Abstract
Background
The Uigur ethnic minority is the largest ethnic group in the Xinjiang Uygur Autonomous Region of China, and valuable resource for the study of ethnogeny. The objective of this study was to estimate the genetic diversities and forensic parameters of 30 insertion-deletion loci in Uigur ethnic group from Xinjiang Uigur Autonomous Region of China and to analyze the genetic relationships between Xinjiang Uigur group and other previously published groups based on population data of these loci.
Results
All the tested loci were conformed to Hardy–Weinberg equilibrium after Bonferroni correction. The observed and expected heterozygosity ranged from 0.3750 to 0.5515; and 0.4057 to 0.5037, respectively. The combined power of discrimination and probability of exclusion in the group were 0.99999999999940 and 0.9963, respectively. We analyzed the DA distance, interpopulation differentiations and population structure, conducted principal component analysis and neighbor-joining tree based on our studied group and 21 reference groups. The present results indicated that the studied Xinjiang Uigur group (represented our samples from the whole territory of Xinjiang Uigur Autonomous Region) had a close relationships with Urumchi Uigur (represented previously reported samples from Urumchi of Xinjiang) and Kazak groups.
Conclusions
The present study may provide novel biological information for the study of population genetics, and can also increase our understanding of the genetic relationships between Xinjiang Uigur group and other groups.
Electronic supplementary material
The online version of this article (doi:10.1186/s40064-016-2730-3) contains supplementary material, which is available to authorized users.
Keywords: Forensic genetics, Population structure, Uigur ethnic group, Genetic relationship
Background
The short tandem repeats (STRs) are commonly used genetic makers in the field of forensic sciences, and single nucleotide polymorphisms (SNPs) are considered as alternative and supplementary markers to STRs (Gill 2001; Kidd et al. 2005; Tan et al. 2015; Ye et al. 2014). SNPs can be captured in smaller amplicons than STRs without stutter in the profile. Insertion-deletion polymorphisms (InDels) as biallelic polymorphic markers are considered to have potential values in forensic application due to number of advantages properties shared with the similar binary variation of SNPs, for example, smaller amplicons, lower mutation rates than STRs and widely distribute in the human genome (Phillips et al. 2007; Fondevila et al. 2012; Shi et al. 2015; Romanini et al. 2012). At present, InDels have been applied in forensic genetic applications including individual identification (Pereira et al. 2009), inferring biogeographic ancestry (Yang et al. 2005) and population genetic studies et al. (Zaumsegel et al. 2013).
The Investigator DIPplex® kit (Qiagen, Hilden, Germany) contains the following components for the simultaneous amplification of Amelogenin and 30 autosomal InDels (the genomic information regarding chromosomal localization of the 30 InDel loci was shown in Table 1). The allele length variations of the InDels range from 4 to 22 bp and all amplicons are shorter than 160 bp, which make them more suitable for highly degraded DNA samples in forensic caseworks. To date, several populations’ genetic data have been published based on this kit, e.g. Japanese, Poland and Korean groups, and so on (Nunotani et al. 2015; Pepinski et al. 2013; Kim et al. 2014).
Table 1.
Allele frequencies and forensic parameters for 30 InDels in Uigur group from Xinjiang Uigur Autonomous Region (n = 136)
| HLD | rs# | Chromosome localization | DIP+ | DIP− | MP | PD | PIC | PE | TPI | Ho | He | HWE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 1610905 | 16q13 | 0.5184 | 0.4816 | 0.3539 | 0.6461 | 0.3747 | 0.1463 | 0.9067 | 0.4485 | 0.5030 | 0.2196 |
| 39 | 17878444 | 1p22.1 | 0.2978 | 0.7022 | 0.4303 | 0.5697 | 0.3308 | 0.1359 | 0.8831 | 0.4338 | 0.4213 | 0.7398 |
| 40 | 2307956 | 1p32.3 | 0.5515 | 0.4485 | 0.3803 | 0.6197 | 0.3723 | 0.1875 | 1.0000 | 0.5000 | 0.4984 | 0.9354 |
| 45 | 2307959 | 2q31.1 | 0.6066 | 0.3934 | 0.3874 | 0.6126 | 0.3634 | 0.1689 | 0.9577 | 0.4779 | 0.4808 | 0.9798 |
| 48 | 28369942 | 2q11.2 | 0.4007 | 0.5993 | 0.3729 | 0.6271 | 0.3650 | 0.1463 | 0.9067 | 0.4485 | 0.4839 | 0.4338 |
| 56 | 2308292 | 4q25 | 0.6581 | 0.3419 | 0.3859 | 0.6141 | 0.3488 | 0.0994 | 0.8000 | 0.3750 | 0.4533 | 0.0724 |
| 58 | 1610937 | 5q14.1 | 0.4154 | 0.5846 | 0.3729 | 0.6271 | 0.3677 | 0.1573 | 0.9315 | 0.4632 | 0.4893 | 0.5714 |
| 64 | 1610935 | 5q12.3 | 0.7206 | 0.2794 | 0.4437 | 0.5563 | 0.3216 | 0.1308 | 0.8718 | 0.4265 | 0.4057 | 0.5961 |
| 67 | 1305056 | 5q33.2 | 0.6140 | 0.3860 | 0.4047 | 0.5953 | 0.3617 | 0.1940 | 1.0149 | 0.5074 | 0.4775 | 0.4608 |
| 70 | 2307652 | 6q16.1 | 0.5699 | 0.4301 | 0.3965 | 0.6035 | 0.3701 | 0.2075 | 1.0462 | 0.5221 | 0.4939 | 0.4839 |
| 77 | 1611048 | 7q31.1 | 0.4890 | 0.5110 | 0.3716 | 0.6284 | 0.3749 | 0.1811 | 0.9855 | 0.4926 | 0.5035 | 0.8346 |
| 81 | 17879936 | 7q21.3 | 0.6471 | 0.3529 | 0.3991 | 0.6009 | 0.3524 | 0.1517 | 0.9189 | 0.4559 | 0.4601 | 0.9524 |
| 83 | 2308072 | 8p22 | 0.3640 | 0.6360 | 0.4417 | 0.5583 | 0.3558 | 0.2366 | 1.1148 | 0.5515 | 0.4664 | 0.0425 |
| 84 | 3081400 | 8q24.12 | 0.6213 | 0.3787 | 0.3654 | 0.6346 | 0.3598 | 0.0994 | 0.8000 | 0.3750 | 0.4740 | 0.0230 |
| 88 | 8190570 | 9q22.32 | 0.4632 | 0.5368 | 0.3854 | 0.6146 | 0.3736 | 0.2007 | 1.0303 | 0.5147 | 0.5010 | 0.7164 |
| 92 | 17174476 | 11q22.2 | 0.4816 | 0.5184 | 0.4054 | 0.5946 | 0.3747 | 0.2366 | 1.1148 | 0.5515 | 0.5030 | 0.2407 |
| 93 | 2307570 | 12q22 | 0.5257 | 0.4743 | 0.3881 | 0.6119 | 0.3743 | 0.2075 | 1.0462 | 0.5221 | 0.5024 | 0.6153 |
| 97 | 17238892 | 13q12.3 | 0.3382 | 0.6618 | 0.3949 | 0.6051 | 0.3475 | 0.1212 | 0.8500 | 0.4118 | 0.4510 | 0.3787 |
| 99 | 2308163 | 14q23.1 | 0.6801 | 0.3199 | 0.4181 | 0.5819 | 0.3404 | 0.1463 | 0.9067 | 0.4485 | 0.4383 | 0.7809 |
| 101 | 2307433 | 15q26.1 | 0.5000 | 0.5000 | 0.3616 | 0.6384 | 0.3750 | 0.1630 | 0.9444 | 0.4706 | 0.5037 | 0.4660 |
| 111 | 1305047 | 17p11.2 | 0.3346 | 0.6654 | 0.4080 | 0.5920 | 0.3461 | 0.1463 | 0.9067 | 0.4485 | 0.4486 | 0.9695 |
| 114 | 2307581 | 17p13.3 | 0.3199 | 0.6801 | 0.4134 | 0.5866 | 0.3404 | 0.1359 | 0.8831 | 0.4338 | 0.4383 | 0.9461 |
| 118 | 16438 | 20p11.1 | 0.6066 | 0.3934 | 0.3941 | 0.6059 | 0.3634 | 0.1811 | 0.9855 | 0.4926 | 0.4808 | 0.7505 |
| 122 | 8178524 | 21q22.11 | 0.4485 | 0.5515 | 0.3803 | 0.6197 | 0.3723 | 0.1875 | 1.0000 | 0.5000 | 0.4984 | 0.9354 |
| 124 | 6481 | 22q12.3 | 0.6360 | 0.3640 | 0.3902 | 0.6098 | 0.3558 | 0.1463 | 0.9067 | 0.4485 | 0.4664 | 0.7053 |
| 125 | 16388 | 22q11.23 | 0.4890 | 0.5110 | 0.3487 | 0.6513 | 0.3749 | 0.1359 | 0.8831 | 0.4338 | 0.5035 | 0.1139 |
| 128 | 2307924 | 1q31.3 | 0.4338 | 0.5662 | 0.4087 | 0.5913 | 0.3706 | 0.2291 | 1.0968 | 0.5441 | 0.4949 | 0.2336 |
| 131 | 1611001 | 7q36.2 | 0.4890 | 0.5110 | 0.4050 | 0.5950 | 0.3749 | 0.2366 | 1.1148 | 0.5515 | 0.5035 | 0.2448 |
| 133 | 2067235 | 3p22.1 | 0.4449 | 0.5551 | 0.3848 | 0.6152 | 0.3719 | 0.1940 | 1.0149 | 0.5074 | 0.4976 | 0.7865 |
| 136 | 16363 | 22q13.1 | 0.4779 | 0.5221 | 0.3689 | 0.6311 | 0.3745 | 0.1750 | 0.9714 | 0.4853 | 0.5027 | 0.7164 |
HLD human locus deletion/insertion polymorphism, DIP− frequency of short allele, DIP+ frequency of long allele, Ho observed heterozygosity, He expected heterozygosity, MP matching probability, PD power of discrimination, PE probability of exclusion, PIC Polymorphic information contents, TPI typical paternity index, HWE probability value of the exact test for Hardy–Weinberg equilibrium, p the short arm of a chromosome, q the long arm of a chromosome
Xinjiang Uigur Autonomous Region is located in the northwest border of China with the land of 1.6649 million square kilometers and account for one-sixth of China’s total area (Fig. 1). It lies in the heart of the ancient Silk Road which has historically experienced migration of many groups of Eastern and Western Eurasians. The Uigur, as the main nationality of Xinjiang Uigur Autonomous Region, has a population of 10.06 million in 2010 (http://www.stats.gov.cn/tjsj/pcsj/rkpc/6rp/indexch.htm). The Uigurs mainly live in Kashi which is located in the south of Tianshan Mountain, and others are scattered in Ili and Urumchi area. Uigurs have their own language and words and their language belongs to the Turkic branch of Altaic language family. The belief of the Uigurs is Islamism which has a great influence on Uigurs’ culture and custom (Shan and Deng 2012). In the present study, we obtained the population genetic data and calculated the forensic parameters of 30 InDels in the studied Xinjiang Uigur group. We also collected the population data from previously reported groups to analysis their genetic relationships including Uigurs living in different area, other groups in China, Asian, European and Amerindian groups.
Fig. 1.
A map showing the geographic location of the Xinjiang Uigur Autonomous Region, China
Methods
Sample collection and DNA extraction
A total of 136 bloodstain samples were collected from Xinjiang Uigur Autonomous Region. All volunteers resided in Xinjiang Uigur Autonomous Region for more than three generations and signed the informed consents before being involved in the study. This study was approved by Institutional Ethics Committee, Xinjiang Medical University, China. Genomic DNA was extracted from bloodstained samples using the Chelex-100 method according to Walsh et al. (1991).
Amplification and genotyping
Amplification of 30 InDel loci was performed using the Investigator DIPplex® kit on GeneAmp PCR System 9700 Thermal Cycler (Applied Biosystems, Foster City, CA, USA) according to the Investigator DIPplex handbook instruction. Amplification products were separated via capillary electrophoresis on an ABI3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) according to manufacturer’s instruction. The control DNA 9948 (Promega, Madison, WI, USA) was analyzed as positive control. Genotyping results were obtained using the software GeneMapper v3.2 (Applied Biosystems, Foster City, CA, USA) by comparing to allelic ladder.
Reference groups
InDel data from 21 previously published groups including 9 Chinese groups: Beijing Han, Tibetan, Kazak, Urumchi Uigur (represented previously reported samples from Urumchi of Xinjiang) (Wei et al. 2014), Guangdong Han (Hong et al. 2013), Shanghai Han, She (Wang et al. 2014), Yi (Zhang et al. 2015) and Xibe (Meng et al. 2015); 6 Mexican groups: Chihuahua Mexican, Mexico Mexica, Jalisco Mexican, Veracruz Mexican, Yucatan Mexican and Mexican Amerindian (Martínez-Cortés et al. 2015); South Korean (Korea) (Seong et al. 2014); Dane (Denmark) (Friis et al. 2012); Two Spanish groups: Basque and Central Spanish (Martín et al. 2013); Uruguayan (Uruguay) (Saiz et al. 2014); and Hungarian (Hungary) (Kis et al. 2012) were collected for population genetic analysis.
Quality control
We strictly followed International Society for Forensic Genetics (ISFG) recommendations on the analysis of the DNA polymorphisms (Schneider 2007).
Statistical analysis
Allele frequencies and forensic parameters including observed heterozygosity (Ho), Hardy–Weinberg equilibrium (HWE), match probability (MP), polymorphic information content (PIC), power of exclusion (PE), discrimination power (PD) and typical paternity index (TPI) were estimated by the modified Powerstat v1.2 spreadsheet (Promega, Madison, WI, USA). Expected heterozygosity (He) was calculated according to the formula: (Nei 1978), pi was the allele frequency of allele i, k was the number of alleles and n was the number of samples. The pairwise Fst and p values were calculated by Arlequin statistical software v.3.5 (Excoffier and Lischer 2010). Principal component analysis (PCA) based on allele frequencies was evaluated in MATLAB2007a (MathWorks Inc., USA). Linkage disequilibrium (LD) analysis was performed using the SNP Analyzer V2.0 (Istech, South Korea) (Yoo et al. 2008). The DA distances were obtained using the DISPAN program (Ota 1993). According to the DA distances the neighbor-joining (NJ) tree was conducted. Population structure analysis was conducted by the STRUCTURE program (version 2.2) using Admixture Model with parameters adjusted to: burn-in-period, 100,000; run time, 100,000 steps in the Markov Chain; K values, 2–7; and iteration time, 15 (Pritchard et al. 2003; Jakobsson and Rosenberg 2007).
Results and discussion
Forensic parameter analysis
All studied loci were found to be in accordance with HWE in Xinjiang Uigur group after Bonferroni correction when the significance level was adjusted to 0.0017 (p = 0.05/30). The allele frequencies and forensic parameters of 30 InDel loci in Xinjiang Uigur group were shown in Table 1; and the raw genotyping data were shown in Additional file 1: Table S1. The Ho and He ranged from 0.3750 (HLD56 and HLD84) to 0.5515 (HLD83, HLD92 and HLD131); 0.4057 (HLD64) to 0.5037 (HLD101), respectively. The PIC, TPI, PD and PE values ranged from 0.3216 to 0.3750; 0.8000 to 1.1148; 0.5563 to 0.6513 and 0.0994 to 0.2366, respectively. The highest and lowest MP were 0.4437 (HLD64) and 0.3487 (HLD125), respectively. The combined power of discrimination (CPD) and probability of exclusion (CPE) in the group were 0.99999999999940 and 0.9963, respectively. The high CPD value demonstrates that the panel of 30 InDel loci had potential in forensic individual identification.
Linkage disequilibrium analysis
Linkage disequilibrium has been tested for all possible combinations between each locus. The linkage disequilibrium pattern revealed by r2 values between each locus was shown in Additional file 2: Table S2, The results showed that there was no linkage disequilibrium observed among all the loci with the values of r2 less than 0.1, which indicated those genetic markers were relatively independent for subsequent comparison among 22 groups.
Clustering analysis
Before conducting the comparison, we had re-read the references and made sure that loci in all reference populations showed no deviation from HWE and linkage equilibrium. We analyzed the population structures of Xinjiang Uigur group (represented our samples from the whole territory of Xinjiang Uigur Autonomous Region) and 21 referenced groups and the results were shown in Fig. 2. The Asian groups were separated from both Amerindian groups and European groups at K = 2, the 5 European groups and 6 Amerindian groups constituted almost entirely by green component while 8 Asian groups by red; The Kazak, Urumchi Uigur and Xinjiang Uigur groups displayed admixture constitution of both green and red components. At K = 4, we could clearly separate Amerindian groups from European groups. Uigurs and Kazaks were much better separated from both Europeans and Asians by K = 6.
Fig. 2.
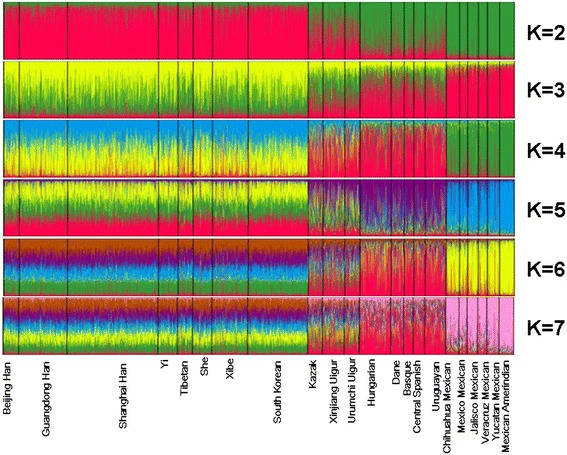
The cluster analysis of 22 groups using the STRUCTURE program based on the genotyping data of 30 InDel loci
Principal component analysis
A PCA was constructed to analyze the relationships between the Xinjiang Uigur group and other 21 groups. The result was shown in Fig. 3. The first and second component accounted for 58.95 and 23.23 %, respectively; and the cumulative contribution of the first two principal components defined 82.18 % of the total variance. In the plot figure, 5 European groups and 6 Amerindian groups located in the left part, while the 8 Asian groups located in the right part and the 3 Eurasian groups (Kazak, Urumchi Uigur and Xinjiang Uigur groups) in the central part. The Xinjiang Uigur group had short distance with the Urumchi Uigur and Kazak groups in PCA plot, which indicated the Xinjiang Uigur group had closer genetic relationships with those two groups.
Fig. 3.
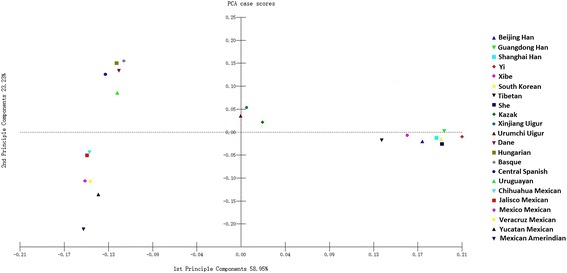
PCA based on population data of 30 InDel loci of Xinjiang Uigur group and 21 reference groups
Interpopulation differentiations
We estimated pairwise Fst and p-values utilizing analysis of molecular variance method between Uigur group and previously published groups at the 30 InDel loci, which were given in Additional file 3: Table S3. The results showed that the least differences were found between the Xinjiang Uigur group and the Urumchi Uigur and Kazak groups, with significant differences at one and three loci, respectively; whereas differences were observed between Xinjiang Uigur group and other groups at 5–20 loci. The results indicated that the distribution of allele frequencies in different groups were different. Therefore, InDel would be a useful tool to study the migration patterns, geneflow, admixture and ancestry with the discovery of more available loci (Hefke et al. 2015).
DA distance
The DA distance was calculated to elucidate the genetic distance. The DA distances between Xinjiang Uigur group and other reference groups were shown in Table 2. According to the DA distances, the Xinjiang Uigur group was closest to the Urumchi Uigur group (DA = 0.0012), and followed by the Kazak (DA = 0.0019) group, both of them belongs to Altaic language family. The greatest distance was detected when comparing the Xinjiang Uigur group with Yucatan Mexican (DA = 0.0353) and Mexican Amerindian (DA = 0.0473) groups.
Table 2.
The D A distances among the 22 groups based on 30 InDel loci
| Populations | Beijing Han | Guangdong Han | Shanghai Han | Yi | Xibe | South Korean | Tibetan | She | Kazak | Urumchi Uigur | Dane | Hungarian | Basque | Central Spanish | Uruguayan | Chihuahua Mexican | Jalisco Mexican | Mexico Mexican | Veracruz Mexican | Yucatan Mexican | Mexican Amerindian |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Guangdong Han | 0.0019 | ||||||||||||||||||||
| Shanghai Han | 0.0011 | 0.0006 | |||||||||||||||||||
| Yi | 0.0054 | 0.0038 | 0.0040 | ||||||||||||||||||
| Xibe | 0.0022 | 0.0023 | 0.0015 | 0.0052 | |||||||||||||||||
| South Korean | 0.0024 | 0.0017 | 0.0008 | 0.0042 | 0.0016 | ||||||||||||||||
| Tibetan | 0.0029 | 0.0055 | 0.0038 | 0.0066 | 0.0037 | 0.0038 | |||||||||||||||
| She | 0.0023 | 0.0015 | 0.0019 | 0.0051 | 0.0032 | 0.0028 | 0.0065 | ||||||||||||||
| Kazak | 0.0083 | 0.0100 | 0.0096 | 0.0133 | 0.0068 | 0.0115 | 0.0074 | 0.0112 | |||||||||||||
| Urumchi Uigur | 0.0100 | 0.0118 | 0.0114 | 0.0163 | 0.0092 | 0.0135 | 0.0093 | 0.0133 | 0.0013 | ||||||||||||
| Dane | 0.0251 | 0.0265 | 0.0264 | 0.0315 | 0.0227 | 0.0288 | 0.0226 | 0.0275 | 0.0093 | 0.0083 | |||||||||||
| Hungarian | 0.0255 | 0.0275 | 0.0271 | 0.0325 | 0.0231 | 0.0295 | 0.0222 | 0.0289 | 0.0084 | 0.0068 | 0.0026 | ||||||||||
| Basque | 0.0270 | 0.0268 | 0.0270 | 0.0328 | 0.0236 | 0.0287 | 0.0258 | 0.0288 | 0.0111 | 0.0096 | 0.0048 | 0.0045 | |||||||||
| Central Spanish | 0.0262 | 0.0269 | 0.0268 | 0.0323 | 0.0226 | 0.0288 | 0.0231 | 0.0285 | 0.0085 | 0.0069 | 0.0030 | 0.0022 | 0.0033 | ||||||||
| Uruguayan | 0.0230 | 0.0244 | 0.0240 | 0.0286 | 0.0203 | 0.0258 | 0.0199 | 0.0255 | 0.0067 | 0.0057 | 0.0039 | 0.0021 | 0.0043 | 0.0023 | |||||||
| Chihuahua Mexican | 0.0445 | 0.0471 | 0.0465 | 0.0522 | 0.0441 | 0.0500 | 0.0422 | 0.0525 | 0.0278 | 0.0241 | 0.0183 | 0.0156 | 0.0212 | 0.0200 | 0.0207 | ||||||
| Jalisco Mexican | 0.0437 | 0.0450 | 0.0448 | 0.0512 | 0.0420 | 0.0484 | 0.0417 | 0.0502 | 0.0260 | 0.0225 | 0.0170 | 0.0137 | 0.0197 | 0.0181 | 0.0189 | 0.0017 | |||||
| Mexico Mexican | 0.0534 | 0.0548 | 0.0544 | 0.0602 | 0.0517 | 0.0576 | 0.0502 | 0.0606 | 0.0345 | 0.0313 | 0.0222 | 0.0210 | 0.0256 | 0.0258 | 0.0274 | 0.0048 | 0.0041 | ||||
| Veracruz Mexican | 0.0498 | 0.0500 | 0.0502 | 0.0566 | 0.0473 | 0.0535 | 0.0468 | 0.0555 | 0.0301 | 0.0263 | 0.0192 | 0.0160 | 0.0209 | 0.0197 | 0.0223 | 0.0040 | 0.0024 | 0.0030 | |||
| Yucatan Mexican | 0.0588 | 0.0599 | 0.0598 | 0.0675 | 0.0569 | 0.0633 | 0.0567 | 0.0655 | 0.0391 | 0.0351 | 0.0267 | 0.0230 | 0.0277 | 0.0279 | 0.0300 | 0.0048 | 0.0046 | 0.0041 | 0.0031 | ||
| Mexican Amerindian | 0.0718 | 0.0724 | 0.0732 | 0.0798 | 0.0698 | 0.0770 | 0.0698 | 0.0796 | 0.0519 | 0.0477 | 0.0365 | 0.0317 | 0.0389 | 0.0393 | 0.0427 | 0.0093 | 0.0090 | 0.0087 | 0.0064 | 0.0053 | |
| Xinjiang Uigur | 0.0101 | 0.0115 | 0.0108 | 0.0156 | 0.0088 | 0.0125 | 0.0086 | 0.0127 | 0.0019 | 0.0012 | 0.0073 | 0.0055 | 0.0090 | 0.0062 | 0.0051 | 0.0246 | 0.0226 | 0.0310 | 0.0262 | 0.0353 | 0.0473 |
Xinjiang Uigur represented our samples from the whole territory of Xinjiang Uigur Autonomous Region. Urumchi Uigur represented previously published samples from Urumchi of Xinjiang
Phylogenetic analysis
A NJ-tree was constructed based on DA distances as presented in Fig. 4, the NJ tree showed that the Xinjiang Uigur group was first clustered with the Urumchi Uigur and Kazak groups. The result was consistent with the above mentioned results of STRUCTURE, DA distance and PCA. According to the relevant historical records, Uigurs were the descendants of ancient Uighur and with large proportion of the descent from Caucasian. Uigurs and Kazaks have common religious belief which indicated that they were likely having the same or similar origin in the process of the formation and development (Palstra et al. 2015; Xu et al. 2006). Therefore, the genetic distances could be relatively close among them. Yuan et al. (2015) studied the genetic polymorphism of 38 STR loci in Uigur group from Southern Xinjiang of China; their Fst distance results (21 loci) indicated the Uigur group was closest to Kazak, and our result was similar to theirs.
Fig. 4.
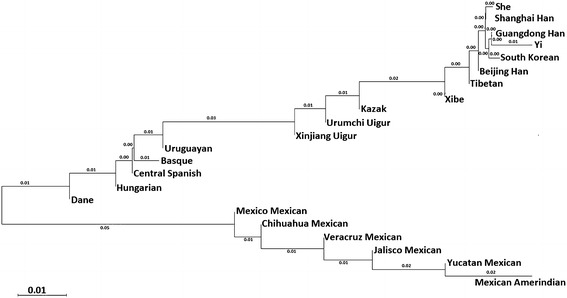
The neighbor-joining tree based on population data of 30 InDel loci of Xinjiang Uigur group and 21 referenced groups
Conclusions
In summary, the 30 InDel loci showed relatively high forensic-efficacy in the Xinjiang Uigur group and could be used in forensic individual identification, and also be used as complement for STR loci in forensic paternity testing. The result of DA distance, STRUCTURE, PCA and NJ tree indicated that the studied Xinjiang Uigur group had a close relationship with Urumchi Uigur and Kazak groups. This study provided valuable data for analysis of genetic relationship and forensic application.
Authors’ contributions
TM and CS wrote the main manuscript text, BZ and LZ designed the study and modified the manuscript. YL, YZ, YG and QD did the sample preparation, HM, XW and JY conducted the data processing. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This project was supported by the Natural Science Foundation of Xinjiang Uygur Autonomous Region (2013211A065). The authors would like to thank the scientists who produced relevant baseline work.
Competing interests
All authors approved and agree with the contents of the manuscript and the authors declare that they have no competing interests.
Additional files
10.1186/s40064-016-2730-3 The genotyping results of the 30 InDel loci from Uigur ethnic group living in Xinjiang Uigur Autonomous Region, China (n = 136).
10.1186/s40064-016-2730-3 The linkage disequilibrium pattern revealed by r 2 values at 30 InDel loci.c.
10.1186/s40064-016-2730-3 Pairwise Fst and p-values between Xinjiang Uigur group and other groups at 30 InDel loci (n = 136).
Footnotes
Ting Mei and Chun-Mei Shen contributed equally to this work
Bo-Feng Zhu and Li-Ping Zhang contributed equally to this work
Contributor Information
Ting Mei, Email: 417387482@qq.com.
Chun-Mei Shen, Email: cmshen2004@126.com.
Yao-Shun Liu, Email: 1132533029@qq.com.
Hao-Tian Meng, Email: menghaotian0803@126.com.
Yu-Dang Zhang, Email: zhangyudang1@163.com.
Yu-Xin Guo, Email: guoyuxin004@163.com.
Qian Dong, Email: dq517087@foxmail.com.
Xin-Xin Wang, Email: 282018327@qq.com.
Jiang-Wei Yan, Email: yanjw@big.ac.cn.
Bo-Feng Zhu, Phone: +86-29-82655117, Email: zhubofeng7372@126.com.
Li-Ping Zhang, Email: zhanglp7684@163.com.
References
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fondevila M, Phillips C, Santos C, et al. Forensic performance of two insertion-deletion marker assays. Int J Legal Med. 2012;5:725–737. doi: 10.1007/s00414-012-0721-7. [DOI] [PubMed] [Google Scholar]
- Friis SL, Børsting C, Rockenbauer E, et al. Typing of 30 insertion/deletions in Danes using the first commercial indel kit–Mentype® DIPplex. Forensic Sci Int Genet. 2012;6:e72–e74. doi: 10.1016/j.fsigen.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Gill P. An assessment of the utility of single nucleotide polymorphisms (SNPs) for forensic purposes. Int J Legal Med. 2001;4–5:204–210. doi: 10.1007/s004149900117. [DOI] [PubMed] [Google Scholar]
- Hefke G, Davison S, D’Amato ME. Forensic performance of Investigator DIPplex indels genotyping kit in native, immigrant, and admixed populations in South Africa. Electrophoresis. 2015;36:3018–3025. doi: 10.1002/elps.201500243. [DOI] [PubMed] [Google Scholar]
- Hong L, Wang XG, Liu SJ, et al. Genetic polymorphisms of 30 Indel loci in Guangdong Han population. J SUN Yat-sen Univ (Med Sci) 2013;34:299–304. [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;15(23):1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kidd KK, Pakstis AJ, Speed WC, et al. Developing a SNP panel for forensic identification of individuals. Forensic Sci Int. 2005;1:20–32. doi: 10.1016/j.forsciint.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Kim EH, Lee HY, Yang IS, et al. Population data for 30 insertion-deletion markers in a Korean population. Int J Legal Med. 2014;128:51–52. doi: 10.1007/s00414-013-0851-6. [DOI] [PubMed] [Google Scholar]
- Kis Z, Zalán A, Völgyi A, et al. Genome deletion and insertion polymorphisms (DIPs) in the Hungarian population. Forensic Sci Int Genet. 2012;6:e125–e126. doi: 10.1016/j.fsigen.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Martín P, García O, Heinrichs B, et al. Population genetic data of 30 autosomal indels in Central Spain and the Basque Country populations. Forensic Sci Int Genet. 2013;7:e27–e30. doi: 10.1016/j.fsigen.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Martínez-Cortés G, García-Aceves M, Favela-Mendoza AF, et al. Forensic parameters of the Investigator DIPplex kit (Qiagen) in six Mexican populations. Int J Legal Med. 2015;130:683–685. doi: 10.1007/s00414-015-1242-y. [DOI] [PubMed] [Google Scholar]
- Meng HT, Zhang YD, Shen CM, et al. Genetic polymorphism analyses of 30 InDels in Chinese Xibe ethnic group and its population genetic differentiations with other groups. Sci Rep. 2015;5(5):8260. doi: 10.1038/srep08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunotani M, Shiozaki T, Sato N, et al. Analysis of 30 insertion-deletion polymorphisms in the Japanese population using the Investigator DIPplex® kit. Leg Med (Tokyo) 2015;17:467–470. doi: 10.1016/j.legalmed.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Ota T. DISPAN: genetic distance and phylogenetic analysis. University Park: Pennsylvania State University; 1993. [Google Scholar]
- Palstra FP, Heyer E, Austerlitz F. Statistical inference on genetic data reveals the complex demographic history of human populations in central Asia. Mol Biol Evol. 2015;32:1411–1424. doi: 10.1093/molbev/msv030. [DOI] [PubMed] [Google Scholar]
- Pepinski W, Abreu-Glowacka M, Koralewska-Kordel M, et al. Population genetics of 30 INDELs in populations of Poland and Taiwan. Mol Biol Rep. 2013;40:4333–4338. doi: 10.1007/s11033-013-2521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R, Phillips C, Alves C, et al. A new multiplex for human identification using insertion/deletion polymorphisms. Electrophoresis. 2009;21:3682–3690. doi: 10.1002/elps.200900274. [DOI] [PubMed] [Google Scholar]
- Phillips C, Fang R, Ballard D, et al. Evaluation of the Genplex SNP typing system and a 49plex forensic marker panel. Forensic Sci Int Genet. 2007;2:180–185. doi: 10.1016/j.fsigen.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Wen X, Falush D (2003) Documentation for structure software: Version 2. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.323.9675&rep=rep1&type=pdf
- Romanini C, Catelli ML, Borosky A, et al. Typing short amplicon binary polymorphisms: supplementary SNP and Indel genetic information in the analysis of highly degraded skeletal remains. Forensic Sci Int Genet. 2012;6:469–476. doi: 10.1016/j.fsigen.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Saiz M, André F, Pisano N, et al. Allelic frequencies and statistical data from 30 INDEL loci in Uruguayan population. Forensic Sci Int Genet. 2014;9:e27–e29. doi: 10.1016/j.fsigen.2013.07.013. [DOI] [PubMed] [Google Scholar]
- Schneider PM. Scientific standards for studies in forensic genetics. Forensic Sci Int. 2007;165:238–243. doi: 10.1016/j.forsciint.2006.06.067. [DOI] [PubMed] [Google Scholar]
- Seong KM, Park JH, Hyun YS, et al. Population genetics of insertion-deletion polymorphisms in South Koreans using Investigator DIPplex kit. Forensic Sci Int Genet. 2014;8:80–83. doi: 10.1016/j.fsigen.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Shan RZ, Deng MB (2012) Analysis the formation of xinjiang uygur gens source and religious beliefs. Academic forum 237. (Article in Chinese)
- Shi M, Liu Y, Bai R, et al. Population data of 30 insertion-deletion markers in four Chinese populations. Int J Legal Med. 2015;129:53–56. doi: 10.1007/s00414-014-1091-0. [DOI] [PubMed] [Google Scholar]
- Tan GK, Tee SF, Tang PY. Genetic association of single nucleotide polymorphisms in dystrobrevin binding protein 1 gene with schizophrenia in a Malaysian population. Genet Mol Biol. 2015;38:138–146. doi: 10.1590/S1415-4757382220140142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Wang Z, Zhang S, Zhao S, et al. Population genetics of 30 insertion-deletion polymorphisms in two Chinese populations using Qiagen Investigator® DIPplex kit. Forensic Sci Int Genet. 2014;11:e12–e14. doi: 10.1016/j.fsigen.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Wei YL, Qin CJ, Dong H, et al. A validation study of a multiplex INDEL assay for forensic use in four Chinese populations. Forensic Sci Int Genet. 2014;9:e22–e25. doi: 10.1016/j.fsigen.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Xu MY, Hong KX, Ma J, et al. Analysis of HLA-B locus gene polymorphism in Sichuan Yi ethnic group and Xinjiang Uygur ethnic group. Yi Chuan. 2006;28:913–917. [PubMed] [Google Scholar]
- Yang N, Li H, Criswell LA, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;3–4:382–392. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- Ye Y, Luo H, Liao L, et al. A case study of SNPSTR efficiency in paternity testing with locus incompatibility. Forensic Sci Int Genet. 2014;9:72–75. doi: 10.1016/j.fsigen.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Yoo J, Lee Y, Kim Y, et al. SNPAnalyzer 2.0: a web-based integrated workbench for linkage disequilibrium analysis and association analysis. BMC Bioinformatics. 2008;23(9):290. doi: 10.1186/1471-2105-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Liu H, Liao Q, et al. Genetics analysis of 38 STR loci in Uygur population from Southern Xinjiang of China. Int J Legal Med. 2015;130:687–688. doi: 10.1007/s00414-015-1270-7. [DOI] [PubMed] [Google Scholar]
- Zaumsegel D, Rothschild MA, Schneider PM. A 21 marker insertion deletion polymorphism panel to study biogeographic ancestry. Forensic Sci Int Genet. 2013;7:305–312. doi: 10.1016/j.fsigen.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Zhang YD, Shen CM, Jin R, et al. Forensic evaluation and population genetic study of 30 insertion/deletion polymorphisms in a Chinese Yi group. Electrophoresis. 2015;36:1196–1201. doi: 10.1002/elps.201500003. [DOI] [PubMed] [Google Scholar]


