Abstract
Background
CUG-BP, Elav-like family member 1 (CELF1) is a multifunctional RNA binding protein found in a variety of adult and embryonic tissues. In the heart, CELF1 is found exclusively in the myocardium. However, the roles of CELF1 during cardiac development have not been completely elucidated.
Results
Myofibrillar organization is disrupted and proliferation is reduced following knockdown of CELF1 in cultured chicken primary embryonic cardiomyocytes. In vivo knockdown of Celf1 in developing Xenopus laevis embryos resulted in myofibrillar disorganization and a trend towards reduced proliferation in heart muscle, indicating conserved roles for CELF1 orthologs in embryonic cardiomyocytes. Loss of Celf1 also resulted in morphogenetic abnormalities in the developing heart and gut. Using optical coherence tomography, we showed that cardiac contraction was impaired following depletion of Celf1, while heart rhythm remained unperturbed. In contrast to cardiac muscle, loss of Celf1 did not disrupt myofibril organization in skeletal muscle cells, although it did lead to fragmentation of skeletal muscle bundles.
Conclusions
CELF1 is required for normal myofibril organization, proliferation, morphogenesis, and contractile performance in the developing myocardium.
Keywords: heart, development, myofibrillogenesis, CELF1, chicken, Xenopus, optical coherence tomography
INTRODUCTION
CUG-BP, Elav-like family member 1 (CELF1) is a versatile RNA binding protein present in a variety of embryonic and adult tissues (Dasgupta and Ladd, 2012). CELF1 regulates alternative splicing (Philips et al., 1998; Savkur et al., 2001; Suzuki et al., 2002), stability (Paillard et al., 1998; Moraes et al., 2006; Lee et al., 2010), or translation (Ezzeddine et al., 2002; Baldwin et al., 2004; Timchenko et al., 2005) of its targets. In the human muscular dystrophy, Myotonic Dystrophy Type 1 (DM1; OMIM#160900), CELF1 is aberrantly up-regulated and hyperactivated in adult striated muscle (Timchenko et al., 1996; Timchenko et al., 2001a; Timchenko et al., 2004), resulting in inappropriate splicing of target transcripts in heart and skeletal muscle (Philips et al., 1998; Savkur et al., 2001; Mankodi et al., 2002). These splicing changes are recapitulated in mouse models of CELF1 over-expression, along with histological, physiological, and functional deficits in heart and skeletal muscle typical of DM1 (Timchenko et al., 2004; Ho et al., 2005; Koshelev et al., 2010; Ward et al., 2010). Repression of nuclear CELF activity (i.e., regulation of alternative splicing) in the mouse heart postnatally results in profound dilated cardiomyopathy, contractile defects, cardiac hypertrophy, and cardiac fibrosis (Ladd et al., 2005b), but its roles in the embryonic myocardium have not been studied.
In Xenopus laevis, Celf1 (formerly called Eden-bp) has been studied in its capacity as a regulator of transcript stability. Both knockdown of Celf1 and antibody-mediated inhibition of Celf1 activity were shown to lead to somite segmentation defects, while differentiation of myotomes (somite-derived muscle precursor structures) was not affected (Gautier-Courteille et al., 2004). Somitogenesis is also obstructed in zebrafish embryos following Celf1 knockdown or abrogation of activity, resulting in failure of two important patterning processes, establishment of bilateral symmetry and establishment of right-left asymmetry (Matsui et al., 2012). In the heart, CELF1 is expressed exclusively in the myocardium, where its expression peaks during cardiac morphogenesis (Brimacombe and Ladd, 2007; Blech-Hermoni et al., 2013).
In this study, we investigated the effects of CELF1 depletion in primary embryonic cardiomyocytes in culture, and in the developing heart in Xenopus laevis embryos. Knockdown of CELF1 in cultured cardiomyocytes led to myofibrillar disorganization and reduced proliferation. In Xenopus, we show that myocardial expression and alternative splicing activity of Celf1 are conserved in the embryonic heart. Moreover, knockdown of Celf1 resulted in defects in myofibril organization, cardiac morphogenesis, and heart function. Consistent with previous reports, loss of Celf1 also led to disruption of skeletal muscle fibers, but surprisingly myofibril structure was not affected. Together, these data indicate that CELF1 is required for normal myofibrillogenesis and morphogenesis in the developing myocardium, but plays a distinct role in developing skeletal muscle.
RESULTS
Knockdown of CELF1 in primary embryonic cardiomyocytes leads to myofibrillar disorganization and reduced proliferation
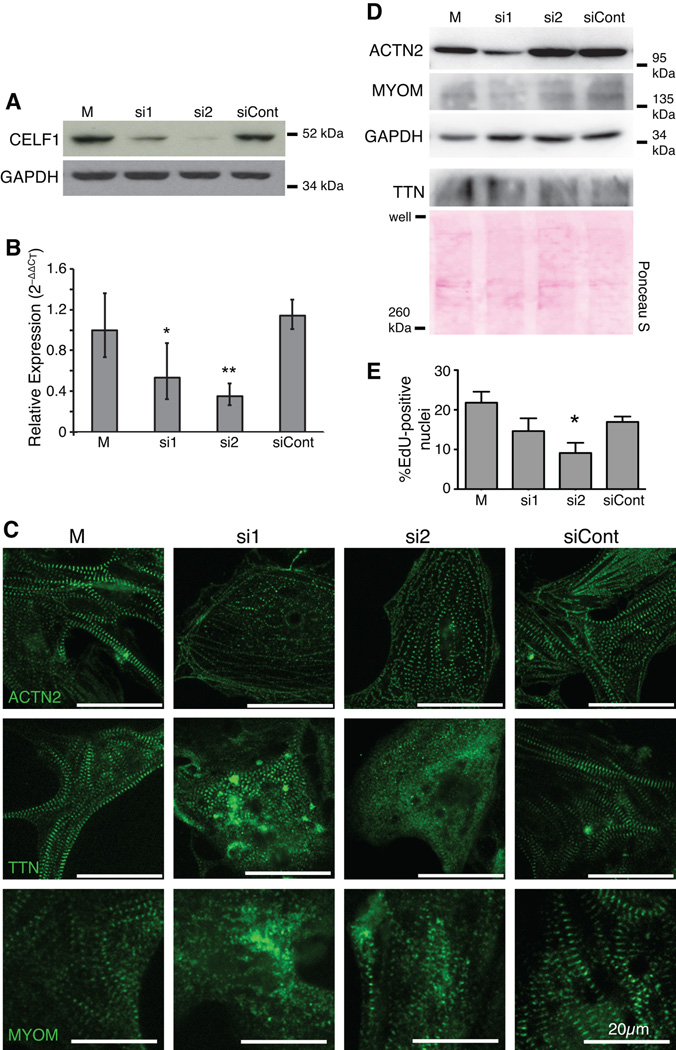
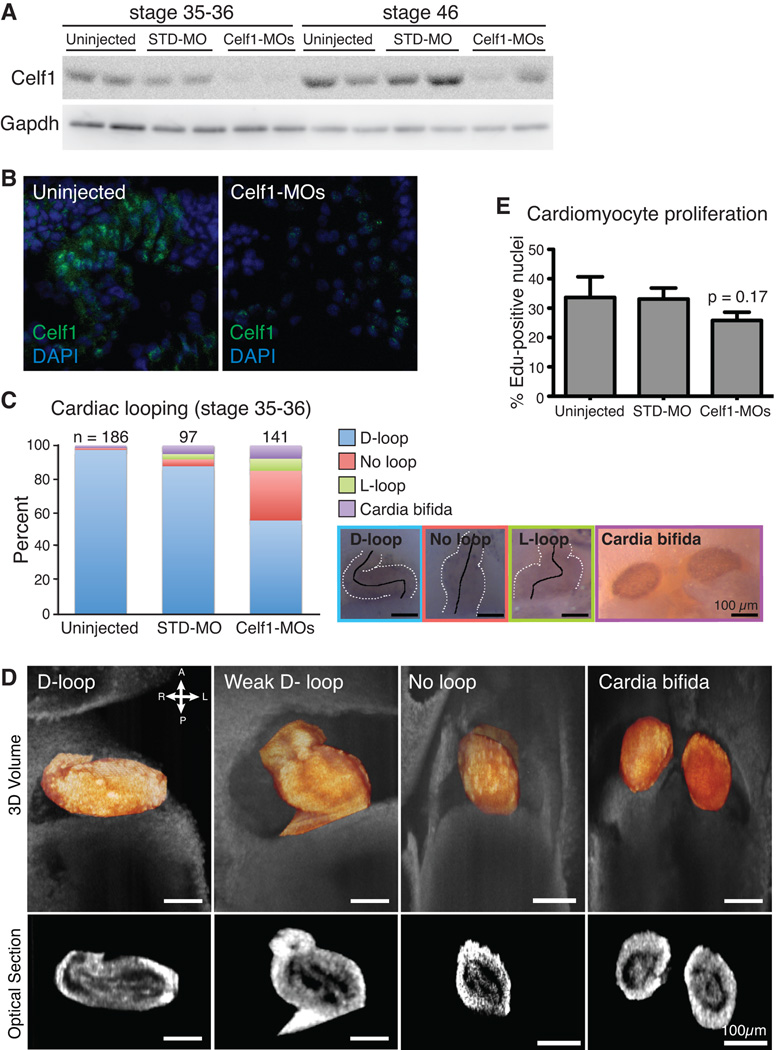
We previously demonstrated that in the embryonic heart, CELF1 is expressed exclusively in the myocardium, and is strongly present in the nucleus in cardiomyocytes (Brimacombe and Ladd, 2007; Blech-Hermoni et al., 2013). In order to investigate the roles of CELF1 in embryonic heart muscle cells, CELF1 was knocked down in chicken primary embryonic cardiomyocytes. As in vivo, CELF1 is expressed predominantly in the nuclei of these cells (Fig. 1). Transient transfection with either of two independent siRNAs (si1 and si2) designed to target the chicken CELF1 transcript resulted in robust knockdown of CELF1 protein in cultured embryonic primary cardiomyocytes by western blot analysis (Fig. 2A) and immunofluorescence (data not shown), as well as reduced levels of CELF1 RNA (Fig. 2B). One siRNA (si2) yielded greater knockdown, but both siRNAs were used in all experiments. A control (non-targeting) siRNA (siCont) did not result in changes to CELF1 protein or transcript levels (Fig. 2A,B). No overt toxicity was observed following transfection with any of the siRNAs at the concentrations tested. Following knockdown of CELF1, the organization of the contractile apparatus was evaluated by immunofluorescence using antibodies against components of the Z-disc (ACTN2 and a C-terminal epitope of titin, TTN) and the M-line (myomesin, MYOM) (Fig. 2C). All sarcomeric markers tested revealed disorganization following knockdown of CELF1: while mock- and siCont-treated cells exhibited aligned, thick myofibrils indicated by broad striations, si1- and si2-treated cells had thin, often web-like myofibrils with small, globular striations accompanied in some cases by diffuse cytoplasmic staining (Fig. 2C). Although cell size was not directly measured, the cells did not appear markedly different under bright-field microscopy (not shown). Despite this disorganization, spontaneous beating of large, synchronized foci across the plate was observed following CELF1 knockdown, and beating in si1- or si2-transfected cells was indistinguishable from beating in mock- or siCont-transfected cells by visual inspection of video microscopy (not shown). Total ACTN2, TTN, and MYOM levels were evaluated by western blot (Fig. 2D). Although ACTN2 levels were consistently reduced in si1-treated cells, they remained normal in si2-treated cells, suggesting this change does not underlie the disorganization of thin filaments. TTN and MYOM levels were not affected by CELF1 knockdown.
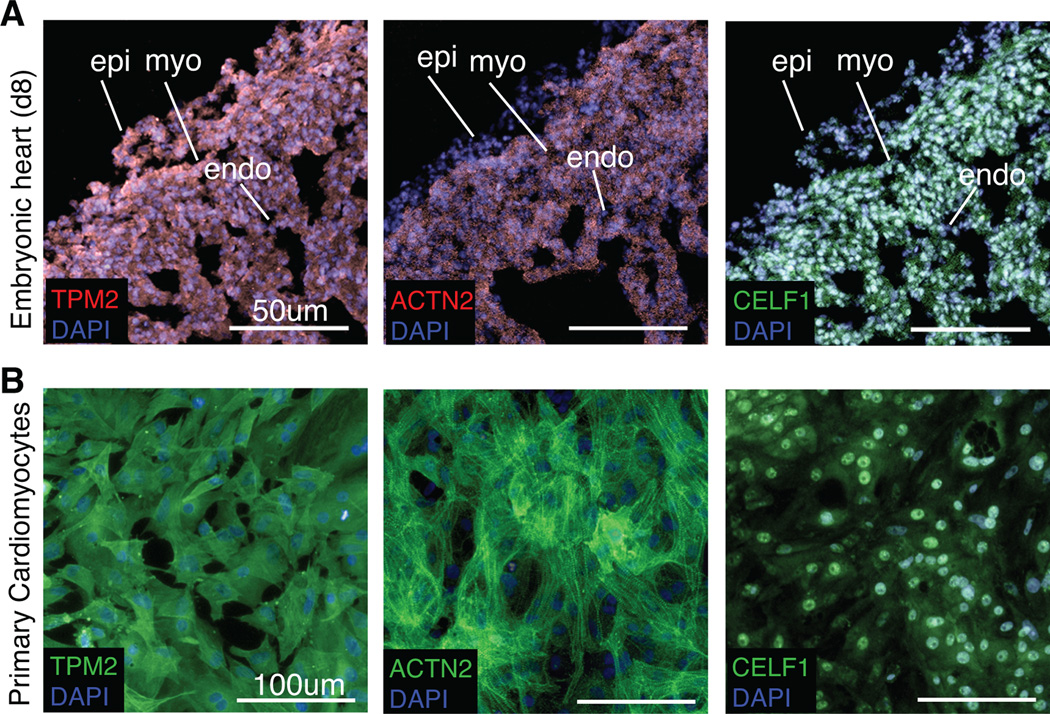
Fig. 1. Cultured chicken primary embryonic cardiomyocytes recapitulate in vivo expression of myofibrillar proteins and CELF1.
Cardiomyocytes in the embryonic day 8 chicken heart in vivo (A) and in cultured chicken primary embryonic cardiomyocytes isolated from day 8 hearts (B) express Tropomyosin (TPM2), sarcomeric alpha-Actinin (ACTN2), and CELF1 as determined by immunofluorescence. epi, epicardium; myo, myocardium; endo, endocardium.
Fig. 2. Knockdown of CELF1 leads to disorganization of myofibrillar structure and reduced proliferation in primary embryonic cardiomyocytes.
(A) Depletion of CELF1 with two independent siRNAs (si1 and si2) but not a control siRNA (siCont) is seen by western blot at 72 hours post-transfection; M, mock transfection. Equivalent loading was confirmed by reprobing for GAPDH. (B) Transfection of cells with si1 and si2, but not siCont, resulted in robust knockdown of CELF1 transcripts as measured by real-time RT-PCR. Values shown are means ± 95% confidence intervals. *, p ≤ 0.05; **, p ≤ 0.01 versus mock-transfected cardiomyocytes as evaluated by Student’s t-test. (C) Knockdown of CELF1 at 24 hrs post-plating leads to disruption of myofibril organization as visualized by ACTN2, TTN, and MYOM immunofluorescence at 72 hrs post-transfection. (D) Western blots for ACTN2, TTN, and MYOM protein were performed on mock-, si1-, si2-, and siCont-transfected cardiomyocytes at 72 hrs post-transfection. Protein integrity, equivalent loading, and successful transfer were confirmed by reprobing for GAPDH or Ponceau S staining. (E) Proliferation was measured at 48 hrs post-transfection by EdU incorporation. Values shown are means + standard errors of the mean. *, p ≤ 0.05 versus siCont as evaluated by Student’s t-test.
CELF1 has been implicated in cell-cycle arrest (Xiao et al., 2011; Chang et al., 2012; Talwar et al., 2013; Liu et al., 2014). To evaluate proliferation in cardiomyocytes, incorporation of a nucleoside analog, 5-ethynyl-2’-deoxyuridine (EdU), during DNA replication in the S stage of the cell cycle was evaluated. Knockdown of CELF1 resulted in a dose-dependent reduction in proliferation, with a robust effect seen following treatment with si2 (Fig. 2E) and a trend toward reduction with the less effective siRNA (si1) that did not attain statistical significance.
CELF1 is required for early events in myofibril assembly in cultured cells
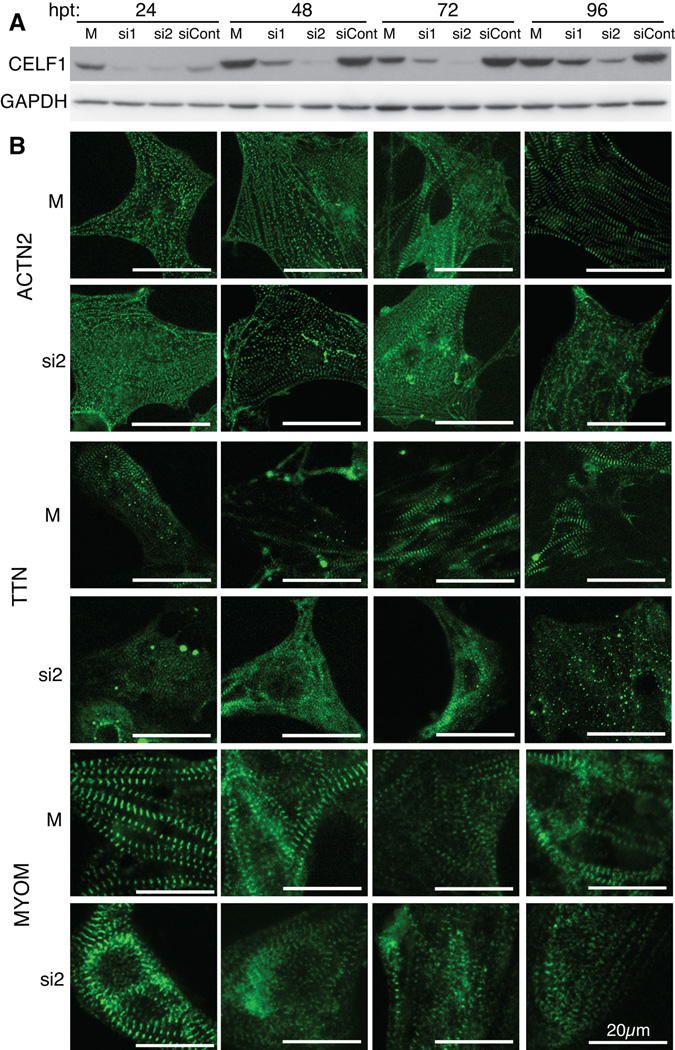
During isolation of the cardiomyocytes, myofibrils disassemble and must be reassembled upon establishment of contacts with the plate surface and with other cells (Lin et al., 1989). The myofibril assembles in a stepwise process (Gregorio and Antin, 2000). In order to determine whether the knockdown of CELF1, which is seen as early as 24 hours post-transfection [hpt] (Fig. 3A), prevents or delays the assembly of the myofibril, or leads to the disassembly of assembled structures, we imaged the myofibrils over consecutive days in culture and compared the appearance of the normally assembling myofibrils to that of the myofibrils following knockdown of CELF1 (Fig. 3B). In control cells, different components appeared to become organized at different time points. Myofibrils visualized using markers of the Z-discs (ACTN2 and TTN) became progressively thicker and more organized between 24 and 72 hpt, after which time no further change was observed. These observations are in agreement with earlier studies of the progression of myofibril reassembly in cultured primary avian cardiomyocytes (Terai et al., 1989; Komiyama et al., 1990). By contrast, a marker of the M-lines (MYOM) already appeared well organized by 24 hpt and no further change was observed. Following knockdown of CELF1, ACTN2 and TTN staining appeared punctate at 24 hpt, similar to the mock-treated cells. However, while the myofibrils in the mock-treated cells became progressively more assembled over time, the myofibrils in the si2-treated cells remained thin, and sometimes assumed a web-like appearance (Fig. 3B). In fact, ACTN2 and TTN failed to fully attain an assembled appearance even by 96 hpt. By contrast, myofibrils stained for MYOM appeared assembled 24 hrs following treatment with si2, but this organization was lost in many, although not all, cells over time (Fig. 3B). Surprisingly, CELF1 protein levels (and transcript levels; not shown) did not recover to normal levels by 96 hpt (Fig. 3A), or even by 144 hpt (not shown). Thus, we were unable to determine whether recovery of CELF1 levels to normal would result in re-organization of myofibrils. Nonetheless, these results indicate that CELF1 is required for both the assembly and maintenance of components in the myofibril.
Fig. 3. CELF1 is required for myofibril assembly and maintenance.
(A) Western blot analysis shows that knockdown of CELF1 protein is detectable by 24 hrs post-transfection (hpt) and persists through 96 hpt. Protein integrity and equivalent loading were confirmed by Ponceau S staining (not shown) and reprobing for GAPDH. (B) Myofibrillar organization was monitored by immunofluorescence using sarcomeric markers (ACTN2, TTN, and MYOM) over 96 hrs following mock (M) or siRNA (si2) transfection. Cells were transfected at 24 hrs post-plating and fixed at 24, 48, 72, and 96 hrs post-transfection; panels match the time points indicated in (A).
Celf1 expression and splicing activity are conserved in the embryonic frog heart
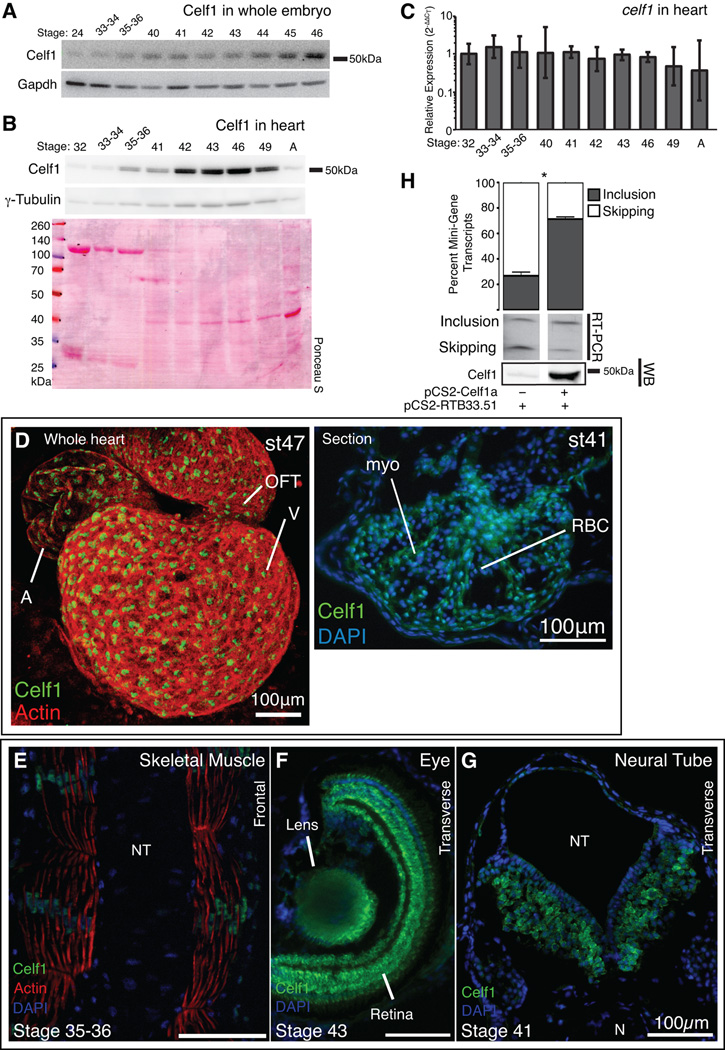
In order to investigate the role of CELF1 in the embryonic heart in vivo, we used African Clawed Frog (Xenopus laevis) embryos, which are amenable to in vivo knockdown. The sequence of CELF1 is highly conserved and Xenopus Celf1-a is 86% identical to the mouse and human CELF1 proteins. Levels of celf1 transcripts have been previously evaluated in X. laevis whole embryos (Wu et al., 2010). As protein levels have never been measured, and neither transcript nor protein has been measured in the developing Xenopus heart, we compared protein levels in whole embryos and in isolated hearts from a series of stages by western blot analysis. Equal amounts of total protein from each stage were loaded in each lane. We found that Celf1 levels in whole embryo gradually increased during development (while Gapdh levels declined slightly; Fig. 4A). This is consistent with celf1 transcript levels measured in the whole embryo, at least between stage 11 and stage 43 (Wu et al., 2010). In isolated hearts, Celf1 levels increased during cardiac morphogenesis, peaking at stage 46, and then declining and remaining low in adult (Fig. 4B). Gapdh expression is highly dynamic during heart development (data not shown), so γ-tubulin was used as a control. The slight gradual increase in γ-tubulin between stages 32 and 41 is likely due to the depletion of intracellular yolk stores, which dilute other proteins in the heart lysates. Indeed, Ponceau S staining revealed the loss of highly abundant proteins corresponding to the molecular weights of vitellogenin species between stages 32 and 41. After stage 41, however, these yolk proteins are depleted and γ-tubulin levels remain relatively constant as Celf1 protein levels rise sharply and then decline. In contrast, while protein levels rose and fell celf1 transcript levels remained steady during heart development (Fig. 4C). A similar transient up-regulation of Celf1 protein that peaks during cardiac morphogenesis without concomitant changes in transcript levels has also been observed in the embryonic chicken heart (Blech-Hermoni et al., 2013), and a postnatal decline in CELF1 protein levels without loss of mRNA expression has been previously reported in both chicken and mouse hearts (Ladd et al., 2005a; Kalsotra et al., 2008). Together, these data suggest that post-transcriptional regulation of CELF1 expression in the heart is highly conserved across vertebrate species.
Fig. 4. Developmental expression and alternative splicing activity of Celf1 are conserved in Xenopus laevis.
Celf1 protein was evaluated in whole embryo extracts (A) and isolated hearts (B) by western blot analysis. Blots were stripped and reprobed for Gapdh or γ-Tubulin. Protein integrity and equivalent loading were also confirmed by Ponceau S staining. (C) celf1 transcript levels were evaluated in embryonic and adult hearts (n = 3–4) by real-time RT-PCR. Error bars indicate 95% confidence intervals. A one-way analysis of variance found no significant differences in celf1 between the stages. Celf1 was visualized by immunofluorescence in the whole embryonic heart (D, left) and heart sections (D, right), skeletal muscle (E), eye (F), and neural tube (G). A, atrium; V, ventricle; OFT, outflow tract; myo, myocardium; RBC, red blood cell; N, notochord; NT, neural tube. (H) The CELF-responsive RTB33.51 mini-gene was co-expressed in COSM6 cells with celf1-a, RNA was collected 72 hrs later, and alternative splicing was assessed by radiolabeled RT-PCR (top). Celf1 expression was confirmed by western blot analysis (WB). Lanes shown are from the same gels; intervening lanes were removed for clarity. *, p ≤ 0.01; versus mini-gene alone as evaluated by Student’s t-test.
In the Xenopus embryo, Celf1 was detected in striated muscle tissues (both cardiac and skeletal muscle), the eye, and the neural tube by immunofluorescence (Fig. 4D–G), similar to chicken and mouse embryos (Blech-Hermoni et al., 2013). Interestingly, Celf1 is predominantly nuclear in the heart muscle and in skeletal muscle of the tail (Fig. 4D,E), while expression in the eye and neural tube was predominantly cytoplasmic (Fig. 4F,G).
Celf1 has been shown to regulate transcript stability in the early Xenopus embryo (Gautier-Courteille et al., 2004), an activity that is conserved in mammals (Vlasova and Bohjanen, 2008). CELF1 orthologs in mammals, birds, and flies have been shown to regulate pre-mRNA alternative splicing (Ladd et al., 2001; Ladd et al., 2005a; Spletter et al., 2015), but Xenopus Celf1 has not yet been shown to possess splicing activity. In order to ascertain whether Xenopus Celf1 can regulate alternative splicing, a CELF-responsive mini-gene reporter (pCS2-RTB33.51) was transfected with or without a Celf1-a expression plasmid into COSM6 cells, which have low levels of endogenous CELF1. While the mini-gene alone exhibited mostly exon skipping, co-expression with Celf1-a strongly promoted exon inclusion (Figure 4H), similar to its human ortholog (Ladd et al., 2001). Thus, both the expression and activity of Celf1 are conserved in Xenopus.
Loss of Celf1 activity in Xenopus embryos results in cardiac looping defects and cardiac dysmorphia
In order to evaluate the effects of Celf1 knockdown on the developing heart, we microinjected 2- to 4-cell stage Xenopus laevis embryos with a mixture of two antisense morpholino oligonucleotides (MOs) designed to target the translation start sites of both Xenopus CELF1 orthologs (celf1-a and celf1-b; previously described as EDEN-BP MOs in (Gautier-Courteille et al., 2004)). Microinjection of these celf1-targeting MOs (Celf1-MOs) resulted in knockdown of Celf1 at stages 35–36 that persisted until at least stage 46, as measured by western blot analysis (Fig. 5A). Either Celf1-MO alone resulted in some reduction of Celf1 protein, while co-injection with both MOs led to a more robust knockdown (data not shown). A standard control MO (STD-MO) had no significant effect on Celf1 levels. Persistent loss of Celf1 expression specifically within the heart at stage 46 was confirmed by immunofluorescence (Fig. 5B).
Fig. 5. Morpholino-mediated Celf1 knockdown in Xenopus laevis causes cardiac looping defects at stage 35–36.
(A) Celf1 protein levels were evaluated by western blot analysis (STD-MO, control morpholino; Celf1-MOs, mix of two celf1-targeting morpholinos). (B) Representative images of Celf1 staining in stage 46 heart sections (5–10 sections were stained per embryo for 2 embryos per group from each of 2 injection sets, n = 4 total). Control and Celf1-MOs-injected sections were processed simultaneously and imaged at the same exposure settings and time. STD-MO-injected embryos looked similar to uninjected controls (not shown). (C) Cardiac looping and cardia bifida were evaluated at stage 35–36. Hearts are outlined with broken white lines; a solid black line indicates the middle of the heart tube. D-loop, dextro-loop; L-loop, levo-loop. (D) Tissue overlaying the heart was removed and hearts were imaged in situ by optical coherence tomography (OCT). Three-dimensional reconstructions (top; heart in orange) and optical sections (bottom) are shown. (E) The percentage of cardiomyocytes with EdU-positive nuclei was determined. Data are means + the standard errors of the means of 5–6 embryos from 2–3 injection sets. Pairwise comparisons of the means were performed via Student’s t-test; p value shown for Celf1-MOs-injected group is versus uninjected.
Disruption of celf1 levels in zebrafish (Danio rerio) was shown to lead to defects in symmetry, affecting somitogenesis and cardiac looping (Matsui et al., 2012). To evaluate cardiac looping following Celf1 knockdown, embryos were collected at stage 35–36 and immunostained using antibodies either against a myosin heavy chain (Meromyosin; MF20) or Tnnt2 in order to easily visualize the heart. Uninjected embryos predominantly exhibited a normally right-looped heart (D-loop), and STD-MO-injected embryos showed few instances of cardiac abnormalities. Hearts in uninjected and STD-MO-injected embryos were rarely unlooped (2/185 and 4/97, respectively) or incorrectly looped (0/185 and 3/97, respectively). In contrast, a substantial fraction of Celf1-MOs-injected embryos exhibited abnormal cardiac looping (Fig. 5C). Unlooped and incorrectly looped hearts were quite commonly seen in Celf1-MOs-injected embryos (41/139 and 10/139, respectively; p ≤ 0.01 for both phenotypes, versus uninjected; Fisher’s exact test). In addition, while about half the hearts observed in Celf1-MOs-injected embryos were looped in the correct direction (D-looped), the degree of looping appeared less pronounced than observed in D-looped hearts of uninjected or STD-MO-injected embryos. The correctly looped hearts in Celf1-MOs-injected embryos appeared stouter than normal hearts suggesting an intermediate state between correctly looped and unlooped hearts (Fig. 5D). In both D-looped and un-looped hearts, the tissue organization of the heart remains intact (myocardial and endocardial layers appear unchanged) as seen by optical coherence tomography (OCT; Fig. 5D). Furthermore, the incidence of cardia bifida, which is not a looping defect but rather a failure of fusion of the bilateral heart fields (Brand, 2003), was also higher following Celf1 knockdown (11/141 in Celf1-MOs-injected embryos versus 3/186 in uninjected embryos; p ≤ 0.05; Fisher’s exact test). To evaluate proliferation of cardiomyocytes following Celf1 depletion, embryos injected at the 2–4-cell stage with Celf1-MOs or STD-MO, as well as uninjected controls, were pulsed with EdU at stage 28, and assayed at stage 35–36. In uninjected and STD-MO-injected embryos, roughly a third of cardiomyocytes were undergoing DNA replication during this period. In Celf1-MOs-injected embryos, a trend was seen toward reduced proliferation, although this difference did not attain statistical significant (Figure 5E).
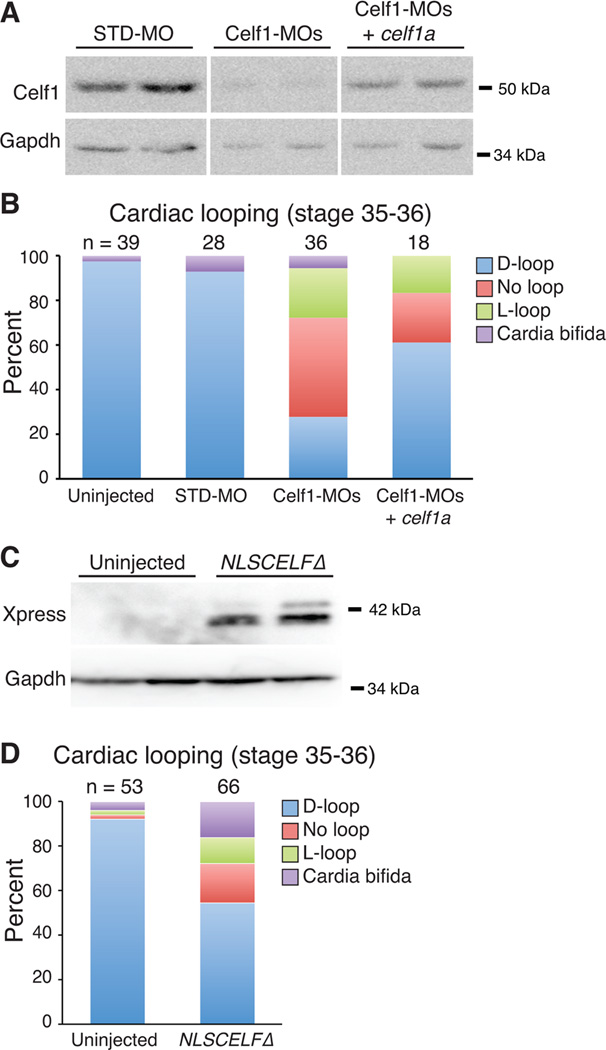
To confirm that the morphogenetic defects are attributable to loss of Celf1, we performed a rescue experiment in which Celf1-MOs were co-injected with an in vitro transcribed celf1a mRNA. Indeed, co-injection with the celf1a transcript increased Celf1 protein levels and ameliorated the cardiac morphogenesis defects (Fig. 6A,B). Significantly more hearts were looped correctly than in Celf1-MOs-injected embryos that did not receive the mRNA (p = 0.04 versus Celf1-MOs-injected; Fisher’s exact test). Celf1-MOs+celf1a-injected embryos still exhibited a higher fraction of defects than controls (p ≤ 0.01 versus uninjected; Fisher’s exact test), likely because co-injection with celf1a mRNA did not completely restore Celf1 protein to normal levels (Fig. 6A). Attempts to co-inject a higher dose of celf1a mRNA resulted in Celf1 over-expression and worsening of the defects (data not shown), a result that is consistent with previous studies investigating Celf1-dependent somite segmentation defects (Gautier-Courteille et al., 2004).
Fig. 6. Morpholino-mediated knockdown of Celf1 can be ameliorated by restoration of Celf1 expression, and mimicked by repression of nuclear Celf activity.
Xenopus laevis embryos were injected either with morpholino oligonucleotides (STD-MO, control morpholino; Celf1-MOs, a mix of two celf1-targeting morpholinos) ± celf1a RNA (A, B), or with RNA encoding a dominant negative CELF protein (NLSCELFΔ; C, D) at the 2-4-cell stage and compared to uninjected controls at stage 35–36. (A) Celf1 protein levels were determined by western blotting. Two embryos per group are shown. Lanes shown are from the same blot. (B) Cardiac looping and the incidence of cardia bifida following knockdown and restoration of Celf1 were evaluated by whole-mount immunohistochemistry using antibodies against meromyosin (MF20) or Tnnt2 (CT3). (C) Dominant negative protein expression was confirmed at stage 26 by western blotting for its Xpress epitope tag. Two embryos per group are shown. (D) Cardiac looping and fusion defects were evaluated in embryos expressing the dominant negative CELF protein by whole mount immunohistochemistry against meromyosin. D-loop, dextro-loop; L-loop, levo-loop.
To test whether morphogenetic defects are caused by disruption of Celf1-mediated alternative splicing, we injected embryos with an in vitro transcribed RNA encoding a dominant negative CELF protein, NLSCELFΔ. NLSCELFΔ is a truncated form of CELF4 fused to an N-terminal Xpress epitope tag and the SV40 large T antigen nuclear localization signal. NLSCELFΔ localizes to the nucleus and represses the splicing activities of all members of the CELF family, but has no effect on alternative splicing in the absence of CELF proteins (Singh et al., 2004; Ladd et al., 2005b). In transgenic mice, NLSCELFΔ expression has been shown to specifically disrupt the splicing of CELF targets in adult heart and skeletal muscle in vivo (Ladd et al., 2005b; Berger et al., 2011; Berger and Ladd, 2012). In the Xenopus embryo, NLSCELFΔ expression (confirmed by western blot, Fig. 6C) also induced a high fraction of morphogenetic defects (Fig. 6D; p ≤ 0.01 versus uninjected; Fisher’s exact test). This suggests that the effects on cardiac looping and fusion observed in Celf1 morphants are likely attributable to disruption of Celf1-mediated alternative splicing.
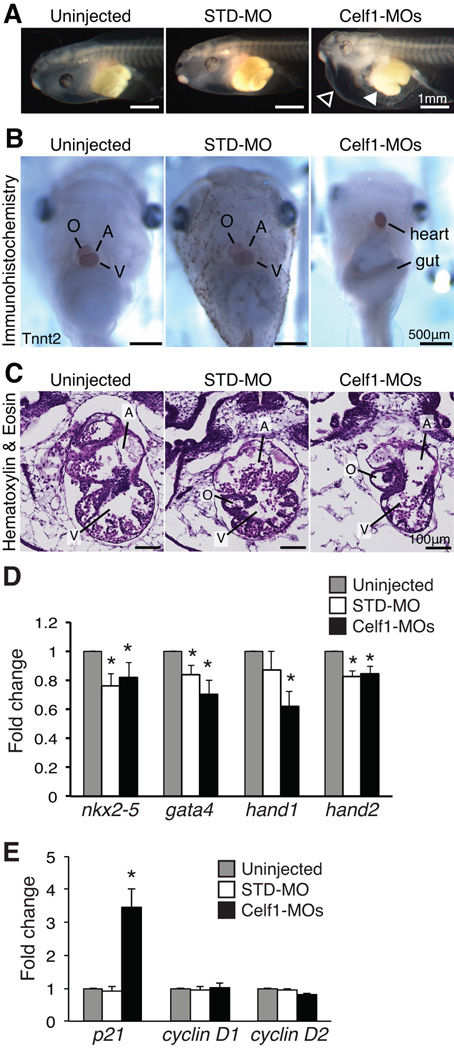
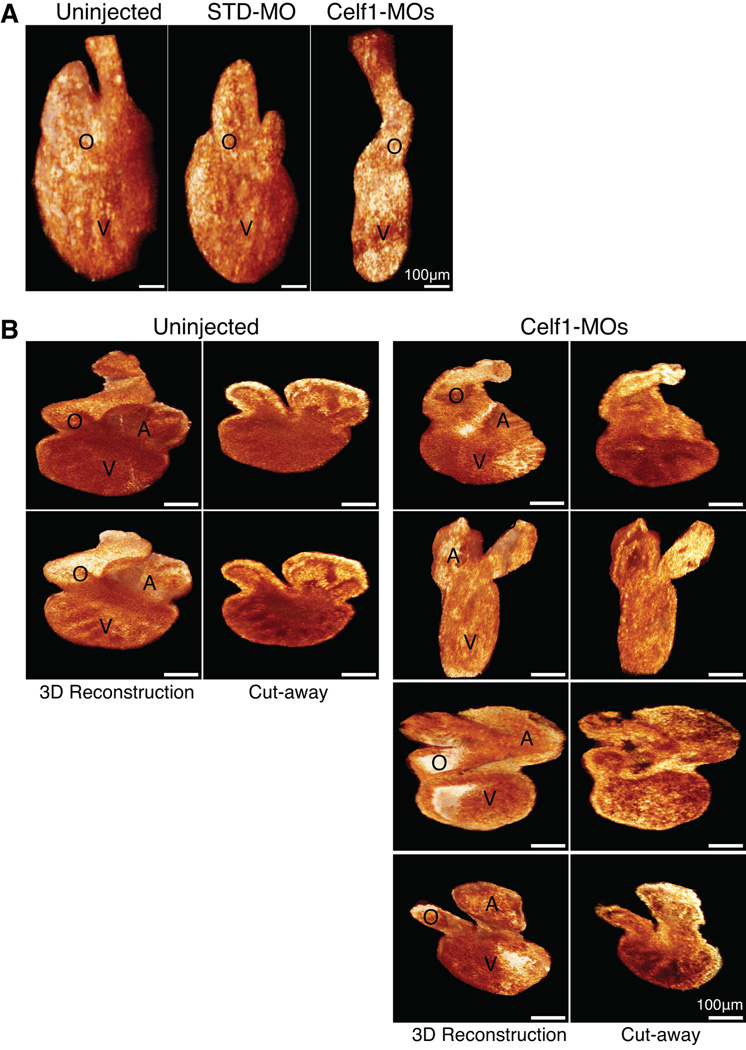
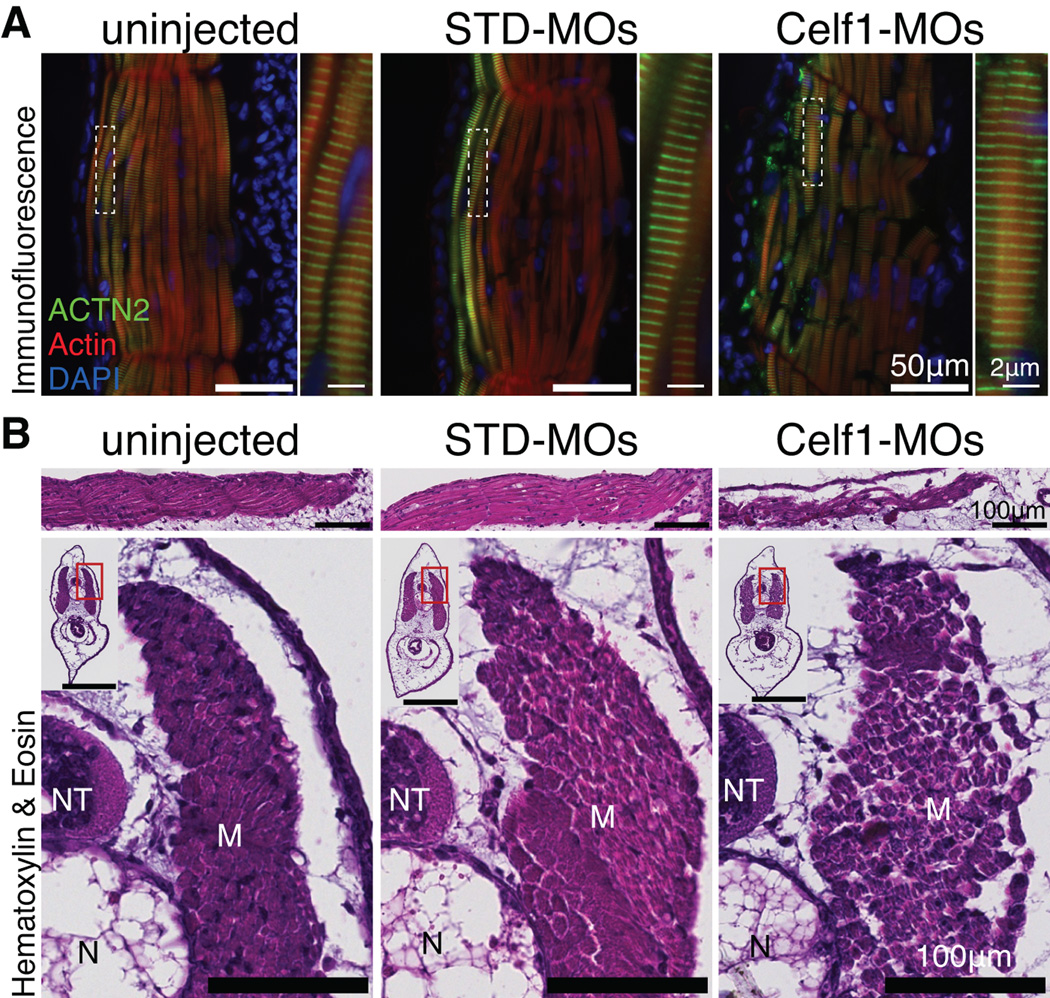
In order to evaluate how aberrant morphogenesis may affect the heart at a later stage of development, embryos were allowed to grow to stage 46, by which time cardiac morphogenesis is complete in uninjected embryos. On gross inspection, most embryos in which Celf1 was knocked down exhibited mild to profound ventral edema (Fig. 7A) compared to very few control embryos (2/218 uninjected embryos, 5/159 STD-MO-injected embryos, and 164/172 Celf1-MOs-injected embryos; p = 0.14 for STD-MO-injected and p < 0.0001 for Celf1-MOs-injected versus uninjected; Fisher’s exact test). Hearts were visualized by immunohistochemistry for cardiac markers and photographed at stage 46 (Fig. 7B), and histological analysis was performed on sections (Fig. 7C). The hearts of Celf1-MOs-injected embryos exhibited various cardiac abnormalities, including chamber dilation, thinning of the chamber wall, and reduced trabeculation (Fig. 7C). In addition, stage 46 hearts were imaged in situ by OCT (Fig. 8A), then removed and re-imaged by optical coherence microscopy (OCM), a high-resolution modification of OCT technology (Fig. 8B). While uninjected embryos and embryos injected with the non-targeting control showed little variability in appearance, hearts in Celf1-MOs-injected embryos exhibited a range of morphogenetic defects. Most hearts were mis-oriented relative to neighboring tissues and/or dramatically reduced in size (Fig. 8). When viewed by light microscopy, some Celf1-MOs-injected hearts appear to beat less vigorously than uninjected or STD-MO-injected embryos (Movies S1–S3). In addition to the cardiac phenotype, a large fraction of Celf1-depleted embryos exhibited profound gut dysmorphia (Fig. 7A,B) compared to controls (0/73 uninjected embryos, 1/57 STD-MO-injected embryos, and 42/48 Celf1-MOs-injected embryos; p = 0.44 for STD-MO-injected and p < 0.0001 for Celf1-MOs-injected versus uninjected; Fisher’s exact test). It is unclear, however, whether this is a primary defect or a consequence of edema.
Fig. 7. Morpholino-mediated knockdown of Celf1 in Xenopus laevis leads to ventral edema, gut malformation, and cardiac dysmorphia at stage 46.
Embryos were injected with morpholino oligonucleotides at the 2-4-cell stage and evaluated at stage 46 (STD-MO, control morpholino; Celf1-MOs, a mix of celf1a- and celf1b-targeting morpholinos). (A) Following depletion of Celf1, most Celf1-MOs-injected embryos exhibited ventral edema (open arrowhead) and many had a malformed gut (filled arrowhead). (B) In embryos stained for Tnnt2, both cardiac and gut dysmorphia can be seen, as well as the aberrant orientation of the heart within the Celf1-MOs-injected embryos. (C) The heart was evaluated by hematoxylin and eosin staining of frontal sections. Serial sections from five embryos per group taken from three separate injection sets were evaluated with similar results. Sections were chosen for similar planes through the heart, since both gross morphology and heart orientation were abnormal in Celf1-MOs-injected embryos. A, atrium; O, outflow tract; V, ventricle. Real-time RT-PCR was performed on RNA from hearts isolated at stage 46 for several cardiac transcription factors (D) and cell cycle genes (E) following Celf1 knockdown (n = 6–7). Error bars indicate standard error of the mean. *, p ≤ 0.05; versus uninjected; Student’s t-test.
Fig. 8. Cardiac dysmorphia following morpholino-mediated knockdown of Celf1 in Xenopus laevis was imaged in situ and ex vivo at stage 46.
The hearts of uninjected (n = 9), STD-MO-injected (n = 7), and Celf1-MOs-injected (n = 8) embryos at stage 46 were imaged in situ by optical coherence tomography (OCT), and then removed and imaged by optical coherence microscopy (OCM). (A) Representative examples of three-dimensional reconstructions of OCT sections through the hearts are shown. General dysmorphia of the heart and outflow tract can be seen following Celf1 depletion; the atria cannot be resolved in these reconstructions. (B) Representative examples of hearts imaged ex vivo using OCM, in intact three-dimensional reconstructions (left) and in cut-away views (right). Multiple hearts from Celf1-MOs-injected embryos are presented to demonstrate the range of dysmorphia observed. Heart reconstructions are shown at equivalent orientations for the purpose of comparison; this orientation does not necessarily match the orientation of the hearts in situ. A, atrium; O, outflow tract; V, ventricle.
Several cardiac transcription factors, including NKX2-5, GATA4, and the HAND proteins, have been shown to play important roles in cardiac morphogenesis and myocardial growth (Brewer and Pizzey, 2006; Bartlett et al., 2010; Vincentz et al., 2011). To determine whether the expression of these factors is altered by Celf1 knockdown, levels of nkx2-5, gata4, hand1, and hand2 transcripts were measured in hearts isolated from control and Celf1-MOs-injected embryos at stage 46 (Fig. 7D). Although mRNA levels of all four cardiac transcription factors were mildly reduced in Celf1-MOs-injected hearts compared to uninjected controls, they were similarly reduced in STD-MO-injected hearts (i.e., levels in Celf1-MOs-injected hearts did not differ significantly from STD-MO-injected hearts, p > 0.05 for each). This suggests the slight decline in their expression is due to a non-specific effect of morpholino injection, and does not underlie the Celf1-dependent defects.
Since knockdown of CELF1 reduced proliferation in primary chicken embryonic cardiomyocytes (Fig. 2E) and showed a trend towards reduced proliferation in the Xenopus embryonic myocardium in vivo (Fig. 5E), we also evaluated the levels of cdkn1, which encodes the cell cycle inhibitor p21, and cyclin D1 (ccnd1) and cyclin D2 (ccnd2), which control entry into the G1 phase of the cell cycle (Lim and Kaldis, 2013). Although the levels of transcripts encoding cyclin D1 and D2 were unaffected, transcripts encoding p21 were significantly elevated specifically in Celf1-MOs-injected hearts (Fig. 7E). We were unable to detect transcripts encoding cyclin E2 (ccne2), which regulates G1 to S progression, and cyclin A1 (ccna1), which regulates G2 to M progression, in any of the heart samples (data not shown). This may be because only a small percentage of cells are actively dividing within the embryonic heart at any given moment in time, and thus the steady state levels of these transcripts are very low. To rule out confounding effects on myocardial growth from changes in cell death, apoptosis was also evaluated in stage 46 hearts by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining. No difference in apoptosis was observed between Celf1-MOs-injected and uninjected hearts, with both exhibiting less than one half of a percent of cells with TUNEL-positive staining (0.49% ± 0.30% in Celf1-MOs-injected hearts, n = 4 versus 0.09%± 0.09% in uninjected hearts, n = 3; p = 0.14; Student’s t-test).
Knockdown of Celf1 results in myofibrillar disorganization in cardiac muscle in vivo
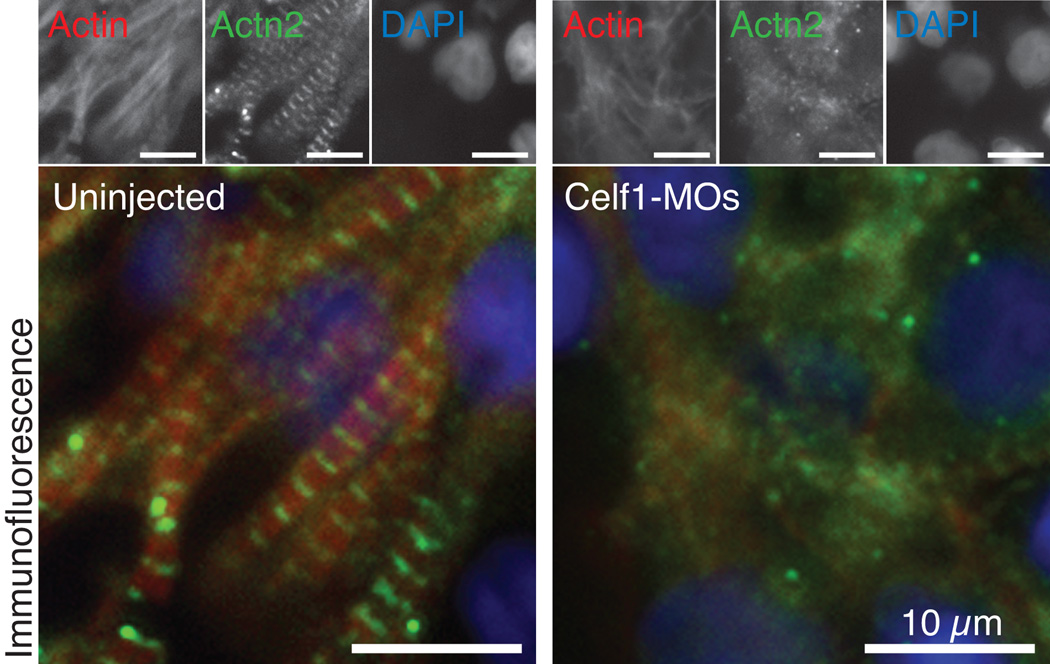
Since CELF1 knockdown in cultured chicken embryonic cardiomyocytes results in myofibrillar disorganization, we evaluated myofibrillar organization in cardiomyocytes of the embryonic Xenopus heart. Embryos were collected at stage 46 and cryosections were processed for immunofluorescence against Actn2 and filamentous Actin (F-Actin). In the heart wall of uninjected embryos, myofibrils are readily visible and striations are prominent, with Actn2 and F-Actin interdigitating along the length of the myofibrils (Fig. 9). By contrast, in Celf1-MOs-injected embryos, myofibrils were sparser (although not absent) within the cardiomyocytes, and although some striations were visible, overall Actn2 staining was more diffuse or punctate (Fig. 9). Actn2 and F-Actin staining in the myocardium of STD-MO-injected embryos was similar to that seen in uninjected embryos (not shown). These findings support our observations that myofibril organization is disrupted by loss of CELF1 in cultured chicken primary embryonic cardiomyocytes (Fig. 2,3). As in the cultured cells, beating is not completely abrogated in the hearts of Celf1-MOs-injected embryos despite this disorganization.
Fig. 9. Morpholino-mediated knockdown of Celf1 in Xenopus laevis leads to myofibril disorganization in the developing heart.
Myofibrillar organization was evaluated in heart wall of stage 46 embryos by immunofluorescence. Sections from eight or more embryos per group from six different injection sets were evaluated with similar results; representative images are shown. In uninjected embryos, mature myofibrils are visible as interdigitating Actn2 and F-Actin staining, whereas Actn2 staining is more diffuse in Celf1-MOs-injected embryos, and does not consistently associate with F-Actin striations in the myofibrils. Myofibrils in STD-MO-injected hearts look similar to those in uninjected embryos (not shown). Top panels show the separate channels for the merged images shown below. F-Actin was detected with fluorophore-conjugated phalloidin.
Knockdown of Celf1 impairs contractile performance, but not rhythm, of the heart
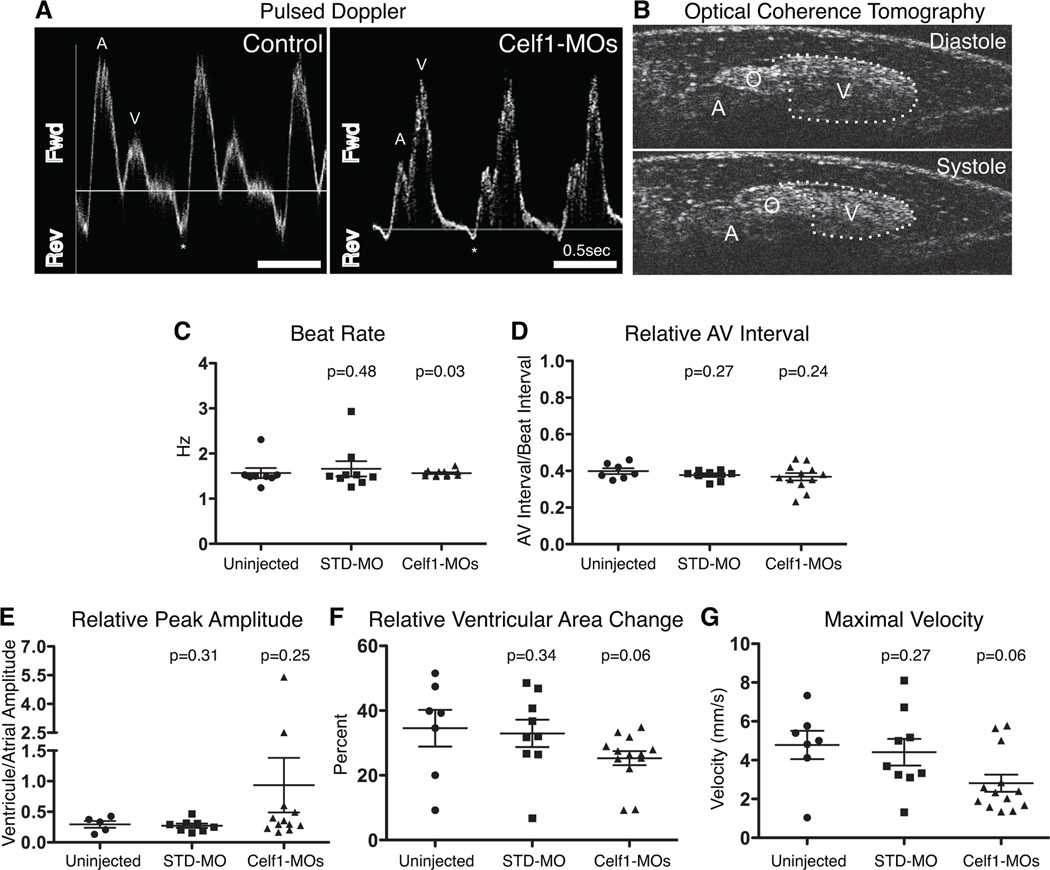
Since hearts of Celf1-MOs-injected embryos appeared grossly malformed or mis-oriented with disorganized myofibrils, cardiac function was evaluated in live Celf1 morphants and control embryos using OCT. OCT has previously been used to evaluate cardiac function in numerous animal models, including Xenopus laevis (Boppart et al., 1997; Yelin et al., 2007; Jenkins et al., 2012; Garcia et al., 2015). Here, we employed OCT to acquire Doppler and pulsed Doppler signals at the level of the right aortic branch (Fig. 10A), as well as video-speed recordings of the heart at the level of the atrial-ventricular axis (Fig. 10B), in stage 46 Xenopus embryos. In characterizing the periodicity of contraction, both beat rate and the relationship between atrial and ventricular contractions were evaluated. By comparing cardiac contraction in multiple OCT movies with the aortic pulsed Doppler traces for the respective embryos, atrial and ventricular contraction could be ascribed to the first and second positive-flow pulsed Doppler peaks within each contractile cycle (Fig. 10A). From these traces, neither the average beat rate (1.57 ± 0.11 Hz in uninjected embryos versus 1.57 ± 0.02 Hz in Celf1-MOs-injected embryos; mean ± standard error of the mean; Fig. 10C) nor the relative length of the A-V interval (as a proportion of a complete contractile cycle; Fig. 10D) was observed to change following Celf1 depletion. Although a significant p value was obtained when the beat rates of Celf1-MOs-injected embryos were compared to those of uninjected embryos using a Mann-Whitney U test (i.e., a non-parametric test used to compare median values), it is important to note that if the sample size is large enough the Mann-Whitney test can also detect differences in spread or shape of the data even when the medians are very similar (Hart, 2001). In this case, Celf1-MOs-injected embryos had less variation in beat rate than the control embryos (variances differed significantly by F test, p < 0.0001).
Fig. 10. Morpholino-mediated knockdown of Celf1 in Xenopus laevis leads to cardiac dysfunction.
Uninjected (n = 8), STD-MO-injected (n = 9), and Celf1-MOs-injected (n = 13) embryos were collected at stage 46, immobilized in methylcellulose gel, and imaged by optical coherence tomography (OCT). Doppler signal was measured at the level of the right aortic branch and both Doppler (not shown) and pulsed Doppler traces (A) were generated for live, beating hearts. Fwd, forward flow (positive Doppler shift); Rev, reverse flow (negative Doppler shift); A, atrial peak; V, ventricular peak; *, regurgitation; horizontal line indicates zero-shift. (B) Video-speed OCT movies were recorded in order to evaluate ventricular contraction. A, Atrium; O, OFT; V, ventricle. (C–G) Contractile parameters were measured and compared to uninjected controls using a one-tailed Mann-Whitney U test. Bars indicate mean ± standard error of the mean. Two control embryos that exhibited a single peak on pulsed Doppler were excluded from the analyses in panels (D) and (E).
While cardiac rhythm was not impaired, parameters associated with the strength of contraction indicated reduced function. In two Celf1-MOs-injected embryos, ventricle-associated flow exceeded atrium-associated flow, a feature never observed in control embryos. This characteristic was associated with increased variability in relative peak amplitude in Celf1-MOs-injected embryos (variances differed significantly by F test, p = 0.0002; Fig. 10E). This difference was in agreement with a trend toward reduction in the change in ventricular area (Fig. 10B,F), a parameter analogous to fractional shortening, suggesting reduced contraction of the ventricle following Celf1 knockdown (34.6 ± 5.7% in uninjected versus 25.3 ± 2.2% in Celf1-MOs-injected embryos; mean ± standard error of the mean; p = 0.06; Fig. 10F). Unfortunately, we could not evaluate changes in atrial area during cardiac contraction, as optical hindrance and the thin aortic wall interfered with the ability to fully visualize the atrium throughout the contractile cycle. Doppler measurements through the right aortic branch were used to calculate the maximal velocity, and this parameter exhibited a strong trend towards being reduced following depletion of Celf1 (4.78 ± 0.73 mm/s in uninjected embryos versus 2.81 ± 0.44 mm/s in Celf1-MOs-injected embryos; mean ± standard error of the mean; p = 0.06; Fig. 10G). Importantly, this difference was not due to a difference in vessel diameter following Celf1 depletion (31.46 ± 2.28 µm in uninjected embryos versus 28.98 ± 1.98 µm in Celf1-MOs-injected embryos; mean ± standard error of the mean; p = 0.21; not shown).
Knockdown of Celf1 in Xenopus embryos results in gross disorganization of muscle fibers, but not myofibrils, in skeletal muscle
It was previously reported that knockdown of Celf1 in Xenopus embryos results in defective somite segmentation and reduced organization of myotomal cells at stage 39 (Gautier-Courteille et al., 2004; Cibois et al., 2010). To compare skeletal muscle to cardiac muscle, we visualized the arrangement of F-Actin and Actn2 within the tail muscle fibers at stage 46 by immunofluorescence. In skeletal muscle, the organization of the myofibrils appeared unperturbed, while the organization of the muscle fibers within muscle bundles was disrupted (Fig. 11A). Similar to previous reports, fragmentation of the myotome-derived fibers was observed in Celf1-MOs-injected embryos and muscle fibers appeared less densely packed (Fig. 11B). Despite these defects, Celf1-MOs-injected embryos were active and their swimming ability was indistinguishable from uninjected embryos (not shown).
Fig. 11. Morpholino-mediated knockdown of Celf1 in Xenopus laevis leads to disorganization of muscle fibers, but not sarcomeric structure, in skeletal muscle.
Embryonic skeletal muscle was evaluated in tails of stage 46 embryos (STD-MO, control morpholino, Celf1-MOs, a mix of two celf1-targeting morpholinos). (A) Representative images of frontal sections stained for Actn2, F-Actin, and DAPI by immunofluorescence are shown (n = 4). In uninjected and STD-MO-injected embryos, muscle bundles are tightly organized, while in Celf1-MOs-injected embryos, muscle bundles exhibit fragmentation and loss of cohesion. Regions indicated with dashed boxes are enlarged in adjoining panels. (B) Representative hematoxylin and eosin-stained frontal (top) and transverse (bottom) sections through stage 46 tails are shown. While the appearance of individual muscle fibers was similar in uninjected, STD-MO-injected, and Celf1-MOs-injected embryos, muscle bundles were looser and more fragmented in tails of Celf1-MOs-injected embryos. M, muscle; NT, neural tube; N, notochord. Insets show whole-section views (scale bar = 100 µm); enlarged regions indicated by red boxes.
DISCUSSION
In this study, we investigated the role of CELF1, a multifunctional RNA binding protein, during heart development by depleting it in cultured primary embryonic cardiomyocytes and in the developing embryo. Our findings indicate essential roles for CELF1 during cardiac morphogenesis (e.g., cardiac looping) in vivo and myofibrillar organization in cardiomyocytes both in the embryo and in culture. Cardiac defects have not been reported in Celf1-knockout mice (Celf1−/−), but heart structure and function have not been evaluated in this mouse model, which is characterized by high perinatal lethality (Kress et al., 2007; Cibois et al., 2012). The cause of death in Celf1−/− mice is not known, but one could speculate that if they have congenital heart defects, these would be exacerbated by the change in circulation and increase in cardiac workload that occurs at birth. We previously reported that the repression of cardiac CELF activity in young mice by transgenic expression of the NLSCELFΔ dominant negative CELF protein in the heart shortly after birth (MHC-CELFΔ) results in dilated cardiomyopathy (Ladd et al., 2005b; Terenzi et al., 2009). Most MHC-CELFΔ mice survive to adulthood, but exhibit profound defects in contractile function, consistent with a role for CELF proteins in the regulation of the contractile apparatus postnatally. Our findings in cultured embryonic cardiomyocytes and in developing Xenopus embryos suggest that CELF1 is important for the assembly of the myofibril in cardiac muscle cells, and that this is a cell autonomous role in cardiomyocytes. It is worth noting that the disorganization of the sarcomeres did not abrogate beating of embryonic heart muscle cells in culture or in vivo. Nonetheless, in Xenopus both gross morphology and functional measures suggest that knockdown of Celf1 perturbs normal cardiac function. The specific targets of Celf1 that underlie these defects are unknown, but CELF1 has been implicated in regulating the splicing or levels of other RNA binding proteins, transcription factors, and contractile proteins in embryonic chicken and mouse hearts (Kalsotra et al., 2008; Blech-Hermoni and Ladd, 2015; Blech-Hermoni et al., 2016).
By contrast, Celf1 does not appear to have an important role in myofibrillar organization of developing skeletal muscle, although its depletion leads to fragmentation of skeletal muscle fibers. Severe fragmentation was recently reported in indirect flight muscles (IFMs) following depletion of a Drosophila CELF1 ortholog, Bruno/Arrest (Bru), resulting in flightless adults (Spletter et al., 2015). Spletter and colleagues proposed that muscle fragmentation in IFMs likely resulted from hypercontraction as the sarcomeres were significantly shorter following Bru depletion (Spletter et al., 2015), but in our study no overt differences in sarcomere length were evident. Drosophila has two other CELF family orthologs, Bru-2 and Bru-3. Phylogenetic analyses suggest that Bru-2 comes from a lineage-specific duplication of Bru, while Bru-3 is likely an ortholog of CELF3-6 (Good et al., 2000). It has been suggested, however, that Bru-3 may be more functionally similar to CELF1 than either Bru or Bru-2 (Delaunay et al., 2004). Thus comparisons between the roles of CELF1 orthologs in Drosophila and vertebrate species should be approached with caution.
In zebrafish and Xenopus embryos, Celf1 has been implicated in aspects of symmetry patterning. In both organisms, bilateral symmetry is disrupted following perturbation of Celf1 levels (Gautier-Courteille et al., 2004; Cibois et al., 2010; Matsui et al., 2012; Cibois et al., 2013). In the Xenopus embryo, this role of Celf1, illustrated by a defect of somite segmentation following Celf1 depletion, was shown to result from the de-repression of su(h)/rbpj, a target of Celf1-mediated transcript degradation and a component of the Notch signaling pathway (Cibois et al., 2010). During early development, bilateral symmetry must be established and maintained, but local symmetry must also be broken (e.g., left-right asymmetry) for proper development of the heart. In zebrafish embryos, disruption of both the establishment of bilateral symmetry and development of left-right asymmetry are observed following perturbation of Celf1 levels, manifested as somite asymmetry and cardiac looping defects (Matsui et al., 2012). We find that knockdown of Celf1 results in cardiac looping defects and other morphological defects in the embryonic Xenopus heart as well, suggesting that the involvement of Celf1 in this process is evolutionarily conserved. Although Celf1 is not expressed in endoderm-derived organs in zebrafish, depletion of Celf1 in zebrafish embryos affects not only mesoderm-derived tissues, but also leads to defects of endoderm-derived organs such as liver and pancreas, including failure of gut looping (Tahara et al., 2013). In our study in the developing Xenopus embryo, we also observe gut morphological abnormalities following knockdown of Celf1, but have not further investigated this phenotype.
Knockdown of CELF1 in primary embryonic cardiomyocytes reduced their proliferation, a trend that was also seen in vivo. Consistent with this, recent network analysis of an RNA-seq dataset found a strong association between CELF1 and cell cycle in embryonic cardiomyocytes (Blech-Hermoni and Ladd, 2015; Blech-Hermoni et al., 2016). CELF1 has been shown to bind to the 5’ UTR of transcripts encoding p21, a cyclin-dependent kinase inhibitor that restricts re-entry into the cell cycle (Lim and Kaldis, 2013), and promote its translation in skeletal muscle cells (Timchenko et al., 2001b; Iakova et al., 2004). Increased relocalization of CELF1 to the nucleus in patients with myotonic dystrophy type 1 leads to reduced p21 protein levels, enhanced proliferation, and impaired differentiation in skeletal muscle cells (Timchenko et al., 2001b; Iakova et al., 2004). Here we report that loss of Celf1 leads to elevated p21 transcript levels in the embryonic heart. This is consistent with reduced cardiomyocyte proliferation, though we do not know if the up-regulation of p21 is due to loss of Celf1 binding to the p21 mRNA or to indirect effects on p21 transcription. While proliferation and differentiation are intimately (and inversely) related during skeletal muscle development, with myocytes leaving the cell cycle as they fuse into syncytial myotubes, this is not the case in cardiac muscle. Cardiomyocytes remain proliferative throughout embryonic and early postnatal development. A gradual switch from a hyperplastic to a hypertrophic state occurs shortly after birth and is influenced by different factors, including nutrient availability and hemodynamic load (Li et al., 1996). Notably, CELF1 undergoes a pronounced decline postnatally (Kalsotra et al., 2008) that is concomitant with the hyperplastic-to-hypertrophic transition. Knockdown of Celf1 was also shown to lead to reduced proliferation in endodermal cells during zebrafish embryonic development (Tahara et al., 2013), and competition between CELF1 and calreticulin has been suggested to drive a biological switch between senescence and proliferation in hepatocytes and fibroblasts (Iakova et al., 2004; Timchenko et al., 2005; Barreau et al., 2006). Together, these data suggest a role for CELF1 in the regulation, directly or indirectly, of proliferation in multiple germ layers.
EXPERIMENTAL PROCEDURES
Animal use
Fertilized White Leghorn Hy-line W-36 chicken eggs were purchased from the Department of Animal Sciences at The Ohio State University or from Charles River Laboratories. Eggs were kept at 15°C (59°F) for up to one week until use. Pre-hatch chicken embryos are not subject to federal regulation and do not require government or institutional approval. Chicken embryos were euthanized by decapitation immediately upon removal from the egg, which is consistent with the recommendations of the American Veterinary Medical Association Panel on Euthanasia for euthanasia of birds.
Frog husbandry and euthanasia were performed in compliance with federal guidelines for the ethical care and use of laboratory animals, and under the approval of the Cleveland Clinic Institutional Animal Care and Use Committee (protocol number 2011-0495).
Chicken primary embryonic cardiomyocytes
Fertilized chicken eggs were incubated at 38°C (100°F), 40–60% humidity, with auto-turning for 8 days. Cardiomyocytes were isolated as previously described (Ladd and Cooper, 2004) and plated at a density of 250,000 cells per 35 mm plate unless otherwise noted. Plates or coverslips (BD Biosciences) were coated with fibronectin (50 µg/mL working solution; Sigma) for at least 1 hour prior to plating. Cells were observed daily and medium was changed every 2–3 days.
Cardiomyocytes were transfected 24 hours post-plating (hpp) using Lipofectamine 2000 transfection reagent (Life Technologies) and a final concentration of 100 nM siRNA; this concentration was determined to be the lowest dose that provided maximal protein knockdown. siRNAs were purchased from Dharmacon (now GE Healthcare). Two independent siRNA duplexes against CELF1 (Vajda et al., 2009) were used (referred to here as “si1” and “si2”). siGLO Green or siGLO Red (referred to in the text as “siCont”) was used as non-targeting control siRNAs. Unless otherwise noted, protein or RNA samples were collected at 72 hours post-transfection (hpt).
For 5-ethynyl-2’-deoxyuridine (EdU) experiments, cardiomyocytes were plated at a density of 250,000 cells per plate in 60 mm culture dishes and transfected at 24 hpp. At 48 hpp, EdU reagent (Click-iT EdU Alexa Fluor 488 Imaging Kit; Life Technologies) was added to the medium, as per the manufacturer’s instructions (final concentration of 10 µM), and cells were incubated for 2 hrs at 37°C followed by EdU conjugation. VECTASHIELD hard-set mounting medium with DAPI (Vector Labs) wash added to each plate and a coverslip was used to cover each sample. EdU (green channel) and DAPI (blue channel) were imaged using an inverted epifluorescence microscope (Olympus) and positive cells were counted at 48 hpt.
Celf1 knockdown, rescue, and repression in Xenopus embryos
Xenopus laevis embryos were generated and maintained following established practices (Sive, 2000). Embryos were staged according to the Nieuwkoop and Faber system (Nieuwkoop and Faber, 1994) and collected into different fixatives, depending on the application (see below).
Celf1-targeting morpholino oligos (MOs) were previously described (XB-MORPHOLINO-17249279 and XB-MORPHOLINO-17249280; (Gautier-Courteille et al., 2004)). Celf1-targeting and a control MO (STD-MO) were purchased from GeneTools. Embryos were microinjected with MOs at the 2- or 4-cell stage into all blastomeres using a pulled glass microinjection needle. For Celf1-MOs, a total of 6.4 pmol/embryo was used [equivalent to 25 ng used in (Gautier-Courteille et al., 2004)]; an equal amount of total STD-MO was injected per embryo in control-treated embryos. Each experiment included Celf1-MOs-injected embryos, STD-MO-injected embryos, and uninjected embryos.
The Xenopus celf1-a sequence (GenBank accession number: KP995066) was amplified from cDNA generated from stage 45 isolated hearts (primers: 5’-AATGAATGGCACAATGGACC-3’ and 5’-TCAGTAGGGTTTGCTGTCATTC-3’) using Pfx50 high-fidelity polymerase (Life Technologies), cloned into pCR-Blunt II-TOPO using the Zero Blunt TOPO PCR Cloning Kit (Life Technologies), and confirmed by sequencing. Compared to the curated Xenopus laevis celf1-a (NM_001090727.1), the cloned sequence contained an inserted stretch between RRM1 and RRM2 that is predicted to result in a change at position p.104 from an A to an evolutionarily conserved G, as well as an insertion of 13 amino acids, an insertion within the divergent domain that has been reported previously in several species (the “LYLQ” variant; (Takahashi et al., 2000; Takahashi et al., 2001)), and one synonymous substitution (c.G1299C; p.G433G) within RRM3. To create the pCS2-celf1a construct, the celf1-a insert subcloned into the pCS2+ plasmid. To create a pCS2-NLSCELFΔ construct, the open reading frame of NLSCELFΔ (Berger and Ladd, 2012) was subcloned into the pCS2+ plasmid. The plasmids were linearized and gel isolated, and celf1a or NLSCELFΔ RNA was transcribed from these templates using an mMESSAGE mMACHINE Kit (Life Technologies) according to the manufacturer’s instructions. Successful in vitro transcription was confirmed by running aliquots on an agarose gel, then the RNAs were extracted with phenol:chloroform, and the concentrations were measured using a Nanodrop ND-1000 spectrophotometer. For rescue experiments, 0.5 fmol (383 pg) of celf1a RNA was co-injected with the Celf1-MOs. It is noteworthy that we had to carefully titrate NLSCELFΔ RNA, as high doses interfered with the function of maternal Celf proteins. High doses of NLSCELFΔ RNA (2 to 8 ng) resulted in strongly dorsalized embryos, while an intermediate dose (800 pg) was weakly dorsalizing (i.e., shortened tail, enlarged head). The latter embryos also exhibited cardiac looping and fusion defects similar to those observed following Celf1 knockdown (data not shown). The experiments shown here were performed with a very low dose of NLSCELFΔ RNA (400 pg), which resulted in cardiac morphogenetic defects (Fig. 6D) in the absence of overt dorsalization.
Immunofluorescence (IF) and immunohistochemistry (IHC)
Embryos were prepared and IF on cryosections was performed as previously described (Blech-Hermoni et al., 2013). Cryosections were probed with mouse monoclonal primary antibodies against CELF1 (1:500 in chicken, 1:100 in Xenopus; 3B1; Santa Cruz Biotechnologies), sarcomeric alpha-Actinin (ACTN2; 1:800 in chicken, 1:50 in Xenopus; EA53; Developmental Studies Hybridoma Bank [DSHB]), Z-disc Titin (TTN; 1:1000; T11; Sigma-Aldrich), or Myomesin (MYOM; 1:100; B4; DSHB). Secondary antibodies used were FITC-conjugated sheep anti-mouse (1:200; 515-095-003; Jackson ImmunoResearch), Alexa Fluor 488-conjugated goat anti-rabbit (1:400; A11008; Life Technologies), Alexa Fluor 594-conjugated goat anti-mouse (1:400; A11005; Life Technologies), and Alexa Fluor 594-conjugated goat anti-rabbit (1:400; A11037; Life Technologies). Nuclei were visualized using DAPI (0.5 µg/mL) and filamentous Actin (F-Actin) was visualized using Alexa Fluor 586-conjugated phalloidin (1:500; Life Technologies). Sections were imaged using upright epifluorescence microscopes (Olympus and Leica). For whole mount IF, Xenopus embryos were collected into 4% paraformaldehyde (PFA), incubated at 4°C overnight, washed briefly in PBS, and transferred into PBSw (0.1% Tween 20 in PBS). Embryos were incubated in PBSw for 2 × 1 hr at room temperature, and then incubated in hybridization solution (3% BSA in PBSw) for 2 hrs at 4°C. Primary antibody was added to the embryos (1:100; 3B1; Santa Cruz Biotechnologies) and they were incubated overnight at 4°C, with rocking. Embryos were washed 4 × 1 hr in PBSw, then incubated for 2 hrs in hybridization solution (3% BSA in PBSw). Secondary antibody was added (1:200; FITC-conjugated sheep anti-mouse; 515-095-003; Jackson ImmunoResearch) and the embryos were incubated at 4°C overnight, with rocking, in the dark. Finally, embryos were washed 5 × 1 hr with PBSw. Embryos were adhered to a culture dish using cyanoacrylate adhesive, overlaid with water, and imaged using an upright laser confocal microscope (Leica) equipped with a water immersion objective lens. Images were processed for publication using Adobe Photoshop CS2 (v.9.0.2), GIMP (v.2.8; http://libguides.library.cofc.edu/gimp), ImageJ (v.1.46; NIH; (Schneider et al., 2012); or Fiji v.2.0.0), and Volocity (v.6.3).
Whole mount IHC was performed as described above for IF with the following modifications: embryos were fixed in Dent’s Solution (20% dimethyl sulfoxide in methanol), then washed twice in methanol before being transferred into PBSw through a graded methanol-PBSw series. Primary antibodies used were monoclonal mouse anti-cardiac Troponin T (TNNT2; 1:100; CT3; DSHB) or monoclonal mouse anti-Meromyosin (1:100; MF20; DSHB). Secondary antibody was HRP-conjugated goat anti-mouse (1:1000; DC02L; Millipore). Finally, signal was detected using the DAB-Plus Substrate Kit (Life Technologies) for 5–30 min, hearts were scored under a dissecting microscope, and embryos were stored in PBS at 4°C.
For Xenopus EdU experiments, embryos were grown to stage 28, anesthetized briefly in 0.05% tricaine mesylate (MS222), microinjected with 8 nL of EdU reagent (as above) near the heart using a pulled glass needle, and incubated until stage 35–36. Embryos were then collected and treated as previously described (Romaker et al., 2012) with minor modifications. Frontal sections (10 µm thickness) were collected and the following antibodies were used in combination to identify cardiomyocytes by immunofluorescence: anti-alpha Actin (ACTA1; 1:50; NBP1-35265SS; Novus Biologicals) and anti-Meromyosin (1:50; MF20; DSHB) primary antibodies, and Alexa Fluor 594-conjugated goat anti-mouse and goat anti-rabbit [R37121 and R37117; Life Technologies] secondary antibodies. All sections were imaged and quantitation of proliferation was based on counts from every other section. EdU-positive cells were counted within ACTA1/MF20-positive cells and compared to the total number of (DAPI-positive) nuclei within these cells. Embryos (5–6 per treatment) were evaluated from multiple (2–3) injection sets.
For Xenopus TUNEL experiments, embryos were collected at stage 46, fixed in 4% PFA, paraffin-embedded, sectioned (frontal, 10 µm thickness), and stained using the DeadEnd Fluorometric TUNEL System (Promega) according to manufacturer’s instructions. TUNEL-stained sections were counter-stained with DAPI and mounted using ProLong Gold Antifade Mountant (Thermo Scientific). The total number (DAPI-positive) and number of TUNEL-positive cells were counted in four heart-containing sections per embryo. Embryos (3–4 per treatment) were evaluated from two independent sets of injections.
Histology
At the appropriate stages, embryos were collected into Bouin’s Fixative Solution (picric acid-formalin-acetic acid; Ricca Chemical Company) and incubated at room temperature with agitation overnight. Embryos were incubated for several additional days at room temperature with agitation, and transferred into fresh 70% ethanol daily until picric acid residue was no longer visible. Embryos were paraffin-embedded and sectioned (transverse and frontal; 7 µm thickness) as described above. Sections were stained with hematoxylin and eosin and imaged using an SCN400 Slide Scanner (Leica Microsystems).
Western blotting
Western blot analyses were performed as previously described (Ladd et al., 2005b; Blech-Hermoni et al., 2013), except as noted. Equal amounts of protein were loaded in each lane. Protein integrity, successful transfer, and equivalent loading were confirmed on all blots by Ponceau S staining (data not shown). Each experiment was carried out at least three separate times. For ACTN2, MYOM, and TTN2, the same primary antibodies were used as for IF; secondary antibodies used were horseradish peroxidase-conjugated goat anti-rabbit (1:20,000; 401393; Calbiochem) or goat anti-mouse (1:4,000–5,000; DC02L; Calbiochem). For TTN blots, samples were run on 5% SDS polyacrylamide separating gels without a stacking gel at a constant 100V for 4.5 to 5 hrs until the highest molecular weight band of the protein ladder (260 kDa) had migrated approximately halfway down the gel, and all other ladder bands had run off the bottom; due to its large size (>1,000 kDa), TTN ran at the top of the gel just below the wells. For primary cardiomyocyte experiments, membranes were stripped using Restore Western Blot Stripping Buffer (Thermo Scientific) and reprobed for GAPDH as previously described (Dasgupta et al., 2013). For western blot analyses of Xenopus proteins, the following modifications were performed: membranes were probed overnight with each antibody, and higher working concentrations of antibody were used when reprobing for Gapdh (1:10,000 primary antibody and 1:5,000 secondary antibody). Since Gapdh was highly dynamic during embryonic heart development (data not shown), heart developmental stage series blots were reprobed for γ-Tubulin (1:1000; T6657; Sigma) using horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:2000; 1010-05; Southern Biotech). For Xenopus developmental series, each embryonic heart sample represents a pool of at least 5 hearts at that stage. Adult Xenopus hearts were cut in half and each protein sample represents one half heart from a unique frog; both male and female adult frogs were used with similar results.
Real-time RT-PCR
Real-time analysis of chicken transcript levels was carried out using TaqMan reagents (Life Technologies) as previously described (Blech-Hermoni et al., 2013) using the following probes: Gg03340922_m1 (CELF1), Gg03346982_m1 (GAPDH), and Gg03358465_m1 (GUSB). Real-time analysis of Xenopus transcript levels was carried out using Power SYBR Green reagents (Life Technologies) as follows: reactions consisted of 10 µL Power SYBR Green Master Mix (Life Technologies), 150 pmol of each primer, and 5 ng cDNA. Xenopus primers are shown in Table 1; celf1 primers were designed to amplify both celf1-a and celf1-b. All samples were run at least in triplicate (technical replicates) and at least three samples (biological replicates) were included in each experiment. For embryonic heart samples, each biological replicate was generated from a pool of several hearts. Data were analyzed by the ΔΔCT method (Livak and Schmittgen, 2001), normalizing target gene values to endogenous control values (GAPDH or GUSB in the chicken; odc1 in Xenopus). Where developmental stages were compared, no endogenous control was found that did not also change during development, so equal amounts of cDNA templates were used and ΔCT values were used for calculations instead. Where indicated, a method for scaling of data from multiple biological replicates was applied (Willems et al., 2008); in these cases, error bars represent 95% confidence intervals.
Table 1.
Primers used for real-time RT-PCR
| Gene | Primer sequencesa |
|---|---|
| celf1 | F: ATGGAGGGCTGTTCCTCAC R: ATTTGCTGCTGAAGTTGCTG |
| nkx2-5 | F: CAGTCCTGGTAAGAGATGGG R: GGCTGGGTTATTGTAGTTGG |
| gata4 | F: CCTATGCTGTGCCAACTGTC R: CTTTCGTGTCTGGATCCCTTC |
| hand1 | F: CTGAGGAGGTGCCAGAAGTTG R: GGATCAGGCATCATGTGGTG |
| hand2 | F: GAACACACGACACCAGCACTG R: GTCATGGTGAACCACTGGGT |
| cdkn1 (p21) | F: TGCAGTCTGCCTTAGCCATC R: GGTGTTTCAGTGGCAAAGCC |
| ccnd1 | F: CAGTGTCTTCTCCACCCAAC R: CTTGGCGCAAACTGGACTCTAG |
| ccnd2 | F: GTGTAGGAGCGGCGATATGTG R: CTCACCAGCACCGACTCAATC |
| odc | F: CCTCGATGGGTACAGATTTCG R: GCTTCAGTCTCACTCCCAAG |
F = forward, R = reverse
Alternative splicing
The pCS2-RTB33.51 construct was created by subcloning the RTB33.51 mini-gene (Ryan and Cooper, 1996) out of the KS+ backbone into the pCS2+ backbone. Cultured COSM6 cells (kindly provided by Dr. Thomas Cooper, Baylor College of Medicine) were grown in COSM6 Growth Medium (DMEM supplemented with 10% FBS, 2 mM L-glutamine, 1% Pen/Strep) in 60 mm culture dishes. When cells reached 50–60% confluence, they were transfected using Lipofectamine 2000 (Life Technologies). All plates were transfected with 100 ng pCS2-RTB33.51, half the plates were co-transfected with 1 µg pCS2-celf1a, and an empty expression plasmid was added to a total of 3 µg DNA per plate. RNA and protein were collected from parallel plates at 48 hrs. Celf1 over-expression was confirmed by western blot, as described above. Semi-quantitative RT-PCR and quantification of splicing products were performed as previously described (Ladd et al., 2001).
Bright-field microscopy
Bright-field images of injected embryos were acquired with a MicroPublisher 5.4 RTV camera (Q-Imaging) mounted onto a Leica dissecting microscope, using the QCapture Pro (v.5.1.1.14) software. Real-time video was acquired using an ocular-mounted PowerShot G6 digital camera (Canon).
Doppler optical coherence tomography (DOCT), optical coherence tomography (OCT), and optical coherence microscopy (OCM)
Embryos were imaged using both a custom-built Fourier domain OCT system and an OCM system. The OCT system (center wavelength 1300 nm) was equipped with a quasi-telecentric scanner, linear-in-wavenumber spectrometer, and a line-scan camera with a line rate of 47 kHz (Hu and Rollins, 2007; Karunamuni et al., 2014). The axial resolution as well as the lateral resolution is approximately 10 µm in tissue, while providing good penetration depth (1–3 mm in cardiac tissues). The OCT system is also capable of quantifying blood velocity within the vessels by measuring Doppler shifts (Jenkins et al., 2010; Jenkins et al., 2012). To measure slower velocities, the line-rate of the system could be lowered below 47 kHz. OCM is similar to OCT except with a higher numerical aperture objective lens giving higher lateral resolution (~2 µm), and more limited penetration depth (< 1 mm). The OCM system had a center wavelength of 870 nm, a lateral resolution of 2 µm and an axial resolution of 5 µm. Thus OCT/OCM imaging offers high spatial resolution, good penetration depth and Doppler capability for phenotyping embryonic hearts (Garita et al., 2011; Jenkins et al., 2012). Custom MATLAB programs (Mathworks; Natick MA) were utilized to create OCT/OCM images from the raw data. Amira software (FEI Visualization Sciences Group; Burlington, MA) was employed to visualize OCT/OCM data.
Each embryo was first prepared for Doppler OCT (DOCT) functional imaging. Embryos were briefly placed into anesthetic (MS222), transferred and immobilized in 3% methylcellulose gel (ventral surface up), and positioned as to maximize Doppler signal at the right aortic branch (Yang et al., 2003). Doppler signal was recorded over a 0.5 mm span at 250 lines/structure (35 frames/second; line rate of 10 kHz; line averaging of 5) and visualized using in-house MATLAB scripts. For calculation of maximal velocity through the right aortic branch, maximal Doppler signal and the angle of vessel deflection were measured manually from the Doppler and structural OCT images. Velocity was calculated following previously published methods (Davis et al., 2009). For pulsed Doppler traces, M-scans were taken at the right aortic branch and visualized using in-house MATLAB scripts. Uninjected (n = 7), STD-MO-injected (n = 8), and Celf1-MOs-injected (n = 12) embryos from two independent injection sets were analyzed. Beat rates were calculated from the pulsed Doppler traces. Relative AV intervals were calculated by measuring the time delay between the atrial and ventricular peaks and then normalizing to the period of the heartbeat (values were averaged for 3–5 beats). Relative peak amplitudes were calculated by dividing the height of the ventricular peak by the height of the atrial peak (values were averaged for 3–5 beats). Following Doppler recordings, short video-speed movies of the beating heart were acquired at an orientation allowing for the visualization of the atrial-ventricular axis (2 mm at 500 lines/frame; 82 frames/second; line rate of 47 kHz). Relative ventricular area change was calculated by measuring ventricular area from frames at which the heart was in diastole and frames at which the heart was in systole, and the following formula was used: (Areadiastole − Areasystole) ÷ Areadiastole × 100 (values were averaged for 3–5 beats).
Embryos were then fixed in situ with 4% PFA and volume stacks (2 × 1 × 3.2 mm at 500 × 200 × 512 pixels) were acquired of the fixed heart. Uninjected (n = 8), STD-MO-injected (n = 10), and Celf1-MOs-injected (n = 14) embryos from two independent injection sets were analyzed. To visualize hearts in greater detail, an OCM system was used. Stage 46 embryos were anesthetized and hearts were removed under a dissecting microscope. Hearts were fixed overnight in 4% PFA at 4°C before being transferred to PBS for imaging.
Statistics
In primary cardiomyocytes, pairwise comparisons between siRNA-treated groups and the mock were performed via one-tailed t-tests assuming unequal variances using Microsoft Excel software. For the Xenopus developmental series, celf1 transcript levels were compared via a one-way analysis of variance performed using the StatPlus:mac (AnalystSoft) extension package for Microsoft Excel. For alternative splicing data, statistical comparisons of means were performed via one-tailed t-tests assuming unequal variances using Microsoft Excel software. Differences in cardiac looping, cardia bifida, edema, and gut dysmorphia were compared between groups using a Fisher’s exact test. Cardiomyocyte proliferation and apoptosis in the developing Xenopus heart were evaluated by performing pairwise comparisons using one-tailed t-tests assuming unequal variances using Microsoft Excel software. Differences in cardiac function were evaluated by performing pairwise comparisons via a one-tailed Mann-Whitney U test using GraphPad Prism software. Comparisons of variance were performed via F test using Microsoft Excel software. For all tests, differences were considered statistically significant when p ≤ 0.05.
Supplementary Material
Magnification: 6X; ventral view.
Magnification: 6X; ventral view.
Magnification: 6X; ventral view.
Key findings.
Strong nuclear CELF1 expression in the embryonic myocardium is conserved between mouse, chicken, and Xenopus embryos.
CELF1 is required for normal myofibrillar organization in embryonic cardiomyocytes.
Celf1 depletion causes cardiac looping defects during embryonic development.
Celf1 depletion impairs contractile function in the embryonic heart.
Celf1 depletion disrupts myofibril organization in cardiac but not skeletal muscle.
Acknowledgments
We thank Dr. Debora Malta Cerquiera Santos, Uyen Tran, and Jessie M. Hassey for their help with maintaining the Xenopus laevis and for technical assistance with Xenopus embryo experiments. We thank Dr. Judith Drazba and Dr. John Peterson for their help with confocal imaging. We thank Dr. Xiaoyong Fu, Dr. Shi Gu, and Dr. Lindsy Peterson for their assistance with acquisition and processing of OCT/OCM data, and Dr. Andrew Rollins for the use of the OCT/OCM equipment. The MF20 antibody was developed by Dr. Donald A. Fischman, the CT3 antibody was developed by Mr. Jim Jung-Ching Lin, the B4 antibody was developed by Mr. Jean-Claude Perriard, and all were obtained from the Developmental Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa (Department of Biology, Iowa City, IA 52242). This work was supported by a grant to A.N.L. from the National Institutes of Health (1R01HL089376), a grant to O.W. from the National Institutes of Health (5R01DK080745-05), and grants to M.W.J. from the National Institutes of Health (R21HL115373 and R01HL083048). Y.B-H. was supported in part by a National Institutes of Health training grant (5T32GM008056).
REFERENCES
- Baldwin BR, Timchenko NA, Zahnow CA. Epidermal growth factor receptor stimulation activates the RNA binding protein CUG-BP1 and increases expression of C/EBPbeta-LIP in mammary epithelial cells. Mol Cell Biol. 2004;24:3682–3691. doi: 10.1128/MCB.24.9.3682-3691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Bartlett H, Veenstra GJ, Weeks DL. Examining the cardiac NK-2 genes in early heart development. Pediat Cardiol. 2010;31:335–341. doi: 10.1007/s00246-009-9605-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger DS, Ladd AN. Repression of nuclear CELF activity can rescue CELF-regulated alternative splicing defects in skeletal muscle models of myotonic dystrophy. PLoS Curr. 2012;4:RRN1305. doi: 10.1371/currents.RRN1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger DS, Moyer M, Kliment GM, van Lunteren E, Ladd AN. Expression of a dominant negative CELF protein in vivo leads to altered muscle organization, fiber size, and subtype. PLoS One. 2011;6:e19274. doi: 10.1371/journal.pone.0019274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blech-Hermoni Y, Dasgupta T, Coram RJ, Ladd AN. Identification of Targets of CUG-BP, Elav-Like Family Member 1 (CELF1) Regulation in Embryonic Heart Muscle. PLoS One. 2016;11:e0149061. doi: 10.1371/journal.pone.0149061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blech-Hermoni Y, Ladd AN. Identification of transcripts regulated by CUG-BP, Elav-like family member 1 (CELF1) in primary embryonic cardiomyocytes by RNA-seq. Genomics Data. 2015;6:74–76. doi: 10.1016/j.gdata.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blech-Hermoni Y, Stillwagon SJ, Ladd AN. Diversity and conservation of CELF1 and CELF2 RNA and protein expression patterns during embryonic development. Dev Dyn. 2013;242:767–777. doi: 10.1002/dvdy.23959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boppart SA, Tearney GJ, Bouma BE, Southern JF, Brezinski ME, Fujimoto JG. Noninvasive assessment of the developing Xenopus cardiovascular system using optical coherence tomography. Proc Natl Acad Sci U S A. 1997;94:4256–4261. doi: 10.1073/pnas.94.9.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- Brewer A, Pizzey J. GATA factors in vertebrate heart development and disease. Expert Rev Mol Med. 2006;8:1–20. doi: 10.1017/S1462399406000093. [DOI] [PubMed] [Google Scholar]
- Brimacombe KR, Ladd AN. Cloning and embryonic expression patterns of the chicken CELF family. Dev Dyn. 2007;236:2216–2224. doi: 10.1002/dvdy.21209. [DOI] [PubMed] [Google Scholar]
- Chang ET, Donahue JM, Xiao L, Cui Y, Rao JN, Turner DJ, Twaddell WS, Wang JY, Battafarano RJ. The RNA-binding protein CUG-BP1 increases survivin expression in oesophageal cancer cells through enhanced mRNA stability. Biochem J. 2012;446:113–123. doi: 10.1042/BJ20120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibois M, Boulanger G, Audic Y, Paillard L, Gautier-Courteille C. Inactivation of the Celf1 gene that encodes an RNA-binding protein delays the first wave of spermatogenesis in mice. PLoS One. 2012;7:e46337. doi: 10.1371/journal.pone.0046337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibois M, Gautier-Courteille C, Kodjabachian L, Paillard L. A gene regulation network controlled by Celf1 protein-rbpj mRNA interaction in Xenopus somite segmentation. Biol Open. 2013;2:1078–1083. doi: 10.1242/bio.20135629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibois M, Gautier-Courteille C, Vallee A, Paillard L. A strategy to analyze the phenotypic consequences of inhibiting the association of an RNA-binding protein with a specific RNA. RNA. 2010;16:10–15. doi: 10.1261/rna.1742610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, Ladd AN. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley interdisciplinary reviews. RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, Stillwagon SJ, Ladd AN. Gene expression analyses implicate an alternative splicing program in regulating contractile gene expression and serum response factor activity in mice. PLoS One. 2013;8:e56590. doi: 10.1371/journal.pone.0056590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A, Izatt J, Rothenberg F. Quantitative measurement of blood flow dynamics in embryonic vasculature using spectral Doppler velocimetry. Anat Rec. 2009;292:311–319. doi: 10.1002/ar.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay J, Le Mee G, Ezzeddine N, Labesse G, Terzian C, Capri M, Ait-Ahmed O. The Drosophila Bruno paralogue Bru-3 specifically binds the EDEN translational repression element. Nucleic Acids Res. 2004;32:3070–3082. doi: 10.1093/nar/gkh627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Paillard L, Capri M, Maniey D, Bassez T, Ait-Ahmed O, Osborne HB. EDEN-dependent translational repression of maternal mRNAs is conserved between Xenopus and Drosophila. Proc Natl Acad Sci U S A. 2002;99:257–262. doi: 10.1073/pnas.012555499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MD, Lopez AL, 3rd, Larin KV, Larina IV. Imaging of cardiovascular development in mammalian embryos using optical coherence tomography. Methods Mol Biol. 2015;1214:151–161. doi: 10.1007/978-1-4939-1462-3_8. [DOI] [PubMed] [Google Scholar]
- Garita B, Jenkins MW, Han M, Zhou C, Vanauker M, Rollins AM, Watanabe M, Fujimoto JG, Linask KK. Blood flow dynamics of one cardiac cycle and relationship to mechanotransduction and trabeculation during heart looping. American journal of physiology. Heart Circ Physiol. 2011;300:H879–H891. doi: 10.1152/ajpheart.00433.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier-Courteille C, Le Clainche C, Barreau C, Audic Y, Graindorge A, Maniey D, Osborne HB, Paillard L. EDEN-BP-dependent post-transcriptional regulation of gene expression in Xenopus somitic segmentation. Development. 2004;131:6107–6117. doi: 10.1242/dev.01528. [DOI] [PubMed] [Google Scholar]
- Good P, Chen Q, Warner S, Herring D. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J Biol Chem. 2000;275:28583–28592. doi: 10.1074/jbc.M003083200. [DOI] [PubMed] [Google Scholar]
- Gregorio CC, Antin PB. To the heart of myofibril assembly. Trends Cell Biol. 2000;10:355–362. doi: 10.1016/s0962-8924(00)01793-1. [DOI] [PubMed] [Google Scholar]
- Hart A. Mann-Whitney test is not just a test of medians: differences in spread can be important. BMJ. 2001;323:391–393. doi: 10.1136/bmj.323.7309.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- Hu Z, Rollins AM. Fourier domain optical coherence tomography with a linear-in-wavenumber spectrometer. Optics Lett. 2007;32:3525–3527. doi: 10.1364/ol.32.003525. [DOI] [PubMed] [Google Scholar]
- Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. Embo J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MW, Peterson L, Gu S, Gargesha M, Wilson DL, Watanabe M, Rollins AM. Measuring hemodynamics in the developing heart tube with four-dimensional gated Doppler optical coherence tomography. J Biomed Optics. 2010;15:066022. doi: 10.1117/1.3509382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MW, Watanabe M, Rollins AM. Longitudinal Imaging of Heart Development with Optical Coherence Tomography. IEEE J Sel Topics Quantum Electron. 2012;18:1166–1175. doi: 10.1109/JSTQE.2011.2166060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunamuni G, Gu S, Doughman YQ, Peterson LM, Mai K, McHale Q, Jenkins MW, Linask KK, Rollins AM, Watanabe M. Ethanol exposure alters early cardiac function in the looping heart: a mechanism for congenital heart defects? Amer J Physiol Heart Circ Physiol. 2014;306:H414–H421. doi: 10.1152/ajpheart.00600.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama M, Maruyama K, Shimada Y. Assembly of connectin (titin) in relation to myosin and alpha-actinin in cultured cardiac myocytes. J Muscle Res Cell Motil. 1990;11:419–428. doi: 10.1007/BF01739762. [DOI] [PubMed] [Google Scholar]
- Koshelev M, Sarma S, Price RE, Wehrens XH, Cooper TA. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:1066–1075. doi: 10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress C, Gautier-Courteille C, Osborne HB, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol Cell Biol. 2007;27:1146–1157. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd A, Charlet-B N, Cooper T. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd A, Cooper T. Multiple domains control the subcellular localization and activity of ETR-3, a regulator of nuclear and cytoplasmic RNA processing events. J Cell Sci. 2004;117:3519–3529. doi: 10.1242/jcs.01194. [DOI] [PubMed] [Google Scholar]
- Ladd A, Stenberg M, Swanson M, Cooper T. A dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev Dyn. 2005a;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- Ladd A, Taffet G, Hartley C, Kearney D, Cooper T. Cardiac-specific repression of CELF activity disrupts alternative splicing and causes cardiomyopathy. Mol Cell Biol. 2005b;25:6267–6278. doi: 10.1128/MCB.25.14.6267-6278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One. 2010;5:e11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- Lin ZX, Holtzer S, Schultheiss T, Murray J, Masaki T, Fischman DA, Holtzer H. Polygons and adhesion plaques and the disassembly and assembly of myofibrils in cardiac myocytes. J Cell Biol. 1989;108:2355–2367. doi: 10.1083/jcb.108.6.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Huang H, Yuan B, Luo T, Li J, Qin X. Suppression of CUGBP1 inhibits growth of hepatocellular carcinoma cells. Clin Invest Med. 2014;37:E10–E18. doi: 10.25011/cim.v37i1.20864. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- Matsui T, Sasaki A, Akazawa N, Otani H, Bessho Y. Celf1 regulation of dmrt2a is required for somite symmetry and left-right patterning during zebrafish development. Development. 2012;139:3553–3560. doi: 10.1242/dev.077263. [DOI] [PubMed] [Google Scholar]
- Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P, Faber J. Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development From the Fertilized Egg Till the End of Metamorphosis. New York: Garland Publishing, Inc; 1994. [Google Scholar]
- Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne H. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips A, Timchenko L, Cooper T. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Romaker D, Zhang B, Wessely O. An immunofluorescence method to analyze the proliferation status of individual nephron segments in the Xenopus pronephric kidney. Methods Mol Biol. 2012;886:121–132. doi: 10.1007/978-1-61779-851-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Cooper T. Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur R, Phillips A, Cooper T. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Charlet BN, Han J, Cooper TA. ETR-3 and CELF4 protein domains required for RNA binding and splicing activity in vivo. Nucleic Acids Res. 2004;32:1232–1241. doi: 10.1093/nar/gkh275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Spletter ML, Barz C, Yeroslaviz A, Schonbauer C, Ferreira IR, Sarov M, Gerlach D, Stark A, Habermann BH, Schnorrer F. The RNA-binding protein Arrest (Bruno) regulates alternative splicing to enable myofibril maturation in Drosophila flight muscle. EMBO Rep. 2015;16:178–191. doi: 10.15252/embr.201439791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Jin Y, Otani H, Yasuda K, Inoue K. Regulation of alternative splicing of a-actinin transcript by Bruno-like proteins. Genes to Cells. 2002;7:133–141. doi: 10.1046/j.1356-9597.2001.00506.x. [DOI] [PubMed] [Google Scholar]
- Tahara N, Bessho Y, Matsui T. Celf1 is required for formation of endoderm-derived organs in zebrafish. Int J Mol Sci. 2013;14:18009–18023. doi: 10.3390/ijms140918009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Sasagawa N, Suzuki K, Ishiura S. The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybird system. Biochem Biophys Res Comm. 2000;277:518–523. doi: 10.1006/bbrc.2000.3694. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Sasagawa N, Usuki F, Kino Y, Kawahara H, Sorimachi H, Maeda T, Suzuki K, Ishiura S. Coexpression of the CUG-binding protein reduces DM protein kinase expression in COS cells. J Biochem. 2001;130:581–587. doi: 10.1093/oxfordjournals.jbchem.a003022. [DOI] [PubMed] [Google Scholar]
- Talwar S, Balasubramanian S, Sundaramurthy S, House R, Wilusz CJ, Kuppuswamy D, D’Silva N, Gillespie MB, Hill EG, Palanisamy V. Overexpression of RNA-binding protein CELF1 prevents apoptosis and destabilizes pro-apoptotic mRNAs in oral cancer cells. RNA Biol. 2013;10:277–286. doi: 10.4161/rna.23315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai M, Komiyama M, Shimada Y. Myofibril assembly is linked with vinculin, alpha-actinin, and cell-substrate contacts in embryonic cardiac myocytes in vitro. Cell Motil Cytoskel. 1989;12:185–194. doi: 10.1002/cm.970120402. [DOI] [PubMed] [Google Scholar]
- Terenzi F, Brimacombe KR, Penn MS, Ladd AN. CELF-mediated alternative splicing is required for cardiac function during early, but not later, postnatal life. J Mol Cell Cardiol. 2009;46:395–404. doi: 10.1016/j.yjmcc.2008.10.030. [DOI] [PubMed] [Google Scholar]
- Timchenko L, Miller J, Timchenko N, Devore D, Datar K, Lin L, Roberts R, Caskey C, Swanson M. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nuc Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko N, Cai Z-J, Welm A, Reddy S, Ashizawa T, Timchenko L. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001a;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- Timchenko N, Iakova P, Cai Z-J, Smith J, Timchenko L. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol Cell Biol. 2001b;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Wang GL, Timchenko LT. RNA CUG-binding protein 1 increases translation of 20-kDa isoform of CCAAT/enhancer-binding protein beta by interacting with the alpha and beta subunits of eukaryotic initiation translation factor 2. J Biol Chem. 2005;280:20549–20557. doi: 10.1074/jbc.M409563200. [DOI] [PubMed] [Google Scholar]
- Vajda NA, Brimacombe KR, LeMasters KE, Ladd AN. Muscleblind-like 1 is a negative regulator of TGF-beta-dependent epithelial-mesenchymal transition of atrioventricular canal endocardial cells. Dev Dyn. 2009;238:3266–3272. doi: 10.1002/dvdy.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, Firulli AB. Hand factors as regulators of cardiac morphogenesis and implications for congenital heart defects. Birth Defects Res A Clin Mol Teratol. 2011;91:485–494. doi: 10.1002/bdra.20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E, Leyns L, Vandesompele J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal Biochem. 2008;379:127–129. doi: 10.1016/j.ab.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Wu J, Li C, Zhao S, Mao B. Differential expression of the Brunol/CELF family genes during Xenopus laevis early development. Int J Dev Biol. 2010;54:209–214. doi: 10.1387/ijdb.082685jw. [DOI] [PubMed] [Google Scholar]
- Xiao L, Cui YH, Rao JN, Zou T, Liu L, Smith A, Turner DJ, Gorospe M, Wang JY. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell. 2011;22:3055–3069. doi: 10.1091/mbc.E11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang VX, Gordon M, Seng-Yue E, Lo S, Qi B, Pekar J, Mok A, Wilson B, Vitkin I. High speed, wide velocity dynamic range Doppler optical coherence tomography (Part II): Imaging in vivo cardiac dynamics of Xenopus laevis. Optics Express. 2003;11:1650–1658. doi: 10.1364/oe.11.001650. [DOI] [PubMed] [Google Scholar]
- Yelin R, Yelin D, Oh WY, Yun SH, Boudoux C, Vakoc BJ, Bouma BE, Tearney GJ. Multimodality optical imaging of embryonic heart microstructure. J Biomed Optics. 2007;12:064021. doi: 10.1117/1.2822904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Magnification: 6X; ventral view.
Magnification: 6X; ventral view.
Magnification: 6X; ventral view.