Abstract
Staphylococcus epidermidis is frequently implicated in human infections associated with indwelling medical devices due to its ubiquity in the skin flora and formation of robust biofilms. The accessory gene regulator (agr) quorum sensing (QS) system plays a prominent role in the establishment of biofilms and infection by this bacterium. Agr activation is mediated by the binding of a peptide signal (or autoinducing peptide, AIP) to its cognate AgrC receptor. Many questions remain about the role of QS in S. epidermidis infections, as well as in mixed-microbial populations on a host, and chemical modulators of its agr system could provide novel insights into this signaling network. The AIP ligand provides an initial scaffold for the development of such probes; however, the structure-activity relationships (SARs) for activation of S. epidermidis AgrC receptors by AIPs are largely unknown. Herein, we report the first SAR analyses of an S. epidermidis AIP by performing systematic alanine and D-amino acid scans of the S. epidermidis AIP-I. Based on these results, we designed and identified potent, pan-group inhibitors of the AgrC receptors in the three S. epidermidis agr groups, as well as a set of AIP-I analogs capable of selective AgrC inhibition in either specific S. epidermidis agr groups or in another common staphylococcal species, S. aureus. In addition, we uncovered a non-native peptide agonist of AgrC-I that can strongly inhibit S. epidermidis biofilm growth. Together, these synthetic analogs represent new and readily accessible probes for investigating the roles of QS in S. epidermidis colonization and infections.
Graphical abstract
INTRODUCTION
Staphylococcus epidermidis is a ubiquitous commensal colonizer of human epithelia (1). Previously thought to be innocuous, S. epidermidis recently has been identified as an important nosocomial pathogen (2, 3). In particular, S. epidermidis is a leading causative agent in infections associated with indwelling medical devices, which is directly related to its ubiquity and propensity to form biofilms on abiotic surfaces (3–6). The barrier provided by this biofilm lifestyle, combined with growing resistance to antibiotics, make S. epidermidis infections especially recalcitrant to traditional antimicrobial treatments and, consequently, a serious problem to public health (2, 3, 6–8).
Playing an important role in the virulence of S. epidermidis is its quorum sensing (QS) system (9–13). QS is a cell-cell communication process that allows bacteria to sense population density and coordinate gene expression to control group behavior at high cell numbers (14, 15). The primary QS circuit in S. epidermidis is the accessory gene regulator (agr) system, which was first characterized and has been most studied to date in the related human pathogen Staphylococcus aureus (16–18). The agr system consists of four components (AgrA–D) that are encoded by the agr locus (Figure 1) (16–19). AgrD is a propeptide containing the sequence of the mature autoinducing peptide (or AIP) signal, and it is processed and secreted by the integral membrane endopeptidase AgrB (20, 21). Once reaching a threshold extracellular concentration (produced by a “quorate” population of cells), the AIP is sensed by a two-component system comprised of AgrC and AgrA. Productive binding of the AIP to AgrC, a transmembrane receptor histidine kinase, triggers its subsequent dimerization and trans-autophosphorylation (22, 23); the activated AgrC then phosphorylates the response regulator AgrA. Phosphorylated AgrA next dimerizes, binds the P2 and P3 promoters, and upregulates the transcription of the agr locus and RNAIII, respectively. RNAIII serves as the main effector of the agr system, and thereby, many virulence phenotypes in S. epidermidis (24). Phosphorylated AgrA also directly activates the production of a group of small amphipathic peptides known as phenol-soluble modulins (PSMs), which are key virulence factors for both the biofilm life cycle and survival in infected hosts (11, 25).
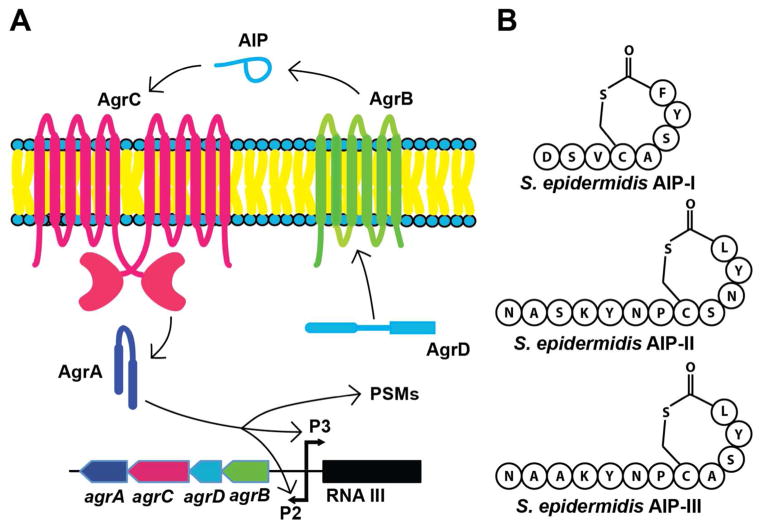
Figure 1.
The agr QS system in S. epidermidis and associated AIP signals. (A) Simplified schematic of the agr system. Propeptide AgrD is processed and secreted by the integral membrane endopeptidase AgrB to generate the mature autoinducing peptide (AIP) signal. Once a threshold extracellular concentration of AIP is reached, the AgrC receptor binds to its cognate AIP and homodimerizes. Activation of AgrC, a transmembrane histidine kinase, by AIP leads to AgrC trans-autophosphorylation, which then phosphorylates the response regulator AgrA. The phosphorylated AgrA subsequently activates transcription from the P2, P3, and PSM promoters. P2 encodes the four components of the agr system, whereas P3 drives the expression of RNAIII, the main effector molecule of the agr system. (B) Primary structures of the three AIP signals (I–III) corresponding to the three S. epidermidis agr Groups (I–III).
The formation of a robust biofilm is the primary mechanism by which S. epidermidis causes infections on medical implants (3). A number of studies have identified the polysaccharide intercellular adhesin (PIA) as being a critical structural component of the biofilm (26). In S. epidermidis strains that produce PIA (but also in some strains that do not), the agr system is involved in many of the distinct stages of biofilm formation (10, 27, 28). Specifically, the agr system negatively regulates the expression of the surface-attached AtlE protein (29), an important adhesion factor in the attachment phase of biofilm growth, and positively regulates the production of PSMs and proteases that facilitate the detachment of bacteria from biofilm (11, 30, 31). Consequently, intentional activation of the agr system could provide an alternative approach of limiting S. epidermidis infection by reducing biofilm growth (32), especially considering that S. epidermidis, unlike S. aureus, does not possess a vast repertoire of damaging toxins under QS control (3, 18). Furthermore, this approach could be combined with administration of antibiotics to potentiate the effectiveness of such drugs (4, 32). In contrast, other studies have proposed that inhibition, as opposed to activation, of the agr system can also attenuate virulence, despite the concern over the enhanced accumulation of biofilm observed in S. epidermidis agr mutants (28, 33, 34). For example, Otto and co-workers, using agr-null strains of S. epidermidis, have demonstrated that the presence of an active agr system offers the bacterium significantly more protection against antimicrobial peptides and oxidative stress, both of which are defense mechanisms against infections employed by the innate immune system (10, 13). The agr system has also been shown to promote bacteria dissemination and tissue infiltration during device-related infections (13, 30, 31, 35). Further, a recent report by Olson et al. revealed the agr system to be necessary for optimal skin colonization (36). Lastly, there is growing appreciation that many S. epidermidis clinical isolates from implant infections do not have the genes to make PIA (37–39), but these strains still build a biofilm (40). The role that the agr system plays in the early stages of biofilm development in these PIA-negative strains is unclear. Combined, these studies and others highlight the complex roles played by the agr system in the lifestyle of S. epidermidis. Therefore, while the agr system is an attractive target for controlling the virulence of S. epidermidis, further fundamental studies are required to assess when and how the agr system should be modulated (either via activation or inhibition) to attenuate infection.
Similar to S. aureus, the agr system is not identical within the S. epidermidis species. Based upon the primary structures of the three reported AIP signals (I–III) (Figure 1B), S. epidermidis can be classified into three agr specificity groups referred to as Groups-I–III (36, 41). Interestingly, the agr systems of these groups are known to interact with each other, a phenomenon called agr interference that was first described in S. aureus (18, 42–44). In S. epidermidis, the agr system of Group-I is significantly inhibited by the AIP-II and –III signals, whereas Groups-II and -III are inhibited by AIP-I (36). In addition, agr interference has also been identified between S. epidermidis Group-I and S. aureus Groups-I–IV, two species that colonize similar environmental niches on the human body (45–47). These cross-inhibitory activities of the agr systems between different specificity groups and species have been hypothesized to play a role in competition for colonization and establishing infections (18, 46, 48). However, in vivo studies have not provided conclusive support for such hypothesis (48, 49). Additional studies are therefore necessary to elucidate the biological significance of agr interference, if any.
Chemical methods to modulate QS in bacteria have attracted considerable interest over the past decade, due in part to the temporal and spatial control they can provide and their often ready applicability to biologically relevant environments (50–52). The development of chemical probes capable of modulating the S. epidermidis agr system could provide valuable tools for better understanding the role of the agr system in colonization and infection by this common bacterium. Both agr agonists and antagonists are of interest, given the dynamic and complex role played by QS during S. epidermidis infection. Moreover, identification of modulators that can selectively target a specific agr group or staphylococcal species in a mixed bacteria population would provide promising tools to investigate the role of QS in the interplay between different agr groups or species. Prior studies by our lab and others have focused on the structure-activity relationships (SARs) for AIP:AgrC interactions in S. aureus, and have uncovered a number of potent, non-native S. aureus AgrC inhibitors in the process (43, 53–61). Little is known, however, about the SARs of AIP:AgrC interactions in S. epidermidis. Earlier work by Otto and co-workers has shown that the thioester linkage and the three-amino acid exocyclic tail of AIP-I (Figure 1B) are required for AgrC-I activation, as they found the hydrolyzed acyclic AIP-I, AIP-I analogs with exocyclic tails either one amino acid shorter or longer, and AIP-I analogs with a lactam or a lactone in place of the thioester were all inactive in cell-based AgrC-I activation assays (47, 62). However, to our knowledge, no non-native AgrC agonists and antagonists have been reported for S. epidermidis to date.
Herein, we report a systematic analysis of the S. epidermidis AIP-I structure and delineate key SARs for AgrC-I activation by this ligand. We used these SAR data to develop a series of AIP-I analogs that strongly antagonize or agonize the AgrC-I receptor. Several of these ligands were capable of either pan-group or group-selective AgrC receptor inhibition in S. epidermidis. In turn, other peptides were found to be species selective, and could be applied to selectively modulate either S. epidermidis or S. aureus AgrC receptors. Notably, one of our non-native AgrC-I agonists was found to strongly inhibit S. epidermidis biofilm growth, with a higher potency and efficacy than that of native AIP-I. These new compounds represent the first synthetic modulators of AgrC in S. epidermidis and constitute powerful new tools to elucidate the complex roles of the agr QS system in this emerging pathogen.
RESULTS AND DISCUSSION
AIP-I SARs revealed through alanine and D-amino acid scans
Our SAR investigation began by systematically replacing each amino acid of the S. epidermidis AIP-I with either alanine or the corresponding D-amino acid to explore the roles of each side chain and stereocenter in the interaction between AIP-I and AgrC-I (note, Cys4 was not mutated to Ala due to its essentiality for macrocycle formation, and Ala5 did not require alanine replacement). The initial set of 14 analogs (listed in Table 1) was synthesized using solid-phase peptide chemistry and purified to homogeneity using our previously established HPLC methods (59). We tested each analog for its ability to modulate the activity of the AgrC-I receptor using a wild-type S. epidermidis Group-I fluorescence reporter strain (see Methods). This strain contained a reporter plasmid encoding a gfp gene fused to the P3 promoter found upstream of RNAIII (36). Under high cell-density conditions, activation of the AgrC-I receptor by the endogenously produced AIP-I leads to phosphorylation of AgrA, which then binds the P3 promoter and activates gfp expression. The level of GFP fluorescence can therefore be quantified as a measurement of the extent of AgrC-I activation (note, this strain then fluoresces in the absence of added AIP-I at high densities). AgrC-I antagonism was measured by testing AIP-I analogs alone, while AgrC-I agonism was measured in the presence of a known strong AgrC-I inhibitor, S. epidermidis AIP-II, to block activation of AgrC-I by endogenously produced AIP-I. For comparison, we also evaluated the activities of the native S. epidermidis AIPs (I–III) and S. aureus AIPs (I–IV), as well as S. aureus AIP-III D4A, the most potent pan-group inhibitor of S. aureus agr systems that we have reported to date (59). The native AIPs served as important controls for our fluorescence reporter assays, as their effects on AgrC-I activation by AIP-I in S. epidermidis have been described previously (36, 45); the activity of the lead S. aureus agr inhibitor, AIP-III D4A, in S. epidermidis was unknown. Compounds demonstrating appreciable antagonism or agonism of AgrC-I in these assays were subjected to dose-response analyses, which permitted calculation of IC50 and EC50 values. The assay data for the alanine and D-amino acid AIP-I analogs and the control AIPs in AgrC-I are summarized in Table 1.
Table 1.
IC50 and EC50 values for alanine and D-amino acid scan analogs of S. epidermidis AIP-I in AgrC-I determined using a fluorescence reporter strain.a Shaded cells represent control peptides.
| Peptide Name | Sequence | IC50 (nM) | EC50b (nM) |
|---|---|---|---|
|
| |||
| AIP-I D1A | A-S-V-(C-A-S-Y-F) | - | 49.3 |
| AIP-I S2A | D-A-V-(C-A-S-Y-F) | - | >1000 |
| AIP-I V3A | D-S-A-(C-A-S-Y-F) | 51.9 | - |
| AIP-I S6A | D-S-V-(C-A-A-Y-F) | - | 71.0 |
| AIP-I Y7A | D-S-V-(C-A-S-A-F) | Inactivec | |
| AIP-I F8A | D-S-V-(C-A-S-Y-A) | Inactivec | |
| AIP-I D-D1 | DD-S-V-(C-A-S-Y-F) | - | >1000 |
| AIP-I D-S2 | D-DS-V-(C-A-S-Y-F) | 192d | |
| AIP-I D-V3 | D-S-DV-(C-A-S-Y-F) | >1000 | - |
| AIP-I D-C4 | D-S-V-(DC-A-S-Y-F) | Inactivec | |
| AIP-I D-A5 | D-S-V-(C-DA-S-Y-F) | Inactivec | |
| AIP-I D-S6 | D-S-V-(C-A-DS-Y-F) | - | >1000 |
| AIP-I D-Y7 | D-S-V-(C-A-S-DY-F) | Inactivec | |
| AIP-I D-F8 | D-S-V-(C-A-S-Y-DF) | - | >1000 |
| AIP-I | D-S-V-(C-A-S-Y-F) | - | 196 |
| AIP-II | N-A-S-K-Y-N-P-(C-S-N-Y-L) | 9.64 | - |
| AIP-III | N-A-A-K-Y-N-P-(C-A-S-Y-L) | 34.3 | - |
| S. aureus AIP-I | Y-S-T-(C-D-F-I-M) | Inactivec | |
| S. aureus AIP-II | G-V-N-A-(C-S-S-L-F) | 62.9 | - |
| S. aureus AIP-III | I-N-(C-D-F-L-L) | Inactivec | |
| S. aureus AIP-IV | Y-S-T-(C-Y-F-I-M) | >1000 | - |
| S. aureus AIP-III D4A | I-N-(C-A-F-L-L) | Inactivec | |
See Methods for details of the reporter strain and assay procedures. See Supplementary Information for peptide MS and HPLC characterization data, dose response curves, and 95% confidence intervals (CI) for IC50 and EC50 values.
Activation dose response curves were performed in the presence of 50 nM S. epidermidis AIP-II (an inhibitor) to block activation of AgrC-I by endogenously produced AIP-I.
Dose response curves revealed neither agonism nor antagonism over the concentration range tested.
Maximum inhibition did not exceed 60%.
The activities of the native S. epidermidis and S. aureus AIPs were congruent with previous reports, with the exception of S. aureus AIP-II. S. aureus AIP-II was previously reported to be inactive toward S. epidermidis AgrC-I (46), but behaved as a relatively strong AgrC-I antagonist in our assay. This difference may be attributable to variances in the assay protocol and the strain used (46). Interestingly, we found S. aureus AIP-III D4A, our potent pan-group inhibitor in S. aureus, to be largely inactive in S. epidermidis Group-I, suggesting that this peptide could be used as a species-specific agr inhibitor in S. aureus. We return to this possibility below.
Turning to the alanine and D-amino acid scans of AIP-I, several interesting trends in the SAR of the AIP-I:AgrC-I interaction were revealed (Table 1). Previous SAR studies of S. aureus AIPs have shown that activation of an AgrC receptor by its cognate AIP likely involves two main steps: (i) initial binding of the AIP to AgrC (i.e., AIP “recognition”) that is mediated largely by the macrocycle and bulky, hydrophobic endocyclic residues, and (ii) subsequent activation of AgrC via interactions with the exocyclic tail region of the bound AIP (55, 61, 63). Consistent with these previous studies, replacement of either of the two hydrophobic residues (Tyr7 and Phe8) in the macrocycle of the S. epidermidis AIP-I with alanine resulted in a complete loss of activity in AgrC-I. Furthermore, the substitution of Tyr7 with the corresponding D-amino acid (i.e., AIP-I D-Y7) also caused a complete loss of activity, demonstrating the stringent requirement of this residue for AIP-I recognition by AgrC-I. AIP-I D-F8, however, maintained agonistic activity, albeit with lower potency relative to AIP-I, suggesting that the spatial configuration at this position plays less of a role in AIP-I:AgrC-I interactions in comparison to the composition of the side chain. Also in agreement with prior observations of AgrC receptor activation by receptor bound AIPs (16, 55), alanine or D-amino acid replacements at residue Val3 in the exocyclic tail region converted AIP-I into a moderate or weak antagonist, respectively, demonstrating that Val3 likely plays an important role in AgrC-I activation. Replacing the nearby residue Ser2 with an alanine caused a decrease in agonistic activity, indicating that Ser2 contributes to AgrC-I activation by AIP-I. In addition, substitution with the D-amino acid counterpart at Ser2 yielded a weak AgrC-I antagonist, suggesting that the orientation of the residue at this position may influence the presentation of the key activating residue, Val3.
Converting both Cys4 and Ala5 residues to their D-isomers caused a complete loss of activity. Given the locations of these two stereocenters relative to the key thioester linkage forming the macrocycle, this loss of activity could be the result of either a stringent requirement for the correct local stereochemistry at these two positions for AIP-I recognition or a global shift in the macrocycle conformation that prevents proper interaction between the key hydrophobic residues and AgrC-I. Additional structural studies are needed to provide a more definitive explanation for these observed effects. Perhaps the most intriguing finding in these initial AIP-I scans was that substitution of either Asp1 in the exocyclic tail or Ser6 in the macrocycle with alanine increased the agonistic activity of AIP-I on AgrC-I (by 2–4-fold). This observation is, to our knowledge, the first report that two residues in a native AIP allow for suboptimal activation of its cognate AgrC receptor. Previous SAR studies in S. aureus have identified only one residue in a single native AIP (S. aureus AIP-II) where a mutation yielded an analog with an increased agonistic activity relative to the starting structure (the extent of this increase was not reported) (16). Replacement of Asp1 and Ser6 with their D-Amino acid counterparts reduced, but did not abolish, the agonistic activity of AIP-I. Together, these SAR trends suggest that the spatial configuration of these two residues contribute to the productive binding of AIP-I to its receptor, while the side chain composition of these residues appear to hinder optimal AIP-I:AgrC-I interactions (relative to alanine analogs).
SAR studies direct the design of second-generation AIP-I analogs
A second set of S. epidermidis AIP-I analogs was next designed and synthesized to further examine the SAR trends revealed by the alanine and D-Amino acid analogs, and to potentially generate more potent modulators of AgrC-I. This set of analogs included peptides with substitutions at Ala5 (a position inaccessible through the alanine scan), combinatorial alanine modifications, and truncations to the exocyclic tail. In addition, a series of S. epidermidis AIP-II analogs with decreasing length of the exocyclic tail were also synthesized. These latter peptides were designed to explore the use of the AIP-II scaffold for AgrC-I inhibition, as the native AIP-II demonstrated the strongest inhibitory potency of our first-generation analogs and control peptides (see Table 1). The structures of the second-generation analogs are shown in Table 2, and their associated activities in S. epidermidis AgrC-I were assessed using the fluorescence reporter assay introduced above.
Table 2.
IC50 and EC50 values of second-generation AIP analogs in S. epidermidis AgrC-I determined using a fluorescence reporter strain.a
| Peptide Name | Sequence | IC50 (nM) | EC50b (nM) |
|---|---|---|---|
|
| |||
| AIP-I A5Y | D-S-V-(C-Y-S-Y-F) | 95.4 | - |
| AIP-I A5S | D-S-V-(C-S-S-Y-F) | 135c, d | |
| AIP-I V3AA5Y | D-S-A-(C-Y-S-Y-F) | 59.6 | - |
| AIP-I V3AA5S | D-S-A-(C-S-S-Y-F) | 29.4 | - |
| AIP-I D1AS6A | A-S-V-(C-A-A-Y-F) | - | 10.3 |
| tAIP-I | Ac-(C-A-S-Y-F)e | 192 | - |
| AIP-I D1AS6AV3A | A-S-A-(C-A-A-Y-F) | 2.84 | - |
| AIP-I D1AS6AV3AA5S | A-S-A-(C-S-A-Y-F) | 2.08 | - |
| AIP-II 11aa | A-S-K-Y-N-P-(C-S-N-Y-L) | 5.43 | - |
| AIP-II 10aa | S-K-Y-N-P-(C-S-N-Y-L) | 5.29 | - |
| AIP-II 9aa | K-Y-N-P-(C-S-N-Y-L) | 2.83 | - |
| AIP-II 8aa | Y-N-P-(C-S-N-Y-L) | 6.08 | - |
| AIP-II 7aa | N-P-(C-S-N-Y-L) | 721 | - |
| tAIP-II | Ac-(C-S-N-Y-L)e | >1000 | - |
See footnotes in Table 1.
Maximum inhibition did not exceed 75%.
Two different AIP-I A5S analogs were isolated that exhibited differing activities; IC50 value listed is derived from the antagonism dose response curves for A5S-1 (see text).
Ac = acetylated N-terminus.
To assess the role of the Ala5 side chain in the AIP-I:AgrC-I interaction, we replaced Ala5 with either serine or tyrosine. These modifications were selected based on the primary structures of the native S. aureus and S. epidermidis AIPs that were found to exhibit inhibitory activities against AgrC-I (Table 1). Specifically, with the exception of S. epidermidis AIP-III having an alanine, all other native AIPs active in AgrC-I possess either a serine or a tyrosine at the second endocyclic position. We found substitution of Ala5 with a tyrosine yielded a weak AgrC-I antagonist (Table 2), whereas substitution with a serine surprisingly produced two peptide products of the correct mass with differing activities (A5S-1 and A5S-2). Analog A5S-1 behaved as a weak inhibitor against AgrC-I, while A5S-2 displayed a non-monotonic dose response that indicated very weak inhibition at low concentrations yet activation at higher concentrations (see Supporting Information). When either the A5Y or A5S modification was combined with the inhibitory modification V3A, the resulting analogs, AIP-I V3AA5Y and AIP-I V3AA5S, exhibited similar inhibitory activities as the parent compound AIP-I V3A. One possible explanation for these observations is that the modifications at Ala5 influence the orientation of the activating residue Val3, leading to varying degrees of AgrC-I activation. When the activating valine is replaced with an alanine, the mutations of Ala5 have little to no effect on the activity of the resulting analogs. The chemical difference between the two AIP-I A5S analogs remains unclear, but we speculate that the two peptides are conformational isomers; efforts to elucidate the structural differences are ongoing.
Next, we tested additional AIP-I analogs with multiple alanine modifications to examine whether their modulatory effects on AgrC-I could be additive. Replacing both Asp1 and Ser6 residues with alanine generated an analog with enhanced agonistic activity in AgrC-I relative to the parent single alanine mutants (Table 2). This observation led to us testing the hypothesis that introduction of the inhibitory modification V3A to the D1AS6A analog, an AIP-I analog that presumably associates strongly with AgrC-I, could produce an AgrC-I antagonist with increased potency. Indeed, the resulting analog – AIP-I D1AS6AV3A – was one of the most potent AgrC-I antagonists identified in this study (IC50 = ~2 nM). A similar analog AIP-I D1AS6AV3AA5S, which contained an additional mutation (A5S) that could potentially further enhance AgrC-I binding, exhibited analogous potency as the parent triple mutant. Overall, the activities of these AIP-I analogs with combinatorial alanine mutations strengthened the SAR trends delineated in our first set of AIP-I analogs. Specifically, their activities confirmed that Val3 is the essential residue for activation of AgrC-I and that the side chains of residues Asp1 and Ser6 play a negative role in the AIP-I:AgrC-I interaction. The importance of the exocyclic tail portion of AIP-I (which includes Val3) for AgrC-I activation was further confirmed by the weak inhibitory activity observed for the truncated analog of AIP-I (tAIP-I; Table 2). This observation was again consistent with prior SAR studies in S. aureus, where AIP macrocycles without exocyclic tails were shown to be sufficient for receptor recognition but not receptor activation, and thus acted as competitive AgrC inhibitors (16, 55, 64).
The last group of peptides examined was the AIP-II analogs with varying lengths of the exocyclic tail (Table 2). Notably, both the native S. epidermidis AIP-II and AIP-III signals have markedly longer exocyclic tails relative to other structurally characterized AIPs, and understanding the role of this structural feature, for receptor activation or inhibition, is of fundamental interest (36). The AIP-II analogs in this study were designed to determine whether the primary structure of AIP-II could be reduced, while maintaining the high potency of its inhibitory activity toward AgrC-I. Removing one amino acid at a time from the N-terminus of AIP-II (Table 2, Rows 9–13), we found that an AIP-II analog with only three amino acids left in the exocyclic tail (AIP-II 8aa) still exhibited inhibitory activity toward AgrC-I with potency similar to the parent AIP-II. Further removal of amino acids from the N-terminus (as in AIP-II 7aa and tAIP-II) caused a dramatic reduction in inhibitory activity. Since AIP-II 8aa has the same number of residues as the most potent AIP-I analogs introduced above, optimization of this AIP-II scaffold using the SAR trends uncovered herein could yield interesting AgrC-I modulators exhibiting strong activities.
Agr group-selective inhibition in S. epidermidis by certain AIP analogs
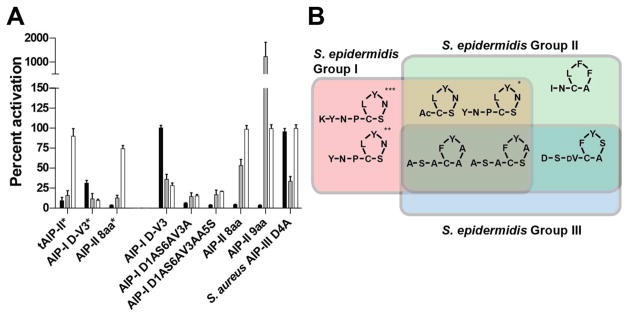
As shown in Table 1, we found the native AIP-II and AIP-III signals of S. epidermidis both inhibit AgrC-I, congruent with the results of Olson et al. (36). However, the biological significance of this agr interference remains unclear, although it has been hypothesized to potentially play a role in competition between different groups in a mixed bacteria milieu (18). Developing chemical modulators that target a specific agr group and are inactive toward the others could provide valuable tools for investigating the hypothesis of agr interference. To this end, we screened all of the AIP analogs introduced above in GFP reporter strains of Groups-II and -III S. epidermidis to complement the Group-I S. epidermidis data in Tables 1 and 2 (see Methods; Figures S-1 and S-2). The only AIP-I analog to show moderate group selectivity was AIP-I Y7A, which was inactive against Group-I but inhibited AgrC-II and –III to ~50% (Figures S-1 and S-2). More interestingly, two AIP-II analogs, tAIP-II and AIP-II 8aa, were shown to be strongly selective against Group-I and -II at 10 μM (inhibiting to ~75%) but were inactive against Group-III (Figure 2A).
Figure 2.
Pan-group activity profiles of selected S. epidermidis AIP-I analogs in S. epidermidis. (A) Effects of AIP-I analogs on AgrC-I–III as determined by fluorescence reporter strains (see Methods for details of strains and assay procedures). Compounds were tested at either 10 μM (indicated with *) or 100 nM. Black: Group-I; Gray: Group-II; White: Group-III. (B) Venn diagram summarizing agr group selectivity profiles for AgrC inhibition by AIP analogs in S. epidermidis. AIP-II 8aa shows differential selectivity at *10 μM and at **100 nM. ***Compound is an agonist of AgrC-II (see text).
We next screened a subset of peptides in these cross-group screens at a lower concentration to better gauge their relative activities (1 μM or 100 nM; Figure 2A, Figure S-3). At the lower concentration, AIP-I D-V3 remained an antagonist against Group-II and Group-III but lost its inhibitory activity toward Group-I (Figure 2A). Conversely, AIP-II 8aa remained active toward Group-I at 100 nM, whereas its inhibitory activity against Group-II decreased, making the activity of this analog more Group-I specific. AIP-II 9aa and S. aureus AIP-III D4A also exhibited group-selective activities, with AIP-II 9aa being inactive toward Group-III and S. aureus AIP-III D4A inhibiting exclusively Group-II. We note, however, that while AIP-II 9aa only inhibited Group-I, AIP-II 9aa was also a highly active agonist for Group-II. These selectively profiles are significant, as few agr group selective probes have been reported in any staphylococcal species (60), and none have been reported in S. epidermidis. Accordingly, these peptides represent a valuable new suite of chemical probes, and hold promise for examining the role of agr-based QS competition between any two agr-groups of S. epidermidis grown together. For example, by using AIP-II 8aa to target Group-I, S. aureus AIP-III D4A to target Group-II, or AIP-I D-V3 to target Group-III, a single agr system can be selectively inhibited in a bacterial population containing two different specificity groups (Figure 2B). We predict that further SAR studies of the AIP-II and AIP-III scaffolds may generate additional peptides with complementary group-selective activities.
Potent pan-group inhibitors of the agr system in S. epidermidis
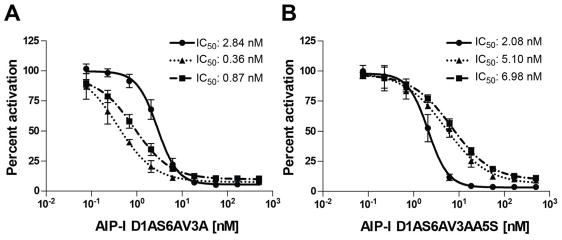
During our efforts to identify agr-group selective modulators, we noticed that AIP-I D1AS6AV3A and AIP-I D1AS6AV3AA5S, two of the most potent inhibitors that we identified in AgrC-I, also strongly inhibited AgrC-II and AgrC-III at 100 nM (Figure 2). To determine whether these two analogs were equally potent against AgrC-II and -III, we performed dose response antagonism assays on these analogs. The IC50 values obtained for AIP-I D1AS6AV3A and AIP-I D1AS6AV3AA5S against AgrC-II and -III were both in the subnanomolar to low nanomolar range and comparable to their IC50 values in AgrC-I (Figure 3). These two analogs represent, to the best of our knowledge, the first potent pan-group inhibitors of the agr system in S. epidermidis. Complementary to the selective probes described above, AIP-I D1AS6AV3A and AIP-I D1AS6AV3AA5S should prove useful in a range of fundamental and applied studies, perhaps most so in those where agr inhibition in all three groups of S. epidermidis is required.
Figure 3.
AgrC antagonism dose response curves and IC50 values for (A) AIP-I D1AS6AV3A and (B) AIP-I D1AS6AV3AA5S in the S. epidermidis Group-I–III GFP reporter strains. Circles: Group-I; triangles: Group-II; squares: Group-III. See Methods for strains and assay procedures.
Species-specific agr inhibition between S. epidermidis Group-I and S. aureus
The S. epidermidis AIP-I is also known to inhibit the agr system in S. aureus. Similar to the cross-group inhibition observed between different agr groups of the same species, this cross-species inhibitory effect has no known biological role but is hypothesized to provide a competitive advantage on a host (18, 46). Thus, we sought to examine the AIP analogs described herein to identify potential species-specific inhibitors that target either S. epidermidis Group-I or S. aureus Groups-I–IV. As noted in the discussions above, S. aureus AIP-III D4A, our most potent pan-group inhibitor against S. aureus, was inactive against S. epidermidis Group-I at 100 nM (Figure 4). We expanded our search and screened all of the AIP analogs reported herein capable of inhibiting the agr system in S. epidermidis Group-I for activity against the agr systems of S. aureus Groups-I–IV using analogous GFP reporter constructs (see Methods). We identified three peptides (Figure 4 and Figure S-4), chief among them AIP-II 8aa, that selectively inhibited the agr system in S. epidermidis Group-I without significantly affecting agr activity in S. aureus Groups-I–IV. Another analog, AIP-II 9aa, was shown to be largely an inhibitor specific to S. epidermidis Group-I, with only weak inhibitory activity in S. aureus Group-III. These two analogs, together with S. aureus AIP-III D4A, represent the first set of species-specific, chemical modulators targeting the agr systems of either S. epidermidis or S. aureus.
Figure 4.
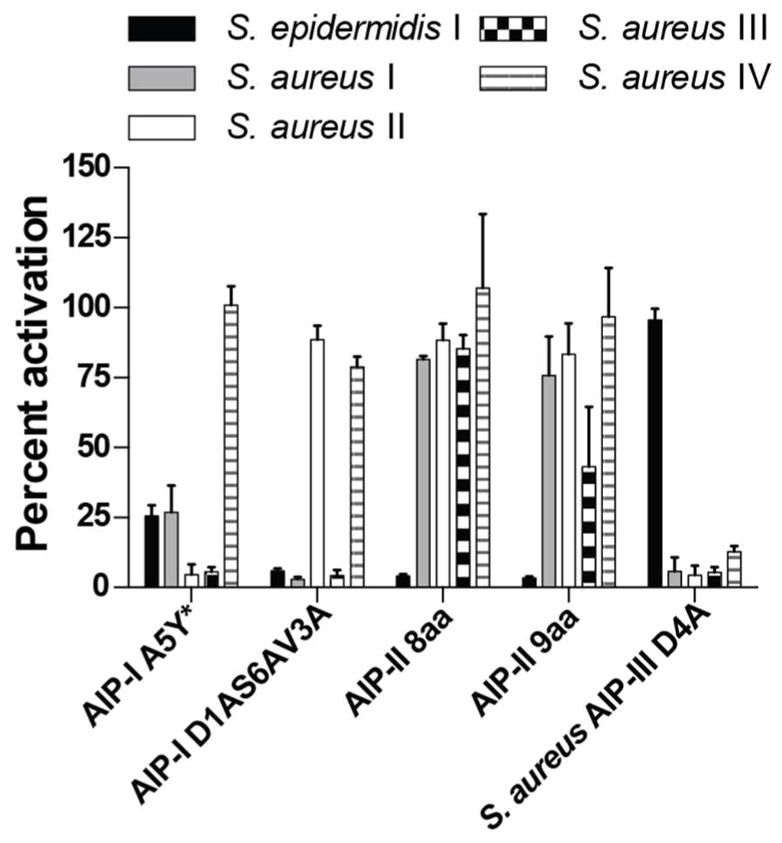
Effects of selected AIP analogs on the AgrC-I receptor in S. epidermidis Group-I and the AgrC I–IV receptors in S. aureus Groups-I–IV. Compounds tested at 100 nM unless otherwise noted. * Compound tested at 1 μM. See Methods for details of strains and assay procedures.
Modulation of biofilm formation in S. epidermidis Group-I using non-native AIPs
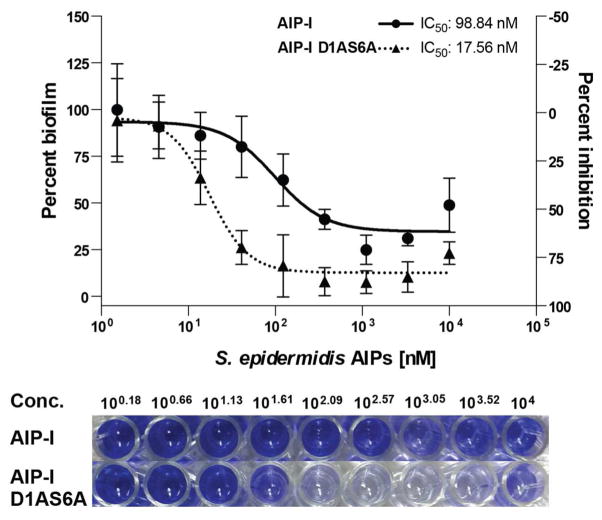
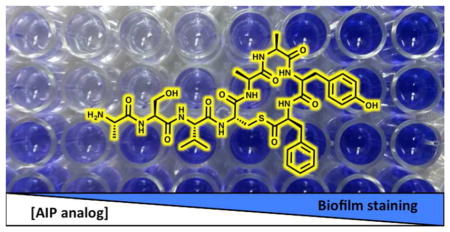
As described above, the use of fluorescence reporter strains allowed for the rapid screening of AIP analogs and the identification of AgrC modulators with varying activities. However, the responses from an artificial reporter gene system may not translate to effects on relevant QS phenotypes. As a validation for the physiological relevance of the new AgrC modulators uncovered in this study, we assessed the ability of select non-native AIPs to modulate biofilm formation by S. epidermidis Group-I using the PIA-producing strain RP62A in a crystal violet (CV) static biofilm assay (see Methods). Again, biofilm formation is the most important virulence phenotype associated with infection by S. epidermidis, and previous reports have established that its agr system plays an important role in biofilm formation (see above) (10, 35). In particular, activation of the agr system is believed to disperse biofilm, thereby preventing the accumulation of sessile bacteria on a surface. We therefore hypothesized that addition of native AIP-I (an AgrC agonist) would inhibit S. epidermidis biofilm growth, and in agreement with a similar study in S. aureus (32), static biofilm assays in 96-well microtiter plates in the presence of exogenous AIP-I showed substantial biofilm reduction (by ~60%, IC50 = 99 nM; Figure 5). We then tested AIP-I D1AS6A, the AIP-I analog identified with enhanced agonistic activity in AgrC-I relative to AIP-I, for modulation of S. epidermidis biofilm formation, and in agreement with its effect on the fluorescent reporter strain, this analog also decreased biofilm formation under similar conditions. In fact, it had overall enhanced efficacy and potency relative to AIP-I (~80% inhibition, IC50 = 18 nM; Figure 5). These matching activity trends provide support for the use of fluorescent reporter strains in identifying S. epidermidis AgrC modulators with physiological relevance. Moreover, our results with AIP-I D1AS6A provide the first non-native AIP analog, to our knowledge, that can strongly inhibit S. epidermidis biofilm growth.
Figure 5.
(Top) Biofilm antagonism dose response curves for S. epidermidis AIP-I and AIP-I D1AS6A against S. epidermidis Group-I RP62A strain as determined by CV staining (see Methods for assay procedure). Dose response curves shown represent the average of at least 4 biological replicates. (Bottom) Representative image of the CV assay with S. epidermidis AIP-I and AIP-I D1AS6A over a range of concentrations. Blue color correlates with biofilm growth.
While agonists of the agr system can decrease biofilm growth, agr system antagonists are generally expected to promote biofilm accumulation. Indeed, as discussed above, this relationship between the agr system and biofilm has raised concern over the merit of inhibiting QS in S. epidermidis (and related staphylococcal species) as a method of reducing virulence (28, 34). To our surprise, however, exposing S. epidermidis Group-I to our nanomolar AgrC-I inhibitor (AIP-I D1AS6AV3AA5S) at 10 μM did not increase biofilm growth as expected in the static biofilm assay (Figure S-6). Instead, the inhibitor did not alter, and perhaps even decreased, biofilm growth. While the CV assay is a relatively rudimentary method to monitor biofilm formation and the results obtained could be specific to our experimental conditions, we believe these results reflect the complex roles played by the agr system during biofilm formation in S. epidermidis. We suspect that the lack of stimulation in biofilm accumulation by the antagonist is a result of the proficiency of S. epidermidis RP62A in PIA production, which could have maximized the formation of biofilm even in the absence of an exogenous AgrC antagonist. Inhibition of the agr system in a PIA-negative S. epidermidis strain could yield a more dramatic impact on biofilm formation. Additional investigation is certainly needed to better assess this hypothesis and the general consequences of modulating QS during S. epidermidis colonization. The new modulators reported in the current study could be useful tools for such an endeavor.
SUMMARY AND CONCLUSIONS
Infections by S. epidermidis have become a significant public health problem (2, 3, 6–8). Current treatment options are limited, as like many other common pathogens, S. epidermidis is increasingly antibiotic resistant and can establish robust biofilms (3–6, 8). The agr QS system is extensively involved in the pathogenesis of S. epidermidis infection, making this system a potentially attractive target for an alternative anti-infective strategy. However, the roles of the agr system in the infection process and biofilm formation are complex and dynamic. Chemical probes that interfere with the S. epidermidis agr system could pave a route toward a deeper understanding of how and when QS should be modulated to attenuate virulence. We report the first such compounds herein.
In this study, we prepared a series of non-native analogs of S. epidermidis AIP-I and performed the first systematic SAR analysis of this AIP scaffold for activation of AgrC-I (key SARs highlighted in Figure 6). This analysis had several important outcomes. First, it revealed two hydrophobic, endocyclic residues (Tyr7 and Phe8) that, in addition to the thioester linkage and Ala5, are essential for AgrC-I recognition. We also found that the exocyclic residue Val3 is critical for AgrC-I activation, and more interestingly, the Asp1 and Ser6 residues allow for submaximal activation of AgrC-I and actually could be mutated to augment the agonistic potencies of AIP-I analogs.
Figure 6.
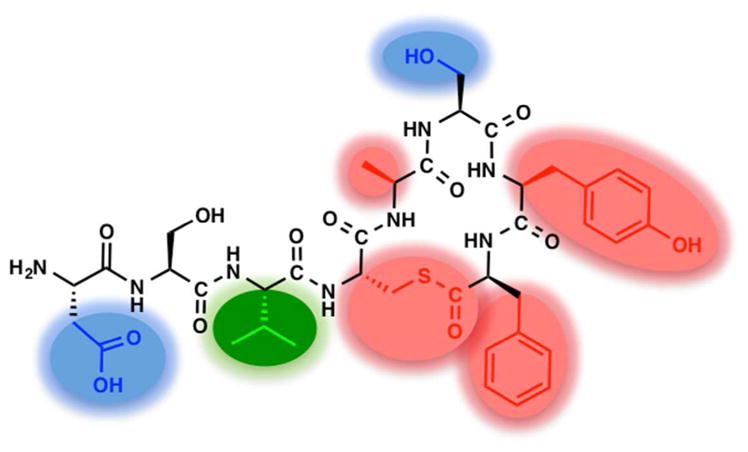
Summary of the key SAR trends for activation of S. epidermidis AgrC-I by AIP-I as revealed by the reporter assays in this study. Red: essential for AgrC receptor recognition. Green: essential for AgrC receptor activation. Blue: detrimental to AgrC receptor activation. Remaining residues contribute but are not critical to receptor recognition.
Second, our studies revealed several highly potent, non-native AgrC modulators, including the first pan-group AgrC inhibitors for S. epidermidis (AIP-I D1AS6AV3A and AIP-I D1AS6AV3AA5S). We also discovered a non-native agonist of AgrC-I (AIP-I D1AS6A) with improved potency over the native AIP-I and demonstrated that this analog was capable of strongly inhibiting S. epidermidis biofilm growth, an important virulence phenotype (at least) partially controlled by the agr system. This result suggests that the AgrC-I modulators reported herein can be used to alter QS-controlled virulence phenotypes in S. epidermidis. Moreover, AIP-I D1AS6A, to the best of our knowledge, represents the first non-native AIP analog with anti-biofilm activity in any staphylococcal species. Co-administration of this peptide with various antibiotics to examine whether S. epidermidis biofilm removal can improve the effectiveness of antibiotics would be an interesting experimental path forward, and is currently underway in our laboratories. As our most potent AgrC-I inhibitor (AIP-I D1AS6AV3AA5S) also slightly inhibited biofilm formation, examining its effects on biofilm growth and morphology in the presence of antibiotics would also be valuable. Looking to the future, applying AIP-I D1AS6AV3AA5S in various infection models at different stages of colonization could better illuminate the role of agr-type QS in S. epidermidis infections and allow us to assess the viability of targeting QS in this pathogen as a possible anti-infective treatment.
Third and to close, this study has provided the first set of AgrC inhibitors that, at the concentrations tested, are selective either for particular S. epidermidis specificity groups or for a staphylococcal species (i.e., S. epidermidis or S. aureus). These new group and species selective AgrC inhibitors now can be utilized to selectively target an agr system in a mixed bacteria population and investigate the hypothesis of agr interference (18, 42–44). Preliminary efforts toward this goal are ongoing in our laboratories and will be reported in due course. Overall, we predict that these peptide tools and the insights that they provide will open new avenues of investigation for QS in Staphylococci and could provide a pathway, with further development, toward new chemical strategies to attenuate infection.
METHODS
Biological reagents, strains, and general methods
All standard reagents were purchased from commercial sources and used according to enclosed instructions. The S. epidermidis fluorescence reporter strains AH3408 (Group-I), AH3567 (Group-II), and AH3409 (Group-III) were cultured in Tryptic Soy Broth (TSB, from Sigma) augmented with 10 μg mL−1 of erythromycin (36). The S. epidermidis wild-type strain RP62A (Group-I) was cultured in TSB (from Sigma or EMD Millipore). The S. aureus fluorescence reporter strains AH1677 (Group-I), AH430 (Group-II), AH1747 (Group-III), and AH1872 (Group-IV) were cultured in Brain Heart Infusion (BHI, from Teknova) augmented with 10 μg mL−1 of chloramphenicol (65). All cultures were grown at 37 °C with shaking (200 rpm) unless noted otherwise. Absorbance and fluorescence measurements were obtained using a Biotek Synergy 2 microplate reader operating Gen5 data analysis software. IC50 values and EC50 values were determined from sigmoidal curve fits using GraphPad Prism software (v. 6.0).
Peptide synthesis
S. epidermidis AIP analogs were synthesized on solid-phase resin, purified by preparative high performance liquid chromatography (HPLC), and characterized using our previously reported methods (59). See Supplementary Information for mass spectrometry (MS) and HPLC characterization data for the peptides in this study, as well as additional information on reagents and instrumentation used in peptide synthesis.
Fluorescence reporter assay protocol
For AgrC antagonism assays, peptide stock solutions in DMSO (1 mM) were serially diluted with DMSO (either 1:3 or 1:10), and 2 μL aliquots of the diluted solutions were added to each of the wells in a black 96-well polystyrene microtiter plate (Costar). Each peptide solution was tested in triplicate. As a negative control, 2 μL of DMSO was included. An overnight culture of the S. epidermidis or S. aureus fluorescence reporter strain was diluted 1:50 with fresh media, and 198 μL of the diluted culture were added to each of the wells of the microtiter plate containing peptide. Plates were then incubated at 37 °C for 24 h. Fluorescence (EX 500 nm / EM 540 nm) and OD600 of each well was measured using a plate reader, and the fluorescence values were normalized to the DMSO control. For S. epidermidis AgrC-I agonism assays, an additional 2 μL of S. epidermidis AIP-II stock solution was added to the wells to a final concentration of 50 nM, in order to block AgrC-I activation by endogenously produced AIP-I. The remaining steps of the fluorescence assay were the same as above.
For single-concentration fluorescence reporter assays, 2 μL of the peptide stock solutions were added to the wells in a black 96-well microtiter plate to a final concentration of 10 μM, 1 μM, or 100 nM. The remaining steps of the fluorescence assay were the same as above.
Crystal violet biofilm assay protocol
Peptide stock solutions were serially diluted with DMSO (1:3), and 2 μL of the diluted solutions were added to each of the wells in a clear, flat-bottom, 96-well polystyrene microtiter plate (Costar). Each solution was tested in triplicate. As a negative control, 2 μL of DMSO was included. An overnight culture of S. epidermidis RP62A was diluted 1:100 with fresh TSB (EMD Millipore) augmented with 0.5% (w/v) glucose, and 198 μL of the diluted culture were added to each of the wells of the microtiter plate containing peptides. Bacteria were incubated at 37 °C for 24 h under static conditions. Quantification of the amount of biofilm formed using crystal violet (CV) assays was performed according to previously reported protocols (66). For single-concentration CV assays, 2 μL of the peptide stock solutions were added to the wells to a final concentration of 10 μM. The remaining steps of the biofilm assay were the same as above.
Supplementary Material
Acknowledgments
Financial support for this work was provided by the Office of Naval Research (N00014-14-1-0791) and the Burroughs Wellcome Fund (to H.E.B) and a Merit Award (I01 BX002711) from the Department of Veteran Affairs (to A.R.H). T.Y. was supported in part by the UW–Madison NIH Biotechnology Training Program (T32 GM08349), and A.E.P was supported by an American Heart Association predoctoral fellowship. MS instrumentation at UW–Madison is supported by the NIH (1S10 0D020022) and a generous gift from P. Bender.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Vuong C, Otto M. Staphylococcus epidermidis infections. Microb Infect. 2002;4:481–489. doi: 10.1016/s1286-4579(02)01563-0. [DOI] [PubMed] [Google Scholar]
- 2.von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis. 2002;2:677–685. doi: 10.1016/s1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- 3.Otto M. Staphylococcus epidermidis--the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCann MT, Gilmore BF, Gorman SP. Staphylococcus epidermidis device-related infections: pathogenesis and clinical management. J Pharm Pharmacol. 2008;60:1551–1571. doi: 10.1211/jpp/60.12.0001. [DOI] [PubMed] [Google Scholar]
- 5.Mertens A, Ghebremedhin B. Genetic determinants and biofilm formation of clinical Staphylococcus epidermidis isolates from blood cultures and indwelling devises. Eur J Microbiol Immunol (Bp) 2013;3:111–119. doi: 10.1556/EuJMI.3.2013.2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers KL, Fey PD, Rupp ME. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am. 2009;23:73–98. doi: 10.1016/j.idc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 8.Claessens J, Roriz M, Merckx R, Baatsen P, Van Mellaert L, Van Eldere J. Inefficacy of vancomycin and teicoplanin in eradicating and killing Staphylococcus epidermidis biofilms in vitro. Int J Antimicrob Agents. 2015;45:368–375. doi: 10.1016/j.ijantimicag.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Cheung GY, Joo HS, Chatterjee SS, Otto M. Phenol-soluble modulins - critical determinants of staphylococcal virulence. FEMS Microbiol Rev. 2014;38:698–719. doi: 10.1111/1574-6976.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong KF, Vuong C, Otto M. Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol. 2006;296:133–139. doi: 10.1016/j.ijmm.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 11.Vuong C, Durr M, Carmody AB, Peschel A, Klebanoff SJ, Otto M. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell Microbiol. 2004;6:753–759. doi: 10.1111/j.1462-5822.2004.00401.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Li M, Dong D, Wang J, Ren J, Otto M, Gao Q. Role of ClpP in biofilm formation and virulence of Staphylococcus epidermidis. Microb Infect. 2007;9:1376–1383. doi: 10.1016/j.micinf.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Yao Y, Vuong C, Kocianova S, Villaruz AE, Lai Y, Sturdevant DE, Otto M. Characterization of the Staphylococcus epidermidis accessory-gene regulator response: quorum-sensing regulation of resistance to human innate host defense. J Infect Dis. 2006;193:841–848. doi: 10.1086/500246. [DOI] [PubMed] [Google Scholar]
- 14.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyon GJ, Novick RP. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides. 2004;25:1389–1403. doi: 10.1016/j.peptides.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 17.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 18.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George EA, Muir TW. Molecular mechanisms of agr quorum sensing in virulent staphylococci. ChemBioChem. 2007;8:847–855. doi: 10.1002/cbic.200700023. [DOI] [PubMed] [Google Scholar]
- 20.Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kavanaugh JS, Thoendel M, Horswill AR. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol Microbiol. 2007;65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Zhao A, Novick RP, Muir TW. Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Mol Cell. 2014;53:929–940. doi: 10.1016/j.molcel.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George Cisar EA, Geisinger E, Muir TW, Novick RP. Symmetric signalling within asymmetric dimers of the Staphylococcus aureus receptor histidine kinase AgrC. Mol Microbiol. 2009;74:44–57. doi: 10.1111/j.1365-2958.2009.06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otto M. Phenol-soluble modulins. Int J Med Microbiol. 2014;304:164–169. doi: 10.1016/j.ijmm.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buttner H, Mack D, Rohde H. Structural basis of Staphylococcus epidermidis biofilm formation: mechanisms and molecular interactions. Front Cell Infect Microbiol. 2015;5:14. doi: 10.3389/fcimb.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otto M. Virulence factors of the coagulase-negative staphylococci. Front Biosci. 2004;9:841–863. doi: 10.2741/1295. [DOI] [PubMed] [Google Scholar]
- 28.Mack D, Davies AP, Harris LG, Rohde H, Horstkotte MA, Knobloch JK. Microbial interactions in Staphylococcus epidermidis biofilms. Anal Bioanal Chem. 2007;387:399–408. doi: 10.1007/s00216-006-0745-2. [DOI] [PubMed] [Google Scholar]
- 29.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188:706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 30.Wang R, Khan BA, Cheung GY, Bach TH, Jameson-Lee M, Kong KF, Queck SY, Otto M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121:238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le KY, Dastgheyb S, Ho TV, Otto M. Molecular determinants of staphylococcal biofilm dispersal and structuring. Front Cell Infect Microbiol. 2014;4:167. doi: 10.3389/fcimb.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boles BR, Horswill AR. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Path. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis. 2004;190:1498–1505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]
- 34.Otto M. Quorum-sensing control in Staphylococci -- a target for antimicrobial drug therapy? FEMS Microbiol Lett. 2004;241:135–141. doi: 10.1016/j.femsle.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 35.Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med. 2013;64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 36.Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, Buttner H, Dunman PM, Rohde H, Cech NB, Fey PD, Horswill AR. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J Bacteriol. 2014;196:3482–3493. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klug D, Wallet F, Kacet S, Courcol RJ. Involvement of adherence and adhesion Staphylococcus epidermidis genes in pacemaker lead-associated infections. J Clin Microbiol. 2003;41:3348–3350. doi: 10.1128/JCM.41.7.3348-3350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hellmark B, Soderquist B, Unemo M, Nilsdotter-Augustinsson A. Comparison of Staphylococcus epidermidis isolated from prosthetic joint infections and commensal isolates in regard to antibiotic susceptibility, agr type, biofilm production, and epidemiology. Int J Med Microbiol. 2013;303:32–39. doi: 10.1016/j.ijmm.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Lianhua Y, Yunchao H, Guangqiang Z, Kun Y, Xing L, Fengli G. The effect of iatrogenic Staphylococcus epidermidis intercellar adhesion operon on the formation of bacterial biofilm on polyvinyl chloride surfaces. Surg Infect (Larchmt) 2014;15:768–773. doi: 10.1089/sur.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaeffer CR, Woods KM, Longo GM, Kiedrowski MR, Paharik AE, Buttner H, Christner M, Boissy RJ, Horswill AR, Rohde H, Fey PD. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect Immun. 2015;83:214–226. doi: 10.1128/IAI.02177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carmody AB, Otto M. Specificity grouping of the accessory gene regulator quorum-sensing system of Staphylococcus epidermidis is linked to infection. Arch Microbiol. 2004;181:250–253. doi: 10.1007/s00203-003-0644-2. [DOI] [PubMed] [Google Scholar]
- 42.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 43.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright JS, 3rd, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otto M. Staphylococcus aureus and Staphylococcus epidermidis peptide pheromones produced by the accessory gene regulator agr system. Peptides. 2001;22:1603–1608. doi: 10.1016/s0196-9781(01)00495-8. [DOI] [PubMed] [Google Scholar]
- 46.Otto M, Echner H, Voelter W, Gotz F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69:1957–1960. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otto M, Sussmuth R, Vuong C, Jung G, Gotz F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999;450:257–262. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- 48.Fleming V, Feil E, Sewell AK, Day N, Buckling A, Massey RC. Agr interference between clinical Staphylococcus aureus strains in an insect model of virulence. J Bacteriol. 2006;188:7686–7688. doi: 10.1128/JB.00700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial Competition for Human Nasal Cavity Colonization: Role of Staphylococcal agr Alleles. Appl Environ Microbiol. 2003;69:18–23. doi: 10.1128/AEM.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amara N, Krom BP, Kaufmann GF, Meijler MM. Macromolecular inhibition of quorum sensing: enzymes, antibodies, and beyond. Chem Rev. 2011;111:195–208. doi: 10.1021/cr100101c. [DOI] [PubMed] [Google Scholar]
- 51.Galloway WR, Hodgkinson JT, Bowden S, Welch M, Spring DR. Applications of small molecule activators and inhibitors of quorum sensing in Gram-negative bacteria. Trends Microbiol. 2012;20:449–458. doi: 10.1016/j.tim.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 52.Praneenararat T, Palmer AG, Blackwell HE. Chemical methods to interrogate bacterial quorum sensing pathways. Org Biomol Chem. 2012;10:8189–8199. doi: 10.1039/c2ob26353j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.PM, Affas Z, Reynolds C, Holden MT, Wood SJ, Saint S, Cockayne A, Hill PJ, Dodd CE, Bycroft BW, Chan WC, Williams P. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol Microbiol. 2001;41:503–512. doi: 10.1046/j.1365-2958.2001.02539.x. [DOI] [PubMed] [Google Scholar]
- 54.Lyon GJ, Mayville P, Muir TW, Novick RP. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc Natl Acad Sci USA. 2000;97:13330–13335. doi: 10.1073/pnas.97.24.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lyon GJ, Wright JS, Muir TW, Novick RP. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry. 2002;41:10095–10104. doi: 10.1021/bi026049u. [DOI] [PubMed] [Google Scholar]
- 56.Scott RJ, Lian LY, Muharram SH, Cockayne A, Wood SJ, Bycroft BW, Williams P, Chan WC. Side-chain-to-tail thiolactone peptide inhibitors of the staphylococcal quorum-sensing system. Bioorg Med Chem Lett. 2003;13:2449–2453. doi: 10.1016/s0960-894x(03)00497-9. [DOI] [PubMed] [Google Scholar]
- 57.George EA, Novick RP, Muir TW. Cyclic peptide inhibitors of staphylococcal virulence prepared by Fmoc-based thiolactone peptide synthesis. J Am Chem Soc. 2008;130:4914–4924. doi: 10.1021/ja711126e. [DOI] [PubMed] [Google Scholar]
- 58.Fowler SA, Stacy DM, Blackwell HE. Design and synthesis of macrocyclic peptomers as mimics of a quorum sensing signal from Staphylococcus aureus. Org Lett. 2008;10:2329–2332. doi: 10.1021/ol800908h. [DOI] [PubMed] [Google Scholar]
- 59.Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J Am Chem Soc. 2013;135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- 60.Tal-Gan Y, Stacy DM, Blackwell HE. N-Methyl and peptoid scans of an autoinducing peptide reveal new structural features required for inhibition and activation of AgrC quorum sensing receptors in Staphylococcus aureus. Chem Commun (Camb) 2014;50:3000–3003. doi: 10.1039/c4cc00117f. [DOI] [PubMed] [Google Scholar]
- 61.Johnson JG, Wang B, Debelouchina GT, Novick RP, Muir TW. Increasing AIP Macrocycle Size Reveals Key Features of agr Activation in Staphylococcus aureus. ChemBioChem. 2015;16:1093–1100. doi: 10.1002/cbic.201500006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otto M, Sussmuth R, Jung G, Gotz F. Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 1998;424:89–94. doi: 10.1016/s0014-5793(98)00145-8. [DOI] [PubMed] [Google Scholar]
- 63.Khan BA, Yeh AJ, Cheung GY, Otto M. Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin Investig Drugs. 2015;24:689–704. doi: 10.1517/13543784.2015.1019062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tal-Gan Y, Ivancic M, Cornilescu G, Cornilescu CC, Blackwell HE. Structural characterization of native autoinducing peptides and abiotic analogues reveals key features essential for activation and inhibition of an AgrC quorum sensing receptor in Staphylococcus aureus. J Am Chem Soc. 2013;135:18436–18444. doi: 10.1021/ja407533e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirchdoerfer RN, Garner AL, Flack CE, Mee JM, Horswill AR, Janda KD, Kaufmann GF, Wilson IA. Structural basis for ligand recognition and discrimination of a quorum-quenching antibody. J Biol Chem. 2011;286:17351–17358. doi: 10.1074/jbc.M111.231258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kratochvil MJ, Tal-Gan Y, Yang T, Blackwell HE, Lynn DM. Nanoporous Superhydrophobic Coatings that Promote the Extended Release of Water-Labile Quorum Sensing Inhibitors and Enable Long-Term Modulation of Quorum Sensing in Staphylococcus aureus. ACS Biomater Sci Eng. 2015;1:1039–1049. doi: 10.1021/acsbiomaterials.5b00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.