Abstract
Purpose
MYC is a critical driver oncogene in many cancers, and its deregulation in the forms of translocation and overexpression has been implicated in lymphomagenesis and progression of diffuse large B-cell lymphoma (DLBCL). The MYC mutational profile and its roles in DLBCL are unknown. This study aims to determine the spectrum of MYC mutations in a large group of DLBCL patients, and to evaluate the clinical significance of MYC mutations in DLBCL patients treated with R-CHOP immunochemotherapy.
Experimental Design
We identified MYC mutations in 750 DLBCL patients using Sanger sequencing and evaluated the prognostic significance in 602 R-CHOP-treated patients.
Results
The frequency of MYC mutations was 33.3% at the DNA level (mutations in either the coding sequence or the untranslated regions), and 16.1% at the protein level (nonsynonymous mutations). Most of the nonsynonymous mutations correlated with better survival outcomes; in contrast, T58 and F138 mutations (which were associated with MYC rearrangements), as well as several mutations occurred at the 3´ untranslated region, correlated with significantly worse survival outcomes. However, these mutations occurred infrequently (only in approximately 2% of DLBCL). A germline single nucleotide polymorphism encoding the Myc-N11S variant (observed in 6.5% of the study cohort) was associated with significantly better patient survival, and resulted in reduced tumorigenecity in mouse xenografts.
Conclusions
Various types of MYC gene mutations are present in DLBCL and show different impact on Myc function and clinical outcomes. Unlike MYC gene translocations and overexpression, most MYC gene mutations may not have a role in driving lymphomagenesis.
Keywords: MYC, DLBCL, mutations, N11S, driver mutations, passenger mutations
INTRODUCTION
MYC is a proto-oncogene encoding the Myc protein, a transcription factor critical for cell proliferation, metabolism, differentiation, apoptosis, microenvironment remodeling and immune responses. MYC-IGH chromosomal rearrangement, resulted from aberrant class-switch recombination during germinal center (GC) reaction and leading to Myc overexpression, underlies the pathogenesis of Burkitt lymphoma, and the poorer prognosis of ∼10% of diffuse large B-cell lymphoma (DLBCL) associated with MYC translocation (1). Paradoxically, Myc overexpression is also a potent inducer of apoptosis through the modulation of both p53-dependent and p53-independent pathways, including the activation of TP53, ARF, CD95/FAS, and BAX, and the inhibition of BCL2, BCLX, and CFLAR/FLIP (2). Therefore, in tumors deregulation of MYC is often concomitant with other abnormalities (e.g., Bcl-2 overexpression) that cooperate with Myc during tumor onset, progression and chemoresistance (3–5).
In addition to MYC rearrangement, MYC mutation is another form of genetic abnormality found in Burkitt lymphoma. Multiple nonsynonymous mutations in the coding sequence (CDS) of the MYC gene have been found in approximately 40–70% of Burkitt lymphoma leading to a mutated Myc protein with amino acid changes (6–9). These Myc mutations cluster in the Myc transactivation domain with hotspots in the Myc box I (MBI) motif (44–63aa, Figure 1A), and have been proposed to have a role in lymphomagenesis by enhancing the oncogenicity of Myc (9–12). Functional studies indicated that Myc T58 mutants had increased transforming ability, increased Myc stability, and decreased proapoptotic ability, owing to alterations in posttranslational modifications of Myc. In contrast, S62 mutations, which are also frequent in the MBI motif and are associated with increased Myc expression, lead to decreased transforming ability without affecting apoptosis (10,13–17); the F138C mutation in the Myc box II (MBII) motif (128–143aa) decreases both transformation and apoptosis (18); and deletion of residues 188–199 in the Myc box III (MBIII) motif correlates with increased response to apoptosis and decreased tumorigenic ability in vivo (19). Moreover, somatic mutations exist in noncoding MYC exon I (5´ untranslated region [UTR]) (20,21) and within intron 1 near the exon 1 boundary (22), which may represent another pathogenic mechanism by deregulating MYC expression (20). For example, in Burkitt lymphoma, mutations at the 3´ border of MYC exon I remove a block to transcriptional elongation (23), and in multiple myeloma, mutations in the MYC internal ribosome entry segment lead to enhanced translation initiation (24).
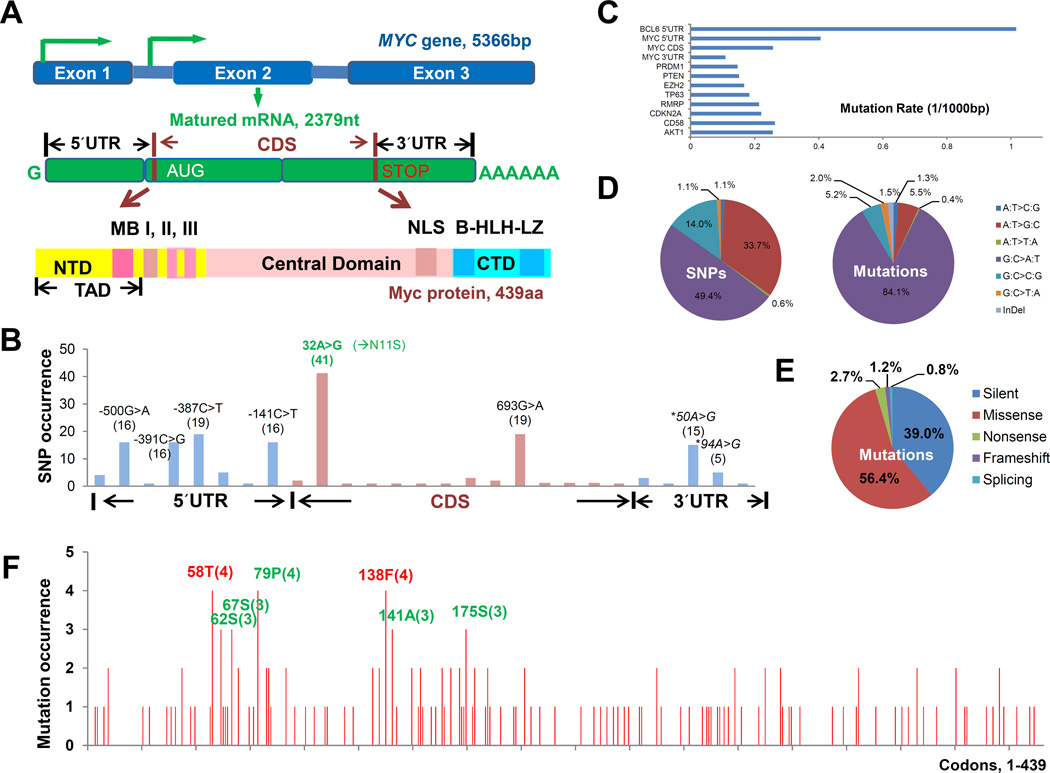
Figure 1.
Schematic illustration of the structure of MYC gene and Myc protein, and the composition and occurrence of MYC mutations. (A) Three MYC exons (top) are transcribed into an mRNA (middle) with untranslated regions (UTR) and the coding sequence (CDS), and then translated into the Myc protein with MYC box I (MBI, 44–63 aa) and MYC box II (MBII, 128–143 aa) in the N-terminal domain (NTD), MYC box III (MBIII including A and B), nuclear localization sequence (NLS), and the basic helix-loop-helix leucine zipper motif (B-HLH-LZ, 355–439 aa, involved in the dimerization with MAX and interacting with other HLH proteins) motif in the C-terminal domain (CTD). TAD indicates transactivation domain. (B) Occurrence of the SNPs (indicated in parentheses) in the 5´UTR, CDS and 3´UTR found in the DLBCL cohort. The SNP nucleotide positions are according to the translation start site resulting in the canonical Myc protein (439 aa). (C) Comparison of the mutation rate of 10 genes we sequenced for the DLBCL cohort. (D) Patterns of the MYC variations (SNPs and somatic mutations) found in the DLBCL cohort. (E) Proportions of silent, missense, nonsense, frame-shift, and splicing mutations in the MYC CDS found in the DLBCL cohort. (F) Frequencies of missense and nonsense Myc mutations. Numbers in parentheses indicate occurrence in the DLBCL cohort.
DLBCL also harbors MYC mutations, as shown by several studies (25–27). Sanger sequencing found DLBCL-specific MYC mutations (absent in GC-derived follicular lymphoma, pre-GC mantle-cell lymphoma, post-GC multiple myeloma and VH-mutated chronic lymphocytic leukemia as well as normal tissues) in the 5´UTR and CDS regions of the MYC gene harbored by 12 (32%) of 37 DLBCL patients; these mutations were proposed to originate from aberrant somatic hypermutation processes during DLBCL lymphomagenesis (25). Through next-generation sequencing, six of 111 DLBCL biopsies were found to have MYC mutations (26). However, the clinical relevance of MYC mutations in DLBCL has not been addressed.
To fill this knowledge gap, this study aims to profile the spectrum and frequency of MYC mutations in a large cohort of DLBCL patients, to study the functional consequences and to evaluate the prognostic significance of these MYC mutations.
PATIENTS AND METHODS
Patients
The study cohort consists of 750 patients with de novo DLBCL between 2000 and 2010 according to the World Health Organization classification criteria as a part of the International DLBCL R-CHOP Consortium Program. Patients with transformed DLBCL, primary mediastinal, cutaneous, or central nervous system large B-cell lymphomas, or human immunodeficiency virus infection were excluded. Cell-of-origin classification by either gene expression profiling or immunohistochemical algorithms have been described previously (1,28). Survival analysis was performed for 602 patients treated with standard rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (i.e., R-CHOP) chemotherapy whose follow-up data were available, randomly divided into a training set (n = 368) and a validation set (n = 234). At last follow-up, 208 of the 602 patients had died. The rest (394) patients were censored and had a median follow-up time of 54 months (range, 3–187 months). This study was conducted in accordance with the Declaration of Helsinki and was approved either as minimal to no risk or as exempt from review by the Institutional Review Boards of all participating centers.
The clinicopathologic features of the patients with or without mutations at the time of presentation were compared using the Fisher’s exact test. Overall survival (OS) was calculated from the date of diagnosis to the date of death from any cause or last follow-up. Progression-free survival (PFS) was calculated from the date of diagnosis to the date of disease progression, disease relapse, or death from any cause. Patients who were alive or had no disease progression were censored at the last follow-up. Survival analysis was performed using the Kaplan–Meier method with GraphPad Prism 6, and survival was compared between groups using the log-rank test. Multivariate survival analysis was performed using the Cox proportional hazards regression model with SPSS statistics software (version 19.0; IBM Corporation). All differences with P ≤ 0.05 were considered statistically significant (4,28–30).
Gene Expression Profiling
For patients in the training set, total RNAs extracted from formalin-fixed, paraffin-embedded tissues were subjected to gene expression profiling (GEP) using the Affymetrix GeneChip Human Genome U133 Plus 2.0 as previously described (28). Totally 350 patients in the training sets have GEP achieved and the CEL files have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus repository (GSE31312). Normalized microarray data underwent univariate analysis using a t-test to identify genes that were differentially expressed between various groups. The P values obtained by multiple t-tests were corrected for false discovery rates using the beta-uniform mixture method.
The mRNA expression levels of selected genes of interest were also compared between DLBCL groups by unpaired t-tests using GraphPad Software.
Detection of MYC Mutations and Rearrangements, Assessment of Myc Expression, and Functional Studies of Myc Mutants in vitro and in vivo
Details of Sanger sequencing for MYC gene (in all patients), functional studies of Myc mutants in vitro and in vivo , fluorescence in situ hybridization for MYC rearrangement detection (successful in 455 patients), and Myc expression evaluation by immunohistochemistry (successful in 556 patients) performed on tissue microarrays using formalin-fixed, paraffin-embedded samples are in the supplementary documents or have been described previously (1,4,29,31).
RESULTS
MYC Gene Resequencing Results Overview
The MYC gene variants found in the 750 patients were predominantly single-nucleotide substitutions of the canonical MYC sequence. The single nucleotide variations (SNVs) from the MYC reference sequence (NG_007161.1) (wild-type [WT] MYC) were herein referred as either germline single nucleotide polymorphisms (SNPs, variations in the dbSNP database [Build 132]), or somatic mutations (MUT, the rest of SNVs). Fourteen SNPs were found in the MYC CDS. Of these, two SNPs were most prevalent: rs4645959 (32A>G) which results in Myc-11S protein, and rs2070582 (693G>A) which is synonymous (Figure 1B). After exclusion of SNPs, MYC gene mutations were found in 250 DLBCL patients (33.3% of the DLBCL cohort), mainly in the 5´UTR and CDS regions (Supplementary Table S1); mutations in the 3´UTR were much less frequent. Mutations at the splicing sites were rare (n = 2). Most (71.4%) of the mutations were heterozygous.
Compared with the other nine genes we sequenced, MYC showed an elevated mutation rate in the 5´UTR although the mutation rate was significantly lower than that of BCL6 5´UTR (Figure 1C). Compared with MYC SNP variants, MYC mutations were predominated by C>T and G>A transitions, thus had a higher transition/transversion ratio than the MYC SNP variants (10.1 versus 4.9, Figure 1D).
Mutations in the MYC Coding Sequence
Mutation Profile
Among the 750 DLBCL patients, 254 point mutation events (Supplementary Table S1) were found in the MYC CDS region harbored by 161 patients (21.5% of the DLBCL cohort). However, 39% of these CDS mutations were synonymous mutations, and nonsynonymous mutations resulting in mutated Myc proteins (MUT-Myc) were found in only 121 patients (16.1% of the DLBCL cohort), 75% of which were heterozygous.
Most of these nonsynonymous mutations were missense mutations (Figure 1E). According to the in silico functional prediction models, 77% of the missense and nonsense mutations had the potential to affect Myc function.
These nonsynonymous mutations were scattered throughout the 439 codons of Myc with one to four occurrences of each mutation (Figure 1F). The frequency of hotspot mutations within or near MBI, for example T58 mutations found in four DLBCL patients, was much lower than that found in Burkitt lymphoma (8,14), and there was another mutation cluster near MBII extending to residue 185. F138 mutations were found in four DLBCL patients including two patients carried concurrent T58A mutations.
Impact of Nonsynonymous MYC Mutations and SNPs on Patient Survival
No clinical parameters significantly differed between the MUT-Myc and WT-Myc groups of the training set, except that MUT-Myc patients with germinal center B-cell–like (GCB) DLBCL had significantly higher frequency of primary nodal (versus extranodal) origin (Table 1). Molecularly, the MUT-Myc group compared with the WT-Myc group had significantly higher frequencies of MYC 5´UTR mutations (P < 0.0001), CD10 (P = 0.0052) and PI3K expression (P = 0.048), but less frequently nuclear p52 expression (P = 0.0044). Moreover, MUT-Myc patients with GCB-DLBCL more frequently had MYC rearrangements (36.8% versus 11.7% in the WT-Myc group, P = 0.011) and less frequently expressed CD30, whereas MUT-Myc patients with activated B-cell–like (ABC) DLBCL more frequently had BCL6 rearrangements (68.8% versus 36.8%, P = 0.015) and p63 expression (Supplementary Table S2).
Table 1.
Clinical characteristics of the 368 patients with diffuse large B-cell lymphoma (DLBCL) (the training set) with wild-type (WT) or mutated MYC
| Variables | MUT-Myc | WT-Myc | P | MUT-3'UTR | WT-3'UTR | P | MUT-5'UTR | WT-5'UTR | P |
|---|---|---|---|---|---|---|---|---|---|
| Age | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | |||
| < 60 y | 27 (52.9) | 129 (40.7) | 0.13 | 7 (35) | 149 (42.8) | 0.49 | 30 (43.5) | 126 (42.1) | 0.84 |
| ≥ 60 y | 24 (47.1) | 188 (59.3) | 13 (65) | 199 (57.2) | 39 (56.5) | 173 (57.9) | |||
| Sex | |||||||||
| Female | 18 (35.3) | 137 (43.2) | 0.29 | 4 (20) | 151 (43.4) | 0.039 | 25 (36.2) | 130 (43.5) | 0.28 |
| Male | 33 (64.7) | 180 (56.8) | 16 (80) | 197 (56.6) | 44 (63.8) | 169 (56.5) | |||
| Stage | |||||||||
| I - II | 21 (41.2) | 135 (44.4) | 0.67 | 10 (52.6) | 146 (43.5) | 0.43 | 31 (45.6) | 125 (43.6) | 0.76 |
| III - IV | 30 (58.8) | 169 (55.6) | 9 (47.4) | 190 (56.5) | 37 (54.4) | 162 (56.4) | |||
| B-symptoms | |||||||||
| No | 31 (63.3) | 193 (64.8) | 0.84 | 13 (68.4) | 211 (64.3) | 0.72 | 42 (63.6) | 182 (64.8) | 0.86 |
| Yes | 18 (36.7) | 105 (35.2) | 6 (31.6) | 117 (35.7) | 24 (36.4) | 99 (35.2) | |||
| LDH level | |||||||||
| Normal | 20 (41.7) | 117 (40.9) | 0.92 | 9 (47.4) | 128 (40.6) | 0.56 | 32 (50.8) | 105 (38.7) | 0.08 |
| Elevated | 28 (58.3) | 169 (59.1) | 10 (52.6) | 187 (59.4) | 31 (49.2) | 166 (61.3) | |||
| No. of extranodal sites | |||||||||
| 0 – 1 | 38 (76.0) | 227 (75.9) | 0.99 | 17 (89.5) | 248 (75.2) | 0.16 | 50 (78.1) | 215 (75.4) | 0.65 |
| ≥ 2 | 12 (24.0) | 72 (24.1) | 2 (10.5) | 82 (24.8) | 14 (21.9) | 70 (24.6) | |||
| ECOG performance status | |||||||||
| 0 – 1 | 39 (83.0) | 230 (82.1) | 0.89 | 17 (89.5) | 252 (81.8) | 0.40 | 49 (83.1) | 220 (82.1) | 0.86 |
| ≥ 2 | 8 (17.0) | 50 (17.9) | 2 (10.5) | 56 (18.2) | 10 (16.9) | 48 (17.9) | |||
| Size of largest tumor | |||||||||
| < 5cm | 21 (60.0) | 146 (59.8) | 0.98 | 9 (64.3) | 158 (59.6) | 0.73 | 28 (54.9) | 139 (61) | 0.42 |
| ≥ 5cm | 14 (40.0) | 98 (40.2) | 5 (35.7) | 107 (40.4) | 23 (45.1) | 89 (39) | |||
| IPI score | |||||||||
| 0 – 2 | 32 (64.0) | 184 (60.5) | 0.64 | 14 (73.7) | 202 (60.3) | 0.24 | 41 (61.2) | 175 (61) | 0.97 |
| 3 – 5 | 18 (36.0) | 120 (39.5) | 5 (26.3) | 133 (39.7) | 26 (38.8) | 112 (39) | |||
| Therapy response | |||||||||
| CR | 36 (70.6) | 235 (74.1) | 0.59 | 16 (80) | 255 (73.3) | 0.50 | 49 (63.6) | 231 (77.3) | 0.015 |
| PR | 6 | 47 | 2 | 51 | 13 | 40 | |||
| SD | 0 | 16 | 0 | 16 | 0 | 14 | |||
| PD | 9 | 19 | 2 | 26 | 15 | 14 | |||
| Primary origin | |||||||||
| Nodal | 39 (76.5) | 193 (62.5) | 0.05 | 11 (55) | 221 (35.0) | 0.40 | 42 (61.8) | 190 (65.1) | 0.61 |
| Extranodal | 12 (23.5) | 116 (37.5) | 9 (45) | 119 (65.5) | 26 (38.2) | 102 (34.9) | |||
| Cell-of-origin | |||||||||
| GCB | 30 (58.8) | 159 (50.5) | 0.29 | 11 (55) | 178 (51.4) | 0.82 | 41 (59.4) | 148 (49.8) | 0.18 |
| ABC | 21 (41.2) | 156 (49.5) | 9 (45) | 168 (48.6) | 28 (40.6) | 149 (50.2) | |||
Abbreviations: LDH, lactate dehydrogenase; ECOG, Eastern Cooperative Oncology Group; IPI, International Prognostic Index; CR, complete remission; PR, partial response; SD, stable disease; PD, progressive disease; GCB, germinal center B-cell–like; ABC, activated B-cell–like. For therapy response, we calculated P values as CR versus other responses. Some data for certain cases were not available.
Compared with the WT-Myc group, the MUT-Myc group showed trends toward better OS (P = 0.08) and PFS (P = 0.05), and patients with nonsynonymous SNPs had significantly better OS (P = 0.015, Figure 2A) and PFS (P = 0.01). When analyzed in GCB-DLBCL and ABC-DLBCL separately, the better survival of MUT-Myc and SNP-Myc groups than the WT-Myc group remained significant or with border-line P values, except that MUT-Myc ABC-DLBCL versus WT-Myc ABC-DLBCL had only slightly better OS [P = 0.43] and PFS [P = 0.44]) (Supplementary Figure S1A–B). Trends for better survival rates were also associated with Myc SNVs (mutations or SNPs) in the validation set (Figure 2B). Between homozygous and heterozygous mutations or SNPs, no significant difference in patient survival was observed (Supplementary Figure S1C–D). Patients with the Myc-11S germline variant had significantly better survival than those with the canonical WT-Myc-11N in the entire (combined training and validation) cohort (Figure 2C). Lists of discovered Myc mutations and SNPs and the associated GEP accession codes and clinical outcomes are shown in Supplementary Table S3.
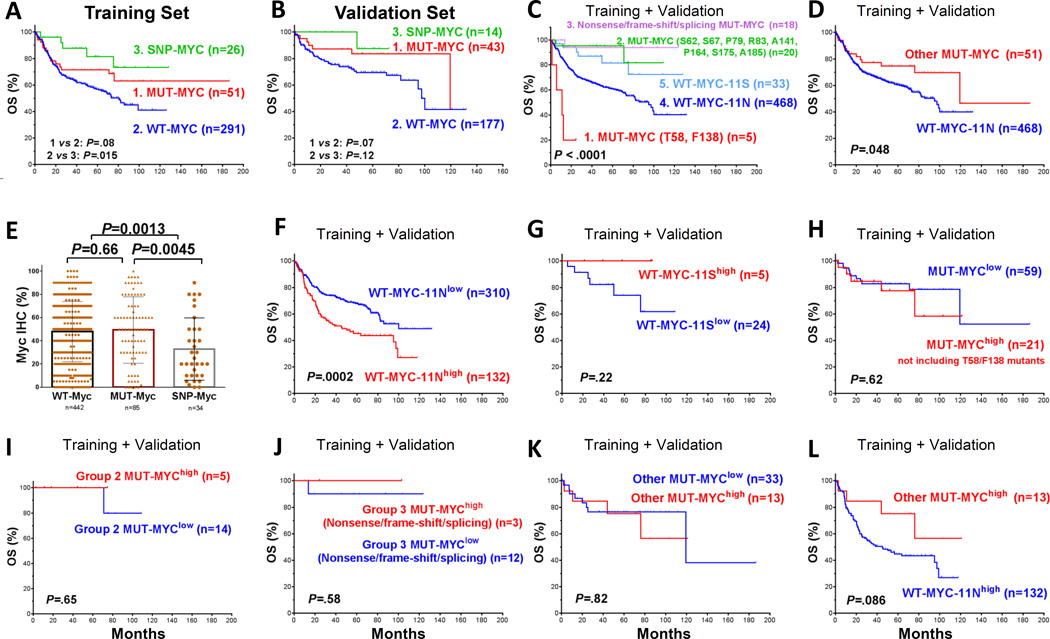
Figure 2.
Impact of nonsynonymous Myc variants on patient survival. (A–B) Overall survival of patient groups with wild-type (WT), mutated (MUT) or polymorphic (SNP) Myc in the training and validation sets. (C–D) Different types of Myc variants were associated with differential prognosis. (E) Comparison of Myc expression levels between groups with WT-, MUT- or SNP-Myc. (F) Overexpression of WT-Myc-11N correlated with significantly poorer survival. (G–H) Expression of the Myc-11S variant and non-T58/F138 MUT-Myc did not impact survival significantly. (I–K) Expression of different types of MUT-Myc (group of recurrent non-T58/F138 mutants; frame-shift or nonsense mutants; and other MUT-Myc) did not impact survival significantly. (L) After exclusion of Myc mutants in Figures I–J, the MUT-Mychigh group continued to show better survival compared with the group with overexpressed WT-Myc-11N with a marginal P value.
However, multivariate survival analysis including clinical parameters and Myc mutation and expression status indicated that Myc protein expression levels but not Myc mutation status independently predicted poorer OS and PFS, although the presence of Myc mutations trended toward conferring better OS (hazard ratio [HR]: 0.61; P = 0.11) and PFS (HR: 0.57; P = 0.057) (Supplementary Table S4).
Prognostic Impact and Heterogeneity of Myc Mutations
Among the R-CHOP-treated patients for survival analysis (n=602), missense mutations at T58, S62, S67, P79, R83, F138, A141, P164, S175 and A185 occurred in at least two patients. We found these recurrent Myc mutations (defined as n ≥ 2 occurred at a same AA) were associated with differential patient survival independently of Myc expression. Mutations at T58 and F138, which have been correlated with increased Myc stability, gain-of-function and reduced response to apoptosis in vitro (13,14,18) , had relative high occurrence in our cohort compared with other mutations, were all overexpressed, and were associated with significantly poor survival (Figure 2C). In contrast, group of other recurrent mutations (S62, S67, P79, R83, A141, S175 and A185 mutations) was associated with significantly better survival than WT-Myc (Figure 2C). Among these mutations, S62 mutations have been associated with impaired transforming ability and normal apoptosis function in vitro and in vivo (15,16) . According to the in silico functional prediction models, all the mutations at these recurrent spots except those at P79 had functional impact.
Nonsense, frame-shift and splicing mutations leading to a truncated Myc protein or substantial amino acid changes were also found associated with significantly better survival than WT-Myc (Figure 2C). The rest of MUT-Myc which have not been functionally characterized in the literature were still associated with significantly better OS in combined training and validation sets (Figure 2D) but not PFS (P = 0.15) compared with the WT-Myc cases.
Prognostic impact of Wild-type and Mutated Myc Overexpression
Myc expression levels were significantly lower in the SNP-Myc group compared with the WT-Myc and MUT-Myc groups (Figure 2E). There was no significant difference in Myc levels between the overall MUT-Myc and WT-Myc groups, but we did observe a higher mean level of Myc expression in the MUT-Myc GCB-DLBCL group compared with the WT-Myc GCB-DLBCL group in the training set only (P = 0.047).
High expression level of the canonical Myc (i.e., WT-Myc-11N) correlated with significantly poorer patient survival; Figure 2F shows the OS curve in overall DLBCL using a 70% cutoff for Mychigh, i.e., ≥70% of tumor cells staining positive on immunohistochemistry analysis (30). This adverse prognostic effect was significant in both GCB-DLBCL (P=0.0019) and ABC-DLBCL (P=0.039) (Figures not shown). In contrast, high level of the Myc-11S germline variant showed trends toward conferring better survival (Figure 2G). Myc overexpression did not have significant prognostic effect in the overall MUT-Myc group (for OS, P = 0.22). After the exclusion of patients with T58 and F138 mutants, which were all expressed at high levels and correlated with significantly poorer survival (Figure 2C), patients with high expression levels of non-T58/F138 MUT-Myc did not have significantly poorer survival with those with low MUT-Myc expression levels (P = 0.62 in overall DLBCL, Figure 2H; P = 0.97 in GCB-DLBCL and P = 0.99 in ABC-DLBCL, Supplementary Figure S1E–F), but significantly better overall survival than patients with overexpressed WT-Myc (P = 0.031 in overall DLBCL). Breaking down into different types of MUT-Myc in Figures 2C–D, analysis showed similar results: expression levels of MUT-Myc with recurrent non-T58/F138 mutations (Figure 2I), nonsense, frame-shift, or splicing mutations (Figure 2J), or other uncharacterized Myc mutations (Figure 2K) did not show prognostic effects. Patients with high expression levels of these Myc mutants showed significant or trends for better survival than those with overexpressed WT-Myc (Figures 2F, I–L).
Prognostic Analysis in the Presence or Absence of MYC Rearrangement
Since approximately 27.3% of the MUT-Myc group had MYC rearrangements (significantly higher compared with the 10% of the WT-Myc group, P = 0.00094, Supplementary Table S2) which has been shown as a significant adverse prognostic factor, we compared the survival outcomes of the WT-Myc and MUT-Myc groups within the MYC rearranged (MYC-R+) and MYC non-rearranged (MYC-R−) DLBCL patients separately. In both the training and validation sets, the MUT-Myc group showed trends toward better survival outcomes compared with the WT-Myc group only in the absence of MYC rearrangements (i.e., MUT-Myc/MYC-R− versus WT-Myc/MYC-R− but not MUT-Myc/MYC-R+ versus WT-Myc/MYC-R+, Supplementary Figure S1G–J). MYC rearrangements correlated with significant poorer prognosis in both WT-Myc and MUT-Myc GCB-DLBCL groups (Supplementary Figure S1K–L, MYC-R+ versus MYC-R−). Among the 13 MUT-Myc/MYC-R+ cases, 4 cases had T58 and/or F138 mutations (totally only five T58/F138-MUT-Myc cases had MYC rearrangement status available) with significantly poorer survival. After excluding these cases from the MUT-Myc/MYC-R+ group, there were still no significant difference in survival outcomes between the MUT-Myc/MYC-R+ and WT-Myc/MYC-R+ groups. Comparison of Myc expression levels between the MUT-Myc, WT-Myc, and SNP-Myc groups within the MYC-R− and MYC-R+ subsets are shown in Supplementary Figure S2A–C.
Mutations in the Untranslated Regions
5´UTR mutations
Compared with the MYC CDS and 3´UTR, the MYC 5´UTR had a higher mutation rate in our cohort (Supplementary Table S1), with the mutations distributed widely starting from the 9th nucleotide of the first exon (Figure 3A).
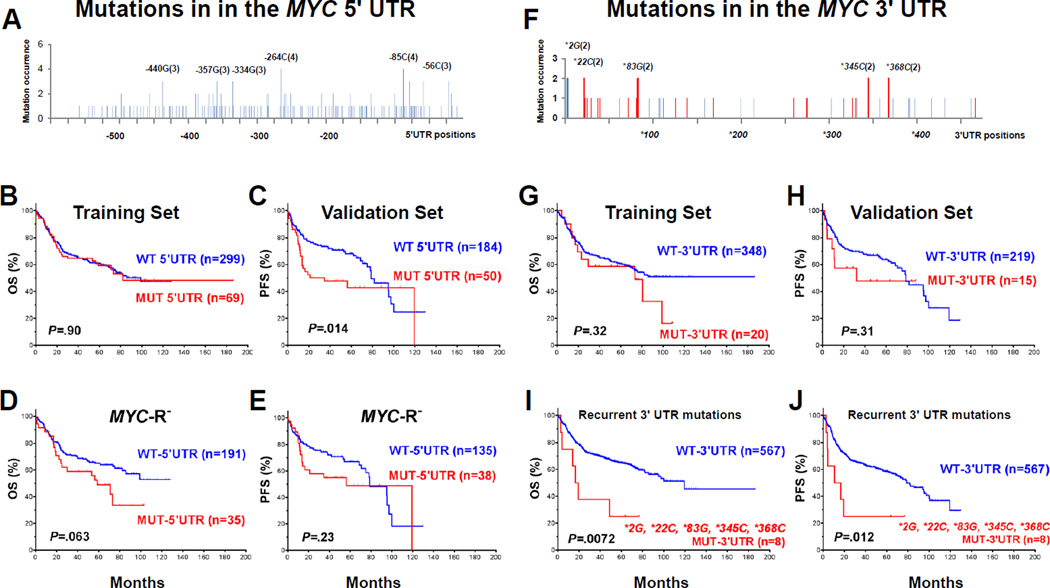
Figure 3.
Mutations in the MYC untranslated regions (UTR). (A) Distribution of mutations in the MYC 5´UTR. The nucleotide positions shown before the parentheses are in relation to the translation start site for canonical Myc protein (439aa). Numbers in parentheses indicate occurrence frequency in our cohort. (B–C) MYC 5´UTR mutations did not correlate with survival in the training set, but did correlate with significantly poorer PFS in the validation set. (D–E) In patients without MYC rearrangements, MYC 5´UTR mutations trended toward conferring poorer OS in the training set and poorer PFS in the validation sets. (F) Distribution of mutations in the MYC 3´UTR. Numbers in parentheses indicate occurrence. Mutations disrupting the known microRNA targeting sites (according to TargetScan) are highlighted in red. (G–H) The overall MUT-MYC-3´UTR group did not show significant poorer survival in the training and validation sets. (I–J) 3´UTR mutations recurrently (n ≥ 2) occurred at *2G, *22C, *83G, *345C, and *368C were associated with significantly poorer survival.
MYC-5´UTR mutations harbored by 139 (19.8%) of the DLBCL cohort were associated with MYC-CDS mutations, Bcl-6 expression, a lower complete remission rate (Supplementary Table S5, Table 1), and differential prognostic impact in the training (no impact) and validation (significantly poorer PFS) cohorts (Figures 3B–C). Multivariate survival analysis indicated that MYC-5´UTR mutation was not a significant prognostic factor. However, in patients without MYC rearrangements, MYC-5´UTR mutations trended toward conferring poorer OS in the training set and poorer PFS in the validation cohort (Figures 3D–E).
3´UTR mutations
Compared with mutations in the MYC CDS and 5´UTR, MYC-3´UTR mutations (Figure 3F) were less frequent (Supplementary Table S1) occurring in 5.8% of DLBCL patients. Half of these mutations occurred in the microRNA targeting sites according to TargetScan. However, MYC-3´UTR mutation status did not correlate with Myc expression levels (Supplementary Table S5). These mutations, the affected microRNA targeting sites, and associated clinical outcomes are listed in Supplementary Table S6.
The MUT-MYC-3´UTR group had a higher proportion of men than the WT-MYC-3´UTR group (Table 1). The overall MUT-MYC-3´UTR group did not have significantly poorer survival than patients with WT-MYC-3´UTR in the training and validation sets (Figures 3G–H). However, multivariate survival analysis adjusting clinical parameters indicated that MYC-3´UTR mutation was an independent prognostic factor for poorer OS (HR: 2.23; P = 0.024) but not PFS (HR: 1.85; P = 0.079) (Supplementary Table S4). MYC-3´UTR mutations were found recurrently at *2G, *22C, *83G, *345C and *368C which were associated with significant poorer survival than WT-MYC-3´UTR (Figures 3I–J), although these mutations were not concurrent with MYC rearrangements.
Gene Expression Profiling Analysis
Comparisons between WT-Myc and MUT-Myc
By supervised clustering analysis, no genes showed significantly differential expression between the MUT-Myc and WT-Myc groups (overall cohort or only Mychigh subcohort), or between MUT-MYC-5´/3´UTR and WT-MYC-5´/3´UTR groups. Individual analysis of particular mutation types showed differential expressed genes involved in proliferation, metabolism and apoptosis (Supplementary Figure S2D–G, Supplementary Table S7), but the significance of these analyses was hindered by small numbers and the heterogeneity of the MUT-Myc cases likely because some patients carried multiple mutations. Notably these signatures included genes involved in Ras/Rho GTPase signaling which interacts with the Myc T58 residue (10,13) and can cooperate with Myc during tumorigenesis (32).
Comparisons between WT- or MUT- Mychigh and Myclow
We further identified the GEP signatures of Myc overexpression (Mychigh GEP signatures) in the WT-Myc and MUT-Myc groups separately, and compared these GEP signatures (Figures 4A–D, Table 2). Differentially expressed genes were shown between WT-Mychigh and WT-Myclow in overall DLBCL, GCB-DLBCL, and ABC-DLBCL, and between MUT-Mychigh and MUT-Myclow in overall DLBCL and GCB-DLBCL but not in ABC-DLBCL even with a high false discovery rate (FDR) threshold of 0.50. These GEP signatures include MYC, some genes which have important oncogenic roles in transformation by Myc (for example, CDCA7L, UVBL2, MKI67IP, NOP16, MINA, and DDX18), and genes which regulate MYC/Myc (for example, PRKDC, PURB, SKP2, NME2, CSNK2A2, APEX1, and AIMP2).
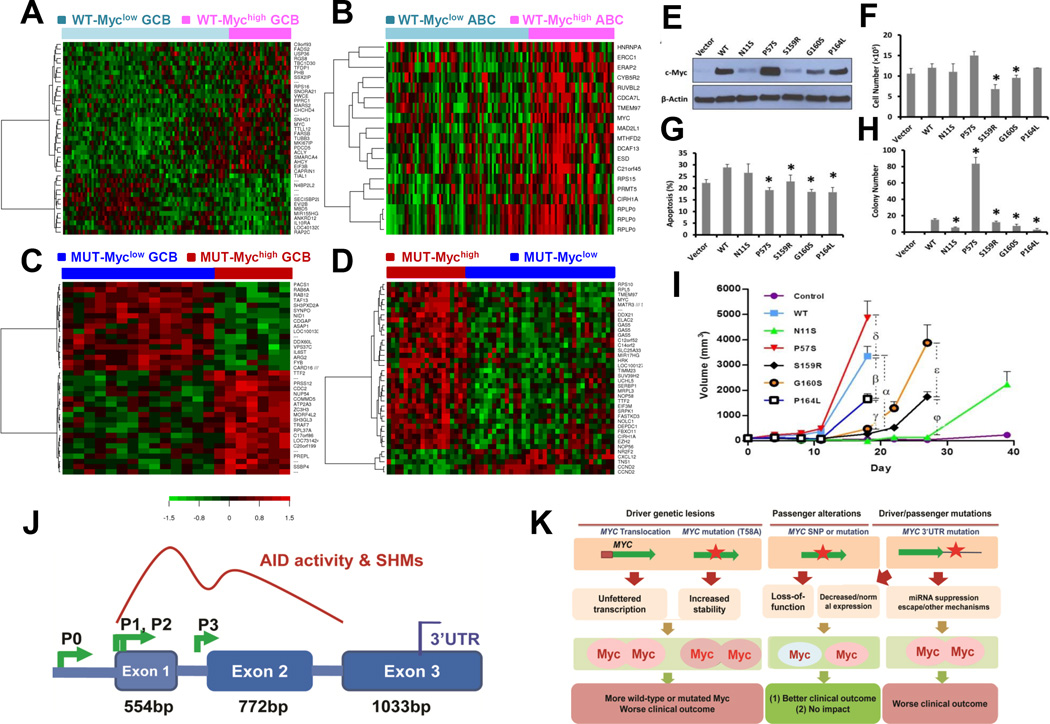
Figure 4.
Gene expression profiling (GEP) analysis, and functional studies of Myc variants in Rat1a cells. (A) GEP signatures for high levels (≥ 70%) of Myc expression (Mychigh) in WT-Myc patients with germinal center B-cell–like (GCB) DLBCL (false discovery rate [FDR] < 0.20). (B) GEP signatures for Mychigh in WT-Myc patients with activated B-cell–like (ABC) DLBCL (FDR < 0.05) with a cutoff of 1.65 for fold change of differential expression. (C) GEP signatures for Mychigh in MUT-Myc GCB-DLBCL patients (FDR < 0.20). (D) GEP signatures for Mychigh in overall MUT-Myc patients (FDR < 0.01). (E) Western blot analysis of expression of wild-type Myc and Myc variants in Rat1a cells transduced with retroviral vector expressing wild type Myc and Myc variants. (F) Cell proliferation analysis of wild-type Myc and Myc variants. Cells with the parental vector were used as control. Error bars show SEM. (G) Cell apoptosis analysis of wild type Myc and Myc variants using serum withdrawal. (H) An anchorage-independent colon formation assay of wild-type Myc and Myc variants. (I) Tumorigenecity of cells expressing wild type Myc or its mutants. Note: Statistical analysis was performed using one-way ANOVA in (G) to (J) with * indicating significant difference (P ≤ 0.05) between wild type Myc and a Myc variant, or two-away ANOVA in (K) with α-φ indicating significant difference (P ≤ 0.05) between the marked two groups. (J) Schematic illustration for the possible mechanism of MYC mutation and rearrangement origin, i.e., the activities of activation-induced cytidine deaminase (AID), which depend on MYC transcription activation and can affect up to ∼2kb downstream DNA from the transcription initiation site. Abbreviations: SHM, somatic hypermutation; UTR, untranslated region; P0, P1, P2, and P3 indicate multiple promoters of the MYC gene. (K) A hypothetical model for origin of MYC genetic lesions and effects on Myc expression, Myc function and clinical outcomes.
Table 2.
Significantly differentially expressed genes between Mychigh and Myclow DLBCL (wild-type [WT] or mutated [MUT])
| WT-Mychigh versus WT-Myclow | MUT-Mychigh versus MUT-Myclow | ||||
|---|---|---|---|---|---|
| Functional categories |
Overall DLBCL (FDR < 0.01) |
GCB-DLBCL (FDR < 0.25) |
ABC-DLBCL (FDR < 0.05) | Overall DLBCL (FDR < 0.05) |
GCB-DLBCL (FDR < 0.25) |
| Signaling |
PAK1IP1 ↑ DIP2A ↓ |
VWCE ↑ RGS8 ↑ RANBP1 ↑ LRP5 ↑ TBC1D30 ↑ SLC6A2 ↑ SOSTDC1 ↑SKI ↓ IL10RA ↓ RAP2C ↓ ATXN1 ↓ IQSEC1 ↓ SOS2 ↓ TNFRSF25 ↓ |
ARHGAP24 ↑ VSNL1 ↑ ZDHHC21 ↑ PTPN11 ↑ LYN ↑ C9orf100 ↑ PAK1IP1 ↑ YWHAG ↑ TRIB3 ↑ GNB2L1 ↑ INVS ↑ TTC1 ↑ CSNK2A2 ↑ PIK3R2 ↑ ARHGAP25 ↑ FYN ↓ PRKCH ↓ CMTM3 ↓ DLG3 ↓ PSG1 ↓ TRPM8 ↓ |
DEPDC1 ↑ RITA1 ↑CXCL12 ↓ |
RITA1 ↑ PRSS12 ↑ SAFB ↑ TNKS ↑ MAP3K4 ↑TRAF7 ↑IL6ST ↓ CDGAP ↓ FYB ↓ PLXND1 ↓ |
|
Cell proliferation, cell cycle, DNA repair, gene expression regulation, ribosome biogenesis, DNA/RNA metabolisms, microRNA, differentiation |
MYC ↑ RUVBL2 ↑ CIRH1A ↑ RPLP0 ↑ MIR17HG ↑ BZW2 ↑ MATR3 ↑ NOP16 ↑ WDR12 ↑ APITD1 ↑ ANP32A ↑ MRPL3 ↑ EEF1B2 ↑ POLR1B ↑ EEF1E1 ↑ RPS21 ↑ FARSB ↑ MRPS33 ↑ GAR1 ↑ POLR1C ↑ STAT3 ↑ DDX21 ↑ NOP2 ↑ UTP20 ↑ MRPS12 ↑ RCL1 ↑ POLR3G ↑ MKI67IP ↑ MINA ↑ GTF2E2 ↑ PRMT1 ↑ NAT10 ↑ PRMT5 ↑ EXOSC2 ↑ APEX1 ↑ PA2G4 ↑ EIF3B ↑ PRPF4 ↑PHF1 ↓ |
MYC ↑ SMARCA4 ↑ RPS21 ↑ TTF2 ↑ BZW2 ↑ MKI67IP ↑ MARS2 ↑ PHB ↑ CAPRIN1 ↑ RPS16 ↑ TFDP1 ↑ EIF3B ↑ EXOSC2 ↑ EIF5A ↑ PPRC1 ↑ SLC29A2 ↑ MIR155HG ↓ TIAL1 ↓ ZBTB4 ↓ CDKN1B ↓ MBD5 ↓ ANKRD12 ↓ |
CDCA7L ↑ HNRNPA1 ↑ RUVBL2 ↑ MAD2L1 ↑ MIS18A ↑ ERCC1 ↑ RPLP0 ↑ RPS15 ↑ DCAF13 ↑ PRMT5 ↑ MYC ↑ CIRH1A ↑ RPSA ↑ EEF1E1 ↑ GINS1 ↑ LRPPRC ↑ ZNF410 ↑ RPLP1 ↑ ZWINT ↑ PABPC1 ↑ ADSL ↑ SPIB ↑ PDCD2L ↑ TOP2A ↑ TRMT10C ↑ IARS2 ↑ MRPL47 ↑ PRKDC ↑ GAR1 ↑ NOP16 ↑ MKI67IP ↑ RPL13A/RPL13AP5/RPL13AP6 ↑ URI1 ↑ KARS ↑ TLK1 ↑ ARPP19 ↑ ESPL1 ↑ POLR1B ↑ DDX21 ↑ TRIP13 ↑ POLR3E ↑ PRIM1 ↑ RPS19 ↑ ZNF22 ↑ RPL13A ↑ HELLS ↑ MATR3/SNHG4 ↑ TAF5 ↑ WDR12 ↑ TFAM ↑ ANP32A ↑ BZW2 ↑ NOP56 ↑ MRPL32 ↑ POLG ↑ UTP20 ↑ EIF3M ↑ CHEK1 ↑ NOLC1 ↑ GTF2E2 ↑ RPL17 ↑ HAUS6 ↑ NOP2 ↑ IRF2BP2 ↑ DENR ↑ PURB ↑ MIS18A ↑ SFRS1 ↑ HIVEP1 ↑ MTPAP ↑ EIF2A ↑ CNP ↑ CDCA5 ↑ RPL24 ↑ HNRNPA0 ↑ FANCI ↑ CDK1 ↑ ZNF511 ↑ MRPS12 ↑ GUF1 ↑ FARSB ↑ RPS3A ↑ MINA ↑ NAP1L1 ↑ EIF4B ↑ MRPL4 ↑ TARBP1 ↑ PRMT1 ↑ RPS16 ↑ NUP93 ↑ POLR1C ↑ HNRNPL ↑ BXDC2 ↑ SPAG5 ↑ NUP205 ↑ FTSJ1 ↑ RPA3 ↑ LUC7L3 ↑ DDX39 ↑ NR2C2AP ↑ DDX18 ↑ RPL37A ↑ DENR ↑ PQBP1 ↑ RPS2 ↑ RAD50 ↑ MTA1 ↑ RPS29 ↑ BARD1 ↑ MIS18A ↑ EXOSC8 ↑ EIF4B ↑ RPL4 ↑ ZNF274 ↑ XPOT ↑ MRPS16 ↑ RPS6KB1 ↑ APEX1 ↑ NAT10 ↑ PTBP1 ↑ POLB ↑ ZNF131 ↑ RPS17 ↑ TEFM ↑ TRMT61B ↑ NCBP2 ↑ DCP1A ↑ RPL38 ↑ MRPS22 ↑ DBF4 ↑ RPS13 ↑ DIMT1L ↑ KANSL2 ↑ RPS28 ↑ PLRG1 ↑ RPP40 ↑ ZNF805 ↑ HAUS4 ↑ CEP295 ↑ KIF23 ↑ EEF2 ↑ RFC1 ↑PATL2 ↓ ARNT ↓ NFATC3 ↓ PHF1 ↓ C19orf40 ↓ ASXL3 ↓ |
MYC ↑ ELAC2 ↑ SRPK1 ↑ EZH2 ↑ CIRH1A ↑ MATR3 ↑ RPS10 ↑ SLC25A33 ↑ RPL5 ↑ DDX21 ↑ EIF3M ↑ TTF2 ↑ SUV39H2 ↑ NOLC1 ↑ MRPL3 ↑ NOP56 ↑ SERBP1 ↑ MIR17HG ↑ NOP58 ↑ ZNF131 ↑ RPS10 ↑ CBX3 ↑ PMS1 ↑ HELLS ↑ EIF2S1 ↑ ZNF138 ↑ RBMX ↑ RRS1 ↑ BUB3 ↑ RPL36A ↑ RPS13 ↑ FARSB ↑ RP9 ↑ RPL5 ↑ RPS10 ↑ TIMELESS ↑ PTCD1 ↑ HNRNPA1 ↑ CCND2 ↓ NR2F2 ↓ JUND ↓ |
BACH2 ↑ MORF4L2 ↑ ZNF675 ↑ ZBTB3 ↑ TTF2 ↑ ZNF331 ↑ CDK1 ↑ ZNF138 ↑ CNOT1 ↑ RBMX ↑ ZNF780B ↑ RPL15 ↑ SKA3 ↑ MRGBP ↑ ZNF480 ↑ HMGA1 ↑ DDX51 ↑ RPS2 ↑ ATRX ↑ RRP36 ↑ STRA13 ↑ COMMD5 ↑ ZBED3 ↑ KHSRP ↑ MRPL19 ↑ NUP54 ↑ ZNF215 ↑ RPL37A ↑ TAF13 ↓ STAT3 ↓ PARP14 ↓ CCND1 ↓ MARCA2 ↓ ZNF205 ↓ CSRP2 ↓ |
| Metabolism |
TMEM97 ↑ CYB5R2 ↑ LDHB ↑ NME1 ↑ NDUFA8 ↑ ATIC ↑ QDPR ↑ MAT2A ↑ SLC7A1 ↑ |
SHMT2 ↑ ARSB ↑ ACLY ↑ PPAT ↑ FADS2 ↑ AHCY ↑ SRXN1 ↑ ATAD3A ↑ |
CYB5R2 ↑ MTHFD2 ↑ ESD ↑ TMEM97 ↑ HINT1 ↑NDUFA12 ↑ USMG5 ↑ NDUFA8 ↑ ATP5S ↑ ADK ↑ NOC3L ↑ LDHB ↑ PIGW ↑ LYPLA1 ↑ DTYMK ↑ PFKM ↑ SFXN4 ↑ LDHB ↑ NME1 ↑ PLA2G5 ↑ ATP5C1 ↑ NDUFB6 ↑ CISD1 ↑ NDUFA9 ↑ QDPR ↑ GOT1 ↑ ATIC ↑ MAT2A ↑ NME1/NME1- NME2/NME2 ↑ PASK ↑ COX7C ↑ PUS1 ↑ PMM2 ↑ COQ3 ↑ PTCD2 ↑ATP6V1G3 ↓ IDH1 ↓ |
TMEM97 ↑ LRPPRC ↑ BDH1 ↑ DCTPP1 ↑ ATIC ↑ MAT2A ↑ AHCY ↑ ATP5J2 ↑ ST3GAL3 ↑ |
BCAT2 ↑ ATP2A3 ↑ DIO3 ↑ HPDL ↑ UGCG ↓ RNASEK ↓ ARG2 ↓ SLC39A6 ↓ |
| Protein folding. chaperon, trafficking, degradation |
PPIA ↑ FKBP4 ↑ CCT2 ↑ CHCHD4 ↑ |
HSPD1 ↑ NUP35 ↑ CHCHD4 ↑ USP36 ↑ TIMM8B ↑ IPO11 ↑ |
TIMM23 ↑ CLNS1A ↑ C15orf63 ↑ CUEDC2 ↑ CCT4 ↑ HSPA14 ↑ COMMD2 ↑ PCBP2 ↑ IPO7 ↑ SLC25A3 ↑ UBA2 ↑ SNX5 ↑ SACS ↑ CSE1L ↑ UBLCP1 ↑ DNAJA3 ↑ TIMM9 ↑ PSME3 ↑ SKP2 ↑ GRPEL1 ↑ IPO4 ↑ TEX10 ↑ PFDN4 ↑ TUBGCP5 ↑ LARP4 ↑ AIMP2 ↑ BAG2 ↑ TOMM40 ↑ NSF ↑ CBLL1 ↑ PPIA ↑USP7 ↓ RNPEP ↓ |
UCHL5 ↑ TIMM23 ↑ FBXO11 ↑ NPM3 ↑ IPO4 ↑ AKAP1 ↑ RAD23A ↑ UBE2G2 ↑ UBQLN4 ↑TNS1 ↓ |
GORASP2 ↑ FBXO41 ↑ TIMM22 ↑ UBQLN4 ↑ SH3GL3 ↑PACS1 ↓ |
| Apoptosis | PDCD5 ↑ | PDCD5 ↑ | BID ↑ GNL3 ↑ |
HRK ↑ FASTKD3 ↑ |
CARD16/CASP1 ↓ RASSF4 ↓ DAPK1 ↓ |
| Adhesion, cell interaction with extracellular matrix, cytoskeleton, transport, motility, membrane/vesicular trafficking and exocytosis |
C14orf104 ↑ SYNE1 ↓ |
SSX2IP ↑ TUBB3 ↑ITM2A ↓ ITM2B ↓ |
DCTN5 ↑ MTMR12 ↑ TUBB ↑ RTTN ↑ RAB3GAP2 ↑ DNAAF2 ↑APOL6 ↓ LAMA3 ↓ PAPPA ↓ |
C21orf59 ↑ |
PICK1 ↑ MYH7 ↑ ITGB1 ↓ DPYSL3 ↓ PALLD ↓ MARCKS ↓ CALD1 ↓ TPM1 ↓ ANXA4 ↓ ANXA7 ↓ NID1 ↓ RAB13 ↓ RAB12 ↓ RAB6A ↓ ASAP1 ↓ SH3PXD2A ↓ TNS1 ↓ SYNPO ↓ MYL12A/MYL12B ↓ VPS37C ↓ SLC39A13 ↓ SYNPR ↓ |
| Immune responses, tumor microenvironment |
NCR3LG1 ↑ |
ERAP2 ↑ C1QBP ↑GIMAP1 ↓ TRA@ ↓ FOXP3 ↓ CD4 ↓ |
MS4A2 ↑IL18BP ↓ | ||
| RNA gene; unknown function |
SNHG4 ↑ SNHG1 ↑ RBM26 ↑ KIAA0114 ↑ C4orf43 ↑ GAS5 ↑ C21orf45 ↑ WDR43 ↑ CCDC58 ↑ GRPEL1 ↑ LYAR ↑ NOC3L ↑ KIAA0020 ↑ SNHG8 ↑ C12orf24 ↑ SNORA21 ↑ FAM86A ↑ TRBC1 ↓ GLCCI1 ↓ LOC100233209 ↓ |
SNHG1 ↑ SNHG12 ↑ SNORA21 ↑ TTLL12 ↑ KIAA0020 ↑ RMND5A ↑ UBXN10 ↑ C9orf93 ↑ ZNRD1-AS1 ↑ TRBC1 ↓ EVI2B ↓ SECISBP2L ↓ KIAA1551 ↓ LOC401320 ↓ ZBED2 ↓ CCDC92 ↓ LOC440944 ↓ LOC651250 ↓ N4BP2L2 ↓ |
KLHL23 ↑ LOC100291837/RPS21 ↑ PUS7 ↑ CCDC86 ↑ RBM26 ↑ C4orf43 ↑ SNHG1 ↑ KIAA0114 ↑ LRRC58 ↑ GAS5 ↑ C3orf26 ↑ METTL9 ↑ZFAS1 ↑ URB1 ↑ WDR75 ↑ SNHG8 ↑ METTL5 ↑ WDR43 ↑ KBTBD8 ↑ CCDC58 ↑ C18orf19 ↑ NOC4L ↑ DYRK4 ↑ LYAR ↑ ATP5SL ↑ C12orf73 ↑ TMEM128 ↑ C19orf48 ↑ LSM12 ↑FAM13AOS ↓ C5orf4 ↓ LOC100233209 ↓ LAMB2L ↓ |
GAS5 ↑ SNHG4 ↑ LOC100127980 ↑ LOC100294028 ↑ C14orf2 ↑ SNHG1 ↑ RPS10P5 ↑ SNHG10 ↑ MARS2 ↑ LOC100288418 ↑ LOC440552 ↑ RRP1B ↑ NOC3L ↑ NUDCD1 ↑ CCDC93 ↑WWC2 ↓ |
ZFAS1 ↑ MATR3/SNHG4 ↑ MPHOSPH9 ↑ ZC3H3 ↑ C19orf43 ↑ C17orf86 ↑ CCDC51 ↑ PREPL ↑ SSBP4 ↑ LOC148189 ↑ LY6H ↑ FLJ40125 ↑ UBE2NL ↑ LOC731424 ↑ DDX60L ↓ EPSTI1 ↓ MEIS3P1 ↓ LOC100133790 ↓ C22orf42 ↓ |
Abbreviations: FDR, false discovery rate. GCB, germinal center B-cell–like; ABC, activated B-cell–like. Note: the genes are listed in the order according to the fold changes in Mychigh versus Myclow group.
All the WT-Mychigh and MUT-Mychigh GEP signatures are characterized by strong proliferation and growth signatures (especially for WT-Mychigh ABC-DLBCL) resembling those identified by previous studies (33,34). However, MUT-Mychigh GEP signatures also included downregulation of CCND2 (cyclin D2) and JUND in overall DLBCL, and downregulation of CCND1 (cyclin D1) and TAF13 RNA polymerase II whereas upregulation of COMMD5 (which negatively regulates cell cycle transition and proliferation) in GCB-DLBCL. In contrast, in WT-Mychigh GCB-DLBCL (versus WT-Myclow GCB-DLBCL), CDKN1B (inhibitor of cell cycle progression) and ANKRD12 (which inhibit transactivation) were downregulated. STAT3 was significantly upregulated in WT-Mychigh DLBCL but downregulated in MUT-Mychigh GCB-DLBCL.
Expression of apoptotic genes also showed differences between WT-Mychigh and MUT-Mychigh GEP signatures. Proapoptotic HRK was significantly upregulated in MUT-Mychigh but not in WT-Mychigh DLBCL, which instead had upregulation of PDCD5 (which promotes p53-mediated apoptosis) (in DLBCL and GCB-DLBCL), BID, and GNL3 (which stabilize MDM2) (in ABC-DLBCL). Other upregulated genes having roles in regulating the p53 pathway included EEF1E1, HINT1, PRKDC, CHEK1, YWHAG, DNAJA3, HIVEP1, PSME3, MTA1, CSNK2A2, AIMP2, USP7, and PHF1 in WT-Mychigh ABC-DLBCL, EIF5A in WT-Mychigh GCB-DLBCL, APITD1 in WT-Mychigh DLBCL, and FBXO11 in MUT-Mychigh DLBCL. In contrast, In MUT-Mychigh GCB-DLBCL, proapoptotic RASSF4 and DAPK1 were downregulated compared with MUT-Myclow GCB-DLBCL.
Moreover, in WT-Mychigh ABC-DLBCL (versus WT-Myclow ABC-DLBCL), several T-cell marker genes (CD4, GIMAP1, TRA@, and FOXP3) were downregulated but C1QBP (which inhibits the complement subcomponent C1) was upregulated. NCR3LG1 (which triggers natural killer cell activation) was upregulated in WT-Mychigh (versus WT-Myclow) GCB-DLBCL. In MUT-Mychigh GCB-DLBCL, MS4A2 (FCER1B, important for mast cell responses) was upregulated whereas IL18BP (which encodes an inhibitor of the proinflammatory cytokine IL18) was downregulated. Other potentially important signatures included downregulation of IL10RA, TNFRSF25, MIR155HG and ATXN1 whereas upregulation of EXOSC2 (components of the RNA exosome complex), HSPD1 and SMARCA4 in WT-Mychigh GCB-DLBCL, upregulation of LYN, PIK3R2 and EXOSC8 whereas downregulation of FYN in WT-Mychigh ABC-DLBCL, and downregulation of IL6ST and FYB whereas upregulation of BACH2, MAP3K4, RITA1 and ZBED3 in MUT-Mychigh GCB-DLBCL. GAS5 and MIR17HG were upregulated in both WT-Mychigh and MUT-Mychigh versus Myclow DLBCL.
Comparisons between WT-Myc and MUT-Myc groups Using Unpaired t Test
By unpaired t test, the MUT-Myc group compared with the WT-Myc group had significantly (P < 0.05) higher levels of MDM2, TP63, CD10/MME and CD22, and significantly lower levels of CD44, ICAM1, JAK3, STAT3, STAT5A, TNFSF13B/BAFF, CTLA4 and ICOS mRNA, as well as subtle changes in HLA, PMAIP1/NOXA, TP53, CDKN2A, BCL2, BCL2L11/BIM (which mediates the proapoptotic function of Myc (17,35)), BID, CHUK/IKK1, IKBKB, NFKBIA and NFKBIZ expression, but not in MIR17HG (which mediates the oncogenic function of Myc (36)), E2F1 and EZH2 levels (Supplementary Figures S3–S4). Genes encoding regulators of Myc degradation/stability according to the literature, such as FBXW7, SKP1/2, PIN1, GSK3, PP2A subunits (13), did not show significantly differential expression between the MUT-Myc and WT-Myc groups. Nonetheless, the MUT-Myc group had significant lower FBXW9 expression compared with both WT-Myclow and WT-Mychigh groups.
Comparison of Protein Expression Levels between WT-Myc and MUT-Myc groups Using Unpaired t Test
The MUT-Myc group compared with the WT-Myc group had significantly increased CD10 and decreased nuclear p52 levels. Expression of p53, Ki-67, pAKT, c-Rel, Bcl-2 and p63 levels may also be affected by Myc mutation status (Supplementary Figure S5).
Functional Studies of MYC Mutations and SNPs
We made MYC expression constructs for three mutants (S159R, G160S, and P164L) and two germline variants including N11S and P57S (by SNP rs28933407), and introduced them into MYC-null Rat1a fibroblasts. We first determined the expression of Myc in Rat1a cells and found that all three mutants and the germline variant N11S resulted in lower Myc protein levels, whereas P57S variant had higher Myc expression, in line with a previous report (P57S was considered as a Myc mutant) (17) (Figure 4E). We seeded 5×104 cells in 6-well plates and 72 h later, adherent cells were enumerated. Cells with Myc-P57S grew fastest and cells with WT-Myc grew modestly faster than did the controls with the parental vector (Figure 4F). Cells with Myc-N11S and Myc-P164L proliferated at similar rates as WT-Myc, yet cells with Myc-S159R and Myc-G160S had a significantly slower rate than WT-Myc. We next asked whether any of the mutations altered the well-known ability of Myc overexpression to sensitize cells to apoptosis induced by serum withdrawal. Cells expressing WT and N11S Myc were sensitized to serum withdrawal-induced apoptosis, while cells with P57S, S159R, G160S and P164L Myc showed apoptosis resistance (Figure 4G). In anchorage-independent colony formation assay, Myc-P57S greatly enhanced the transformation ability compared to WT-Myc, consistent with the previous report (17); however, N11S, S159R, G160S and P164L Myc had compromised transformation ability compared with WT-Myc (Figure 4H). To further assess the tumorigenesis of these Myc mutants in vivo, xenograft in nude mice was applied. Rat1a cells stably expressing WT-Myc or its mutants were subcutaneously implanted to 8 weeks old male nude mice (10 implantation each). Eighteen days post inoculation, significant tumors were visible in mice injected with cells expressing P57S, WT, and P164L Myc with different tumor volumes; cells with G160S, S159R and N11S were able form tumors at day 27, 27 and 39 respectively compared to cells with the parental vector (Figure 4I). Tumorigenesis effect (P57S >WT >P164L >G160S >S159R >N11S, Figure 4I) was more correlated with Myc expression levels (P57S >WT >P164S >G160S >S159R and N11S, Figure 4E) than colony formation (P57S >WT >S159R >G160S >N11S >P164L, Figure 4H), cell proliferation (P57S >WT, P164L, and N11S >G160S >S159R, Figure 4F), or apoptosis (WT >N11S >S159R >P164L, G160S, and S159R, Figure 4G). Nonetheless, these data suggest that a substantial number of MYC CDS mutations are of “loss-of-function” rather than “gain-of-function”.
DISCUSSION
In 750 de novo DLBCL patients, we found somatic mutations either in the MYC CDS or UTR in 33.3% of DLBCL patients; 71.4% of these mutations were heterozygous. Nonsynonymous mutations were found in 16.1% of DLBCL patients. The mutation frequency in DLBCL by previous studies was 32% for MYC exon 1 and 2 areas by Sanger sequencing, 6.3% for MYC-CDS by next generation sequencing (25,26), and 29% for BCL2 nonsynonymous mutation by RNA-seq and Sanger sequencing methods (37). We acknowledge the limitation of our data lacking paired normal DNA for each patient, and the potential false mutations due to sample and sequencing limitations. However, the frequency (6.5%) of the Myc-11S germline variant found in our cohort was comparable to the 7.2% and 8.1% by two previous studies (38) (38,39). Myc-N11S was associated with better survival in our cohort, and was controversially associated with, or not associated with breast cancer risk by the two previous studies.
Supplementary Table S8 and Figures 4J–K summarize the major findings by this study. The effects of Myc mutations on Myc stability, function, and apoptosis have been inconsistent in previous studies (8,13). Contrary to the notion that tumor-derived MYC mutations are associated with gain-of-function (10,12) and poorer clinical outcomes, our study showed that most Myc mutants (resulted from nonsynonymous MYC-CDS mutations), carried by approximately 6% to 15% (i.e., the frequencies for group 2 and 3 mutations in Figure 2C and all non-T58/F138 mutants in Figures 2C–D) of DLBCL patients, were often associated with better patient survival compared with WT-Myc (NG_007161.1), regardless of Myc expression levels. This correlation and functional study results may suggest that many Myc mutations attenuated Myc oncogenic function, potentially due to functional changes or haploinsufficiency effects (40). Attenuated pro-apoptosis function may paly roles in tumorigenesis (17,18,41). Moreover, identified Mychigh GEP signatures may suggest that to a certain extent, there were differences in tumor survival, proliferation, and microenvironment between the WT-Myc and MUT-Myc groups.
In contrast, T58 mutations, which are frequent in BL and have gain-of-function in vitro and in vivo (13), were associated with significantly poorer survival than other DLBCL patients. However, the occurrence was low in DLBCL (0.8% of the training set, and 0.5% of the combined training and validation cohorts) and associated with MYC rearrangement (an independent prognostic factor for adverse survival). In addition, the recurrent MYC-3´UTR mutations in 1.3% of DLBCL patients were associated with significant poorer survival than WT-MYC-3´UTR. TargetScan indicated that *22C, *83G, *345C of MYC-3´UTR are targeted by miR-196b, miR-33b, and miR-429, respectively. Intriguingly, 3´UTR mutations were not associated with Myc overexpression, suggesting the presence of MYC suppression by multiple microRNAs (42) and posttranslational regulations, and that the molecular mechanisms underlying the adverse prognostic impact of these 3´UTR mutations may not be simply Myc activation due to the disruption of microRNA-mediated MYC suppression. GEP analysis showed that TNRC6B; which plays important roles in microRNA-mediated suppression, was significantly downregulated in patients harboring recurrent MYC-3´UTR mutations. In addition, MAGEA2/MAGEA2B and ZNF415 which inhibit p53 transcription activities were upregulated, and RAD1 which plays a role in DNA repair was downregulated (Supplementary Table S7).
In this study, MYC mutations were not associated with TP53 mutations which increase genomic instability. It has been proposed that MYC mutations originated from aberrant somatic hypermutation initiated by activation-induced cytidine deaminase (AID) (25). In the present study, the following features of MYC mutations resemble those of AID activities, which may support this origin hypothesis: (a) the predominance of single nucleotide mutations; (b) the elevated transition/transversion mutation ratios compared with SNP variants (Figure 1D) (25); (c) the high to low mutation rate ranged from the 5´UTR, CDS, to 3´UTR, consistent with the AID action pattern (Figure 4J); (d) the association between CDS and 5´UTR mutations; (e) the higher frequency of MYC mutations in MYC and BCL6 rearranged cases (MYC rearrangement is thought to be mediated by AID activities (43)) (supplemental Table S2); and (f) the elevated mutation rate in the MYC 5´UTR compared with other genes we sequenced concurrently (except BCL6 5´UTR) (Figure 1C) (25,44–46). MYC translocation break-points in DLBCL may have affected the mutation rate in 5´UTR and CDS (47,48). The predominance of heterozygous instead of homozygous mutations suggested that most mutations happen during or after the translocation event. Moreover, the association between MUT-5´UTR and Bcl-6 expression, and between MYC-CDS mutation and BCL6 rearrangement in ABC-DLBCL (Supplementary Table S2 and Supplementary Table S5), as well as the high transition/transversion ratio of MYC mutations may suggest that genome instability caused by Bcl-6 expression (49), and loss of protective high-fidelity repair (50) may be additional mutation mechanisms during lymphomagenesis. As most MYC-CDS and MYC-5´UTR mutations did not appear to adversely impact prognosis, our results suggest that most MYC mutations were just passenger mutations acquired during tumorigenesis driven by other oncogenic mechanism. This may also explain why the ABC subtype (post-GC) having high AID expression (43) also had MYC mutations (with a lower frequency than the GCB subtype, Table 1) which did not show significant prognostic effect or Mychigh GEP signatures. These MYC mutations might be insufficient for tumor onset, and occurred either during GC reaction or concurrently with other post-GC transforming events.
In summary, MYC mutations are also present in DLBCL (in addition to Burkitt lymphoma) and have differential functional and clinical effects. Particular mutations such as T58/F138 mutations and some MYC-3´UTR mutations, were found in approximately 2% of DLBCL cases and associated with significantly poorer prognosis. In contrast with these infrequent mutations which may have roles in lymphomagenesis, most MYC-CDS mutations (in approximately 15% of DLBCL) were associated with better clinical outcomes compared with the canonical WT-Myc-11N and probably passenger mutations during lymphomagenesis. The Myc-N11S germline variant (in 6.5% of DLBCL) was also associated with better clinical outcomes compared with the canonical WT-Myc-11N. This study provides knowledge of MYC mutations and variations in DLBCL, supports the oncogenic role of the canonical WT-Myc, and has important clinical and therapeutic implications.
Supplementary Material
Translational Relevance.
MYC mutations in diffuse large B-cell lymphoma (DLBCL) are not as well studied as MYC translocations, another form of MYC genetic aberrations. This study fills in this knowledge gap by profiling MYC gene mutations and germline variations in a large group of DLBCL patients, and attempted to understand their impact on Myc function and clinical outcomes. We found a wide range of single nucleotide variations of MYC genes in DLBCL which correlated with different clinical outcomes. Mutations known to have gain-of-functions implicated in the pathogenesis of Burkitt lymphoma by previous studies were not frequent in DLBCL, whereas most MYC mutations were associated with better clinical outcomes. These results suggested that most MYC mutations in DLBCL were probably passenger mutations instead of driver mutations during lymphomagenesis. This study showed, for the first time, the clinical significance of MYC mutations in DLBCL, and supports the oncogenic role of MYC.
Acknowledgments
ZYXM is the recipient of the Harold C. and Mary L. Daily Endowment Fellowship and Shannon Timmins Fellowship for Leukemia Research.
FUNDING SUPPORT
This study is supported by National Cancer Institute and National Institutes of Health grants (R01CA138688 and R01CA187415, YL and KHY). KHY is also supported by The University of Texas MD Anderson Cancer Center Institutional Research Grant Award, an MD Anderson Lymphoma Specialized Programs of Research Excellence (SPORE) Research Development Program Award, an MD Anderson Myeloma SPORE Research Developmental Program Award, MD Anderson Collaborative Research Funds with High-Throughput Molecular Diagnostics, Gilead Pharmaceutical and Roche Molecular Systems. The study is also partially supported by P50CA136411 and P50CA142509, and the MD Anderson Cancer Center Support Grant CA016672.
Footnotes
DISCLOSURE OF CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Tzankov A, Xu-Monette ZY, Gerhard M, Visco C, Dirnhofer S, Gisin N, et al. Rearrangements of MYC gene facilitate risk stratification in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Mod Pathol. 2014;27:958–971. doi: 10.1038/modpathol.2013.214. [DOI] [PubMed] [Google Scholar]
- 2.Albihn A, Johnsen JI, Henriksson MA. MYC in oncogenesis and as a target for cancer therapies. Adv Cancer Res. 2010;107:163–224. doi: 10.1016/S0065-230X(10)07006-5. [DOI] [PubMed] [Google Scholar]
- 3.Johnson NA, Slack GW, Savage KJ, Connors JM, Ben-Neriah S, Rogic S, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu S, Xu-Monette ZY, Tzankov A, Green T, Wu L, Balasubramanyam A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021–4031. doi: 10.1182/blood-2012-10-460063. quiz 4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horn H, Ziepert M, Becher C, Barth TF, Bernd HW, Feller AC, et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121:2253–2263. doi: 10.1182/blood-2012-06-435842. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith-Sorensen B, Hijmans EM, Beijersbergen RL, Bernards R. Functional analysis of Burkitt’s lymphoma mutant c-Myc proteins. J Biol Chem. 1996;271:5513–5518. doi: 10.1074/jbc.271.10.5513. [DOI] [PubMed] [Google Scholar]
- 9.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 10.Wasylishen AR, Chan-Seng-Yue M, Bros C, Dingar D, Tu WB, Kalkat M, et al. MYC phosphorylation at novel regulatory regions suppresses transforming activity. Cancer Res. 2013;73:6504–6515. doi: 10.1158/0008-5472.CAN-12-4063. [DOI] [PubMed] [Google Scholar]
- 11.Yano T, Sander CA, Clark HM, Dolezal MV, Jaffe ES, Raffeld M. Clustered mutations in the second exon of the MYC gene in sporadic Burkitt’s lymphoma. Oncogene. 1993;8:2741–2748. [PubMed] [Google Scholar]
- 12.Chakraborty AA, Scuoppo C, Dey S, Thomas LR, Lorey SL, Lowe SW, et al. A common functional consequence of tumor-derived mutations within c-MYC. Oncogene. 2015;34:2406–2409. doi: 10.1038/onc.2014.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16:288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- 15.Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 2011;71:925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pulverer BJ, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett JR. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 17.Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuttler F, Ame P, Clark H, Haughey C, Mougin C, Cahn JY, et al. c-myc box II mutations in Burkitt’s lymphoma-derived alleles reduce cell-transformation activity and lower response to broad apoptotic stimuli. Oncogene. 2001;20:6084–6094. doi: 10.1038/sj.onc.1204827. [DOI] [PubMed] [Google Scholar]
- 19.Herbst A, Hemann MT, Tworkowski KA, Salghetti SE, Lowe SW, Tansey WP. A conserved element in Myc that negatively regulates its proapoptotic activity. EMBO Rep. 2005;6:177–183. doi: 10.1038/sj.embor.7400333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabbitts TH, Forster A, Hamlyn P, Baer R. Effect of somatic mutation within translocated c-myc genes in Burkitt’s lymphoma. Nature. 1984;309:592–597. doi: 10.1038/309592a0. [DOI] [PubMed] [Google Scholar]
- 21.Taub R, Moulding C, Battey J, Murphy W, Vasicek T, Lenoir GM, et al. Activation and somatic mutation of the translocated c-myc gene in burkitt lymphoma cells. Cell. 1984;36:339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- 22.Yu BW, Ichinose I, Bonham MA, Zajac-Kaye M. Somatic mutations in c-myc intron I cluster in discrete domains that define protein binding sequences. J Biol Chem. 1993;268:19586–19592. [PubMed] [Google Scholar]
- 23.Cesarman E, Dalla-Favera R, Bentley D, Groudine M. Mutations in the first exon are associated with altered transcription of c-myc in Burkitt lymphoma. Science. 1987;238:1272–1275. doi: 10.1126/science.3685977. [DOI] [PubMed] [Google Scholar]
- 24.Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, et al. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- 25.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 26.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat.Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Jima D, Moffitt AB, Liu Q, Czader M, Hsi ED, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123:2988–2996. doi: 10.1182/blood-2013-07-517177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Li Y, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26:2103–2113. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visco C, Tzankov A, Xu-Monette ZY, Miranda RN, Tai YC, Li Y, et al. Patients with diffuse large B-cell lymphoma of germinal center origin with BCL2 translocations have poor outcome, irrespective of MYC status: a report from an International DLBCL rituximab-CHOP Consortium Program Study. Haematologica. 2013;98:255–263. doi: 10.3324/haematol.2012.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu-Monette ZY, Dabaja BS, Wang X, Tu M, Manyam GC, Tzankov A, et al. Clinical features, tumor biology and prognosis associated with MYC rearrangement and overexpression in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Mod Pathol. 2015;28:1555–1573. doi: 10.1038/modpathol.2015.118. [DOI] [PubMed] [Google Scholar]
- 31.Xu-Monette ZY, Tu M, Jabbar KJ, Cao X, Tzankov A, Visco C, et al. Clinical and biological significance of de novo CD5+ diffuse large B-cell lymphoma in western countries. Oncotarget. 2015;6:5615–5633. doi: 10.18632/oncotarget.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 33.Seitz V, Butzhammer P, Hirsch B, Hecht J, Gutgemann I, Ehlers A, et al. Deep sequencing of MYC DNA-binding sites in Burkitt lymphoma. PLoS One. 2011;6:e26837. doi: 10.1371/journal.pone.0026837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muthalagu N, Junttila MR, Wiese KE, Wolf E, Morton J, Bauer B, et al. BIM is the primary mediator of MYC-induced apoptosis in multiple solid tissues. Cell Rep. 2014;8:1347–1353. doi: 10.1016/j.celrep.2014.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26:262–272. doi: 10.1016/j.ccr.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuetz JM, Johnson NA, Morin RD, Scott DW, Tan K, Ben-Nierah S, et al. BCL2 mutations in diffuse large B-cell lymphoma. Leukemia. 2012;26:1383–1390. doi: 10.1038/leu.2011.378. [DOI] [PubMed] [Google Scholar]
- 38.Wirtenberger M, Hemminki K, Forsti A, Klaes R, Schmutzler RK, Grzybowska E, et al. c-MYC Asn11Ser is associated with increased risk for familial breast cancer. Int J Cancer. 2005;117:638–642. doi: 10.1002/ijc.21225. [DOI] [PubMed] [Google Scholar]
- 39.Figueiredo JC, Knight JA, Cho S, Savas S, Onay UV, Briollais L, et al. Polymorphisms cMyc-N11S and p27-V109G and breast cancer risk and prognosis. BMC Cancer. 2007;7:99. doi: 10.1186/1471-2407-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baudino TA, McKay C, Pendeville-Samain H, Nilsson JA, Maclean KH, White EL, et al. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes & Development. 2002;16:2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang DW, Claassen GF, Hann SR, Cole MD. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol Cell Biol. 2000;20:4309–4319. doi: 10.1128/mcb.20.12.4309-4319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bueno MJ, Gomez de Cedron M, Gomez-Lopez G, Perez de Castro I, Di Lisio L, Montes-Moreno S, et al. Combinatorial effects of microRNAs to suppress the Myc oncogenic pathway. Blood. 2011;117:6255–6266. doi: 10.1182/blood-2010-10-315432. [DOI] [PubMed] [Google Scholar]
- 43.Lenz G, Nagel I, Siebert R, Roschke AV, Sanger W, Wright GW, et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J Exp Med. 2007;204:633–643. doi: 10.1084/jem.20062041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han L, Masani S, Yu K. Overlapping activation-induced cytidine deaminase hotspot motifs in Ig class-switch recombination. Proc Natl Acad Sci U S A. 2011;108:11584–11589. doi: 10.1073/pnas.1018726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelicci PG, Knowles DM, 2nd, Magrath I, Dalla-Favera R. Chromosomal breakpoints and structural alterations of the c-myc locus differ in endemic and sporadic forms of Burkitt lymphoma. Proc Natl Acad Sci U S A. 1986;83:2984–2988. doi: 10.1073/pnas.83.9.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy W, Sarid J, Taub R, Vasicek T, Battey J, Lenoir G, et al. A translocated human c-myc oncogene is altered in a conserved coding sequence. Proc Natl Acad Sci U S A. 1986;83:2939–2943. doi: 10.1073/pnas.83.9.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2:920–932. doi: 10.1038/nri953. [DOI] [PubMed] [Google Scholar]
- 50.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.