Abstract
Purpose
Promising results in the treatment of NSCLC have been seen with agents targeting immune checkpoints, such as PD-1 or PD-L1. However, only a select group of patients respond to these interventions. The identification of biomarkers that predict clinical benefit to immune checkpoint blockade is critical to successful clinical translation of these agents.
Methods
We conducted an integrated analysis of three independent large datasets, including The Cancer Genome Atlas (TCGA) of lung adenocarcinoma and two datasets from MD Anderson Cancer Center, Profiling of Resistance patterns and Oncogenic Signaling Pathways in Evaluation of Cancers of the Thorax (named PROSPECT) and Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination (named BATTLE-1). Comprehensive analysis of mRNA gene expression, reverse phase protein array (RPPA), immunohistochemistry and correlation with clinical data were performed.
Results
Epithelial-mesenchymal transition (EMT) is highly associated with an inflammatory tumor microenvironment in lung adenocarcinoma, independent of tumor mutational burden. We found immune activation co-existent with elevation of multiple targetable immune checkpoint molecules, including PD-L1, PD-L2, PD-1, TIM-3, B7-H3, BTLA and CTLA-4, along with increases in tumor infiltration by CD4+Foxp3+ regulatory T cells in lung adenocarcinomas that displayed an EMT phenotype. Furthermore, we identify B7-H3 as a prognostic marker for NSCLC.
Conclusions
The strong association between EMT status and an inflammatory tumor microenvironment with elevation of multiple targetable immune checkpoint molecules warrants further investigation of using EMT as a predictive biomarker for immune checkpoint blockade agents and other immunotherapies in NSCLC and possibly a broad range of other cancers.
Keywords: Epithelial-mesenchymal transition, PD-1, PD-L1, Immune suppression, Lung adenocarcinoma
Introduction
Non-small cell lung cancer (NSCLC), long thought to be a ‘non-immunogenic’ tumor, has shown responses to immunotherapeutic approaches targeting immune checkpoints programmed cell death 1 (PD-1), programmed death-ligand 1 (PD-L1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), even in heavily pretreated patients (1–3). These findings clearly indicate the crucial role of immune checkpoint pathways in mediating immune tolerance in NSCLC. The PD-1/PD-L1 pathway has emerged as a critical inhibitory pathway that regulates T cell response and maintains immune suppression (1, 2, 4, 5). Although anti-PD-1/PD-L1 treatment can produce durable responses, it appears to benefit only a subset of patients. While the expression levels of PD-L1 on tumor cells and tumor infiltrating immune cells have recently been shown to correlate with clinical response to anti-PD-1 therapy (6–8), only a subset of patients with PD-L1 expressing tumors had clinical response and others without PD-L1 staining demonstrate clinical benefit, indicating that additional factors in the tumor microenvironment exist which define the subgroup of patients who derive benefit (6, 9).
EMT is a key process that drives cancer metastasis, drug resistance and has been associated with poor prognosis in multiple cancers including NSCLC (10–12). However, most studies that investigate the impact of EMT in cancer have mainly focused on metastasis and drug resistance (12, 13). The impact of EMT on reprogramming the tumor immune microenvironment is largely unknown. Our group has previously developed a robust EMT gene signature that correlates with cellular phenotypes and predicts in vitro and in vivo NSCLC resistance to EGFR or PI3K/Akt inhibitors, highlighting differential patterns of drug responsiveness for “epithelial” and “mesenchymal” NSCLC (14). We have also recently demonstrated a molecular link between EMT and intratumoral CD8+ T cell suppression, through regulation of PD-L1 in both animal models and human cell lines (15). To further explore the impact of EMT on tumor immune microenvironment and identify potential biomarkers for selecting patients who might preferentially benefit from PD-1/PDL-1 blockade or immunotherapies more broadly, we conducted an integrated analysis of three independent large patient datasets.
Patients and methods
Clinical datasets and patient sample characteristics
A total of 439 patients from three clinical datasets including The Cancer Genome Atlas (TCGA, n = 230), Profiling of Resistance patterns and Oncogenic Signaling Pathways in Evaluation of Cancers of the Thorax (named PROSPECT hereafter, n = 152) and the Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination (named BATTLE-1 hereafter, n = 57) were included in this study, but analyzed as independent datasets (16). Clinical and demographic information for 230 early-stage, surgically resected lung adenocarcinomas included in the Cancer Genome Atlas (TCGA) dataset was obtained from the TCGA portal (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). The PROSPECT dataset includes tumor tissue collected from patients who underwent surgical resection of NSCLC with curative intent between 1996 and 2008 at MD Anderson Cancer Center. 152 tumors classified as adenocarcinoma resected from patients who had not received neo-adjuvant chemotherapy or radiation therapy and had detectable tumor by H&E immunostaining were included in the current study. Case selections were made by two independent pathologists who evaluated the amount of tumor tissue present in H&E slides corresponding to the paraffin blocks from surgically resected lung specimens available for this study. Cases with at least one representative tumor tissue block containing > 30% viable tumor cells, were selected for further IHC analysis. BATTLE-1 was a randomized, biomarker-based clinical trial for patients with recurrent or metastatic NSCLC in the second-line setting (Trial registration ID: NCT00409968). Core-needle biopsies were required for enrollment. Samples with detectable adenocarcinoma by H&E immunostaining were included in our analysis (n = 57). Clinical characteristics of these samples are described in supplementary table 1. The study was approved by the institutional review boards of MD Anderson Cancer Center.
mRNA expression profiling
Experimental details regarding lung adenocarcinoma from TCGA dataset including RNA extraction from tumors, mRNA library preparation, sequencing (Illumina HiSeq platform), quality control, data processing and quantification of gene expression have been previously reported (17). For the PROSPECT samples, the mRNA was extracted from frozen tumor tissue corresponding to the same specimen from which the formalin fixed, paraffin embedded blocks were made. The frozen tumor tissue was harvested by a pathologist who examined and sampled the gross tissue specimen and only tumor tissues were included for mRNA extraction. Array-based expression profiling of PROSPECT tumors was performed using the Illumina Human WG-6 v3 BeadChip, according to the manufacturer’s protocol. BATTLE-1 adenocarcinoma was profiled using the GeneChip®Human Gene 1.0 ST Array from Affymetrix. Gene expression data for the PROSPECT and BATTLE-1 dataset have been previously deposited in the GEO repository (GSE42127) and (GSE33072) (18, 19), respectively. For each sample, an EMT score was computed using an averaging scheme based on the mRNA expression of 76 genes previously published by our group and originally derived from analysis of NSCLC(14),(15). The scores were calculated as the average expression level of “mesenchymal” genes minus the average expression level of “epithelial” genes. Tumor samples were then classified by EMT score as EMT-low (defined by EMT scores ≤ lowest 1/3) or EMT-high (defined by EMT scores ≥ highest 1/3) in both TCGA and PROSPECT datasets. By comparing the tumor samples with either relatively high or low EMT score, but not intermediate, we expected to increase the likelihood of identifying the immune markers associated with either “epithelial” or “mesenchymal” lung adenocarcinomas. In the BATTLE-1 dataset of metastatic tumor specimens the sample size was much more limited (n = 57) and a narrowed spectrum of EMT scores was observed, with generally more “mesenchymal” phenotypes. This is likely due to the nature of advanced stage/metastatic disease in this patient population and the small amount of sampled tissue for each case versus resection specimens. Based upon this narrowed distribution, a different EMT threshold was used in the BATTLE-1 dataset, defining EMT score < 0 as EMT-low (“epithelial”) and EMT score ≥ 0 as EMT high (“mesenchymal”). The mRNA expression profiles of immune related genes in each sample were then analyzed. The raw data files of transcriptomes were analyzed using Bioconductor R packages.
Non-synonymous mutational burden per Mb of DNA and C >A transversion rate in TCGA tumors were calculated using the MutSig2CV algorithm as published previously (17). This algorithm takes into account recurrence of mutations, nucleotide context, gene-expression, replication time, and somatic background mutation rate. Genes with a Bonferroni-corrected p-value less than 0.025 were deemed significant. It has been previously established that transversion low samples represent tumors from lifelong never-smokers and transversion high samples as tumors from patients with 60 or more pack years (17). A linear discriminant analysis based on GC>AT frequency, GC>TA frequency, and total mutation count by using the R MASS library2 was performed based on previously published methods to classify all samples as belonging to either transversion high or transversion low categories.
Immunohistochemistry staining and reverse phase protein array (RPPA) analysis
For the detailed IHC and RPPA analysis of PD- L1, see the Supplementary Methods section. Briefly, formalin fixed, paraffin embedded tumor blocks from the PROSPECT dataset were selected for immunohistochemistry (IHC) staining. Thirty-five tumor tissues with “mesenchymal” lung adenocarcinomas and 33 tumor tissues with “epithelial” lung adenocarcinomas, as defined by the mRNA expression, from the PROSPECT tissue bank were found to have sufficient quality material for additional analyses and were blinded to the pathologists who performed the IHC study. The markers assessed in this study included: PD-L1, PD1, CD3, CD4, CD8, CD45RO, CD57, CD68, Granzyme B and FOXP3 (see Supplementary Methods for antibody details). All antibodies were detected with the Leica Bond Polymer Refine detection kit (Leica Microsystems), including diaminobenzidine reaction to detect the antibody labeling and hematoxylin counterstaining. The stained slides were digitally scanned using the Aperio® ScanScope Turbo slide scanner (Leica Microsystems). The digital images were captured at ×20 magnification.
For RPPA analysis of TCGA and PROSPECT specimens, the tumors were lysed as we have previously published (20). Five serial dilutions of each protein lysate were printed on nitrocellulose-coated slides using an Aushon Biosystems 2470 arrayer (Burlington, MA) and stained sequentially with primary and secondary antibodies in an autostainer (BioGenex), prior to signal detection using a signal amplification system and DAB-based colorimetric reaction. MicroVigene Software (VigeneTech) as well as an in-house R package was used to assess spot intensities and the SuperCurve method was applied to estimate protein levels in each sample. For comparisons, data were log transformed (to the base of 2) and median-centered across antibodies to correct for protein loading. Rabbit polyclonal antibody to PD-L1 (Cat: ab174838, Abcam, MA) at 1:250 dilution was used to detect PD-L1 expression by RPPA. Differences in protein expression between “epithelial” and “mesenchymal” lung adenocarcinoma tumor samples were compared by t-test. Pearson correlation between CD274 (PD-L1) mRNA expression and PD-L1 protein expression by RPPA were assessed, along with E-cadherin protein RPPA and PD-L1 RPPA expression. All statistical analyses were performed using R packages (version 2.10.0).
Statistical Analysis
Statistical analyses were conducted using GraphPad Prism version 6.00 (GraphPad Software, La Jolla) or alternatively, the R system (version 2.10.0) for statistical computing. The un-paired t-test was used for comparisons between two-group means, where the data could be assumed to have been sampled from populations with normal (or approximately normal) distributions. Mann-Whitney U test was used to compare the mean ranks between two groups. All P values are two-tailed and for all analyses, P ≤ 0.05 is considered statistically significant, unless otherwise specified.
Results
PD-1, PD-L1 and PD-L2 are significantly elevated in “mesenchymal” lung adenocarcinoma
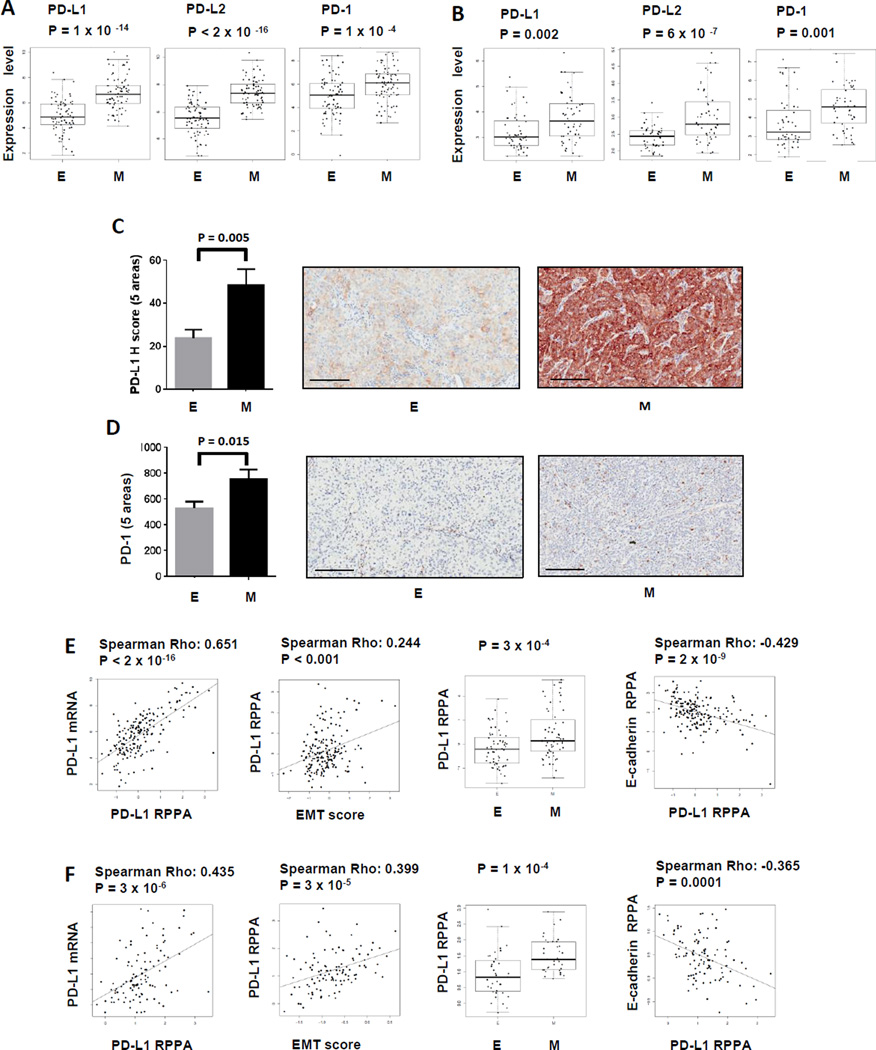
The PD-1:PD-L1/PD-L2 pathway has been implicated in tumor escape from immune destruction in various cancers, including NSCLC (21). Although the expression of PD-L1 in NSCLC is an area of active study, whether specific subgroups of NSCLC patients display distinct PD-L1 expression patterns is unknown. Our recent report demonstrated that “epithelial” and “mesenchymal” NSCLC have distinct patterns of drug responsiveness to EGFR and PI3K/Akt inhibitors. To study the expression of the PD-1:PD-L1/PD-L2 axes in “epithelial” and “mesenchymal” lung adenocarcinomas, mRNA expression data from TCGA and PROSPECT datasets were analyzed. As shown in Fig 1A and 1B, the expression levels of PD-L1, PD-L2 and PD-1 were significantly elevated in “mesenchymal” versus “epithelial” tumors in both TCGA and PROSPECT, consistent with our recent findings (15). To confirm whether the elevated mRNA expression of PD-L1 and PD-1 correlated to protein levels, immunohistochemical (IHC) staining and scoring of PD-L1 (on tumor cells) and PD-1 (on tumor infiltrating immune cells) were performed for full tumor sections from a subset of cases in the PROSPECT dataset for which we have sufficient tumor tissues of high quality for IHC analysis (n=68). For these analyses we validated the specificity of the antibody E1L3N (Cell Signaling Technology), which we employed for the IHC PD-L1 staining (Supplementary Figure 1A–G). As shown in Fig 1C and 1D, we found significantly higher expression of PD-L1 and PD-1 in “mesenchymal” than in “epithelial” adenocarcinoma cases (P = 0.005 and P = 0.015, respectively). We also addressed this question by using reverse phase protein arrays (RPPA), an independent technique to detect and quantify the protein levels. We first validated the specificity of the antibody (Abcam 174838) used for RPPA versus the widely used 5H1 antibody, by Western blot of cell line lysates, IHC of cell pellets, IHC of control placenta samples, and tumor samples with a range of PD-L1 expression on tumor cells and infiltrates (Supplementary Figure 2A–H). The PD-L1 protein levels measured by RPPA correlated strongly with the PD-L1 mRNA levels (CD274) in both independent datasets (Fig. 1E & F). We further demonstrated a positive correlation between PD-L1 protein by RPPA and EMT score, when assessed as a continuous variable, or significantly higher levels of PD-L1 in “mesenchymal” as compared to “epithelial” tumors, when analyzed by group (Fig 1E & F). The expression of E-cadherin, an epithelial marker that is also present on the RPPA panel, strongly negatively correlated with PD-L1 protein expression in TCGA samples (Fig 1E) and the independent PROSPECT dataset (Fig 1F).
Figure 1. Elevated PD-1:PD-L1/PD-L2 axis in “mesenchymal” versus “epithelial” lung adenocarcinoma.
“Epithelial” lung adenocarcinoma is defined by EMT scores ≤ lowest 1/3 as described in methods and represented as E. Similarly, “mesenchymal” lung adenocarcinoma is defined by EMT scores ≥ highest 1/3 and represented as M. Gene expression levels of PD-1: PD-L1/PD-L2 axis in TCGA dataset (A), PROSPECT dataset (B). PD-L1 (C) and PD-1 expression (D) by IHC in tumors from PROSPECT dataset are shown. 35 and 33 tumor tissues with “mesenchymal” or “epithelial” lung adenocarcinomas were used for the IHC study. Five random regions (1 mm2) in the core of each tumor in each group were analyzed. Unpaired T test was performed. 200 µm scale bar is shown in each representative IHC picture. PD-L1 RPPA correlated to PD-L1 mRNA or EMT score, RPPA in “epithelial” versus “mesenchymal” lung adenocarcinomas, and versus E-cadherin RPPA in TCGA (E) and PROSPECT (F) samples.
A distinct tumor microenvironment immune profile with elevation of multiple immune checkpoint molecules is revealed in “mesenchymal” lung adenocarcinoma
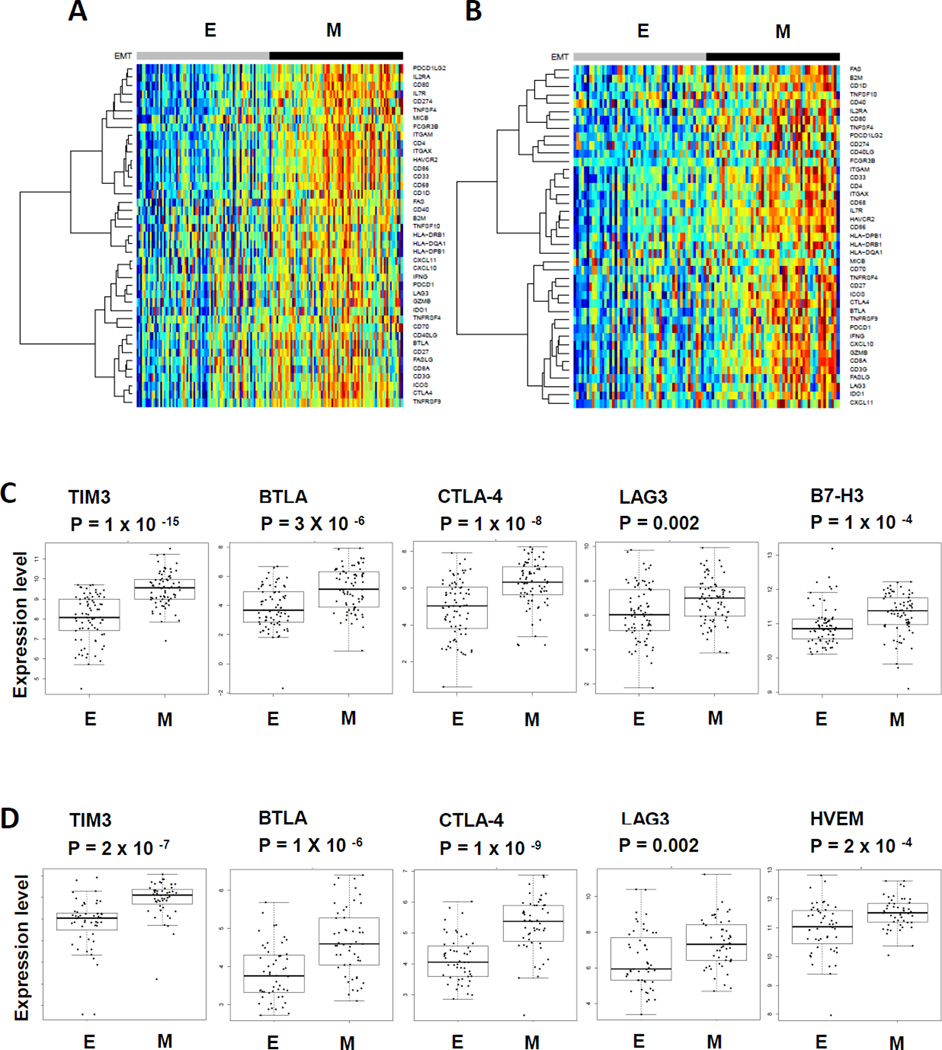
The findings of elevated PD-1:PD-L1/PD-L2 axis in “mesenchymal” lung adenocarcinoma prompted us to investigate whether there is an association with a broader immunosuppressed phenotype beyond the PD-1:PD-L1/PD-L2 axis. To address this question, a comprehensive list of 89 immune-related genes including co-stimulatory molecules, immune checkpoints, cytokines, chemokines, MHC class I and II, and genes highly expressed on dendritic cells, T cells, NK cells, myeloid cells and macrophages was generated from the literature (22) (Supplementary Table 2). There is no overlap between the immune-related gene list and the previously identified EMT gene signature (19). The mRNA expression of each immune-related molecule was tested in the “mesenchymal” versus “epithelial” adenocarcinomas in TCGA and PROSPECT datasets. Strikingly, profound immune-related phenotypic changes were found in “mesenchymal” lung adenocarcinoma in contrast to much lower expression of immune-related molecules in “epithelial” lung adenocarcinoma (Fig 2A and 2B). Notably, multiple immune checkpoint molecules were significantly elevated in TCGA and PROSPECT, including T cell Immunoglobulin and Mucin protein-3 (TIM-3), B- and T-lymphocyte attenuator (BTLA), cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), lymphocyte-activation protein 3 (LAG-3) and B7-H3 (Fig 2C and 2D). Similar to PD-1, TIM-3, BTLA, CTLA-4, LAG-3 and B7-H3 all negatively regulate T cell function through relatively unique and potentially non-overlapping molecular mechanisms (23). Co-expression of multiple immune checkpoint molecules has been frequently found on exhausted T cells in tumors and chronic infections (23). Although the expression pattern of elevated immune checkpoint molecules was largely consistent between TCGA and PROSPECT, some differences were observed. For example, B7-H3 was significantly elevated in lung adenocarcinoma in TCGA dataset, but not in the PROSPECT dataset. By contrast, herpes virus entry mediator (HVEM) was elevated in PROSPECT, but not in TCGA. Although both datasets comprised patients with mainly surgically resectable disease, more patients with stage IV (4%) were included in TCGA than in PROSPECT (0%), indicating that potential differences in the patient population might contribute to the observed molecular differences. Of note, although the association between this distinct tumor microenvironment immune profile and “mesenchymal” lung adenocarcinoma was most readily observed when the data was analyzed as the top versus bottom 1/3 group of cases, we observed similar results when analyzed in either a continuous manner or categorized into EMT low, EMT intermediate and EMT high groups (Supplementary Figure 3A–D).
Figure 2. Elevation of multiple immune checkpoint molecules in “mesenchymal” as compared to “epithelial” lung adenocarcinoma.
Supervised cluster heatmap of immune related molecules in “epithelial” lung adenocarcinoma versus “mesenchymal” lung adenocarcinoma from TCGA (A), and PROSPECT (B) respectively. Expression levels of immune checkpoint molecules in “epithelial” lung adenocarcinoma versus “mesenchymal” lung adenocarcinoma in tumor tissues from TCGA (C) and PROSPECT (D) respectively.
Significant increase in tumor infiltrating CD4+FOXP3+ regulatory T cells is found in “mesenchymal” lung adenocarcinoma
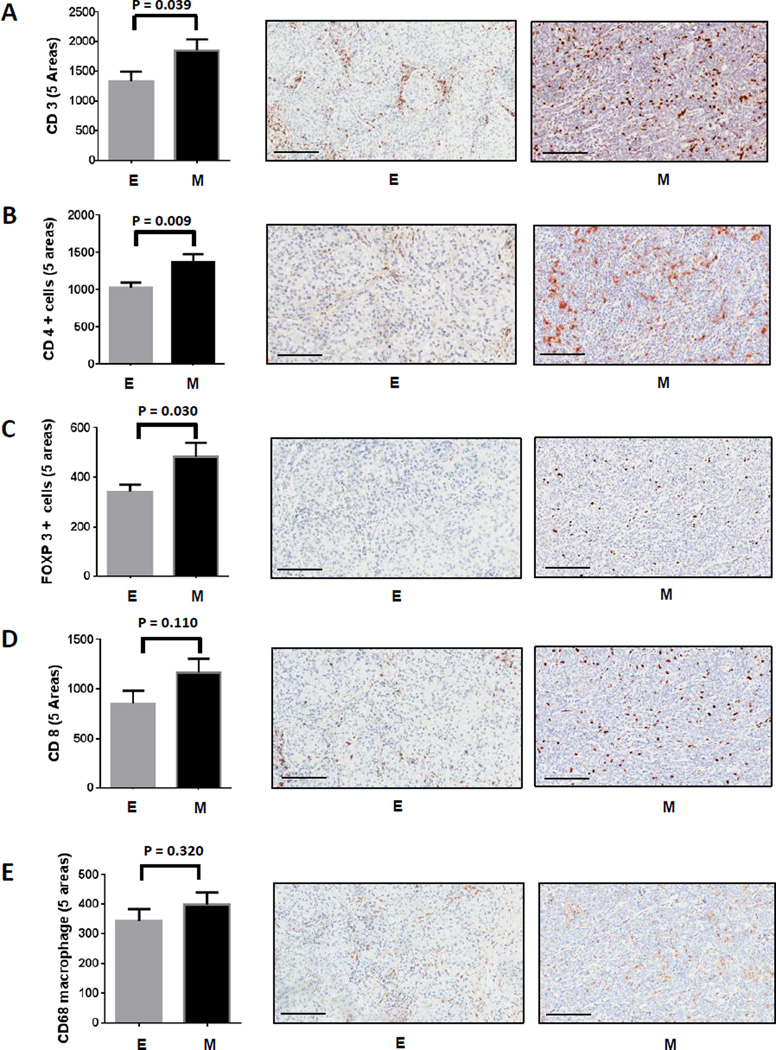
The expression of multiple immune suppressive molecules in “mesenchymal” lung adenocarcinoma suggested the possibility of a complex immunosuppressive tumor microenvironment. To explore the cellular consequence of this observation, “mesenchymal” and “epithelial” lung adenocarcinoma specimens from the PROSPECT study were stained and scored in a blinded fashion for CD3, CD4, CD8, FOXP3, CD68, CD45RO, CD57 and Granzyme B, well-established IHC markers for T cell subsets, regulatory T cells, macrophages and natural killer cells, respectively. As shown in Fig 3A, a robust increase in tumor infiltrating CD3+ T cells was found in “mesenchymal” as compared to “epithelial” lung adenocarcinoma (P = 0.039). We found much higher levels of tumor infiltration by CD4+ and FOXP3+ T cells in “mesenchymal” lung adenocarcinomas (P = 0.009 and P = 0.030 respectively) (Fig 3B and 3C). These likely represent CD4+FOXP3+ regulatory T cells, although we are unable to confirm that interpretation since methodology for double staining cells was not used. Although there was a trend toward more tumor infiltrating CD8+ T cells in “mesenchymal” samples, the difference was not statistically significant. As shown in Fig 3E, we observed no significant difference in tumor infiltration of total CD68+ macrophages, suggesting that the CD4+ and FOXP3+ T cells rather than tumor-associated macrophages likely played a greater role in this EMT-associated immunosuppressive phenotype. However, additional staining to assess M1 vs M2 macrophages was not evaluated in this study. No significant difference was detected in CD45RO, CD57 or Granzyme B between the “mesenchymal” and “epithelial” lung adenocarcinoma samples (data not shown).
Figure 3. Increased tumor infiltrating CD3+ T cells and CD4+FOXP3+ regulatory T cells, but not CD8+ T cells and macrophages in “mesenchymal” versus “epithelial” lung adenocarcinoma.
IHC staining and scoring of CD3 (A), CD4 (B), FOXP3 (C), CD8 (D) and CD68 (E) were performed in tumors from PROSPECT. 35 and 33 tumor tissues with “mesenchymal” or “epithelial” lung adenocarcinomas were used for the IHC study, respectively. The staining and density of each marker were analyzed using the cell membrane staining algorithm, except for FOXP3, which used a nuclear staining algorithm. Five random regions (1 mm2) in the core of each tumor at the same region in each group were analyzed. 200 µm scale bar is shown in each representative IHC picture.
Elevation of multiple immune-stimulatory molecules and interferon gamma signals are found in “mesenchymal” lung adenocarcinoma
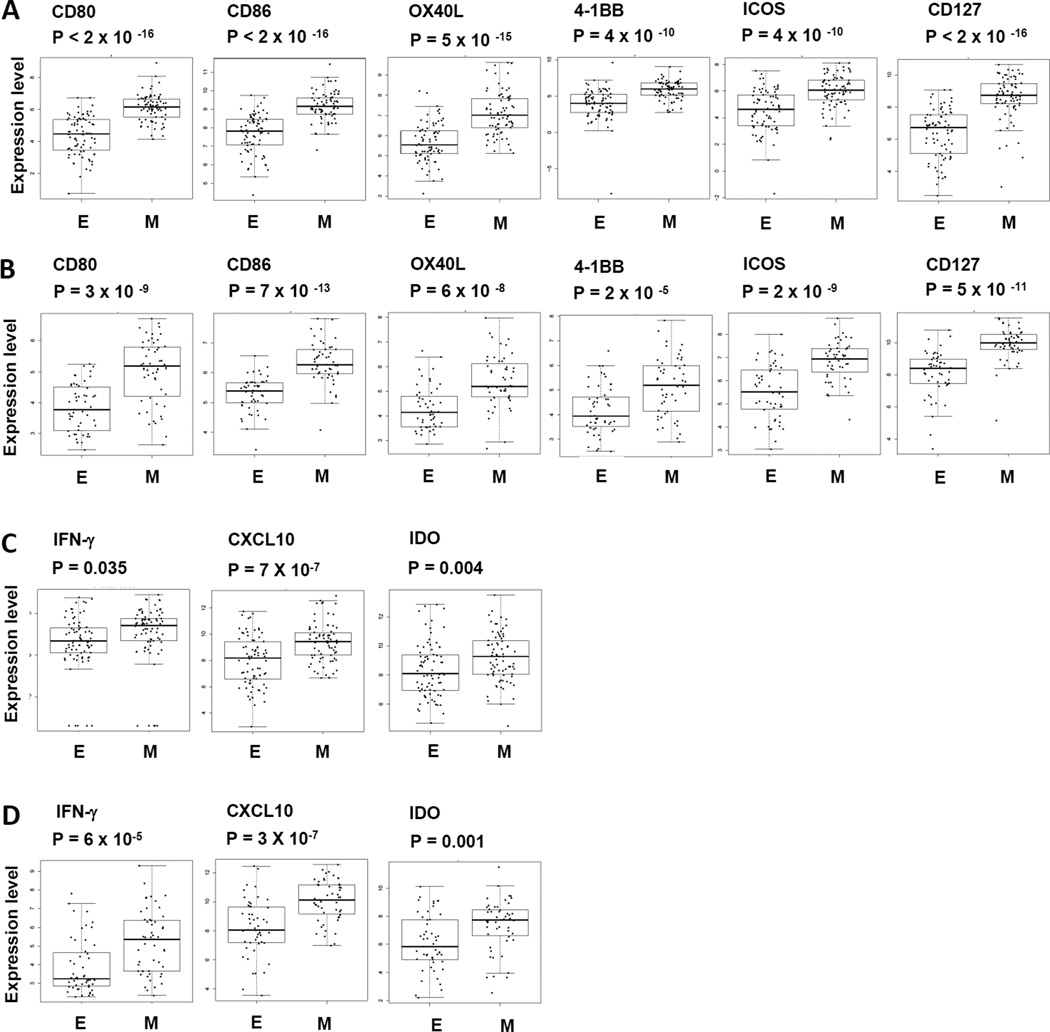
Although PD-L1 is generally viewed as an immune inhibitory molecule, its expression has been reported to reflect an ongoing anti-tumor immune response (9, 24, 25). To test if the immune suppressive profile we found in “mesenchymal” lung adenocarcinoma is associated with the infiltrating immune response, analysis of gene expression was performed on an extensive panel of immune stimulatory molecules. As shown in Fig 4A–B, multiple immune stimulatory molecules including CD80, CD86, OX40L, 4-1BB, ICOS and CD127 were significantly elevated in “mesenchymal” as compared to “epithelial” lung adenocarcinoma. In addition, as shown in Fig 4C–D, interferon gamma (IFN-γ), IFN-γ-induced protein 10 (CXCL10) and an IFN-γ inducible enzyme indoleamine 2,3-dioxygenase (IDO) were also significantly elevated in “mesenchymal” lung adenocarcinoma as compared to the “epithelial” samples. Together, these data indicate an ongoing immune response in the tumor microenvironment of lung adenocarcinomas with “mesenchymal” phenotype. The elevated IFN-γ released from ongoing host immune response most likely contributed to the increased PD-L1 expression and activation of other immune suppressive molecules observed in “mesenchymal” lung adenocarcinoma. Once again, the results from the independent TCGA and PROSPECT datasets were highly consistent with one another.
Figure 4. Evidence of immune activation in “mesenchymal” compared to “epithelial” adenocarcinoma.
Gene expression levels of multiple immune stimulatory molecules including CD80, CD86, OX40L, 4-1BB, ICOS, CD127 were analyzed in “mesenchymal” lung adenocarcinoma versus “epithelial” lung adenocarcinoma in tumor tissues from (A) TCGA, (B) PROSPECT. Gene expression of IFN-γ and IFN-γ inducible genes including CXCL10 and IDO were also analyzed in “mesenchymal” in comparison to “epithelial” lung adenocarcinoma in tumor tissues from (C) TCGA, (D) PROSPECT.
EMT is not associated with tumor mutational burden, indicating an independent factor mediating an inflammatory tumor microenvironment
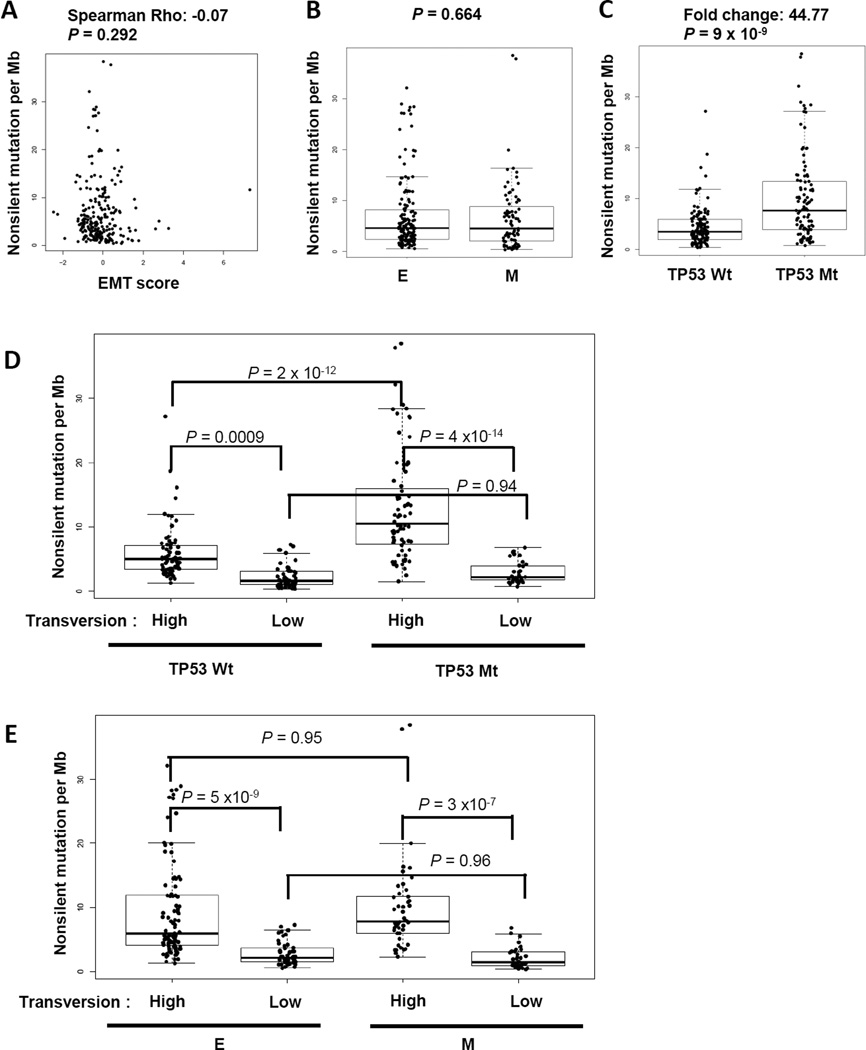
Genomic mutational burden has been posited as a marker for response to immunotherapy (26, 27). To investigate if the profound inflammatory tumor microenvironment we observed in “mesenchymal” lung adenocarcinoma is confounded by increases in mutational burden, we studied the correlation of EMT and tumor mutation frequency. Using the TCGA dataset, for which we have DNA/RNA sequencing data and can assess mutational burden, there was no correlation between tumor mutational burden, as measured by non-synonymous mutation per Mb, and EMT status (Fig 5A–B). By contrast, and as expected, there was a significantly higher mutational burden found in NSCLC tumors with TP53 mutation as compared to tumors with wild-type TP53 (~45-fold higher) (Fig 5C). Smoking is known to cause a high rate of mutations, including TP53, and high frequency of cytosine to adenine (C >A) nucleotide transversions has been identified as a surrogate marker for amount of smoking exposure (17, 28). We next tested the possible impact of EMT status on mutational burden, using nucleotide tranversion as a surrogate measure for amount of tobacco exposure. Patients with high frequency of transversion (C >A) carried significantly higher mutational burden than patients with low transversion, regardless of the status of TP53 (Fig. 5D). However, the presence of both TP53 mutation and high transversion frequency led to even higher mutational burden than the combination of transversion high and wild-type TP53, indicating the additional or synergistic effects between smoking and TP53 mutation. In contrast, no impact of EMT status on mutational burden was found, regardless of smoking exposure measured by transversion frequency (Fig 5E). This data indicates that EMT is not a surrogate for mutational burden and is an independent mediator driving changes in the tumor immune microenvironment. Interestingly, although no correlation was seen between total mutational burden and EMT, we investigated the correlation between commonly-identified individual mutations in lung adenocarcinoma and EMT, which revealed a significantly higher frequency of STK11 (p = 0.001) or KEAP1 (3.9 × 10−5) mutations in “epithelial” than “mesenchymal” lung adenocarcinomas (Supplementary Table 3). None of the other studied mutations, e.g. TP53, KRAS, EGFR, or BRAF, were associated with a difference in the “epithelial” or “mesenchymal” groups (Supplementary Table 3).
Figure 5. EMT is not associated with tumor mutational burden.
Non-silent mutational rate, transversion rate and TP53 mutation of TCGA lung adenocarcinoma samples were included in the analysis. (A) Spearman correlation between non-silent mutational rate per Mb and EMT score. (B) Non-silent mutational rate in “epithelial” versus “mesenchymal” lung adenocarcinoma. (C) Non-silent mutational rate in TP53 mutant versus TP53 wild-type lung adenocarcinoma. (D) Non-silent mutational rate in TP53 mutant versus wild-type lung adenocarcinoma further separated by molecular smoking exposure via transversion high and low. (E) Non-silent mutational rate in “epithelial” versus “mesenchymal” lung adenocarcinoma further separated by molecular smoking exposure via transversion high and low.
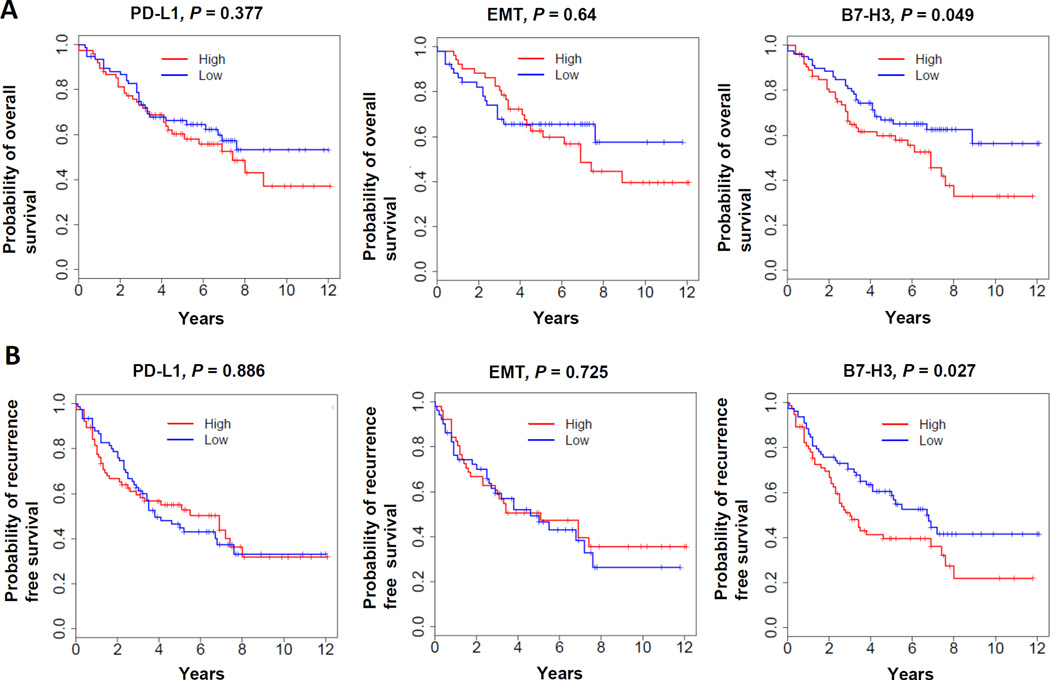
B7-H3 is associated with poor OS and RFS in lung adenocarcinoma
We next analyzed the correlation between immune checkpoint molecules, EMT status and overall survival (OS) or recurrence free survival (RFS). Neither PD-L1 nor EMT status was prognostic of OS or RFS. Among all the immune checkpoint molecules, only B7-H3 was found to negatively correlate with OS and RFS (Figure 6 and Supplementary Figure 4 using 1/2 or 1/3 cut-off, respectively). This may indicate a potential pathway to explore in therapeutic targeting of lung adenocarcinoma with immunotherapy.
Figure 6. B7-H3 is associated with poor OS and RFS in lung adenocarcinoma.
The probability of overall survival and recurrence free survival of patients from PROSPECT were analyzed by dividing the patients into either high or low group based on the expression levels of each immune checkpoint molecule or EMT score. Patients with gene expression levels higher or lower than average expression level of each gene or EMT score are considered as high or low respectively. (A) Overall survival, (B) Recurrence free survival. Both are from PROSPECT.
Analysis of an independent dataset of advanced lung adenocarcinoma confirms the association of EMT with inflammatory tumor microenvironment
Our analyses using the two independent datasets, TCGA and PROSPECT, demonstrated consistent conclusions in surgically-resected early-stage lung adenocarcinoma samples. To test whether the association between EMT and a profound inflammatory/immunosuppressive tumor microenvironment is limited to early-stage lung adenocarcinoma or more broadly applicable to all clinical stages, we next analyzed the gene expression profile of biopsy specimens from the BATTLE-1 study (16). This dataset consists only of core-needle biopsy specimens from patients with advanced, treatment refractory NSCLC. For the sake of consistency with our TCGA and PROSPECT analyses, we only included the lung adenocarcinoma samples for this study. As shown in Supplementary Fig 5A, we observed a similar distinction in the immune-related marker profiles. Multiple immune checkpoint molecules, including PD-L1, PD-L2, TIM3 and CTLA-4 were significantly elevated in “mesenchymal” as compared to “epithelial” lung adenocarcinoma specimens (Supplementary Fig 5B). Furthermore, multiple immune stimulatory molecules, including CD80, CD86, OX40L, 4-1BB, CD127, IFN-γ-induced protein 10 and IFN-γ inducible enzyme IDO were also significantly elevated in “mesenchymal” compared to the “epithelial” samples (Supplementary Fig 5C and 5D). Taken together, our data demonstrate that the association of EMT with elevation of inflammatory signals and multiple immune checkpoints is a phenomenon broadly observed across all stages of lung adenocarcinoma.
Discussion
Therapies targeting the immune checkpoint molecules PD-1 and PD-L1 have achieved remarkable clinical responses in multiple types of cancer including NSCLC. Unfortunately, durable responses have been observed in only a subset of patients with chemotherapy-refractory metastatic disease. Identification of biomarkers that predict clinical benefit to immune-based approaches is needed. Accumulating data have suggested that immune checkpoint agents are most effective in patients in which an endogenous immune response co-exists with elevation of immune checkpoints (7, 27, 29, 30). However, biomarkers to identify this subgroup of patients who carry both endogenous immune response and elevation of immune checkpoints are essentially lacking. By using integrated gene expression analysis of three independent NSCLC datasets, we demonstrated that lung adenocarcinoma with a “mesenchymal” phenotype is associated with distinct tumor microenvironment changes. It is constituted by endogenous immune activation, such as elevation of immune co-stimulatory molecules, IFN-γ and CXCL10, along with simultaneous elevation of multiple immune checkpoint molecules including elevated PD-1 and PD-L1, as compared to lung adenocarcinoma with an “epithelial” phenotype. Consistent with gene analysis, high expression of PD-L1 was confirmed by IHC and RPPA in “mesenchymal” lung adenocarcinoma. Enhanced tumor infiltration by CD4+Foxp3+ regulatory T cells and CD3+ T cells was also demonstrated in patients with “mesenchymal” lung adenocarcinoma in contrast to those with an “epithelial” phenotype.
EMT, a biologic program associated with loss of cell adhesion and increased invasive behavior has been established as a major mechanism for metastasis and drug resistance in several types of epithelial cancers including NSCLC (10, 11, 13, 31–33). Studies have demonstrated that expression of transcription factors such as snail or neu can induce EMT, and are associated with activation of immunosuppressive cytokines and T lymphocyte resistance in pre-clinical models of melanoma, pancreatic and breast cancer (34–37). Our current study provides evidence in NSCLC that EMT is associated with much broader inflammatory changes in the tumor microenvironment, with immune activation co-existent with elevation of multiple targetable immune checkpoint molecules. These data suggest a previously under-recognized role of tumor cell EMT. That is, EMT might accelerate cancer growth and metastasis not only by direct reprogramming of cancer cells, but also by reprogramming the immune response in the local tumor microenvironment. Furthermore, our data indicate that therapies targeting immune checkpoints might have a therapeutic impact on tumor metastases and drug resistance mediated via EMT.
PD-L1 expression on tumor cells or tumor infiltrating immune cells has recently been studied as a potential single predictive biomarker for clinical activity to anti-PD-1/PD-L1-directed therapy (7, 38). Although studies have suggested that patients with over-expression of PD-L1 have improved clinical outcomes to anti-PD-1–directed therapy, some patients with over-expression of PD-L1 do not derive benefit and other patients with low level PD-L1 expression demonstrate robust responses, indicating that PD-L1 is not an exclusionary biomarker and suggesting that a broader measure of the tumor microenvironment is needed. PD-L1 can be up-regulated through different mechanisms such as PTEN silencing, AKT activation or inflammatory immune responses (24),(39, 40). IFN-γ, secreted by activated T cells, is known to be the primary cytokine driving PD-L1 expression. The patients who clinically respond to ipilimumab and anti-PD-1 have been found to have tumors with pre-existing immunity or inflamed tumors where activated T cells are present (7, 29, 41). Gene expression suggestive of T cell activation, such as elevated expression of IFN-γ and CXCL10, were also found in responders as compared to non-responders (7, 41). These findings suggest that up-regulation of PD-L1 expression and subsequent development of immune suppression in the tumor microenvironment were most likely driven by an ongoing immune response as an adaptive immune escape mechanism (25, 29, 42, 43). Although the association of PD-L1 and increased tumor infiltrating immune cells is not well-understood, it was shown in murine models that elevation of PD-L1 in the tumor microenvironment is dependent on the presence of CD8+ T cells secreting IFN-γ24. This data emphasizes that PD-L1 expression is likely not the cause, but rather the consequence, of increased tumor infiltration by immune cells. Our recent study demonstrated that the transcription factor ZEB1, a known EMT driver, regulates the micro-RNA-dependent expression of PD-L1 on tumor cells and enhances the tumor response to IFN-γ (15). However, the redundant mechanisms underlying the association between EMT and the tumor immune microenvironment require further study. One possibility is that the cytokine milieu generated upon EMT increases tumor infiltration by immune cells. The selective clinical response to immunotherapies targeting the PD-1/PD-L1 pathway is most likely restricted to patients whose PD-L1 expression is associated with pre-existing immunity where immune activation is augmented with immune checkpoint blocking agents (24),(7, 25, 29, 39, 40, 42, 43). Biomarker panels or a collective scoring system, such as the EMT score or the recently developed PD-L1 score that incorporates infiltrating immune cells, to identify this subgroup of patients who have characteristic features of high levels of PD-L1 signals of T cells activation and an inflammatory tumor microenvironment, will help us to select the best candidate patients for immunotherapies targeting the PD-1/PD-L1pathway (7).
Consistent with these features, our data demonstrate a strong association between an ongoing immune response and elevation of immune checkpoints in the tumor microenvironment when lung adenocarcinomas undergo EMT. Increased gene expression of CD80, CD86, OX40L, 4-1BB, ICOS, CD127, IFN-γ and CXCL10 were found in “mesenchymal” compared to “epithelial” tumors. This ongoing immune response likely promoted the elevation of multiple immune checkpoint molecules. One could question why tumors were not rejected if active immune cell infiltration existed in the tumor microenvironment. Several lines of evidence have demonstrated that cytotoxic CD8 T cells isolated from tumor showed functional impairment despite initial proper T cell activation (41, 44), indicating that immune suppressive mechanisms inhibit the T cell function in the tumor microenvironment. As revealed in our current study, in addition to the PD-1:PD-L1/PD-L2 axis, several other immune checkpoint molecules were elevated, such as TIM-3, BTLA, B7-H3 and CTLA-4. This redundant suppressive tumor environment might imply the necessity of combinatorial strategies in the design of clinical trials in future.
Although expression of PD-L1 protein or mRNA has been associated with longer survival and better clinical outcome in patients with NSCLC, such a prognostic correlation was not found in our study (9). The use of variable techniques such as RNAscope assay versus Illumina sequencing, as well as different cutoffs and processing variability, or the types of samples utilized likely explain the differences. Interestingly, B7-H3, one of the immune checkpoint molecules commonly up-regulated on NSCLC was found significantly associated with poor OS and RFS and may represent a novel target for immunotherapy in NSCLC in the future. Although all three independent datasets shared the same trend in elevated immune checkpoints such as PD-L1, PD-L2, CTLA-4, TIM-3 along with elevated co-stimulatory molecules, including CD80, CD86, OX40L, 4-1BB, CD127, some differences were noticed in comparison of late-stage to early-stage lung adenocarcinoma samples. For example, BTLA, LAG3, PD-1, ICOS and IFN-γ were significantly elevated in “mesenchymal” as compared to “epithelial” lung adenocarcinomas in patients with early, but not late-stage of disease. Although we cannot completely explain this observation, it may indicate the potential presence of different tumor immune microenvironments in advanced stage lung cancers. Further investigation of the host immune system and tumor microenvironment in advanced versus early-stage lung cancer will better elucidate the optimal strategy for personalization of immunotherapies.
Recent studies in NSCLC and melanoma revealed that the clinical response to anti-PD-1 or CTLA-4 blockade correlates with genomic mutational burden (27, 45). A high mutational load is postulated to generate non-self-antigens that can be recognized by the immune system and trigger a mutation-reactive immune response. Although both smoking and TP53 mutation are highly associated with increased tumor mutational burden in lung adenocarcinoma, our data demonstrated no association between EMT and mutational burden (46). Our study therefore indicates that EMT is an independent mediator driving inflammatory and immunosuppressive tumor microenvironment changes in addition to total mutational burden, and suggests that patients with tumors carrying both high EMT score and high mutation burden are more likely to benefit from immune checkpoint therapy than those who have either alone. Interestingly, investigating the correlation between common individual mutations and EMT revealed a significantly higher frequency of STK11 and KEAP1 mutations in “epithelial” lung adenocarcinomas. This is consistent with our recent study showing that KRAS mutant lung adenocarcinomas with concurrent STK11 or KEAP1 mutations appeared largely “immune-inert” and expressed low level of immune markers (47). Further study to investigate the underlying mechanisms behind the association of specific driver mutations and immune suppression will facilitate the implementation of personalized therapy.
In summary, our data demonstrate a strong association between EMT and an inflammatory tumor microenvironment with expression of multiple immune checkpoint molecules and immune activation. Along with using the difference between “epithelial” and “mesenchymal” groups to explore the mechanistic biology driving treatment response and resistance, further validation of potential utility of using EMT as a predictive biomarker to select patients for immune checkpoint blockade and other immunotherapies in NSCLC is needed. Because implementing an mRNA-based gene signature will be clinically challenging, other simplified testing schema will need to be devised. We are currently exploring the use of simplified methodologies that could be implemented in a clinical testing environment.
Supplementary Material
Translational relevance.
Therapeutic agents targeting immune checkpoints, such as PD-1 have achieved encouraging clinical activity and been granted recent approval in the treatment of NSCLC. However, it appears that only a select group of patients respond to these interventions. The identification of potential biomarkers that predict clinical benefit to immune checkpoint blockade is critical to successful clinical application of these agents. By analyzing three large independent datasets, we report that lung adenocarcinomas displaying a “mesenchymal” phenotype are associated with distinct tumor microenvironment changes, including elevated expression of multiple immune checkpoints, such as PD-1 and PD-L1, along with evidence of pre-existing immunity and increases in tumor infiltration by CD3+ T cells and CD4+Foxp3+ regulatory T cells. Our data warrant further investigation of using EMT as a potential predictive biomarker to guide selection of patients who are likely to benefit from immune checkpoint blockade agents in NSCLC and possibly a range of other cancers.
Acknowledgments
Grant Support: This study is supported by the Department of Defense-supported PROSPECT Grant and NCI-funded Lung SPORE (P50 CA070907, J.V.H. & I.I.W.), Conquer Cancer Foundation of ASCO Young Investigator Award 2014 (Y.L.), NIH–T32 Research Training in Academic Medical Oncology (Y.L.), NIH R01 grant (R01CA1668484, J.V.H), LUNGevity Foundation Research Award (D.L.G. & L.A.B.), Uniting Against Lung Cancer/Lung Cancer Research Foundation (D.L.G.), Rexanna’s Foundation for Fighting Lung Cancer (D.L.G), The Stading Lung Cancer Research Fund (J.V.H.), MD Anderson Cancer Center Physician Scientist Award (D.L.G.), K08-CA151651 (D.L.G.), and CPRIT RP150405 (D.L.G.). D.L.G. is an R. Lee Clark Fellow of the University of Texas MD Anderson Cancer Center, supported by the Jeane F. Shelby Scholarship Fund. The work was also supported by the generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shots Program.
Footnotes
Disclosure: Part of this study has been presented as a poster presentation in 2014 ASCO annual meeting, 2015 AACR annual meeting, 2015 ASCO annual meeting, and the 2015 WCLC meeting.
References
- 1.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bondarenko I, Luft A, Serwatowski P, Barlesi F, Chacko R, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line treatment in stage IIIB/IV non-small-cell lung cancer: results from a randomized, double-blind, multicenter phase II study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:2046–2054. doi: 10.1200/JCO.2011.38.4032. [DOI] [PubMed] [Google Scholar]
- 4.Topalian SL, Drake CG, Pardoll DM. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Current opinion in immunology. 2012;24:207–212. doi: 10.1016/j.coi.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. The Journal of experimental medicine. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 Ligands, and Other Features of the Tumor Immune Microenvironment with Response to Anti-PD-1 Therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014 doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the Treatment of Non-Small-Cell Lung Cancer. The New England journal of medicine. 2015 doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 9.Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Laboratory investigation; a journal of technical methods and pathology. 2014;94:107–116. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prudkin L, Liu DD, Ozburn NC, Sun M, Behrens C, Tang X, et al. Epithelial-to-mesenchymal transition in the development and progression of adenocarcinoma and squamous cell carcinoma of the lung. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2009;22:668–678. doi: 10.1038/modpathol.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zavadil J, Haley J, Kalluri R, Muthuswamy SK, Thompson E. Epithelial-mesenchymal transition. Cancer research. 2008;68:9574–9577. doi: 10.1158/0008-5472.CAN-08-2316. [DOI] [PubMed] [Google Scholar]
- 12.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 13.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer research. 2005;65:9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 14.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An Epithelial-Mesenchymal Transition Gene Signature Predicts Resistance to EGFR and PI3K Inhibitors and Identifies Axl as a Therapeutic Target for Overcoming EGFR Inhibitor Resistance. Clin Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nature communications. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR, Jr, Tsao A, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow CW, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. 2013;19:1577–1586. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nanjundan M, Byers LA, Carey MS, Siwak DR, Raso MG, Diao L, et al. Proteomic profiling identifies pathways dysregulated in non-small cell lung cancer and an inverse association of AMPK and adhesion pathways with recurrence. J Thorac Oncol. 2010;5:1894–1904. doi: 10.1097/JTO.0b013e3181f2a266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blank C, Gajewski TF, Mackensen A. Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion: implications for tumor immunotherapy. Cancer immunology, immunotherapy : CII. 2005;54:307–314. doi: 10.1007/s00262-004-0593-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Nirschl CJ, Drake CG. Molecular pathways: coexpression of immune checkpoint molecules: signaling pathways and implications for cancer immunotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:4917–4924. doi: 10.1158/1078-0432.CCR-12-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Science translational medicine. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. The New England journal of medicine. 2014 doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbons DL, Byers LA, Kurie JM. Smoking, p53 mutation, and lung cancer. Molecular cancer research : MCR. 2014;12:3–13. doi: 10.1158/1541-7786.MCR-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer immunology, immunotherapy : CII. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nature genetics. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 32.Bremnes RM, Veve R, Hirsch FR, Franklin WA. The E-cadherin cell-cell adhesion complex and lung cancer invasion, metastasis, and prognosis. Lung Cancer. 2002;36:115–124. doi: 10.1016/s0169-5002(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 33.Gibbons DL, Lin W, Creighton CJ, Zheng S, Berel D, Yang Y, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One. 2009;4:e5401. doi: 10.1371/journal.pone.0005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 35.cxKudo-Saito C, Shirako H, Ohike M, Tsukamoto N, Kawakami Y. CCL2 is critical for immunosuppression to promote cancer metastasis. Clinical & experimental metastasis. 2013;30:393–405. doi: 10.1007/s10585-012-9545-6. [DOI] [PubMed] [Google Scholar]
- 36.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Lu H, Knutson KL, Gad E, Disis ML. The tumor antigen repertoire identified in tumor-bearing neu transgenic mice predicts human tumor antigens. Cancer research. 2006;66:9754–9761. doi: 10.1158/0008-5472.CAN-06-1083. [DOI] [PubMed] [Google Scholar]
- 38.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. The New England journal of medicine. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 39.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nature reviews Immunology. 2004;4:336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- 40.Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nature medicine. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 41.Gajewski TF, Louahed J, Brichard VG. Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. Cancer journal. 2010;16:399–403. doi: 10.1097/PPO.0b013e3181eacbd8. [DOI] [PubMed] [Google Scholar]
- 42.Gajewski TF, Meng Y, Harlin H. Immune suppression in the tumor microenvironment. Journal of immunotherapy. 2006;29:233–240. doi: 10.1097/01.cji.0000199193.29048.56. [DOI] [PubMed] [Google Scholar]
- 43.Gajewski TF, Fuertes M, Spaapen R, Zheng Y, Kline J. Molecular profiling to identify relevant immune resistance mechanisms in the tumor microenvironment. Current opinion in immunology. 2011;23:286–292. doi: 10.1016/j.coi.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harlin H, Kuna TV, Peterson AC, Meng Y, Gajewski TF. Tumor progression despite massive influx of activated CD8(+) T cells in a patient with malignant melanoma ascites. Cancer immunology, immunotherapy : CII. 2006;55:1185–1197. doi: 10.1007/s00262-005-0118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibbons DL, Byers LA, Kurie JM. Smoking, p53 mutation, and lung cancer. Mol Cancer Res. 2014;12:3–13. doi: 10.1158/1541-7786.MCR-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.