Summary
Interferon gamma (IFN-γ) is an essential mediator of host defense against intracellular pathogens, including the protozoan parasite Toxoplasma gondii. However, prior T. gondii infection blocks IFN-γ-dependent gene transcription, despite the downstream transcriptional activator STAT1 being activated and bound to cognate nuclear promoters. We identify the parasite effector that blocks STAT1-dependent transcription and show it is associated with recruitment of the Mi-2 Nucleosome Remodeling and Deacetylase (NuRD) complex, a chromatin-modifying repressor. This secreted effector, Toxoplasma Inhibitor of STAT1-dependent Transcription (TgIST), translocates to the host cell nucleus where it recruits Mi-2/NuRD to STAT1-dependent promoters, resulting in altered chromatin and blocked transcription. TgIST is conserved across strains, underlying their shared ability to block IFN-γ-dependent transcription. TgIST deletion results in increased parasite clearance in IFN-γ-activated cells and reduced mouse virulence, which is restored in IFN-γ-receptor deficient mice. These findings demonstrate the importance of both IFN-γ responses and the ability of pathogens to counteract these defenses.
Keywords: transcription, acetylation, nuclear transport, phosphorylation, signaling
Graphical Abstract

Introduction
Signal Transducer and Activator of Transcription (STAT) proteins transmit signals from engagement of cytokine and growth factors to control transcription of genes important for immunity, growth, and development (Reich and Liu, 2006; Stark and Darnell, 2012). Binding of IFN-γ to its receptors drives dimerization and recruitment of JAK kinases that phosphorylate STAT1 on residue Y701 (Shuai et al., 1993). Phosphorylation of STAT1 promotes dimerization, nuclear trafficking, and binding to a conserved motif known as the gamma-activated sequence (GAS) resulting in the transcription of many genes, including those important for immunity (Darnell et al., 1994; Schindler and Darnell, 1995). STAT1-mediated transcription is important in control of viral, bacterial, and eukaryotic pathogens and, not surprisingly, many microbes have evolved elaborate mechanisms to block this important defense mechanism (Najjar and Fagard, 2010).
Immune defense against the intracellular parasite Toxoplasma gondii infection is mediated by a potent Th1 response characterized by secretion of IL-12 (Gazzinelli et al., 1994) and induction of IFN-γ (Suzuki et al., 1989), which activates defenses in both hematopoietic and non-hematopoietic cells (Yap and Sher, 1999). Mice deficient in IFN-γ receptors (Deckert-Schlüter et al., 1996) or STAT1 (Gavrilescu et al., 2004; Lieberman et al., 2004) are extremely susceptible to an otherwise nonlethal challenge with T. gondii. IFN-γ induces a number of antimicrobial responses including upregulation of reactive oxygen species (Nathan et al., 1983), degradation of tryptophan (Pfefferkorn, 1984), induction of nitric oxide synthase (NOS2) (Stuehr et al., 1989), and upregulation of a family of immunity related GTPases (IRGs) (Taylor et al., 2007) and guanylate binding proteins (GBPs) (Kim et al., 2012). Not surprising given its widespread success, T. gondii is equipped with potent defenses that block IFN-γ-mediated immunity (Hunter and Sibley, 2012).
Importantly, when cells are infected by T. gondii prior to encountering IFN-γ, the parasite globally blocks STAT1-mediated transcription (Kim et al., 2007; Lang et al., 2012). This block occurs despite normal phosphorylation, dimerization, and translocation of STAT1 to the nucleus (Rosowski et al., 2014; Schneider et al., 2013). Repression of STAT1-mediated transcription is observed following infection with all three clonal types of T. gondii (Kim et al., 2007; Rosowski and Saeij, 2012) in a variety of both mouse and human cell types. (Kim et al., 2007; Lang et al., 2012). This block does not depend on previously characterized virulence determinants (Hunter and Sibley, 2012), suggesting the presence of an effector. Inhibition of STAT1 likely aids parasite survival as it down-modulates important defenses including MHC and inducible NOS2 expression (Luder et al., 2003; Lüder et al., 2001; Lüder et al., 1998). However, the basis of this inhibition has remained elusive despite extensive study (Lang et al., 2012; Rosowski et al., 2014; Rosowski and Saeij, 2012).
Previous studies have shown that STAT1 remains bound to GAS sequences in its cognate promoters in T. gondii infected cells treated with IFN-γ despite a block of transcription (Rosowski et al., 2014). In the present study, we capitalized on the stability of this interaction to identify proteins that are found in complex with activated STAT1 in the nucleus of T. gondii infected cells. We identified the Mi-2 Nucleosome Remodeling and Deacetylase (Mi-2/NuRD) complex in tight association with STAT1, an interaction that is mediated by a secreted parasite effector, which is responsible for the block in STAT1 transcription in murine and human cells. These studies reveal a mechanism by which T. gondii blocks host immunity by turning a normal transcriptional activator into a repressor of immune response genes.
Results
Identification of a repressive complex recruited to activated STAT1 complexes in T. gondii-infected cells
To identify the molecular basis of the transcriptional block induced by T. gondii, we compared STAT1-containing complexes in control vs. infected U3A-STAT1 cells, which express FLAG-tagged STAT1 (Zhang et al., 2005), that were treated with IFN-γ. Following STAT1-FLAG immunoprecipitation (IP) from IFN-γ-treated cells, LC-MS/MS analysis was used to characterize STAT1 interacting complexes in control vs. infected cells (Figure 1A, Table S1). We observed a slight decrease in the abundance of the chromatin modifier BRG1 (Figure 1A, Table S1), which is normally an essential activator of IFN-γ responsive promoters (Ni et al., 2005), consistent with a previous report (Lang et al., 2012). Unexpectedly, a number of STAT1-interacting proteins were enriched in T. gondii infected vs. uninfected cells treated with IFN-γ (Figure 1A, Table S1). In particular, STAT1 was associated with core members of the Mi-2/NuRD complex including CHD4, an ATPase, HDAC1 and HDAC2, two histone deacetylases, and a variety of other subunits (Bowen et al., 2004; Denslow and Wade, 2007). Additionally, STAT1 complexes in infected and IFN-γ activated cells contained the co-repressor C-terminal binding proteins (CTBP) 1 and 2 (Figure 1A, Table S1). However, neither the CTBPs or the Mi-2/NuRD complex were found to be associated with STAT1 in IFN-γ-treated cells in the absence of T. gondii infection (Figure 1A, Table S1).
Figure 1. Identification of T. gondii Inhibitor of STAT1-dependent transcription (TgIST) and Mi-2/NuRD repressor complex.
(A) STAT1-associated host and T. gondii proteins identified by MS analysis. STAT1-FLAG IP from uninfected (−) or type I (RH) parasite (+) infected U3A-STAT1 cells + IFN-γ (100 U/ml). Combination of two experiments. *Increased protein coverage specifically targeting IST.
(B) Western blot analysis of STAT1-FLAG IP from uninfected (−) or type I (RH) parasite (+) infected U3A-STAT1 cells ± IFN-γ (100 U/ml). IP’d samples (20% of total) were resolved by SDS-PAGE, blotted with primary antibodies (listed to right) and imaged with LI-COR specific secondary antibodies. Representative of three or more experiments with similar outcomes. See also Figure S1.
(C) Western blot analysis of TgIST-Ty IP’d from U3A-STAT1 host cells infected with type I (RH) parasites. Host cells were either uninfected or infected with parasites expressing a Ty-tagged copy of TgIST ± IFN-γ (100 units/ml) for 30 min. Nuclear extracts (2% of total) and IP’d samples (20% of total) were resolved on SDS-PAGE gels, blotted with primary antibodies and imaged with LI-COR specific secondary antibodies.
To validate the LC-MS/MS analysis, we performed co-IPs, first immunoprecipitating STAT1-FLAG and then assessing the phosphorylation status of STAT1 and the presence of subunits of the Mi-2/NuRD complex by western blotting. Similar to IFN-γ receptors, infection with wild type T. gondii led to low levels of STAT1-Y701 phosphorylation, as previously reported (Schneider et al., 2013), while the combination of infection and IFN-γ treatment resulted in enhanced levels of STAT1-Y701 phosphorylation (Figure 1B, Figure S1A). Consistent with the LC-MS/MS findings, IPs of STAT1-FLAG from infected plus IFN-γtreated cells led to capture of a number of the Mi-2/NuRD complex members (Figure 1B, Figure S1A). This interaction was specific to infected cells where low levels of some components were detected in association with STAT1 in the absence of IFN-γ activation, including MTA1 and RBBP7 (Figure 1B, Figure S1A). Collectively, these findings reveal the formation of a STAT1-associated Mi-2/NuRD complex in the nucleus of T. gondii-infected cells treated with IFN-γ.
Recruitment of the Mi-2/NuRD complex to STAT1 requires a unique T. gondii effector
To identify putative effector proteins responsible for the STAT1-Mi-2/NuRD interaction, we searched the MS data for parasite proteins associated with STAT1. A single parasite protein was found in IFN-γ-treated and infected cells, referred to as T. gondii Inhibitor of STAT1-dependent Transcription (TgIST), which was represented by several high confidence (>95%) peptides (Figure 1A, Table S1). Although the initial frequency of peptides was low, when the MS spectra were searched for specific matches to peptides derived from this protein, the frequency of matches was highly enriched in infected samples (Figure 1A, Table S1). TgIST is encoded by a single copy gene (TGME49_240060) that is distantly conserved (~16% identical at the amino acid level) in the closely related livestock parasite Neospora caninum, which lacks the ability to block STAT1 transcription (Kim et al., 2007). However, TgIST has no close similarity to other proteins in the NCBI database nor does it contain conserved domain features that would suggest function.
To test the requirement for TgIST in mediating the interaction with the Mi-2/NuRD complex, we generated a TgIST-knockout line of the type I RH strain of T. gondii, and also reinserted the TgIST gene under control of its native promoter to create a complemented line that was epitope tagged (Figure 1B, C, Figure S1B, C). In co-IP experiments, the association of TgIST with STAT1 was dependent on both infection and IFN-γ-treatment, suggesting that it specifically associates with the active phospho-Y701-STAT1 dimer found in the nucleus (Figure 1B). Reciprocal IPs of Ty-epitope tagged TgIST confirmed this interaction by preferentially capturing the phospho-Y701-STAT1 dimer from nuclear extracts (Figure 1C). Infection of U3A-STAT1 cells with the TgIST knockout line no longer led to STAT1-Y701 phosphorylation in untreated cells nor increased phosphorylation following IFN-γ treatment, and phosphorylation was restored during infection with the complemented line (Figure 1B). Likewise, the association of STAT1 with the Mi-2/NuRD complex was abolished during infection with the TgIST knockout line, and this interaction was fully restored in the complemented line (Figure 1B). Taken together, these findings indicate that the association of the Mi-2/NuRD complex with STAT1 is dependent on the T. gondii effector TgIST.
TgIST is necessary to block STAT1-dependent transcription induced by IFN-γ-treatment
We engineered a 3T3 cell line containing an IFN-γ-inducible GAS promoter driving expression of the HaloTag reporter that allows for live cell detection via staining with tetramethyl rhodamine (TMR). Infection of 3T3 reporter cells with wild type T. gondii blocked subsequent IFN-γ-stimulated expression of the GAS-HaloTag reporter as seen by FACS (Figure 2A) or microscopy analysis (Figure 2B). The block in induction of the GAS-HaloTag reporter seen in infected cells treated with IFN-γ was dependent on TgIST as shown quantitatively using FACS analysis (Figure 2A). Moreover, the pattern of TgIST dependent suppression was also observed for IRF1 in human fibroblasts (Figure 2C), NOS2 in mouse RAW macrophages (Figure 2D) and IRF1 and ICAM1 gene expression in HeLa cells (Figure 2E). In all cases, infection with wild type parasites repressed the induction of the protein, while the blockade was lost with knockout parasite and was restored with complemented parasite infection (Figure 2 BE). TgIST is found in a diverse collection of T. gondii strains and shows strong evidence of being under positive selection based on the ratio of non-synonymous to synonymous changes (Figure S2 A, B). Despite this diversity, when TgIST was deleted in type II or type III strains, they also lost the ability to block STAT1-dependent expression of IRF1 in response to IFN-γ (Figure S2 C). Collectively, these findings show that TgIST is necessary for the ability of all three clonal types of T. gondii to block IFN-γ-stimulated, STAT1-dependent gene transcription in mouse and human cells.
Figure 2. TgIST blocks STAT1-dependent transcription.
(A) 3T3 GAS HaloTag reporter cells infected with type I (RH) wild type, TgIST knockout or complemented GFP-expressing parasites were activated with IFN-γ (100 U/ml) for 14 hr, stained with TMR (red), fixed, and analyzed by flow cytometry. Uninfected/unstimulated (UI/US) (grey) and uninfected/IFN-γ stimulated (US/IFN-γ) (black) cells for comparison.
(B) 3T3 GAS HaloTag reporter cells stained with TMR (red).
(C) Human foreskin fibroblast (HFF) cells stained for IRF-1.
(D) RAW264.7 cells stained for NOS-2 (red).
(B–D) GFP-expressing type I (RH), wild type, knockout, or complement parasites were stimulated with IFN-γ (100 U/ml) for 15 hr prior to staining. Scale bar = 20 μm. (A–D) Representative of three or more experiments with similar outcomes.
(E) qRT-PCR analysis of IFN-γ induced transcripts of IRF1 and ICAM1 using RNA isolated from HeLa cells infected with type I (RH) wild type, TgIST knockout, or complemented parasites and either left untreated (−) or activated (+) with IFN-γ (100 U/ml) for 15 hr. Mean ± SEM (n = 3 experiments, each with 3 replicates, n = 9). *P < 0.05 and **P < 0.001, two sample unpaired Student’s t-test. See also Figure S2.
TgIST is found in secretory granules, traffics to the host cell nucleus, and is sufficient to block STAT1-dependent gene expression
The TgIST gene encodes a ~90 kDa protein with several potential nuclear localization sequences and two putative PEXEL-like export sequences (Hsiao et al., 2013) (Figure 3A). Cellular localization studies revealed that TgIST was found in secretory compartments in the parasite that partially overlapped with the dense granule marker GRA2 in extracellular parasites and early (i.e. 20 min) after infection, and then accumulated in the nucleus of infected cells at later time points (i.e. 20 hr) after infection (Figure 3B, Figure S3). Although the above findings indicated that TgIST is necessary for the block in STAT1-dependent gene expression, they do not rule out the possibility that TgIST participates indirectly in this process, for example by affecting another protein that in turn is responsible for blocking STAT1-dependent transcription. To test whether TgIST was sufficient to block STAT1-dependent transcription we expressed TgIST as a transgene in HeLa cells and tested their response to IFN-γ. Transient expression of GFP-tagged, or Ty-epitope-tagged, TgIST in HeLa cells completely blocked IRF1 induction by IFN-γ (Figure 3C, D), establishing that it is fully sufficient on its own and that it operates independently of other effectors. The strong nuclear localization of TgIST in HeLa cells, similar to the pattern seen in infected cells, suggests that it functions in the host cell nucleus, a finding consistent with the co-IP of STAT1 with TgIST from nuclear extracts (Figure 1).
Figure 3. TgIST traffics to the host cell nucleus and blocks IRF1 induction.
(A) Mass spectrometry coverage (yellow bars). Computational analysis identified two PEXEL-like motifs and three potential nuclear localization signals (NLS) as well as an internal duplication site (arrowheads). Sequence boxed in green indicates corrected ME49 (II) strain sequence vs. that found in ToxoDB (TGME_240060).
(B) Extracellular type I (RH) tachyzoites vs. HFF cells infected with type I (RH) parasites and fixed at intervals post-infection, stained with α-Ty (red), α-GRA2 (green), and DAPI (blue). See also Figure S3.
(C) HeLa cells transiently expressing GFP-tagged TgIST.
(D) HeLa cells transiently expressing Ty-tagged TgIST.
(C, D) treated with + IFN-γ (100 U/ml) for 6 hr, stained for α-IRF1 (red) and DAPI (blue).
(B–D) Representative of 3 or more experiments with similar outcomes. Scale bars in microns.
TgIST associates with activated STAT1 and the Mi-2/NuRD complex on GAS sequences
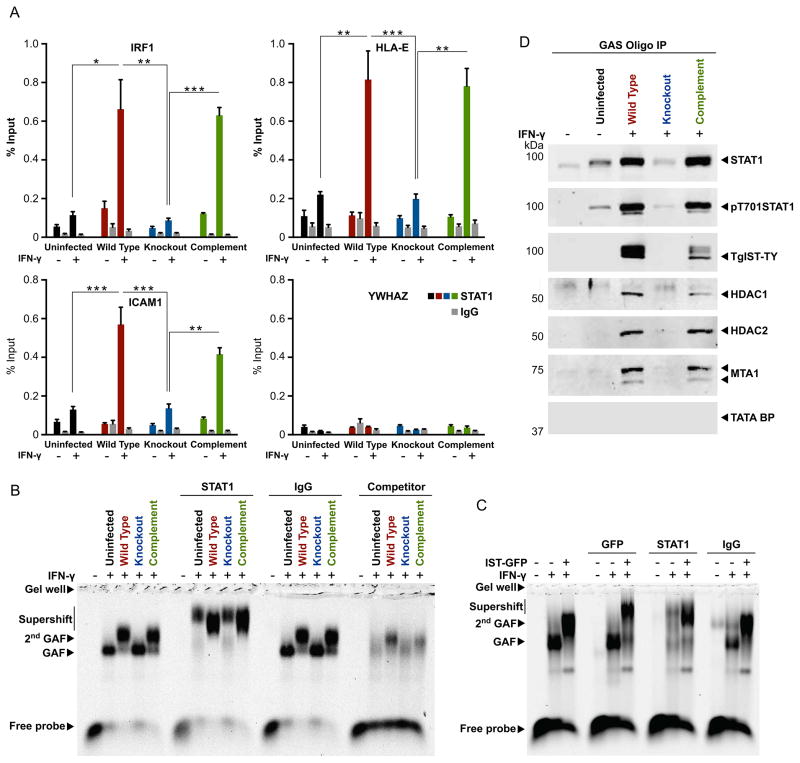
To examine the role of TgIST in binding to GAS sequences, we performed chromatin IP (ChIP) studies in normal and IFN-γ–activated cells to interrogate STAT1 interactions with endogenous GAS-containing promoters. Infection with T. gondii led to enhanced association of STAT1 with GAS sequences in promoters of IFN-γ-regulated genes including IRF-1, HLA-E, ICAM1 but not the control gene YWHAZ (Figure 4A). The enhanced interaction of STAT1 with GAS-containing promoters was dependent on TgIST as it was lost in the knockout line and restored in the complemented line (Figure 4A).
Figure 4. Activated STAT1 protein complexes at GAS sites contain TgIST and the NuRD complex.
(A) ChIP of α-Stat1 from HeLa cells infected for 3 hr with type I (RH) wild type (red), TgIST knockout (blue), and complemented parasites (green), IgG control (gray) ± IFN-γ (100 U/ml) for 30 min. Analyzed by qPCR for the GAS sites in promoter elements of the genes shown. Composite of three independent experiments each with 3 technical replicates, *P < 0.05 and **P < 0.001, ***P ≤ 0.0001, Kruskal-Wallis with Dunn’s correction.
(B) HFF cells infected with type I (RH) wild type (red), TgIST type I knockout (blue), or complemented parasites (green) for 20 hr ± IFN-γ (100 units/ml) for the last 30 min. Nuclear extracts were tested for electrophoretic mobility shift using labeled GAS oligo (Fc gamma receptor). Supershift with α-STAT1 or rabbit IgG (α-Rab IgG). Unlabeled competitor: 200x concentration of unlabeled GAS probe. GAF, gamma activated factor. See also Figure S4 A, B.
(C) HEK293T cells ± transfected with TgIST-GFP ± IFN-γ (100 units/ml) for the last 30 min. Nuclear extracts were tested for electrophoretic mobility shift using labeled GAS oligo (Fc gamma receptor). Supershift with α-GFP, α-STAT1 or rabbit IgG (α-Rab IgG). See also Figure S4C. (B–D) Representative of 3 or more experiments with similar outcomes. See also Figure S4C.
(D) Western blot analysis of biotinylated GAS oligo (Fc gamma receptor) incubated with, and pulled-down from, HFF host cell lysate. HFF cells were either left uninfected or infected with type I (RH) parasites expressing endogenous TgIST-Ty, type I TgIST deletion mutant, or the complemented line for 20 hr. Cells were either left naive (−) or activated (+) with 100 U/ml IFN-γ for the final 30 min of infection. Pull-down was performed on nuclear extracts. Samples (20% of total) were resolved on SDS-PAGE gels, blotted with primary antibodies and imaged with LI-COR specific secondary antibodies. See also Figure S4D.
Following IFN-γ treatment, nuclear extracts containing the active STAT1 complex, referred to as the gamma-activated factor (GAF) (Schindler and Darnell, 1995), can be visualized by electrophoretic mobility shift assay (EMSA). T. gondii infection shifted the normal GAF complex to migrate as a broad, slower migrating band (2nd GAF in Figure 4B), which required the presence of STAT1 (Figure S4 A, B), consistent with a previous report (Lang et al., 2012). Importantly, the slower migrating 2nd GAF form was lost in nuclear extracts from cells infected with the TgIST knockout line and was fully restored in the complement line, demonstrating that the altered mobility of this 2nd GAF species was TgIST-dependent (Figure 4B). Anti-STAT1 antibodies super-shifted both the normal and TgIST-dependent 2nd GAF, indicating the presence of STAT1 at the core of both complexes (Figure 4B, Figure S4 A, B). To verify the presence of TgIST in association with STAT1 on the GAS oligo, we examined nuclear extracts of HEK293T cells transiently transfected with GFP-tagged TgIST by EMSA. The slower migrating second band (2nd GAF), which was only present in TgIST-transfected cells, was super-shifted both with anti-STAT1 and anti-GFP antibodies (Figure 4C, Figure S4C).
The slower migrating 2nd GAF form seen in T. gondii infected cells suggests formation of a larger complex, perhaps not containing TgIST and also members of the Mi-2/NuRD complex. To test this we used a biotinylated GAS oligo to precipitate complexes and then analyzed them by western blot. Consistent with the ChIP studies above, pull-downs with the GAS oligo captured higher levels of pT701-STAT1 in the presence of T. gondii infection (Figure 4D, Figure S4D). This enhanced interaction was lost in the knockout parasite and regained in the complemented parasite showing it is specific to TgIST (Figure 4D, Figure S4D). Similarly, the interaction of the Mi-2/NuRD complex members was strictly dependent on TgIST (Figure 4D). Together, these findings demonstrate that TgIST mediates an interaction between STAT1 and the Mi-2/NuRD complex on GAS containing sequences.
Although Mi-2/NuRD complex members are known to interact with a variety of transcription factors during development (Bowen et al., 2004; Denslow and Wade, 2007), there is no precedent for their interaction with STAT transcription factors. However, it is possible that the Mi-2/NuRD complex has a low affinity interaction with STAT1 that is stabilized by TgIST, or that Mi-2/NuRD is only recruited to the STAT1 complex once bound by TgIST. To test the ability of TgIST to bind directly to the Mi-2/NuRD complex, we conducted co-IPs with a GAS oligo in U3A cells that lacked STAT1 vs. U3A cells expressing STAT1. Immunoprecipitation of TgIST in U3A-STAT1 cells confirmed that TgIST interacts with phospho-Y701-STAT1 (Figure 5A). In addition, TgIST IP’d a number of members of the Mi-2/NuRD complex both in the absence and presence STAT1 and IFN-γ, respectively (Figure 5A). To gain insights into the mechanistic role of the interaction between TgIST and the Mi-2/NuRD complex at active GAS sites we performed ChIP experiments in normal and IFN-γ-activated cells. Infection with T. gondii significantly reduced the acetylation status of histone H3 bound to GAS sequences in promoter regions of HLA-E and GBP1 in response to IFN-γ treatment, but not in the control gene GAPDH (Figure 5B, Table S5). The inhibition of H3 acetylation was dependent on TgIST and was reversed in cells infected with the knockout parasite (Figure 5B).
Figure 5. TgIST associates with members of the Mi2/NuRD complex independently of STAT1 and IFN-γ activation.
(A) Western blot analysis of TgIST-Ty IP from Toxoplasma type I (RH) infected U3A (STAT1 null) or U3A-STAT1 (STAT1+) cells that were either left naive (−) or activated (+) with IFN-γ (100 U/ml) for the final 30 min of infection. Nuclear extracts were precipitated using the GAS oligo, resolved by SDS-PAGE, blotted with primary antibodies shown to the right and imaged with LI-COR specific secondary antibodies. Representative of three or more experiments with similar outcomes.
(B) ChIP of acetylated Histone H3 of HFF cells infected for 3 hr with type I (RH) wild type (red) and TgIST knockout (blue) ± IFN-γ (100 U/ml) for 2 hr. Data analyzed by qPCR with primers for GAS sites in promoter elements of genes shown. GAPDH served as control. Means ± S.D., n=6, *P < 0.05 and **P < 0.001. Student’s t-test.
TgIST is an important component of virulence due to enhanced survival in IFN-γ activated macrophages
IFN-γ is a critical component of innate and adaptive immune defense against T. gondii (Suzuki et al., 1989; Yap and Sher, 1999) and as such the ability to block IFN-γ signaling is likely important for parasite survival. To examine the role of TgIST in virulence, we compared parasite survival in RAW macrophages activated with IFN-γ in vitro. The absence of TgIST led to enhanced clearance and reduced growth of type II (Pru) parasites in cells that were infected first and then activated with IFN-γ (Figure 6 A, B), but not when cells were pre-activated with IFN-γ, nor in naïve cells (Figure S5 A–D). These findings are consistent with TgIST having to act prior to the host cell receiving the IFN-γ signal, and being distinct from effectors that act downstream of activation. Moreover, loss of TgIST in type II (Pru) strain parasites decreased virulence in C57BL/6 mice (Figure 6 C, D, Figure S5 E, F). This defect was evident from failure of the knockout to disseminate beyond the initial site of infection, a greatly decreased tissue burden, and the survival of all infected mice. The decreased virulence of the knockout was restored in IFN-γ-receptor 1 deficient mice (Ifngr1−/−) (Figure 6D), indicating it was not intrinsically impaired in vivo. Collectively these findings indicate that the blocking of IFN-γmediated gene expression by TgIST plays an important role in pathogenesis in vivo.
Figure 6. Deletion of TgIST leads to attenuation in vivo and enhanced susceptibility in IFN-γ activated macrophages.
(A) Survival of type II (Pru) parasites in IFN-γ-activated RAW 264.7 macrophages comparing percentage of cells infected at 24 hr vs. 30 min. (B) Growth of type II (Pru) parasites in IFN-γ-activated RAW 264.7 macrophages at 36 hr. Cells were infected and then activated with IFN-γ (200 U/ml). Mean ± SEM (n = 3 experiments, each with 3 technical replicates, n=9). *P < 0.05 and **P < 0.001, Kruskal-Wallis with Dunn’s correction. See also Figure S5 A–D.
(C) Comparison of C57/BL6 mice infected with 1,000 tachyzoites i.p. of type II (Pru strain) parasites by bioluminescence imaging. Cumulative results of 2 experiments with 5 mice each group (n = 10) shown. *P ≤ 0.05 between wild type, complement or knockout. Representative mice shown in the inset. See also Figure S5 E, F.
(D) Survival curve of mice challenged with type II (Pru) wild type, TgIST knockout, or complemented parasites.
Discussion
Our findings reveal the molecular basis for the ability of T. gondii to block IFN-γ signaling by identifying the effector TgIST that is secreted by the parasite and which binds to activated STAT1 dimers in the nucleus of IFN-γ treated cells. TgIST also binds to the Mi-2/NuRD complex and recruits it to activated STAT1 dimers bound to GAS sites. The formation of this TgIST-Mi2/NuRD-STAT1 complex explains the slower migration of STAT1-containing GAF complexes detected by EMSA gels, as previously reported (Lang et al., 2012; Schneider et al., 2013) and also observed here. Collectively, our findings suggest that TgIST recruits the Mi-2/NuRD to STAT1 on GAS sites of its cognate regulated genes in IFN-γ activated cells thus blocking transcription of IFN-stimulated genes (ISGs).
Using genetic depletion studies, TgIST was shown to be essential for the interaction between STAT1 and Mi-2/NuRD, and for the repression of IFN-γ-dependent gene expression. Moreover, expression of TgIST in HeLa cells blocked STAT1-induced expression of IRF1. Hence, TgIST is both necessary and sufficient to block IFN-γ-mediated gene expression. Furthermore, we show that the function of TgIST is conserved among different T. gondii strains, consistent with their ability to block STAT1-mediated transcription (Kim et al., 2007; Rosowski and Saeij, 2012). Further studies using ChIP will be necessary to establish the complete occupancy of TgIST on STAT1-dependent promoters. However, the consistent effect on the ISGs that we tested here suggests that TgIST mediates the widespread disruption of IFN-γ-dependent gene expression described previously (Kim et al., 2007; Lang et al., 2012).
Our studies also provide direct evidence that the ability of T. gondii to block STAT1-mediated transcription is important for parasite survival. Although TgIST was not required for normal growth, when infected cells were treated with IFN-γ after infection, type II TgIST deletion mutants were significantly more susceptible to clearance. As well, type II TgIST mutants were also highly attenuated in the mouse model, likely as a result of impaired ability to survive in macrophages and other cells where IFN-γ is important for control. In contrast, type I mutants of TgIST did not show attenuation in vivo (unpublished data), presumably because they possess potent pathways for blocking downstream mediators of IFN-γ (Hunter and Sibley, 2012). Nonetheless, TgIST may act in concert with other mediators to enhance the virulence of type I strains. Importantly the function of TgIST requires that it is delivered into the host cell prior to activation by IFN-γ, in contrast to a variety of other effectors that block downstream immune mediators once they are turned on (Hunter and Sibley, 2012). Hence, TgIST may be especially important for parasite infection of naïve cells during the acute dissemination phase into tissues, or sites of immune privilege, where immune responses may be delayed.
TgIST is found is secreted into the parasitophorous vacuole and then traffics out of the vacuolar space to ultimately reside in the host cell nucleus. This cellular distribution of TgIST is similar to other recently described T. gondii effectors such as GRA16 and GRA24 that affect p38 MAPK signaling (Braun et al., 2013) and p53 (Bougdour et al., 2013), respectively. TgIST contains features that suggest it might be processed by the aspartic protease TgASP5, which has recently been shown to be important for export of a number of exported dense granule effectors (Curt-Varesano et al., 2015; Hammoudi et al., 2015). Moreover, the absence of the parasite effector MYR1, a distinct protein involved in export of dense granule effectors (Franco et al., 2016), phenocopies the defect of TgIST in blocking IRF1 induction by IFN-γ, suggesting it may affect export of TgIST. Defining the domains of TgIST that are involved in export and trafficking to the nucleus may be useful for future studies designed to block the delivery of effectors like TgIST, thereby restoring normal host cell function.
Our findings reveal a complex of Mi-2/NuRD in association with STAT1, an interaction mediated by the effector TgIST. The Mi-2/NuRD complex is most often associated with repression of transcription during development, and not normally involved in STAT-mediated transcription (Bowen et al., 2004; Denslow and Wade, 2007). Nonetheless, this interaction suggests several possible mechanisms to explain the block in STAT1 transcription. For example, the HDAC activity in the Mi-2/NuRD complex may underlie the previous observation that T. gondii infection blocks IFN-γ-induced acetylation of core histones in the CIITA promoter (Lang et al., 2012), and our findings also corroborate this pattern for the genes HLA-E and GBP1 genes. Hence, deacetylation via the activities of HDAC1 and HDAC2 may be important in the reduced induction of STAT1-regulated genes, consistent with previous reports that HDAC inhibitors reverse this phenotype (Lang et al., 2012). However, it has also been argued that IRF1 is not dependent in acetylation status, but rather the enhanced association of STAT1 with chromatin prevents normal recycling and dampens expression (Rosowski et al., 2014). Thus, the tight binding of the Mi-2/NuRD may prevent recycling by inhibiting normal de-phosphorylation of STAT1, a process that is needed for maximum expression (Reich and Liu, 2006). The Mi2-/NuRD complex also contains CHD3 and CHD4 ATPases implicated in nucleosome remodeling and these domains are known to associate with co-repressors (Bowen et al., 2004; Denslow and Wade, 2007). Finally, the presence of a large and highly altered STAT1 complex on the chromatin may displace normal co-activators such as CBP/p300 and BRG1 that are important for activation (Ni et al., 2005; Ramana et al., 2000). It is interesting to note that inhibition of STAT1 by T. gondii infection can be reversed by over-expression of some co-activators, including the recently described orphan nuclear receptor TLX (Beiting et al., 2015). As such, it may be possible to reverse the action of TgIST by disrupting its interaction with either STAT1 or Mi-2/NuRD or by augmenting other activating factors such as p300/CBP (Zhang et al., 2005) and other histone modifiers (e.g., PARP9-DTX3L ubiquitin ligase) that increase ISG expression (Zhang et al., 2015).
Given the importance of epigenetic modification, it is not surprising that microbes have found a variety of ways to alter chromatin and affect host gene expression (Silmon de Monerri and Kim, 2014). TgIST represents an example of a pathogen hijacking the Mi-2/NuRD complex to regulate host gene expression and dampen immunity, although there are several examples of viral proteins interacting with Mi-2/NuRD to regulate their own gene expression (Cismasiu et al., 2008; Dembowski and DeLuca, 2015; Terhune et al., 2010). In contrast, TgIST acts by recruitment of the chromatin-modifying Mi-2/NuRD complex to STAT1 transcriptional complexes, resulting in chromatin remodeling and decreased ISG expression. TgIST also underlies the previously described defect in type I IFN signaling (Rosowski et al., 2014) that is mediated by STAT1/STAT2 heterodimers (unpublished data). Moreover, the tethering of chromosomal modifying complexes like Mi-2/NuRD, which participates in regulation of a variety of pathways (Bowen et al., 2004; Denslow and Wade, 2007), to transcription factors may be a general mechanism employed by pathogens to manipulate host gene expression to their advantage.
EXPERIMENTAL PROCEDURES
In Vivo Infections Studies
Animals were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care approved facility and the Institutional Care Committee at Washington University in St. Louis, School of Medicine, approved all protocols. Female outbred CD-1 (Charles River), C57BL/6, and IFN-γ receptor 1 knockout mice (Ifngr11-1) (Jackson Laboratories) (8–10 weeks) (n=5 per group) were challenged by intraperitoneal (ip) injection of 1,000 T. gondii tachyzoites. Cumulative mortality was determined at 30 days post injection.
Bioluminescence Imaging
C57BL/6 mice infected i.p. with 103 luciferase expressing parasites and imaged day 0 through day 10 using a Xenogen IVIS200 machine and analyzed with the Xenogen Living Image software (Caliper Life Sciences). Mice were anaesthetized with 2% isoflurane and injected ip with D-luciferin (Biosynth AG) (150 mg/kg), prior to imaging.
Parasite and Cell Culture
T. gondii tachyzoites were serially passaged in human foreskin fibroblast (HFF) monolayers. Strains used in this study are listed in Table S2. Gene disruptants and complemented lines were generated using CRISPR/Cas9 (Shen et al., 2014), as described in the Supplemental materials. Stable transgenic parasites were selected in mycophenolic acid (25 μg/ml) and xanthine (50 μg/ml) (MPA/Xa) or pyrimethamine (Pyr) (3 mM) (Sigma). Cultures were negative for Mycoplasma contamination using the e-Myco plus mycoplasma PCR detection kit (Boca Scientific). U3A and U3A-STAT1 cells (Zhang et al., 2005), HeLa cells (ATCC CCL-2), 3T3 cells (ATCC CCL-163) and RAW 264.7 mouse macrophages (ATCC TIB-71) were maintained in complete media without gentamicin at 37°C in 5% CO2.
Construction of HaloTag reporter
Primers used for construction of plasmids and testing transgenic parasites are listed in Table S3 and plasmids are listed in Table S4. The CMV promoter of the pHTN HaloTag CMV-neo vector was replaced by a minimal (mCMV) promoter fused with five tandem GAS sequences (5′-AGTTTCATATTACTCTAAATC-3′) and the construct was transfected with Lipofectamine 2000 (Life Technologies) into mouse 3T3 cells, selected with 600 μg/ml G418 (Sigma) and single clonal colonies were tested for response to 100 U/ml IFN-γ.
Flow cytometry
The stable HaloTag reporter 3T3 cell line was either left uninfected or infected with parasites for 3 hr followed by activation with 100 U/ml IFN-γ for the final 15 hr. Cells were trypsinized, fixed with 4% formaldehyde, and analyzed using an iCyt Synergy cell sorter (Sony Biotech) and FlowJo software (Tree star).
Reverse transcription quantitative PCR (RT-qPCR)
HeLa cells were either left uninfected or were infected with parasites for 2 hr followed by activation with 100 U/ml IFN-γ for 4 hr. RNA was purified using the RNeasy Mini Kit (Qiagen), and reverse transcribed using the iScript Reverse Transcription Supermix (Biorad). Primers for references genes (Table S5) were analyzed in triplicate using SYBR Green PCR Master Mix (Applied Biosystems) and the CFX96 Real-Time PCR Detection System (Biorad), and analyzed as described in the Supplemental materials.
Immunofluorescence Assays
Infected cells were fixed with 4% formaldehyde in PBS for 10 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked in 10% FBS for 20 min. Samples were incubated with primary antibodies (Table S6), washed in PBS, and incubated with secondary antibodies (goat anti-mouse IgG or goat anti-rabbit IgG) conjugated to Alexa fluor dyes (Invitrogen). Slides were mounted with Prolong Gold with DAPI (Life technologies). Samples were examined with a Zeiss Axioskop 2 MOT Plus epifluorescence microscope equipped with an AxioCam MRm camera (Carl Zeiss, Inc.,) and images acquired using Axiovision v4.6. Images were processed with Photoshop CS4. For confocal analysis, samples were examined with a Zeiss LSM 880 Confocal Laser Scanning Microscope with Airyscan using Z-Sectioning and further analyzed with ZEN software. For immunoEM, samples were processed as described in the Supplemental materials.
Chromatin Immunoprecipitation (ChIP)
Cells were left uninfected or infected with parasites for 3 hr followed by activation with 100 U/ml IFN-γ for 30 min. Cells were cross-linked with 1% formaldehyde for 10 min, stopped by adding 0.125 M glycine for 5 min, washed twice with cold PBS and resupended in 0.5% SDS cell lysis buffer (Majumder et al., 2015). Lysates were sonicated using a Diagenode Biorupter and immunoprecipitated using antibodies (Table S6) and complexes isolated with protein G magnetic Dynabeads (Life technologies). Following elution, chromatin-antibody complexes and input DNA were reverse crosslinked overnight at 65°C. The DNA was purified after proteinase K treatment using a DNA purification kit (Qiagen) and analyzed by qPCR using SYBR Green (Life Technologies) on the CFX96 Real-Time PCR Detection System (Biorad) and primer combinations shown in Table S5.
Electromobility Shift Assay (EMSA)
Infected or control cells were cultured for 20 hr followed by activation with 100 U/ml IFN-γ for the final 30 min. Nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction kit (Life Technologies). Sense and antisense DNA strand probes to the GAS sequence in the Fc gamma receptor were labeled at the 5′ end with LICOR IRDye 700 fluorophores (Integrated DNA Technologies Inc.,) and annealed to produce double stranded DNA probes. Probes (5 pmoles/reaction) were combined with nuclear extracts in 10 mM Tris pH 7.5, 100 mM KCl, 3.5 mM DTT, 0.25% Tween20, Poly (dI:dC) 50 μg/ml, and 10 mM EDTA (EMSA buffer). For supershift assays antibodies (Table S6) were added at 2 μg/μl final. Samples were diluted with LICOR 10X Orange Loading Dye, resolved by agarose gel electrophoresis and visualized using the ODYSSEY infrared imager (LI-COR Biosciences).
Biotinylated GAS Oligo Pull-Down
Samples were prepared as for EMSA only the GAS oligo (from Fc gamma receptor) was labeled at the 5′ end with biotin (Integrated DNA Technologies Inc.,). Annealed oligos were incubated for 4 hr at 4°C with streptavidin magnetic beads (Thermo Scientific) followed by washing with PBS. Nuclear extracts were incubated with Poly (dI:dC) 25 μg/ml for 30 min at 4°C, then incubated with oligo-coated beads overnight at 4°C. Beads were washed with PBS, samples were eluted using denaturing SDS-PAGE sample buffer, resolved on 8–16% acrylamide gels, and Western blotted using antibodies (Table S6).
Cell Transfection
For heterologous expression, the coding sequence of TgIST from the GT1 strain of T. gondii was cloned in frame with GFP or the Ty-epitope and placed into the pcDNA™4/TO Mammalian Expression Vector (Life Technologies) using a CMV promoter. The plasmid (500 ng) was transfected using the Lipofectamine LTX Reagent (Life Technologies) according to the manufacturers protocol into HeLa or HEK293T cells.
Clearance and Growth Restriction Assay
RAW 264.7 mouse macrophages were infected with GFP-expressing type II Pru parasites. Macrophages were either pre-activated with IFN-γ (200 U/ml) (R&D Systems) and LPS (10 ng/ml) derived from E. coli 055:B5 (Sigma) for 16 hr prior to infection, activated with IFN-γ (200 U/ml) 3 hr post-infection or were left untreated. RAW cells were challenged with parasites in complete medium at 37°C 5% CO2 for 10 min, washed and fixed with 4% formaldehyde either at 30 min or 24 hr for the clearance assay, or 36 hr for the growth restriction assay. The percentage of infected cells remaining, and average vacuole area, were assessed by imaging of GFP (parasite) and host cell nuclei (Hoechst dye) on the Cytation 3 Cell Imaging Multi-Mode Reader (BioTek). Experiments were repeated three times with 2 to 3 technical replicates per experiment.
Immunoprecipitation (IP) and Western Blotting
U3A-STAT1 cells were either left uninfected or were infected for 16 hr followed by activation with 100 U/ml IFN-γ for 30 min. Nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagent kit (ThermoFisher) on ice at 4°C. Anti-Flag bound agarose (Sigma) beads or anti-Ty mAb BB2 bound to Protein G Dynabeads (Life Technologies) beads were precleared and incubated with cell lysates. Beads were washed and samples were eluted using denaturing SDS-PAGE sample buffer, resolved on 8–16% acrylamide gels, and Western blotted. Blots were probed with primary antibodies (Table S6) incubated in 3% milk in PBS for 1 hr, washed in PBS-Tween (0.05%), incubated with secondary antibodies conjugated to IRDye 680CW (goat anti-rabbit or anti-mouse IgG) or IRDye 800CW (goat anti-rabbit or anti-mouse IgG) (LI-COR Biosciences) and signals detected using the ODYSSEY infrared imager (LI-COR Biosciences).
MS Analyses
U3A-STAT1 cells were left uninfected or infected with the T. gondii type I strain RH and subsequently activated by IFN-γ (100 U/ml, 3 hr). Immunoprecipitations of STAT1-FLAG from U3A-STAT1 cells were eluted from beads with RapiGest (Waters), trypsinized and peptides purified on C18 columns. Peptides were analyzed using an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) coupled with a nanoLC Ultra (Eksigent). Data was processed using Mascot Distiller v2.2 and searched using Mascot Daemon 2.2 (Matrix Science). Searches were performed against the Toxoplasma gondii database (http://ToxoDB v10; 8345 entries) and NCBI nr database (March 2014; 276756 human protein entries). MS spectra were submitted to Scaffold 4.3 (Proteome Software) to validate identifications.
Statistical Analyses
Statistical analyses were conducted with Prism 5 (GraphPad) and the test for significance for groups of three or more was performed using the nonparametric one-way analysis of variance Kruskal-Wallis test with Dunn’s post-test for correction. Groups of two were analyzed using the unpaired Student’s t-test. *P ≤ 0.05 was considered the minimum cutoff for significance.
Supplementary Material
Acknowledgments
We are grateful to Jenifer Barks for cell culture, Qiuling Wang for assistance with animal studies, Bryan Anthony for assistance with confocal microscopy, Wandy Beatty for immunoEM studies, and Kinjal Majumder and Patrick Collins for advice on ChIP studies. Supported in part by grants from the National Institutes of Health (AI 118426 and AI 070489). P.O. was supported by a fellowship from the Leopoldina Foundation (LPDS 2012-10).
Footnotes
Author Contributions
P.O. and R.D.E. designed and performed the experiments, analyzed the data, and generated the figures. Y.Z. and M.J.H provided key biological materials and insight into experimental design. L.D.S. wrote the manuscript with input from all authors.
The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beiting DP, Hidano S, Baggs JE, Geskes JM, Fang Q, Wherry EJ, Hunter CA, Roos DS, Cherry S. The Orphan Nuclear Receptor TLX Is an Enhancer of STAT1-Mediated Transcription and Immunity to Toxoplasma gondii. PLoS Biol. 2015;13:e1002200. doi: 10.1371/journal.pbio.1002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Durandau E, Brenier-Pinchart MP, Ortet P, Barakat M, Kieffer S, Curt-Varesano A, Curt-Bertini RL, Bastien O, Coute Y, et al. Host cell subversion by Toxoplasma GRA16, an exported dense granule protein that targets the host cell nucleus and alters gene expression. Cell Host Microbe. 2013;13:489–500. doi: 10.1016/j.chom.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Braun L, Brenier-Pinchart MP, Yogavel M, Curt-Varesano A, Curt-Bertini RL, Hussain T, Kieffer-Jaquinod S, Coute Y, Pelloux H, Tardieux I, et al. A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation. J Exp Med. 2013;210:2071–2086. doi: 10.1084/jem.20130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu VB, Paskaleva E, Suman Daya S, Canki M, Duus K, Avram D. BCL11B is a general transcriptional repressor of the HIV-1 long terminal repeat in T lymphocytes through recruitment of the NuRD complex. Virology. 2008;380:173–181. doi: 10.1016/j.virol.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curt-Varesano A, Braun L, Ranquet C, Hakimi MA, Bougdour A. The aspartyl protease TgASP5 mediates the export of the Toxoplasma GRA16 and GRA24 effectors into host cells. Cell Microbiol. 2015 doi: 10.1111/cmi.12498. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Deckert-Schlüter M, Rang A, Weiner D, Huang S, Wiestler OD, Hof H, Schlüter D. Interferon-g receptor-deficiency renders mice highly susceptible to toxoplasmosis by decreased macrophage activation. Laboratory Investigation. 1996;75:827–841. [PubMed] [Google Scholar]
- Dembowski JA, DeLuca NA. Selective recruitment of nuclear factors to productively replicating herpes simplex virus genomes. PLoS Pathog. 2015;11:e1004939. doi: 10.1371/journal.ppat.1004939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Franco M, Panas MW, Marino ND, Lee MC, Buchholz KR, Kelly FD, Bednarski JJ, Sleckman BP, Pourmand N, Boothroyd JC. A Novel Secreted Protein, MYR1, Is Central to Toxoplasma’s Manipulation of Host Cells. MBio. 2016;7 doi: 10.1128/mBio.02231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilescu LC, Butcher BA, Del Rio L, Taylor GA, Denkers EY. STAT1 is essential for antimicrobial effector function but dispensable for gamma interferon production during Toxoplasma gondii infection. Infect Immun. 2004;72:1257–1264. doi: 10.1128/IAI.72.3.1257-1264.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzinelli R, Wysocka M, Hayashi S, Denkers E, Hieny S, Caspar P, Trinchieri G, Sher A. Parasite-induced IL-12 stimulates early INF-gamma synthesis and resistance during acute infection with Toxoplasma gondii. Journal of Immunology. 1994;153:2533–2543. [PubMed] [Google Scholar]
- Hammoudi PM, Jacot D, Mueller C, Di Cristina M, Dogga SK, Marq JB, Romano J, Tosetti N, Dubrot J, Emre Y, et al. Fundamental Roles of the Golgi-Associated Toxoplasma Aspartyl Protease, ASP5, at the Host-Parasite Interface. PLoS Pathog. 2015;11:e1005211. doi: 10.1371/journal.ppat.1005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao CH, Luisa Hiller N, Haldar K, Knoll LJ. A HT/PEXEL motif in Toxoplasma dense granule proteins is a signal for protein cleavage but not export into the host cell. Traffic. 2013;14:519–531. doi: 10.1111/tra.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CA, Sibley LD. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat Rev Microbiol. 2012;10:766–778. doi: 10.1038/nrmicro2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Fouts AE, Boothroyd JC. Toxoplasma gondii dysregulates IFN-g inducible gene expression in human fibroblasts: insights from a genome-wide transcriptional profiling. J Immunol. 2007;178:5154–5165. doi: 10.4049/jimmunol.178.8.5154. [DOI] [PubMed] [Google Scholar]
- Lang C, Hildebrandt A, Brand F, Opitz L, Dihazi H, Luder CG. Impaired chromatin remodelling at STAT1-regulated promoters leads to global unresponsiveness of Toxoplasma gondii-infected macrophages to IFN-gamma. PLoS Pathog. 2012;8:e1002483. doi: 10.1371/journal.ppat.1002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman LA, Banica M, Reiner SL, Hunter CA. STAT1 plays a critical role in the regulation of antimicrobial effector mechanisms, but not in the development of Th1-type responses during toxoplasmosis. J Immunol. 2004;172:457–463. doi: 10.4049/jimmunol.172.1.457. [DOI] [PubMed] [Google Scholar]
- Luder CG, Algner M, Lang C, Bleicher N, Gross U. Reduced expression of the inducible nitric oxide synthase after infection with Toxoplasma gondii facilitates parasite replication in activated murine macrophages. Int J Parasitol. 2003;33:833–844. doi: 10.1016/s0020-7519(03)00092-4. [DOI] [PubMed] [Google Scholar]
- Lüder CG, Walter W, Beuerle B, Maeurer MJ, Gross U. Toxoplasma gondii down-regulates MHC class II gene expression and antigen presentation by murine macrophages via interference with nuclear translocation of STAT1a. Eur J Immunol. 2001;31:1475–1484. doi: 10.1002/1521-4141(200105)31:5<1475::AID-IMMU1475>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Lüder CGK, Lang T, Beuerle B, Gross U. Down-regulation of MHC class II molecules and inability to up-regulate class I molecules in murine macrophages after infection with Toxoplasma gondii. Clinical and Experimental Immunology. 1998;112:308–316. doi: 10.1046/j.1365-2249.1998.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder K, Koues OI, Chan EA, Kyle KE, Horowitz JE, Yang-Iott K, Bassing CH, Taniuchi I, Krangel MS, Oltz EM. Lineage-specific compaction of Tcrb requires a chromatin barrier to protect the function of a long-range tethering element. J Exp Med. 2015;212:107–120. doi: 10.1084/jem.20141479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar I, Fagard R. STAT1 and pathogens, not a friendly relationship. Biochimie. 2010;92:425–444. doi: 10.1016/j.biochi.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan CF, Murray HW, Weibe ME, Rubin BY. Indentification of interferon gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med. 1983;158:670–689. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Karaskov E, Yu T, Callaghan SM, Der S, Park DS, Xu Z, Pattenden SG, Bremner R. Apical role for BRG1 in cytokine-induced promoter assembly. Proc Natl Acad Sci U S A. 2005;102:14611–14616. doi: 10.1073/pnas.0503070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn ER. Interferon-gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host to degrade tryptophan. Proc Natl Acad Sci (USA) 1984;81:908–912. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana CV, Chatterjee-Kishore M, Nguyen H, Stark GR. Complex roles of Stat1 in regulating gene expression. Oncogene. 2000;19:2619–2627. doi: 10.1038/sj.onc.1203525. [DOI] [PubMed] [Google Scholar]
- Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol. 2006;6:602–612. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- Rosowski EE, Nguyen QP, Camejo A, Spooner E, Saeij JP. Toxoplasma gondii Inhibits gamma interferon (IFN-gamma)- and IFN-beta-induced host cell STAT1 transcriptional activity by increasing the association of STAT1 with DNA. Infect Immun. 2014;82:706–719. doi: 10.1128/IAI.01291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosowski EE, Saeij JP. Toxoplasma gondii clonal strains all inhibit STAT1 transcriptional activity but polymorphic effectors differentially modulate IFNgamma induced gene expression and STAT1 phosphorylation. PLoS One. 2012;7:e51448. doi: 10.1371/journal.pone.0051448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Darnell JE., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- Schneider AG, Abi Abdallah DS, Butcher BA, Denkers EY. Toxoplasma gondii triggers phosphorylation and nuclear translocation of dendritic cell STAT1 while simultaneously blocking IFNgamma-induced STAT1 transcriptional activity. PLoS One. 2013;8:e60215. doi: 10.1371/journal.pone.0060215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Brown KM, Lee TD, Sibley LD. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio. 2014;5(3):e01114–14. doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- Silmon de Monerri NC, Kim K. Pathogens hijack the epigenome: a new twist on host-pathogen interactions. The American journal of pathology. 2014;184:897–911. doi: 10.1016/j.ajpath.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuehr D, Gross S, Levi R, Nathan C. Activated murine macrophages secrete a metabolite of arginine with the bioactivity of endothelial-derived relaxing factor and the chemical reactivity of nitric oxide. Journal of Experimental Medicine. 1989;169:1011. doi: 10.1084/jem.169.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Conley FK, Remington JS. Importance of endogenous IFN-gamma for prevention of toxoplasmic encephalitis in mice. Journal of Immunology. 1989;143:2045–2050. [PubMed] [Google Scholar]
- Taylor GA, Feng CG, Sher A. Control of IFN-gamma-mediated host resistance to intracellular pathogens by immunity-related GTPases (p47 GTPases) Microb Infect. 2007;9:1644–1651. doi: 10.1016/j.micinf.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Terhune SS, Moorman NJ, Cristea IM, Savaryn JP, Cuevas-Bennett C, Rout MP, Chait BT, Shenk T. Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA. PLoS Pathog. 2010;6:e1000965. doi: 10.1371/journal.ppat.1000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap GS, Sher A. Effector cells of both nonhemopoietic and hemopoietic origin are required for interferon (IFN)-gamma- and tumor necrosis factor (TNF)-alpha- dependent host resistance to the intracellular pathogen, Toxoplasma gondii. J Exp Med. 1999;189:1083–1091. doi: 10.1084/jem.189.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mao D, Roswit WT, Jin X, Patel AC, Patel DA, Agapov E, Wang Z, Tidwell RM, Atkinson JJ, et al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat Immunol. 2015;16:1215–1227. doi: 10.1038/ni.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Takami K, Lo MS, Huang G, Yu Q, Roswit WT, Holtzman MJ. Modification of the Stat1 SH2 domain broadly improves interferon efficacy in proportion to p300/CREB-binding protein coactivator recruitment. J Biol Chem. 2005;280:34306–34315. doi: 10.1074/jbc.M503263200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.