Abstract
Aim:
This study aims to determine the serogroup distribution and molecular diagnosis, as well as antimicrobial resistance profiles among Shigella spp. isolated from patients with diarrhea in Kerman, southeast of Iran.
Background:
Shigella species are frequent cause of bacterial dysentery worldwide. Previous studies have been reported that S. sonnei and S. flexneri are the most prevalent serogroups in various parts of Iran.
Patients and methods:
A total of 624 stool samples were randomly collected from patients with diarrhea from June 2013 to August 2014. Biochemical and serological characterizations were performed for identifying Shigella spp. In addition, the multiplex PCR assay was carried out for the detection and differentiation of three pathogenic Shigella spp. Antibiotic susceptibility testing was performed according to the Clinical Laboratory Standards Institute (CLSI) guidelines.
Results:
Fifty six (9%) Shigella strains were isolated from stool samples. The most common species were S. flexneri 31(55.4%), followed by S .sonnei 18(32.1%) and S. boydii 7(12.5%). S. dysentery was not detected in the present study. All the isolates that identified by serological test as Shigella spp. were confirmed by the multiplex PCR method. The highest rate of resistance was observed for ampicillin and trimethoprim-sulphamethoxazole antibiotics with 52(92.9%) resistant, followed by tetracycline 44(78.6%) and cefotaxime 33(58.9%). All Shigella isolates were susceptible to ciprofloxacin. A significant relationship was found between the Shigella species and cefotaxime resistance (p<0.05).
Conclusion:
S. flexneri was found as the most prevalent serogroup causing shigellosis. The high rate of resistance to third-generation cephalosporins limits the treatment options available for the management of shigellosis in Kerman, Iran.
Key Words: Multiplex PCR, Shigella flexneri, Shigella sonnei, third-generation cephalosporins
Introduction
Shigellosis is an acute gastroenteritis infection and one of the most common causes of morbidity and mortality, especially in children with diarrhea in developing countries (1). Four species of Shigella, namely, Shigella dysenteriae, S. flexneri, S. boydii, and S. sonnei are responsible for shigelosis in humans (2). The most prevalent Shigella species in developed and developing countries are S. sonnei and S. flexneri, respectively (2). S. flexneri had been recognized as the mainspring of shigellosis in Tehran until 2003 (3, 4), then it was replaced by S. sonnei (2, 5). Nowadays, there are several methods for detection of Shigella species, including conventional culture and molecular techniques (6). Conventional methods have some limitations due to problems in processing and relying on the viable organisms (7). Moreover, these methods require several days to give reliable results (8). In various studies, molecular techniques have been used for detecting Shigella spp. (9-12). Multiplex PCR is one of the most popular techniques has been used due to rapidity and its capability to detect and differentiate Shigella spp. in a single reaction (7, 13).
Antibiotic treatment for shigellosis is usually recommended to reduce the severity of symptoms and potentially lethal complications and shorten the duration of the fecal excretion of the Shigella (14). However, due to the emergence of multi-drug resistant strains, empirical treatment with antibiotics such as sulfonamides, tetracycline, ampicillin and trimethoprim-sulphamethoxazole were no longer recommended (15). Previous studies have been indicated that, antimicrobial resistance patterns among Shigella spp. are varying between the countries and even within a country. Therefore, it is necessary to determine the local anti-microbial resistance pattern for selecting a proper antibiotic to treat shigellosis (1, 16). The present study aims to determine the serogroup distribution and molecular diagnosis, as well as antimicrobial resistance pattern among Shigella spp. isolated from patients with diarrhea in Kerman, southeast of Iran.
Patients and Methods
Sample collection and Shigella identification
A total of 624 stool samples were randomly collected since June 2013 to August 2014 from patients with diarrhea admitted to the hospitals of Afzalipour and Ayatollah Kashani in Kerman, Iran. The stools were immediately cultured on xylose lysine deoxycholate agar (XLD) and MacConkey agar media. Then, lactose-negative colonies were selected and identified as Shigella by standard biochemical methods (17) and were grouped serologically by slide agglutination with specific antisera (MAST Group LTD, Merseyside, UK). The isolates were kept in the TSB broth containing 30% glycerol at -70°C until testing.
Identification of the Shigella species through PCR
For PCR amplification, DNA template was obtained as previously described by Ranjbar et al. (18). Shigella species were identified using specific primers of each species. The sequences of the primers are shown in Table 1. PCR reactions were performed in a final volume of 20 µl using the 3µl DNA solution, 10 µl Master mix (Amplicon, Brighton, UK), 10 pmol primer and water. Amplification was carried out in the thermo cycler (Biometra-T gradient, Biometra GmbH, Gottingen, Germany). The PCR conditions were as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles, including denaturation at 95°C for 45 seconds, annealing at 56 for 35 seconds and extension at 72°C for 45 seconds and a single final extension at 72°C for 5 minutes.
Table 1.
Sequences of primers used for the multiplex PCR
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing for 11 antibiotics, including: chloramphenicol (30μg), ampicillin (10μg), trimethoprim-sulphamethoxazole (1.25/23.75μg), azteronam (30μg), gentamicin (10μg), cefotaxime (30μg), ceftriaxone (30μg), tetracycline (30μg), nalidixic acid (30μg), ofloxacin (5μg) and ciprofloxacin (5μg) was determined by disk diffusion method according to the Clinical Laboratory Standards Institute guidelines (CLSI) (19). E. coli ATCC 25922 was used as the anti-biogram control.
Statistical analysis
The SPSS (Statistical Product and Service Solutions, version 20.0) (SPSS Inc., Chicago, IL, USA) was used for the analysis of data. We used the chi-square test to compare proportions and Fisher’s exact test when appropriate. A value of p<0.05 was considered statistically significant.
Results
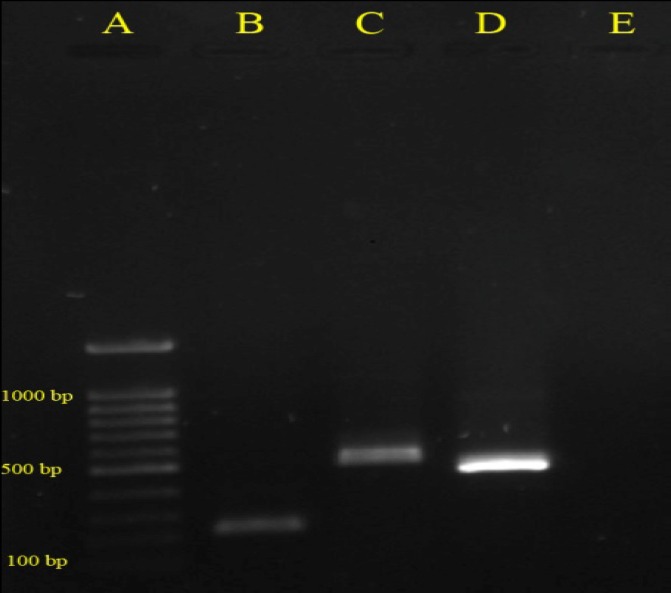
Fifty six (9%) Shigella strains were isolated from 624 stool samples. S. flexneri was the most frequently isolated serogroup 31(55.4%), followed by S. sonnei 18(32.1%), and S. boydii 7(12.5%). S. dysentery was not detected in the present study. All isolates that identified by serological test as Shigella spp. were confirmed by the multiplex PCR method. Multiplex PCR was successfully developed and optimized for rapid, accurate and simultaneous detection of Shigella species (Fig.1). As shown in Table 2, the highest resistance rate was observed for ampicillin and trimethoprim-sulphamethoxazole 52(92.9%), followed by tetracycline 44(78.6%) and nalidixic acid 12(21.4%). Notably, Shigella spp. were also resistant to the third generation cephalosporins, so that 27 (48.2%) and 33 (58.9%) of isolates were resistant to ceftriaxone and cefotaxime, respectively. Despite observed resistance towards the third generation cephalosporins in all serogroups, S. sonnei strains were highly resistant to these antibiotics, so that 16(88.9%) of its isolates were cefotaxime resistant and 13(72.2%) isolates were ceftriaxone resistant. A significant relationship was found between the shigella species and cefotaxime resistance (p<0.05). Although resistance to quinolones and chloramphenicol was mainly found in S. flexneri strains, overall analysis showed higher resistance to antibiotics in the S. sonnei more than other species (Table 2). All Shigella isolates were susceptible to the ciprofloxacin.
Figure 1.
Detection of specific Shigella species genes by multiplex PCR, A: Marker- 100 bp, B: S. boydii- 248 bp, C: S. flexneri- 537 bp, D: S. sonnei- 503 bp, E: Negative Control
Table 2.
Antibiotic resistance patterns of the Shigella spp
| Total No. (%) |
S. boydii
No. (%) |
S. sonnei
No. (%) |
S. flexneri
No. (%) |
Antibiotic |
|---|---|---|---|---|
| 52(92.9) | 7(100) | 18(100) | 27(87.1) | Trimethoprim-sulphamethoxazole |
| 52(92.9) | 6(85.7) | 17(94.4) | 29(93.5) | Ampicillin |
| 44(78.6) | 5(71.4) | 17(94.4) | 22(71) | Tetracycline |
| 23(41.1) | 2(28.6) | 1(5.6) | 20(64.5) | Chloramphenicol |
| 9(16.1) | 0(0) | 8(44.4) | 1(3.2) | Gentamicin |
| 27(48.2) | 3(42.9) | 13(72.2) | 11(35.5) | Ceftriaxone |
| 33(58.9) | 4(57.1) | 16(88.9) | 13(41.9) | Cefotaxime |
| 19(33.9) | 2(28.6) | 6(33.3) | 11(35.5) | Aztreonam |
| 12(21.4) | 1(14.3) | 3(16.7) | 8(25.8) | Nalidixic-acid |
| 3(5.4) | 0(0) | 0(0) | 3(9.7) | Ofloxacin (5μg) |
| 0(0) | 0(0) | 0(0) | 0(0) | Ciprofloxacin (5μg) |
Discussion
The present study investigated the prevalence of diarrhea caused by Shigella isolates and their antibiotic resistance in Kerman, southeast of Iran. In Tehran, Iran, S. flexneri was the predominant serogroup up to 2003 (3, 4). But recent studies indicated that it was replaced with S. sonnei (2, 5). In a study in Shiraz, Iran, S. sonnei was found to be the main cause of shigellosis (74.39%) (20). In the present study, however, S. flexneri was the most common serogroup causing shigellosis in Kerman. The results of this study are consistent with several other studies in different parts of Iran (21, 22). Differences in the dominant serogroup in Tehran and Shiraz compared to this study may be related to the higher levels of hygiene and development in those two cities (3).
Due to the speed, sensitivity and specificity, molecular methods have offered advantages over the conventional methods. However, some molecular techniques used to identify and differentiate the Shigella are relatively expensive, difficult to perform, and requiring special equipment. Some of these molecular techniques include: PCR-ELISA (11), seminested PCR (12), PCR-RFLP (9) and PCR-Nonradioactive labeling (10). Without these constraints, multiplex PCR method can serve as the valuable detective tool for diagnosis of Shigella species. Several previous studies have used multiplex PCR for rapid and simultaneous detection of Shigella species (7, 13). But it should be considered that molecular techniques complement rather than replace conventional methods.
The emergence of multidrug resistant (MDR) Shigella isolates has complicated the selection of empirical antibiotics for treatment of shigellosis (1). As many previous studies in Iran and other countries, high rate of resistance to ampicillin, trimethoprim-sulphamethoxazole and tetracycline was observed (3, 21-24). Due to the high resistance to these antibiotics, WHO suggested the ciprofloxacin as a drug of choice for the treatment of shigellosis in 1990 (14). However, the third generation cephalosporins are used as the alternative treatment of shigellosis because of the concerns about the side effects of ciprofloxacin in children and an increased resistance to it (25). In the present study, resistance to ciprofloxacin was not found. Therefore, it can be used as the treatment of choice in patients with shigellosis in this area. However, resistance to third-generation cephalosporins is really concerning because of their high importance in the shigellosis treatment especially in children. Despite previous reports on the resistance to third-generation cephalosporins in various parts of the world and Iran, This resistance rate is surprisingly high, and the findings of the current study do not support the previous studies (3, 5, 21-24). This result may be due to excessive using of beta-lactam antibiotics in treatment of infections, resulting in selecting the resistant strains and propagation of beta-lactamase genes in the normal intestinal flora. Reports indicate the easy transferring of the resistance genes from the commensal members of the Enterobacteriaceae family to the enteropathogens (23, 26). In consistent with our results, Farshad, et al. in Shiraz, Iran showed higher resistance rates to antibiotics in the S. sonnei strains than other serogroups (20). High resistance of the S. sonnei strains to the antibiotics of beta-lactam families and gentamicin will pose challenges in the treatment of infections caused by this serogroup.
In conclusion, S. flexneri was recognized as the dominant serogroup causing shigellosis in Kerman, Iran. The results showed that multiplex PCR can be used for identification of Shigella species with high speed and appropriate sensitivity and specificity. Empirical treatment for shigellosis entails the knowledge of the antimicrobial resistance pattern of local Shigella strains. Despite the high sensitivity to ciprofloxacin, the Shigella isolates showed resistance to the antibiotics like Trimethoprim-sulphamethoxazole and ampicillin in the present study. The high rate of resistance to third-generation cephalosporins limits the treatment options available for the management of shigellosis in Kerman, Iran. Continuous monitoring of antimicrobial resistance is necessary for appropriate antibiotic treatment of Shigellosis.
Acknowledgments
This study was a part of the PhD thesis of medical microbiology performed by first author. The research was supported by Kerman University of Medical Sciences. The authors acknowledge Kerman University of Medical Sciences for the financial support.
References
- 1.Yang H, Chen G, Zhu Y, Liu Y, Cheng J, Hu L, et al. Surveillance of antimicrobial susceptibility patterns among Shigella species isolated in China during the 7-year period of 2005-2011. Ann Lab Med. 2013;33:111–15. doi: 10.3343/alm.2013.33.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranjbar R, Dallal MMS, Talebi M, Pourshafie MR. Increased isolation and characterization of Shigellasonnei obtained from hospitalized children in Tehran, Iran. J Health Popul Nutr. 2008;26:426. doi: 10.3329/jhpn.v26i4.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MoezArdalan K, Zali MR, Dallal MMS, Hemami MR, Salmanzadeh-Ahrabi S. Prevalence and pattern of antimicrobial resistance of Shigella species among patients with acute diarrhoea in Karaj, Tehran, Iran. J Health Popul Nutr. 2003;21:96–102. [PubMed] [Google Scholar]
- 4.Nikkah J, Mehr-Movahead A. Antibiotic resistance among Shigella species isolated in Tehran, Iran. Ann Trop Med Parasitol. 1988;82:481–83. doi: 10.1080/00034983.1988.11812280. [DOI] [PubMed] [Google Scholar]
- 5.Tajbakhsh M, Migura LG, Rahbar M, Svendsen CA, Mohammadzadeh M, Zali MR, et al. Antimicrobial-resistant Shigella infections from Iran: an overlooked problem? J Antimicrob Chemother. 2012;67:1128–33. doi: 10.1093/jac/dks023. [DOI] [PubMed] [Google Scholar]
- 6.Thiem VD, Sethabutr O, von Seidlein L, Van Tung T, Chien BT, Lee H, et al. Detection of Shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in Nha Trang, Vietnam. J Clin Microbiol. 2004;42:2031–5. doi: 10.1128/JCM.42.5.2031-2035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranjbar R, Afshar D, Tavana AM, Najafi A, Pourali F, Safiri Z, et al. Development of multiplex PCR for simultaneous detection of three pathogenic Shigella species. Iran J Pub Health. 2014;43:1657–63. [PMC free article] [PubMed] [Google Scholar]
- 8.Villalobo E, Torres A. PCR for detection of Shigella spp. in mayonnaise. Appl Environ Microbiol. 1998;64:1242–45. doi: 10.1128/aem.64.4.1242-1245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bin Kingombe CI, Cerqueira-Campos M-L, Farber JM. Molecular strategies for the detection, identification, and differentiation between enteroinvasive Escherichia coli and Shigella spp. J Food Prot. 2005;68:239–45. doi: 10.4315/0362-028x-68.2.239. [DOI] [PubMed] [Google Scholar]
- 10.Jackson MP. Detection of Shiga toxin-producing Shigelladysenteriae type 1 and Escherichia coli by using polymerase chain reaction with incorporation of digoxigenin-11-dUTP. J Clin Microbiol. 1991;29:1910–14. doi: 10.1128/jcm.29.9.1910-1914.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sethabutr O, Venkatesan M, Yam S, Pang LW, Smoak BL, Sang WK, et al. Detection of PCR products of the ipaH gene from Shigella and enteroinvasive Escherichia coli by enzyme linked immunosorbent assay. Diagn Microbiol Infect Dis. 2000;37:11–6. doi: 10.1016/s0732-8893(00)00122-x. [DOI] [PubMed] [Google Scholar]
- 12.Theron J, Morar D, Du Preez M, Brözel V, Venter S. A sensitive seminested PCR method for the detection of Shigella in spiked environmental water samples. Water Res. 2001;35:869–74. doi: 10.1016/s0043-1354(00)00348-1. [DOI] [PubMed] [Google Scholar]
- 13.Ojha SC, Yean Yean C, Ismail A, Banga Singh K-K. A pentaplex PCR assay for the detection and differentiation of Shigella species. Biomed Res Int. 2013;2013:412370. doi: 10.1155/2013/412370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taneja N, Mewara A, Kumar A, Verma G, Sharma M. Cephalosporin-resistant Shigellaflexneri over 9 years (2001–09) in India. J Antimicrob Chemother. 2012;67:1347–53. doi: 10.1093/jac/dks061. [DOI] [PubMed] [Google Scholar]
- 15.Xia S, Xu B, Huang L, Zhao J-Y, Ran L, Zhang J, et al. Prevalence and characterization of human Shigella infections in Henan Province, China, in 2006. J Clin Microbiol. 2011;49:232–42. doi: 10.1128/JCM.01508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pazhani GP, Niyogi SK, Singh AK, Sen B, Taneja N, Kundu M, et al. Molecular characterization of multidrug-resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. J Med Microbiol. 2008;57:856–63. doi: 10.1099/jmm.0.2008/000521-0. [DOI] [PubMed] [Google Scholar]
- 17.Bopp CA, Brenner F, Fields P, Wells J, Strockbine N. Escherichia, Shigella, and Salmonella. Manual Clin Microbiol. 2003;8:654–71. [Google Scholar]
- 18.Ranjbar R, Memariani M. Multilocus variable-number tandem-repeat analysis for genotyping of Shigella sonnei strains isolated from pediatric patients. Gastroenterol Hepatol Bed Bench. 2015;8:225. [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing: 20th Informational Supplement M100-S20. CLSI, Wayne, PA, USA: 2010. [Google Scholar]
- 20.Farshad S, Sheikhi R, Japoni A, Basiri E, Alborzi A. Characterization of Shigella strains in Iran by plasmid profile analysis and PCR amplification of ipa genes. J Clin Microbiol. 2006;44:2879–83. doi: 10.1128/JCM.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jomezadeh N, Babamoradi S, Kalantar E, Javaherizadeh H. Isolation and antibiotic susceptibility of Shigella species from stool samples among hospitalized children in Abadan, Iran. Gastroenterol Hepatol Bed Bench. 2014;7:218. [PMC free article] [PubMed] [Google Scholar]
- 22.Savadkoohi RB, Ahmadpour-Kacho M. Prevalence of Shigella species and their antimicrobial resistance patterns at Amirkola children hospital, North of Iran. Iran J Pediatr. 2007;17:118–22. [Google Scholar]
- 23.Fulla N, Prado V, Duran C, Lagos R, Levine MM. Surveillance for antimicrobial resistance profiles among Shigella species isolated from a semirural community in the northern administrative area of Santiago, Chile. Am J Trop Med Hyg. 2005;72:851–54. [PubMed] [Google Scholar]
- 24.Peirano G, Souza FdS, Rodrigues DdP. Frequency of serovars and antimicrobial resistance in Shigella spp. from Brazil. Mem Inst Oswaldo Cruz. 2006;101:245–50. doi: 10.1590/s0074-02762006000300003. [DOI] [PubMed] [Google Scholar]
- 25.Lee W, Chung H-S, Lee H, Yum JH, Yong D, Jeong SH, et al. CTX-M-55-type extended-spectrum β-lactamase-producing Shigellasonnei isolated from a Korean patient who had travelled to China. Ann Lab Med. 2013;33:141–44. doi: 10.3343/alm.2013.33.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blake D, Hillman K, Fenlon D, Low J. Transfer of antibiotic resistance between commensal and pathogenic members of the Enterobacteriaceae under ileal conditions. J Appl Microbiol. 2003;95:428–36. doi: 10.1046/j.1365-2672.2003.01988.x. [DOI] [PubMed] [Google Scholar]