crystallin causes aberrant Desmin and B-Crystallin aggregation and cardiomyopathy in mice. Circ. Res. 89, 84–91 [DOI] [PubMed] [Google Scholar]
- 29. Forrest K. M., Al-Sarraj S., Sewry C., Buk S., Tan S. V., Pitt M., Durward A., McDougall M., Irving M., Hanna M. G., Matthews E., Sarkozy A., Hudson J., Barresi R., Bushby K., et al. (2011) Infantile onset myofibrillar myopathy due to recessive CRYAB mutations. Neuromuscul. Disord. 21, 37–40 [DOI] [PubMed] [Google Scholar]
- 30. Takahashi K., Okita K., Nakagawa M., and Yamanaka S. (2007) Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2, 3081–3089 [DOI] [PubMed] [Google Scholar]
- 31. Christians E. S., Banerjee Mustafi S., and Benjamin I. J. (2014) Chaperones and cardiac misfolding protein diseases. Curr. Protein Pept. Sci. 15, 189–204 [DOI] [PubMed] [Google Scholar]
- 32. Yusa K., Rashid S. T., Strick-Marchand H., Varela I., Liu P. Q., Paschon D. E., Miranda E., Ordóñez A., Hannan N. R., Rouhani F. J., Darche S., Alexander G., Marciniak S. J., Fusaki N., Hasegawa M., et al. (2011) Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature 478, 391–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amrani N., Sachs M. S., and Jacobson A. (2006) Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 7, 415–425 [DOI] [PubMed] [Google Scholar]
- 34. Lee D. H., and Goldberg A. L. (1998) Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8, 397–403 [DOI] [PubMed] [Google Scholar]
- 35. Bloom J., Amador V., Bartolini F., DeMartino G., and Pagano M. (2003) Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115, 71–82 [DOI] [PubMed] [Google Scholar]
- 36. Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., and Tashiro Y. (1998) Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23, 33–42 [DOI] [PubMed] [Google Scholar]
- 37. Nawrocki S. T., Carew J. S., Dunner K. Jr., Boise L. H., Chiao P. J., Huang P., Abbruzzese J. L., and McConkey D. J. (2005) Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res. 65, 11510–11519 [DOI] [PubMed] [Google Scholar]
- 38. Flores-Jasso C. F., Arenas-Huertero C., Reyes J. L., Contreras-Cubas C., Covarrubias A., and Vaca L. (2009) First step in pre-miRNAs processing by human Dicer. Acta Pharmacol. Sin. 30, 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jackstadt R., and Hermeking H. (2015) MicroRNAs as regulators and mediators of c-MYC function. Biochim. Biophys. Acta 1849, 544–553 [DOI] [PubMed] [Google Scholar]
- 40. Perng M. D., Wen S. F., van den Ijssel P., Prescott A. R., and Quinlan R. A. (2004) Desmin aggregate formation by R120G αB-crystallin is caused by altered filament interactions and is dependent upon network status in cells. Mol. Biol. Cell 15, 2335–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vabulas R. M., Raychaudhuri S., Hayer-Hartl M., and Hartl F. U. (2010) Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kondrat F. L., Struwe W., and Benesch J. P. (2015) in Structural Proteomics (Owens R. J., ed.) pp. 349–371, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 43. Aquilina J. A., Benesch J. L., Bateman O. A., Slingsby C., and Robinson C. V. (2003) Polydispersity of a mammalian chaperone: mass spectrometry reveals the population of oligomers in αB-crystallin. Proc. Natl. Acad. Sci. U.S.A. 100, 10611–10616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peroutka R. J., Elshourbagy N., Piech T., and Butt T. R. (2008) Enhanced protein expression in mammalian cells using engineered SUMO fusions: secreted phospholipase A(2). Protein Sci. 17, 1586–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Panavas T., Sanders C., and Butt T. (2009) SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. Methods Mol. Biol. 497, 303–317 [DOI] [PubMed] [Google Scholar]
- 46. Békés M., Prudden J., Srikumar T., Raught B., Boddy M. N., and Salvesen G. S. (2011) The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J. Biol. Chem. 286, 10238–10247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lian X., Zhang J., Azarin S. M., Zhu K., Hazeltine L. B., Bao X., Hsiao C., Kamp T. J., and Palecek S. P. (2013) Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 8, 162–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hosoyama T., McGivern J. V., Van Dyke J. M., Ebert A. D., and Suzuki M. (2014) Derivation of myogenic progenitors directly from human pluripotent stem cells using a sphere-based culture. Stem Cells Transl. Med. 3, 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang H., Rajasekaran N. S., Orosz A., Xiao X., Rechsteiner M., and Benjamin I. J. (2010) Selective degradation of aggregate-prone CryAB mutants by HSPB1 is mediated by ubiquitin-proteasome pathways. J. Mol. Cell. Cardiol. 49, 918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.