Abstract
To determine the effects of single nucleotide polymorphisms (SNPs) identified in a genome-wide association study of progressive supranuclear palsy (PSP), we tested their association with brain gene expression, CpG methylation and neuropathology. In 175 autopsied PSP subjects, we performed associations between seven PSP risk variants and temporal cortex levels of 20 genes in-cis, within ±100 kb. Methylation measures were collected using reduced representation bisulfite sequencing in 43 PSP brains. To determine whether SNP/expression associations are due to epigenetic modifications, CpG methylation levels of associated genes were tested against relevant variants. Quantitative neuropathology endophenotypes were tested for SNP associations in 422 PSP subjects. Brain levels of LRRC37A4 and ARL17B were associated with rs8070723; MOBP with rs1768208 and both ARL17A and ARL17B with rs242557. Expression associations for LRRC37A4 and MOBP were available in an additional 100 PSP subjects. Meta-analysis revealed highly significant associations for PSP risk alleles of rs8070723 and rs1768208 with higher LRRC37A4 and MOBP brain levels, respectively. Methylation levels of one CpG in the 3′ region of ARL17B associated with rs242557 and rs8070723. Additionally, methylation levels of an intronic ARL17A CpG associated with rs242557 and that of an intronic MOBP CpG with rs1768208. MAPT and MOBP region risk alleles also associated with higher levels of neuropathology. Strongest associations were observed for rs242557/coiled bodies and tufted astrocytes; and for rs1768208/coiled bodies and tau threads. These findings suggest that PSP variants at MAPT and MOBP loci may confer PSP risk via influencing gene expression and tau neuropathology. MOBP, LRRC37A4, ARL17A and ARL17B warrant further assessment as candidate PSP risk genes. Our findings have implications for the mechanism of action of variants at some of the top PSP risk loci.
Introduction
Progressive supranuclear palsy (PSP) is a neurodegenerative parkinsonian disorder pathologically characterized by the presence of neurofibrillary tangles and tau-positive glial lesions referred to as tufted astrocytes and oligodendroglial coiled bodies[12]. Although PSP is largely considered a sporadic disorder, families with an autosomal dominant pattern of inheritance have been reported[38]. Furthermore PSP patients are more likely to report a family history of parkinsonism or dementia, implicating a genetic component to this disease[13,20]. Indeed, a genome-wide association study (GWAS) of a PSP case-control cohort identified common variants at four loci that associate with disease risk significantly and three others with suggestive association[19]. The most significant of these are the H1 haplotype (tagged by rs8070723) on chromosome 17q21 that encompasses the tau encoding MAPT gene, and a variant of the H1 haplotype termed H1c (partially tagged by rs242557)[33].
While case-control disease association studies have been successful for identifying novel disease risk variants, frequently the common variants that mark the association at these loci are located within introns or in intergenic regions. Consequently, neither the identity of the disease gene nor the disease mechanism of the risk variant(s) are immediately clear. Assessment of disease relevant endophenotypes can provide further insights into the putative function of the risk variants for neurodegenerative diseases (reviewed[15]).
We previously determined that common variants at 3 of the 7 PSP risk loci associate with brain expression of nearby genes (rs1768208(A)-MOBP, rs11568563(C)-SLCO1A2, rs242557(A)-MAPT and rs8070723(G)-MAPT)[48,1]. Others have reported similar findings, including association of the STX6 locus SNP rs1411478(A) with lower STX6 expression levels in white matter[16] and MAPT locus risk variants with altered MAPT brain expression[29,36]. Collectively these findings implicate transcriptional regulation as a plausible molecular mechanism for common PSP risk variants and importantly indicate the likely impacted gene.
Gene expression can also be influenced by epigenetic modifications, such as DNA methylation changes, which have been implicated in the etiology of multiple neurodegenerative diseases including Alzheimer’s[9], Parkinson’s[8,22] and Huntington’s disease[30]. Most recently association of DNA methylation levels with the PSP risk haplotype (H1) has been reported, adding PSP to the growing list of neurodegenerative disorders that may be affected by epigenetic mechanisms[25].
Here we evaluate association of the PSP risk GWAS variants with three relevant endophenotypes. We previously assessed these variants for association with brain gene expression in 100 PSP autopsied-confirmed cases. Herein, we expand this assessment to an independent cohort of 175 additional PSP subjects. We pursue the variants implicated in our expression analysis, for association with DNA methylation as a putative disease mechanism. Finally we combine this analysis with an assessment of PSP neuropathological measures to determine the specific neuropathological features most influenced by these variants. This combined endophenotype approach can provide valuable additional insights into disease mechanisms affected by these variants that can inform future studies aimed at therapeutic interventions.
Methods
Subjects and samples
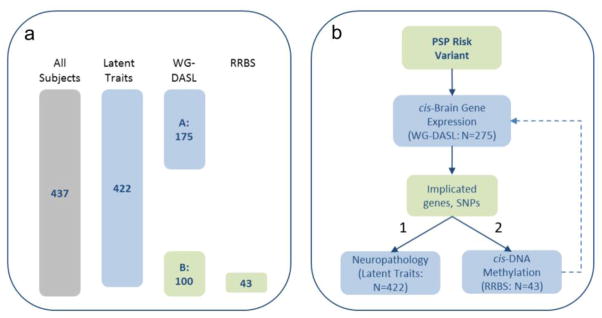
All brain samples were obtained from the Mayo Clinic Brain Bank and received a pathological diagnosis of PSP at autopsy[18]. Following quality control (QC) described below, 437 PSP subjects were evaluated in this study, all of whom had genome-wide genotypes and at least one of the three investigated endophenotype measures: gene expression, DNA methylation and/or neuropathology latent traits. For expression analysis, samples fall into one of two cohorts: Cohort A comprises 175 PSP temporal cortex (TCX) samples that have not previously been assessed for eQTL. Cohort B comprises 100 PSP TCX samples that were included in our prior gene expression study[8], 43 of whom also have DNA methylation data. A total of 422 PSP subjects have quantitative neuropathology measures transformed into latent traits as described below. All subjects in expression cohort A, 71/100 subjects in cohort B and 32/43 from the subset with DNA methylation measures have latent traits (Figure. 1). Demographics for subjects utilized in the various endophenotype analyses are shown in Table. 1. This study was approved by the appropriate institutional review board.
Figure 1. Outline of analyses.
a. Number of subjects within each of the analyses. b. Analytic paradigm. Endophenotypes of brain gene expression measures, neuropathology latent traits and DNA CpG methylation levels were evaluated for association with PSP risk GWAS index SNPs (primary analysis, solid arrows). Correlations between brain gene expression and DNA methylation were also assessed when relevant (secondary analysis, dashed arrows).
Table 1.
Sample demographics.
| Phenotye | Approach | N | Females (%) | Age at death (SD) | Mean RIN (SD) | Genotyping Array |
|---|---|---|---|---|---|---|
| Neuropathology | Latent Traits | 422 | 210 (50%) | 75.0 (7.7) | NA | Illumina 660W-Quad Infiniumac |
| Gene Expression | WG-DASL | 175a | 78 (45%) | 73.4 (7.2) | 7.35 (0.83) | Illumina 660W-Quad Infiniumc |
| Gene Expression | WG-DASL | 100b | 41 (41%) | 71.8 (5.3) | 7.01 (1.04) | Illumina HumanHap 300d |
| DNA methylation | RRBS | 43b | 22 (51%) | 73.8 (4.9) | NA | Illumina HumanHap 300d |
WG-DASL = Whole genome cDNA-mediated Annealing, Selection, extension, and Ligation;; RRBS = reduced representation bisulfite sequencing;
= cohort A;
= cohort B;
= genotypes from PSP GWAS5;
= genotypes from Mayo Clinic AD GWAS18.
Genotyping
DNA was extracted from frozen post-mortem brain tissue. Genotypes for the 8 single nucleotide polymorphisms (SNPs) that represent the strongest variants at the 7 PSP risk GWAS loci assessed in this study (rs8070723, rs242557, rs1411478, rs7571971, rs1768208, rs6687758, rs2142991 and rs11568563) were extracted from two published genome-wide datasets[7,19] (Table. 2). Subjects in the latent trait cohort and expression cohort A were genotyped using Human 660W-Quad Infinium BeadChips as part of the PSP risk GWAS[19]. Subjects in expression cohort B, a subset of which also has DNA methylation measures, were genotyped using the HumanHap300-Duo Genotyping BeadChips as part of the Mayo Clinic AD risk GWAS, where the control group included PSP subjects[7]. QC for the Mayo Clinic AD GWAS is already published[7,8]. QC was performed for the 498 PSP subjects from the Mayo Clinic Brain Bank that were genotyped as part of the PSP risk GWAS[19]. Unless otherwise stated all QC procedures were carried out using PLINK[35]. All subjects met a QC threshold of >98% call rate. Twenty-two subjects were removed due to mismatched sex and 2 subjects were removed due to suspected contamination (pi-hat > 0.05 with >10 other subjects). Genotypes were available for 561,882 SNPs. We removed SNPs with a call rate <98% and a minor allele frequency (MAF)< 2%. 476 PSP cases and 526,351 SNPs remained. We did not implement a Hardy-Weinberg threshold due to all subjects being cases. We performed pruning of the dataset based on linkage disequilibrium (LD) between the SNPs (r2=0.5, 50 SNP windows) reducing the number of SNPs to 233,597 prior to running the EIGENSOFT package[32,34] to evaluate the cohort for population stratification. Two subjects were removed that were determined to be population outliers (>6 standard deviations (SD) from mean) after 5 iterations. Principal components were recalculated for the remaining 474 subjects. We removed an additional 37 subjects that were > 2 standard deviations (SD) from the mean based on the first two principal components. The first 4 principal components for the remaining 437 samples were retained for inclusion as covariates in regression analyses.
Table 2.
PSP risk GWAS loci.
| rs# | Chr | Position bp (hg19) | Major Allelea | Minor Allelea | MAFa | Location | Genes in cisb | PSP risk Associationc | |
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | ||||||||
| rs8070723 | 17q21 | 44,081,064 | A | G | 0.208 | Intron MAPT | MAPT, KANSL1, STH. CRHR1*, PLEKHM1*, LRRC37A4*, ARL17A*, ARL17B* | 0.18 (0.16–0.21) | 1.50E-116 |
| rs242557 | 17q21 | 44,019,712 | G | A | 0.407 | Intron MAPT | MAPT, KANSL1, STH. CRHR1*, PLEKHM1*, LRRC37A4*, ARL17A*, ARL17B* | 1.96 (1.82–2.13) | 4.20E-70 |
| rs1411478 | 1q25 | 180,962,282 | G | A | 0.473 | Intron STX6 | STX6, KIAA1614d, MR1, IER5 | 1.27 (1.18–1.35) | 2.30E-10 |
| rs7571971 | 2p11 | 88,895,351 | C | T | 0.267 | Intron EIF2AK3 | EIF2AK3, TEX37d, RPIA | 1.33 (1.23–1.45) | 3.20E-13 |
| rs1768208 | 3p22 | 39,523,003 | C | T | 0.295 | Intron MOBP | MOBP, SLC25A38, RPSA, SNORA6, SNORA62d | 1.39 (1.28–1.49) | 1.00E-16 |
| rs6687758 | 1q41 | 222,164,948 | A | G | 0.208 | Intergenic | no genes +/− 100kb | 1.27 (1.15–1.37) | 2.80E-07 |
| rs2142991 | 10q11 | 43,341,105 | A | G | 0.164 | 10kb 3′ of BMS1 | BMS1 | 0.77 (0.69–0.85) | 4.90E-07 |
| rs11568563 | 12p12 | 21,457,434 | A | C | 0.083 | Exon SLCO1A2 | SLCO1A2e, SLCO1B1f, IAPP | 1.47 (1.27–1.67) | 1.90E-07 |
Minor Allele based on frequency information in HapMap-CEU population phase 3.
cis defined as +/− 100kb from index SNP position.
PSP risk GWAS5 results from Stage1+2 analysis. Odds ratios (OR) and 95% confidence intervals (95%CI) are the inverse of published PSP risk GWAS results to show effects for minor alleles.
These genes did not have expression probes.
Probes for this gene either harbored a SNP or were below our expression threshold.
This gene had expression probes but did not meet our criteria of expression above background in >50% of subjects.
These genes are outside of the cis-boundary but given the extended LD of the MAPT locus, all genes within the inversion region were included.
Gene expression measures
For both expression cohorts, total RNA was extracted from frozen post-mortem TCX tissue, using the Ambion RNAqueous kit. RNA integrity number (RIN) was measured using the Agilent Technologies 2100 Bioanalyzer. A minimum RIN of 5.5 was required for inclusion in either cohort. The majority of samples (Cohort A: 85%; Cohort B: 65%) had a RIN of ≥ 6.5 (Online Resource Figure 1). Illumina Whole Genome DASL (WG-DASL) microarray (Illumina, San Diego, CA) was used to collect gene expression measures at the Mayo Clinic Medical Genome Facility. For expression cohort A, expression measures were collected for 192 PSP TCX samples using the WG-DASL HT assay which has 29,285 probes. RNA samples were randomized and seven samples were included in duplicate as biological replicates. Raw probe levels were exported from GenomeStudio software (Illumina Inc.). Lumi package (Bioconductor) was used for preprocessing with background subtraction, variance stabilizing transformation, quantile normalization and probe filtering[14]. Following background subtraction, principal components analysis (PCA) was performed, 1 outlier sample was identified and was removed from further analysis. Probes were annotated according to human genome build19 (GRCh37), and screened for the presence of common variants (>1%) within the probe sequence according to dbSNP138. Probes with such SNPs within their sequence were removed from further analyses. For probes with nominally significant association results, probe specificity was assessed using the UCSC Genome Browser Blat tool (GRCh37) for visualization. All hits that aligned to autosomes with 100% identity were assessed and alignments that matched with exons of RefSeq transcripts were recorded. (Online Resource Table 2). One hundred seventy-five out of 191 of the subjects in expression cohort A had genome-wide genotypes from the PSP risk GWAS[19]. Gene expression measures for expression cohort B were previously collected from the TCX of 106 PSP subjects. This cohort used the prior version of the WG-DASL array with 24,526 probes. Similar QC measures were applied as for cohort A and are described elsewhere[48]. 100/106 of the subjects in expression cohort B had genome-wide genotypes[7].
A subset of subjects from each expression cohort (Cohort A, N= 78; Cohort B, N= 85) also had expression measures collected using next-generation RNA sequencing (RNASeq) [Online Resource Methods]. For those genes with significant expression level-cisSNP associations, WG-DASL expression levels were validated by performing correlations of WG-DASL vs. RNAseq measurements. WG-DASL levels were correlated with both RNAseq levels of the corresponding gene, as well as the RNAseq levels of the specific exon which binds the WG-DASL probe sequence.
To compare expression measures collected using microarray and RNAseq approaches, gene expression residuals were generated, using multivariable linear regression accounting for key covariates. All expression measures were adjusted for age at death, sex, RIN, and adjusted RIN ((RINsample-RINmean)2). Additionally, WG-DASL adjustment also included microarray PCR plates; and RNASeq adjustment included sequencing flowcell. All pairwise comparisons (WG-DASL vs. RNAseq Gene Counts and WG-DASL vs. RNAseq Exon Counts) were assessed for correlation using the Spearman rank correlation test. All analysis were executed using R Statistical Software (R Foundation for Statistical Computing, version 3.2.3).
We also compared the results for significant expression level-cisSNP associations in our study with publically available eQTL data from different diagnostic groups and/or tissue regions [Online Resource Methods].
DNA methylation RRBS measures
DNA was isolated from frozen post-mortem TCX tissue using an AutoGenFlex STAR instrument (AutoGen). Reduced representation bisulfite sequencing (RRBS) was used to collect genome-wide DNA methylation measures for 55 PSP samples, a subset of cohort B samples (Figure. 1; Table. 1), at the Mayo Clinic Medical Genome Facility. Bisulfite-converted DNA libraries were prepared using Msp1 DNA digest followed by end-repair, dA-tailing and column purification prior to ligation with Illumina adapters using T4 DNA ligase. Ligated DNA was purified and size selected using Pippin Prep (Sage Science) followed by bisulfite modification. Modified DNA was PCR amplified and purified using Ampure beads. The RRBS library DNA was run on the Illumina HiSeq2000, indexing 4 samples per lane, using a standard operating procedure based on Illumina’s protocol. RRBS data was analyzed using the workflow of Mayo Bioinformatics Core (BIC). Briefly, FASTQ data were trimmed to remove adaptor sequences, and reads with less than 15bp were discarded; the remaining reads were aligned against the reference genome hg19 using BSMAP. Samtools was used to get mpileup and custom scripts were used to determine CpG methylation and bisulfite conversion ratios[43].
QC included removal of subjects and CpGs with low call rates. A sample call rate threshold of 90% led to removal of 7 samples after merging common CpGs greater than 5X coverage across all samples with CpG call rate >90%. Principal components analysis identified an additional 2 PSP samples that were outliers, which were removed. Following QC, 46 PSP samples were retained for further analysis. Approximately 2 million CpG sites (forward + reverse strand) were retained for analysis. Density plots of the overall methylation patterns indicated that the majority of the identified CpG sites were either un-methylated or fully methylated in most samples. 570,982 CpG sites that were either 0% or 100% methylated in all samples were removed from further analysis as these are uninformative for statistical models. Forty-three out of 46 samples also have genome-wide genotypes[7].
Neuropathology Latent Traits
In order to generate continuous quantitative neuropathology measures from semi-quantitative neuropathology scores, latent trait analysis was applied in R using the ltm package[37] (URL: http://www.jstatsoft.org/v17/i05/) on all subjects on whom neuropathology data was available (n=848). Semi-quantitative tau pathology measures (none, mild, moderate, severe) were assessed from CP13 immunostained sections in 19 different anatomical structures for the following four lesions: tau neurofibrillary tangles (NFT), oligodendroglial coiled bodies (CB), tufted astrocytes (TA), and tau neuropil threads (TAUTH). Brain regions were selected based on those affected in PSP, which include: basal nucleus, caudate putamen, globus pallidus, hypothalamus, motor cortex, subthalamic nucleus, thalamic fasciculus, ventral thalamus, cerebellar white matter, dentate nucleus, inferior olive, locus ceruleus, medullary tegmentum, midbrain tectum, oculomotor complex, pontine base, pontine tegmentum, red nucleus, and substantia nigra.
The neuropathology measures were each on a 0 to 3 scale and were used to create continuous scores for the degree of pathology, for each of the four pathological lesions (NFT, CB, TA, TAUTH), based on a latent trait approach[6,37]. An overall latent variable was also calculated by using the semi-quantitative scores for all four lesion types in all regions. These scores are an estimate of an assumed underlying level of pathology severity that all individual scores are dependent on, or correlated with. Four hundred twenty-two out of 848 subjects with neuropathology latent traits also had genome-wide genotypes available.
Statistical Analysis
Of the 8 index SNPs from the seven PSP risk GWAS loci with significant or suggestive association (Table. 2), all but rs6687758 had genes within ±100kb for which cisSNP/gene expression associations could be tested. Multivariable linear regression analyses were utilized to test for these associations using PLINK[35], assuming an additive model for the SNP minor allele, and adjusting for age at death, sex, microarray PCR plates, RIN, and adjusted RIN ((RINsample-RINmean)2). For cohort A, the first four principal components were also included as covariates. Cohort B cisSNP/gene expression association results were previously obtained[48] and used to conduct meta-analysis with the top results from cohort A. Meta-analysis was conducted in METAL[44].
To determine whether any of the significant cisSNP/gene expression associations were driven by changes in methylation, CpGs within the body±5kb of these genes were tested for association with the index SNP, assuming an additive model. Given that CpG methylation often assumes a bimodal, semi-quantitative distribution, we used the non-parametric Kruskal-Wallis test for cisSNP/CpG methylation analysis. To determine correlations between gene expression and CpG methylation levels, we performed multivariable linear regression with the former as the dependent variable and the latter as the independent variable, correcting for all gene expression covariates mentioned above.
The PSP index SNPs were also tested for their association with neuropathology latent traits, using multivariable linear regression analysis in PLINK[35] with an additive model and including age at death, sex and the first four principal components as covariates. Kernel density plots were generated as previously described[2] for the most significant SNP associations with gene expression and neuropathology latent traits, using R.
Results
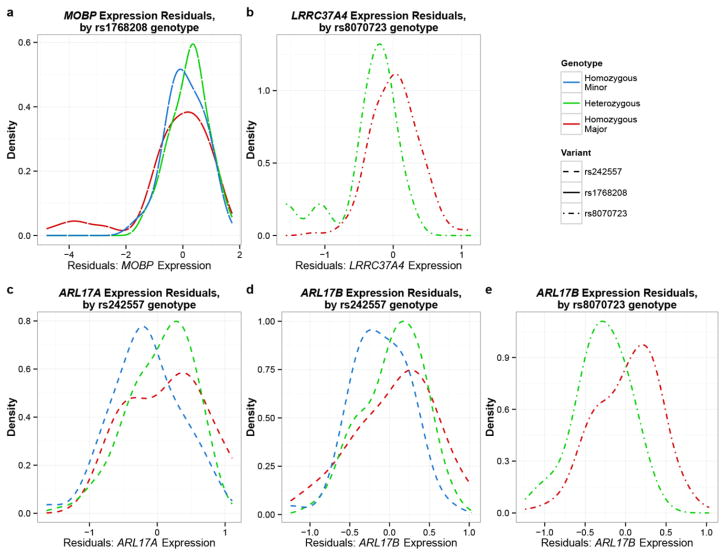
In total, there were 20 unique genes (30 transcript probes) for which cis-associations could be tested with the PSP risk GWAS index SNPs[19] (Table. 2; Online Resource Table 1.). Following QC, there were 41 cisSNP/expression association results of which five were nominally significant (Table. 3). Although only rs8070723/LRRC37A4 would achieve study-wide significance by stringent Bonferroni corrections for 41 tests, we retained all five nominally significant findings for further assessments. Of these five associations, four were at the MAPT locus and one was at the MOBP locus. At the MAPT locus, TCX levels of three genes, ARL17A, ARL17B and LRRC37A4 were associated with one or both of the PSP risk SNPs, rs8070723 and rs242557. MAPT region H2-haplotype tagging rs8070723 SNP G allele (rs8070723-G), which has association with lower risk of PSP (Table. 2) is associated with lower brain levels of LRRC37A4 and ARL17B. Rs242557-A, which partially tags MAPT H1c-haplotype and confers PSP risk is associated with lower brain levels of ARL17A and ARL17B. MOBP locus SNP rs1768208-T with genome-wide significant PSP risk association, associates with higher brain MOBP levels. Visualization of these associations by kernel density plots (Figure. 2) clearly demonstrate the shift for MAPT H2-haplotype carriers to lower brain LRRC37A4 (Figure. 2b) and ARL17B levels (Figure. 2e). Both ARL17A (Figure. 2c) and ARL17B (Figure. 2d) brain expression levels are lower in rs242557-A minor homozygote carriers, although there appears to be greater overlap in expression levels of subjects who are heterozygote and major homozygotes for this allele. While this might suggest a recessive mode of transmission, applying this model did not lead to significant results, which may be due to lack of power. A subset of the rs1768208-T major homozygote carriers had the lowest brain MOBP levels. Further, there was a modest shift to higher brain MOBP levels for minor allele carriers (Figure. 2a).
Table 3.
Significant PSP risk GWAS SNP associations with genes in cis
| Locus | Chr | SNP | Tested Allele | Gene | Probe | Cohort A | Cohort B | Meta-analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||
| N | BETA | CI | P | N | BETA | CI | P | N | BETA | CI | P | ||||||
| MAPT | 17 | rs8070723 | G | LRRC37A4 | ILMN_2393693 | 175 | −0.45 | −0.67 to −0.22 | 1.60E-04 | 100 | −0.41 | −0.67 to −0.15 | 2.93E-03 | 275 | −0.43 | −0.60 to −0.26 | 8.01E-07 |
| MOBP | 3 | rs1768208 | T | MOBP | ILMN_2298464 | 175 | 0.30 | 0.06 to 0.54 | 1.68E-02 | 99 | 0.36 | 0.07 to 0.66 | 1.84E-02 | 274 | 0.32 | 0.14 to 0.51 | 6.94E-04 |
| MAPT | 17 | rs8070723 | G | ARL17B | ILMN_3231952 | 175 | −0.35 | −0.59 to −0.10 | 6.43E-03 | NA | NA | ||||||
| MAPT | 17 | rs242557 | A | ARL17B | ILMN_3231952 | 175 | −0.12 | −0.02 to −0.21 | 2.04E-02 | NA | NA | ||||||
| MAPT | 17 | rs242557 | A | ARL17A | ILMN_1698680 | 175 | −0.18 | −0.06 to −0.30 | 5.35E-03 | NA | NA | ||||||
Abbreviations: NA: Not Available (No probes available for indicated genes). Cohort B results are from the PSP subset within the published Mayo Clinic eGWAS7 and based on an earlier version of WG-DASL expression microarray than Cohort A measurements. Locus name is given by the PSP risk GWAS and is usually the gene name nearest the index SNP. Gene: Transcript levels of the genes in-cis tested for associations with the index SNP. Probe: WG-DASL probe measured.
Figure 2. Kernel density plots for significant PSP risk SNP associations with brain gene expression levels.
Distribution of brain gene expression level residuals from PSP patients obtained after adjustment for all covariates is shown. Blue line indicates distribution of gene expression residuals for homozygous minor individuals; green line indicates the same for heterozygotes; red line indicates the same for major homozygotes. Different line types were used for the different SNPs.
Since a subset of 100 subjects in the published Mayo Clinic brain eGWAS[8] had PSP diagnosis, we pursued meta-analysis between the top associations in the current study with this published dataset. Due to the differences in the probe content for the expression microarrays between this former study and the current study, we were able to meta-analyze only two results, rs8070723/LRRC37A4 and rs1768208/MOBP brain level associations. Both of these associations were also nominally significant in the PSP subjects from our former study. Importantly, there was remarkable similarity in the SNP allele effect sizes between these two independent studies (Table. 3). As expected, meta-analysis revealed highly significant results for both of these associations, which achieved study-wide significance for both.
To ensure that the WG-DASL probes for the significant results (Table. 3) are specific to the transcripts of the specified genes, we performed sequence alignments. We confirmed that all four probes had 100% sequence identity only for the specified gene with the exception of ILMN_3231952, which had alignment to ARL17B, as expected, but also an isoform of ARL17A (Online Resource Table 2).
To validate the WG-DASL microarray-based expression measures, we evaluated MOBP, LRRC37A4, ARL17A and ARL17B levels for correlation with RNAseq expression measures available in a subset of subjects from cohort A; and MOBP in a subset of subjects from cohort B. For each gene we investigated the correlation of microarray measures with both overall gene counts (Online Resource Figure 2a–f) and counts for the same exon targeted by the microarray probe (Online Resource Figure 3a–f) [Online Resource Methods]. For all assessments the Spearman rank correlation coefficient indicated a positive correlation between each pair of variables. All of the correlation tests were likewise nominally significant (p<0.05) with the exception of MOBP for cohort A (Microarray vs Gene Counts, p=0.09; Microarray vs Exon Counts, p=0.09) and LRRC37A4 (Microarray vs Exon count, p=0.22) for cohort A (Online Resource Table 3).
We also compared the results from Table 3, with publically available data, where available and determined that rs1768208/MOBP, rs8070723/ARL17B and rs8070723/LRRC37A4 associations could also be detected in brain tissue of subjects with different diagnoses and/or in a different brain region (Online Resource Table 4).
To determine whether the PSP risk SNP associations with gene expression were driven by their influence on CpG methylation, we next tested their effects on these endophenotypes. There were no CpGs, captured in our study, within the gene body±5 kb of LRRC37A4. There were 11 such CpGs for MOBP which were tested for their associations with rs1768208; 12 for ARL17A tested with rs242557 and 8 for ARL17B tested with both rs242557 and rs8070723. There were four nominally significant associations (Table. 4), of which ARL17B-3′ region CpG methylation/rs8070723 achieved study-wide significance after corrections for 39 multiple tests. Visualization of the CpG methylation levels by genotype using box plots revealed lower levels of methylation at this ARL17B-3′ CpG for carriers of rs8070723-G allele in comparison to major homozygotes (data not shown). Box plots for the next significant association ARL17A intronic CpG/rs242557 demonstrated higher methylation for rs242557-A carriers. Likewise, carriers of this allele were associated with higher methylation levels of a CpG in the ARL17B-3′ region. At the MOBP locus an intronic CpG with significant rs1768208 association had higher levels of methylation with one copy of rs1768208-T, but not with two copies. Of these 3 genes, only MOBP had both gene expression and CpG methylation measurements in the same subset of subjects (Figure. 1, n=43). To determine whether epigenetic changes may be driving gene expression changes, we correlated MOBP levels with CpG methylation in this cohort and did not observe significant correlations.
Table 4.
Significant PSP risk GWAS SNP associations with CpG methylation across nominated genes
| Gene | Index SNP | N | Number of CpGs tested | Position (hg19) | Location | % Methylation, Mean (Range) | Chi Sq | P-value |
|---|---|---|---|---|---|---|---|---|
| MOBP | rs1768208 | 43 | 11 | 39,539,967 | Intron MOBP | 0.13 (0.0 – 0.39) | 6.37 | 4.15E-02 |
| ARL17A | rs242557 | 40 | 13 | 44,583,458 | Intron ARL17A | 0.94 (0.71 – 1.0) | 13.30 | 1.29E-03 |
| ARL17B | rs242557 | 40 | 8 | 44,346,670 | 4.9kb 3′ ARL17B | 0.87 (0.19 – 1.0) | 8.03 | 1.81E-02 |
| ARL17B | rs8070723 | 43 | 8 | 44,346,670 | 4.9kb 3′ ARL17B | 0.88 (0.19 – 1.0) | 16.14 | 5.87E-05 |
Genes with significant expression associations were assessed. CpGs within the body +/− 5 kb of these genes were tested for association of their methylation with the PSP risk index SNPs at the relevant loci. LRRC37A4 did not have such CpGs. Significant associations are shown. N: number of subjects tested. Additive model was assumed for the tested allele.
We investigated the CpG methylation/SNP association in the entire inversion region of chromosome 17 (Online Resource Table 5). We determined that rs8070723 had the strongest association with the ARL17B-3′ CpG methylation levels, although this variant also had associations with other CpGs including those near MAPT. The association with rs242557 and the intronic ARL17A CpG at position 44,583,458 had the 3rd strongest significance behind those for CpGs near CRHR1 and MAPT. Our CpG results across the chromosome 17 inversion region are annotated for presence of SNPs (dbSNP135) at the tested CpG positions (Online Resource Table 5). The CpG at position 44,346,670 that has the strongest association with rs8070723 has a SNP (rs139480590 aka rs2696560) annotated at this position, however the evidence for this variant is limited to reports from just two subjects (dbSNP) and was not identified by the 1000 genomes project, indicating that it is unlikely to account for the altered methylation we observe in this study.
To ascertain whether any of the top PSP risk SNPs with expression associations influenced levels of neuropathology and tau lesion subtypes, we tested them for associations with the latent traits. MAPT locus variants rs242557 and rs8070723 were tested in addition to MOBP locus variant rs1768208 (Table. 5). There were nominally significant associations between PSP risk allele rs242557-A and increased overall neuropathology latent trait; and suggestive results for rs1768208-T for the same endophenotype. When subtypes of neuropathology latent traits were evaluated, rs242557-A had the strongest associations with tufted astrocytes (TA), followed by coiled bodies (CB). Tau threads (TAUTH) were the most strongly associated neuropathology subtype with rs1768208-T, followed by coiled bodies that have suggestive results. Given that these analyses are conducted in only PSP subjects, H2-haplotype tagging rs8070723-G is rare with only 4% allele frequency amongst the 422 subjects tested. Despite this, all neuropathology latent traits had trends towards lesser severity associated with rs8070723-G, where neurofibrillary tangles (NFT) had a suggestive p value of association. Kernel density plots for the SNP/neuropathology trait associations are shown for each of the SNPs and the overall latent trait (Figures. 3a–c) and for the significant neuropathology subtype associations (Figures. 3d–f). These plots depict the shifts towards greater neuropathology severity (higher residuals) for carriers of rs242557-A (Figures. 3a, d, e) and rs1768208-TT minor homozygotes (Figures. 3b, f). Likewise, a shift towards lower overall neuropathology is observed for rs8070723-G carriers (Figure. 3c).
Table 5.
Association of Neuropathological Latent Traits with PSP risk variants.
|
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Locus_SNP (Allele; MAF; N) | |||||||||
|
|
|||||||||
| MAPT_rs242557 (A; 0.56; 422) | MOBP_rs1768208 (T; 0.36; 421) | MAPT_rs8070723 (G; 0.04; 422) | |||||||
|
| |||||||||
| Phenotype | BETA | 95% CI | P | BETA | 95% CI | P | BETA | 95% CI | P |
| Overall | 0.15 | 0.02 – 0.27 | 2.11E-02 | 0.13 | 0.00 – 0.25 | 5.24E-02 | −0.19 | −0.49 to 0.11 | 2.07E-01 |
|
| |||||||||
| Neurofibrillary tangles (NFT) | 0.02 | −0.10 – 0.13 | 7.98E-01 | 0.01 | −0.10 – 0.13 | 6.70E-01 | −0.23 | −0.50 to 0.04 | 9.96E-02 |
| Coiled Bodies (CB) | 0.17 | 0.06 – 0.29 | 3.12E-03 | 0.10 | −0.01 – 0.22 | 8.27E-02 | −0.16 | −0.44 to 0.11 | 2.41E-01 |
| Tufted Astrocytes (TA) | 0.24 | 0.12 – 0.37 | 1.20E-04 | 0.03 | −0.10 – 0.15 | 6.79E-01 | −0.12 | −0.41 to 0.18 | 4.46E-01 |
| Tau threads (TAUTh) | 0.04 | −0.07 – 0.16 | 4.85E-01 | 0.16 | 0.04 – 0.27 | 9.00E-03 | −0.13 | −0.41 to 0.14 | 3.44E-01 |
Locus, SNP (tested allele, minor allele frequency=MAF; number of subjects tested=N) are shown in the header row. BETA = direction and magnitude of effect of each copy of the minor allele on pathological measure; CI = 95% confidence interval; P = p-value. Nominally significant results <0.05 are bolded, suggestive results with p<0.1 are italicized.
Figure 3. Kernel density plots for PSP risk SNP associations with neuropathology latent traits.
Results for overall neuropathology trait residual distributions by genotype is shown for the three PSP risk SNPs evaluated (a–c) and for the significant neuropathology subtype associations (d–f). All symbol definitions are per Figure. 2.
Discussion
With the advent of genome-wide association studies (GWAS), many disease risk loci have been identified, though discovery of the actual disease genes, the functional genetic variants and their mechanism of action require alternative approaches[15]. In this study, we evaluated the most significant PSP risk variants identified in a GWAS[19] for their effects on brain gene levels in a sizable cohort of 175 autopsied PSP subjects, followed by meta-analysis with data from an independent cohort of 100 PSP brains. To our knowledge this study constitutes the largest gene expression association study conducted in brains from subjects with PSP. The goals of this analysis were twofold: To assess whether any of the PSP risk SNPs were markers of functional, regulatory variants; and to identify genes with influenced brain levels as potential candidate PSP risk genes. In order to further explore the potential functional consequence of PSP risk variants, we next tested all of the SNPs with nominally significant brain gene expression associations for epigenetic effects. For this, we used CpG methylation levels measured by RRBS in 43 PSP subjects. We also tested the effects of PSP risk SNPs with expression associations, on quantitative PSP neuropathology phenotypes obtained from 422 subjects. These multi-endophenotype associations revealed plausible functional effects for some of the PSP risk variants on brain gene expression, CpG methylation and severity of neuropathology.
The expression associations for cis-genes implicate LRRC37A4, ARL17A and ARL17B at the MAPT locus and MOBP as candidate PSP risk genes. We previously identified PSP risk SNP/expression associations for rs1768208/MOBP, rs11568563/SLCO1A2 and both rs8070723 and rs242557 with MAPT temporal cortex and/or cerebellar levels in a smaller cohort of PSP subjects[8]. In the current study, we took a more stringent approach and excluded all gene expression probes with a known polymorphism within their sequence, due to their potential for false positive SNP associations[42]. Consequently, SLCO1A2 ILMN_2381020 and MAPT ILMN_1710903 probes which harbor a polymorphism within their sequence and which yielded expression associations with PSP risk SNPs in the prior study were not assessed in this current study. These genes require further investigations for PSP risk SNP/expression associations using alternative methods not prone to such artifacts, such as next generation RNA sequencing. In the current study of 175 temporal cortex samples from PSP subjects and after applying such stringent exclusion criteria, we identified nominally significant associations with rs1768208/MOBP as in our previous study, and also associations with rs8070723/LRRC37A4, rs242557/ARL17A and both rs8070723 and rs242557 with ARL17B brain levels. ARL17A and ARL17B could not be evaluated in the prior study due to their absence from the older version of the microarray applied. The remarkable similarities in the direction of SNP effects for MOBP and LRRC37A4 associations in our two independent studies and meta-analysis that yield strong significance underscore the authenticity of the expression results for these two genes.
In another study[19], MAPT region H1-tagging variant rs8070723 was shown to associate with LRRC37A4 brain expression of control subjects, consistent with our results. The same study determined strong expression associations with ARL17A brain levels and another MAPT region variant rs8079215. This SNP is in LD with both rs8070723 and rs242557, but is not as strongly associated with PSP risk. Rs8079215 does not confer PSP risk after accounting for H1/H2 haplotype status. Given this, rs8079215 was not a focus of our study, although we confirmed that this variant has a stronger effect than rs242557 on brain levels of ARL17A in our cohort, as well (data not shown). We also showed expression associations with both rs242557 and rs8070723 on ARL17B, which is likewise influenced by rs8079215 to an even greater extent. Whether changes in LRRC37A4, ARL17A and ARL17B brain levels contribute to pathogenesis of PSP or are merely coincidental outcomes of regulatory variants in LD with the actual PSP risk variants is an important question that warrants addressing.
The extent of LD in the MAPT region poses great difficulty in discerning the gene(s) and functional variants that impose risk for PSP and other neurodegenerative diseases that associate with SNPs in this region, such as Alzheimer’s (AD)[24,31,1] and Parkinson’s disease(PD)[11,40]. The hallmark of tau pathology in AD, PSP and other tauopathies and presence of deterministic MAPT mutations that lead to neurodegeneration (reviewed[17]) strongly implicate MAPT as the culprit in these disease associations. Nevertheless, the complex interactions between disease risk variants and expression levels of a number of genes in the MAPT region should at least raise the possibility that additional genes at this locus may play a role in disease pathogenesis either independent of the MAPT gene or as effect modifiers.
LRRC37A4, which is at the boundary of the MAPT locus inversion, pertains to the family of “leucine rich repeat containing 37 genes” which have evolved through segmental duplication on chromosome 17[3]. Through evolution, LRRC37 genes acquired new promoters leading to increased expression in the cerebellum and fetal brain. LRRC37 proteins are predicted to have a transmembrane domain and some isoforms were shown to undergo cleavage, followed by extracellular release[3]. Other LRR-containing proteins have been implicated in neurodegenerative diseases[28,26,23]. Early transfection studies of LRRC37A showed induction of filopodia formation[3]. ARL17A and ARL17B pertain to the sub-family of ADP-ribosylation factor-like (ARF-like) genes, which belong to the ARF family that regulates membrane trafficking and vesicular transport[39]. Mutations in ARF family genes can lead to a number of neurologic diseases, including autosomal recessive periventricular heterotopia, X-linked intellectual disability, and Joubert’s syndrome ([reviewed [39]), which is not surprising given the dependence of synaptic transmission on vesicular trafficking. Collectively, these findings suggest that LRRC37A4, ARL17A and ARL17B may be additional plausible candidate PSP risk genes at the MAPT locus which should be explored further.
Our expression analyses showed lower levels of LRRC37A4 and ARL17B associated with the MAPT region H2 haplotype. Although LRRC37 gene levels previously analyzed in lymphoblastoid cells showed the opposite association, i.e. higher levels with H2 haplotype[10,3], this discrepancy may arise from differences in tissue, subject diagnosis, and analysis of multiple vs. single LRRC37 gene(s). Partial H1c-tagging rs242557 also associates with lower levels of ARL17A and ARL17B. The association of both the protective H2 and risky H1c variants with lower ARL17B levels may be due to the presence of allelic heterogeneity on these haplotypic backgrounds and/or collective effect of susceptibility variants which may confer either risk or protection depending on their relative frequencies and levels of expression. Sequencing and haplotype analysis in conjunction with detailed in vitro studies are needed to explore these results further. It also remains possible that one or more of these associations reflect false positives, though this seems especially unlikely for LRRC37A4 and MOBP, which have remarkable consistency in two independent cohorts.
Another important level of complexity in the chromosome 17 inversion region is the presence of copy-number variations (CNVs), which have been shown to arise independently on the H1 and H2 haplotypes[5,41]. A 205 bp long duplication CNV, which has arisen on the H1 and a shorter 155 bp duplication CNV, which has arisen on the H2 haplotype both involve 5′ exons of KANSL1. Brain levels of KANSL1 do not associate with either rs8070723 or rs242557 in our study. While this result does not rule out KANSL1 as a candidate risk gene for PSP, it suggests that the PSP risk conferred by rs8070723 or rs242557 variants are unlikely to be due to their effects on gene dosage of KANSL1. Alternatively, rs8070723 or rs242557 may not sufficiently distinguish the haplotypic complexity in this region that associates with KANSL1 levels.
There is another 210 bp CNV in the inversion region, which harbors LRRC37A, ARL17A, ARL17B and NSF genes. This CNV can occur as 1–2 copies on the H2 haplotype and as 1–4 copies on the H1 haplotype. In our study, we did not have gene expression measurements for LRRC37A or NSF. It should be noted that LRRC37A4, which resides upstream of all three CNVs and for which there is a significant eQTL is a different gene than LRRC37A. Importantly, the probe for LRRC37A4 does not bind LRRC37A. Hence, the eQTL result for LRRC37A4 is unlikely to be driven by the CNVs in the inversion region.
In contrast, ARL17A and ARL17B gene expression associations could be driven by the 210 bp CNV. ARL17B levels are lower in H2-haplotype carriers. Given that H1-haplotype carriers can have as many as 4 copies of this 210 bp CNV, whereas H2-carriers have only 1–2 copies, it is plausible that the rs8070723/ARL17B level association is driven by this CNV. On the other hand, the association of rs242557, which partly tags H1c, with lower ARL17A and ARL17B levels could also be theoretically explained by CNVs. The H1c haplotype was found to be less prone to the proximal duplication events[41]. If this is also true for the distal 210 bp CNV, this can explain the lower ARL17A and ARL17B level associations with partial H1c-tagging rs242557. Since we have not directly measured CNVs in our samples, the influence of these structural variants on the expression associations detected in our study is speculative. Nevertheless, these collective results should encourage combined CNV and SNP studies to fully explore the role of these variations in gene expression and risk of PSP.
Higher levels of MOBP are associated with the PSP risk allele (rs1768208-T) in both of our expression cohorts of PSP subjects (N=275). The PSP risk allele at this locus has also been reported to associate with expression of SLC25A38, in a cohort of control brains (N=387) assessed with microarray expression chips [19]and a mixed cohort of control and PSP brains (N=38) assessed by quantitative RT-PCR[47]. In the larger cohort of control brains, SNPs at the MOBP locus associated with levels of both MOBP and SLC25A38, but more strongly for the latter. In that study, the SLC25A38 gene expression associations were stronger in the cerebellum than in the frontal cortex. In the smaller study, frontal cortex RNA was assessed from 19 subjects who were carriers of rs1768208-T vs. 19 rs1768208-C allele carriers, where the authors detected higher SLC25A38 but not MOBP levels in the rs1768208-T carriers. In our study of 175 PSP brains with RNA isolated from the temporal cortex we do not observe an association of rs1768208-T with SLC25A38 levels (p = 0.23: data not shown).
The identification of brain MOBP expression associations with MOBP locus SNPs in the 387 control brains from another study using the microarray gene expression measurement approach is consistent with our findings[19]. The lack of MOBP level associations in the study of 38 brains measured with qRT-PCR [47] could be due to the limited power of this study or difference in methodology. The absence of SLC25A38 level associations with rs1768208-T in our study of PSP brains could be due to our assessment of disease subjects vs. control or mixed cohorts or differences in the tissue regions assessed. It is possible that some expression associations may be context dependent and only be observed in certain brain regions, cell types or diseased subjects. This may be due to differences in transcription factors, other cis-acting elements, interactions with environmental factors or compensatory mechanisms in control tissue. Both MOBP and SLC25A38 are plausible candidate genes based on their known functions. SLC25A38 is located ~70kb 5′ of MOBP, encodes for apoptosin and has been implicated in apoptosis and neurodegeneration [46]. MOBP, which encodes myelin oligodendrocytic basic protein is exclusively expressed in the central nervous system[45] and also represents a plausible candidate PSP risk gene, particularly given the abundance of oligodendroglial tau neuropathology in PSP[12]. Indeed, the PSP risk allele at this locus is associated with a higher burden of oligodendroglial and white-matter tau lesions (Coiled Bodies and Tau Threads) but not neurofibrillary tangles (Table 5), further supporting the relevance of this variant in cell types in which MOBP is expressed.
Given that epigenetic modifications can influence gene expression changes[27], we sought to determine whether any of the PSP risk loci with SNP/expression associations also harbored SNP associations with CpG methylation. Although CpG methylation is not the only form of epigenetic modification, it is the one most commonly evaluated[27,4]. Although most epigenetic studies evaluate DNA CpG methylation using microarray-based approaches, we chose to utilize a sequencing-based approach, since microarrays cover only a relatively small percentage of CpGs typically targeting promoters. Using RRBS, we were able to assess CpGs in both the body and ±5kb flanking regions of the genes of interest. We identified one CpG in the 3′ region of ARL17B that showed study-wide significant associations with rs8070723, where carriers of H2 haplotype had lower levels of methylation. The same CpG had higher levels of methylation in rs242557-A carriers. There were two additional CpG/SNP associations of nominal significance in the vicinity of ARL17A and MOBP. The epigenetic associations in the chromosome 17 region raise the question about the role of CNVs in these findings. We have assessed this indirectly by evaluation of rs8070723 and rs242557 associations with CpG methylation levels in the entire chromosome 17 inversion region. Although, associations with ARL17A and ARL17B region CpGs were some of the strongest in this region, strong associations were also noted for other genic regions, including MAPT, which should not be influenced by the CNVs. Thus, the epigenetic associations in chromosome 17 are unlikely to be entirely explained by CNVs. Epigenetic studies in neurodegenerative disorders are an emerging theme with only one published study focused on the differential DNA methylation in blood from subjects with PSP, in addition to frontotemporal dementia against controls[1]. This study, which assessed 43 PSP samples against 185 controls identified differentially methylated probes especially at the MAPT locus on chromosome 17 and associations of methylation levels at this region with H1 haplotype. Methylation changes were also observed in post-mortem AD[21] and PD[8] brains in the MAPT region. Given the differences in methodology, regions and extent of CpGs evaluated in these studies, a direct comparison with our results is not possible, other than to indicate that our study and others identified H1 haplotype associations with CpG methylation at the MAPT locus.
To our knowledge, our findings constitute the first evaluation of epigenetic changes in brain tissue from PSP subjects. Although intriguing, they must be considered preliminary and replication must be sought due to the relatively small sample size of 43 subjects with RRBS DNA methylation measurements. Further, these results require functional validation by joint assessment of methylation and gene expression levels using samples from the same tissue region of the same subjects. This type of validation is essential to propose a role for CpG methylation changes in modifying expression levels of particular genes. In our study, we had joint gene expression and methylation data only for MOBP, where correlations were not observed. This may imply that the methylation changes are not the major drivers of expression for this gene, that we are underpowered to detect such correlations or that the SNP/CpG association for this region may be a false positive. Gene expression and methylation correlations for ARL17A and ARL17B need to be assessed in future studies. Further, assessment of both whole transcriptome and methylome of PSP brains are needed to characterize the full extent of transcriptional and epigenetic modifications that may be driving pathophysiology of this condition.
Finally, our study evaluated the PSP risk SNPs that show expression associations, for their influence on levels of neuropathology based on the postulate that aberrations in gene levels may influence disease pathology. The latent trait[6] approach using which we converted semi-quantitative pathologic scores to quantitative traits enabled these analyses for the overall pathology as well as subtypes of tau lesions. PSP is characterized by neuropathology involving not only neuronal tau accumulation (NFT), but also glial lesions in the form of tufted astrocytes (TA) and oligodendroglial coiled bodies (CB) and tau threads (TAUTH) in white matter. In our study, the risk variants rs242557-A and rs1768208-T associated with increased overall neuropathology. The rs8070723-G allele, which tags the protective H2 haplotype, was rare, as expected, in our PSP cohort. Despite limited power due to this protective allele’s rarity in this cohort, there were trends towards lower neuropathology association with this variant.
Interestingly, the most significantly associated neuropathologic lesion differed for the various risk variants. Whereas rs242557 had the most significant associations with TA followed by CB, rs1768208 was significantly associated with TAUTH and CB. These findings may have implications for the specific cell types and neuropathologic lesions influenced by the different genes and variants. Given the oligodendroglial expression of MOBP and our collective results, we hypothesize that regulatory variants, which increase MOBP levels, drive the oligodendroglial neuropathology and thus contribute to PSP risk. Though this seems the most parsimonious explanation of these collective data, it requires functional validation in vitro and in animal models.
In summary, in this investigation of PSP risk variants for their effects on multiple endophenotypes, we find replicable brain expression associations with LRRC37A4 and MOBP and nominate ARL17A and ARL17B as additional candidate PSP genes to evaluate in follow-up studies. In a preliminary assessment of PSP brain CpG methylation, several associations emerge in the vicinity of ARL17A, ARL17B and MOBP, which require replication in larger cohorts and validation with gene expression correlative studies. Although our findings strongly support a role for misregulation of LRRC37A4, ARL17A, ARL17B and MOBP in PSP pathophysiology, the precise regulatory changes that promote disease risk cannot be discerned until all genetic variants in these regions, including CNVs can be assessed concurrently with levels of all genes in their vicinity and their isoforms. Given the known functions of the genes with expression associations in our study, we can postulate that disruption of pathways implicated in synaptic transmission, neurodevelopment and/or oligodendroglial biology could underlie the risk for PSP. Assessing the top PSP risk and expression variants for their effects on neuropathology subtypes can provide insight into the differential effects of these variants on pathophysiology. Our findings should be helpful in providing guidance for fine mapping efforts at the PSP risk loci and also for downstream biological studies.
Supplementary Material
Acknowledgments
For the brain donations, we thank all patients and their families, without whom this work would not have been possible. This work is supported in part by grants from National Institutes of Health, National Institute on Aging (R01 AG032990 and U01 AG046139 to N.E.T); National Institute of Neurological Disorders and Stroke (R01 NS080820 to N.E.T); Cure PSP (to N.E.T and D.W.D); Mayo Clinic Center for Individualized Medicine, Epigenetics Program Grant (to N.E.T and M.A.).
Footnotes
Potential Conflicts of Interest: None
References
- 1.Allen M, Kachadoorian M, Quicksall Z, Zou F, Chai HS, Younkin C, et al. Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels. Alzheimers Res Ther. 2014;6:39. doi: 10.1186/alzrt268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen M, Kachadoorian M, Carrasquillo MM, Karhade A, Manly L, Burgess JD, et al. Late-onset Alzheimer disease risk variants mark brain regulatory loci. Neurology:Genetics. 2015;1:e15. doi: 10.1212/NXG.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bekpen C, Tastekin I, Siswara P, Akdis CA, Eichler EE. Primate segmental duplication creates novel promoters for the LRRC37 gene family within the 17q21.31 inversion polymorphism region. Genome Res. 2012;22:1050–1058. doi: 10.1101/gr.134098.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett DA, Yu L, Yang J, Srivastava GP, Aubin C, De Jager PL. Epigenomics of Alzheimer’s disease. Transl Res. 2015;165:200–220. doi: 10.1016/j.trsl.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boettger LM, Handsaker RE, Zody MC, McCarroll SA. Structural haplotypes and recent evolution of the human 17q21.31 region. Nature genetics. 2012;44:881–885. doi: 10.1038/ng.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollen KA. Latent variables in psychology and the social sciences. Annu Rev Psychol. 2002;53:605–634. doi: 10.1146/annurev.psych.53.100901.135239. [DOI] [PubMed] [Google Scholar]
- 7.Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coupland KG, Mellick GD, Silburn PA, Mather K, Armstrong NJ, Sachdev PS, et al. DNA methylation of the MAPT gene in Parkinson’s disease cohorts and modulation by vitamin E in vitro. Mov Disord. 2014;29:1606–1614. doi: 10.1002/mds.25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong S, Chepelev I, Janson E, Strengman E, van den Berg LH, Veldink JH, et al. Common inversion polymorphism at 17q21.31 affects expression of multiple genes in tissue-specific manner. BMC Genomics. 2012;13:458. doi: 10.1186/1471-2164-13-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desikan RS, Schork AJ, Wang Y, Witoelar A, Sharma M, McEvoy LK, et al. Genetic overlap between Alzheimer’s disease and Parkinson’s disease at the MAPT locus. Mol Psychiatry. 2015;20:1588–1595. doi: 10.1038/mp.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickson DW, Rademakers R, Hutton ML. Progressive supranuclear palsy: pathology and genetics. Brain Pathol. 2007;17:74–82. doi: 10.1111/j.1750-3639.2007.00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donker Kaat L, Boon AJ, Azmani A, Kamphorst W, Breteler MM, Anar B, et al. Familial aggregation of parkinsonism in progressive supranuclear palsy. Neurology. 2009;73:98–105. doi: 10.1212/WNL.0b013e3181a92bcc. [DOI] [PubMed] [Google Scholar]
- 14.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 15.Ertekin-Taner N, De Jager PL, Yu L, Bennett DA. Alternative Approaches in Gene Discovery and Characterization in Alzheimer’s Disease. Curr Genet Med Rep. 2013;1:39–51. doi: 10.1007/s40142-013-0007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari R, Ryten M, Simone R, Trabzuni D, Nicolaou N, Hondhamuni G, et al. Assessment of common variability and expression quantitative trait loci for genome-wide associations for progressive supranuclear palsy. Neurobiol Aging. 2014;35:1514 e1511–1512. doi: 10.1016/j.neurobiolaging.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goedert M. NEURODEGENERATION. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Abeta, tau, and alpha-synuclein. Science. 2015;349:1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- 18.Hauw JJ, Daniel SE, Dickson D, Horoupian DS, Jellinger K, Lantos PL, et al. Preliminary NINDS neuropathologic criteria for Steele-Richardson-Olszewski syndrome (progressive supranuclear palsy) Neurology. 1994;44:2015–2019. doi: 10.1212/wnl.44.11.2015. [DOI] [PubMed] [Google Scholar]
- 19.Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im SY, Kim YE, Kim YJ. Genetics of Progressive Supranuclear Palsy. J Mov Disord. 2015;8:122–129. doi: 10.14802/jmd.15033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwata A, Nagata K, Hatsuta H, Takuma H, Bundo M, Iwamoto K, et al. Altered CpG methylation in sporadic Alzheimer’s disease is associated with APP and MAPT dysregulation. Human molecular genetics. 2014;23:648–656. doi: 10.1093/hmg/ddt451. [DOI] [PubMed] [Google Scholar]
- 22.Jowaed A, Schmitt I, Kaut O, Wullner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson’s disease patients’ brains. J Neurosci. 2010;30:6355–6359. doi: 10.1523/JNEUROSCI.6119-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kett LR, Dauer WT. Leucine-rich repeat kinase 2 for beginners: six key questions. Cold Spring Harb Perspect Med. 2012;2:a009407. doi: 10.1101/cshperspect.a009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouri N, Ross OA, Dombroski B, Younkin CS, Serie DJ, Soto-Ortolaza A, et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun. 2015;6:7247. doi: 10.1038/ncomms8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Chen JA, Sears RL, Gao F, Klein ED, Karydas A, et al. An epigenetic signature in peripheral blood associated with the haplotype on 17q21.31, a risk factor for neurodegenerative tauopathy. PLoS Genet. 2014;10:e1004211. doi: 10.1371/journal.pgen.1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lincoln S, Allen M, Cox CL, Walker LP, Malphrus K, Qiu Y, et al. LRRTM3 interacts with APP and BACE1 and has variants associating with late-onset Alzheimer’s disease (LOAD) PLoS One. 2013;8:e64164. doi: 10.1371/journal.pone.0064164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lunnon K, Mill J. Epigenetic studies in Alzheimer’s disease: current findings, caveats, and considerations for future studies. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:789–799. doi: 10.1002/ajmg.b.32201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majercak J, Ray WJ, Espeseth A, Simon A, Shi XP, Wolffe C, et al. LRRTM3 promotes processing of amyloid-precursor protein by BACE1 and is a positional candidate gene for late-onset Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17967–17972. doi: 10.1073/pnas.0605461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers AJ, Pittman AM, Zhao AS, Rohrer K, Kaleem M, Marlowe L, et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis. 2007;25:561–570. doi: 10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Ng CW, Yildirim F, Yap YS, Dalin S, Matthews BJ, Velez PJ, et al. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci U S A. 2013;110:2354–2359. doi: 10.1073/pnas.1221292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastor P, Moreno F, Clarimon J, Ruiz A, Combarros O, Calero M, et al. MAPT H1 Haplotype is Associated with Late-Onset Alzheimer’s Disease Risk in APOEvarepsilon4 Noncarriers: Results from the Dementia Genetics Spanish Consortium. J Alzheimers Dis. 2015;49:343–352. doi: 10.3233/JAD-150555. [DOI] [PubMed] [Google Scholar]
- 32.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pittman AM, Myers AJ, Abou-Sleiman P, Fung HC, Kaleem M, Marlowe L, et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J Med Genet. 2005;42:837–846. doi: 10.1136/jmg.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 35.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rademakers R, Melquist S, Cruts M, Theuns J, Del-Favero J, Poorkaj P, et al. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum Mol Genet. 2005;14:3281–3292. doi: 10.1093/hmg/ddi361. [DOI] [PubMed] [Google Scholar]
- 37.Rizopoulos D. ltm: An R Package for Latent Variable Modeling and Item Response Theory Analyses. Journal of Statistical Software. 2006;17:1–25. [Google Scholar]
- 38.Rojo A, Pernaute RS, Fontan A, Ruiz PG, Honnorat J, Lynch T, et al. Clinical genetics of familial progressive supranuclear palsy. Brain. 1999;122( Pt 7):1233–1245. doi: 10.1093/brain/122.7.1233. [DOI] [PubMed] [Google Scholar]
- 39.Seixas E, Barros M, Seabra MC, Barral DC. Rab and Arf proteins in genetic diseases. Traffic. 2013;14:871–885. doi: 10.1111/tra.12072. [DOI] [PubMed] [Google Scholar]
- 40.Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinberg KM, Antonacci F, Sudmant PH, Kidd JM, Campbell CD, Vives L, et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nature genetics. 2012;44:872–880. doi: 10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Z, Baheti S, Middha S, Kanwar R, Zhang Y, Li X, et al. SAAP-RRBS: streamlined analysis and annotation pipeline for reduced representation bisulfite sequencing. Bioinformatics. 2012;28:2180–2181. doi: 10.1093/bioinformatics/bts337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto Y, Mizuno R, Nishimura T, Ogawa Y, Yoshikawa H, Fujimura H, et al. Cloning and expression of myelin-associated oligodendrocytic basic protein. A novel basic protein constituting the central nervous system myelin. J Biol Chem. 1994;269:31725–31730. [PubMed] [Google Scholar]
- 46.Zhang H, Zhang YW, Chen Y, Huang X, Zhou F, Wang W, et al. Appoptosin is a novel pro-apoptotic protein and mediates cell death in neurodegeneration. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:15565–15576. doi: 10.1523/JNEUROSCI.3668-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y, Tseng IC, Heyser CJ, Rockenstein E, Mante M, Adame A, et al. Appoptosin-Mediated Caspase Cleavage of Tau Contributes to Progressive Supranuclear Palsy Pathogenesis. Neuron. 2015;87:963–975. doi: 10.1016/j.neuron.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.