Summary
Autophagy, a cellular waste disposal process, has well-established tumor suppressive properties. New studies indicate that in addition to its cell autonomous anti-tumorigenic functions, autophagy inhibits cancer development by orchestrating inflammation and immunity. While attenuating tumor-promoting inflammation, autophagy enhances the processing and presentation of tumor antigens and thereby stimulates anti-tumor immunity. Although cancer cells can escape immunosurveillance by tuning down autophagy, certain chemotherapeutic agents with immunogenic properties may enhance anti-tumor immunity by inducing autophagic cell death. Understanding the intricate and complex relationships within this troika and how they are affected by autophagy enhancing drugs should improve the efficacy of cancer immunotherapy.
Introduction
Homeostasis and rapid adaptation to environmental changes are key to organismal health and survival. Autophagy, a “self-eating” process that clears intracellular waste attenuates cell stress and keeps pro-carcinogenic processes at bay. These tumor suppressive functions of autophagy are well-recognized and recently reviewed (Mah and Ryan, 2012). While many previous studies had focused on the cell autonomous nature of autophagy, it has become clear that autophagy-dependent tumor suppression is also executed through downregulation of chronic tumor-promoting inflammation and/or enhancement of anti-tumor immunity. By surveying the relationships between autophagy, inflammation and immunity (the “AII Troika”) this review aims to explain how they shape the immune landscape that modulates malignancy. We will discuss how these basic concepts can be translated to improve cancer immunotherapy.
Although “autophagy” was described in the late 1950’s, its mechanism was elucidated much later using yeast genetics (Harding et al., 1995; Tsukada and Ohsumi, 1993). Autophagosome formation requires three main steps: initiation, nucleation and expansion (Figure 1), which have been extensively reviewed elsewhere (Choi et al., 2013). Autophagy is classified as microautophagy, chaperone-mediated autophagy and macroautophagy (Cuervo and Wong, 2014; Mizushima and Komatsu, 2011). Microautophagy involves invagination of either lysosomal or endosomal membranes, resulting in direct engulfment of cytoplasmic cargo. Chaperone-mediated autophagy entails selective degradation of proteins containing a KFERQ-like motif (Cuervo and Wong, 2014). Macroautophagy consists of classic double membrane autophagosomes that recognize and sequester cellular cargo that has been tagged by autophagy adaptors [e.g. sequestosome 1 (p62/SQSTM1), neighbor of Brca 1 (NBR1) and optineurin] (Stolz et al., 2014). Cargo recognition often depends on ubiquitylation, but under certain circumstances non-ubiquitinated cargo is also cleared by autophagy (Zhang and Ney, 2009). Successfully encapsulated cargo is eventually degraded by lysosomal hydrolases.
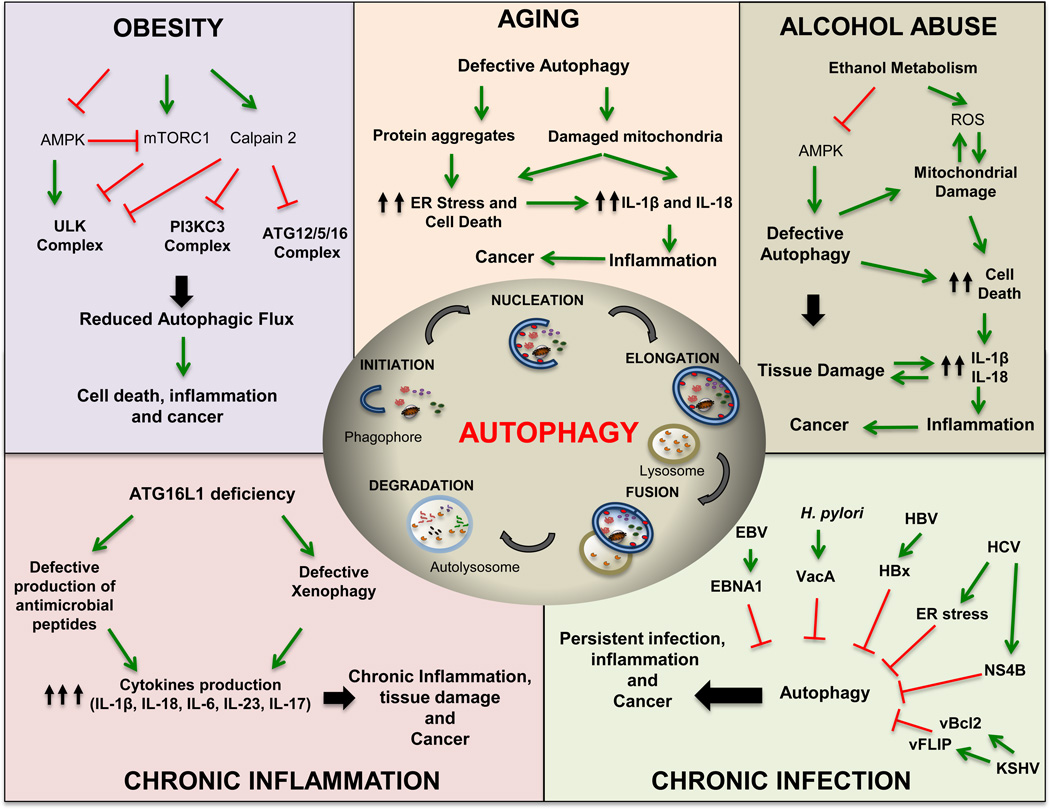
Figure 1. Speculative Models of How Cancer Risk Factors Compromise Autophagy.
Cancer risk factors, including obesity, aging, alcohol abuse, chronic inflammation and infection interfere with either the initiation or termination of autophagy to promote cancer development. ULK, unc51-like kinase; PI3KC, class III phosphatidylinositol (PI) 3’ kinase; ATG, autophagy related gene; AMPK AMP-activated protein kinase; mTORC1, mammalian target of rapamycin complex 1; ER, endoplasmic reticulum; IL-1β, interleukin 1β; ROS, reactive oxygen species; HCV, hepatitis C virus; HBV, hepatitis B virus; EBV, Epstein-Barr virus; KSHV, Kaposi’s sarcoma-Assocaited Herpesvirus; cFLIP, viral FLICE inhibitory protein; vBcl-2, viral B-cell lymphoma 2; EBNA1, EBV nuclear antigen 1; VacA, vacuolating cytotoxin A; HBx, HBV×protein; H. pylori, Helicobacter pylori. Mechanistic details are provided in the main text.
Relative to other autophagy types, macroautophagy (hereafter referred as autophagy) is most important for maintaining homeostasis, because it regulates the turnover and functionality of key cellular organelles (Levine, 2005; Mizushima and Komatsu, 2011). For instance, tissue damage and stress cause the release of danger-associated molecular patterns (DAMP) that are sensed by resident tissue macrophages, which mount an acute inflammatory response, whose goal is to get rid of cell corpses and initiate tissue repair and regeneration (Karin and Clevers, 2016). However, macrophage overactivation can result in hyperplasia and tumor promotion. Therefore, the inflammatory response needs to be properly terminated, a task mediated by autophagy (Gross et al., 2011). DAMP-induced macrophage activation often involves mitochondrial stress which results in release of mitochondrial signals that affect secretion of inflammasome-dependent cytokines to fight infections and promote tissue repair (Lamkanfi and Dixit, 2012). However, uncontrolled macrophage activation results in self-inflicted death, which subsequently triggers extensive neutrophil recruitment, thereby causing severe immunopathologies. This dangerous process is counteracted by mitochondrial autophagy (mitophagy) in macrophages (Zhong et al., 2016). In addition, mitophagy in epithelial and mesenchymal cells is important for preventing mitochondrial reactive oxygen species (mtROS)-induced tissue damage and tumor initiation (Antonucci et al., 2015; Chourasia et al., 2015). Moreover, autophagy is required for optimal induction of protective adaptive immunity (Ma et al., 2013). Hence, in addition to its well-recognized cell-autonomous anti-tumorigenic properties, autophagy, as outlined below, also controls important non-cell-autonomous functions that counteract tumorigenesis, mainly through activation of adaptative immunity and inhibition of chronic inflammation.
Autophagy and Cancer: A Two Way Street
Numerous cancer risk factors (e.g. aging, obesity and chronic inflammation) interfere with the proper functioning of the autophagic machinery (Figure 1). In epithelial cells, defective autophagy can promote tumor initiation by enhancing oxidative stress and genomic instability, as well as by activating transcription factor NRF2 (nuclear factor erythroid 2-related factor 2), which paradoxically induces expression of genes encoding anti-oxidant and drug metabolizing enzymes (Umemura et al., 2016). Defective autophagy also interferes with oncogene-induced senescence, an important tumor suppressive mechanism, and thereby leads to uncontrolled proliferation of cancer progenitor cells (Dou et al., 2015). Conversely, once the malignant phenotype has been established, autophagy serves as a survival mechanism that provides rapidly proliferating cancer cells with nutrients (Perera et al., 2015). Nonetheless, cancers in which autophagy is upregulated, as indicated by accumulation of microtubule-associated protein 1A/1B light chain 3 (LC3) puncta, exhibit higher density of CD8+ T cells and lower number of Foxp3+ T regulatory cells (Treg) in the tumor bed (Ladoire et al., 2016). Thus, enhanced autophagy correlates with activation of anti-tumor immunity and its downregulation may allow malignant growths to avoid immune surveillance. Indeed, oncogene activation can inhibit autophagy, in part through a mechanism similar to one used for inhibition of apoptosis (Degenhardt et al., 2006). Further complicating the intricate power balance within the “AII Troika”, rapid tumor growth results in hypoxia and necrosis at the tumor core, leading to DAMP release and recruitment and activation of macrophages and dendritic cells (Ma et al., 2013). Stimulation of autophagy may suppress this inflammatory response that drives tumor growth by promoting the survival of hypoxic and nutrient-starved cancer cells and by clearing damaged mitochondria (Figures 2 and 3).
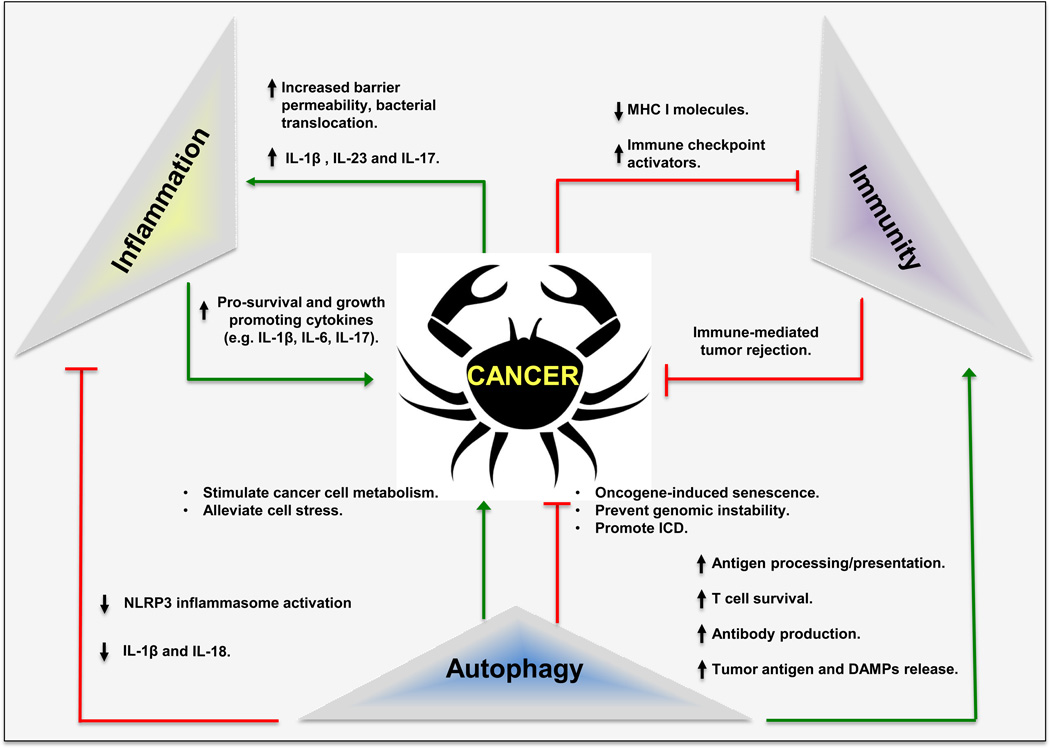
Figure 2. The “Autophagy-Inflammation-Immunity” (AII) troika in Cancer.
Overview of the cancer governing “AII” troika and how its function is modified by certain cancer-associated processes. MHC I, major histocompatibility complex I; ICD, immunogenic cell death; DAMP, damage-associated molecular patterns; NLRP3, nod-like receptor pyrin domain containing protein 3.
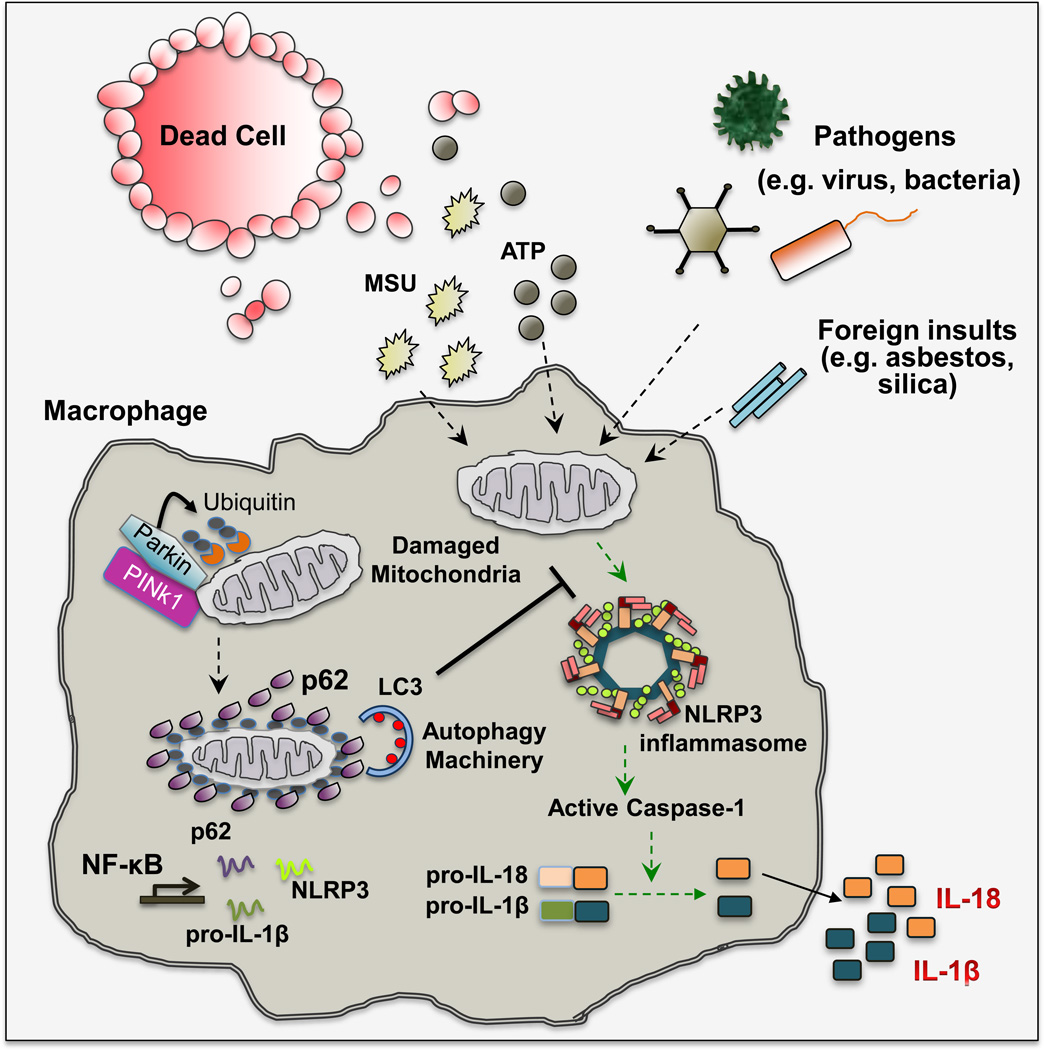
Figure 3. Mitophagy/Autophagy Dials Down Inflammation.
Stimulation of macrophages with DAMP (e.g. ATP and uric acid), carcinogenic particles (asbestos and silica microfibers/crystals) or PAMP (e.g. bacterial toxins) results in mitochondrial damage that is characterized by loss of mitochondrial membrane potential and subsequent release of mtDNA and mtROS. These mitochondrial signals in turn activate the NLRP3 inflammasome to induce IL-1β and IL-18 secretion. Loss of mitochondrial membrane potential (ψm) activates PINK1, a mitochondrial protein kinase that phosphorylates ubiquitin chains attached to mitochondrial outer membrane proteins. Phosphorylated ubiquitin interacts with and activates Parkin, an E3 ubiquitin ligase that further ubiquitinates mitochondrial outer membrane proteins. Ubiquitin-tagged mitochondria are recognized by the UBA domain of p62, whose expression is induced upon NF-κB activation. p62 also binds to LC3 and targets ubiquitinated mitochondria to autophagosomal clearance. By eliminating signal-emitting mitochondria, macrophage limits the extent of NLRP3-inflammasome activation. MSU, monosodium urate crystal; PINK1, PTEN-induced putative kinase 1; LC3, microtubule-associated protein 1A/1B light chain 3; NF-κB, nuclear factor-kappa B; UBA, ubiquitin association domain.
In addition to cell autonomous modulation of cancer cell physiology, systemic and local autophagy defects, caused by various cancer risk factors, also affect cancer-associated inflammation and immune responses (Figures 1 and 2). As described in detail below, defective autophagy in myeloid cells enhances tumor promoting inflammation while compromising antigen presentation. Conversely, stimulation of autophagy suppresses tumor-promoting inflammation and enhances anti-cancer immunity (Figure 2). The interaction and crosstalk between members of the “AII Troika”, their effects on cancer development/progression and the response to immunotherapy are the focal points for this review.
Corruption of Autophagy by Cancer Risk Factors
Age-related, metabolic and environmental factors as well as infectious agents that increase cancer risk may do so by interfering with the initiation or completion of autophagy (Figure 1). Unlike laboratory-induced ablation of autophagy related genes (ATG) defects, cancer risk factors do not block autophagy completely, but over time even a partial decrease in autophagic flux will result in chronic pathologies and elevated cancer risk due to accumulation of cellular waste. Cancer-causing infectious agents, including Helicobacter pylori, Epstein-Barr virus (EBV), Hepatitis B virus (HBV) and Kaposi’s sarcoma-associated Herpesvirus (KSHV), have evolved multiple strategies to avoid xenophagic elimination (Silva and Jung, 2013; Zhang et al., 2014). These pathogens produce bacterial and viral proteins that bind lysosomes and alter their acidification and ability to degrade autophagosome-delivered cargo. For instance, H. pylori produces vacuolating cytotoxin A (VacA) that blocks autolysosome function to promote its own survival and protect the cytotoxin-associated gene A (CagA) protein from degradation (Tsugawa et al., 2012). Stabilized CagA spreads to other host cells, where it activates signaling pathways that stimulate motility and proliferation and induce gastric mucosal metaplasia (Hayashi et al., 2013). Although it is not entirely clear how H. pylori promotes gastric cancer development, CagA-induced changes in the behaviour of gastric epithelial cells and accumulation of p62/SQSTM1 and reactive oxygen species (ROS) in the gastric mucosa of patients infected with virulent VacA-producing strains are thought to be of importance (Tsugawa et al., 2012). In a similar vein, EBV nuclear antigen 1 (EBNA1) blocks autophagy by inhibiting lysosome acidification thereby evading xenophagy, a “selective” form of autophagy that eliminates cell-invading microbes. Defective autophagy also impairs EBNA1 presentation by major histocompatibility class II (MHC-II) molecules, an autophagy-dependent process (Paludan et al., 2005). Furthermore, dysregulation of autophagy results in cytosolic accumulation of the EBV oncoprotein latent membrane protein 1, LMP1, which activates signaling pathways that control cell proliferation and survival, and inhibits tumor suppressors (Fathallah et al., 2010; Zhao et al., 2013). In contrast, HBV, which greatly increases liver cancer risk, produces the small surface protein (HBs) that triggers the unfolded protein response (UPR), resulting in stimulation of autophagy (Silva and Jung, 2013). Moreover, another HBV viral protein, HBx (HBV X protein), binds to class III PI 3-kinase, VPS34, and causes persistent stimulation of autophagy initiation that promotes HBV core protein maturation (Silva and Jung, 2013). However, HBx also interacts with V-ATPase and impairs lysosome acidification and proteolysis (Liu et al., 2014). This results in accumulation of p62/SQSTM1 and viral proteins in infected hepatocytes, increasing their likelihood to die (Liu et al., 2014). Enhanced hepatocyte death and subsequent compensatory proliferation strongly enhance hepatocellular carcinogenesis (Maeda et al., 2005). By activating NRF2, p62 accumulation protects hepatocellular carcinoma (HCC) initiating cells from ROS induced death, allowing them to accumulate numerous oncogenic mutations (Umemura et al., 2016). Hepatitis C virus (HCV), on the other hand, induces endoplasmic reticulum (ER) stress to interfere with autophagy (Malhi and Kaufman, 2011). In addition, HCV viral protein NS4B interacts with VPS34 and Rab5 to inhibit autophagosome maturation and autolysosome formation, although the precise mechanism remains unknown (Silva and Jung, 2013). Interestingly, certain viruses encode viral proteins that are homologs of important regulators of autophagy. For instance, KSHV encoded-protein vFLIP (viral FLICE inhibitory protein) and vBcl-2 (viral B-cell lymphoma 2), that interacts with Atg3 and Beclin1 to block autophagosome formation or vesicle nucleation, respectively (Silva and Jung, 2013).
Infectious agents that do not produce oncoproteins stimulate cancer development by inducing chronic tumor promoting inflammation. Inflammatory bowel diseases (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), greatly increase the risk of colorectal cancer (CRC) due to elevated expression of inflammatory cytokines, such as IL-6, TNF and IL-1β (Grivennikov et al., 2010). Autophagy related 16-like 1 (ATG16L1) maps to a CD susceptibility locus that is associated with an elevated CRC risk (Hampe et al., 2007). ATG16L1 deficiency afffects the initiation of xenophagy (Maurer et al., 2015), and enhances expression of Laptm5, a lysosomal transmembrane protein that promotes A20 degradation, thereby potentiating nuclear factor-kappa B (NF-κB) signaling and dendritic cell (DC) activation (Hubbard-Lucey et al., 2014). Due to enhanced inflammasome activity, ATG16L1-deficient macrophages produce high amounts of IL-1β and IL-18 (Saitoh et al., 2008), two cytokines known to increase CRC risk. ATG16L1 deficiency also compromises Paneth cell maturation, resulting in decreased production of antimicrobial peptides (Cadwell et al., 2008), which together with defective xenophagy contribute to high microbial load in the lamina propia, which can stimulate CRC development and progression by increasing IL-23 and IL-17 production (Grivennikov et al., 2012).
Another life threatening inflammatory disease associated with insufficient autophagy is chronic pancreatitis (Clemens et al., 2014). Chronic pancreatitis greatly increases pancreatic cancer risk and so do other pancreatitis and pancreatic cancer risk factors, such as alcohol consumption and high caloric intake, all of which interfere with autophagy activity (Clemens et al., 2014). Genetic manipulations that attenuate with autophagy lead to chronic pancreatitis in mice (Antonucci et al., 2015; Li et al., 2013). Alcohol abuse also causes alcoholic hepatitis (ASH), which is accompanied by lipid doplet accumulation and a marked increase in HCC risk. Ethanol metabolism interferes with AMP-activated protein kinase (AMPK) activity, thereby compromising autophagy (You et al., 2004). Moreover, ethanol metabolism elicits mitochondrial damage, causing ROS production and hepatocyte death (Louvet and Mathurin, 2015). Dying hepatocytes release DAMP that trigger NOD-like receptor family pyrin domain-containing 3 (NLRP3) inflammasome activation in resident liver macrophages, resulting in neutrophil infiltration and ASH development (Szabo and Csak, 2012). Defective myeloid cell autophagy enhances NLRP3 inflammasome activation thereby augmenting IL-1β production, neutrophil infiltration and liver damage (Zhong et al., 2016).
Obesity causes nonalcoholic fatty liver disease (NAFLD) that can progress to nonalcoholic steatohepatitis (NASH), which increases HCC risk (Starley et al., 2010). Ongoing autophagy reduces accumulation of lipid droplets in hepatocytes, thus providing a safeguard against NAFLD and NASH (Schneider and Cuervo, 2014). Conversely, obesity suppresses autophagy by multiple mechanisms, including activation of calcium-dependent protease calpain-2 that leads to ATG7 degradation (Yang et al., 2010), activation of mammalian target of rapamycin complex 1 (mTORC1) and inhibition of unc-51 like autophagy activating kinase 1 (ULK1) activity (Tremblay et al., 2005) or defective autophagosome-lysosome fusion, due to changes in membrane lipid composition and ER stress (Yang et al., 2010). Obesity also compromises macrophage autophagy (Liu et al., 2015), which can result in enhanced IL-1β and IL-18 production (Zhong et al., 2016). Consistent with this notion, myeloid specific Atg5 ablation greatly increases the likelihood of NASH development upon consumption of high fat diet (Liu et al., 2015). Steatotic hepatocytes release linoleic acid that causes depletion of liver resident CD4+ T cells, thereby contributing to the dismanteling of antitumor immunity (Ma et al., 2016).
The ultimate cancer risk factor is old age, which has a detrimental effect on autophagy (Martinez-Lopez et al., 2015). Defective autophagy in aged individuals or animals results in aberrant clearance of damaged mitochondria, leading to elevated inflammation and accumulation of ROS and protein aggregates that cause ER stress (Bujak et al., 2015; Komatsu et al., 2006). These cellular defects contribute to different degenerative diseases and enhance tumor initiation and malignant progression (Martinez-Lopez et al., 2015). Aging is accompanied by indolent inflammation and parainflammation manifested by increased basal production of IL-1β, IL-18, TNF and IL-6. (Chung et al., 2009; Licastro et al., 2005). All of these cytokines enhance cancer development and progression (Grivennikov et al., 2010). Chronic inflammation also compromises anti-cancer immunity (Shalapour and Karin, 2015).
Autophagy turns off the inflammatory fire
In addition to xenophagy that reduces tumor promoting inflammation by eliminating pathogen-associated molecular patterns (PAMP)-producing intracellular microbes, other forms of autophagy also suppress tumor-promoting inflammation. In addition to increasing cancer risk, sustained and/or unresolved inflammation causes collateral tissue damage, doing more harm than good. Conversely, a properly mounted, focused and transient inflammation promotes tissue repair and regeneration (Karin and Clevers, 2016). As discussed below, autophagy ensures a well-balanced inflammatory response that is accompanied by restoration of homeostasis.
The ability of autophagy to prevent excesive inflammation was first observed in mice rendered deficient in Atg16l1, which produce significantly more IL-1β than wild-type (WT) controls and are more vulnerable to septic shock (Saitoh et al., 2008). Further studies demonstrated that anything that blocks autophagy, be it Atg gene ablation or pharmacological intervention, results in enhanced caspase-1 activation and increased IL-1β production and secretion (Nakahira et al., 2011; Zhou et al., 2011). Caspase-1 is activated after its incorporation into large protein assemblies called inflammasomes, which also contain a sensor protein that belongs to the NLR (Nod-like receptor) family and the adaptor protein ASC (apoptosis-associated speck-like). Production of biologically active IL-1β, one of the two most potent inflammatory cytokines (the other being TNF), depends on two steps. First, IL-1β gene transcription is induced upon NF-κB activation caused by PAMP or DAMP binding to Toll-like receptors (TLR) or engagement of TNF receptors. Translation of IL-1β mRNA results in production of precursor pro-IL-1β, which cannot be secreted by macrophages. The second step is inflammasome dependent and is initiated by NLR-specific stimuli (Martinon et al., 2009). For instance, NLRC4 (NLR family CARD domain-containing protein 4) senses cytosolic flagelin, whereas NLRP1b (NLR Family, Pyrin Domain Containing 1) detects anthrax lethal toxin. The sensor protein with the broadest sensitivity is NLRP3, which responds to a panoply of stimuli including ATP, toxins (e.g. nigericin), uric acid, cholesterol, alum, silica and asbestos microcrystals and lipid particles. Notably, none of these stimuli is directly recognized by NLRP3. Instead they either open potassium channel(s), disrupt membrane integrity or function through other indirect mechanisms that somehow culminate in mitochondria damage (Elliott and Sutterwala, 2015). Damaged mitochondria release or present substances such as mtROS, oxidized mitochondrial DNA (mtDNA) or cardiolipin, which are presumed to be direct NLRP3 inflammasome activators (Elliott and Sutterwala, 2015). Although the identity of the ultimate NLRP3 ligand and how it activates the inflammasome remains to be determined, it is quite well established that macrophages encountering NLRP3 activating stimuli display mitochondrial damage and that elimination of mitochondria or mtROS specifically prevents NLRP3 inflammasome activation and production of mature IL-1β and IL-18 (Zhong et al., 2016; Zhou et al., 2011) (Figure 3).
Given its dependence on mitochondrial damage, NLRP3 inflammasome activation is negatively regulated by autophagy which keeps IL-1β and IL-18 production and subsequent inflammation in check (Nakahira et al., 2011; Zhong et al., 2016). Mitochondria that have been damaged upon macrophage encounter with NLRP3 activating stimuli, undergo Parkin-dependent ubiquitylation, followed by recruitment of the ubiquitin-binding autophagy adaptor p62/SQSTM1 (Figure 3). By recognizing LC3 on phagophore membranes, p62 ensures the mitophagic elimination of damaged mitochondria and termination of NLRP3 inflammasome activation (Zhong et al., 2016). Of note, p62 gene transcription is induced by NF-κB, the same process that controls the first step in IL-1β production and accounts for upregulation of NLRP3 itself. Genetic ablation of p62 or inhibition of IκB kinase β (IKKβ), which attenuates p62 induction, results in excessive secretion of IL-1β and IL-18 by macrophages that have been presented with NLRP3 activating stimuli, despite the concomitant decrease in Il1b gene transcription (Greten et al., 2007; Zhong et al., 2016). Enhanced IL-1β secretion results in excessive inflammation and neutrophilia (Greten et al., 2007; Hsu et al., 2011; Zhong et al., 2016). Of note, the NLRP3 inflammasome likely plays an important and rather general role in the onset of tumor promoting inflammation, as it is activated by carcinogenic asbestos and silica microparticles (Dostert et al., 2008), as well as by lipids and cholesterol, which stimulate NAFLD progression to NASH and thereby increase HCC risk.
Another mechanism through which autophagy squelches the tumor-promoting inflammatory fire is xenophagy. For instance, following infection of phagocytes with H. pylori, Salmonella, Listeria or Shigella, PAMP recognition by TLR and NLR stimulates xenophagy, which by reducing pathogen load attenuates microbe-induced inflammation (Levine, 2005). Like damaged mitochondria, intracellular bacteria are recognized by autophagy receptors such as p62, NBR1 and NDP52, which promote their autophagic clearance (Manzanillo et al., 2013). One trigger of xenophagy could be energy imbalance, caused by competition between the invading microorganism and the host cell for nutrients, which results in AMPK activation and inhibition of mTORC1, stimulating the initiation of autophagy through modulation of ULK1/2 phosphorylation (Mizushima and Komatsu, 2011). Intriguingly, xenophagy and mitophagy are evolutionarily related as mitochondria are thought to have originated from endosymbiotic bacteria (Randow and Youle, 2014). Thus the control of inflammation, by either exogenous or endogenous insults may be an ancient function of autophagy in multicellular organisms. Given the importance of inflammation in tumor development (Grivennikov et al., 2010), there is little doubt that the anti-inflammatory function of autophagy makes a key contribution to its tumor suppressive ability.
Autophagy as an immune stimulator
Adaptive immunity relies on recognition of extracellular (exogenous) or intracellular (endogenous) peptide epitopes presented by MHC class II and I molecules that are recognized by CD4+ and CD8+ T cells, repectively (Shibutani et al., 2015). Autophagy in antigen-presenting cells (APC) can promote antigen presentation by both MHC class II and I molecules. For instance, upon uptake of extracellular antigens (e.g. microbial or tumor antigens) by APC, autophagy promotes trafficking of the engulfed antigens to endosomes, where they are digested by cathepsins and loaded onto MHC class II molecules that eventually translocate to the plasma membrane and present antigens to CD4+ T cells. Although the precise mechanism remains to be further investgated, autophagy also facilitates “cross-presentation” of exogenous constituents by facilitating their loading onto MHC class I molecules that ultimately activate antigen specific CD8+ T cells. Notably, autophagy-mediated cross-presentation is important for mounting T cell responses under various stressed conditions, when proteasome function is compromised and antigenic peptides cannot be imported to the ER for loading onto MHC class I molecules. In support of this notion, in tumor-bearing mice or cancer patients, tumor infiltrating APC found in the draining lymph nodes are often functionally compromised. To ensure the generation of an effective antitumor cytotoxic T cells (CTL) response, APC autophagy then becomes of utmost importance and needs to be stimulated to facilitate processing and cross-presentation of tumor antigenic peptides by MHC class I molecules (Ma et al., 2013; Shibutani et al., 2015). Furthermore, autophagy also induces upregulation of MHC class I molecules in response to IFN-γ (Li et al., 2010), further enhancing the “cross-presentation” of extracellular antigens (Shibutani et al., 2015). Last but not least, in addition to its roles in APC, autophagy in cancer cells can indirectly promote “cross-presentation” of tumor antigens by facilitating their release from dying cells, thereby increasing extracellular antigen availability (Ma et al., 2013).
Autophagy also stimulates thymic “negative selection” of autoreactive CD4+ T cells, thereby maintaining central T cell tolerance (Aichinger et al., 2013; Schmid et al., 2007). Autophagy further regulates lymphocyte development and functional diversification. Naïve T cell number is dramatically reduced in the absence of mitophagy, and mature T cells require autophagy for survival (Pua et al., 2007). Additionally, autophagy indirectly influences Th17 cell polarization by restraining IL-1β production by innate immune cells. Since IL-1β promotes Th17 lineage commitment together with IL-6 and TGFβ (Zhou and Littman, 2009), defective autophagy should enhance IL-17 production and tumorigenesis (Grivennikov et al., 2012). As mentioned above, defective hepatocyte autophagy results in accumulation of lipid doplets, which may promote the release of linoleic acid, causing the depletion of hepatic CD4+ T cells and creating an immunosuporessive microenvironment that promotes cancer growth (Ma et al., 2016). The generation of T cell-dependent and – independent antibody responses also requires functional autophagy as its absence results in ER stress and consequent plasma cell death (Pengo et al., 2013). In summary, autophagy tunes down inflammation while boosting adaptive immunity capable of curtailing tumor growth and progression.
Autophagy and its split personality: modulation of immunotherapy
Concomitant with the improved understanding of tumor biology and immunology, great progress has been made towards development of immunological therapies for cancer. Cancer immunotherapies include vaccines, chimeric antigen receptor (CAR) expressing T cells, bispecific antibodies and immune checkpoint inhibitors (Sharma and Allison, 2015). By-and-large, all of these strategies unleash the killing power of T cells and focus it on malignant cells. Nonetheless, cancer cells often adopt various ways to evade immune destruction, an ability that is critical for tumor progression (Figure 2). In addition to MHC downregulation, common immune escape mechanisms include recruitment and expansion of Foxp3+ Treg cells and induction of T cell anergy or exhaustion through direct and indirect interactions between cancer cells and infiltrating T cells (Sharma and Allison, 2015). Fortunately, a better understanding in tumor immunology has led to effective strategies for overcoming some of these immune escape mechanisms. Especially important were the discoveries of cytotoxic T lymphocyte associated protein 4 (CTLA-4) and programmed death 1 (PD-1), which are inhibitory receptors expressed by activated T cells (Ishida et al., 1992; Krummel and Allison, 1995). CTLA-4 competes with the CD28 co-stimulatory receptor for binding B7 molecules on APC, but unlike CD28 which acts positively, CTLA-4 engagement generates a regulatory signal that inhibits T cell proliferation and causes cell death (Krummel and Allison, 1995). CTLA-4 blockade relieves this negative input and potentiates antigen-mediated activation of T cells, including T cells directed against tumor antigens but also autoreactive T cells (Sharma and Allison, 2015). PD-1 is engaged by two ligands appropriately called PD-1 ligands 1 and 2 (PD-L1 and PD-L2) that are expressed by different cell types, including cancer cells, myeloid cells and immunosuppressive plasmocytes (Shalapour et al., 2015). PD-1 engagement switches activated CD8+ T cells to an exhausted phenotype, characterized by expression of specific markers and inability to mount a CTL response (Freeman et al., 2000). Correspondingly, PD-1 or PD-L1 blockade allows activation of CTL responses, including those that are directed against tumor antigens (Dong et al., 2002). Although checkpoint inhibitors targeting CTLA-4 or PD-1 are highly effective in treatment of metastatic cutaneous melanoma, non-small cell lung cancer and renal cell carcinoma, quite a few cancer types have not shown a significant response when treated with such drugs as monotherapies (Sharma and Allison, 2015). Even in melanoma, an immunogenic cancer that is highly responsive to checkpoint inhibitors, only a fraction of patients, usually not exceeding 50%, show a complete response to a single agent. The causes of treatment failure are not clear, but it has been suggested that insufficient release or presentation of tumor antigens is a major factor (Sharma and Allison, 2015). One way to overcome this limitation is to combine checkpoint inhibitors with inducers of so-called immunogenic cell death (ICD), a unique response that is thought to depend on autophagy (Bezu et al., 2015) (Figure 4).
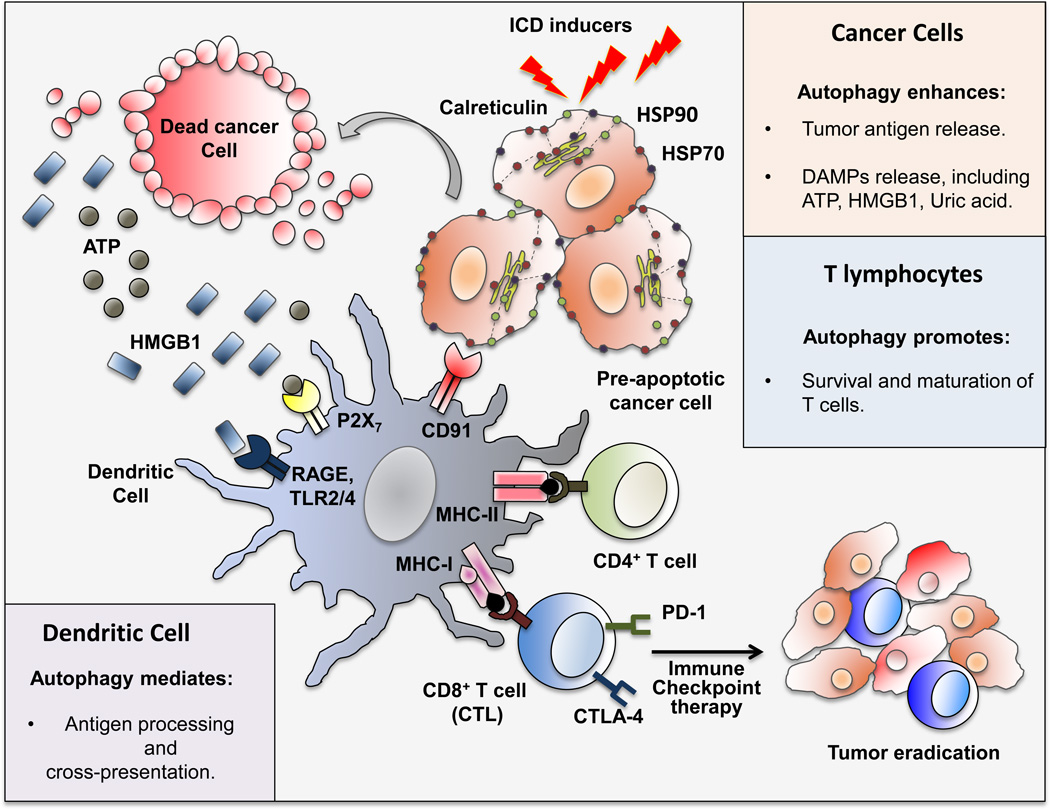
Figure 4. Autophagy Fuels Immunogenic Cell Death.
Overview of the roles of autophagy in ICD. Ionizing radiation, oncolytic viruses and certain chemotherapeutic agents induce autophagy-dependent release of DAMP and tumor antigens by cancer cells. DAMP lead to maturation and advanced activation of APC, which after engulfing tumor antigens, use autophagy to process antigens and cross-present them to T cells, leading to activation of CTL survival and maturation are also stimulated by autophagy. CTL activation can be limited upon engagement of the inhibitory receptors CTLA-4 and PD-1 by ligands that are present within the tumor bed. Immune checkpoints inhibitors overcome this inhibition and promote effective CTL-mediated tumor eradication, especially when combined with ICD inducers. HSP70/90, heat shock protein 70/90; HMGB1, high mobility group box 1 protein; RAGE, receptor for advanced glycation endproducts; P2X7, purinergic receptor P2X, ligand-gated ion channel 7; TLR2/4, toll-like receptor 2/4; CTL, cytotoxic T-lymphocytes; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; PD-1, programmed cell death protein 1.
Unlike immunologically silent or even tolerogenic apoptosis, ICD is a non-silent form of cell death that results in immune system activation. In cancer cells, ICD is postulated to induce the release of tumor antigens which are taken up and processed by APC, ultimately resulting in a tumor eradicating CTL response (Kroemer et al., 2013). Of note, a number of conventional cancer therapeutic agents/strategies are capable of inducing cancer cell ICD. These include anthracyclines (i.e., doxorubicin, epirubicin, and idarubicin), mitoxantrone, oxaliplatin and cyclophosphamide, especially when used at a low non-myeloablative dose (Kroemer et al., 2013). Radiotherapy also induces ICD, resulting in CTL activation and systemic tumor rejection, even in mice bearing multiple tumors in which only a single tumor has been irradiated (Twyman-Saint Victor et al., 2015). Certain oncolytic viruses (e.g. Coxsackievirus B3, Parvovirus and Herpes simplex virus) are also potent ICD inducers (Kaufman et al., 2015). Although their mechanism of action is not entirely clear, ICD inducers were suggested to trigger ER stress in cancer cells, causing pre-apoptotic calreticulin translocation to the cell surface. Calreticulin translocation involves inhibition of protein synthesis due to phosphorylation of eukaryotic initiation factor 2α (eIF2α), activation of pro-apoptotic molecules, including caspase-8, BAX (BCL2-associated X protein) and BAK1 (BCL2 antagonist/killer 1), and activation of intracellular vesicular trafficking (Kepp et al., 2014). Surface exposed calreticulin is one of the three main DAMP produced by dying cancer cells along with ATP and high mobility group box 1 protein (HMGB1). By binding CD91, calreticulin generates an “eat-me” signal that triggers the uptake of dying cancer cells by recruited phagocytes, leading to CTL activation in the tumor bed (Basu et al., 2001). Other DAMP, such as heat shock proteins 70 (HSP70) and 90 (HSP90), further enhance APC maturation and antigen cross-presentation (Krysko et al., 2012)(Figure 4).
ICD also results in ATP release, a process considered to depend on activation of autophagy. Extracellular ATP functions as a “find-me” signal that by engaging the purinergic receptor P2Y2 recruits monocytes that upon maturation into macrophages/DCs engulf dying cancer cells. By binding a related purinergic receptor, ligand-gated ion channel 7 (P2X7), extracellular ATP activates the NLRP3 inflammasome to induce IL-1β secretion, a process critical for a successful CTL response (Ghiringhelli et al., 2009). Of note, the preferential engagement of distinct purinergic receptors depends on ATP concentration: at <1 uM ATP predominantly binds P2Y2 to induce monocyte infiltration; whereas at > 100 uM it engages P2X7 to activate the NLRP3 inflammasome (Krysko et al., 2012). Last but not least, HMGB1 is also required for ICD induction by chemotherapeutic agents and subsequent activation of anti-tumor immunity as shown in colorectal cancer (Apetoh et al., 2007). HMGB1 released from cancer cells undergoing ICD is thought to engage TLR4 on APC, resulting in enhanced CD8+ T cell infiltration, which in combination with immune checkpoint inhibitors, leads to a strong antitumor response (Pfirschke et al., 2016). In addition, HMGB1-TLR4 signaling can prime the NLRP3 inflammasome, which further enhances induction of an antitumor CTL response (Ghiringhelli et al., 2009).
Autophagy plays a key role in promoting release of DAMP and antigens. Hence, stimulation of cancer cell autophagy is an important requisite for ICD (Martins et al., 2014). During ICD, intact autophagosomes, loaded with multiple tumor antigens, HSP and DAMP, are released to the extracellular space to be taken up by APC, inducing their maturation, activation and antigen cross-presentation (Ma et al., 2013). In support of this notion, ablation of autophagy genes (ATG5, ATG7, and BECN1) inhibited DAMP release from cancer cells killed by mitoxanthrone or oxaliplatin, and subsequently attenuated induction of anti-tumor immunity (Martins et al., 2012; Michaud et al., 2011). It was also suggested that cisplatin, which can induce apoptosis as effectively as oxaliplatin, does not trigger ICD because it does not stimulate autophagy in prostate cancer cells (Shalapour et al., 2015). Autophagy also controls exposure of phosphatidylserine (PS), another “eat-me” signal that promotes uptake of cancer cell corpses and subsequent presentation of tumor antigens (Ma et al., 2013). However, the precise molecular mechanism that drives autophagy-dependent DAMP and antigen release remains largely unknown. It is also unknown why only certain chemotherapeutic agents can trigger ICD.
Recruiting autophagy to enhance cancer immunotherapy
As discussed above, autophagy is tumor suppressive during early tumorigenesis, but its role in advanced cancer remains controversial. It was suggested that survival of established cancers depends on autophagy and therefore autophagy inhibitors should be useful as cancer therapeutics (White, 2015). However, an extensive examination of this proposal, using a large number of human cancer cell lines, failed to detect regression of tumor xenografts on inhibition of autophagy (Eng et al., 2016). Although the autophagy inhibitor chloroquine inhibited growth of some cancer cell lines, its effect was autophagy-independent.
Conversely, since autophagy mediates the release of tumor antigens and DAMP, it appears that downregulation of autophagy will allow advanced tumors to escape immunosurveillance. Indeed, tumors that retained elevated autophagic flux contain more CD8+ T cells than low autophagy tumors (Ladoire et al., 2016). Given these findings, it seems that the concept of treating cancer with autophagy inhibitors is ill conceived and likely to result in immune escape and accelerated progression of premalignant lesions and early tumors. Thus, rather than inhibit cancer cell autophagy, we should consider stimulating it, in order to enhance anti-tumor immunity. However, the direct manipulation of autophagy is not an easy task, as most autophagy enhancers do not target the autophagy machinery directly or selectively and may affect multiple targets. Indirect autophagy activators include agents that inhibit energy generation and metabolic pathways, including caloric restriction mimetics (e.g. blockers of cytosolic acetyl coenzyme A), inhibitors of autophagy-repressive acetyltransferases and activators of autophagy-stimulatory deacetylases, metformin and related biguanides (weak inhibitors of mitochondrial complex I and AMPK activators), and rapalogs (inhibitors of mTORC1, a negative regulator of autophagy initiation). Many of these agents exhibit anti-aging and anti-tumor properties (Bhullar and Hubbard, 2015; Foretz et al., 2014; Lamming et al., 2013; Morselli et al., 2010). Given that cancer is an age-related disease, it is not surprising that dietary or pharmacological caloric restriction, which extends lifespan, can enhance antitumor immunity and delay cancer development (Pietrocola et al., 2016). But whether such treatments alone will affect established malignancies is questionable. To achieve significant therapeutic efficacy, caloric restriction or starvation mimetics should be considered in combination with immunogenic chemotherapeutics capable of inducing ICD. It is also important that starvation mimetics will not exert any immunosuppressive effects that are typical of rapamycin and similar agents. In addition, ICD inducers need to be used at low, non myelosuppressive doses and their combination with starvation mimetics should not augment immunosuppression. Another potential complication associated with autophagy enhancers is the ability of autophagy to attenuate activation of the NLRP3 inflammasome by DAMP released upon ICD induction. Since IL-1β is required for ICD-mediated DC maturation and antigen presentation (Ghiringhelli et al., 2009), inhibition of its production could be counterproductive. One way to circumvent this problem is to combine autophagy inducers with inhibitors of p62 induction, such as IKKβ antagonists (Zhong et al., 2016). IKKβ inhibitors have the added benefit of sensitizing cancer cells to chemotherapy- or radiation-induced cell death. Future studies should be directed at studying the effect of such drug combinations on anti-tumor immunity and its stimulation by checkpoint inhibitors.
Autophagy is also critical for mounting antigen-specific T cell responses (Bezu et al., 2015). But again, the effect of autophagy enhancement on T cell mediated immune responses is complex, even in the context of cancer vaccines. Most vaccines rely on adjuvants, such as alum and complete Freund's adjuvant (CFA), which were proposed to exert their immunomostimulatory effect through NLRP3-inflammasome activation (Eisenbarth et al., 2008; Li et al., 2008), which is attenuated by autophagy (Zhong et al., 2016). Thus, systemic use of autophagy enhancers may reduce the efficacy of cancer vaccines, unless combined with agents that inhibit p62 induction. Alternatively, autophagy inducers should be delivered directly and specifically into cancer cells, perhaps as antibody-drug conjugates or ligand-coated nanoparticles.
Concluding Remarks
Autopahgy is a key tumor suppressive process and many of its effects are exerted in a cell non-autonomous manner through inhibition of tumor-promoting inflammation and activation of anti-tumor immunity. As our appreciation and understanding of cancer-related autophagy grow, it is time to start putting basic knowledge into practice. We propose that, together with immunogenic chemotherapeutics and immune checkpoint blockade, autophagy enhancers should expand the pharmacological arsenal and augment the efficacy of cancer immunotherapy.
Acknowledgments
Z.Z. was supported by Cancer Research Institute (CRI) Irvington postdoctoral fellowship; E.S.-L. was supported by Sara Borrell fellowship under ISCIII/MICINN program; Research was supported by grants from the NIH (AI043477 and CA163798) and a Leukemia and Lymphoma Society SCOR (20132569) to M.K, who is an American Cancer Society Research Professor and holder of the Ben and Wanda Hildyard Chair for Mitochondrial and Metabolic Diseases. We apologize to the authors, that due to the space constraints, their work could not be cited.
Abbreviation List
- AMPK
AMP-activated protein kinase
- APC
Antigen-presenting cells
- ASC
Apoptosis-associated speck-like
- ASH
Alcoholic hepatitis
- ATG
Autophagy related gene
- ATG16L1
Autophagy related 16-like 1
- BAK1
BCL2 antagonist/killer 1
- BAX
BCL2-associated X protein
- CAR
Chimeric antigen receptor
- CagA
Cytotoxin-associated gene A
- CD
Crohn’s disease
- CFA
Complete Freund's adjuvant
- CRC
Colorectal cancer
- CTL
Cytotoxic T lymphocytes
- CTLA-4
Cytotoxic T lymphocyte associated protein 4
- DAMP
Danger-associated molecular patterns
- DC
Dendritic cells
- EBNA1
EBV nuclear antigen 1
- EBV
Epstein - Barr virus
- eIF2α
Eukaryotic initiation factor 2α
- ER
Endoplasmic reticulum
- HBs
Hepatitis B virus small surface protein
- HBV
Hepatitis B virus
- HBx
HBV X protein
- HCC
Hepatocellular carcinoma
- HCV
Hepatitis C virus
- HMGB1
High mobility group box 1 protein
- H. pylori
Helicobacter pylori
- HSP70/90
Heat shock proteins 70/90
- IBD
Inflammatory bowel diseases
- ICD
Immunogenic cell death
- IKKβ
IκB kinase β
- KSHV
Kaposi’s sarcoma-Assocaited Herpesvirus
- LC3
Microtubule-associated protein 1A/1B light chain 3
- LMP1
Latent membrane protein 1
- MHC-I/II
Major histocompatibility class I/II
- MSU
Monosodium urate crystal
- mtDNA
Mitochondrial DNA
- mTORC1
mammalian target of rapamycin complex 1
- mtROS
Mitochondrial reactive oxygen species
- NAFLD
Nonalcoholic fatty liver disease
- NASH
Nonalcoholic steatohepatitis
- NBR1
Neighbor of Brca 1
- NF-κB
Nuclear factor-κB
- NLR
Nod-like receptors
- NLRC4
NLR family CARD domain-containing protein 4
- NLRP1b
NLR Family, Pyrin Domain Containing 1
- NLRP3
NOD-like receptor family pyrin domain-containing 3
- NRF2
Nuclear factor erythroid 2-related factor 2
- P2X7
Purinergic receptor P2X, ligand-gated ion channel 7
- P62/SQSTM1
Sequestosome 1
- PAMP
Pathogen-associated molecular patterns
- PD-1
Programmed death 1
- PD-L1/2
PD-1 ligands 1/2
- PI3KC
Class III phosphatidylinositol (PI) 3’ kinase
- PINK1
PTEN-induced putative kinase 1
- PS
Phosphatidylserine
- ROS
Reactive oxygen species
- TLR
Toll-like receptors
- Treg
T regulatory cells
- UBA
Ubiquitin association domain
- UC
Ulcerative colitis
- ULK1
Unc-51 like autophagy activating kinase 1
- UPR
Unfolded protein response
- VacA
Vacuolating cytotoxin A
- vBcl-2
Viral B-cell lymphoma 2
- vFLIP
Viral FLICE inhibitory protein
- VPS34
Class III PI 3-kinase
- WT
Wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aichinger M, Wu C, Nedjic J, Klein L. Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance. J Exp Med. 2013;210:287–300. doi: 10.1084/jem.20122149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonucci L, Fagman JB, Kim JY, Todoric J, Gukovsky I, Mackey M, Ellisman MH, Karin M. Basal autophagy maintains pancreatic acinar cell homeostasis and protein synthesis and prevents ER stress. Proc Natl Acad Sci U S A. 2015;112:E6166–E6174. doi: 10.1073/pnas.1519384112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- Basu S, Binder RJ, Ramalingam T, Srivastava PK. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity. 2001;14:303–313. doi: 10.1016/s1074-7613(01)00111-x. [DOI] [PubMed] [Google Scholar]
- Bezu L, Gomes-de-Silva LC, Dewitte H, Breckpot K, Fucikova J, Spisek R, Galluzzi L, Kepp O, Kroemer G. Combinatorial strategies for the induction of immunogenic cell death. Front Immunol. 2015;6:187. doi: 10.3389/fimmu.2015.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar KS, Hubbard BP. Lifespan and healthspan extension by resveratrol. Biochim Biophys Acta. 2015;1852:1209–1218. doi: 10.1016/j.bbadis.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Bujak AL, Crane JD, Lally JS, Ford RJ, Kang SJ, Rebalka IA, Green AE, Kemp BE, Hawke TJ, Schertzer JD, et al. AMPK activation of muscle autophagy prevents fasting-induced hypoglycemia and myopathy during aging. Cell Metab. 2015;21:883–890. doi: 10.1016/j.cmet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- Chourasia AH, Tracy K, Frankenberger C, Boland ML, Sharifi MN, Drake LE, Sachleben JR, Asara JM, Locasale JW, Karczmar GS, et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16:1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HY, Cesari M, Anton S, Marzetti E, Giovannini S, Seo AY, Carter C, Yu BP, Leeuwenburgh C. Molecular inflammation: underpinnings of aging and age-related diseases. Ageing Res Rev. 2009;8:18–30. doi: 10.1016/j.arr.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Wells MA, Schneider KJ, Singh S. Molecular mechanisms of alcohol associated pancreatitis. World J Gastrointest Pathophysiol. 2014;5:147–157. doi: 10.4291/wjgp.v5.i3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527:105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott EI, Sutterwala FS. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol Rev. 2015;265:35–52. doi: 10.1111/imr.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CH, Wang Z, Tkach D, Toral-Barza L, Ugwonali S, Liu S, Fitzgerald SL, George E, Frias E, Cochran N, et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci U S A. 2016;113:182–187. doi: 10.1073/pnas.1515617113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathallah I, Parroche P, Gruffat H, Zannetti C, Johansson H, Yue J, Manet E, Tommasino M, Sylla BS, Hasan UA. EBV latent membrane protein 1 is a negative regulator of TLR9. J Immunol. 2010;185:6439–6447. doi: 10.4049/jimmunol.0903459. [DOI] [PubMed] [Google Scholar]
- Foretz M, Guigas B, Bertrand L, Pollak M, Viollet B. Metformin: from mechanisms of action to therapies. Cell Metab. 2014;20:953–966. doi: 10.1016/j.cmet.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, Vermaelen K, Panaretakis T, Mignot G, Ullrich E, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med. 2009;15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- Greten FR, Arkan MC, Bollrath J, Hsu LC, Goode J, Miething C, Goktuna SI, Neuenhahn M, Fierer J, Paxian S, et al. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell. 2007;130:918–931. doi: 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross O, Thomas CJ, Guarda G, Tschopp J. The inflammasome: an integrated view. Immunol Rev. 2011;243:136–151. doi: 10.1111/j.1600-065X.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Tsujii M, Wang J, Kondo J, Akasaka T, Jin Y, Li W, Nakamura T, Nishida T, Iijima H, et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut. 2013;62:1536–1546. doi: 10.1136/gutjnl-2011-301625. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Enzler T, Seita J, Timmer AM, Lee CY, Lai TY, Yu GY, Lai LC, Temkin V, Sinzig U, et al. IL-1beta-driven neutrophilia preserves antibacterial defense in the absence of the kinase IKKbeta. Nat Immunol. 2011;12:144–150. doi: 10.1038/ni.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard-Lucey VM, Shono Y, Maurer K, West ML, Singer NV, Ziegler CG, Lezcano C, Motta AC, Schmid K, Levi SM, et al. Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity. 2014;41:579–591. doi: 10.1016/j.immuni.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature. 2016;529:307–315. doi: 10.1038/nature17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov. 2015;14:642–662. doi: 10.1038/nrd4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepp O, Senovilla L, Vitale I, Vacchelli E, Adjemian S, Agostinis P, Apetoh L, Aranda F, Barnaba V, Bloy N, et al. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology. 2014;3:e955691. doi: 10.4161/21624011.2014.955691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol. 2013;31:51–72. doi: 10.1146/annurev-immunol-032712-100008. [DOI] [PubMed] [Google Scholar]
- Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. 1995;182:459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer. 2012;12:860–875. doi: 10.1038/nrc3380. [DOI] [PubMed] [Google Scholar]
- Ladoire SE, Senovilla L, Ghiringhelli F, Poirier-Colame V, Chaba K, Semeraro M, Chaix M, Penault-Llorca F, Arnould L, Arveux P, et al. The presence of LC3B puncta and HMGB1 expression in malignant cells correlate with the immune infiltrate in breast cancer. Autophagy. 2016 doi: 10.1080/15548627.2016.1154244. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol. 2012;28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Sabatini DM, Baur JA. Rapalogs and mTOR inhibitors as anti-aging therapeutics. J Clin Invest. 2013;123:980–989. doi: 10.1172/JCI64099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Li B, Lei Z, Lichty BD, Li D, Zhang GM, Feng ZH, Wan Y, Huang B. Autophagy facilitates major histocompatibility complex class I expression induced by IFN-gamma in B16 melanoma cells. Cancer Immunol Immunother. 2010;59:313–321. doi: 10.1007/s00262-009-0752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Willingham SB, Ting JP, Re F. Cutting edge: inflammasome activation by alum and alum's adjuvant effect are mediated by NLRP3. J Immunol. 2008;181:17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wu X, Holzer RG, Lee JH, Todoric J, Park EJ, Ogata H, Gukovskaya AS, Gukovsky I, Pizzo DP, et al. Loss of acinar cell IKKalpha triggers spontaneous pancreatitis in mice. J Clin Invest. 2013;123:2231–2243. doi: 10.1172/JCI64498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licastro F, Candore G, Lio D, Porcellini E, Colonna-Romano G, Franceschi C, Caruso C. Innate immunity and inflammation in ageing: a key for understanding age-related diseases. Immun Ageing. 2005;2:8. doi: 10.1186/1742-4933-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, et al. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014;10:416–430. doi: 10.4161/auto.27286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, Tanaka KE, Czaja MJ. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy. 2015;11:271–284. doi: 10.1080/15548627.2015.1009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet A, Mathurin P. Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat Rev Gastroenterol Hepatol. 2015;12:231–242. doi: 10.1038/nrgastro.2015.35. [DOI] [PubMed] [Google Scholar]
- Ma C, Kesarwala AH, Eggert T, Medina-Echeverz J, Kleiner DE, Jin P, Stroncek DF, Terabe M, Kapoor V, ElGindi M, et al. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253–257. doi: 10.1038/nature16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Galluzzi L, Zitvogel L, Kroemer G. Autophagy and cellular immune responses. Immunity. 2013;39:211–227. doi: 10.1016/j.immuni.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Mah LY, Ryan KM. Autophagy and cancer. Cold Spring Harb Perspect Biol. 2012;4:a008821. doi: 10.1101/cshperspect.a008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N, Athonvarangkul D, Singh R. Autophagy and aging. Adv Exp Med Biol. 2015;847:73–87. doi: 10.1007/978-1-4939-2404-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- Martins I, Michaud M, Sukkurwala AQ, Adjemian S, Ma Y, Shen S, Kepp O, Menger L, Vacchelli E, Galluzzi L, et al. Premortem autophagy determines the immunogenicity of chemotherapy-induced cancer cell death. Autophagy. 2012;8:413–415. doi: 10.4161/auto.19009. [DOI] [PubMed] [Google Scholar]
- Martins I, Wang Y, Michaud M, Ma Y, Sukkurwala AQ, Shen S, Kepp O, Metivier D, Galluzzi L, Perfettini JL, et al. Molecular mechanisms of ATP secretion during immunogenic cell death. Cell Death Differ. 2014;21:79–91. doi: 10.1038/cdd.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer K, Torres VJ, Cadwell K. Autophagy is a key tolerance mechanism during Staphylococcus aureus infection. Autophagy. 2015;11:1184–1186. doi: 10.1080/15548627.2015.1058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. Transcriptional control of autophagylysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfirschke C, Engblom C, Rickelt S, Cortez-Retamozo V, Garris C, Pucci F, Yamazaki T, Poirier-Colame V, Newton A, Redouane Y, et al. Immunogenic Chemotherapy Sensitizes Tumors to Checkpoint Blockade Therapy. Immunity. 2016;44:343–354. doi: 10.1016/j.immuni.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F, Pol J, Vacchelli E, Rao S, Enot DP, Baracco EE, Levesque S, Castoldi F, Jacquelot N, Yamazaki T, et al. Caloric restriction mimetics reinforce anticancer immunosurveillance. Cancer Cell. 2016 doi: 10.1016/j.ccell.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JL, Cuervo AM. Liver autophagy: much more than just taking out the trash. Nat Rev Gastroenterol Hepatol. 2014;11:187–200. doi: 10.1038/nrgastro.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalapour S, Font-Burgada J, Di Caro G, Zhong Z, Sanchez-Lopez E, Dhar D, Willimsky G, Ammirante M, Strasner A, Hansel DE, et al. Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature. 2015;521:94–98. doi: 10.1038/nature14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125:3347–3355. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani ST, Saitoh T, Nowag H, Munz C, Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat Immunol. 2015;16:1014–1024. doi: 10.1038/ni.3273. [DOI] [PubMed] [Google Scholar]
- Silva LM, Jung JU. Modulation of the autophagy pathway by human tumor viruses. Semin Cancer Biol. 2013;23:323–328. doi: 10.1016/j.semcancer.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- Szabo G, Csak T. Inflammasomes in liver diseases. J Hepatol. 2012;57:642–654. doi: 10.1016/j.jhep.2012.03.035. [DOI] [PubMed] [Google Scholar]
- Tremblay F, Krebs M, Dombrowski L, Brehm A, Bernroider E, Roth E, Nowotny P, Waldhausl W, Marette A, Roden M. Overactivation of S6 kinase 1 as a cause of human insulin resistance during increased amino acid availability. Diabetes. 2005;54:2674–2684. doi: 10.2337/diabetes.54.9.2674. [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Suzuki H, Saya H, Hatakeyama M, Hirayama T, Hirata K, Nagano O, Matsuzaki J, Hibi T. Reactive oxygen species-induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem-like cells. Cell Host Microbe. 2012;12:764–777. doi: 10.1016/j.chom.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, Benci JL, Xu B, Dada H, Odorizzi PM, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520:373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Duran A, Linares J, et al. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell. 2016 doi: 10.1016/j.ccell.2016.04.006. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Matsumoto M, Pacold CM, Cho WK, Crabb DW. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology. 2004;127:1798–1808. doi: 10.1053/j.gastro.2004.09.049. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Sung JJ, Yu J, Ng SC, Wong SH, Cho CH, Ng SS, Chan FK, Wu WK. Xenophagy in Helicobacter pylori- and Epstein-Barr virus-induced gastric cancer. J Pathol. 2014;233:103–112. doi: 10.1002/path.4351. [DOI] [PubMed] [Google Scholar]
- Zhao J, Liang Q, Cheung KF, Kang W, Dong Y, Lung RW, Tong JH, To KF, Sung JJ, Yu J. Somatostatin receptor 1, a novel EBV-associated CpG hypermethylated gene, contributes to the pathogenesis of EBV-associated gastric cancer. Br J Cancer. 2013;108:2557–2564. doi: 10.1038/bjc.2013.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, et al. NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Littman DR. Transcriptional regulatory networks in Th17 cell differentiation. Curr Opin Immunol. 2009;21:146–152. doi: 10.1016/j.coi.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]