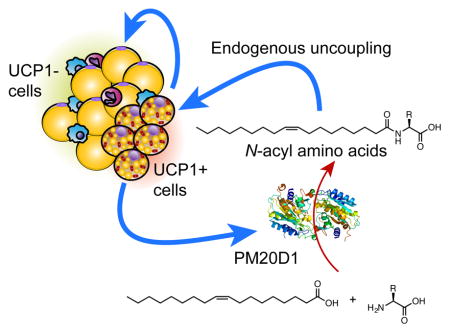
SUMMARY
Brown and beige adipocytes are specialized cells that express UCP1 and dissipate chemical energy as heat. These cells likely possess alternative UCP1-independent thermogenic mechanisms. Here we identify a secreted enzyme, peptidase M20 domain containing 1 (PM20D1), that is enriched in UCP1+ versus UCP1- adipocytes. We demonstrate that PM20D1 is a bidirectional enzyme in vitro, catalyzing both the condensation of fatty acids and amino acids to generate N-acyl amino acids and also the reverse hydrolytic reaction. N-acyl amino acids directly bind mitochondria and function as endogenous uncouplers of UCP1-independent respiration. Mice with increased circulating PM20D1 have augmented respiration and increased N-acyl amino acids in blood. Lastly, administration of N-acyl amino acids to mice improves glucose homeostasis and increases energy expenditure. These data identify an enzymatic node and a family of metabolites that regulate energy homeostasis; this pathway might be useful for treating obesity and associated disorders.
Graphical Abstract
eTOC PARAGRAPH
Beige fat cells secrete an enzyme that tacks lipids onto amino acids responsible for directly licensing mitochondria for thermogenesis.
INTRODUCTION
Adaptive thermogenesis has gained increasing attention as a process to fight the epidemic of obesity and type 2 diabetes. Mammals have brown and beige fat, two tissues that express uncoupling protein 1 (UCP1) and that are specialized to dissipate stored chemical energy in the form of heat (Cannon and Nedergaard, 2004; Harms and Seale, 2013). Either ablation of brown or beige cells (Cohen et al., 2014; Lowell et al., 1993) or knockout (KO) of the Ucp1 gene (Feldmann et al., 2009) predisposes mice to obesity and diabetes. Conversely, increasing the number or activity of brown and beige cells is protective against weight gain and metabolic disease (Seale et al., 2011).
Most studies of adaptive thermogenesis and thermogenic fat have centered upon the expression and function of UCP1. This protein catalyzes a “proton leak” whereby protons that are pumped out of the mitochondrial matrix in the electron transport chain (ETC) are transported back across the inner mitochondrial membrane (Nicholls et al., 1978; Rousset et al., 2004). This results in oxidative metabolism with no production of ATP, a process referred to as uncoupled respiration. While UCP1 is a very important part of adaptive thermogenesis, in principle, any biochemical process that requires energy and is not linked to energy storage or work can function as a thermogenic event. Indeed, data have emerged indicating that UCP1 is not the only mediator of this process (Kazak et al., 2015; Ukropec et al., 2006). Moreover, other carriers of the mitochondrial SLC25 family, of which UCP1 is only one member (SLC25A7), also have the ability catalyze a proton leak across the inner mitochondrial membrane (Brand et al., 2005).
In addition to storing chemical energy, adipose cells are now recognized to be important sensors of energy balance and secrete many bioactive proteins, including adipsin, leptin, and adiponectin (Kershaw and Flier, 2004). Proteins secreted by brown and beige fat cells in particular have not been extensively or systematically studied (Svensson et al., 2016; Wang et al., 2014). We recently developed the UCP1-TRAP mouse to identify the gene expression signature of brown and beige cells in vivo, regardless of their anatomical localization (Long et al., 2014). These initial experiments elucidated a smooth muscle-like origin for beige adipocytes, while also providing a comprehensive molecular inventory of the thermogenic fat cell in vivo.
Here, by combining the UCP1-TRAP molecular inventory with other datasets, we have compiled a core thermogenesis gene set that is co-expressed in vivo with Ucp1. We hypothesized that such a gene set might be used to identify secreted proteins that play a significant role in adaptive thermogenesis. This analysis led to the identification of a previously unstudied secreted enzyme, peptidase M20 domain containing 1 (PM20D1). We demonstrate that PM20D1 is a biosynthetic enzyme for a class of N-lipidated amino acids in vivo, and these metabolites function as endogenous uncouplers of mitochondrial respiration in a UCP1-independent manner.
RESULTS
Peptidase M20 domain containing 1 (PM20D1) is expressed in UCP1+ adipocytes and promotes energy expenditure in vivo
To identify secreted factors from brown and beige adipocytes, we first generated a list of “core thermogenesis” genes that are co-expressed with Ucp1 in vivo. To this end, we identified the overlapping gene set from the following publicly available microarray/RNAseq datasets: 1) enrichment in the classical brown adipose tissue (BAT) versus the epididymal white fat (eWAT); 2) equivalent expression in both brown and beige cells in vivo using the TRAP method; and 3) induction in the cold in the subcutaneous inguinal white fat (iWAT) following 1 or 5 weeks cold exposure (Long et al., 2014; Seale et al., 2007; Xue et al., 2009) (Figure 1A). 32 genes passed these filters (Table S1). Consistent with our original search strategy, cross-referencing of these 32 genes with the Universal Protein Resource (UniProt) demonstrated that half of these candidates (16 out of 32) were mitochondrial in subcellular localization (Table S1). Of the remainder, only one, peptidase M20 domain containing 1 (PM20D1), contained a signal peptide without any transmembrane domains, two features characteristic of a classically secreted protein. We validated the original microarray/RNAseq datasets in a new cohort of mice, and found that Pm20d1 mRNA was higher in BAT versus the other fat depots and cold-inducible in the iWAT depot (Figure S1A and B). Further supporting Pm20d1 co-expression with Ucp1 in vivo, Pm20d1 and Ucp1 mRNA were coordinately upregulated regulated in the iWAT following treatment of mice with the β-adrenergic receptor agonist CL-316,243 (Figure S1C), and coordinately downregulated in the eWAT following high fat diet (Figure S1D). Shotgun proteomics with tandem mass tag (TMT) labeling confirmed the presence of PM20D1 in blood, though its circulating levels were unchanged following 10 days of cold exposure (Figure S1E and F, and Table S2). This is presumably because, in addition to UCP1+ fat cells, liver and kidney also highly express PM20D1 (Figure S1A). To confirm that PM20D1 can be secreted from intact cells, we generated a C-terminal flag-tagged PM20D1 cDNA construct and transfected this plasmid into HEK293A cells. PM20D1 was detected both in cells and in conditioned media, whereas flag-tagged GFP was found exclusively in the cellular fraction (Figure S1G). These data demonstrate that PM20D1 is a bona fide secreted factor enriched in UCP1+ versus UCP1- adipocytes and induced in adipose tissues by cold exposure.
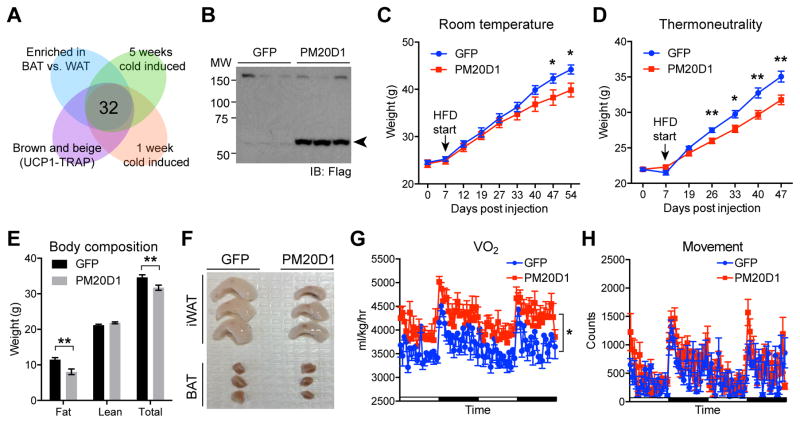
Figure 1. Increased circulating PM20D1 augments whole body energy expenditure.
(A) Schematic diagram of search strategy to identify factors expressed by UCP1+ cells. The following publicly available datasets were used for the comparisons: UCP1-TRAP (GSE56248), brown versus white adipose tissues (GSE8044), and inguinal fat following 1 or 5 weeks cold exposure (GSE13432).
(B and C) Anti-flag Western blot of plasma 40 days post injection (B) and whole body weight curves (C) from male C57BL/6 mice after tail vein injection of AAV-GFP or AAV-PM20D1 fed high fat diet (HFD) at room temperature. Mice were 7 weeks old at the time of injection, HFD was started 7 days post injection, and mice were maintained at room temperature for the duration of the experiment. For (B), arrow indicates band corresponding to PM20D1-flag. For (C), n=8–10/group, mean ± SEM, * p<0.05.
(D–F) Whole body weight curves (D), MRI analysis of total body composition (E), and representative images of adipose tissues (F) from male C57BL/6 mice after tail vein injections of AAV-GFP or AAV-PM20D1 fed HFD at thermoneutrality. Mice were placed into thermoneutrality (30°C) at 6 weeks old, injected with virus at 7 weeks old, and HFD was started 7 days post injection. Mice were maintained at 30°C for the duration of the experiment. For (E) and (F), data are from 47 days post injection. n=8–10/group, mean ± SEM, * p<0.05, ** p<0.01.
(G and H) VO2 (G) and movement (H) of male C57BL/6 mice over a period of two days. Mice were 7 weeks old at the time of injection, high fat diet (HFD) was started 7 days post injection, and mice were maintained at room temperature for the duration of the experiment. For (G) and (H), measurements were taken at 4 weeks post injection, a time point prior to any sigificant divergence of body weight (body weight means ± SEM: GFP 33.3±1.3 g, PM20D1 31.6±1.3 g, p>0.05). n=8/group, mean ± SEM, * p<0.05.
To assess the functions of PM20D1 in vivo, mice were tail vein-injected with adeno-associated viral vectors (serotype AAV8) expressing PM20D1 or GFP. These vectors are primarily taken up and expressed by the liver, though other tissues may also express them (Zincarelli et al., 2008). One week following the injections, the mice were placed on high fat diet (HFD). Increased circulating PM20D1 was observed by western blots of the plasma at 40 days post-injection (Figure 1B). At room temperature (22°C), mice with augmented circulating PM20D1 showed blunted weight gain (final weight means ± SEM: GFP, 44.2±1.0 g; PM20D1, 39.8±1.5 g; 10% weight difference) compared to the control animals (Figure 1C). A similar blunting of weight gain was observed at thermoneutralty (30°C) (final weight means ± SEM: GFP, 35.0±0.8 g; PM20D1, 31.8±0.7 g; 9% weight difference; Figure 1D), where the sympathetic nervous input to adipose tissues is decreased. Body composition analysis by MRI at the end of the experiment at thermoneutrality revealed that the weight difference was due exclusively to a 30% reduction in fat mass in those animals receiving PM20D1 compared to GFP; there were no effects on lean mass (Figure 1E and F). In a separate cohort of mice treated with AAV-PM20D1 or AAV-GFP at room temperature, we performed whole body indirect calorimetry measurements at a time point prior to the divergence in weights (4 weeks HFD, Figure S2A). These analyses revealed significantly augmented VO2 (Figure 1G) and VCO2 (Figure S2B), indicative of increased energy expenditure. Most importantly, this occurred with no changes in movement (Figure 1H) or food intake (Figure S2C).
PM20D1 regulates N-lipidated amino acids in vivo
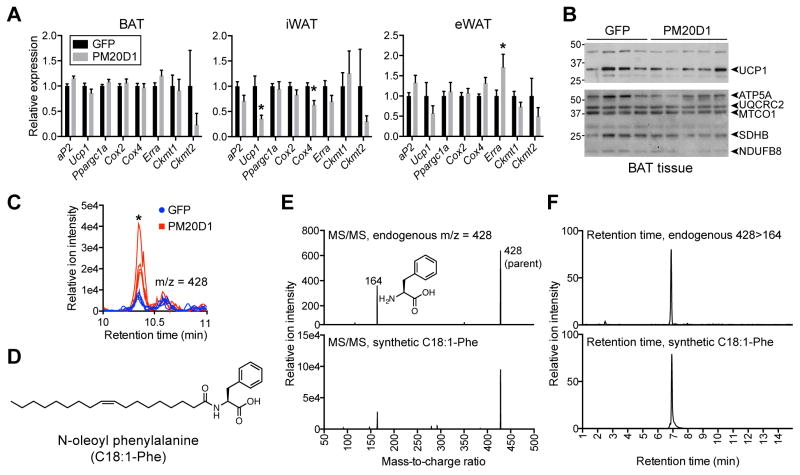
Changes in energy expenditure with no change in physical movement are usually indicative of activation of a thermogenic gene program in the classical brown fat, subcutaneous iWAT (termed browning), or both. In addition, recent work has indicated the presence of an additional thermogenic pathway based upon a futile cycle of creatine phosphorylation (Kazak et al., 2015). Surprisingly, the increased metabolic rates observed here were not accompanied by any broad molecular change corresponding to these processes, such as an induction of UCP1 (Figure 2A and B), or changes in gene expression related to creatine metabolism (Figure 2A). These data suggest that PM20D1 increases whole body energy expenditure through an alternative mechanism.
Figure 2. Lack of classical browning and identification of increased N-oleoyl phenylalanine in mice injected with AAV-PM20D1.
(A and B) mRNA expression of the indicated genes in BAT, iWAT, and eWAT (A) and Western blot of UCP1 and mitochondrial proteins (B) from male C57BL/6 mice at thermoneutrality after tail vein injections of AAV-GFP or AAV-PM20D1. Mice were placed into thermoneutrality (30°C) at 6 weeks old, injected with virus at 7 weeks old, and high fat diet (HFD) was started 7 days post injection. Mice were maintained at 30°C for the duration of the experiment. For (A) and (B), data are from 47 days post injection. For (A), n=8/group, mean ± SEM, * p<0.05. For (B), n=4–5/group.
(C) Chromatogram at m/z = 428 from plasma of male C57BL/6 mice after tail vein injection of AAV-GFP or AAV-PM20D1. For (C), mice were 7 weeks old at the time of injection, high fat diet (HFD) was started 7 days post injection, and mice were maintained at room temperature for the duration of the experiment. The comparative metabolomics was performed on plasma harvested 54 days post injection. n=4/group, * p<0.05.
(D) Chemical structure of N-oleoyl phenylalanine (C18:1-Phe).
(E and F) MS/MS spectra (E) and retention time (F) of endogenous (top) or synthetic (bottom) C18:1-Phe.
See also Table S3.
To further investigate how PM20D1 augmented energy expenditure, we examined the detailed molecular consequences of PM20D1 actions in the blood. PM20D1 is one of five members of the mammalian M20 peptidase family, but remains entirely uncharacterized with respect to its endogenous substrates and products. Despite their annotation as metallopeptidases, the other four other mammalian M20 family members possess peptidase activity on a variety of small molecule substrates (Teufel et al., 2003; Van Coster et al., 2005; Veiga-da-Cunha et al., 2014). We therefore performed untargeted polar small molecule liquid chromatography-mass spectrometry (LC-MS) profiling of plasma from mice injected with AAV-GFP or AAV-PM20D1 viral vectors (Smith et al., 2006). Manual inspection of differential peaks revealed the most robust difference occurred in a metabolite with an m/z = 428, which was increased in mice injected with AAV-PM20D1 versus AAV-GFP (Figure 2C and Table S3). We identified this m/z = 428 peak as the [M-H]− ion of N-oleoyl phenylalanine (C18:1-Phe, chemical formula C27H42NO3−, expected m/z = 428) (Figure 2D). This identification was confirmed by MS/MS experiments, which revealed a predominant product ion of m/z = 164 corresponding to the phenylalanine anion (Figure 2E, chemical formula C9H10NO2−, expected m/z = 164). Further supporting this identification, both synthetic C18:1-Phe and the endogenous m/z = 428 peak eluted at similar retention times by LC (Figure 2F).
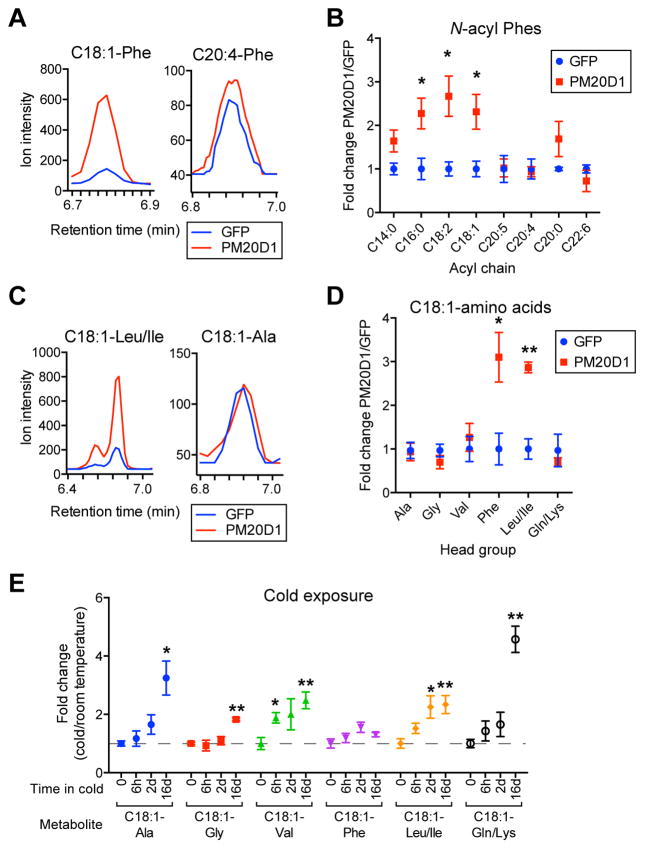
We then developed a targeted multiple reaction monitoring (MRM) program to assess the scope of N-acyl amino acids regulated by PM20D1 in plasma in vivo. By absolute quantitation using a C15-Phe internal standard, the basal plasma concentrations of specific members of the N-acyl amino acids in AAV-GFP injected mice were found to be in the 1–100 nM range (Table S4). PM20D1 predominantly elevates medium and long N-acyl Phes (C14–18, Figure 3A and B) and also N-oleoyl amino acid metabolites with large and hydrophobic head groups (C18:1-Phe and C18:1-Leu/Ile, Figure 3C and D). Similar changes in N-acyl amino acids were observed for mice injected with AAV-PM20D1 at room temperature (Figure S3). Since Pm20d1 mRNA is induced in the iWAT following cold exposure (Figure 1A and S1A), we also assessed plasma levels of the N-oleoyl amino acids following cold exposure by targeted MS. 6 hours or 2 days cold exposure increased specific members of this class (C18:1-Leu/Ile and C18:1-Val, Figure 3E). Long-term cold exposure (16 days) significantly elevated the levels of most N-oleoyl amino acids that were measured (Figure 3E). Therefore, both PM20D1 and its N-acyl amino acid products are physiologically co-regulated by cold exposure.
Figure 3. PM20D1 regulates the levels of N-acyl amino acids in vivo.
(A–D) Chromatograms (A and C) and quantitation of fold change (B and D) of various N-acyl Phes (A and B) or various C18:1-amino acids (C and D) in plasma of male C57BL/6 mice in thermoneutrality after tail vein injection of AAV-GFP or AAV-PM20D1 by targeted MRM. Mice were placed into 30°C at 6 weeks old, injected with virus at 7 weeks old, and high fat diet (HFD) was started 7 days post injection. Mice were maintained at 30°C for the duration of the experiment. The comparative targeted metabolomics was performed on plasma harvested 47 days post injection. For (A) and (C), chromatograms are from one representative mouse per group. For (B) and (D), the absolute quantitation in AAV-GFP versus AAV-PM20D1 is as follows: C16-Phe, 6 versus 15 nM, respectively; C18:2-Phe, 2 versus 7 nM, respectively; C18:1-Phe, 4 versus 10 nM, respectively; C18:1-Leu, 6 versus 29 nM, respectively.
(E) Fold change (cold/room temperature) of the indicated plasma N-acyl amino acids following cold exposure for the indicated times by targeted MRM.
For (B) and (D–E), n=4–5/group, mean ± SEM, * p<0.05, ** p<0.01.
PM20D1 is a bidirectional N-acyl amino acid synthase and hydrolase in vitro
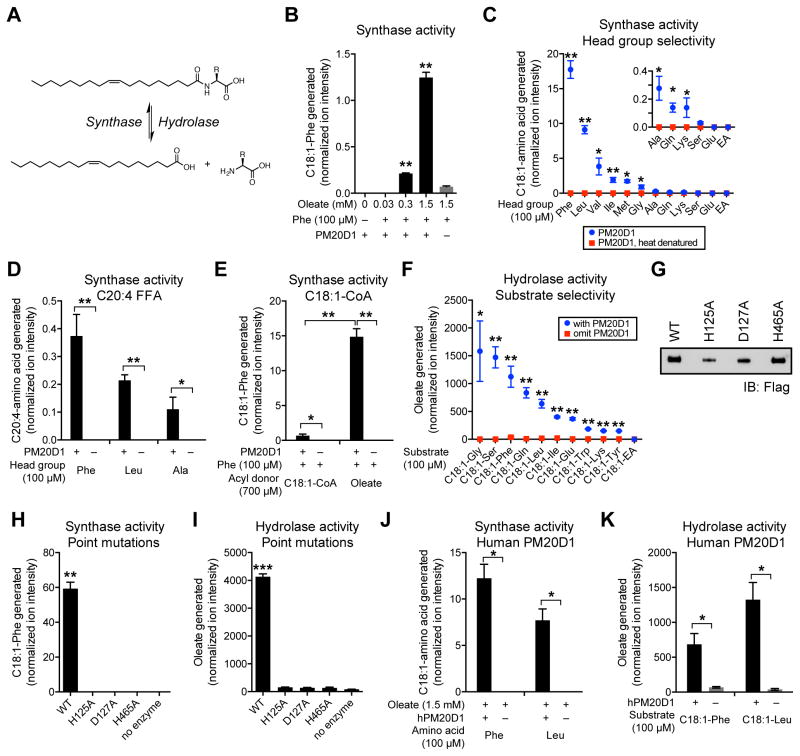
Although the endogenous presence of N-acyl amino acids has been previously described (Tan et al., 2010), their biosynthesis has remained a long-standing mystery. The increase of these metabolites in AAV-PM20D1 mice suggested that PM20D1 might be an enzyme responsible for N-acyl amino acid biosynthesis in vivo. To investigate this process in vitro, we generated purified, mammalian recombinant mouse PM20D1 for enzymatic assays. As expected, purified mouse PM20D1 migrated as a ~60kDa band by Coomassie staining (Figure S4A). Significant formation of C18:1-Phe was observed by LC-MS when recombinant PM20D1 was incubated in the reaction mixture with physiologically relevant concentrations of free oleate and Phe (300 μM and 100 μM, respectively; Figure 4A and B). Among different amine head groups, Phe was the amino acid most efficiently converted to its corresponding N-acyl amino acid product (Figure 4C). PM20D1 could also condense other amino acids with oleate, although less efficiently than Phe (Figure 4C). The negatively charged amino acid glutamate, as well as ethanolamine (EA), were not substrates for the synthase reaction (Figure 4C). PM20D1 was also capable of using arachidonate as a fatty acid donor (Figure 4D), and showed strong preference for free oleate over oleoyl-coenzyme A (C18:1-CoA, Figure 4E). Thus free fatty acids and free amino acids are substrates for PM20D1, and its N-acyl amino acid synthase activity shows selectivity for specific amino groups and acyl donors.
Figure 4. Enzymatic activity of PM20D1 in vitro.
(A) Schematic of synthase and hydrolase reaction of free fatty acid and free amino acid to N-acyl amino acid.
(B) Relative levels of C18:1-Phe generated in vitro from Phe (100 μM), oleate (0.03–1.5 mM), and purified mouse PM20D1-flag.
(C–E) Relative levels of C18:1-amino acid generated in vitro from the indicated head group (100 μM) and purified mouse PM20D1-flag using either oleate (1.5 mM, C), arachidonate (1.5 mM, D) or oleoyl-coenzyme A (C18:1-CoA, 0.7 mM, E). For (C), EA, ethanolamine.
(F) Relative levels of oleate generated in vitro from the indicated N-acyl amide substrates (100 μM) and purified mouse PM20D1-flag. C18:1-EA, N-oleoyl ethanolamine.
(G) Anti-flag Western blot of immunoaffinity purified mouse PM20D1-flag or the indicated point mutants.
(H) Relative levels of C18:1-Phe generated in vitro from Phe (100 μM), oleate (1.5 mM), and the indicated wild-type (WT) or mutant PM20D1-flag protein.
(I) Relative levels of oleate generated in vitro from C18:1-Phe (100 μM) and the indicated wild-type (WT) or mutant PM20D1-flag protein.
(J) Relative levels of C18:1-amino acid generated in vitro from the indicated head group (100 μM), oleate (1.5 mM), and purified human PM20D1-flag.
(K) Relative levels of oleate generated in vitro from the indicated N-acyl amide substrate (100 μM) and purified human PM20D1-flag.
For (B–F) and (H–K), enzymatic assays were carried out in PBS at 37°C for 1.5 hours, n=3/group, mean ± SEM, * p<0.05, ** p<0.01, for reaction with PM20D1 versus reaction omitting PM20D1, or reaction with PM20D1 versus reaction with heat-denatured PM20D1. Y-axes indicates relative ion intensity normalized to 1 nmol of a D3,15N-serine internal standard that was doped in during the extraction process prior to MS analysis.
See also Figure S4.
We also observed that incubation of C18:1-Phe with PM20D1 liberated free oleate, indicating that PM20D1 can also act as an N-acyl amino acid hydrolase (Figure 4A and F). The hydrolase activity was apparently more promiscuous than the synthase activity since PM20D1 hydrolyzed all N-oleoyl amino acids tested (Figure 4F). In contrast, N-oleoyl ethanolamine (C18:1-EA), a well-characterized substrate for fatty acid amide hydrolase (FAAH) (Cravatt et al., 1996; Saghatelian et al., 2004), was not a PM20D1 substrate under these conditions (Figure 4F).
Based on sequence homology with the other members of the mammalian M20 family and Uniprot annotations, we generated three point mutations in PM20D1 that we anticipated would disrupt catalytic activity. Due to differences in protein stability and expression, following flag-immunoaffinity purification, each construct was titrated such that approximately equivalent protein amounts were used in the subsequent enzymatic assays (Figure 4G). Under synthase activity reaction conditions using oleate and Phe as substrates, or hydrolase activity reaction conditions using C18:1-Phe, only wild-type (WT) PM20D1 possessed enzymatic activity, whereas no activity was observed for any of the mutants (Figure 4H and I). In these in vitro assays, we observe 1.2±0.1% and 94.0±0.8% conversion (means ± SEM, n=3) in the synthase and hydrolase direction, respectively for the wild-type enzyme. Therefore the synthase and hydrolase activities are embodied within the PM20D1 polypeptide and not a co-purifying protein. PM20D1 appears to require residues predicted to coordinate divalent cations, and both synthase and hydrolase activities are coordinately disrupted by point mutations in the cation binding sites.
Finally, we generated purified, recombinant human PM20D1, which shares 71% identity and 86% similarity with the mouse enzyme; the human protein also shows complete conservation of the H125, D127, and H465 residues required for catalysis (Figure S4B and C). Human PM20D1 also possessed N-acyl amino acid synthase and hydrolase activities (Figure 4J and K), demonstrating the conservation of this enzymatic activity in the PM20D1 polypeptide from these two mammalian species.
Taken together, these studies indicate that PM20D1 is a bidirectional enzyme that can generate N-acyl amino acids from amino acids and free fatty acids, and can also hydrolyze N-acyl amino acids into amino acids and free fatty acids. The changes in the various species of N-acyl amino acids in plasma from AAV-PM20D1 injected mice are therefore likely to reflect a balance of the relative synthase and hydrolase activities on, as well as the relative concentrations of, the particular substrates and products. Differences between N-acyl amino acids regulated by PM20D1 overexpression and cold exposure (Figure 3) likely reflect different levels of fatty acid or amino acid substrates under various physiologic conditions. However, we cannot exclude other, as of yet unknown enzymes that might perform similar chemical reactions.
N-acyl amino acids are endogenous uncouplers of respiration in cells
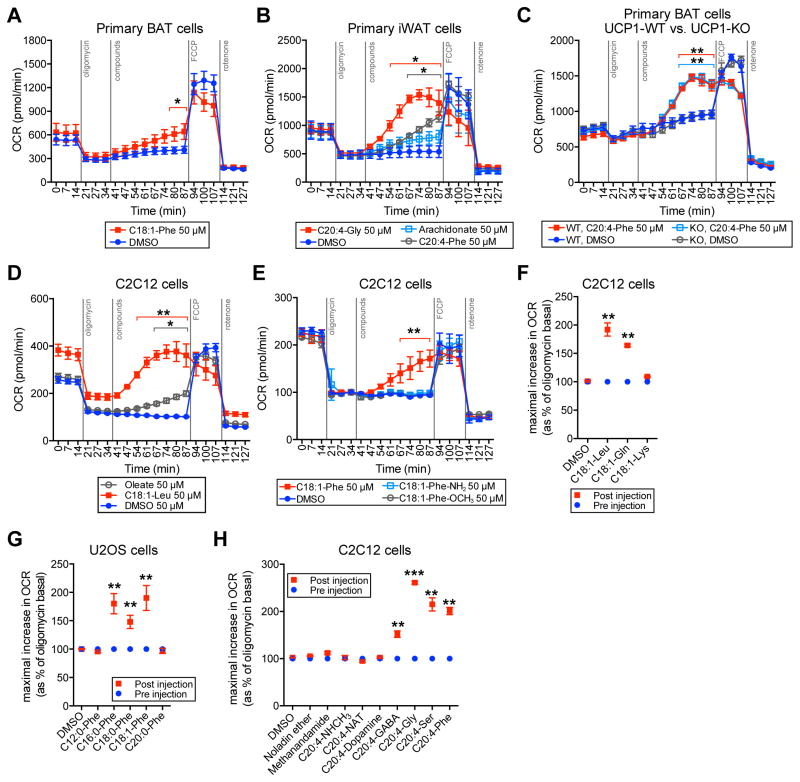
To directly test the effect of N-acyl amino acids on respiration, we acutely treated differentiated primary BAT adipocytes first with the ATP synthase inhibitor oligomycin to block coupled respiration, and then with C18:1-Phe (50 μM). Cellular oxygen consumption was significantly augmented by C18:1-Phe treatment (maximal OCR of 198%, versus oligomycin-treated basal respiration; and 156% versus DMSO at the same time point, Figure 5A). Similarly, in differentiated primary iWAT adipocytes, both C20:4-Gly and C20:4-Phe augmented respiration in the presence of oligomycin (maximal OCR of 295% and 243%, respectively, versus oligomycin-treated basal respiration; and 285% and 214%, respectively, versus DMSO at the same time point, Figure 5B). As a control, arachidonate itself also increased the oxygen consumption rate very modestly, but not nearly as much in magnitude as the N-acyl amino acids (Figure 5B).
Figure 5. Effects of N-acyl amino acids on respiration in cells.
(A–C) Oxygen consumption rates (OCRs) of differentiated primary BAT cells (A), differentiated primary iWAT cells (B), and differentiated primary BAT cells from UCP1-WT or UCP1-KO mice (C), treated with the indicated compounds for the indicated time. For (A–C), adipocytes were differentiated and analyzed on day 5.
(D–H) OCRs of C2C12 cells (D–F, and H) or U2OS cells (G) treated with the indicated compounds for the indicated time. For (D–H), cells were seeded and analyzed the following day. For (F–H), data is shown as the maximal increase in OCR as a percentage of the oligomycin basal OCR, which is normalized to 100%. For (A–H), the following concentrations of compounds were used: oligomycin (1 μM), indicated N-acyl amino acid or fatty acid (50 μM), FCCP (0.2 μM), or rotenone (3 μM). For (E), the following non-standard abbreviations are used: C18:1-Phe-NH2 (N-oleoyl phenylalanine amide), C18:1-Phe-OCH3 (N-oleoyl phenylalanine methyl ester). For (H), the following non-standard abbreviations are used: C20:4-NHCH3 (N-arachidonoyl N-methyl amide), C20:4-NAT (N-arachidonoyl taurine), C20:4-GABA (N-arachidonoyl gamma-amino butyric acid).
For (A–H), n=3–6/group, mean ± SEM, * p<0.05, ** p<0.01, *** p<0.001 for treatment versus DMSO at the same time point.
The major biological mechanism currently understood for inducing uncoupled respiration in adipocytes is through the action of UCP1 (Rousset et al., 2004). However, C20:4-Phe produced virtually identical increases in uncoupled respiration in both UCP1-WT and KO cells (maximal OCR of 221% and 214%, in UCP1-WT and UCP1-KO cells, respectively, versus the oligomycin-treated basal respiration, p>0.05 between genotypes, Figure 5C), establishing that UCP1 is not required for this effect. Consistent with these observations, N-acyl amino acids were also competent inducers of uncoupled respiration in cell types that completely lack UCP1, including the mouse myoblast cell line C2C12 (Figure 5D–F) and the human osteosarcoma cell line U2OS (Figure 5G). These data show that N-acyl amino acids are endogenous chemical uncouplers of mitochondrial respiration in a UCP1-independent manner.
We next explored the structural requirements for N-acyl amino acids to exert their effects on uncoupled respiration. A direct comparison of C18:1-Leu and oleate demonstrated that the N-acyl amino acid conjugate was significantly more potent than the free fatty acid alone (Figure 5D). Modification of the free amino acid carboxylate of C18:1-Phe, either by methyl esterification (C18:1-Phe-OCH3), or by primary amidation (C18:1-Phe-NH2), completely abrogated uncoupling activity (Figure 5E), indicating that the amino acid carboxylate moiety is required for activity. We observed some amino acid head group selectivity to the respiration effects, as C18:1-Gln, but not C18:1-Lys, possessed the uncoupling activity (Figure 5F). Modification of the lipid chain revealed that only C16-, C18-, and C18:1-Phe possessed uncoupling activity, whereas saturated acyl chains that were too short (C12:0-Phe) or too long (C20:0-Phe) completely lacked uncoupling activity (Figure 5G). Finally, testing multiple commercially available fatty acid amides revealed that most did not stimulate uncoupled respiration, unless they possessed these general structural features outlined above (e.g., C20:4-GABA, Figure 5H).
N-acyl amino acids directly uncouple mitochondria and interact with mitochondrial proteins
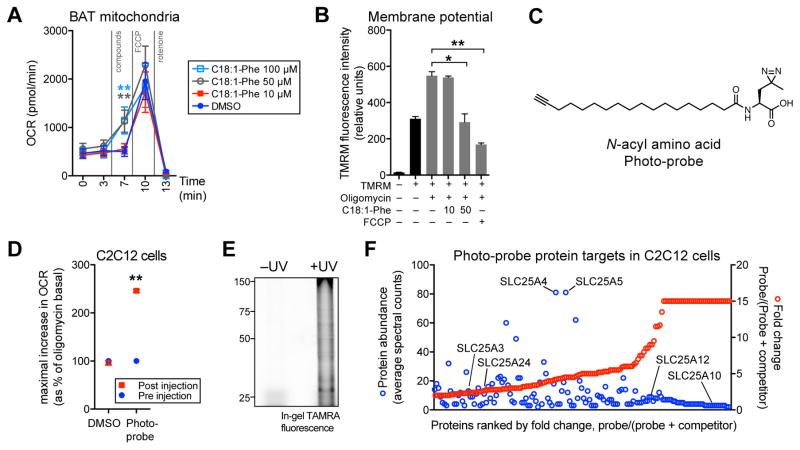
To directly assess the ability of N-acyl amino acids to uncouple mitochondria, we employed two approaches. First, we isolated mitochondria from the BAT tissues and treated these mitochondria with increasing concentrations of C18:1-Phe (10–100 μM). Isolated BAT mitochondria increased respiration in a dose-dependent manner following C18:1-Phe treatment (Figure 6A), indicating that N-acyl amino acids do not require any other cellular components or organelles for their uncoupling effects. Second, we used tetramethylrhodamine methyl ester (TMRM) fluorescence to directly measure the mitochondrial membrane potential in live cells. As expected, treatment of C2C12 cells with oligomycin increased the membrane potential (Figure 6B). Co-treatment of oligomycin with C18:1-Phe (50 μM) decreased the TMRM fluorescence by ~45% (Figure 6B). As a positive control, the well-known chemical uncoupler FCCP (0.4 μM) also reduced TMRM fluorescence by an even larger magnitude (~70% reduction). Therefore N-acyl amino acids can directly augment uncoupled respiration in isolated mitochondria, resulting in a decreased mitochondrial membrane potential.
Figure 6. Effects of N-acyl amino acids in mitochondria and identification of N-acyl amino acid-interacting proteins.
(A) Oxygen consumption rates (OCRs) of freshly isolated BAT mitochondria treated with the indicated compounds for the indicated times. Respiration was measured with 10 mM pyruvate and 5 mM malate as substrates, and FCCP and rotenone were used at 2 μM and 3 μM, respectively. n=4–5/group, ** p<0.01.
(B) Tetramethyl rhodamine methyl ester (TMRM) fluorescence in C2C12 cells following 20 min treatment with oligomycin alone (1 μM), or in combination with C18:1-Phe (10 or 50 μM) or FCCP (0.4 μM). n=3/group, mean ± SEM, * p<0.05, ** p<0.01.
(C) Chemical structure of the N-acyl amino acid photocrosslinkable probe (“photo-probe”).
(D) OCR of C2C12 cells treated with DMSO or the photo-probe (50 μM). For (D), data is shown as the maximal increase in OCR as a percentage of the oligomycin basal OCR, which is normalized to 100%. n=3–4/group, mean ± SEM, ** p<0.01
(E) TAMRA in-gel fluorescence of C2C12 cells treated with the photo-probe (50 μM, 20 min), followed by UV irradiation (on ice, 10 min), cell lysis, and click chemistry with TAMRA-N3. For (E), control cells that were not UV irradiated were kept under ambient light (on ice, 10 min).
(F) Proteins in C2C12 cells that showed C20:4-Phe competeable photoprobe labeling. For (F), C2C12 cells were incubated with 20 μM photo-probe (“probe only”), or 20 μM photo-probe with 100 μM C20:4-Phe competitor (“probe + competitor”). Cells were then UV irradiated, lysed, subjected to click chemistry with biotin-N3, and analyzed by MS (see Methods). Proteins satisfying the following filtering criteria are shown: >50% reduction in peptide counts with competitor present versus without competitor, and detection of at least one peptide in all three probe only samples. Comparisons in which no peptides were detected in “probe + competitor” samples were assigned a fold-change of 15.
See also Table S5.
The structure activity relationships (SAR) observed with N-acyl amino acids-induced uncoupling (Figure 5), and the direct effects of N-acyl amino acids on isolated mitochondria (Figure 6A), are consistent with specific binding interactions between these metabolites and mitochondrial proteins (Niphakis et al., 2015). To identify the proteins that may be mediating the uncoupling by N-acyl amino acid, we synthesized a photo-crosslinkable version (Figure 6C). This molecule, which we term “photo-probe,” contains a modified Met amino acid with a photo-crosslinking diazarine side chain, and a fatty acid-alkyne for downstream click chemistry applications. Photo-probe (50 μM) was a competent inducer of uncoupled respiration in C2C12 cells, demonstrating that the alkyne and diazarine modifications did not affect the bioactivity (Figure 6D). Moreover, photo-probe demonstrated robust, UV-dependent crosslinking as assessed by in-gel TAMRA fluorescence (Figure 6E).
C2C12 cells were selected for LC-MS/MS analysis of photo-probe targets since they demonstrate robust N-acyl amino acid induced uncoupling. To this end, C2C12 cells were incubated with photo-probe (20 μM, “probe only” samples), or co-incubated with both photo-probe (20 μM) and a C20:4-Phe competitor at 5-fold excess (100 μM, “probe + competitor” samples). Cells were then UV irradiated on ice and lysed by sonication. Probe-labeled proteins were conjugated to biotin-N3 by click chemistry, streptavidin-enriched, and subject to LC-MS/MS analysis with spectral counting. In total, 149 proteins were identified that showed >50% competition by C20:4-Phe (Table S5 and Figure 6F). Of these, 31 proteins (21%) are localized to the mitochondria by Uniprot annotation, including 6 members of the SLC25 carrier family (Table S5 and Figure 6F). Notably, the two most abundantly detected proteins in the entire dataset were the mitochondrial SLC25A4 and SLC25A5 (also known as ANT1 and 2). In addition to their ADP/ATP symport activity, these transporters have previously been demonstrated to translocate protons across the inner membrane (Brand et al., 2005). Taken together, these data are consistent with a model where N-acyl amino acids increase uncoupled respiration by liganding SLC25 family members, including SLC25A4 and SLC25A5, and increasing SLC25-mediated proton flux into the matrix.
Administration of N-acyl amino acids to mice increases energy expenditure and improves glucose homeostasis
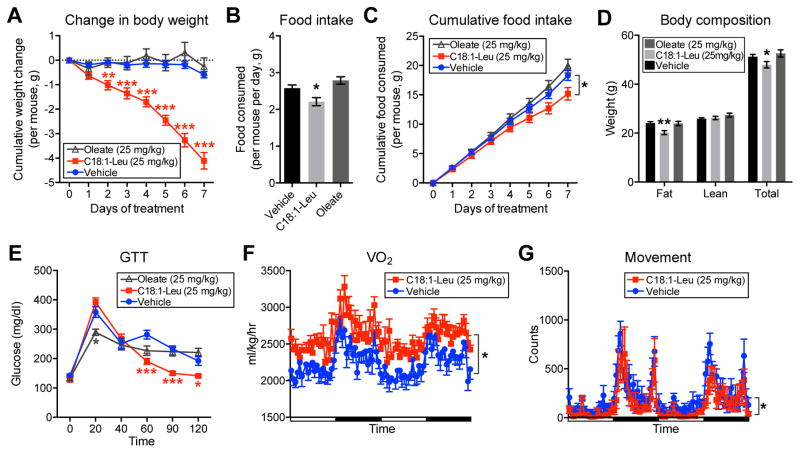
We next sought to determine the physiologic effects of N-lipidated amino acid administration to mice. Diet induced obese (DIO) mice were treated daily with vehicle, oleate, or C18:1-Leu (25 mg/kg, i.p.). After 8 days treatment, mice receiving C18:1-Leu lost 4.1±0.3 g, whereas mice treated with oleate or vehicle only lost 0.3±0.2 g and 0.6±0.1 g, respectively (means ± SEM, Figure 7A). Over this time course, food intake was slightly but significantly reduced in mice receiving C18:1-Leu (17% reduction versus vehicle-treated mice, Figure 7B and C). Body composition analysis at the end of this experiment showed the weight loss induced by C18:1-Leu was entirely accounted for by a difference in fat mass (Figure 7D). Lastly, C18:1-Leu treated mice also showed improvements in GTT versus either vehicle- or oleate-treated mice (Figure 7E).
Figure 7. In vivo effects of chronic C18:1-Leu administration to mice.
(A–E) Change in body weight (A), daily and cumulative food intake (B and C), body composition by MRI (D), and GTT (E) of 21 week DIO mice treated daily with vehicle, or C18:1-Leu (25 mg/kg/day, i.p.), or oleate (25 mg/kg/day, i.p.). For (D), MRI measurements were taken on day 7. For (A–E), the initial weights of the mice were not statistically different (means ± SEM: vehicle, 51.9±0.8 g; 25 mg/kg C18:1-Leu, 52.1±1.1 g; 25 mg/kg oleate, 52.9±1.2 g). For (A–E), n=9/group, for vehicle and C18:1-Leu, and n=5/group for oleate, mean ± SEM, * p<0.05, ** p<0.01, *** p<0.001. For (E), after the last dose on day 7, mice were fasted overnight and the GTT was performed the next morning with glucose at a dose of 1.5 g/kg.
(F and G) VO2 (F) and movement (G) of mice treated with vehicle or C18:1-Leu. For (F and G), measurements were recorded for 2 days following 8 days chronic treatment with vehicle or C18:1-Leu (25 mg/kg/day, i.p.); during this time daily administration of the indicated compounds continued. For (F and G), n=8/group, mean ± SEM, * p<0.05. See also Figure S5, Figure S6, and Figure S7.
To assess the effects of C18:1-Leu on whole body energy expenditure, we performed indirect calorimetry measurements in a separate cohort of mice treated daily with vehicle or C18:1-Leu (25 mg/kg/day, i.p.). After 8 daily injections, mice were placed into metabolic cages and injected for an additional two days (Figure S5). Mice treated with C18:1-Leu showed significantly augmented VO2 (Figure 7F) and VCO2 (Figure S5F) compared with vehicle-treated mice, and also slightly reduced movement (Figure 7G). In this cohort, C18:1-Leu treated mice also showed a reduced food intake over the final two days of the experiment (Figure S5H), but not during the earlier time period (days 0–7, Figure S5C and D). Finally, The RER was significantly lower in mice receiving C18:1-Leu, indicating a switch to fats as a metabolic fuel type (Figure S5G).
The uncoupling activity of N-acyl amino acids appears to be a property of this entire class of metabolites, at least in cells. To explore the generality of N-acyl amino acid bioactivity in vivo, we analyzed additional cohorts of DIO mice treated daily with C18:1-Phe (30 or 50 mg/kg/day, i.p.) or C20:4-Gly (15 mg/kg/day, i.p.). Blood levels of C18:1-Phe were 3.0±0.3 μM and 0.4±0.1 μM (means ± SEM, n=3) at 2 h and 6 h post-injection, respectively, following a single 30 mg/kg i.p. dose. While chronic treatment of mice with C18:1-Phe or C20:4-Gly reduced food intake, these compounds nevertheless significantly augmented VO2, with no effects on movement (Figure S6 and S7). Measurements of plasma AST and ALT, as well as plama cytokines, revealed no sigificant elevations in mice treated with C20:4-Gly versus vehicle (Figure S7G–J). Taken together, these data demonstrate that N-acyl amino acids can directly augment whole body energy expenditure, reduce fat mass, and improve glucose clearance in mice.
DISCUSSION
We have characterized a previously unstudied enzyme, PM20D1, that is enriched in UCP1+ versus UCP1- cells and catalyzes the condensation of fatty acids and certain amino acids to form N-acyl amino acids. While these lipidated metabolites had previously been detected in biological tissues, little was known about their biosynthesis or physiologic functions. Here, we show that N-acyl amino acids function as endogenous uncouplers of mitochondrial respiration, even in cells lacking UCP1. It is important to note that the experiments demonstrating in vitro and in vivo augmentation of respiration by N-acyl amino acids have been performed in two independent laboratories (BMS and PRG). These data thus suggest a new model of brown and beige fat thermogenesis, whereby UCP1+ adipocytes can stimulate uncoupled respiration in neighboring cells that are not specialized to dissipate chemical energy as heat (i.e., UCP1- cells). In this model, brown and beige fat cells are not only the terminal effectors of thermogenesis via UCP1 and a creatine futile cycle, but also are the initiators of a broader cascade of UCP1-independent thermogenic events (Figure S7K). This model also suggests that the thermogenic and metabolic benefits of brown and beige fat extend beyond their own intracellular functions. Based on the N-acyl amino acid concentrations observed in blood, the PM20D1/N-acyl amino acid pathway likely endogenously occurs in a local, paracrine manner. However, our in vivo experiments with viral vectors indicate that this mechanism can potentially function with systemic administration of the protein as well. Future work using tissue-specific knockout mice will definitely establish the contribution of various tissues to circulating PM20D1 levels, and the physiologic roles of PM20D1 in adipose tissues.
These data suggest that either PM20D1, or N-acyl amino acids themselves, might be used therapeutically for the treatment of obesity and other obesity-associated disorders. The therapeutic use of synthetic chemical uncouplers has been limited by untoward and even fatal side effects (Grundlingh et al., 2011). Whether a wider therapeutic window for N-acyl amino acids exists remains to be determined. Although administration of several distinct N-acyl amino acids to mice augments energy expenditure and promotes weight loss, we also observe a small but significant reduction in food intake, at least under our chronic dosing regime. The total weight loss observed in the DIO mice treated with N-acyl amino acids is likely due to a combination of both reduced food intake and augmented energy expenditure. Further exploration of other naturally occurring lipidated amino acids or chemical modifications of such molecules might identify compounds that can dissociate the beneficial from any potential undesired effects. Alternatively, PM20D1 protein injections might also be used to augment N-acyl amino acid levels in vivo.
Besides potential therapeutic applications, these studies on the enzymology of PM20D1 address long-standing questions regarding biosynthetic pathways for N-acyl amino acids. Historically, an enzymatic activity involving the condensation of fatty acids and amino acids has been previously reported in tissues (Fukui and Axelrod, 1961), but its molecular identity has remained unclear. The data here provide strong evidence that PM20D1 is an enzyme responsible for these activities. From an chemical equilibrium point of view, we calculate a thermodynamic equilibrium of ~1% conversion to N-acyl amino acid, based on the in vitro synthase reactant concentrations (1.5 mM oleate and 100 μM Phe) and the equilibrium constant for a related amide condensation and hydrolysis reaction (Katayama et al., 1999). Our experimentally observed N-acyl amino acid generation in the synthase direction (1.2±0.1% conversion; mean ± SEM) are consistent with these calculations. Moreover, such concentrations of fatty acid and amino acid reactants are within the physiologic range (Stegink et al., 1991). The energetic driving force of the synthase reaction in vivo is likely to arise from a disequilibrium of the fatty acid and amino acid reactants and their N-acyl amino acid products, analogous to the proton-motive force that drives ATP production by ATP synthase.
An outstanding issue is the molecular target(s) responsible for the uncoupling activity of N-acyl amino acids. The photo-crosslinking experiments provide direct evidence that N-acyl amino acids engage members of the SLC25 family of inner mitochondrial carriers, including ANT1 and 2. Notably, the function of this photo-probe reagent requires both diazarine and alkyne “ends” of the molecule. Consequently, it must be the intact photo-probe, and not a hydrolyzed product, that is interacting with proteins. The proton conductance activity of the ANTs, or other SLC carriers, might be directly or allosterically activated by N-acyl amino acid binding. Loss-of-function studies will ultimately be required to definitely establish such a role for these carriers.
In summary, these data identify a new enzymatic node and a class of lipidated metabolites that might be used for the treatment of human obesity and diabetes, and to modulate thermogenesis more generally.
EXPERIMENTAL PROCEDURES
Plasmids and viruses
Full-length mouse Pm20d1 cDNA (GE Dharmacon) was subcloned with an in-frame C-terminal 6xHis and Flag tag into pENTR/D-TOPO (Thermo Fisher Scientific) according to the manufacturer’s instructions. This construct was subsequently cloned into the appropriate expression vectors as described in the Supplemental Experimental Procedures. Full-length C-terminal Flag tagged human Pm20d1 cDNA (Origene) was used directly for transient transfection experiments.
Chemicals
The full inventory of commercially available compounds, purchased from Sigma, Cayman Chemical Company, or Santa Cruz Biotechnology, as well as the synthesis of N-acyl amino acids, is described in the Supplemental Experimental Procedures.
General animal information
Animal experiments were performed according to procedures approved by the Beth Israel Deaconess Medical Center IACUC. Mice were maintained in 12 h light-dark cycles at 22°C and fed a standard irradiated rodent chow diet. All experiments on wild-type mice were performed with male C57BL/6 mice purchased from Jackson Laboratories, except for the AAV experiments, which were performed with male C57BL/6 mice from Charles River. UCP1-KO (stock #017476) and 16–20 week DIO mice (stock #380050) were obtained from Jackson. Details of the mouse injections and treatments can be found in the Supplemental Experimental Procedures.
Indirect calorimetry and body composition measurements
Energy expenditure, O2 consumption, CO2 production, respiratory exchange ratio, total locomotor activity and food intake measurements were made with a 16-cage Columbus Instruments Oxymax Comprehensive Lab Animal Monitoring System at ambient room temperature (21–23°C). Mice were acclimated for one day in metabolic cages prior to data collection. Whole-body composition was assessed with an EchoMRI 3-in-1 on conscious mice.
In vitro enzymatic assays
Flag-tagged mouse and human PM20D1 were immunopurified using magnetic Flag-M2 beads (Sigma Aldrich) as detailed in the Supplemental Experimental Procedures. In vitro PM20D1-catalyzed synthesis of N-acyl amino acids was measured by incubating purified PM20D1 protein with oleate (1.5 mM) and phenylalanine (0.1 mM) in PBS (to 100 μl final volume) at 37°C for 1.5 hrs. In vitro PM20D1-catalyzed N-acyl amino acid hydrolysis was measured by incubating purified PM20D1 protein with the indicated N-acyl amino acid (0.1 mM) in PBS (to 100 μl final volume) at 37°C for 1.5 hrs. Reactions were terminated by placing the vials at −80°C and analyzed as described below by LC-MS.
Measurements of metabolites in vivo and enzyme activities in vitro by LC-MS
Frozen serum (30 μl) for polar metabolomic analyses were extracted in 160 μl of 1:1 acetonitrile/methanol with inclusion of internal standard D3,15N-serine (1 nmol). Activity assays (100 μl) were extracted in 400 μl 1:1 acetonitrile/methanol. Following 30 s of thorough vortexing and 1 min of bath sonication, the polar metabolite fraction (supernatant) was isolated by centrifugation at 13,000 × g for 10 min. 10 μl of this supernatant was analyzed by SRM-based targeted LC-MS/MS, or untargeted LC-MS. The Supplemental Experimental Procedures contains details about MS parameters used for these analyses.
Cellular respiration measurements
Cellular oxygen consumption rates were determined using an XF24 Extracellular Flux Analyzer (Seahorse Biosciences) as detailed in the Supplemental Experimental Procedures.
Statistics
The Student’s t-test was used for pair-wise comparisons, and ANOVA was used for indirect calorimetry experiments. Unless otherwise specified, statistical significance was set at p<0.05.
Supplementary Material
ARTICLE HIGHLIGHTS.
PM20D1 is a secreted enzyme that regulates N-acyl amino acids in vivo
N-acyl amino acids are endogenous metabolites that uncouple mitochondria
Increased PM20D1 or N-acyl amino acid administration augments energy expenditure
Acknowledgments
We thank Marc Kirschner for many critical discussions as well as members of the Spiegelman, Nomura, and Griffin laboratories for helpful comments. We are grateful to Julie J. Johnson and Arbansjit Sandhu at the University of Pennsylvania Vector Core for AAV production. This work was supported by grants to JZL (NIH DK105203), KJS (Swedish Research Council Postdoctoral Fellowship PE2013/613), JAP (NIH DK098285), DKN (American Cancer Society Research Scholar Award RSG14-242-01-TBE and NIH R01CA172667), and BMS (NIH DK5447, DK31405 and the JPB Foundation).
Footnotes
CONFLICTS OF INTEREST
BMS is a consultant for Calico, Inc.
AUTHOR CONTRIBUTIONS
Conceptualization, JZL and BMS; Validation, MRC and PRG; Investigation, JZL, KJS, LAB, IAL, JL, RRR, MPJ, JAP, and DKN; Resources, HL, TK, LAB and DKN; Writing, JZL and BMS; Funding Acquisition, JZL, KJS, DKN, and BMS; Supervision, SPG, PRG, DKN, and BMS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brand MD, Pakay JL, Ocloo A, Kokoszka J, Wallace DC, Brookes PS, Cornwall EJ. The basal proton conductance of mitochondria depends on adenine nucleotide translocase content. Biochem J. 2005;392:353–362. doi: 10.1042/BJ20050890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, et al. Ablation of PRDM16 and Beige Adipose Causes Metabolic Dysfunction and a Subcutaneous to Visceral Fat Switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Fukui T, Axelrod B. Enzymatic formation of lipo-amino acids by rat liver preparations and the nature of the product. J Biol Chem. 1961;236:811–816. [PubMed] [Google Scholar]
- Grundlingh J, Dargan PI, El-Zanfaly M, Wood DM. 2,4-dinitrophenol (DNP): a weight loss agent with significant acute toxicity and risk of death. J Med Toxicol. 2011;7:205–212. doi: 10.1007/s13181-011-0162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- Katayama K, Ueda N, Katoh I, Yamamoto S. Equilibrium in the hydrolysis and synthesis of cannabimimetic anandamide demonstrated by a purified enzyme. Biochim Biophys Acta. 1999;1440:205–214. doi: 10.1016/s1388-1981(99)00124-9. [DOI] [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, Vetrivelan R, Lu GZ, Laznik-Bogoslavski D, Hasenfuss SC, et al. A Creatine-Driven Substrate Cycle Enhances Energy Expenditure and Thermogenesis in Beige Fat. Cell. 2015;163:643–655. doi: 10.1016/j.cell.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. J Clin Endocrinol Metab. 2004;89:2548–2556. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. A smooth muscle-like origin for beige adipocytes. Cell Metab. 2014;19:810–820. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell BB, VSS, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Bernson VS, Heaton GM. The identification of the component in the inner membrane of brown adipose tissue mitochondria responsible for regulating energy dissipation. Experientia Suppl. 1978;32:89–93. doi: 10.1007/978-3-0348-5559-4_9. [DOI] [PubMed] [Google Scholar]
- Niphakis MJ, Lum KM, Cognetta AB, 3rd, Correia BE, Ichu TA, Olucha J, Brown SJ, Kundu S, Piscitelli F, Rosen H, et al. A Global Map of Lipid-Binding Proteins and Their Ligandability in Cells. Cell. 2015;161:1668–1680. doi: 10.1016/j.cell.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset S, Alves-Guerra MC, Mozo J, Miroux B, Cassard-Doulcier AM, Bouillaud F, Ricquier D. The biology of mitochondrial uncoupling proteins. Diabetes. 2004;53(Suppl 1):S130–135. doi: 10.2337/diabetes.53.2007.s130. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Stegink LD, Filer LJ, Jr, Brummel MC, Baker GL, Krause WL, Bell EF, Ziegler EE. Plasma amino acid concentrations and amino acid ratios in normal adults and adults heterozygous for phenylketonuria ingesting a hamburger and milk shake meal. Am J Clin Nutr. 1991;53:670–675. doi: 10.1093/ajcn/53.3.670. [DOI] [PubMed] [Google Scholar]
- Svensson KJ, Long JZ, Jedrychowski MP, Cohen P, Lo JC, Serag S, Kir S, Shinoda K, Tartaglia JA, Rao RR, et al. A Secreted Slit2 Fragment Regulates Adipose Tissue Thermogenesis and Metabolic Function. Cell Metab. 2016;23:454–466. doi: 10.1016/j.cmet.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, O’Dell DK, Yu YW, Monn MF, Hughes HV, Burstein S, Walker JM. Identification of endogenous acyl amino acids based on a targeted lipidomics approach. J Lipid Res. 2010;51:112–119. doi: 10.1194/jlr.M900198-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel M, Saudek V, Ledig JP, Bernhardt A, Boularand S, Carreau A, Cairns NJ, Carter C, Cowley DJ, Duverger D, et al. Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J Biol Chem. 2003;278:6521–6531. doi: 10.1074/jbc.M209764200. [DOI] [PubMed] [Google Scholar]
- Ukropec J, Anunciado RP, Ravussin Y, Hulver MW, Kozak LP. UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1−/− mice. J Biol Chem. 2006;281:31894–31908. doi: 10.1074/jbc.M606114200. [DOI] [PubMed] [Google Scholar]
- Van Coster RN, Gerlo EA, Giardina TG, Engelke UF, Smet JE, De Praeter CM, Meersschaut VA, De Meirleir LJ, Seneca SH, Devreese B, et al. Aminoacylase I deficiency: a novel inborn error of metabolism. Biochem Biophys Res Commun. 2005;338:1322–1326. doi: 10.1016/j.bbrc.2005.10.126. [DOI] [PubMed] [Google Scholar]
- Veiga-da-Cunha M, Chevalier N, Stroobant V, Vertommen D, Van Schaftingen E. Metabolite proofreading in carnosine and homocarnosine synthesis: molecular identification of PM20D2 as beta-alanyl-lysine dipeptidase. J Biol Chem. 2014;289:19726–19736. doi: 10.1074/jbc.M114.576579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GX, Zhao XY, Meng ZX, Kern M, Dietrich A, Chen Z, Cozacov Z, Zhou D, Okunade AL, Su X, et al. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med. 2014;20:1436–1443. doi: 10.1038/nm.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Petrovic N, Cao R, Larsson O, Lim S, Chen S, Feldmann HM, Liang Z, Zhu Z, Nedergaard J, et al. Hypoxia-independent angiogenesis in adipose tissues during cold acclimation. Cell Metab. 2009;9:99–109. doi: 10.1016/j.cmet.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.