Abstract
The tight, relative positioning of the nucleus and centrosome in mammalian cells is important for the regulation of cell migration. Under pathophysiological conditions, the purinergic A2b receptor can regulate cell motility, but the underlying mechanism remains unknown. Expression of A2b, normally low, is increased in tissues experiencing adverse physiological conditions, including hypoxia and inflammation. ATP is released from such cells. We investigated whether extracellular cues can regulate centrosome-nucleus positioning and cell migration. We discovered that hypoxia as well as extracellular ATP cause a reversible increase in the distance between the centrosome and nucleus and reduced cell motility. We uncovered the underlying pathway: both treatments act through the A2b receptor and specifically activate the Epac1/RapGef3 pathway. We show that cells lacking A2b do not respond in this manner to hypoxia or ATP but transfection of A2b restores this response, that Epac1 is critically involved, and that Rap1B is important for the relative positioning of the centrosome and nucleus. Our results represent, to our knowledge, the first report demonstrating that pathophysiological conditions can impact the distance between the centrosome and nucleus. Furthermore, we identify the A2b receptor as a central player in this process.
Keywords: ATP, cell migration, guanine nucleotide exchange factor (GEF), hypoxia, microtubule, Ras-related protein 1 (Rap1)
Introduction
In mammalian cells, the centriole organizes pericentriolar material, forming the centrosome that nucleates polar arrays of microtubules. In non-cycling cells, the centriole functions as a basal body and generates the primary cilium. In most mammalian cells, the centrosome is in the center of the cell, in close proximity to the nucleus at all interphase stages, only to separate at the onset of mitosis to establish the poles of the mitotic spindle. The close proximity of the centrosome and nucleus in interphase stages appears to be conserved from unicellular organisms, including fungi, where the spindle pole body is the centrosome equivalent embedded in the nuclear membrane (1), chrysomonads and other flagellates, where a prominent microfibril structure connects the basal body to the nucleus (2), to Caenorhabditis elegans, where microtubules, together with Sun1 and ZYG-12 protein, are required for positioning of the two (3). In mammalian cells, microtubules that are nucleated from the centrosome play a role in the establishment of the relative positioning, as disruption by microtubule-depolymerizing drugs causes an increase in the distance between the centrosome and nucleus (4, 5). Coupling microtubules to the nucleus requires proteins, including Lis1, NudE/NudEL, Ndel1, DCX, the dynein/dynactin complex, LaminB, Nup133, Nup358/RanBP2, BICD2, Asunder, CenpF, the SUN domain proteins SUN1 and SUN2, and the KASH domain proteins Syne-1/Nesprin-1 and Syne-2/Nesprin-2 (4, 6–14). Ablation of their function or down-regulation of their expression result in an increased distance between the two organelles. The tight positioning of the two organelles is required during development. In Drosophila, eye development requires sequential co-migration of the two organelles during cell differentiation (15). Disruption of the positioning causes abnormal eye formation (16–18). Also in Drosophila, loss of the tight positioning is correlated with defective embryonic development (19, 20). In humans, the brain formation disorder lissencephaly is caused by defective neuronal migration during gestation, resulting in a lack of development of brain folds and grooves (21). Several studies have indicated that, in cultured neuronal cells, RNAi-mediated down-regulation of lissencephaly-associated genes can induce an increase in the distance between the centrosome and nucleus, causing defective nuclear translocation and retarded cell migration (4–6). Despite the progresses made in our understanding of the positioning of the centrosome and nucleus and its importance in cell migration, there is still the question of whether positioning can be regulated by extracellular cues. It is possible that, under certain circumstances, e.g. when adverse, extreme conditions are met, temporary separation and, consequently, retarded cell migration may be of overall benefit to the organism. We set out to discover whether such signaling pathways exist and focused on the purinergic receptor A2b for the following reasons. The level of expression of the purinergic A2b receptor is normally low but increases in response to adverse conditions, including necrosis, ischemia, hypoxia, and inflammation (22, 23). ATP is released from damaged or dying cells, in ischemia (24), and in response to gentle mechanical disturbance or hypoxia (25). A2b is activated by extracellular ATP and adenosine (26). Elevated A2b is believed to assist tissues in coping with the extreme condition. Indeed, although A2b receptor knockout mice are viable and fertile (27), organs of A2b knockout mice, including the heart, liver, lung, intestine, brain, and kidney, display increased susceptibility to ischemic and inflammatory injury (28–34). Here we discovered a specific pathway that is activated through the purinergic receptor A2b by either hypoxia or extracellular ATP, triggering a cascade of events culminating in Epac1 and Rap1B activation and movement of the nucleus away from the centrosome. The end result is reduced cell migration.
Results
ATP Affects Cell Migration and Causes an Increase in the Distance between the Centrosome and Nucleus
ATP is released into the extracellular milieu under pathological conditions from damaged cells, potentially acting as an extracellular signaling molecule (25, 35). During injury, released ATP stimulates purinergic receptors, altering cell migration and impacting wound repair (36). To mimic this adverse condition, we first tested the effect of ATP on the migration of two cell types, human retinal epithelial pigment (RPE)3 cells and human foreskin fibroblasts (HS68) using the cell scratch damage assay (37). The results (Fig. 1) show that ATP had no effect on the migration of HS68 cells but significantly reduced RPE cell migration in the scratch assay (Fig. 1b). The inhibition was reversible: removal of ATP from the culture medium allowed RPE cell migration to resume. Other nucleotides had no effect on RPE cell migration (data not shown). To confirm this result, we tested the effect of ATP on cell migration of individual cells using the Transwell® system (38). In these assays, ∼500 untreated cells migrated consistently through the membrane during the test period (1.5 days), whereas fewer than five ATP-treated cells migrated through the membrane over this period, confirming the scratch cell assay results.
FIGURE 1.
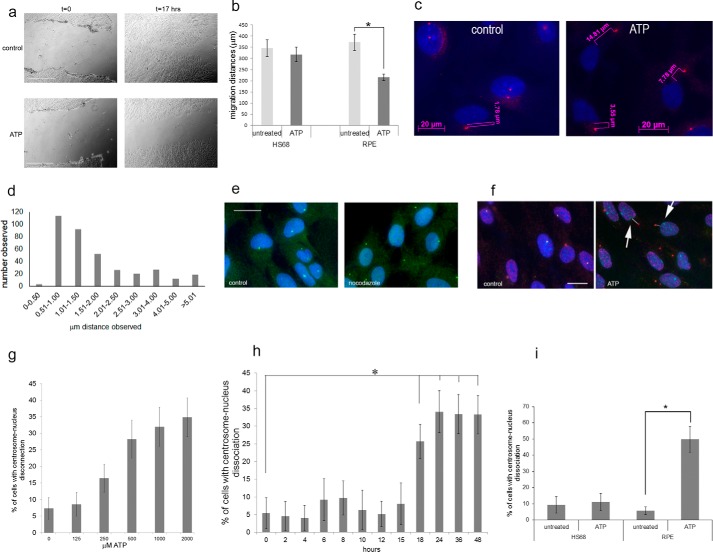
Extracellular ATP inhibits cell migration and causes centrosome-nucleus separation. The effect of ATP on cell migration was studied using the scratch assay. a, examples of phase-contrast images of scratch-damaged monolayer RPE cells immediately and 17 h after the scratch with and without 2 mm ATP treatment. Scale bars = 500 μm. b, quantitation of results from eight scratch assays for each cell line. c, we analyzed centrosome-nucleus separation in untreated RPE cell cultures (control) and in cells treated with ATP (ATP) as follows. Cells were fixed and stained with an antibody to ninein to detect the centrosome (pink) and DAPI to stain the nucleus (blue). Images were obtained with immunofluorescence microscopy. We analyzed centrosome-nucleus distances as shown using Axiovert software. d, the distance between the centrosome and nucleus was measured in RPE cells growing under normal conditions. Data were collected from 360 cells. The distribution of distances was analyzed and is shown. e, nocodazole causes centrosome-nucleus separation. Control, untreated RPE cells; nocodazole, RPE cells treated with 0.5 μm nocodazole for 3 h. Anti-pericentrin was used to show centrosomes, and anti-β-tubulin was used to reveal microtubules. DAPI stains nuclei. Images were obtained with immunofluorescence microscopy. Scale bar = 20 μm. f, RPE cells were analyzed without treatment (control) or after treatment with 2 mm ATP (ATP) for 24 h, fixed, and stained with human autoimmune serum (M4491) and an antibody to ninein, both to visualize the centrosome, and with DAPI to stain nuclei. Images were obtained with immunofluorescence microscopy. Arrows point to examples of cells with distanced centrosomes and nuclei, schematically indicated by the white lines. Distances were measured. Scale bar = 20 μm. g, results of ATP dose-response experiments. RPE cells were treated with the indicated concentrations of ATP for 16 h. Data were collected from three independent assays, for each of which the centrosome-nucleus distance was measured in 300 cells. h, ATP treatment time course. RPE cells were treated with 1 mm ATP for the indicated times, and centrosome-nucleus distances were measured. Data were collected from three independent assays, for each of which at least 300 cells were analyzed. i, HS68 cells and RPE cells were analyzed untreated or after treatment with 1 mm ATP, and distances between the centrosome and nucleus were measured. Data were collected from three independent assays, for each of which at least 300 cells were counted. *, p < 0.05.
We next examined the position of the centrosome and nucleus in ATP-treated RPE cells compared with untreated cells. We first had to establish the distribution of distances between the two organelles in RPE cells under normal culture conditions. As expected, the centrosome and nucleus were in close proximity in the majority of RPE cells (Fig. 1c, control). Examples of such measurements are shown in Fig. 1c. The range observed (Fig. 1d) was between 0.32 and 13.12 μm, with a median of 1.30 μm and an average of 1.91 μm. Our analysis revealed that the distance between the centrosome and nucleus was smaller than 2.15 μm in >95% of cells analyzed. In subsequent experiments, we therefore took an observed distance of greater than 2.15 μm as indicative of enhanced separation between the two organelles. This cutoff was confirmed as an appropriate criterion by measuring centrosome-nucleus distances in RPE cells treated with nocodazole, which causes the nucleus to move away from the centrosome (3, 4, 39, 40). The results (Fig. 1e shows examples) indicate that, in 47% of nocodazole-treated cells, the distance between the two organelles was >2.8 μm.
Next, we analyzed the centrosome-nucleus distance in RPE cells treated for 24 h with 2 mm ATP, which caused an increased distance between the centrosome and nucleus (Fig. 1, c, ATP, and f). The average distance observed for the two organelles was 6.94 μm. ATP-induced centrosome-nucleus separation showed a dose-response effect (Fig. 1g) and was observed with 250 μm. ATP-mediated centrosome-nucleus separation is not restricted to a particular cell cycle stage and reversible. A time course experiment showed that treatment with 1 mm ATP results in significant centrosome-nucleus separation after 18 h (Fig. 1h). To visualize the effect of ATP, we recorded live-cell time-lapse images every 15 min for 16 h using HeLa cells expressing mCherry-H2B, an GFP-centrin. supplemental Movie 1 shows the results. It can be observed that, although the centrosome continues to reside in the center of the cell, the nucleus moves away as a result of ATP treatment. In contrast to RPE cells, ATP does not affect the distance between the centrosome and nucleus in HS68 cells (Fig. 1i). Finally, we also investigated a possible effect of ATP on the cell cycle. There was no significant difference between untreated and ATP-treated RPE cells. In untreated cells, 70.2% were in G1, 9.4% in G2, and 20.4% in S phase, whereas, for ATP-treated cells, 70.7% were in G1, 9.5% in G2, and 18.8% in S phase. Together, these results are the first demonstration that ATP can reversibly affect the distance between the centrosome and nucleus and cause reduced cell migration.
Adenosine Receptor A2b Mediates ATP-induced Centrosome and Nucleus Separation
Other triphosphate nucleotides did not cause separation (data not shown). In contrast, ADP, AMP, and the non-hydrolyzable ATP analog PNP-AMP cause centrosome-nucleus separation (Fig. 2, a and b). Indeed, adenosine also causes centrosome-nucleus separation (Fig. 2c). Adenosine-mediated separation can be detected as early as 3 h after treatment (Fig. 2d). A dose-response experiment is shown in Fig. 2e. These data suggest that one of the adenosine receptors mediates ATP/adenosine-induced centrosome-nucleus separation.
FIGURE 2.
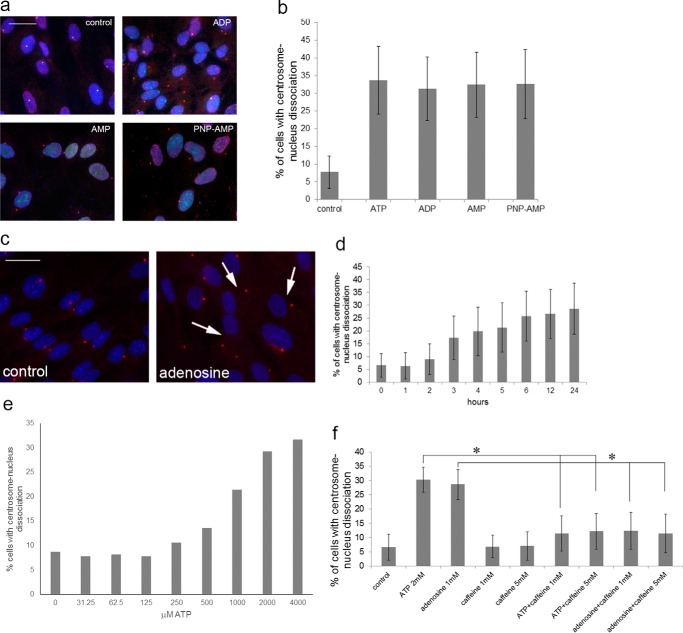
ATP, ADP, AMP, and adenosine induce centrosome-nucleus separation. a, RPE cells were left untreated or treated for 24 h with 2 mm ADP, AMP, or the non-hydrolyzable analog PNP-AMP, fixed, and analyzed as described to visualize the centrosome and nucleus. Control, untreated RPE cells. Scale bar = 20 μm. b, quantitation of the results shown in a. Data were collected from three independent assays, for each of which 300 cells were analyzed. c, RPE cells were treated with 2 mm adenosine for 18 h and analyzed as above. Control, untreated cells. Arrows point to examples of cells with distanced centrosomes and nuclei. Scale bar = 20 μm. d, RPE cells were treated with 2 mm adenosine for the indicated times. Data were collected from three independent assays, for each of which 300 cells were analyzed. e, adenosine dose-response experiment. Data were collected from three independent assays, for each of which 300 cells were analyzed. f, RPE cells were treated with caffeine alone or in combination with ATP or adenosine at the indicated concentrations. Centrosome-nucleus separation was measured as above and quantitated. Data were collected from three independent assays, for each of which 300 cells were analyzed. *, p < 0.05.
Four adenosine receptors, which belong to the P1 class of purinergic receptors, had been described, viz. A1, A2a, A2b, and A3. Caffeine is a non-selective antagonist (41), and we tested its effect first. Caffeine by itself did not affect the position of the centrosome and nucleus, but caffeine efficiently abrogated ATP and adenosine-induced separation (Fig. 2f). To identify the adenosine receptor responsible for the ATP effect, we used specific chemical inhibitors for the adenosine receptors (PSB36 for A1 (42), SCH58261 for A2a (43), MRS1754 for A2b (44), and MRS1523 for A3 (45)). The results are shown in Fig. 3a. Inhibitors alone had no effect on the position of the centrosome and nucleus. In combination with ATP, only the A2b inhibitors MRS1754 and PSB1115 (46) completely abrogated ATP-mediated centrosome-nucleus separation (Fig. 3a).
FIGURE 3.
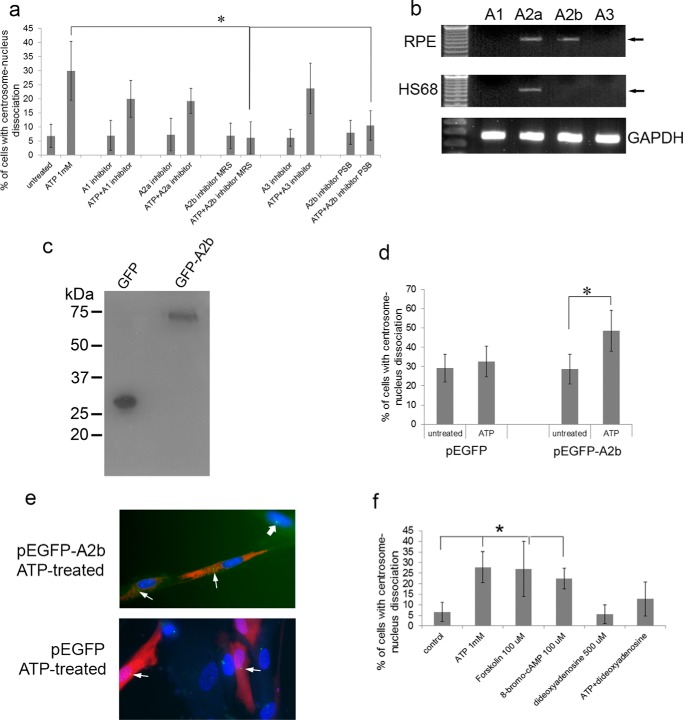
The A2b receptor is required for ATP induced centrosome-nucleus separation. Our data suggested that one of the P1 purinergic adenosine receptors is involved in mediating the observed ATP effect. a, RPE cells were treated for 18 h with the indicated compounds alone or in combination with 1 mm ATP. The concentrations used for the inhibitors were as follows: A1 inhibitor, 40 μm PSB36; A2a inhibitor, 20 μm SCH58261; A2b inhibitor MRS, 40 μm MRS1754; A2b inhibitor PSB, 40 μm PSB1115; and A3 inhibitor, 40 μm MRS1523. Untreated and treated cells were analyzed for centrosome-nucleus separation as described above. Data were collected from three independent assays, for each of which 300 cells were analyzed. b, RT-PCR analysis of A1, A2a, A2b, and A3 mRNA expression in RPE and HS68 cells. c, Western blotting analysis of HS68 cells, which lack A2b, transfected with pEGFP (as a control) and with pEGFP-A2b expressing a GFP-A2b fusion protein. d, HS68 cells transfected with pEGFP or with pEGFP-A2b were left untreated or treated with 2 mm ATP for 17 h, and the distance between the centrosome and nucleus was analyzed as described above. Data were collected from three independent assays, for each of which 300 cells were analyzed. e, example of HS68 cells transfected with EGFP or EGFP-A2b and treated with ATP. Thin arrows point to centrosomes in transfected cells, and the thick arrow points to a centrosome in an untransfected cell. f, RPE cells were treated with two adenylate cyclase activators (100 μm forskolin, 100 μm 8-bromo-cAMP) or with an adenylate cyclase inhibitor (500 μm dideoxyadenosine). ATP alone was used as a control for separation. Untreated and treated cells were analyzed for centrosome-nucleus separation as described above. Data were collected from three independent assays, for each of which 300 cells were analyzed. *, p < 0.05.
To prove that the A2b receptor is critically involved in the observed ATP effect, we carried out the following experiments. We first analyzed adenosine receptor mRNA expression in RPE cells and HS68 cells by RT-PCR. The results (Fig. 3b) show that the adenosine receptors A1 and A3 are not expressed in RPE and HS68 cells and that A2a mRNA expression was detected in both. In contrast, A2b mRNA was detected in RPE cells, not in HS68 cells. This suggested that A2b is required for the observed ATP-induced centrosome-nucleus separation. To support this, HS68 cells were transfected with pEGFP-A2b and pEGFP and treated with ATP. Western blotting analysis showed production of both GFP and GFP-A2b proteins (Fig. 3c). Without ATP, pEGFP-A2b did not affect the position of the centrosome and nucleus (Fig. 3d). However, pEGFP-A2b-transfected and ATP-treated HS68 cells showed centrosome-nucleus separation (Fig. 3, d and e), demonstrating a critical role for the adenosine A2b receptor in the ATP effect.
A2b associates with the Gs subunit of heterotrimeric G protein. Upon A2b activation Gs activates adenylate cyclase, producing cAMP. To explore whether adenylate cyclase is involved in A2b-mediated centrosome-nucleus separation, we treated RPE cells with adenylate cyclase activators (forskolin and 8-bromo-cAMP) and an inhibitor (dideoxyadenosine). As shown in Fig. 3f, forskolin and 8-bromo-cAMP promote an increase in the distance between the centrosome and nucleus, but dideoxyadenosine had no effect. However, dideoxyadenosine efficiently reduced ATP-induced separation (Fig. 3f), showing that adenylate cyclase is the downstream effector.
PKA Does Not Mediate ATP-induced Centrosome-Nucleus Separation
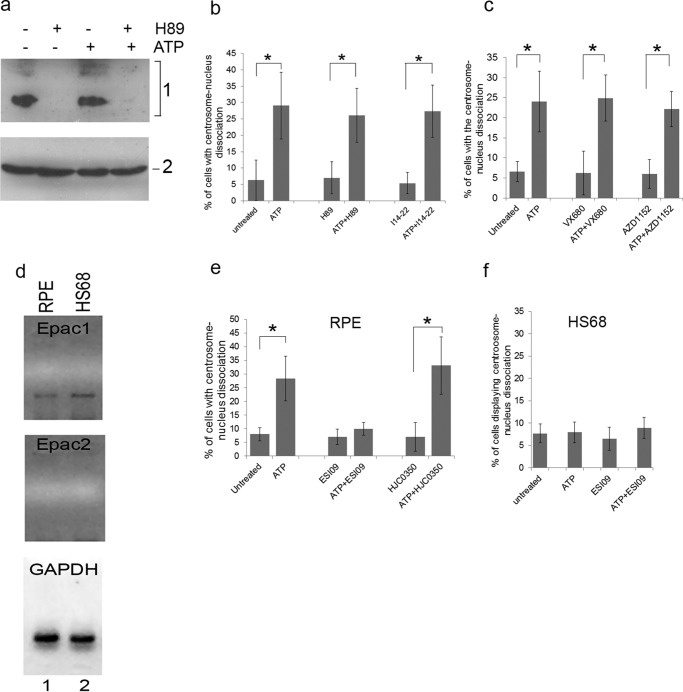
cAMP has been shown to activate two pathways: the cAMP-dependent protein kinase (PKA) pathway, and the exchange protein directly activated by cAMP (Epac) pathway. We tested whether PKA mediates ATP-induced centrosome-nucleus separation using the PKA inhibitors H89 and I14–22 (a plasma membrane-permeable peptide inhibitor). No changes in the overall phosphorylation profiles of PKA substrates with or without ATP treatment were observed by Western blotting (Fig. 4a). PKA inhibitors significantly reduced the levels of PKA phosphosubstrates as expected (Fig. 4a). Neither of the PKA inhibitors exhibited any effect on the positioning of the centrosome and nucleus (Fig. 4b). Importantly, they were unable to block ATP-induced centrosome-nucleus separation (Fig. 4b). To further rule out PKA involvement, we analyzed mitotic kinase Aurora A, a PKA phosphosubstrate (47). Aurora activates Polo-like kinase 1 (Plk-1) (48), which plays a role in controlling centrosome-nucleus separation at the onset of mitosis (19). To analyze the involvement of Aurora in the ATP effect, RPE cells were treated with two Aurora kinase inhibitors: VX-680 with a high potency toward Aurora A and AZD-1152, a selective inhibitor of Aurora kinase B. These inhibitors did not cause centrosome-nucleus separation and did not interfere with ATP-induced centrosome-nucleus separation (Fig. 4c). We conclude that PKA is not involved in the ATP effect.
FIGURE 4.
Epac1 regulates the close positioning of centrosome and nucleus. To distinguish a role for PKA or Epac1 in the observed ATP effect, we treated RPE cells with two different PKA inhibitors: the chemical compound H89 (10 μm) and the plasma membrane-permeable peptide inhibitor I14–22 (10 μm) either alone or in combination with 2 mm ATP. a, top panel, Western blot of RPE cells treated with ATP and/or H89 using anti-PKA phosphorylated substrate antibodies. Bottom panel, Western blot using anti-tubulin antibodies as a loading control. b, RPE cells were treated with the indicated inhibitors with or without ATP, and the distance between the centrosome and nucleus was measured as described above. Data were collected from three independent assays, for each of which 200 cells were analyzed. c, we also tested a possible role for Aurora kinase, which is activated by PKA and involved in centrosome separation normally seen in mitotic diving cells. RPE cells were treated for 17 h with two different Aurora kinase activity inhibitors, VX680 (1 μm) and AZD1152 (10 μm), either alone or in combination with 1 mm ATP. Untreated and treated cells were analyzed for centrosome-nucleus separation as described above. Data were collected from three independent assays, for each of which 300 cells were analyzed. d, we analyzed a possible involvement of Epac in the observed ATP effect. First, we analyzed Epac 1 and Epac 2 mRNA expression in RPE cells (lane 1) and HS68 cells (lane 2) by RT-PCR. GAPDH served as a positive control. e, we tested a functional role for Epac1 as follows. RPE cells were treated for 17 h with 2.5 μm of ESI09 (an inhibitor of both Epac1 and Epac2) or with 10 μm of HJC0350 (which specifically inhibits only Epac2) either alone or in combination with 1 mm ATP. Untreated and treated cells were analyzed for centrosome-nucleus separation as described above. Data were collected from three independent assays, for each of which 250 cells were analyzed. f, we tested whether the Epac inhibitor ESI09 caused any effect in HS68 cells, which lack the A2b receptor but express Epac1. Cells were treated as above and analyzed in the same fashion. *, p < 0.05.
Epac1 Mediates ATP-induced Centrosome-Nucleus Separation
We next investigated a role for Epac, a cAMP-regulated guanine nucleotide exchange factor (49). In mammals, two Epac isoforms, Epac1 and Epac2, are encoded by two distinct genes, RAPGEF3 and RAPGEF4 (50). We determined Epac mRNA expression in RPE and HS68 cells using RT-PCR. Epac1 mRNA is expressed in both cell types (Fig. 4d), whereas Epac2 mRNA was not detectable (Fig. 4d). We next used Epac1 and Epac2 inhibitors. ESI09 is an inhibitor of both Epac isoforms, with an IC50 of 3.2 μm and 1.4 μm for Epac1 and Epac2, respectively (51). At 2.5 μm, ESI09 did not affect the positioning of the centrosome and nucleus in RPE cells (Fig. 4e). However, ESI09 completely abrogated ATP-induced centrosome-nucleus separation (Fig. 4e). ESI09 had no effect on the position of the centrosome and nucleus in untreated and ATP-treated HS68 cells (Fig. 4f). The Epac2 inhibitor HJC0350 did not affect ATP-induced separation (Fig. 4e) as expected because Epac2 is not detectably expressed.
Epac1 localizes to the cytosol and nuclear membrane and translocates to the cell membrane after cAMP activation (52). We show that Epac1 regulates a centrosomal process and thus expected Epac1 to also localize to the centrosome. This was examined before and after ATP treatment using the GFP-Epac1 expression vector typically used in Epac1 localization studies (52). We observed GFP-Epac1 in the cytosol and on the nuclear membrane, as expected, and, importantly, also on the centrosome (Fig. 5, a–c). ATP treatment not only caused a translocation to the plasma membrane, but GFP-Epac1 was also no longer detectable at the centrosome (Fig. 5, d and e). The Epac1 inhibitor ESI09 has no effect on GFP-Epac1 localization in untreated cells (Fig. 5, g–i). However, ESI09, which abrogates ATP-induced centrosome-nucleus separation, blocks translocation of GFP-Epac1 after ATP treatment, which remains localized at the centrosome (Fig. 5, j–l). We conclude that Epac1 is a centrosomal protein and that it mediates ATP-induced centrosome-nucleus separation.
FIGURE 5.
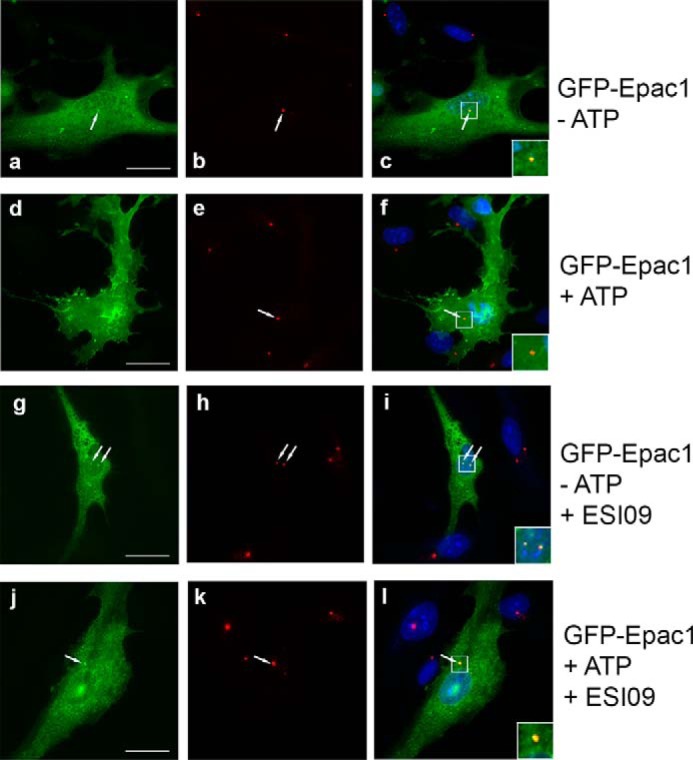
Centrosomal localization of Epac1 is lost upon ATP treatment. To analyze Epac1 localization, we employed GFP-Epac1 (a, d, g, and j), which had been used extensively for localization studies. b, e, h, and k show pericentrin on the centrosome. c, f, i, and l show merged images. a–c, localization of Epac1 in the cytosol, on the nuclear membrane, and on the centrosome (arrow) in untreated cells. Inset, an enlarged image of the centrosome (yellow). d–f, ATP treatment causes significant translocation of Epac1 to the plasma membrane, and Epac1 can no longer be detected at the centrosome (the inset shows a red centrosome). g–i, without ATP, Epac1 localization at the centrosome is not affected by the Epac1 inhibitor ESI09 (the inset shows yellow centrosomes). j–l, the Epac1 inhibitor ESI09 prevents loss of Epac1 at the centrosome (the inset shows a yellow centrosome) and blocks translocation to the plasma membrane. Scale bar = 20 μm.
Rap1B Is Required for Centrosome-Nucleus Positioning
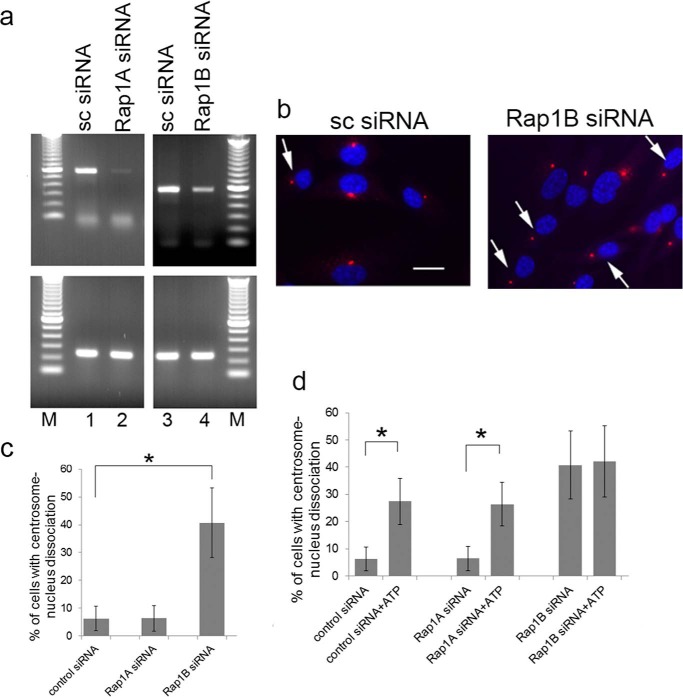
The GTPase Rap 1 is a cytosolic protein whose GTP/GDP-bound state is regulated by cAMP. Epac promotes exchange from GDP to GTP, thereby activating Rap1. Two isoforms, Rap1A and Rap1B, are encoded by separate genes. To determine whether Rap1 may be involved in ATP-mediated centrosome-nucleus separation, we analyzed Rap1A and Rap1B mRNA expression in RPE cells under control conditions (cells treated with a scrambled siRNA) and after knockdown using specific siRNAs. Rap1A and Rap1B are both expressed in RPE cells (Fig. 6a, lanes 1 and 3, respectively). Knockdown using specific Rap1A siRNA and Rap1B siRNA resulted in a reduction of Rap1A and Rap1B expression to 5% and 46%, respectively (Fig. 6a, lanes 2 and 4). Knockdown of Rap1B but not Rap1A caused an increase in the distance between the centrosome and nucleus, whereas a scrambled siRNA control had no effect (Fig. 6, b and c). Quantitation shows that this increase is significant and to a level similar to that observed for cells treated with ATP (Fig. 6c). RPE cells treated with siRNA against Rap1A or a scrambled siRNA are still responsive to the effect of ATP on centrosome-nucleus linkage (Fig. 6d), demonstrating that Rap1A is not involved. There was no measurable additive or synergistic effect for Rap1B siRNA and ATP treatments. Our results suggest that Rap1B is the effector protein of ATP-induced centrosome-nucleus separation.
FIGURE 6.
Rap1B activation is involved in relative positioning of centrosome and nucleus. a, a possible involvement of Rap1 protein(s) in the ATP effect mediated by Epac1 was analyzed as follows. We first determined mRNA expression in RPE cells of the two isoforms Rap1A and Rap1B and the level of knockdown achieved using specific siRNA using RT-PCR. GAPDH was used as a positive control. Lanes 1 and 2, Rap1A expression in cells treated with 25 nm scrambled siRNA (sc siRNA) or 25 nm Rap1A-specific siRNA. Lanes 3 and 4, Rap1B expression in cells treated with 25 nm scrambled siRNA or 25 nm Rap1B-specific siRNA. M, molecular mass marker lane. b, the requirement for Rap1 protein in the ATP effect was analyzed using siRNA. Shown are examples of scrambled siRNA-treated RPE cells and Rap1B siRNA-treated RPE cells (Rap1B siRNA). Images were obtained as described above. Arrows point to examples of cells with separated centrosomes and nuclei. Scale bar = 20 μm. c, quantitation of centrosome-nucleus separation data from experiments of siRNA-treated RPE cells. Data were collected from three independent assays, for each of which 300 cells were analyzed. d, a possible effect of siRNA treatment on ATP-induced centrosome-nucleus separation was analyzed. RPE cells were treated with the indicated siRNA alone or with the indicated siRNA plus 1 mm ATP. Data were collected from three independent assays, for each of which 300 cells were analyzed. *, p < 0.05.
Hypoxia Induces Centrosome-Nucleus Separation and Reduced Cell Migration via the A2b-Epac1 Pathway
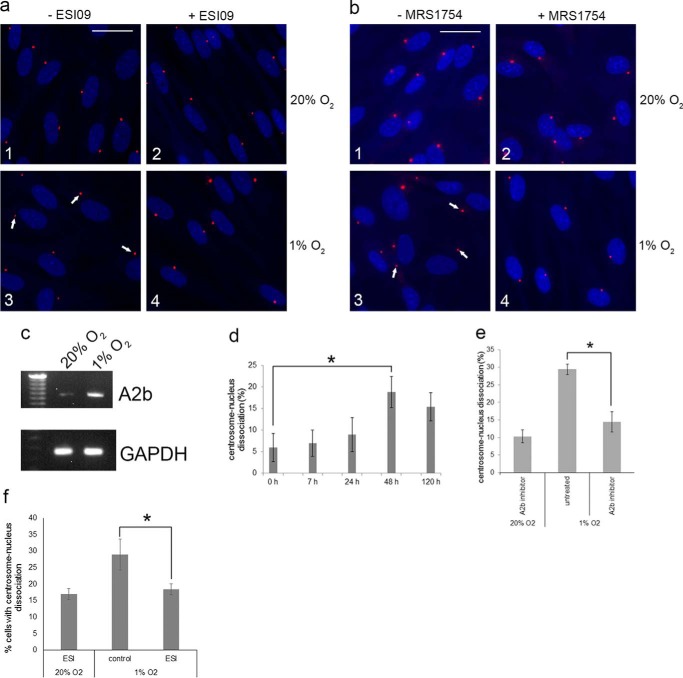
As described above, it was reported that A2b expression is up-regulated under adverse conditions, including hypoxia, ischemia, inflammation, cell injury, and cell stress (22, 23). To analyze whether A2b activation by hypoxia also affects the positioning of the centrosome and nucleus, we carried out the following experiments. First, we analyzed whether hypoxia (1% O2) had any effect on the distance between the centrosome and nucleus in RPE cells. The results show that RPE cells grown in 1% O2 exhibit centrosome-nucleus separation in comparison with cells grown in 20% O2 (Fig. 7, a and b, 1 and 3 show examples). We next analyzed whether hypoxia induced A2b mRNA expression, which, as expected, it did (Fig. 7c). A time course and quantitation of data show significant centrosome-nucleus separation after 48 h under 1% O2 conditions (Fig. 7d). We then investigated the pathway activated by hypoxia that results in centrosome-nucleus separation and reduced cell motility. Cells grown under normoxic and hypoxic conditions were left untreated or treated with the A2b inhibitor MRS1754 or the Epac1 inhibitor ESI09. Both ESI09 and MRS1745 block hypoxia-induced centrosome-nucleus separation (Fig. 7, a and b, 3 and 4 show examples). Quantitation of data indicated that the A2b inhibitor (Fig. 7e) and Epac1 inhibitor (Fig. 7f) efficiently blocked hypoxia-induced centrosome-nucleus separation.
FIGURE 7.
Hypoxia causes increased distance between the centrosome and nucleus and activation of the A2b-Epac1 pathway. We analyzed the effect of hypoxia on RPE cells and the possible role of the A2b-Epac1 pathway. a, in comparison with normoxic conditions (1), hypoxia (1% O2) causes centrosome-nucleus separation (3, arrows point to examples). The Epac1 inhibitor ESI09 had no effect under normoxic conditions (2) but blocked the hypoxia-induced separation (4). b, the A2b inhibitor MRS1754 had no effect under normoxic conditions (1 and 2) but blocked hypoxia-induced centrosome-nucleus separation (3 and 4). c, RT-PCR analysis of A2b mRNA expression in normoxic and hypoxic RPE cells. d, time course experiments of hypoxia-induced centrosome-nucleus separation. Data were collected from three independent assays, for each of which 250 cells were analyzed. e, quantitation of data obtained in the experiments shown in b using the A2b inhibitor MRS1754. Data were collected from three independent assays, for each of which 300 cells were analyzed. f, quantitation of data obtained in the experiments shown in a using the Epac1 inhibitor ESI09. Data were collected from three independent assays, for each of which 300 cells were analyzed. *, p < 0.05.
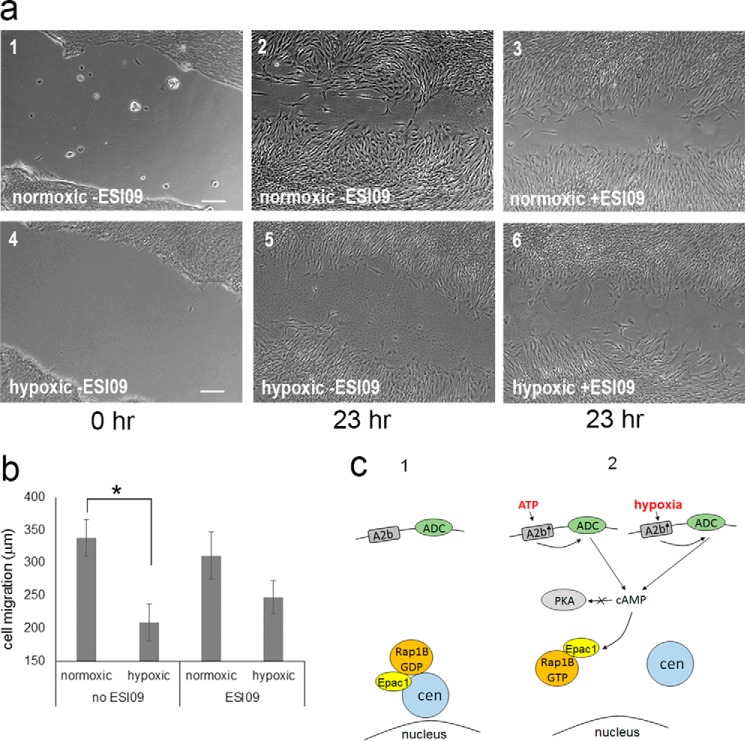
Our results predicted that RPE cells grown under hypoxic conditions should show reduced migration comparable with RPE cells treated with ATP and cultured under normoxic conditions. To measure the effect of hypoxia on cell migration, the cell scratch assay was carried out under normoxic and hypoxic conditions. The results (Fig. 8) show reduced cell migration under hypoxic conditions compared with normoxic conditions (Fig. 8a, 2 and 5 show examples). Quantitation demonstrated that the reduction in migration is significant (Fig. 8b). We next analyzed the effect of the Epac1 inhibitor ESI09 on cell migration under both oxygen level conditions. ESI09 had no effect under normoxic conditions (Fig. 8a, panels 2 and 3). However, inhibition of Epac1 by ESI09 consistently caused a partial reversion of hypoxia-induced cell migration block (Fig. 8a, 5 and 6).
FIGURE 8.
Hypoxia reduces cell migration via the A2b-Epac1 pathway. a, RPE cells were grown under normal conditions (normoxic, 1–3) and reduced oxygen conditions (hypoxic, 4–6). After 48 h, the cell scratch assay was performed, and cells were observed immediately (1 and 4) and after 23 h (2, 3, 5, and 6). Cell migration was measured for untreated cultures (−ESI09; 1, 2, 4, and 5), and cultures were treated with the Epac1 inhibitor ESI09 (3 and 6). b, quantitation of data obtained in scratch assays as shown in a. Data were collected from four independent assays, for each of which 20 distance points were analyzed. c, a model comparing the normal state (1) and the activation of the A2b-Epac1 pathway (2) for ATP treatment and hypoxia. Indicated are the critical players A2b, Epac1, and Rap1B (GDP or GTP-bound). ADC, adenylyl cyclase; cen, centrosome. *, p < 0.05.
Discussion
It has been well documented that pathophysiological conditions activate the purinergic A2b receptor, an event believed to be important in protecting tissues from significant damage. We report here that activation of the purinergic A2b receptor (under pathophysiological conditions) induces an increase in the distance between the centrosome and nucleus and impaired cell migration. We uncovered the responsible signaling pathway starting at the A2b receptor, which stimulates adenylate cyclase and results in Epac1 activation. Disruption of Rap1B expression also causes centrosome-nucleus separation. Thus we demonstrate for the first time that an extracellular cue can affect the relative positioning of the centrosome and nucleus.
Adenosine Receptor A2b Activation
We provide evidence that the A2b receptor, not other P1 purinergic receptors, is critically involved in hypoxia- and ATP-induced centrosome-nucleus separation. This is interesting in light of the known expression patterns and activities of P1 purinergic receptors where A2b is generally linked to pathophysiological conditions. The A2b receptor displays unique features. Although widely expressed, its expression level is normally very low. A2b expression is increased under pathologic conditions, including hypoxia, ischemia, inflammation, cell injury, and cell stress (22, 23). We found that the A2b receptor is dispensable for maintaining the normal positioning of the centrosome and nucleus because positioning is normal in HS68 cells lacking A2b. However, A2b activity is stimulated by high levels of extracellular ATP or adenosine (26), and we show that this results in centrosome-nucleus separation and reduced cell migration. In contrast, activation of A1, A2a, and A3 receptors requires low levels of adenosine. ATP can be released from damaged or dying cells, in ischemia (24), and in response to gentle mechanical disturbance or hypoxia (25). In brain ischemia, the local extracellular adenosine concentration increases, mainly by degradation of ATP by ecto-nucleotidase (53). Here we found that activation of the A2b receptor causing centrosome-nucleus separation requires 250 μm of either ATP or adenosine. Levels of extracellular ATP in damaged tissues have not been accurately measured but are described as significantly elevated (54, 55), ranging from 50–250 μm (56)
It is believed that A2b activation is protective. A2b receptor knockout mice are viable and fertile (27). However, the organs of A2b knockout mice, including the heart, liver, lung, intestine, brain, and kidney, display increased susceptibility to ischemic and inflammatory injury (28–34). It is tempting to speculate that the absence of A2b in A2b knockout mice prevents centrosome-nucleus separation under adverse conditions and an inability to modulate cell migration, which may contribute to the observed organ injury. Indeed, it was shown that adenosine or A2b receptor agonists attenuate cell migration of polymorphonuclear neutrophils (57).
Epac and Rap1B
Our data show that the hypoxia- and ATP-induced increased distance between the centrosome and nucleus involves the Epac-Rap1 pathway. Interestingly, we found that, under normal conditions, Epac1 activity is not required for maintaining the relative positioning of the centrosome and nucleus: inhibition of Epac1 activity by ESI09 does not result in centrosome-nucleus separation. This suggests that Epac1 activity becomes important only under adverse conditions, e.g. higher levels of extracellular ATP or hypoxia, that activate the A2b pathway. Under these conditions, we show that inhibition of Epac activity abolished centrosome-nucleus separation. In addition, we observed that Epac1 inhibition in part reverses the reduction in cell migration induced by hypoxic conditions.
The regulation of Epac activation is complex and largely unknown. Stimulation of the β-adrenergic receptor by isoproterenol or epinephrine causes production of cAMP, which activates Epac, not PKA (58, 59). Compartmentalization in local microdomains was shown for cAMP and PKA (60, 61) and may be involved in Epac activation. It should be noted that activation of PKA activity by adenosine is possible. The adenosine agonist 5′-N-ethylcarboxamidoadenosine causes phosphorylation and delayed prenylation of Rap1B (62) in HEK293 cells, whereas the Epac activator 8-CPT-2-OMe-cAMP did not alter Rap1B prenylation (62).
Epac1 distribution in the cell varies with cell type and has been reported in the cytosol, at the plasma membrane, and in perinuclear regions, the nuclear envelope, mitochondria, and phagosomes (63–65). Its subcellular location changes with the cell cycle stage and with the state of its activation (63). In addition, it appears that Epac1 binding to microtubules stimulates its guanine exchange activity toward Rap1 (66). Disruption of microtubules causes the distance between the centrosome and nucleus to increase (Refs. 3, 4, 39, 40 and this study). This study adds to this list the centrosome as an Epac1 locale in the cell. We show that ATP causes a translocation of Epac1 to the plasma membrane as well as loss from the centrosome, whereas another centrosomal protein, pericentrin, did not change (Fig. 5).
Rap1B is both cytosolic and membrane-associated (67). Its distribution within cells, like Epac1, varies. In HEK293 cells, Rap1B is mainly located at the plasma membrane (62). In human umbilical vein endothelial cells and in a squamous cell carcinoma cell line, Rap1B has a perinuclear distribution (68, 69). In mouse natural killer cells, Rap1B is localized to the centrosome and regulates the proper formation of this organelle (70). In RPE cells, we found that Rap1B protein levels are below detection by Western blotting, and immunofluorescence data are inconclusive (data not shown). In summary, two proteins that we identified in this study as downstream effectors of extracellular pathophysiological conditions are, in part, localized to the centrosome.
Our model (Fig. 8c) shows Epac1/Rap1B at the centrosome under normal conditions. Extracellular ATP (released locally under adverse conditions) or hypoxia activate a pathway specifically from the A2b purinergic receptor to Epac1, which is lost from the centrosome, causing the nucleus to move away from the centrosome. Rap1B plays a critical role. Based on recent work showing that the Rap1B family member Rab6a activates cytoplasmic dynein (71), it is possible that, in our system, Rap1B may also affect dynein function, which is normally required to bring the nucleus in close proximity to the centrosome. The end result is impaired cell migration. Several questions remain, including the mechanism that causes separation. Our experimental findings should allow addressing this in future work.
Rap1B, Centrosome-Nucleus Positioning, and Cell Migration
The centrosome occupies a central position in cells, which requires cytoplasmic microtubules that mediate pushing or pulling forces involving motor molecules. In addition, actomyosin can play a direct or indirect role in this process (72–74). The position of the nucleus is coordinated with that of the centrosome. Factors that contribute to changes in nuclear positioning include cell type, cell cycle stage, cell migration, and cell differentiation (75). The maintenance of a precise position of the nucleus within the cytoplasm is important. Studies in fungi, C. elegans, and Drosophila indicate that the nucleus has to maintain a precise position within the cytoplasm of many cell types for proper cellular and developmental processes (for a review, see Ref. 75).
The positioning of the centrosome and nucleus impacts cell migration. Studies in yeast and filamentous fungi identified many proteins required for both nuclear positioning and migration, including NudA, NudC, NudE, NudF, NudG, and NudI (76). In mammalian cells, orthologs were identified, including cytoplasmic dynein (NudA), dynactin (NudG and NudI), and Lis-1 (NudF). Mutation or deletion of these proteins causes an increase in the distance between the centrosome and nucleus.
Cell migration involves a repetition of basic steps: protrusion formation, adhesion, contraction, and retraction. At the subcellular level, migration requires the formation of the front-rear axis where the orientation of the centrosome and nucleus determines direction. The formation of the centrosome-nucleus axis requires the actin cytoskeleton. In a majority of cell types, the centrosome localizes in front of the nucleus in this front-rear axis (77, 78). In fibroblasts, the centrosome is stationary in the cell center, whereas the nucleus is moved by actin through a mechanism dependent on nesprin and its partners (77, 78). Here we also observed that the centrosome and nucleus are in the front-rear axis orientation (data not shown). In this study, we also found that, after ATP treatment, the nucleus moves away, whereas the centrosome remains where it is (see supplemental Movie 1). Examination of actin microfibers in ATP-treated cells by phalloidin staining fluorescence microscopy showed no observable difference in comparison with untreated cells (data not shown). This suggests that A2b-mediated repositioning of the centrosome and nucleus does not involve actin.
In cells including neurons, the process of cell migration involves two coordinated, asynchronous events: first, centrosome movement into the leading process of cell migration, followed by nuclear translocation (39). Microtubules are required for pulling the nucleus toward the centrosome (7, 40). Par6α signaling controls centrosomal protein localization and cytoskeletal dynamics, which in turn coordinate centrosomal motion and neuronal migration (39). Uncoupling of the positioning of the centrosome and nucleus can severely perturb cell migration (8, 79).
Rap1B is known for its role in controlling cell migration. Lung endothelial cells isolated from Rap1B-deficient mice display decreased migration in vitro (80). Rap1B knockdown by RNAi in endothelial cells and human umbilical vein endothelial cells results in the same phenotype (81, 82). Here we demonstrate that down-regulation of Rap1B by siRNA leads to an increase in the distance between the centrosome and nucleus and reduced migration.
The A2b-Epac1-Rap1B Pathway in the Context of Migration of Cancer Cells
Our discovery of a critical involvement of Rap1B in the positioning of the centrosome and nucleus may explain previous observations that cells isolated from Rap1B-deficient mice display decreased migration (80), that Rap1B knockdown results in impaired cell migration of human endothelial cells (81), and that Rap1B suppression by miR-708 results in inhibition of ovarian cancer cell migration/invasion (83). However, some studies reported a correlation between hypoxia and cell motility in cancer cells, including enhanced metastatic potential (84, 85). Hypoxia-inducible factor plays a significant role in this process, in addition to many other cellular factors. Our results suggest that hypoxia can reduce cell migration in normal cells via the A2b-Epac1 pathway, and it is tempting to speculate that deregulation of the normal A2b-Epac1-Rap1B pathway may be one factor involved in cancer progression, allowing a metastatic phenotype. In support of this possibility, Epac1 activation causes enhanced endothelial barrier function (86), and it can be speculated that deregulating this pathway causes loosening of cell-cell connections. Also, Rap1B knockdown inhibits gastric tumor cell invasion and hypoxia-induced gastric cancer cell invasion (and Rap1B and HIF1 expression) (87), suggesting the possibility that such cells undergo deregulation of the normal pathway. Future work beyond the scope of this study can address such possibilities.
Experimental Procedures
Chemicals
All of the following chemical compounds were purchased from Sigma-Aldrich (St. Louis, MO): ATP, ADP, AMP, PNP-AMP, adenosine, GTP, CTP, TTP, UTP, dATP, dGTP, dCTP, dTTP, nocodazole, forskolin, 8-bromo-cAMP, dideoxyadenosine, and caffeine. The following inhibitors were purchased from EMD Millipore (Billerica, MA): PSB 36, SCH58261, MRS1754, MRS1523, PSB1115, H89, PKA peptide inhibitor 14-22, ESI09, and HJC0350.
Cell Culture
Human foreskin fibroblasts (88) and human RPE cells (ATCC, CRL-4000) were grown in DMEM (Gibco/Thermo Fisher Scientific) supplemented with 10% fetal calf serum (Gibco/Thermo Fisher Scientific). All work was carried out in accordance with Biohazardous Materials regulations of the University of Calgary. The drug treatments, concentrations, and duration of treatments are indicated in the text. For hypoxia experiments, cells were cultured as above but in a chamber with 1% O2, 5% CO2 at 37 °C.
Cell Migration Assays
The cell scratch assay was done using established procedures (37). Briefly, cells were grown to confluence. Then, a micropipette tip was used to make a scratch in the monolayer. Images of the damaged area were taken immediately and after 17 h. The width of the scratch gaps was measured using a Zeiss microscope and the Axiovert software Measurement-Length function. The gap distance reduction parameter was calculated by subtracting the initial scratch gap distance from the final gap distance. The cell migration distance was calculated by dividing the gap distance reduction parameter by 2 (i.e. the migration length from one side of the scratch edge). To measure the effect of ATP on cell migration, cells were pretreated for 5 h with 1 mm ATP before applying the scratch damage. To measure the effect of hypoxia on cell migration, cells were incubated under 1% O2 for 2 days, after which the scratch damage assay was carried out. The Transwell assay was carried out as follows. RPE cells grown on 60-mm culture dishes were cultivated in DMEM supplemented with 10% FBS. At 70% confluence, the cells were treated for 1 day with either fresh medium (control) or medium that contained 500 μm ATP or 250 nm cytochalasin D. After treatment, cells were trypsinized and resuspended in 1.5 ml DMEM without serum. 5 × 105 cells from each treatment were loaded into the upper chamber of the Transwell kit (Corning Inc., Corning, NY). The lower chamber was filled with 2.0 ml DMEM containing 10% FBS. After 1.5 days of incubation at 37 °C to allow cells to migrate through the upper chamber membrane, cells that adhered to the lower chamber substratum were imaged and counted.
Cell Cycle Analysis
RPE cells grown in 60-mm culture dishes were trypsinized, washed, spun at 300 × g for 8 min, and resuspended in 250 μl of Hanks' buffer. The cells were fixed by mixing slowly with 250 μl of cold 95% ethanol and incubating at 4 °C overnight. Fixed cells were stained with propidium iodide using FxCycle PI/RNase staining solution as recommended by the manufacturer (Molecular Probes, Eugene, OR). Stained cells were analyzed at the University of Calgary Flow Cytometry Facility.
Antibodies
Anti-phospho-PKA substrate (RRXS*/T*) (100G7E) antibody was purchased from Cell Signaling Technology, Inc. (rabbit polyclonal, catalog no. 9621S, lot no. 8). Anti-ninein antibody was made in-house and was characterized previously (89). Anti-GFP monoclonal antibody was purchased from Santa Cruz Biotechnology, Inc. (mouse monoclonal, catalog no. SC-9996, lot no. F1115). Anti-pericentrin antibody was purchased from Abcam Inc. (rabbit polyclonal, catalog no. 4448, lot no. GR126417-2). HRP-conjugated secondary antibody and Cy3-, and Alexa488-labeled secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA) and Molecular Probes, respectively.
RNA Isolation and RT-PCR
RNA was isolated from monolayer cells using TRIzol (Invitrogen/Thermo Fisher Scientific) under conditions recommended by the manufacturer. RT-PCR was performed as described previously (90). Briefly, RNA was transcribed into cDNA using the Moloney murine leukemia virus (M-MLV) RT kit (Invitrogen/Thermo Fisher Scientific). For each sample, 100 ng of random primers was used, and a total volume of 20 μl of cDNA was generated. 2.5 μl of cDNA solution was used as template for PCR in a 50-μl reaction volume. For each primer set, 30 cycles of PCR were carried out using Taq polymerase (Invitrogen/Thermo Fisher Scientific). For PCR, the following primers were used: Adenosine receptor A1, ADORA1F (CCG CCC TCC ATC TCA GCT TTC) and ADORA1R (GTC ATC AGG CCT CTC TTC TGG); adenosine receptor A2a, ADORA2aF (ATG CCC ATC ATG GGC TCC TCG G) and ADORA2AR (GGA CAC TCC TGC TCC ATC CTG); adenosine receptor A2b, ADORA2BF (CTC AAG CTT CGA ATT CTG GCC GCC ACC ATG Gtg ctg gag aca cag gac) and ADORA2BR2 (GGC GAC CGG TGG ATC CCG TAG GCC CAC ACC GAG AGC AGG CTG); adenosine receptor A3, ADORA3F (CCC AAC AAC AGC ACT ACT CTG) and ADORA3R (ATT CTT CTC AAT GCT TGT GTC); Epac1, EPAC1F1 (ATG GTG TTG AGA AGG ATG CAC) and EPAC1R (GCC AGA CAT CAC TGT ATA CCG); Epac2, EPAC2F (GCA AAC TTG GAT TTG TTC CTG) and EPAC2R (TCG ACG AGG CTC TAA TCT GTG); Rap1A, Rap1aF (CTC AGA TCT CGA GCT CGT GAG TAC AAG CTA GTG GTC) and RAP1aR1 (GAC TGC AGA ATT CGA GAG CAG CAG ACA TGA TTT CTT TTT AGG); and Rap1B, Rap1bF (CTC AGA TCT CGA GCT CGT GAG TAT AAG CTA GTC GTT C) and RAP1bR (GTC GAC TGC AGA ATT CGA AAG CAG CTG ACA TGA TGA C). PCR fragments were analyzed by 1.2% agarose gel electrophoresis, and the sequence of all PCR fragments was determined at the University of Calgary Core DNA Services.
Plasmid Constructions and DNA Transfection
For construction of the pEGFP-A2b plasmid, a DNA fragment encoding A2b was amplified using the primers ADORA2BF and ADORA2BR2. After restriction digestion with EcoRI and BamHI, the PCR fragment was inserted into the corresponding sites of the pEGFP-C1 vector (Invitrogen/Thermo Fisher Scientific). Plasmid constructs were transfected into the indicated cells using Lipofectamine under conditions recommended by the manufacturer (Invitrogen/Thermo Fisher Scientific).
Protein Analysis and Western Blotting
Gel electrophoresis and Western blotting analyses were carried out as described previously (88). In short, proteins were boiled in SDS loading buffer, separated on 10% acrylamide SDS-PAGE gels, and electrophoretically transferred onto nitrocellulose blotting membranes (Amersham Biosciences/GE Healthcare). Membranes were blocked overnight at 4 °C in blocking buffer (54 mm Tris (pH 7.5), 150 mm NaCl, 0.05% Nonidet P-40, 0.05% Tween 20, and 5% nonfat dry milk), and analyzed using the indicated primary antibodies followed by HRP-conjugated secondary antibody. Prestained protein standards were used as a marker (Bio-Rad). LumiGLO substrate (Kirkegaard & Perry Laboratories, Inc.) was used to develop the blots. Luminescence was captured using Hyperfilm ECL films (Amersham Biosciences/GE Healthcare).
Immunofluorescence Microscopy and Measurement of the Distance between the Centrosome and Nucleus
Immunofluorescence microscopy was performed as described previously (88). Briefly, cells were fixed in cold methanol and stained with the primary and secondary antibodies mentioned in the text at a concentration recommended by the manufacturer. Images were obtained using a Zeiss Observer.Z1 microscope with a ×40 objective (Plan-Apochromat ×40/1.4 oil DIC (UV) VIS-IR M27) and an Axiocam MRm (426509-9901-0000) digital camera in conjunction with the Zeiss software AxioVision Rel.4.9.1. The shortest direct distance between the centrosome (demarcated by ninein immunofluorescence) and the periphery of the nucleus (DAPI) was measured using the Zeiss AxioVision software Measurement and Length functions. The scratch assay was imaged using the Zeiss Observer.Z1 microscope and a ×5 objective (EC Plan-Neofluar 4203319911).
Live Cell Imaging
To visualize the movement of the nucleus relative to the central position of the centrosome, we used time-lapse live cell imaging. HeLa cells that express mCherry-H2B (to visualize the nucleus) and GFP-centrin (to visualize the centrosome) were used. Cells seeded in a 35-mm glass-bottom dish (MatTek Corp. or Fluordish) were placed onto a sample stage within an incubator chamber maintained at a temperature of 37 °C in an atmosphere of 5% CO2. Imaging was performed using a spinning disk confocal (PerkinElmer Life Sciences, UltraView) on an inverted microscope (Axiovert 200 M, Carl Zeiss, with a ×63 objective lens and 1.4 numerical aperture) equipped with a Scientific CMOS camera (ORCA-FLASH-4.0, Hamamatsu Photonics). Cells were treated with 1 mm ATP, and images were recorded every 15 min over a 16-h period using Volocity software (version 6.3.0, PerkinElmer Life Sciences). The time-lapse imaging data were exported from Volocity software as OME tiff image format. The time-lapse movies were subsequently analyzed using Imaris (version 7.6 Bitplane) three-dimensional visualization software. The cell nuclei were surface-rendered with the H2B signal, and the centrosomes were tracked using the spot detection and tracking function of the software. The still tiff-format images from videos were exported in Imaris and processed using Photoshop. Time-lapse movies were recorded at a frame rate of 30 frames/s from Imaris using the quicktime.mov format to record nucleus movement.
siRNA Experiments
All of the following siRNA products were purchased from Qiagen: negative control (scramble) siRNA, catalog no. SI03650325; Rap1A, catalog no. SI02662296; and Rap1B, catalog no. SI02662303. The siRNA products were transfected using RNAi-Max (Invitrogen/Thermo Fisher Scientific), and cells were cultured for 72 h. The cells were next fixed with cold methanol and analyzed by immunofluorescence microscopy or were collected for analysis by semiquantitative RT-PCR.
Statistical Analysis
Data are presented as the mean value ± S.D. of at least three independent experiments (as indicated in the figure legends). Z test was used to determine the p value. p Values of less than 0.05 were considered to indicate a significant difference between measured values.
Author Contributions
Y. O. and F. V. D. H. conceived the project and designed the experiments. Y. O., J. B. R., and F. V. D. H. discussed all data interpretations and build models to allow study progress. Y. O. performed most experiments, with significant contributions of cell treatments and distance measurements by J. Z. Life cell imaging was performed by G. C. Y. O., and F. A. V. D. H. wrote the article with major contributions from G. C.
Supplementary Material
Acknowledgments
We thank Dr. J. L. Bos for the gift of the EGFP-Epac1 expression vector, Dr. D. Slater for the gifts of ESI09 and Epac1 and Epac2 siRNA, and Dr. F. Jirik for use of the hypoxic chamber. We thank J. Pan for technical assistance.
This work was supported by Canadian Institutes of Health Research Grant MOP 111008 (to F. A. V. D. H.) and a summer studentship from Alberta Innovates - Health Solutions (to J. Z.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Movie 1.
- RPE
- retinal epithelial pigment
- PNP-AMP
- adenylyl-imidodiphosphate.
References
- 1. Snyder M. (1994) The spindle pole body of yeast. Chromosoma 103, 369–380 [DOI] [PubMed] [Google Scholar]
- 2. Brugerolle G., and Mignot J. P. (2003) The rhizoplast of chrysomonads, a basal body-nucleus connector that polarises the dividing spindle. Protoplasma 222, 13–21 [DOI] [PubMed] [Google Scholar]
- 3. Malone C. J., Misner L., Le Bot N., Tsai M. C., Campbell J. M., Ahringer J., and White J. G. (2003) The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115, 825–836 [DOI] [PubMed] [Google Scholar]
- 4. Tanaka T., Serneo F. F., Higgins C., Gambello M. J., Wynshaw-Boris A., and Gleeson J. G. (2004) Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 165, 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsai J. W., Chen Y., Kriegstein A. R., and Vallee R. B. (2005) LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 170, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shu T., Ayala R., Nguyen M. D., Xie Z., Gleeson J. G., and Tsai L. H. (2004) Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263–277 [DOI] [PubMed] [Google Scholar]
- 7. Tsai J. W., Bremner K. H., and Vallee R. B. (2007) Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 10, 970–979 [DOI] [PubMed] [Google Scholar]
- 8. Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., Xu R., and Han M. (2009) SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 64, 173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Splinter D., Tanenbaum M. E., Lindqvist A., Jaarsma D., Flotho A., Yu K. L., Grigoriev I., Engelsma D., Haasdijk E. D., Keijzer N., Demmers J., Fornerod M., Melchior F., Hoogenraad C. C., Medema R. H., and Akhmanova A. (2010) Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLOS Biol. 8, e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jodoin J. N., Shboul M., Sitaram P., Zein-Sabatto H., Reversade B., Lee E., and Lee L. A. (2012) Human Asunder promotes dynein recruitment and centrosomal tethering to the nucleus at mitotic entry. Mol. Biol. Cell 23, 4713–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burakov A. V., and Nadezhdina E. S. (2013) Association of nucleus and centrosome: magnet or velcro? Cell Biol. Int. 37, 95–104 [DOI] [PubMed] [Google Scholar]
- 12. Agircan F. G., Schiebel E., and Mardin B. R. (2014) Separate to operate: control of centrosome positioning and separation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng Y., Olson E. C., Stukenberg P. T., Flanagan L. A., Kirschner M. W., and Walsh C. A. (2000) LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 28, 665–679 [DOI] [PubMed] [Google Scholar]
- 14. Sasaki S., Shionoya A., Ishida M., Gambello M. J., Yingling J., Wynshaw-Boris A., and Hirotsune S. (2000) A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28, 681–696 [DOI] [PubMed] [Google Scholar]
- 15. Tomlinson A., and Ready D. F. (1986) Sevenless: a cell-specific homeotic mutation of the Drosophila eye. Science 231, 400–402 [DOI] [PubMed] [Google Scholar]
- 16. Mosley-Bishop K. L., Li Q., Patterson L., and Fischer J. A. (1999) Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 9, 1211–1220 [DOI] [PubMed] [Google Scholar]
- 17. Patterson K., Molofsky A. B., Robinson C., Acosta S., Cater C., and Fischer J. A. (2004) The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell 15, 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kracklauer M. P., Banks S. M., Xie X., Wu Y., and Fischer J. A. (2007) Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly 1, 75–85 [DOI] [PubMed] [Google Scholar]
- 19. Archambault V., Zhao X., White-Cooper H., Carpenter A. T., and Glover D. M. (2007) Mutations in Drosophila Greatwall/Scant reveal its roles in mitosis and meiosis and interdependence with Polo kinase. PLoS Genet. 3, e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Archambault V., D'Avino P. P., Deery M. J., Lilley K. S., and Glover D. M. (2008) Sequestration of Polo kinase to microtubules by phosphopriming-independent binding to Map205 is relieved by phosphorylation at a CDK site in mitosis. Genes Dev. 22, 2707–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dobyns W. B. (1987) Developmental aspects of lissencephaly and the lissencephaly syndromes. Birth Defects Orig. Artic. Ser. 23, 225–241 [PubMed] [Google Scholar]
- 22. Eltzschig H. K., Ibla J. C., Furuta G. T., Leonard M. O., Jacobson K. A., Enjyoji K., Robson S. C., and Colgan S. P. (2003) Coordinated adenine nucleotide phosphohydrolysis and nucleoside signaling in posthypoxic endothelium: role of ectonucleotidases and adenosine A2B receptors. J. Exp. Med. 198, 783–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xaus J., Mirabet M., Lloberas J., Soler C., Lluis C., Franco R., and Celada A. (1999) IFN-γ up-regulates the A2B adenosine receptor expression in macrophages: a mechanism of macrophage deactivation. J. Immunol. 162, 3607–3614 [PubMed] [Google Scholar]
- 24. Dale N., and Frenguelli B. G. (2009) Release of adenosine and ATP during ischemia and epilepsy. Curr. Neuropharmacol. 7, 160–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burnstock G. (2014) Purinergic signalling: from discovery to current developments. Exp. Physiol. 99, 16–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fredholm B. B., Irenius E., Kull B., and Schulte G. (2001) Comparison of the potency of adenosine as an agonist at human adenosine receptors expressed in Chinese hamster ovary cells. Biochem. Pharmacol. 61, 443–448 [DOI] [PubMed] [Google Scholar]
- 27. Yang D., Zhang Y., Nguyen H. G., Koupenova M., Chauhan A. K., Makitalo M., Jones M. R., St Hilaire C., Seldin D. C., Toselli P., Lamperti E., Schreiber B. M., Gavras H., Wagner D. D., and Ravid K. (2006) The A2B adenosine receptor protects against inflammation and excessive vascular adhesion. J. Clin. Invest. 116, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eckle T., Krahn T., Grenz A., Köhler D., Mittelbronn M., Ledent C., Jacobson M. A., Osswald H., Thompson L. F., Unertl K., and Eltzschig H. K. (2007) Cardioprotection by ecto-5′-nucleotidase (CD73) and A2B adenosine receptors. Circulation 115, 1581–1590 [DOI] [PubMed] [Google Scholar]
- 29. Eckle T., Grenz A., Laucher S., and Eltzschig H. K. (2008) A2B adenosine receptor signaling attenuates acute lung injury by enhancing alveolar fluid clearance in mice. J. Clin. Invest. 118, 3301–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frick J. S., MacManus C. F., Scully M., Glover L. E., Eltzschig H. K., and Colgan S. P. (2009) Contribution of adenosine A2B receptors to inflammatory parameters of experimental colitis. J. Immunol. 182, 4957–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schingnitz U., Hartmann K., Macmanus C. F., Eckle T., Zug S., Colgan S. P., and Eltzschig H. K. (2010) Signaling through the A2B adenosine receptor dampens endotoxin-induced acute lung injury. J. Immunol. 184, 5271–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hart M. L., Grenz A., Gorzolla I. C., Schittenhelm J., Dalton J. H., and Eltzschig H. K. (2011) Hypoxia-inducible factor-1α-dependent protection from intestinal ischemia/reperfusion injury involves ecto-5′-nucleotidase (CD73) and the A2B adenosine receptor. J. Immunol. 186, 4367–4374 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Choukèr A., Ohta A., Martignoni A., Lukashev D., Zacharia L. C., Jackson E. K., Schnermann J., Ward J. M., Kaufmann I., Klaunberg B., Sitkovsky M. V., and Thiel M. (2012) In vivo hypoxic preconditioning protects from warm liver ischemia-reperfusion injury through the adenosine A2B receptor. Transplantation 94, 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grenz A., Bauerle J. D., Dalton J. H., Ridyard D., Badulak A., Tak E., McNamee E. N., Clambey E., Moldovan R., Reyes G., Klawitter J., Ambler K., Magee K., Christians U., Brodsky K. S., et al. (2012) Equilibrative nucleoside transporter 1 (ENT1) regulates postischemic blood flow during acute kidney injury in mice. J. Clin. Invest. 122, 693–710 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35. Idzko M., Ferrari D., and Eltzschig H. K. (2014) Nucleotide signalling during inflammation. Nature 509, 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakagawa S., Omura T., Yonezawa A., Yano I., Nakagawa T., and Matsubara K. (2014) Extracellular nucleotides from dying cells act as molecular signals to promote wound repair in renal tubular injury. Am. J. Physiol. Renal Physiol. 307, F1404–F1411 [DOI] [PubMed] [Google Scholar]
- 37. Liang C. C., Park A. Y., and Guan J. L. (2007) In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2, 329–333 [DOI] [PubMed] [Google Scholar]
- 38. Marshall J. (2011) Transwell((R)) invasion assays. Methods Mol. Biol. 769, 97–110 [DOI] [PubMed] [Google Scholar]
- 39. Solecki D. J., Model L., Gaetz J., Kapoor T. M., and Hatten M. E. (2004) Par6α signaling controls glial-guided neuronal migration. Nat. Neurosci. 7, 1195–1203 [DOI] [PubMed] [Google Scholar]
- 40. Tsai L. H., and Gleeson J. G. (2005) Nucleokinesis in neuronal migration. Neuron 46, 383–388 [DOI] [PubMed] [Google Scholar]
- 41. Dunwiddie T. V., and Masino S. A. (2001) The role and regulation of adenosine in the central nervous system. Annu. Rev. Neurosci. 24, 31–55 [DOI] [PubMed] [Google Scholar]
- 42. Weyler S., Fülle F., Diekmann M., Schumacher B., Hinz S., Klotz K. N., and Müller C. E. (2006) Improving potency, selectivity, and water solubility of adenosine A1 receptor antagonists: xanthines modified at position 3 and related pyrimido[1,2,3-cd]purinediones. ChemMedChem 1, 891–902 [DOI] [PubMed] [Google Scholar]
- 43. Zocchi C., Ongini E., Conti A., Monopoli A., Negretti A., Baraldi P. G., and Dionisotti S. (1996) The non-xanthine heterocyclic compound SCH 58261 is a new potent and selective A2a adenosine receptor antagonist. J. Pharmacol. Exp. Ther. 276, 398–404 [PubMed] [Google Scholar]
- 44. Kim Y. C., Ji X., Melman N., Linden J., and Jacobson K. A. (2000) Anilide derivatives of an 8-phenylxanthine carboxylic congener are highly potent and selective antagonists at human A(2B) adenosine receptors. J. Med. Chem. 43, 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li A. H., Moro S., Melman N., Ji X. D., and Jacobson K. A. (1998) Structure-activity relationships and molecular modeling of 3, 5-diacyl-2,4-dialkylpyridine derivatives as selective A3 adenosine receptor antagonists. J. Med. Chem. 41, 3186–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Müller C. E., Sandoval-Ramírez J., Schobert U., Geis U., Frobenius W., and Klotz K. N. (1998) 8-(Sulfostyryl)xanthines: water-soluble A2A-selective adenosine receptor antagonists. Bioorg. Med. Chem. 6, 707–719 [DOI] [PubMed] [Google Scholar]
- 47. Macurek L., Lindqvist A., Lim D., Lampson M. A., Klompmaker R., Freire R., Clouin C., Taylor S. S., Yaffe M. B., and Medema R. H. (2008) Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123 [DOI] [PubMed] [Google Scholar]
- 48. Archambault V., and Carmena M. (2012) Polo-like kinase-activating kinases: Aurora A, Aurora B and what else? Cell Cycle 11, 1490–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Rooij J., Zwartkruis F. J., Verheijen M. H., Cool R. H., Nijman S. M., Wittinghofer A., and Bos J. L. (1998) Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396, 474–477 [DOI] [PubMed] [Google Scholar]
- 50. Bos J. L. (2003) Epac: a new cAMP target and new avenues in cAMP research. Nat. Rev. Mol. Cell Biol. 4, 733–738 [DOI] [PubMed] [Google Scholar]
- 51. Almahariq M., Tsalkova T., Mei F. C., Chen H., Zhou J., Sastry S. K., Schwede F., and Cheng X. (2013) A novel EPAC-specific inhibitor suppresses pancreatic cancer cell migration and invasion. Mol. Pharmacol. 83, 122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ponsioen B., Gloerich M., Ritsma L., Rehmann H., Bos J. L., and Jalink K. (2009) Direct spatial control of Epac1 by cyclic AMP. Mol. Cell Biol. 29, 2521–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Melani A., Corti F., Stephan H., Müller C. E., Donati C., Bruni P., Vannucchi M. G., and Pedata F. (2012) Ecto-ATPase inhibition: ATP and adenosine release under physiological and ischemic in vivo conditions in the rat striatum. Exp. Neurol. 233, 193–204 [DOI] [PubMed] [Google Scholar]
- 54. Cauwels A., Rogge E., Vandendriessche B., Shiva S., and Brouckaert P. (2014) Extracellular ATP drives systemic inflammation, tissue damage and mortality. Cell Death. Dis. 5, e1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bours M. J., Swennen E. L., Di Virgilio F., Cronstein B. N., and Dagnelie P. C. (2006) Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol. Ther. 112, 358–404 [DOI] [PubMed] [Google Scholar]
- 56. Wilhelm K., Ganesan J., Müller T., Dürr C., Grimm M., Beilhack A., Krempl C. D., Sorichter S., Gerlach U. V., Jüttner E., Zerweck A., Gärtner F., Pellegatti P., Di Virgilio F., Ferrari D., et al. (2010) Graft-versus-host disease is enhanced by extracellular ATP activating P2X7R. Nat. Med. 16, 1434–1438 [DOI] [PubMed] [Google Scholar]
- 57. Wakai A., Wang J. H., Winter D. C., Street J. T., O'Sullivan R. G., and Redmond H. P. (2001) Adenosine inhibits neutrophil vascular endothelial growth factor release and transendothelial migration via A2B receptor activation. Shock 15, 297–301 [DOI] [PubMed] [Google Scholar]
- 58. Steininger T. S., Stutz H., and Kerschbaum H. H. (2011) β-Adrenergic stimulation suppresses phagocytosis via Epac activation in murine microglial cells. Brain Res. 1407, 1–12 [DOI] [PubMed] [Google Scholar]
- 59. van Hooren K. W., van Agtmaal E. L., Fernandez-Borja M., van Mourik J. A., Voorberg J., and Bierings R. (2012) The Epac-Rap1 signaling pathway controls cAMP-mediated exocytosis of Weibel-Palade bodies in endothelial cells. J. Biol. Chem. 287, 24713–24720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Baillie G. S. (2009) Compartmentalized signalling: spatial regulation of cAMP by the action of compartmentalized phosphodiesterases. FEBS J. 276, 1790–1799 [DOI] [PubMed] [Google Scholar]
- 61. Edwards H. V., Christian F., and Baillie G. S. (2012) cAMP: novel concepts in compartmentalised signalling. Semin. Cell Dev. Biol. 23, 181–190 [DOI] [PubMed] [Google Scholar]
- 62. Ntantie E., Gonyo P., Lorimer E. L., Hauser A. D., Schuld N., McAllister D., Kalyanaraman B., Dwinell M. B., Auchampach J. A., and Williams C. L. (2013) An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering. Sci. Signal. 6, ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qiao J., Mei F. C., Popov V. L., Vergara L. A., and Cheng X. (2002) Cell cycle-dependent subcellular localization of exchange factor directly activated by cAMP. J. Biol. Chem. 277, 26581–26586 [DOI] [PubMed] [Google Scholar]
- 64. Brock T. G., Serezani C. H., Carstens J. K., Peters-Golden M., and Aronoff D. M. (2008) Effects of prostaglandin E2 on the subcellular localization of Epac-1 and Rap1 proteins during Fcγ-receptor-mediated phagocytosis in alveolar macrophages. Exp. Cell Res. 314, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gloerich M., Vliem M. J., Prummel E., Meijer L. A., Rensen M. G., Rehmann H., and Bos J. L. (2011) The nucleoporin RanBP2 tethers the cAMP effector Epac1 and inhibits its catalytic activity. J. Cell Biol. 193, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gupta M., and Yarwood S. J. (2005) MAP1A light chain 2 interacts with exchange protein activated by cyclic AMP 1 (EPAC1) to enhance Rap1 GTPase activity and cell adhesion. J. Biol. Chem. 280, 8109–8116 [DOI] [PubMed] [Google Scholar]
- 67. Klinz F. J., Seifert R., Schwaner I., Gausepohl H., Frank R., and Schultz G. (1992) Generation of specific antibodies against the rap1A, rap1B and rap2 small GTP-binding proteins: analysis of rap and ras proteins in membranes from mammalian cells. Eur. J. Biochem. 207, 207–213 [DOI] [PubMed] [Google Scholar]
- 68. Mitra R. S., Zhang Z., Henson B. S., Kurnit D. M., Carey T. E., and D'Silva N. J. (2003) Rap1A and rap1B ras-family proteins are prominently expressed in the nucleus of squamous carcinomas: nuclear translocation of GTP-bound active form. Oncogene 22, 6243–6256 [DOI] [PubMed] [Google Scholar]
- 69. Wittchen E. S., Aghajanian A., and Burridge K. (2011) Isoform-specific differences between Rap1A and Rap1B GTPases in the formation of endothelial cell junctions. Small GTPases 2, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Awasthi A., Samarakoon A., Chu H., Kamalakannan R., Quilliam L. A., Chrzanowska-Wodnicka M., White G. C. 2nd, and Malarkannan S. (2010) Rap1b facilitates NK cell functions via IQGAP1-mediated signalosomes. J. Exp. Med. 207, 1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yamada M., Kumamoto K., Mikuni S., Arai Y., Kinjo M., Nagai T., Tsukasaki Y., Watanabe T. M., Fukui M., Jin M., Toba S., and Hirotsune S. (2013) Rab6a releases LIS1 from a dynein idling complex and activates dynein for retrograde movement. Nat. Commun. 4, 2033. [DOI] [PubMed] [Google Scholar]
- 72. Burakov A., Nadezhdina E., Slepchenko B., and Rodionov V. (2003) Centrosome positioning in interphase cells. J. Cell Biol. 162, 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Manneville J. B., and Etienne-Manneville S. (2006) Positioning centrosomes and spindle poles: looking at the periphery to find the centre. Biol. Cell 98, 557–565 [DOI] [PubMed] [Google Scholar]
- 74. Zhu J., Burakov A., Rodionov V., and Mogilner A. (2010) Finding the cell center by a balance of dynein and myosin pulling and microtubule pushing: a computational study. Mol. Biol. Cell 21, 4418–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gundersen G. G., and Worman H. J. (2013) Nuclear positioning. Cell 152, 1376–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Morris N. R. (2000) Nuclear migration. From fungi to the mammalian brain. J. Cell Biol. 148, 1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Luxton G. W., and Gundersen G. G. (2011) Orientation and function of the nuclear-centrosomal axis during cell migration. Curr. Opin. Cell Biol. 23, 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Luxton G. W., Gomes E. R., Folker E. S., Worman H. J., and Gundersen G. G. (2011) TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus 2, 173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Renaud J., Kerjan G., Sumita I., Zagar Y., Georget V., Kim D., Fouquet C., Suda K., Sanbo M., Suto F., Ackerman S. L., Mitchell K. J., Fujisawa H., and Chédotal A. (2008) Plexin-A2 and its ligand, Sema6A, control nucleus-centrosome coupling in migrating granule cells. Nat. Neurosci. 11, 440–449 [DOI] [PubMed] [Google Scholar]
- 80. Chrzanowska-Wodnicka M., Kraus A. E., Gale D., White G. C. 2nd, and Vansluys J. (2008) Defective angiogenesis, endothelial migration, proliferation, and MAPK signaling in Rap1b-deficient mice. Blood 111, 2647–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Carmona G., Göttig S., Orlandi A., Scheele J., Bäuerle T., Jugold M., Kiessling F., Henschler R., Zeiher A. M., Dimmeler S., and Chavakis E. (2009) Role of the small GTPase Rap1 for integrin activity regulation in endothelial cells and angiogenesis. Blood 113, 488–497 [DOI] [PubMed] [Google Scholar]
- 82. Yan J., Li F., Ingram D. A., and Quilliam L. A. (2008) Rap1a is a key regulator of fibroblast growth factor 2-induced angiogenesis and together with Rap1b controls human endothelial cell functions. Mol. Cell Biol. 28, 5803–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lin K. T., Yeh Y. M., Chuang C. M., Yang S. Y., Chang J. W., Sun S. P., Wang Y. S., Chao K. C., and Wang L. H. (2015) Glucocorticoids mediate induction of microRNA-708 to suppress ovarian cancer metastasis through targeting Rap1B. Nat. Commun. 6, 5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Semenza G. L. (2015) The hypoxic tumor microenvironment: a driving force for breast cancer progression. Biochim. Biophys. Acta 1863, 382–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sullivan R., and Graham C. H. (2007) Hypoxia-driven selection of the metastatic phenotype. Cancer Metastasis Rev. 26, 319–331 [DOI] [PubMed] [Google Scholar]
- 86. Cullere X., Shaw S. K., Andersson L., Hirahashi J., Luscinskas F. W., and Mayadas T. N. (2005) Regulation of vascular endothelial barrier function by Epac, a cAMP-activated exchange factor for Rap GTPase. Blood 105, 1950–1955 [DOI] [PubMed] [Google Scholar]
- 87. Yang Y., Li M., Yan Y., Zhang J., Sun K., Qu J. K., Wang J. S., and Duan X. Y. (2015) Expression of RAP1B is associated with poor prognosis and promotes an aggressive phenotype in gastric cancer. Oncol. Rep. 34, 2385–2394 [DOI] [PubMed] [Google Scholar]
- 88. Ou Y., Zhang Y., Cheng M., Rattner J. B., Dobrinski I., and van der Hoorn F. A. (2012) Targeting of CRMP-2 to the primary cilium is modulated by GSK-3β. PLoS ONE 7, e48773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ou Y. Y., Mack G. J., Zhang M., and Rattner J. B. (2002) CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 115, 1825–1835 [DOI] [PubMed] [Google Scholar]
- 90. Fitzgerald C., Sikora C., Lawson V., Dong K., Cheng M., Oko R., and van der Hoorn F. A. (2006) Mammalian transcription in support of hybrid mRNA and protein synthesis in testis and lung. J. Biol. Chem. 281, 38172–38180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.