Abstract
YAP signaling pathway plays critical roles in tissue homeostasis, and aberrant activation of YAP signaling has been implicated in cancers. To identify tractable targets of YAP pathway, we have performed a pathway-based pooled CRISPR screen and identified tankyrase and its associated E3 ligase RNF146 as positive regulators of YAP signaling. Genetic ablation or pharmacological inhibition of tankyrase prominently suppresses YAP activity and YAP target gene expression. Using a proteomic approach, we have identified angiomotin family proteins, which are known negative regulators of YAP signaling, as novel tankyrase substrates. Inhibition of tankyrase or depletion of RNF146 stabilizes angiomotins. Angiomotins physically interact with tankyrase through a highly conserved motif at their N terminus, and mutation of this motif leads to their stabilization. Tankyrase inhibitor-induced stabilization of angiomotins reduces YAP nuclear translocation and decreases downstream YAP signaling. We have further shown that knock-out of YAP sensitizes non-small cell lung cancer to EGFR inhibitor Erlotinib. Tankyrase inhibitor, but not porcupine inhibitor, which blocks Wnt secretion, enhances growth inhibitory activity of Erlotinib. This activity is mediated by YAP inhibition and not Wnt/β-catenin inhibition. Our data suggest that tankyrase inhibition could serve as a novel strategy to suppress YAP signaling for combinatorial targeted therapy.
Keywords: CRISPR/Cas, drug resistance, functional genomics, protein degradation, signal transduction, yes-associated protein (YAP)
Introduction
The evolutionarily conserved Hippo-YAP (yes-associated protein)3 pathway is gaining increasing attention as critical regulators in organ size homeostasis, tissue regeneration, and tumorigenesis (1, 2). Hippo-YAP pathway functions through regulating the activity of transcription cofactor YAP and its paralog TAZ. Central to Hippo pathway is the highly conserved MST1/2-LATS1/2 kinase cascade. MST1/2, in complex with its coregulatory protein Salvador (SAV1), phosphorylate and activate LATS1/2. Hippo kinase cascade is controlled by multiple upstream regulators, most notably tumor suppressor NF2. When Hippo kinase cascade is activated, YAP is phosphorylated by LATS1/2 and sequestered in the cytoplasm. When Hippo kinase cascade is inactivated, YAP enters the nucleus and binds to TEAD family transcription factors to activate the downstream transcriptional program. The Hippo-YAP pathway has emerged as a hub that integrates extra- and intracellular cues, including cell-cell interaction, mechanical and cytoskeletal changes, mitogens, and G-protein-coupled receptor (GPCR) signaling, to control the transcription activity of YAP. Among these regulations, the actin cytoskeleton plays a central role orchestrating extracellular and intracellular stimuli to regulate YAP signaling (2). Angiomotin family of proteins (AMOTs), including AMOT, AMOTL1, and AMOTL2, are key negative YAP regulators that link cytoskeletal changes and YAP signaling through phosphorylation-dependent and -independent mechanisms (3–6).
YAP signaling plays critical roles in cell proliferation and tissue homeostasis, and must be tightly regulated. Hyper-activation of YAP signaling can lead to uncontrolled cell proliferation and is associated with many human cancers (7, 8). Elevated expression of nuclear YAP protein is observed in many types of cancers, such as lung, liver, prostate, breast, colon, and ovary, and it often correlates with bad patient prognosis. Many mechanisms can lead to increased YAP signaling in cancer, including YAP gene amplification mutation or silencing of negative regulators, mutation of upstream GPCRs, or crosstalk with other signaling pathways (9). Elevated YAP signaling has been linked to drug resistance. Breast cancer cells with high YAP/TAZ activity show resistance to drugs such as taxol, doxorubicin, and tamoxifen (10–12). Furthermore, cancer cells with high YAP activity are resistant to RAF- and MEK-targeted therapies and YAP depletion sensitizes cells harboring BRAF-V600E mutations to RAF and MEK inhibitors (13). Therefore, small molecules inhibiting YAP signaling are needed to overcome drug resistance in cancers.
Tankyrase 1 (TNKS1) and tankyrase 2 (TNKS2) are enzymes catalyzing poly(ADP-ribosylation) (PARsylation) of protein substrates (14, 15). Through its ankyrin repeats, tankyrase recognizes substrate proteins and transfers poly-(ADP-ribose) chains to these proteins. Our lab has previously discovered that tankyrase and its associated E3 ligase RNF146 control Wnt/β-catenin signaling through promoting PARsylation-dependent degradation of Axin (16, 17). Over the years, tankyrase has been implicated in many biological processes and has been proposed as a drug target for cancer therapy (18–21).
In this study, we have performed pooled CRISPR screen and identified tankyrase and its associated E3 ligase RNF146 as positive regulators of YAP activity. Using a proteomics approach, we discovered angiomotins as substrates for tankyrases. Inhibition of tankyrase and RNF146 or blocking the interaction between angiomotins and tankyrase leads to accumulation of angiomotins and inhibition of YAP signaling. More importantly, tankyrase inhibition enhances the growth inhibitory activity of EGFR inhibitor in non-small cell lung cancer (NSCLC) through stabilization of angiomotins and inhibition of YAP signaling. Our study represents one of the first examples of pathway-based CRISPR screen for pathway dissection and target identification. Our work uncovers tankyrase/RNF146 as novel regulators of YAP signaling and expands the application of tankyrase inhibitor in combatting YAP-mediated drug resistance.
Results
Identification of RNF146 and Tankyrase as Positive Regulators of YAP Signaling from Pooled CRISPR Screen
YAP signaling is activated in a variety of cancers, and tractable targets are needed to block YAP signaling in cancer. We used a pooled CRISPR screen strategy (22, 23) to identify druggable components of the YAP signaling pathway. NF2, also known as Merlin, is a tumor suppressor and a negative regulator of YAP signaling (24). We chose to perform the screen in a NF2-deficient background since NF2 deficiency activates YAP signaling and bypasses cell density mediated YAP regulation. HEK293T NF2 KO cells were generated by CRISPR (supplemental Fig. S1). GTIIC-GFP, a GFP reporter for YAP/TEAD transcription factor complex (25, 26), was incorporated into HEK293T NF2 KO cells and HEK293A cells. A Cas9 expression cassette was also introduced into these cells to facilitate CRISPR-mediated gene inactivation.
We constructed a lentivirus library encoding 90,000 individual guide RNAs (gRNA) targeting 18,000 human genes. HEK293T NF2 KO GTIIC-GFP cells were infected with the lentiviral gRNA library and subjected to FACS analysis. Cells expressing low GFP signal and high GFP signal were collected for next-generation sequencing (NGS) and subjected to informatics analysis (Fig. 1A). Genes/gRNAs underrepresented in the GFP-high population compared with the GFP-low population are expected to be positive regulators for YAP signaling. As seen in Fig. 1B, YAP and its paralog TAZ (also called WWTR1) scored strongly in the screen, demonstrating the robustness of the screen. Much to our surprise, RNF146, a tankyrase-associated E3 ligase originally cloned in our laboratory, is also among the top hits (Fig. 1B). We previously discovered that tankyrase negatively regulates Wnt/β-catenin signaling through promoting degradation of Axin (16). Our follow-up work revealed that RNF146, a RING domain E3 ligase, binds to PARsylated Axin through its WWE domain and mediates PARsylation-dependent degradation of Axin (17). RNF146 physically associates with tankyrase and is directly activated by PAR (17, 27). RNF146 appears to be an E3 ligase dedicated to tankyrase; it mediates all known tankyrase-dependent protein degradation (AXIN1/2, TNKS1/2, BLZF1, CACS3, 3BP2, PTEN) (16, 17, 20, 28). Because of the close connection between RNF146 and tankyrase, we took a careful look at the screening data and found that TNKS2 also scored as a positive regulator of YAP signaling (Fig. 1B). However, TNKS2 scored as a weaker hit as compared with RNF146, possibly due to the redundant function of TNKS1 (16). These results suggest that tankyrase and RNF146 might function as positive regulators of YAP signaling.
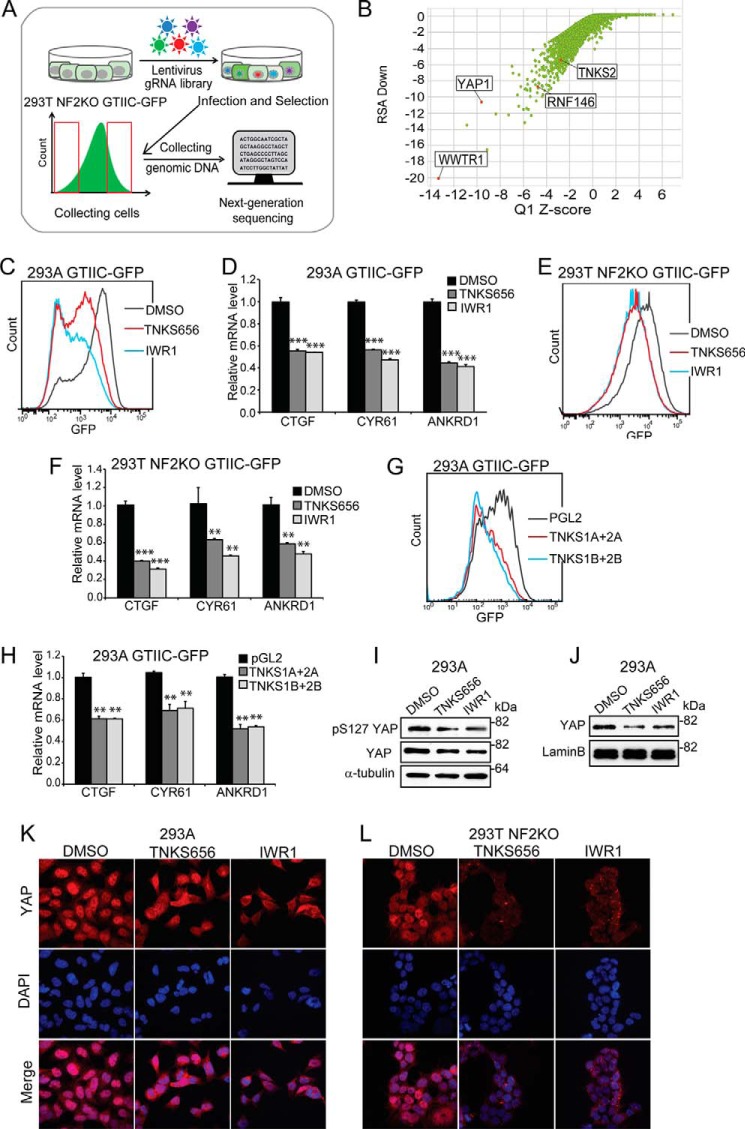
FIGURE 1.
Identification of RNF146 and tankyrase as positive regulators of YAP signaling through pooled CRISPR screen. A, an illustration of FACS-based pooled CRISPR screen of YAP signaling pathway in HEK293T NF2 KO GTIIC-GFP cells. B, scatter plot depicting results of GTIIC-GFP pooled CRISPR screen. Y-axis indicates the RSA down value (the statistical significance of all gRNA targeting each gene being unusually distributed toward the low end of the distribution), and X-axis indicates the Q1 z-score. C, tankyrase inhibitors attenuate GTIIC-GFP YAP reporter in HEK293A cells. For FACS assay, data are representative from at least two independent experiments. D, tankyrase inhibitors decrease the expression of YAP target genes in HEK293A cells. For qPCR assay, error bars are S.D., n = 4. Data are representative from at least two independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not significant. E, tankyrase inhibitors attenuate GTIIC-GFP YAP reporter in HEK293T NF2 KO cells. F, tankyrase inhibitors decrease the expression of YAP target genes in HEK293T NF2 KO cells. G, depletion of TNKS1/2 decreases GTIIC-GFP in HEK293A cells. H, depletion of TNKS1/2 decreases the expression of YAP target genes in HEK293A cells. I, effect of tankyrase inhibitors on the level of pS127 YAP and total YAP in total cell lysates of HEK293A cells. J, tankyrase inhibitors decrease the level of YAP in nuclear extracts of HEK293A cells. K, tankyrase inhibitors decrease nuclear localization of YAP in HEK293A cells. L, tankyrase inhibitors decrease nuclear localization of YAP in HEK293T NF2 KO cells.
To test a potential role of tankyrase in YAP signaling, we determined the effect of two structurally unrelated tankyrase inhibitors TNKS656 (29) and IWR1 (30) on YAP signaling. At 1 μm, both inhibitors suppressed GTIIC-GFP reporter activity in HEK293A cells (Fig. 1C) and HEK293T NF2 KO cells (Fig. 1E). Consistently, both tankyrase inhibitors suppressed the expression of YAP target genes, CTGF, CYR61, and ANKRD1 (Fig. 1, D and F). Together, these data suggest that tankyrase positively regulates YAP signaling and the catalytic activity of tankyrase is required for this function.
We then used siRNA to validate the function of tankyrase in YAP signaling. Since TNKS1 and TNKS2 play redundant function, we used two previously validated pairs of siRNAs (16) to simultaneously deplete TNKS1 and TNKS2 in HEK293A GTIIC-GFP cells (supplemental Fig. S2A). FACS analysis revealed a significant reduction of GFP signal intensity in TNKS1/2-knockdown cells compared with cells transfected with PGL2 control siRNA (Fig. 1G), suggesting that depletion of TNKS1 and TNKS2 suppresses YAP activity. We further showed that co-depletion of TNKS1 and TNKS2 reduced YAP target gene expression (Fig. 1H). These data further consolidate our finding that TNKS1 and TNKS2 are positive regulators of YAP signaling.
We next asked whether tankyrase inhibition influences the upstream kinase cascade that phosphorylates YAP. Interestingly, phosphorylation status of YAP at serine 127 was not significantly affected by tankyrase inhibitors in HEK293A cells (Fig. 1I). Phosphorylation of YAP at serine 127 in HEK293T NF2 KO cells was too low to be detected. These results suggest that tankyrase inhibitors do not have a strong effect on YAP phosphorylation.
Since nuclear translocation of YAP is required for activation of downstream target genes, we examined the effect of TNKS inhibitors on YAP expression in the nucleus. As shown in Fig. 1J, TNKS inhibitors decreased the level of YAP proteins in the nuclear extracts in HEK293A cells. In addition, we performed immunofluorescent staining to assess YAP nuclear localization in response to treatment of tankyrase inhibitors. In HEK293A cells, YAP proteins accumulated in the nucleus at low cell density (Fig. 1K, DMSO panel). However, nuclear staining of YAP was much reduced in tankyrase inhibitor-treated cells (Fig. 1K, TNKS656 and IWR1 panels). Similar results were also obtained in HEK293T NF2 KO cells (Fig. 1L). These results suggest that tankyrase inhibitor reduces YAP signaling through reducing its nuclear localization.
Tankyrase and RNF146 Degrade Angiomotins
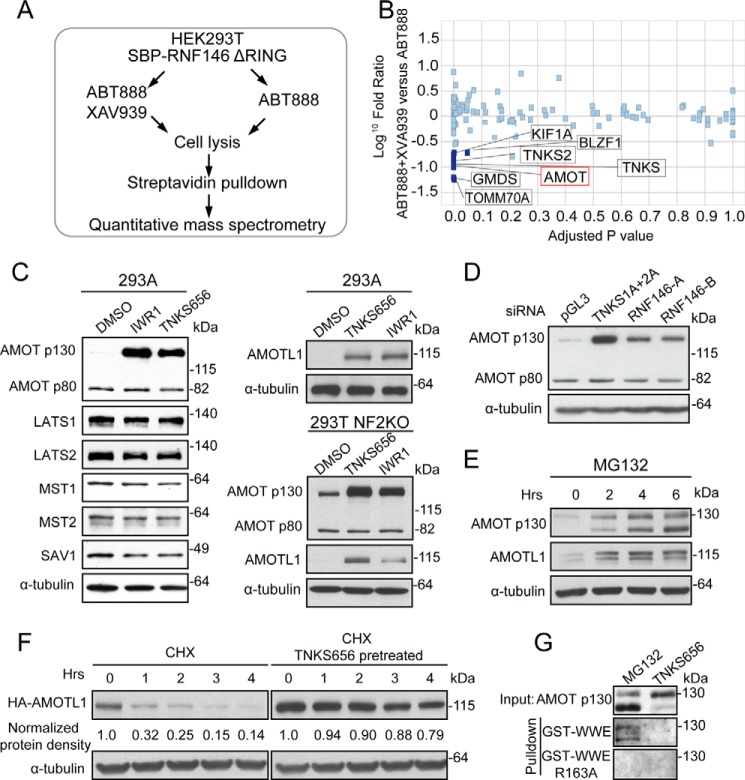
Next, we sought to identify proteins through which tankyrase regulates YAP activity. To this end, we employed an affinity purification approach to isolate PARsylated proteins specifically modified by tankyrase. RNF146 directly interacts with PARsylated proteins through its WWE domain (17). RNF146ΔRING, an RNF146 mutant retaining the WWE domain but lacking the E3 ligase activity, was used as a bait to purify PARsylated proteins. HEK293T cells expressing SBP-tagged RNF146ΔRING were treated with either PARP1/2 inhibitor ABT888 to reduce background PARsylation, or co-treated with ABT888 and tankyrase inhibitor XAV939 (16) (Fig. 2A). Cell lysates were incubated with streptavidin beads and bound proteins were eluted for quantitative mass spectrometry analysis. Proteins reduced in the XAV939-treated sample were candidates of tankyrase substrates. As expected, TNKS1 (also known as TNKS) and TNKS2, which auto-PARsylate themselves, were top hits of this experiment (Fig. 2B). In addition, we found angiomotin (AMOT) in the top hit list (Fig. 2B and supplemental Table S1). Angiomotin family of proteins (AMOT, AMOTL1, AMOTL2) are known negative regulators of YAP signaling (3–6). AMOT exists in two isoforms, a longer p130 isoform and a shorter p80 isoform. We found that tankyrase inhibitors strongly increased the protein level of p130 isoform, but not that of p80 isoform, in HEK293A cells (Fig. 2C). As controls, tankyrase inhibitors did not affect protein levels of other negative regulators of YAP pathway, including LATS1/2, MST1/2, and SAV1 (Fig. 2C). Furthermore, tankyrase inhibitors increased the protein level of AMOTL1, a close paralog of AMOT (Fig. 2C). Similar findings were made in HEK293T NF2 KO cells (Fig. 2C). We also found that siRNA-mediated depletion of tankyrase or RNF146 in HEK293A cells led to accumulation of AMOT p130 protein without affecting AMOT p80 protein (Fig. 2D, supplemental Fig. S2B), suggesting that tankyrase and RNF146 specifically degrade the longer isoform of AMOT proteins. Furthermore, AMOT and AMOTL1 proteins accumulated upon MG132 treatment (Fig. 2E), indicating that angiomotins are degraded by the proteasome. Consistently, after protein translation was blocked by cycloheximide, the half-life of HA-AMOTL1 was increased when cells were pre-treated with tankyrase inhibitor (Fig. 2F). In addition, the GST- WWE domain of RNF146, which specifically binds the poly(ADP-ribose) moiety (17), pulled down endogenous AMOT p130 from MG132-treated cells but not from TNKS656-treated cells (Fig. 2G), indicating that AMOT proteins are PARylated by tankyrase in vivo. As a control, WWE R163A mutant, which cannot bind to poly(ADP-ribose) moiety (17), did not pull down AMOT proteins (Fig. 2G), demonstrating the specificity of pulldown assay. These data are consistent with the idea that tankyrase and RNF146 promote PARsylation-dependent degradation of angiomotins.
FIGURE 2.
Inhibition of tankyrase stabilizes AMOT family proteins. A, proteomics strategy to identify tankyrase substrates. B, scatter plot depicting proteins identified and quantified in a quantitative proteomics experiment. Proteins significantly depleted in XAV939-treated condition are highlighted as potential substrates of tankyrase. C, tankyrase inhibitors increase protein levels of AMOT p130 and AMOTL1. D, depletion of TNKS1/TNKS2 or RNF146 increases the protein level of AMOT p130. E, proteasome inhibitor MG132 increases protein levels of AMOT p130 and AMOTL1. F, tankyrase inhibitor increases half-life of HA-AMOTL1. G, tankyrase inhibitor suppresses PARsylation of AMOT p130.
Angiomotins Bind to Tankyrase through N-terminal Tankyrase Binding Motif
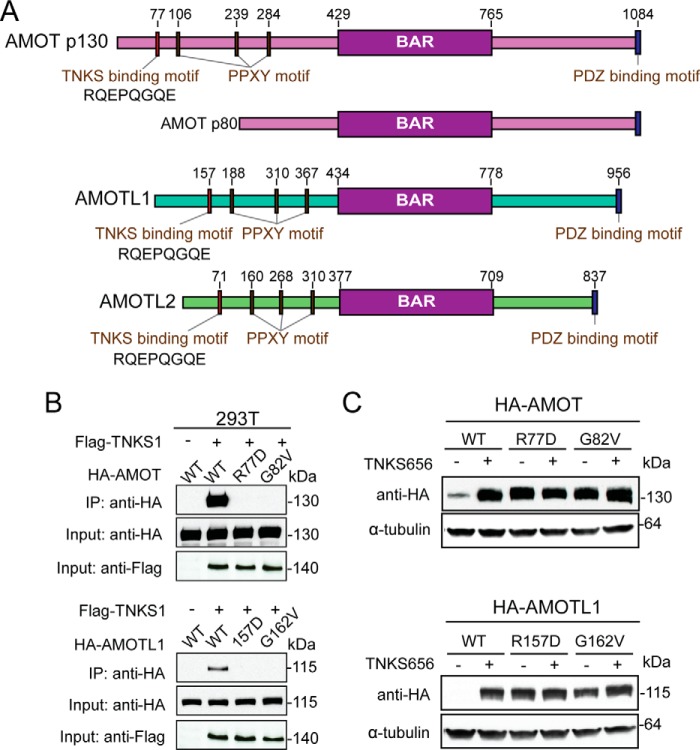
To further characterize the interaction between tankyrase and angiomotins, we searched for tankyrase binding motifs in all three paralogs of AMOT family of proteins. Ankyrin repeats of tankyrase recognize a consensus of motif of RXXG/PXGX(E/D) (31, 32), although our recent work suggested that certain deviations can be tolerated (27, 33). We surveyed the full length AMOT and identified only one motif (RQEPQGQE) that matches perfectly to the consensus tankyrase binding motif (Fig. 3A). Furthermore, this motif is located within the N terminus of AMOT p130, a region missing from AMOT p80, which is consistent with our earlier finding that AMOT p130 but not the p80 isoform is stabilized by tankyrase inhibition (Fig. 2D). Importantly, this motif is highly conserved among AMOT, AMOTL1, and AMOL2 (Fig. 3A). To test the function of this motif, we introduced single point mutations (Arg → Asp or Gly → Val) to AMOT and AMOTL1, and tested the interaction between wild-type or mutant proteins with co-expressed Flag-TNKS1 in a co-immunoprecipitation assay. As expected, Flag-TNKS1 only pulled down wild-type AMOT and AMOTL1 but not mutant proteins bearing the mutated tankyrase binding motif (Fig. 3B). We next assessed the functional consequence of disrupting the physical interaction between tankyrase and angiomotins. If binding between angiomotins and tankyrase is required for degradation of angiomotins, mutation of tankyrase binding motif should lead to stabilization of angiomotins. Indeed, although wild-type HA-AMOT and HA-AMOTL1 were expressed at low basal protein levels that were strongly increased upon treatment of tankyrase inhibitor, HA-AMOT and HA-AMOTL1 mutants bearing the mutated tankyrase binding motif were expressed at much higher basal levels and did not further respond to compound treatment (Fig. 3C).
FIGURE 3.
The tankyrase binding motif is required for tankyrase-induced degradation of angiomotins. A, schematic diagram of the domain structure of angiomotins. The tankyrase binding motif at the N terminus of angiomotins is indicated. B, angiomotins interact with tankyrase through the N-terminal tankyrase binding motif. C, mutation of tankyrase binding motif stabilizes AMOT and AMOTL1.
These data collectively lead to a conclusion that AMOT and AMOTL1, and likely AMOTL2, are substrates of tankyrase and RNF146 and that their degradation is controlled through tankyrase-catalyzed PARsylation and RNF146-dependent degradation.
Angiomotins Are Major Mediators of Tankyrase Inhibitor-induced Down-regulation of YAP Signaling
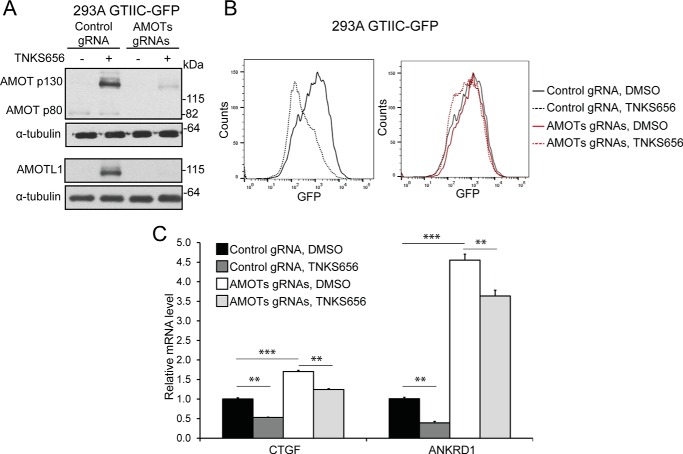
Next, we asked whether angiomotins mediate YAP inhibitory activity of tankyrase inhibitor. To this end, we co-infected HEK293A GTIIC-GFP-Cas9 cells with virus expressing gRNAs against AMOT, AMOTL1, and AMOTL2, and analyzed YAP reporter expression by flow cytometry 10 days after infection. Since suitable AMOTL2 antibodies are not available, we only examined AMOT and AMOTL1 protein levels in AMOTs knock-out cells. As seen in Fig. 4A, expression of both AMOT and AMOTL1 proteins was significantly reduced in AMOTs knock-out cells. In cells infected with control gRNA, the GTIIC-GFP reporter signal was significantly decreased by tankyrase inhibitor TNKS656 (Fig. 4B, left panel). However, this decrease was largely abolished in cells infected with gRNAs targeting angiomotins (Fig. 4B, right panel). Moreover, loss of angiomotins overcame TNKS656-induced suppression of YAP target gene expression (Fig. 4C). These results demonstrate that angiomotins are major mediators of tankyrase in regulation of YAP signaling.
FIGURE 4.
Knock-out of angiomotins abolishes the inhibitory activity of tankyrase inhibitor on YAP signaling. A, protein levels of AMOT and AMOTL1 are decreased in 293A GTIIC-GFP Cas9 cells expressing gRNAs against angiomotins. B, knock-out of angiomotins abolishes the inhibitory effect of tankyrase inhibitor on GTIIC-GFP. C, knock-out of angiomotins attenuates the inhibitory activity of tankyrase inhibitor on YAP target genes.
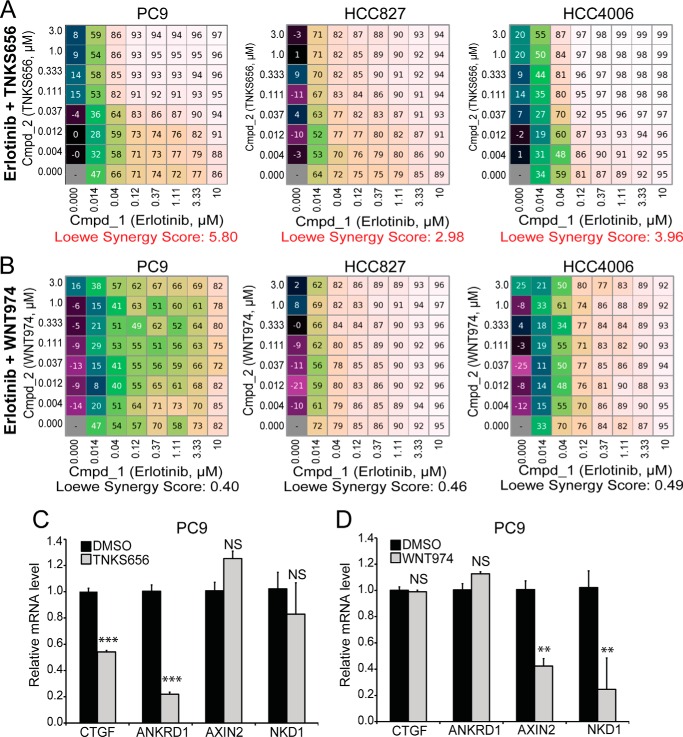
Tankyrase Inhibition Synergizes with Erlotinib through YAP Inhibition
Drug resistance in targeted therapy is a major challenge for many anti-cancer drugs. Increased YAP signaling has been linked to resistance to chemotherapies and targeted cancer therapies. Since tankyrase inhibition is able to inhibit YAP activities, we explored if combining tankyrase inhibitor with targeted therapy could provide additional benefits in targeting cancer cells. Interestingly, a previous study demonstrated that tankyrase inhibition potentiated Erlotinib activity in NSCLC, but this activity was attributed to Wnt/β-catenin inhibition (34). We set out to test whether the growth inhibitory activity of tankyrase inhibition in NSCLC is mediated by inhibition of Wnt/β-catenin signaling or inhibition of YAP signaling. We performed checkbox assays (35) to measure the synergistic effect of tankyrase inhibitor TNKS656 and Erlonitib in three NSCLC cell lines, PC9, HCC827, and HCC4006. Combination of these two drugs displayed synergy in all three cells with a Loewe synergy score above 2 (Fig. 5A). However, WNT974, a porcupine inhibitor that specifically blocks WNT secretion and downstream Wnt/β-catenin signaling (36), displayed no synergistic activity with Erlotinib (Fig. 5B). Importantly, tankyrase inhibitor TNKS656 inhibited expression of YAP target gene CTGF and ANKRD1 with minimal effect on β-catenin target gene AXIN2 and NKD1 (Fig. 5C). On the contrary, porcupine inhibitor WNT974 decreased expression of β-catenin target gene AXIN2 and NKD1 but not that of YAP target genes (Fig. 5C). Together, these results suggest that the observed effect of TNKS656 in synergizing with Erlotinib is not mediated by inhibition of WNT/β-catenin pathway.
FIGURE 5.
Tankyrase inhibitor TNKS656, but not porcupine inhibitor WNT974, sensitizes NSCLC cells to EGFR inhibitor Erlotinib. A and B, checkbox assays were performed to measure growth inhibitory effect of combination of Erlotinib and TNKS656 (A) or WNT974 (B) in PC9, HCC827, and HCC4006 cells, and Loewe synergy scores were calculated. Loewe Synergy score above 2 is considered significant. C, effects of TNKS656 and WNT974 on YAP target genes (CTGF and ANKRD1) and β-catenin target genes (AXIN2 and NKD1).
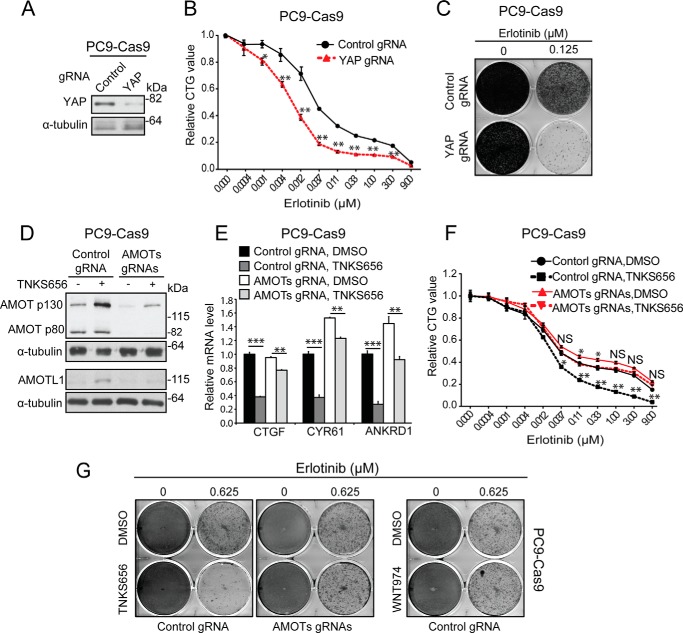
To determine a possible role of YAP signaling in mediating Erlotinib resistance, we tested whether knock-out YAP can enhance the growth inhibitory activity of Erlotnib. PC9 cells stably expressing Cas9 were infected with virus expressing control gRNA or gRNA against YAP. Decrease of YAP expression in the cell pool expressing YAP gRNA was confirmed by Western blot analysis (Fig. 6A). PC9 cells expressing YAP gRNA showed enhanced sensitivity to Erlotnib in CTG assay (Fig. 6B) and colony formation assay (Fig. 6C). To determine a possible role of angiomotins in mediating the effect of tankyrase inhibition in NSCLC, we co-infected PC9 cells stably expressing Cas9 with virus expressing gRNAs against AMOT, AMOL1, and AMOTL2. Guide RNAs against angiomotins decreased but did not abolish the expression of AMOT and AMOTL1 (Fig. 6D), which reflected the heterogeneous nature of infected cell population. Knock-out angiomotins attenuated tankyrase inhibitor-dependent inhibition of YAP target genes (Fig. 6E). Significantly, growth inhibitory activities of TNKS656 in CTG assay and colony formation assay were largely abolished in PC9 cells deficient of angiomotins (Fig. 6, F and G). Similar results were obtained in HCC827 cells (supplemental Fig. S3). Taken together, these results suggest that tankyrase inhibition increases the potency of Erlotinib, and this activity is mostly mediated by stabilization of AMOTs and inhibition of YAP signaling. This finding provides a rationale of further testing tankyrase inhibition in combination with targeted therapies in preclinical tumor models and corresponding early clinical trials.
FIGURE 6.
Stabilization of angiomotins is required for growth inhibitory activity of tankyrase inhibitor in NSCLC. A–C, knock-out of YAP sensitizes cells to the growth inhibitory activity of Erlotinib. PC9 stably expressing Cas9 were infected with lentivirus expressing control gRNA or gRNA against YAP. Cells were subjected to Western blot assay (A), CTG assay (B), or colony formation assay (C) in combination with different doses of Erlotinib. For proliferation assays, error bars are S.E., n = 3. Data are representative from at least three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not significant. D–G, knock-out of angiomotins attenuates YAP inhibitory and growth inhibitory activities of tankyrase inhibitor. PC9-Cas9 cells expressing control gRNA or gRNAs against angiomotins were treated with indicated compound, and subjected to Western blot assay (D), qRT-PCR assay (E), CTG assay (F), or colony formation assay (G).
Discussion
In this study, we performed pooled CRISPR screen using a YAP/TEAD reporter and discovered tankyrase and its associated E3 ligase RNF146 as positive regulators of YAP signaling. Using a proteomics approach, we identified angiomotins as substrates of tankyrase and RNF146 and demonstrated that stabilization of angiomotins mediates YAP inhibitory activity of tankyrase inhibitor. The same notion was put forward in a study (37) that was published during the preparation of this manuscript. Thus, tankyrase plays a key role in controlling angiomotins stability and YAP activation. More importantly, we demonstrate that tankyrase inhibition, but not porcupine inhibition, sensitize NSCLC to Erlotinib, and this activity is dependent on angiomotins. Our data suggest that the growth inhibitory activity of tankyrase inhibitor in NSCLC involves inhibition of YAP signaling but not Wnt/β-catenin signaling.
Over the past decade, RNAi screening has been the dominating genomic screening approach, but the approach is sometimes complicated by RNAi off-target activities and inefficient knockdown. High-throughput CRISPR screen represents a revolutionary technology in the field of functional genomics (38). However, most pooled CRISPR screens published so far are based on cell proliferation and survival. Following a recent study (39), our work represents the second example of pathway-based pooled CRISPR screen and highlights the power of CRISPR screen for pathway dissection and target identification.
Angiomotins bind to F-actin and also localize at tight junctions (40–42). They negatively regulate YAP signaling through different mechanisms: firstly, angiomotins directly interact with NF2, which leads to activation of LATS kinases, increased YAP phosphorylation and reduced YAP nuclear accumulation (3, 43); secondly, angiomotins recruit YAP to tight junctions through PPXY motifs on angiomotins and the WW domain of YAP and prevent it from shuttling to the nucleus (4–6). In this report, we showed that tankyrase inhibition suppressed YAP signaling through stabilizing angiomotins and retaining YAP in the cytoplasm. The function of angiomotins in YAP signaling is dynamically regulated, including phosphorylation by LATS and AMPK, and ubiquitination by NEDD4-like E3 ligases (44–46). Degradation of angiomotins by tankyrase and RNF146 represents another layer of regulation of angiomotins. Whether this regulation itself is controlled should be examined in future studies.
Targeted therapy has been a great success for cancer treatment in the past decades. Despite its success, drug resistance still poses a major threat to cancer patients. Tackling drug resistance has emerged as an increasingly important need in cancer treatment. Interestingly, high YAP/TAZ activity has been correlated to drug resistance (10, 11, 13, 47). Combination of certain targeted therapies with strategies concurrently targeting YAP signaling may alleviate drug resistance. Indeed, we have shown that tankyrase inhibition enhances growth inhibitory activity of Erlotinib in NSCLC. Importantly, this activity requires angiomotins and it does not appear to involve inhibition of β-catenin signaling as one previous study suggested. YAP hyper-activation can also overcome KRAS dependence in mouse tumor models (48, 49). Interestingly, tankyrase inhibition sensitizes KRAS mutant cancer cells to MEK inhibition (50). The exact mechanism of this finding is not clear although it might involve FGFR2 feedback. Whether the synergy between tankyrase inhibitor and MEK inhibitor in KRAS mutant cells involves angiomotins stabilization and YAP inhibition should be examined in the future. Because of the key roles of tankyrase in signaling pathways such as Wnt pathway, several tankyrase inhibitors are being developed for cancer. Testing tankyrase inhibitors in combination with targeted therapies in preclinical models and early clinical trials will likely provide new strategies to block YAP signaling and combat drug resistance.
Experimental Procedures
Pooled CRISPR Screen
We designed five gRNAs against each gene using Illumina Human BodyMap 2.0 and NCBI CCDS data sets. The gRNA library containing 90,000 gRNAs was synthesized using array synthesis and cloned into a lentivirus vector by CELLECTA Inc. HEK293T NF2 KO cells with constitutive Cas9 expression and GTIIC-GFP reporter were transduced at MOI 0.5. Ten days after virus transduction, GFP-high and GFP-low cells were sorted using BD FACSAria Cell Sorter. Genomic DNA was collected and subjected to Illumina DNA sequencing for barcode counts. Raw counts from each sample were normalized before analysis. The number of counts for each barcode in the GFP-high sample was divided by the corresponding number in the GFP-low sample to give the fold change. A robust z-score was calculated using the median and mean-absolute deviation across the log2 fold changes of the library combined results. To summarize the results at the gene level, we evaluated the statistical significance of all gRNAs targeting each gene being unusually distributed toward the high end of the distribution (RSA up) and the low end of the distribution (RSA down) using the Redundant siRNA Activity (RSA) algorithm (51). To visualize the gene significance and result strength, we plotted the RSA down value against the Q1 (the second most depleted gRNA within 5 gRNAs) z-score for each gene for investigating YAP positive regulators. Guide RNA sequences used in this work: Control: GACCGGAACGATCTCGCGTA; AMOT: GAGACGACAAGAGCTGGAAG; AMOTL1: GCAGCCTCAGCAGAACAACG; AMOTL2: GCGGTGCAGGACTGTCCCCG; YAP: TGGGGGCTG TGACGTTCATC.
Cell Culture
HEK293T, HEK293A, PC9, HCC827, and HCC4006 were obtained from ATCC from 2014- 2015 and authenticated using SNP testing in November 2015.
RNA Interference and Transfection
Sequences of siRNAs were described previously (16, 17). Plasmid transfection was done using Fugene 6 (Roche) and siRNA transfection was done using Lipofectamine® RNAiMAX (ThermoFisher Scientific).
Western Blot Analysis, Co-immunoprecipitation, GST Pulldown Assay
Western blot analysis, co-immunoprecipitation, and GST pulldown assay were performed as previously described (17). Nuclear extracts were generated using NE-PERTM nuclear and cytoplasmic extraction reagents (ThermoFisher). Sources of primary antibodies are: anti-YAP, anti-phospho-S127 YAP, anti-NF2, and anti-GAPDH (Cell Signaling Technology); anti-AMOT (Bethyl Laboratories), anti-AMOTL1, anti-FLAG, anti-α tubulin (Sigma); anti-HA (Roche).
Immunofluorescence Staining
Cells were seeded on glass chamber slides and treated with 1 μm TNKS656, 1 μm IWR1, or DMSO control for 24 h. Cells were then fixed with 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100 in PBS. After blocked with 2% BSA/4% goat serum, cells were stained with anti-YAP antibody (Santa Cruz Biotechnology, 1:500) and followed with secondary antibody Alexa Fluor 568 goat anti-mouse IgG (Molecular Probes, 1:2000). Coverslips were mounted on glass slides using ProLong® Gold antifade reagent containing DAPI (Invitrogen), and the images were taken using confocal laser scanning microscope (Carl Zeiss, Germany) with 40× objective.
Colony Formation Assays
For colony formation assays, cells were seeded at 10,000 cells per well into 6-well plates. At the next day, cells were treated with various compounds at the indicated concentration. Medium was replenished every 3 days. After 14 days, plates were washed with PBS, fixed, and stained with crystal violet solution. After washed with water, plates were dried and imaged.
N-SBP-RNF146ΔRING Pulldown and Quantitative Mass Spectrometry
HEK293T cells stably expressing SBP-RNF146 ΔRING construct were treated with 5 μm ABT888 and 3 μm XAV939 or 5 μm ABT-888 alone. ABT888 is a PARP1/2 inhibitor to suppress strong background PARsylation mediated by PARP1/2. Cells were then washed with ice-cold PBS twice and lysed in RIPA buffer supplemented with 5 μm ADP-HPD (Alexis), and pulled down with Streptavidin beads (Sigma). Eluted protein samples were solved by SDS-PAGE. Complete gel lanes were excised and samples were subjected to in-gel tryptic digestion. Peptide extracts from control lane slices (no XAV939 treatment) were labeled with TMT reagent 131 and combined with extracts from corresponding slices of the XAV939 treatment lane which were labeled with TMT reagent 129. Peptide sequencing was performed by liquid chromatography-tandem mass spectrometry. Peptide quantification and normalization were performed as previously described. Protein fold changes were derived as median peptide fold change, p values were calculated using a one-way t test (arbitrarily set to 1 for non-significant single peptide quantitations) and adjusted using the Benjamini-Hochberg False Discovery Rate (FDR). All identified proteins are shown in supplemental Table S1.
Cell Viability Assay
Cell viability was determined by Cell Titer Glo Luminescence Assay (Promega). Cells were seeded in triplicates in 96-well plates and 1 day after drugs are added accordingly. Five days after luminescence was recorded on an EnVision plate reader (PerkinElmer). For checkbox assays the inhibition of viability relative to DMSO-treated cells was calculated and analyzed as previously described (35).
Statistical Analysis
For proliferation assays, error bars are S.E., n = 3. Data are representative from at least three independent experiments. For qRT-PCR assay, error bars are S.D., n = 4. Data are representative from at least two independent experiments. For FACS data, Data are representative from at least two independent experiments. Statistical analysis was carried out using one-way ANOVA. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not significant.
Author Contributions
H. W., B. L., Y. Z., Z. Y., G. M., J. R., J. T., G. H., C. R., W. X., M. S., and F. C. conceived and designed the study. H. W., B. L., J. C., Y. Z., Z. Y., G. M., J. R., G. H., C. R., W. X., M. S., and F. C. designed and implemented experiments. H. W. and F. C. wrote the paper.
Supplementary Material
Acknowledgments
We thank Raffaella Zamponi, Malini Varadarajan, Akos Szilvasi, Alan Ho, Deborah Ahern-Ridlon, Amy Janiak, and Jennifer Kelliher for technical assistance. We also thank Ralph Tiedt, Marion Wiesmann, Tobias Schmelzle, Andreas Bauer, Huaixiang Hao, Xiaomo Jiang, Chen Liu, and Yi Yang for comments and advices.
The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Figs. S1–S3.
- YAP
- yes-associated protein
- TNKS
- tankyrase
- AMOT
- angiomotin
- RSA
- redundant siRNA activity
- PARsylation
- poly(ADP-ribosylation).
References
- 1. Pan D. (2010) The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yu F. X., Zhao B., and Guan K. L. (2015) Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paramasivam M., Sarkeshik A., Yates J. R. 3rd, Fernandes M. J., and McCollum D. (2011) Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol. Biol. Cell 22, 3725–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao B., Li L., Lu Q., Wang L. H., Liu C. Y., Lei Q., and Guan K. L. (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan S. W., Lim C. J., Chong Y. F., Pobbati A. V., Huang C., and Hong W. (2011) Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J. Biol. Chem. 286, 7018–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang W., Huang J., and Chen J. (2011) Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 286, 4364–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harvey K. F., Zhang X., and Thomas D. M. (2013) The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 [DOI] [PubMed] [Google Scholar]
- 8. Plouffe S. W., Hong A. W., and Guan K. L. (2015) Disease implications of the Hippo/YAP pathway. Trends Mol. Med. 21, 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Irvine K. D. (2012) Integration of intercellular signaling through the Hippo pathway. Semin. Cell Dev. Biol. 23, 812–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lai D., Ho K. C., Hao Y., and Yang X. (2011) Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 71, 2728–2738 [DOI] [PubMed] [Google Scholar]
- 11. Cordenonsi M., Zanconato F., Azzolin L., Forcato M., Rosato A., Frasson C., Inui M., Montagner M., Parenti A. R., Poletti A., Daidone M. G., Dupont S., Basso G., Bicciato S., and Piccolo S. (2011) The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 [DOI] [PubMed] [Google Scholar]
- 12. Zhou X., Wang S., Wang Z., Feng X., Liu P., Lv X. B., Li F., Yu F. X., Sun Y., Yuan H., Zhu H., Xiong Y., Lei Q. Y., and Guan K. L. (2015) Estrogen regulates Hippo signaling via GPER in breast cancer. J. Clin. Invest. 125, 2123–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin L., Sabnis A. J., Chan E., Olivas V., Cade L., Pazarentzos E., Asthana S., Neel D., Yan J. J., Lu X., Pham L., Wang M. M., Karachaliou N., Cao M. G., Manzano J. L., Ramirez J. L., Torres J. M., Buttitta F., Rudin C. M., Collisson E. A., Algazi A., Robinson E., Osman I., Muñoz-Couselo E., Cortes J., Frederick D. T., Cooper Z. A., McMahon M., Marchetti A., Rosell R., Flaherty K. T., Wargo J. A., and Bivona T. G. (2015) The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 47, 250–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riffell J. L., Lord C. J., and Ashworth A. (2012) Tankyrase-targeted therapeutics: expanding opportunities in the PARP family. Nat. Rev. Drug Discov. 11, 923–936 [DOI] [PubMed] [Google Scholar]
- 15. Lehtiö L., Chi N. W., and Krauss S. (2013) Tankyrases as drug targets. FEBS J. 280, 3576–3593 [DOI] [PubMed] [Google Scholar]
- 16. Huang S. M., Mishina Y. M., Liu S., Cheung A., Stegmeier F., Michaud G. A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C. J., Mickanin C., Myer V., Fazal A., Tomlinson R., Serluca F., Shao W., Cheng H., Shultz M., Rau C., Schirle M., Schlegl J., Ghidelli S., Fawell S., Lu C., Curtis D., Kirschner M. W., Lengauer C., Finan P. M., Tallarico J. A., Bouwmeester T., Porter J. A., Bauer A., and Cong F. (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., Michaud G. A., Schirle M., Shi X., Hild M., Bauer A., Myer V. E., Finan P. M., Porter J. A., Huang S. M., and Cong F. (2011) RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 13, 623–629 [DOI] [PubMed] [Google Scholar]
- 18. Smith S., Giriat I., Schmitt A., and de Lange T. (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487 [DOI] [PubMed] [Google Scholar]
- 19. Chang P., Coughlin M., and Mitchison T. J. (2005) Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 7, 1133–1139 [DOI] [PubMed] [Google Scholar]
- 20. Li N., Zhang Y., Han X., Liang K., Wang J., Feng L., Wang W., Songyang Z., Lin C., Yang L., Yu Y., and Chen J. (2015) Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev. 29, 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cho-Park P. F., and Steller H. (2013) Proteasome regulation by ADP-ribosylation. Cell 153, 614–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., Heckl D., Ebert B. L., Root D. E., Doench J. G., and Zhang F. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou Y., Wang Y., Tischfield M., Williams J., Smallwood P. M., Rattner A., Taketo M. M., and Nathans J. (2014) Canonical WNT signaling components in vascular development and barrier formation. J. Clin. Invest. 124, 3825–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang N., Bai H., David K. K., Dong J., Zheng Y., Cai J., Giovannini M., Liu P., Anders R. A., and Pan D. (2010) The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shimizu N., Smith G., and Izumo S. (1993) Both a ubiquitous factor mTEF-1 and a distinct muscle-specific factor bind to the M-CAT motif of the myosin heavy chain beta gene. Nucleic Acids Res. 21, 4103–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., and Piccolo S. (2011) Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 [DOI] [PubMed] [Google Scholar]
- 27. DaRosa P. A., Wang Z., Jiang X., Pruneda J. N., Cong F., Klevit R. E., and Xu W. (2015) Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 517, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levaot N., Voytyuk O., Dimitriou I., Sircoulomb F., Chandrakumar A., Deckert M., Krzyzanowski P. M., Scotter A., Gu S., Janmohamed S., Cong F., Simoncic P. D., Ueki Y., La Rose J., and Rottapel R. (2011) Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell 147, 1324–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shultz M. D., Cheung A. K., Kirby C. A., Firestone B., Fan J., Chen C. H., Chen Z., Chin D. N., Dipietro L., Fazal A., Feng Y., Fortin P. D., Gould T., Lagu B., Lei H., Lenoir F., Majumdar D., Ochala E., Palermo M. G., Pham L., Pu M., Smith T., Stams T., Tomlinson R. C., Touré B. B., Visser M., Wang R. M., Waters N. J., and Shao W. (2013) Identification of NVP-TNKS656: the use of structure-efficiency relationships to generate a highly potent, selective, and orally active tankyrase inhibitor. J. Med. Chem. 56, 6495–6511 [DOI] [PubMed] [Google Scholar]
- 30. Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C. W., Wei S., Hao W., Kilgore J., Williams N. S., Roth M. G., Amatruda J. F., Chen C., and Lum L. (2009) Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsiao S. J., and Smith S. (2008) Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 90, 83–92 [DOI] [PubMed] [Google Scholar]
- 32. Guettler S., LaRose J., Petsalaki E., Gish G., Scotter A., Pawson T., Rottapel R., and Sicheri F. (2011) Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell 147, 1340–1354 [DOI] [PubMed] [Google Scholar]
- 33. Morrone S., Cheng Z., Moon R. T., Cong F., and Xu W. (2012) Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc. Natl. Acad. Sci. U.S.A. 109, 1500–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Casás-Selves M., Kim J., Zhang Z., Helfrich B. A., Gao D., Porter C. C., Scarborough H. A., Bunn P. A. Jr., Chan D. C., Tan A. C., and DeGregori J. (2012) Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer Res. 72, 4154–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lehár J., Krueger A. S., Avery W., Heilbut A. M., Johansen L. M., Price E. R., Rickles R. J., Short G. F. 3rd, Staunton J. E., Jin X., Lee M. S., Zimmermann G. R., and Borisy A. A. (2009) Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 27, 659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu J., Pan S., Hsieh M. H., Ng N., Sun F., Wang T., Kasibhatla S., Schuller A. G., Li A. G., Cheng D., Li J., Tompkins C., Pferdekamper A., Steffy A., Cheng J., Kowal C., Phung V., Guo G., Wang Y., Graham M. P., Flynn S., Brenner J. C., Li C., Villarroel M. C., Schultz P. G., Wu X., McNamara P., Sellers W. R., Petruzzelli L., Boral A. L., Seidel H. M., McLaughlin M. E., Che J., Carey T. E., Vanasse G., and Harris J. L. (2013) Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl. Acad. Sci. U.S.A. 110, 20224–20229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang W., Li N., Li X., Tran M. K., Han X., and Chen J. (2015) Tankyrase Inhibitors Target YAP by Stabilizing Angiomotin Family Proteins. Cell Rep. 13, 524–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shalem O., Sanjana N. E., and Zhang F. (2015) High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16, 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parnas O., Jovanovic M., Eisenhaure T. M., Herbst R. H., Dixit A., Ye C. J., Przybylski D., Platt R. J., Tirosh I., Sanjana N. E., Shalem O., Satija R., Raychowdhury R., Mertins P., Carr S. A., Zhang F., Hacohen N., and Regev A. (2015) A Genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishimura M., Kakizaki M., Ono Y., Morimoto K., Takeuchi M., Inoue Y., Imai T., and Takai Y. (2002) JEAP, a novel component of tight junctions in exocrine cells. J. Biol. Chem. 277, 5583–5587 [DOI] [PubMed] [Google Scholar]
- 41. Patrie K. M. (2005) Identification and characterization of a novel tight junction-associated family of proteins that interacts with a WW domain of MAGI-1. Biochim. Biophys. Acta 1745, 131–144 [DOI] [PubMed] [Google Scholar]
- 42. Ernkvist M., Aase K., Ukomadu C., Wohlschlegel J., Blackman R., Veitonmäki N., Bratt A., Dutta A., and Holmgren L. (2006) p130-angiomotin associates to actin and controls endothelial cell shape. FEBS J. 273, 2000–2011 [DOI] [PubMed] [Google Scholar]
- 43. Li Y., Zhou H., Li F., Chan S. W., Lin Z., Wei Z., Yang Z., Guo F., Lim C. J., Xing W., Shen Y., Hong W., Long J., and Zhang M. (2015) Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway. Cell Res. 25, 801–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dai X., She P., Chi F., Feng Y., Liu H., Jin D., Zhao Y., Guo X., Jiang D., Guan K. L., Zhong T. P., and Zhao B. (2013) Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J. Biol. Chem. 288, 34041–34051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang C., An J., Zhang P., Xu C., Gao K., Wu D., Wang D., Yu H., Liu J. O., and Yu L. (2012) The Nedd4-like ubiquitin E3 ligases target angiomotin/p130 to ubiquitin-dependent degradation. Biochem. J. 444, 279–289 [DOI] [PubMed] [Google Scholar]
- 46. DeRan M., Yang J., Shen C. H., Peters E. C., Fitamant J., Chan P., Hsieh M., Zhu S., Asara J. M., Zheng B., Bardeesy N., Liu J., and Wu X. (2014) Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein. Cell Rep. 9, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Touil Y., Igoudjil W., Corvaisier M., Dessein A. F., Vandomme J., Monté D., Stechly L., Skrypek N., Langlois C., Grard G., Millet G., Leteurtre E., Dumont P., Truant S., Pruvot F. R., Hebbar M., Fan F., Ellis L. M., Formstecher P., Van Seuningen I., Gespach C., Polakowska R., and Huet G. (2014) Colon cancer cells escape 5FU chemotherapy-induced cell death by entering stemness and quiescence associated with the c-Yes/YAP axis. Clin. Cancer Res. 20, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kapoor A., Yao W., Ying H., Hua S., Liewen A., Wang Q., Zhong Y., Wu C. J., Sadanandam A., Hu B., Chang Q., Chu G. C., Al-Khalil R., Jiang S., Xia H., Fletcher-Sananikone E., Lim C., Horwitz G. I., Viale A., Pettazzoni P., Sanchez N., Wang H., Protopopov A., Zhang J., Heffernan T., Johnson R. L., Chin L., Wang Y. A., Draetta G., and DePinho R. A. (2014) Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158, 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shao D. D., Xue W., Krall E. B., Bhutkar A., Piccioni F., Wang X., Schinzel A. C., Sood S., Rosenbluh J., Kim J. W., Zwang Y., Roberts T. M., Root D. E., Jacks T., and Hahn W. C. (2014) KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schoumacher M., Hurov K. E., Lehár J., Yan-Neale Y., Mishina Y., Sonkin D., Korn J. M., Flemming D., Jones M. D., Antonakos B., Cooke V. G., Steiger J., Ledell J., Stump M. D., Sellers W. R., Danial N. N., and Shao W. (2014) Inhibiting Tankyrases sensitizes KRAS-mutant cancer cells to MEK inhibitors via FGFR2 feedback signaling. Cancer Res. 74, 3294–3305 [DOI] [PubMed] [Google Scholar]
- 51. Hoffman G. R., Rahal R., Buxton F., Xiang K., McAllister G., Frias E., Bagdasarian L., Huber J., Lindeman A., Chen D., Romero R., Ramadan N., Phadke T., Haas K., Jaskelioff M., Wilson B. G., Meyer M. J., Saenz-Vash V., Zhai H., Myer V. E., Porter J. A., Keen N., McLaughlin M. E., Mickanin C., Roberts C. W., Stegmeier F., and Jagani Z. (2014) Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl. Acad. Sci. U.S.A. 111, 3128–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.