Abstract
The adenosine A2A receptor (A2AR) is a much-studied class A G protein-coupled receptor (GPCR). For biophysical studies, A2AR is commonly purified in a detergent mixture of dodecylmaltoside (DDM), 3-(3-cholamidopropyl) dimethylammoniopropane sulfonate (CHAPS), and cholesteryl hemisuccinate (CHS). Here we studied the effects of CHAPS on the ligand binding activity and stability of wild type, full-length human A2AR. We also tested the cholesterol requirement for maintaining the active conformation of the receptor when solubilized in detergent micelles. To this end, the receptor was purified using DDM, DDM/CHAPS, or the short hydrocarbon chain lipid 1,2-dihexanoylsn-glycero-3-phosphocholine (DHPC, di-6:0PC). After solubilization in DDM, DDM/CHAPS, or DHPC micelles, although A2AR was found to retain its native-like fold, its binding ability was significantly compromised compared to DDM or DDM/CHAPS with CHS. It therefore appears that although cholesterol is not needed for A2AR to retain a native-like, α-helical conformation, it may be a critical component for high affinity ligand binding. Further, this result suggests that the conformational differences between the active and inactive protein may be so subtle that commonly used spectroscopic methods are unable to differentiate between the two forms, highlighting the need for activity measurements. The studies presented in this paper also underline the importance of the protein’s purification history; i.e., detergents that interact with the protein during purification affect the ligand binding properties of the receptor in an irreversible manner.
Keywords: GPCR, lipids, detergents, ligand binding, cholesterol
Introduction
Membrane proteins are essential biomolecules needed for a vast array of cellular processes including cellular signaling, ion and metabolite transport, adhesion, and migration [1], to name a few. G protein-coupled receptors (GPCRs) are integral membrane proteins consisting of seven α-helical segments that span the plasma membrane and respond to different extracellular stimuli (e.g. peptides, neurotransmitters, and small molecules). Upon binding to a ligand, GPCRs transmit a cellular signal mainly through intracellular G proteins and arrestins [2].
Structural studies typically require milligrams of purified protein [3, 4], and unlike water-soluble proteins, membrane proteins require a membrane mimetic system to stabilize their hydrophobic transmembrane regions. However, obtaining substantial amounts of active GPCRs remains a challenge. This is in part due to their low expression in native tissues, structural flexibility, and instability when in detergent solutions [5]. Because of these challenges, structure-function studies of GPCRs using their native, full-length sequence, are not commonly carried out; modifications to their native sequences have been used to stabilize the receptors, facilitating crystal formation [6, 7].
Detergents with a hydrophobic tail of 6–12 carbon atoms are commonly used to solubilize, stabilize and crystallize GPCRs [8]. In the case of GPCR X-ray crystal structures, the membrane protein was first solubilized in detergent prior to crystallization in detergent micelles, lipidic cubic phases, or bilayered micelles (also referred to as bicelles), without exception.
Even though many GPCRs are stable and retain functionality in detergent micelles, GPCRs and other membrane proteins often unfold and aggregate when solubilized in detergents [9], altering their native structure and eliminating or suppressing their biological function(s). At this point, selecting a detergent that retains membrane protein stability and function is typically a matter of trial and error [10]. In the case of the wild type, full-length human A2A adenosine receptor (A2AR), a class A GPCR, purification with the detergent dodecylmatoside (DDM) requires the presence of a cholesterol analog (cholesteryl hemisuccinate, CHS) in order for the receptor to retain its ligand binding activity [11, 12]. It should be pointed out that three cholesterol interaction sites have been identified from molecular dynamic simulations [13], and cholesterol’s presence was observed in one A2AR crystal structure [14]. However, it remains unclear whether cholesterol stabilizes the high affinity conformation of the receptor due to changes to the membrane properties, direct lipid-protein interactions, or a combination of both.
When purifying A2AR, the zwitterionic detergent 3-(3-cholamidopropyl) dimethylammoniopropane sulfonate (CHAPS) is often used to solubilize CHS into micelles. In the current studies, we employed the wild type, full-length human A2AR to investigate the effects of CHAPS on the ligand binding activity and stability of this receptor. We also solubilized A2AR in micelles, with and without CHS, to further test the notion that a cholesterol analog is needed to retain the native fold and ligand binding activity of A2AR. Finally, we tested the efficacy of 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC), a short chain phospholipid that self-assembles into micelles, and which acts as a biologically relevant detergent [9, 15]. DHPC has been shown to minimally perturb membrane proteins [16] and has been studied extensively in conjunction with the long hydrocarbon chain lipid, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC, di-14:0PC) [16–19]. This lipid mixture (i.e., DHPC/DMPC) is tunable, and forms a number of different morphologies (e.g. bilayered micelles, unilamellar vesicles, multilamellar vesicles, perforated lamellae, ribbon-meshed lamellae) depending on the total lipid concentration, the molar ratio of DMPC-to-DHPC, net charge of the system, and temperature [20, 21].
Materials and Methods
Expression and purification of A2AR from yeast membrane preparations
A2AR was expressed in Saccharomyces cerevisiae cells, BJ5464, using the multi-integrating pITy-A2AR-His10 plasmid, as previously described [11]. In order to improve protein purity and reproducibility between purifications, a previous purification protocol [11, 22] was adapted to employ membrane preparations instead of crude cell lysis [23]. Briefly, cell pellets were collected via centrifugation 24 hours post-induction. 600 ml of liquid culture with a total OD600 of 22 was separated into 50 mL aliquots, centrifuged at 3,22×g, and cooled to −80°C. Both lipid and detergent purificat ions started from cell pellets, with cell pellets consistently collected at an OD close to 22, which reduced batch-to-batch variability.
For details regarding the purification protocol refer to [23]. For these studies, additional detergent and lipid mixtures were used as follows. The homogenized membrane preparations were resuspended in 22 ml Buffer A (composed of 10% glycerol, 50 mM sodium phosphate monobasic, and 300 mM sodium chloride at pH 8, supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF) and complete EDTA-free protease inhibitor tablets (Roche Applied Science, Indianapolis, IN)), and the appropriate detergent or lipid: 1) 2% (w/v) DDM+1% (w/v) CHAPS+0.2% (w/v) CHS (all from Anatrace, Maumee, OH); 2) 2% (w/v) DDM+0.2% (w/v) CHS; 3) 2% (w/v) DDM+1% (w/v) CHAPS; or 4) 2% (w/v) DDM, and 5) 6.25% (w/v) DHPC (Avanti Polar Lipids Alabaster, AL). 6.25% (w/v) DHPC corresponds to 138 mM, approximately 10 times the critical micellar concentration (CMC) of DHPC (11–16 mM) [15]. The concentrations of DDM and CHAPS used correspond to approximately 200 and 3 times their CMC (i.e., approximately 0.2 mM and 6 mM, respectively) [24].
The elution buffers contained the appropriate detergent or lipid mixtures: 1) 0.1% (w/v) DDM+0.1% (w/v) CHAPS+0.02% (w/v) CHS; 2) 0.1% (w/v) DDM+0.02% (w/v) CHS; 3) 0.1% (w/v) DDM+0.1% (w/v) CHAPS; 4) 0.1% (w/v) DDM; or 5) 0.8% (w/v) DHPC. Purified A2AR samples were stored at 4 °C and used within one w eek of preparation to ensure maximal ligand binding activity.
Protein purity, concentration and biophysical characterization
Samples were separated via electrophoresis on 12% SDS-PAGE, and protein bands were detected via staining with Sypro Ruby (Life Technologies, Carlsbad, CA). For Western blotting mouse anti-A2A (Santa Cruz Biotechnology, Santa Cruz, CA, catalogue # 32261) primary antibody was used at a 1:5000 dilution. Alexa Fluor 488 goat anti-mouse (Life Technologies, Carlsbad, CA, catalogue # A11029) was used as the secondary antibody at a 1:5000 dilution. Fluorescence was detected using a BioSpectrum Imaging System (UVP, Upland, CA). Protein concentration was determined using UV absorbance at 280 nm as described in [11], and protein purity was quantified from the Sypro Ruby stained gel images using FIJI [25].
For characterizing the protein’s secondary structure, circular dichroism (CD) measurements were conducted using a Jasco J-810 spectropolarimeter (Jasco, Easton, MD), as previously described in [11]. Measurements were performed at 25°C, and spectra collected with 1 nm resolution, with at least 3 integrations per spectrum. Reference spectra containing the appropriate buffer were collected and subtracted from their respective spectra. Fluorescence spectra were collected using a PC-1 spectrofluorimeter (ISS, Champaign, IL), as described previously in [11]. To minimize light scattering effects, measurements were collected with the excitation polarizer set to 90° and the emission polarizer set to 0°. Intrinsic fluorescence was measured at 15°C, and excitation was set to 280 nm. CD and fluorescence measurements were taken at a protein concentration of 0.05 – 0.06 mg/ml.
To estimate particle sizes, Dynamic Light Scattering (DLS) measurements were taken using a Brookhaven Instruments 90Plus Particle Analyzer (Brookhaven Instruments, Holtsville, NY). A 50 µL micelle solution was loaded into Eppendorf UVette small volume cuvettes (Fisher Scientific, Pittsburgh, PA) and mounted on the instrument’s small-volume cuvette adapter pedestal.
Radioactive ligand binding
Ligand binding of purified receptors was carried out as described previously [11], with minor modifications. Briefly, approximately 1 ml of purified receptor was incubated with a Ni-NTA Superflow resin (approximately 15 µl of settled resin) overnight. If necessary, samples were centrifuged briefly to allow the resin to settle, the elution buffer volume was removed, and the sample volume adjusted to achieve a target concentration of 55 µg/ml. 180 µl of the protein/resin mix was added per well, to enable loading of 10 µg of A2AR-His10 per well in a 96-well plate (glass fiber type B filters, Millipore, Billerica, MA) for saturation and point measurement ligand binding analysis. For saturation binding experiments, protein samples were incubated with increasing amounts of [3H] CGS 21680 (Perkin Elmer, Waltham, MA) for 4 hours. Non-specific binding was determined using 10 µM of the cold ligand, CGS 21680, which was added to the samples at each value of titriated ligand. The counts obtained from these controls were subtracted from the sample data. For ligand binding activity point measurements, 100 nM of the titriated ligand was added to each A2AR sample, and 10 µM of cold ligand was added to measure non-specific binding. Binding was measured using a Wallac 1450 Trilux MicroBeta Jet (Perkin Elmer). Error bars reflect effects of variability in both technical and biological replicates. Saturation binding data were fit to a single-site binding model using the Curve Fitting toolbox in Matlab 7.10 (R2010a), and point measurement data were compared to data from A2AR purified and solubilized in DDM/CHAPS/CHS.
Protein thermal denaturation
A thiol-reactive fluorescent probe, 7-(Diethylamino)-3-(4’-maleimidylphenyl)-4-methylcoumarin (CPM) (Invitrogen, Carlsbad, CA) was used to characterize the thermal stability of A2AR in various membrane-mimetic environments. CPM stocks were protected from light and stored at −80 °C as described in [22]. 5.6 µM CPM was added to the protein sample while A2AR was bound to Ni-NTA Superflow resin prior to elution (CPM was allowed to react with accessible free thiols, protected from light, on an end-over-end mixer at 4 °C overnight). The following day the protein was eluted from the Ni-NTA Superflow resin with 500 mM imidazole as described in [23].
Fluorescence measurements were performed on a PC-1 (ISS) spectrofluorimeter, as described in [22] with some modifications. Measurements were taken with the excitation set to 280 nm (intrinsic absorbance of the purified receptor) and the emission at 460 nm, close to the emission maximum of CPM bound to the protein sample. Thus, any fluorescence from CPM close to the emission peak is expected to occur due to tryptophan excitation and energy transfer to the CPM in its vicinity; further details are included in the Supplemental Information. The fluorescence signal is expected to change as the protein unfolds. CPM fluorescence was normalized to the maximal and minimal intensity values.
Results and Discussion
The full-length, wild type human A2A receptor (A2AR) was expressed in yeast BJ5464 cells, and membrane preparations were carried out as described in [23] and the Materials and Methods section. Samples of A2AR were extracted and purified from cellular membranes using different detergent and lipid mixtures: DDM, CHAPS, CHS, and DHPC. To solubilize CHS in DDM in the absence of CHAPS, we used a sonication protocol described by Stevens and the JCIMPT at Scripps Research Institute (jcimpt.usc.edu/protocols/JCIMPT_PreparationofCHSStock.pdf). Samples were separated via electrophoresis, and protein bands were detected using Sypro Ruby staining and Western analysis using anti-A2AR antibodies (Figure 1).
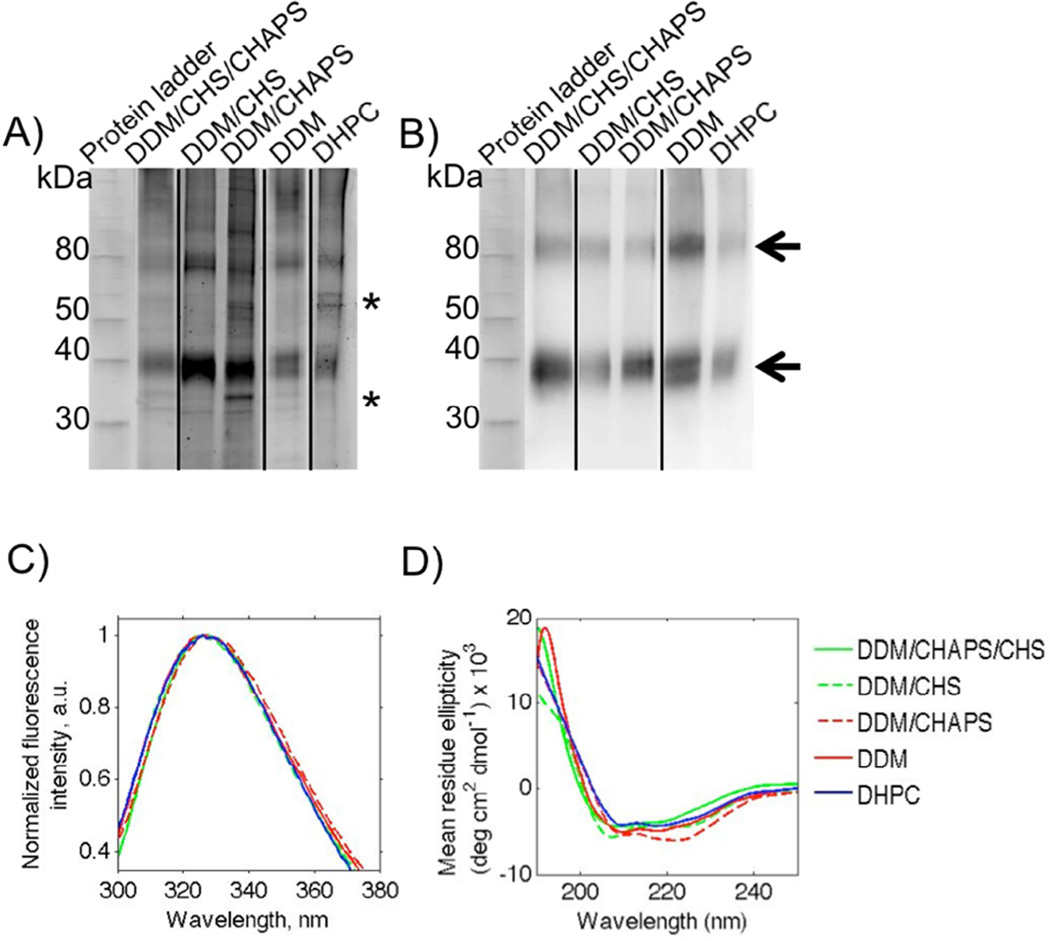
Figure 1.
Wild type, full-length human A2AR bands detected using Sypro Ruby staining (A) and Western blotting (B). Samples were treated with a reducing agent (10 µM TCEP) and separated via 12 % SDS-PAGE, stained with Sypro Ruby (A) and blotted onto a nitrocellulose membrane for Western blot detection with an anti-A2A primary antibody (B). Lanes contain MagicMark protein standards, with molecular weights indicated on the left, and A2AR purified from membrane preparations using various detergents mixtures and DHPC, as shown. Arrows indicate the A2AR monomer and oligomers, and asterisks indicate the main impurities. Results are typical of multiple independent purifications. The contrast and brightness of images in Figure 1A were adjusted in order to highlight the impurities present in each purification; results shown were not used to quantify protein concentration. C) Intrinsic fluorescence spectra of A2AR purified using detergents and lipids following excitation at 280 nm. The data represent the average from at least two independent purifications. Lines represent the fine lowess curve fit to the average in GraphPad Prism. DDM/CHAPS/CHS, solid green; DDM/CHS, dashed green; DDM/CHAPS, solid red; DDM, dashed red; DHPC, solid blue. D) CD spectra of A2AR in different micelle environments. Lines represent the fine lowess curve fit to data in GraphPad Prism. DDM/CHAPS/CHS, solid green; DDM/CHS, dashed green; DDM/CHAPS, solid red; DDM, dashed red; DHPC, solid blue. Intrinsic fluorescence and CD measurements were taken at a protein concentration of 0.05 – 0.06 mg/ml.
Differences between the protein obtained from the detergent and lipid purification protocols are highlighted in Figure 1. Sypro Ruby staining (Figure 1A) revealed a pronounced band at approximately 40 kDa, which corresponds to A2AR monomers, and whose identity was confirmed by Western analysis (Figure 1B) and mass spectrometry of protein isolated from gels (results not shown). A final protein concentration of 0.05 – 0.09 mg/ml in 3.5 ml (corresponding to 3.5 – 6.2 mg/L of culture) was determined for the A2AR purifications using UV absorbance at 280 nm (Table 1). As seen in Figure 1A and Table 1, the detergent and lipid purifications yielded varying degrees of receptor purity, as confirmed by Sypro Ruby stain and densitometry analysis.
Table 1.
Protein purity, concentration and intrinsic fluorescence center of mass (n≥2 independent purifications). Total cell density in the lysate was kept constant as described in the Materials and Methods.
| Purity ± s.d. (%) |
Concentration µg/ml ± s.d. (mg per L of culture) |
Center of mass ± s.d. (nm) |
|
|---|---|---|---|
| DDM/CHAPS/CHS | 90.1 ± 3.2 | 63.0 ± 7.6 (4.4) | 327.9 ± 0.8 |
| DDM/CHS | 96.2 ± 2.1 | 55 ± 8.0 (3.9) | 326.2 ± 0.8 |
| DDM/CHAPS | 64.0 ± 2.1 | 50 ± 2.0 (3.5) | 327.8 ± 1.6 |
| DDM | 91.7 ± 2.0 | 55.0 ± 7.4 (3.9) | 327.0 ± 0.0 |
| DHPC | 73.4 ± 8.6 | 88 ± 46.2 (6.2) | 327.7 ± 1.3 |
Despite the differences in purity, A2AR isolated using these different detergent conditions did not show significant differences in their secondary and tertiary structures, as determined by intrinsic fluorescence and circular dichroism (CD) spectroscopy (Figure 1C and 1D). The intrinsic fluorescence emission spectra of A2AR in the different membrane-mimetic environments had a center of mass ranging from 326–328 nm (Table 1). This result implies that the receptor is in a hydrophobic environment, and has a compact and native-like fold (Figure 1C and Table 1). CD analysis was used as a semi-quantitative measure to confirm that the purified receptor was predominantly α-helical (Figure 1D). We have previously reported that A2AR lost its α-helical content when purified in DDM [26]; we believe that the difference here – i.e., the observation of α-helical structure – may result from the improved purity we obtained using our current membrane preparation purification protocol.
The small shift in the intrinsic fluorescence maximum and CD spectra of the protein in these different detergents (DDM/CHAPS/CHS) and lipid (DHPC) could imply minor differences in the structural conformation of the receptor and its local environment. However, it is unclear whether these subtle changes in secondary and tertiary structure correlate to changes in receptor function.
To determine whether A2AR in these different micelle environments was able to bind ligand, we conducted ligand binding assays using 100 nM of the tritiated agonist CGS 21680 (Figure 2). Results were compared to A2AR in DDM/CHAPS/CHS, whose activity we previously characterized in detail [11, 27, 28].
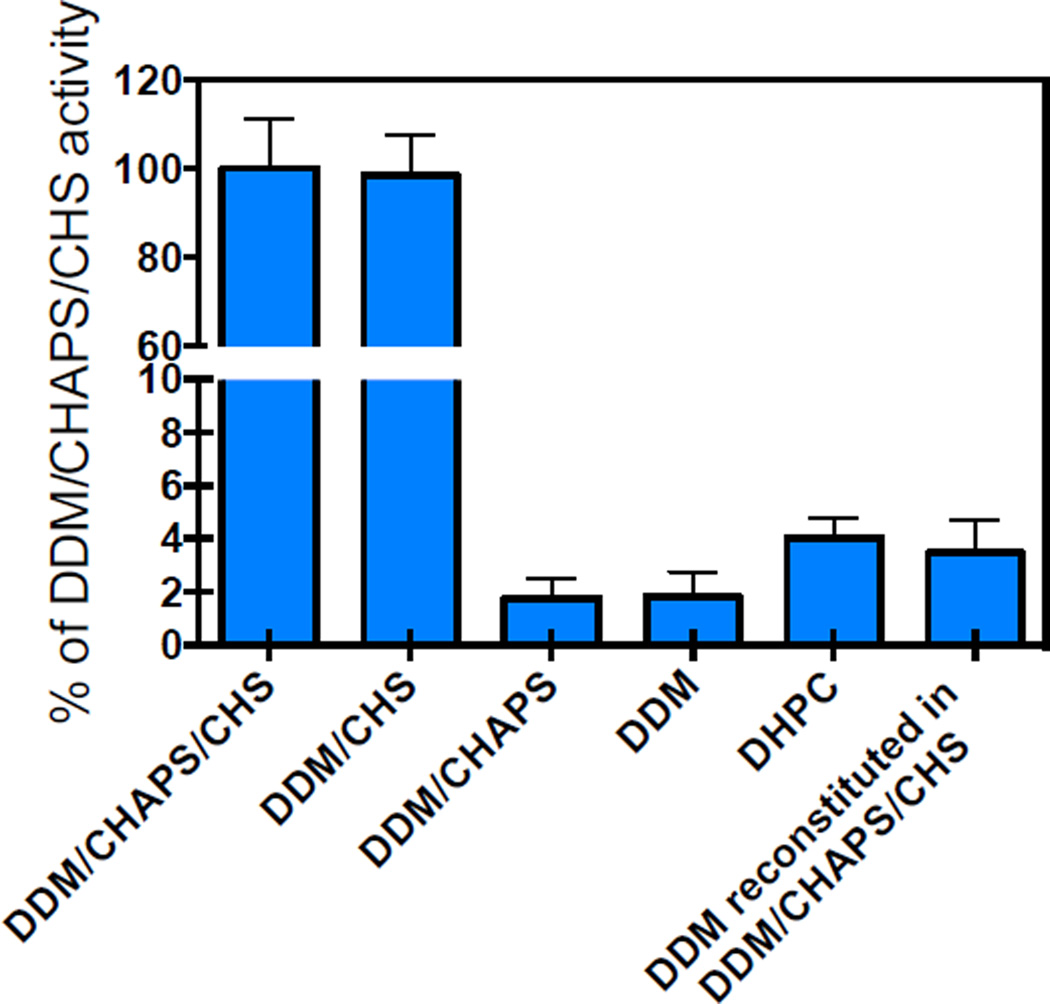
Figure 2.
Specific activity measurements for A2AR solubilized in different micelle environments. 100 nM of the titriated ligand was added to each A2AR sample, and 10 µM of cold ligand was added to measure non-specific binding. The resulting signal (measured in counts per minute) was compared to the signal obtained for A2AR solubilized in DDM/CHAPS/CHS. Results for the sample labeled “DDM reconstituted in DDM/CHAPS/CHS”, correspond to those of the receptor initially purified in DDM only, and subsequently reconstituted in DDM/CHAPS/CHS. Error bars represent the standard deviation from the average of at least two independent purifications, with measurements conducted in triplicate (n=6).
We previously reported ligand binding activity for A2AR in DDM/CHAPS/CHS, but no detectable ligand binding activity was observed for A2AR in DDM [11, 27, 28]. Here, we expand this analysis and report activity for A2AR in DDM/CHS, but almost undetectable activity in DDM/CHAPS (1.8%; Figure 2). Interestingly, we detected ligand binding activity of A2AR purified using the short chain lipid DHPC (4.0%); however the receptor bound with significantly reduced affinity (refer to Fig. S1 in the Supporting Material). It is worth highlighting that if the receptor is first purified in DDM and then reconstituted in DDM/CHAPS/CHS within one day, the receptor does not recover its native ligand binding activity (sample labeled “DDM reconstituted in DDM/CHAPS/CHS” in Figure 2). This result implies that once the receptor is purified in a detergent that does not promote ligand-binding activity (in this case DDM), changing the detergent mixture to one that promotes ligand-binding activity (such as DDM/CHAPS/CHS) does not recover the receptor’s ligand binding ability. The inability to restore ligand binding implies that this system exhibits a detergent related hysteresis, which prevents it from regaining its full activity. Similar results have been reported for the cannabinoid receptor CB2, where in the absence of CHS the receptor did not show any measurable activity. Importantly, in the absence of CHS even a short exposure of the receptor to micelles leads to a rapid and irreversible loss of function [29]. These results have important implications for nanodisc studies, where receptors are often reconstituted in DDM prior to lipid reconstitution.
Dynamic light scattering (DLS) was used to characterize micelle size (Table 2), and to determine if the lack of ligand binding activity correlated to micelle size/protein aggregation. A2AR completely loses its ligand binding activity when solubilized in DDM/CHAPS or DDM, and has low affinity for CGS2180 when solubilized in DHPC. Large micelles were observed when A2AR was solubilized in DDM/CHAPS or DHPC (47 – 145 nm), while A2AR in DDM had a similar size to A2AR in DDM/CHAPS/CHS and DDM/CHS (3.5 – 10 nm), indicating that ligand binding affinity did not correlate with micelle size (p=0.24). The effect of CHAPS was surprising, in contrast to previous reports of the serotonin receptor and other membrane proteins, which reported a decrease in protein association with this detergent [30–33]; this result further emphasizes that this class of membrane proteins have unique detergent interaction properties that are not easy to predict.
Table 2.
Micelle size determined using DLS. Data were collected from at least three independent purifications (n= number of independent purifications and measurements). The size range shown corresponds to ≥ 98% of the particles measured.
| Diameter range (nm) |
n | |
|---|---|---|
| DDM/CHAPS/CHS | 3.9–9.7 | 5 |
| DDM/CHS | 3.5–7.6 | 3 |
| DDM/CHAPS | 46.8–110.9 | 3 |
| DDM | 4.0–9.7 | 3 |
| DHPC | 84.4–144.5 | 4 |
Specific ligand binding point measurements (Figure 2) indicated that A2AR retained high ligand binding affinity only when solubilized in DDM/CHAPS/CHS and DDM/CHS. Even though high affinity ligand binding was lost when A2AR was solubilized in DDM, DDM/CHAPS or DHPC, the receptor retained a native-like fold. These data are strong evidence that cholesterol is not needed to attain an α-helical conformation and native-like fold for A2AR, but may be critical for high affinity ligand binding. Using intrinsic fluorescence and CD we were unable to differentiate between an active and inactive receptor, highlighting the importance of complementary biophysical measurements and functional assays. This result also indicates that the differences between the high affinity state of the receptor and its inactive state are subtle. For example, using atomic force microscopy-based single-molecule force spectroscopy, Zocher et al. found that CHS stabilizes structural segments of the β2-adrenergic receptor. They also concluded that CHS stabilizes the same structural segments in the absence of CHS [34]. This implies that cholesterol enhances the interactions that stabilize the receptor, but cholesterol does not necessarily alter the receptor’s conformation.
Since A2AR retained high ligand binding affinity when solubilized in DDM/CHAPS/CHS and DDM/CHS, these samples were chosen for saturation ligand binding and temperature unfolding experiments to study differences in ligand binding affinity and structural stability when the receptor is solubilized into these different micelle environments (Figure 3).
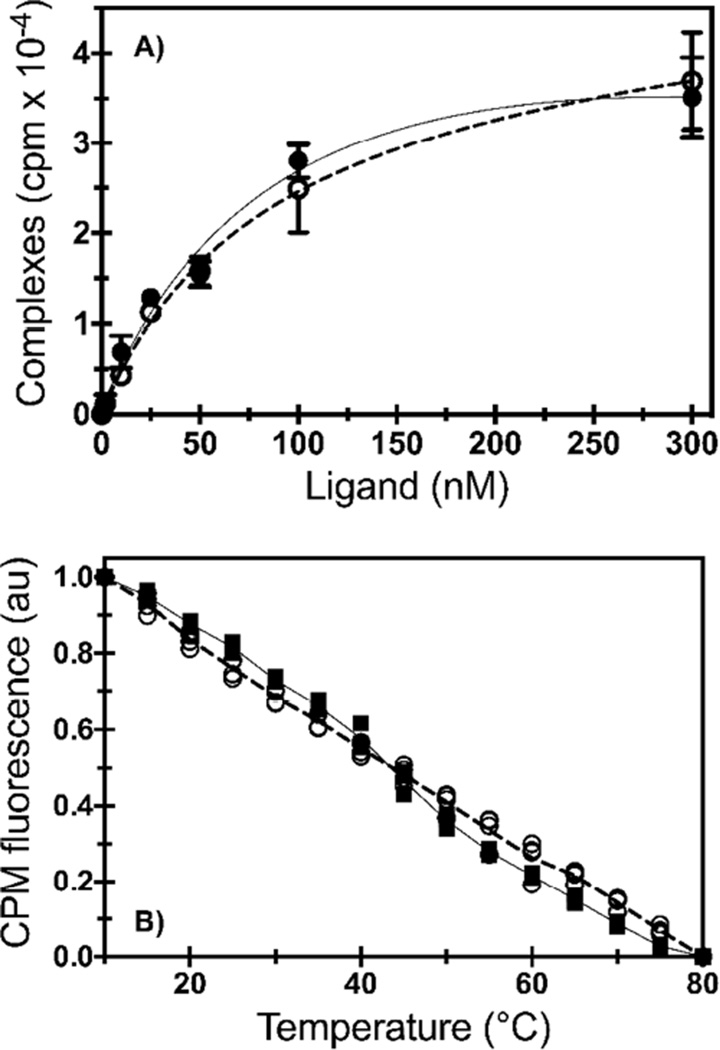
Figure 3.
Ligand binding affinity and temperature unfolding studies. A) Saturation ligand binding of 3H-CGS 21680 to A2AR in DDM/CHAPS/CHS (filled circles; solid line) and DDM/CHS micelles (open circles; dashed line). The data points show the average of two different purifications run in triplicate (n=6). A2AR in DDM/CHAPS/CHS had a KD of 67 nM; we previously reported a KD of 66 ± 4 nM [11]. A2AR in DDM/CHS had a KD of 96 nM with a 95% confidence interval of 77 – 114 nM. Data were fit to a single-site binding model (line). Error bars are denoted as the standard deviation. B) Temperature unfolding of A2AR-CPM solubilized in DDM/CHAPS/CHS (filled squares; solid line) and DDM/CHS micelles (open circles; dashed line) via the fluorescence intensity (at 463 nm) of the thiol-reactive probe CPM. Data points were collected from at least three independent experiments and all replicates are shown. Lines connect the median value of the data points.
Saturation binding data were fit to a single-site binding model exhibiting a typical saturation binding curve with an equilibrium dissociation constant (KD) of 67 nM for A2AR in DDM/CHAPS/CHS, and 96 nM for A2AR in DDM/CHS (Figure 3A). Previously, we have used the thiol-reactive fluorescent probe 7-(Diethylamino)-3-(4’-maleimidylphenyl)-4-methylcoumarin (CPM) to determine changes in thermal unfolding of A2AR in the presence of different ligands [35]. As CPM binds irreversibly, the unfolding is not reversible. Here, we removed unbound CPM prior to the temperature denaturation experiments and use bound CPM fluorescence to determine whether the solubilization conditions would impact receptor thermal unfolding behavior, with only minor differences observed (Figure 3B). The data could not be fit to a single-transition folding model, as there was only a gradual transition from the folded to the unfolded state as temperature increased.
A2AR exhibited high ligand binding affinity when solubilized in DDM/CHAPS/CHS and DDM/CHS micelles. Although there were only subtle differences between the secondary and tertiary structure—based on intrinsic fluorescence and CD data of similar size micelles—it appears that A2AR is slightly more active when solubilized in DDM/CHAPS/CHS micelles.
Conclusion
These results highlight the importance of ligand binding activity screening, as intrinsic fluorescence, CD, and DLS studies were not capable of differentiating between an inactive and an active native-like protein fold. Furthermore, it may be that the conformational differences between an active and inactive protein are so subtle that spectroscopic methods are unable to differentiate between them, or improved methods need to be developed – such as site-specific labeling – in order to determine the subtle differences in protein conformation.
A2AR was minimally active when solubilized in DDM or DDM/CHAPS micelles lacking CHS, and exhibited low affinity when solubilized in DHPC (Figure 2). This low affinity indicates that cholesterol is not needed for the protein to retain its α-helical content and ligand-binding activity, but may be crucial for high affinity binding.
Furthermore, it is important to note that the high affinity conformation was not recovered after purification of A2AR in the absence of cholesterol and its subsequent solubilization in DDM/CHAPS/CHS, indicating that the detergent-protein interaction history is crucial, and alters the ligand binding properties of the receptor in an irreversible matter.
Supplementary Material
Highlights.
Examined effects of lipids on the ligand binding activity and stability of human A2AR
Cholesterol is not needed for A2AR to retain a native-like, α-helical conformation
Cholesterol appears to be a critical component for high affinity ligand binding
Purification history impacts long-term protein activity in irreversible manner
Acknowledgments
This project was supported by grants from the National Institutes of Health (National Center for Research Resources (5P30RR031160-03) and the National Institute of General Medical Sciences (8 P30 GM103519-03), and from the National Science Foundation (1033268/1249200) and NSF Graduate Research Fellowship Program (ANN). JK is supported through the Scientific User Facilities Division of the DOE Office of Basic Energy Sciences (BES), under contract no. DE-AC05 00OR2275. We also thank Dr. K. Dane Wittrup (Massachusetts Institute of Technology, Cambridge, MA) for the pITy plasmid, Dr. Marlene Jacobson (Merck) for the human adenosine hA2AR gene, Dr. Kelvin Lee (University of Delaware) for assistance with mass spectrometry, Dr. William Wimley (Tulane University) for the use of the circular dichroism equipment, and Dr. Robert F. Standaert (Oak Ridge National Laboratory) for discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
ASR, ANN, and JK designed research; ANN and PMM performed the research; ASR, ANN, PMM, and JK analyzed data; ASR, ANN, PMM, and JK wrote and edited the manuscript. ASR oversaw the research.
References
- 1.McNeely PM, Naranjo AN, Robinson AS. Structure-function studies with G protein-coupled receptors as a paradigm for improving drug discovery and development of therapeutics. Biotechnol J. 2012;7:1451–1461. doi: 10.1002/biot.201200076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry SJ, Lefkowitz RJ. Arresting developments in heptahelical receptor signaling and regulation. Trends Cell Biol. 2002;12:130–138. doi: 10.1016/s0962-8924(01)02239-5. [DOI] [PubMed] [Google Scholar]
- 3.Tapaneeyakorn S, Goddard AD, Oates J, Willis CL, Watts A. Solution- and solid-state NMR studies of GPCRs and their ligands. Biochim Biophys Acta. 2011;1808:1462–1475. doi: 10.1016/j.bbamem.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Chiu ML, Tsang C, Grihalde N, MacWilliams MP. Over-expression, solubilization, and purification of G protein-coupled receptors for structural biology. Comb Chem High Throughput Screen. 2008;11:439–462. doi: 10.2174/138620708784911456. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-proteincoupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 6.Tate CG. A crystal clear solution for determining G-protein-coupled receptor structures. Trends Biochem Sci. 2012;37:343–352. doi: 10.1016/j.tibs.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Q, Wu BL. Ice breaking in GPCR structural biology. Acta pharmacologica Sinica. 2012;33:324–334. doi: 10.1038/aps.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao XJ, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang SG. Designer short peptide surfactants stabilize G protein-coupled receptor bovine rhodopsin. Proc Natl Acad Sci U S A. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prive GG. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Robinson AS. Production of membrane proteins : strategies for expression and isolation. Wiley-VCH; 2011. [Google Scholar]
- 11.O'Malley MA, Lazarova T, Britton ZT, Robinson AS. High-level expression in Saccharomyces cerevisiae enables isolation and spectroscopic characterization of functional human adenosine A2a receptor. J Struct Biol. 2007;159:166–178. doi: 10.1016/j.jsb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss HM, Grisshammer R. Purification and characterization of the human adenosine A(2a) receptor functionally expressed in Escherichia coli. European Journal of Biochemistry. 2002;269:82–92. doi: 10.1046/j.0014-2956.2002.02618.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee JY, Lyman E. Predictions for cholesterol interaction sites on the A(2A) adenosine receptor. J Am Chem Soc. 2012;134:16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, AP IJ, Cherezov V, Stevens RC. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauser H. Short-chain phospholipids as detergents. Biochimica Et Biophysica Acta-Biomembranes. 2000;1508:164–181. doi: 10.1016/s0304-4157(00)00008-3. [DOI] [PubMed] [Google Scholar]
- 16.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magnetic resonance in chemistry : MRC. 2006;44 Spec No:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 17.Katsaras J, Donaberger RL, Swainson IP, Tennant DC, Tun Z, Vold RR, Prosser RS. Rarely observed phase transitions in a novel lyotropic liquid crystal system. Phys Rev Lett. 1997;78:899–902. [Google Scholar]
- 18.Katsaras J, Harroun TA, Pencer J, Nieh MP. "Bicellar" lipid mixtures as used in biochemical and biophysical studies. Naturwissenschaften. 2005;92:355–366. doi: 10.1007/s00114-005-0641-1. [DOI] [PubMed] [Google Scholar]
- 19.Sanders CR, Hare BJ, Howard KP, Prestegard JH. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Progress in Nuclear Magnetic Resonance Spectroscopy. 1994;26:421–444. [Google Scholar]
- 20.Harroun TA, Koslowsky M, Nieh MP, de Lannoy CF, Raghunathan VA, Katsaras J. Comprehensive examination of mesophases formed by DMPC and DHPC mixtures. Langmuir : the ACS journal of surfaces and colloids. 2005;21:5356–5361. doi: 10.1021/la050018t. [DOI] [PubMed] [Google Scholar]
- 21.Nieh MP, Dolinar P, Kucerka N, Kline SR, Debeer-Schmitt LM, Littrell KC, Katsaras J. Formation of kinetically trapped nanoscopic unilamellar vesicles from metastable nanodiscs. Langmuir : the ACS journal of surfaces and colloids. 2011;27:14308–14316. doi: 10.1021/la2023314. [DOI] [PubMed] [Google Scholar]
- 22.O'Malley MA, Naranjo AN, Lazarova T, Robinson AS. Analysis of adenosine A(2)a receptor stability: effects of ligands and disulfide bonds. Biochemistry. 2010;49:9181–9189. doi: 10.1021/bi101155r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blocker KM, Britton ZT, Naranjo AN, McNeely PM, Young CL, Robinson AS. Chapter Eight - Recombinant G Protein-Coupled Receptor Expression in Saccharomyces cerevisiae for Protein Characterization. In: Arun KS, editor. Methods Enzymol. Academic Press; 2015. pp. 165–183. [DOI] [PubMed] [Google Scholar]
- 24.Maler L. Solution NMR studies of peptide-lipid interactions in model membranes. Mol Membr Biol. 2012;29:155–176. doi: 10.3109/09687688.2012.683456. [DOI] [PubMed] [Google Scholar]
- 25.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Malley MA. Expression, Purification, and Biophysical Characterization of G-Protein Coupled Receptors Expressed from Saccharomyces cerevisiae. Newark: Chemical Engineering, University of Delaware; 2009. [Google Scholar]
- 27.O'Malley MA, Helgeson ME, Wagner NJ, Robinson AS. The morphology and composition of cholesterol-rich micellar nanostructures determine transmembrane protein (GPCR) activity. Biophys J. 2011;100:L11–L13. doi: 10.1016/j.bpj.2010.12.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Malley MA, Helgeson ME, Wagner NJ, Robinson AS. Toward rational design of protein detergent complexes: determinants of mixed micelles that are critical for the in vitro stabilization of a G-protein coupled receptor. Biophys J. 2011;101:1938–1948. doi: 10.1016/j.bpj.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vukoti K, Kimura T, Macke L, Gawrisch K, Yeliseev A. Stabilization of functional recombinant cannabinoid receptor CB(2) in detergent micelles and lipid bilayers. Plos One. 2012;7:e46290. doi: 10.1371/journal.pone.0046290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chattopadhyay A, Jafurulla M, Kalipatnapu S. Solubilization of serotonin1A receptors heterologously expressed in Chinese hamster ovary cells. Cell Mol Neurobiol. 2004;24:293–300. doi: 10.1023/B:CEMN.0000018623.81954.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee P, Joo JB, Buse JT, Dawson G. Differential solubilization of lipids along with membrane proteins by different classes of detergents. Chem Phys Lipids. 1995;77:65–78. doi: 10.1016/0009-3084(95)02455-r. [DOI] [PubMed] [Google Scholar]
- 32.Jafurulla M, Chattopadhyay A. Solubilization of human serotonin 1A receptors expressed in neuronal cells. Chem Phys Lipids. 2007;150:244–249. doi: 10.1016/j.chemphyslip.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Talmont F, Mouledous L, Mollereau C, Zajac JM. Solubilization and reconstitution of the mu-opioid receptor expressed in human neuronal SH-SY5Y and CHO cells. Peptides. 2014;55:79–84. doi: 10.1016/j.peptides.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Zocher M, Zhang C, Rasmussen SrGF, Kobilka BK, Muller DJ. Cholesterol increases kinetic, energetic, and mechanical stability of the human β2-adrenergic receptor. Proceedings of the National Academy of Sciences. 2012;109:E3463–E3472. doi: 10.1073/pnas.1210373109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Malley MA, Naranjo AN, Lazarova T, Robinson AS. Analysis of adenosine Aa receptor stability: effects of ligands and disulfide bonds. Biochemistry. 2010;49:9181–9189. doi: 10.1021/bi101155r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.