Abstract
Relative to European Americans, type 2 diabetes (T2D) is more prevalent in African Americans (AAs). Genetic variation may modulate transcript abundance in insulin-responsive tissues and contribute to risk; yet published studies identifying expression quantitative trait loci (eQTLs) in African ancestry populations are restricted to blood cells. This study aims to develop a map of genetically regulated transcripts expressed in tissues important for glucose homeostasis in AAs, critical for identifying the genetic etiology of T2D and related traits. Quantitative measures of adipose and muscle gene expression, and genotypic data were integrated in 260 non-diabetic AAs to identify expression regulatory variants. Their roles in genetic susceptibility to T2D, and related metabolic phenotypes were evaluated by mining GWAS datasets. eQTL analysis identified 1,971 and 2,078 cis-eGenes in adipose and muscle, respectively. Cis-eQTLs for 885 transcripts including top cis-eGenes CHURC1, USMG5, and ERAP2, were identified in both tissues. 62.1% of top cis-eSNPs were within ±50kb of transcription start sites and cis-eGenes were enriched for mitochondrial transcripts. Mining GWAS databases revealed association of cis-eSNPs for more than 50 genes with T2D (e.g. PIK3C2A, RBMS1, UFSP1), gluco-metabolic phenotypes, (e.g. INPP5E, SNX17, ERAP2, FN3KRP), and obesity (e.g. POMC, CPEB4). Integration of GWAS meta-analysis data from AA cohorts revealed the most significant association for cis-eSNPs of ATP5SL and MCCC1 genes, with T2D and BMI, respectively. This study developed the first comprehensive map of adipose and muscle tissue eQTLs in AAs (publically accessible at https://mdsetaa.phs.wakehealth.edu) and identified genetically-regulated transcripts for delineating genetic causes of T2D, and related metabolic phenotypes.
Keywords: Expression Quantitative Trait (eQTL), Genotype, Transcript, Single nucleotide polymorphism (SNP), Adipose, Muscle, African American, Genomics, Diabetes, Obesity
INTRODUCTION
The importance of genetic factors in modulating the susceptibility to type 2 diabetes (T2D) is well established (Groop and Pociot, 2014). Relative to European Americans, T2D is twice as prevalent in African Americans (Cowie et al., 2010) and associated risk factors such as insulin resistance and obesity are more prevalent (Cowie et al., 1993). Whether genetic variation modulates molecular processes and contributes to the enhanced susceptibility to T2D in African Americans is unknown. Large-scale linkage, candidate-gene, and genome-wide association studies (GWAS), primarily in European and Asian populations, have identified approximately 88 loci associated with T2D and 83 loci associated with glucose homoeostasis-related phenotypes (Mohlke and Boehnke, 2015). T2D-associated loci identified in GWAS reveal relatively weak effects, together explaining only a small fraction of the heritability in African Americans (Mahajan et al., 2014;Ng et al., 2014). Moreover, most associated variants are located in noncoding genomic regions. Thus, determining how these loci modulate systemic glucose homeostasis at the molecular level remains unclear. Approaches investigating molecular endophenotypes more proximal to gene products may assist in identifying the molecular basis of genetic susceptibility to T2D in African Americans.
Genetic variation can impact transcript abundance. We and others reported that T2D- and related trait-associated variants are enriched for expression-regulatory single nucleotide polymorphisms (eSNPs) in tissues important for glucose homeostasis (Das and Sharma, 2014;GTEx consortium, 2015;Nicolae et al., 2010). Identifying genetic variants associated with transcript expression in metabolically relevant tissues may identify functionally meaningful sets of SNPs involved in T2D, obesity, and related metabolic phenotypes. However, studies on the genetics of gene expression in populations of African ancestry are predominantly limited to lymphocytes/lymphoblastoid cell lines (Storey et al., 2007;Stranger et al., 2012;Zhang et al., 2008). The goal of this study was to identify genetic regulatory variants modulating expression of adipose and muscle transcripts in African Americans at risk for T2D and evaluate their role in susceptibility to T2D, obesity, and related metabolic disorders. A systematic analysis was performed on the genome-wide transcript expression profiles of insulin responsive tissues (subcutaneous adipose and skeletal muscle) and genome-wide SNP genotypes in a metabolically characterized cohort of 260 non-diabetic African Americans from North Carolina. To our knowledge, this is the largest existing cohort of non-diabetic African Americans characterized for gluco-metabolic phenotypes with available biological samples (DNA and tissue) for conducting an integrative multi-omics approach. These data were used to test two hypotheses: 1) expression levels of a subset of transcripts would associate with genotype and manifest as expression quantitative trait loci (eQTL) in both adipose and muscle, while a subset of transcripts would be modulated by tissue-specific eQTLs; and 2) a subset of the expression regulatory SNPs (eSNPs) would associate with glucose homeostasis related phenotypes, obesity and/or T2D in large GWAS, identifying putative causal SNPs in African Americans.
MATERIALS AND METHODS
Human subjects
Participants were healthy, self-reported African Americans residing in North Carolina aged 18–60 years with a body mass index (BMI) between 18 and 42 kg/m2. A total of 260 unrelated non-diabetic individuals completed all study visits and are referred to as the “African American Genetics of Metabolism and Expression” (AAGMEx) cohort; subcutaneous adipose tissue (from the abdomen near the umbilicus) and skeletal muscle (from the vastus lateralis) biopsies were collected from 256 individuals. Studies were performed at the Wake Forest School of Medicine (WFSM) Clinical Research Unit. This study was approved by the WFSM Institutional Review Board and all participants provided written informed consent.
A standard 75-g oral glucose tolerance test (OGTT) was used to exclude individuals with diabetes and results were analyzed by homeostatic model assessment (HOMA; http://mmatsuda.diabetes-smc.jp/MIndex.html) to evaluate insulin sensitivity (Matsuda Index) and insulin resistance (HOMA-IR) (Matsuda and DeFronzo, 1999; Matthews et al., 1985). High quality insulin modified (0.03 U/kg) frequently sampled intravenous glucose tolerance test (FSIGT) data were available in 235 participants. The MINMOD Millennium program was used to analyze FSIGT data to determine insulin sensitivity (SI) and acute insulin response (AIRG) (Bergman et al., 2014). Clinical, anthropometric, and physiological characteristics of the AAGMEx cohort have been described (Sharma et al., 2016).
Gene expression analysis and genotyping
Genome-wide expression data were generated using HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA) whole genome gene expression arrays for quantitative analyses of transcript expression in adipose and muscle samples. Infinium HumanOmni5Exome-4 v1.1 DNA Analysis BeadChips (Illumina) were used to genotype DNA samples based on the manufacturer’s recommendations. Additional technical details of standard gene expression analyses and genotyping methods are described in Supplementary methods.
Quality control
Detailed data quality control methods are presented in Supplementary methods. In brief, measures of glucose homeostasis and obesity were examined for outliers in a univariate fashion and as correlated pairs. Genome-wide gene expression data (probe level) for both the adipose and muscle samples were extracted separately using Illumina GenomeStudio V2011.1. Expression level was log2 transformed, robust multi-array average normalized (RMA, includes quantile normalization) (Irizarry et al., 2003) and batch-corrected using ComBat (Johnson et al., 2007). The HumanHT-12 v4 Expression BeadChip includes 47,231 probes annotated to transcripts. Significant expression (p<0.05) of 16,010 and 13,118 transcript probes was observed in adipose and muscle RNA, respectively, in 90% of participants. Data from these probes were primarily used for analysis. Probes were further filtered out based on bioinformatic criteria described in the supplementary methods. Genotype data were examined to verify sample and SNP quality. Genotype assays of 4,210,443 SNPs passed technical quality filters. The genotype of 2,296,925 autosomal SNP assays (representing 2,210,735 unique high-quality genotyped SNPs with MAF>0.01 and HWE-p value >1×10−6) was used in eQTL analysis.
Statistical and Bioinformatic analyses
To identify expression quantitative trait loci (eQTLs), linear regression was computed with the log2 transformed expression values as the outcome and an additive genetic model for the SNP as implemented in the R-package MatrixEQTL (Shabalin, 2012); age, gender, and African ancestry proportion were covariates. Analyses scanned for both cis and trans eQTLs, but partitioned the overall type 1 error rate of α=0.05 into α=0.04 for cis and α=0.01 for trans. However, we considered as significant any cis- and trans-eSNPs with a false discovery rate (FDR)-corrected p-value (Q-value) <0.01 (or 1.0%). Detailed statistical and bioinformatic data analysis methods are presented in Supplementary methods. Sample sizes in each analysis (Supplementary Table 1) varied based on available data after quality control.
Replication of eQTL data
Adipose cis-eQTL data from the Multiple Tissue Human Expression Resource (MuTHER) project (Grundberg et al., 2012) and muscle cis-eQTL data reported by Keildson et al. (Keildson et al., 2014) were mined to replicate cis-eGenes identified in the AAGMEx cohort (Supplementary Methods). Additionally, replication of adipose and muscle cis-eGenes was tested in publically available tissue eQTL data from GTEx project (GTEx_Analysis_v6 updated) and lymphoblastoid cell line (LCL) eQTL data from both the Geuvadis RNA sequencing project and the SeeQTL data depository.
Integration of GWAS data
Cis-eSNPs identified in adipose and muscle of African Americans represented a prioritized set of SNPs providing statistical evidence for genotype-dependent variation in transcript abundance. The NHGRI Catalog of Published GWAS (Hindorff et al., 2009) and meta-analysis data from the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) (Scott et al., 2012) were mined to identify the role of putatively functional SNPs in T2D susceptibility and gluco-metabolic phenotypes. We also searched for associations of eSNPs with T2D and BMI in the Meta-analysis of Type 2 Diabetes in African Americans (MEDIA) Consortium (Ng et al., 2014) and African Ancestry Anthropometry Genetic Consortium (AAAGC) (Monda et al., 2013) GWAS (Supplementary Methods).
RESULTS
eQTLs identified in adipose and muscle
We identified 1971 and 2078 transcripts with at least one significant cis-eQTL (top eSNP within ±500kb around the expressed transcript at FDR <0.01) in adipose and muscle, respectively (Figure 1A, 1B, Supplementary Tables 2 and 3). These transcripts were considered cis-eGenes.
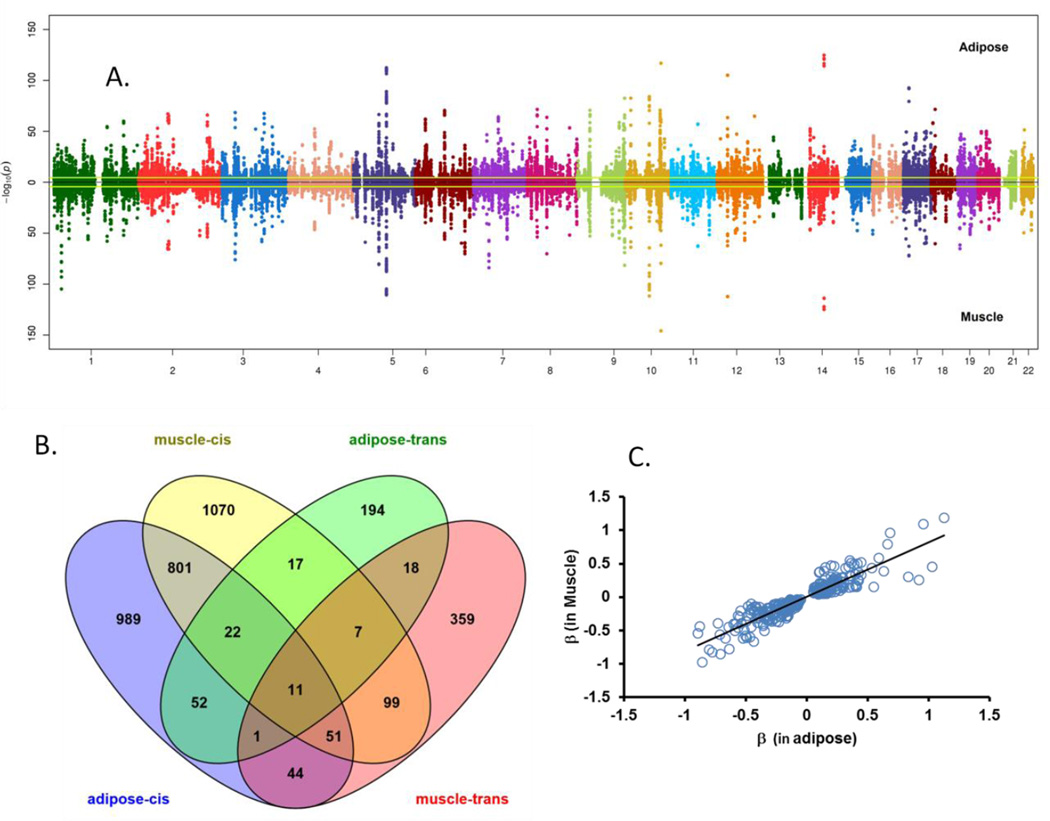
Figure 1. Expression quantitative trait locus (eQTL) analysis identified regulatory polymorphisms for adipose and muscle tissue transcripts in African Americans.
Opposing Manhattan plot showing chromosomal distribution of −log10(p-values) for association of all cis-SNPs (±500 kb of the transcript start and end) tested for transcripts (all probes representing refSeq genes) expressed in adipose and muscle. Significance threshold (Q-value <0.01) is marked by fluorescent yellow color lines. (A). A Venn diagram (B) shows common and tissue specific cis- and trans-eGenes (FDR<0.01 and selected clean probes representing known genes) in adipose and muscle. Top cis-eSNPs for 317 transcripts showed the same direction of effect in both tissues (C)
Overlap between cis-eGenes was identified in adipose and muscle. Cis-eQTLs for 885 transcripts were identified in both insulin-responsive tissues (Figure 1B). Among these, 317 were most strongly associated with the same cis-eSNPs in both tissues and showed the same direction of effect (Figure 1C and Supplementary Table 4). The most significant cis-eGenes (FDR <1×10−100) observed in both tissues included churchill domain containing 1 (CHURC1), up-regulated during skeletal muscle growth-homolog 5 (USMG5), and endoplasmic reticulum aminopeptidase 2 (ERAP2) (Table 1).
Table 1.
Top 20 cis-eQTLs common in adipose and muscle tissue of African Americans
| cis-eSNP | Chr | A1 | MAF | Adipose | Muscle | Probe ID | Entrez Gene ID |
Symbol | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | P- value |
Q- value |
β | P- value |
Q- value |
|||||||
| rs7143432 | 14 | C | 0.391 | 0.96 | 1.97E-125 | 7.97E-119 | 1.10 | 1.58E-125 | 2.23E-119 | ILMN_1798177 | 91612 | CHURC1 |
| rs11191642 | 10 | G | 0.091 | 1.13 | 1.59E-117 | 1.61E-111 | 1.18 | 1.36E-146 | 3.36E-140 | ILMN_1773313 | 84833 | USMG5 |
| rs74345571 | 5 | G | 0.440 | 1.03 | 4.60E-113 | 3.49E-107 | 0.45 | 2.52E-111 | 1.18E-105 | ILMN_1743145 | 64167 | ERAP2 |
| rs1055340 | 10 | A | 0.251 | −0.41 | 2.91E-83 | 8.03E-78 | −0.32 | 1.58E-64 | 1.44E-59 | ILMN_1795336 | 9317 | PTER |
| rs8413 | 9 | G | 0.365 | −0.40 | 3.42E-83 | 9.02E-78 | −0.28 | 2.75E-82 | 4.74E-77 | ILMN_1811301 | 56623 | INPP5E |
| rs200198999 | 18 | G | 0.191 | −0.60 | 3.01E-72 | 5.63E-67 | −0.40 | 4.18E-61 | 3.24E-56 | ILMN_1776515 | 65258 | MPPE1 |
| rs2137471 | 8 | G | 0.396 | −0.40 | 3.45E-72 | 6.34E-67 | −0.23 | 1.79E-54 | 9.46E-50 | ILMN_1720059 | 79618 | HMBOX1 |
| rs59347416 | 9 | T | 0.153 | −0.54 | 2.44E-71 | 4.28E-66 | −0.21 | 1.55E-53 | 7.87E-49 | ILMN_1744980 | 84186 | ZCCHC7 |
| rs2232745 | 2 | T | 0.457 | −0.34 | 8.32E-68 | 1.23E-62 | −0.29 | 2.54E-66 | 2.63E-61 | ILMN_1655340 | 51255 | RNF181 |
| rs6873912 | 5 | A | 0.271 | 0.39 | 3.47E-67 | 5.02E-62 | 0.49 | 5.95E-81 | 9.44E-76 | ILMN_2103295 | 10412 | TINP1 |
| rs7021977 | 9 | A | 0.327 | 0.43 | 3.66E-67 | 5.23E-62 | 0.37 | 2.42E-49 | 9.58E-45 | ILMN_1808661 | 401505 | TOMM5 |
| rs28405687 | 2 | T | 0.375 | 0.40 | 1.11E-66 | 1.50E-61 | 0.34 | 1.73E-54 | 9.22E-50 | ILMN_1770020 | 53938 | PPIL3 |
| rs6663 | 12 | T | 0.165 | 0.55 | 1.75E-65 | 2.13E-60 | 0.15 | 1.18E-39 | 2.66E-35 | ILMN_1719064 | 83892 | KCTD10 |
| rs6873912 | 5 | A | 0.271 | 0.42 | 2.41E-57 | 1.80E-52 | 0.50 | 2.02E-81 | 3.26E-76 | ILMN_1694259 | 10412 | NSA2 |
| rs3796683 | 4 | T | 0.366 | −0.58 | 3.31E-53 | 2.09E-48 | −0.40 | 2.36E-47 | 8.22E-43 | ILMN_1656560 | 25849 | PARM1 |
| rs73992309 | 17 | G | 0.064 | −0.47 | 9.31E-50 | 5.00E-45 | −0.61 | 8.54E-61 | 6.46E-56 | ILMN_1685112 | 51204 | TACO1 |
| rs73366229 | 17 | C | 0.270 | 0.38 | 4.57E-49 | 2.36E-44 | 0.15 | 7.24E-21 | 3.47E-17 | ILMN_1720708 | 1453 | CSNK1D |
| rs17856037 | 14 | T | 0.178 | −0.73 | 1.37E-44 | 5.73E-40 | −0.39 | 1.11E-30 | 1.33E-26 | ILMN_3243744 | 55837 | EAPP |
| rs3211938 | 7 | G | 0.100 | −0.86 | 9.59E-44 | 3.78E-39 | −0.98 | 2.37E-35 | 4.01E-31 | ILMN_1796094 | 948 | CD36 |
| rs2908700 | 12 | C | 0.415 | 0.40 | 2.72E-43 | 1.04E-38 | 0.18 | 1.73E-23 | 1.11E-19 | ILMN_1784608 | 9976 | CLEC2B |
Shown are the cis-eSNPs (genotyped SNP within ±500kb of the 5' and 3' end of the transcript) that are most strongly associated (Q-value < 0.01) and in the same direction for the same transcripts. Chr, Chromosome; A1, Minor Allele; MAF, Minor Allele Frequency; β, effect size of minor allele (A1); P-value, significance in additive model (MatrixEQTL analysis); Q-value, false discovery rate. Results for all 317 cis-eGenes associated with same cis-eSNPs and showing the same allele direction of effect in both tissues are provided in Supplementary Table 4.
SNP–transcript expression-level associations for variants located on other chromosomes or outside the defined cis boundary (±500kb around the transcript) were examined to identify trans-regulatory variants. Associations were identified (FDR<0.01) for expression of 603 and 943 transcripts with a genotype of at least one trans-eSNP in adipose and muscle, respectively (data not shown). Considering the large number of tests performed to identify trans-eQTLs, we conservatively considered a transcript associated with >1 trans-eSNP as statistically significant. Using this conservative criterion, 322 and 591 trans-eGenes were identified in adipose and muscle, respectively. Overlap of these trans-eGenes with cis-eGenes is shown in Figure 1B. Summary statistics of all cis- and trans-eSNPs is publically accessible through a searchable database at https://mdsetaa.phs.wakehealth.edu.
Genomic distribution of cis-eSNPs
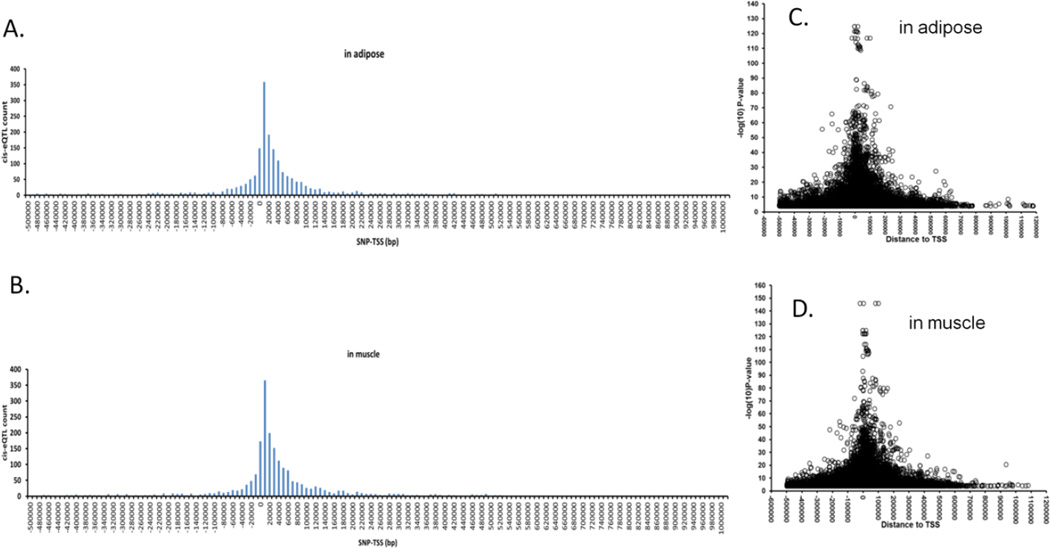
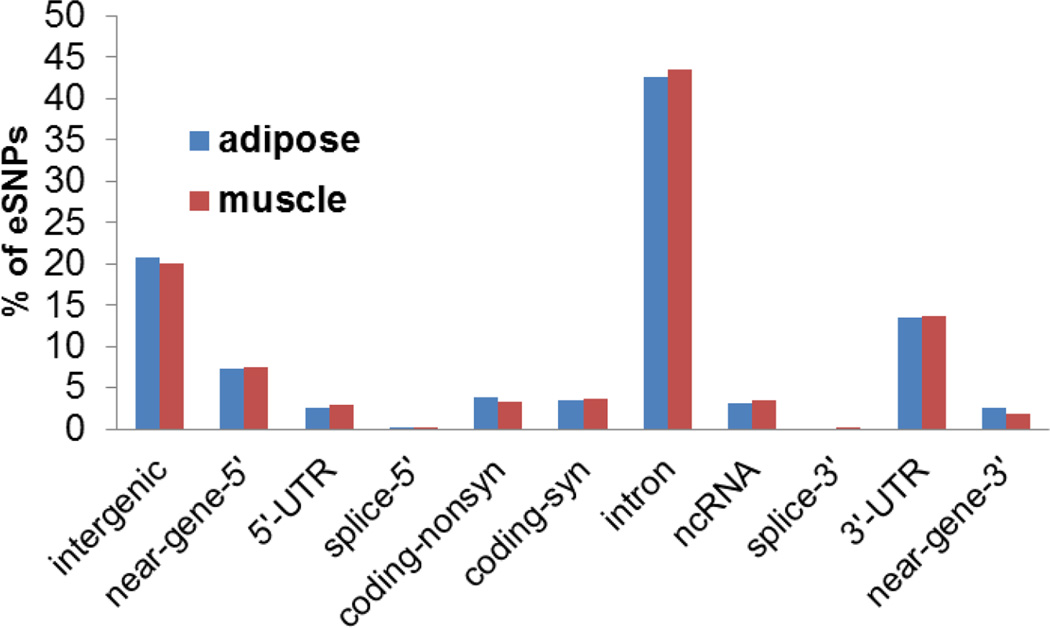
The distribution of the top cis-eSNPs for each transcript was assessed in relation to gene proximity and plotted against the distribution of distances between cis-eSNPs with the lowest p-values and transcription start site (TSS). As reported (Stranger et al., 2005;Stranger et al., 2007;Veyrieras et al., 2008), the majority of top cis-eSNPs in adipose (62.6%) and muscle (61.7%) were located within ±50kb of the TSS (Figure 2A and 2B). For adipose, 80% and 95% of the top cis-eSNPs were within 118.6 kb and 374.4 kb, respectively, of the TSS. For muscle, these distances were 122.8 kb and 371.4 kb, respectively. Cis-eSNPs with larger effect sizes were also overrepresented close to TSS (Figure 2C and 2D). Greater than 80% (80.8% in adipose; 84% in muscle) of the highly significant cis-eSNPs (P-value <1×10−10) were located within ±100 kb of the TSS. To assess potential functional significance, we annotated the genomic locations (based on the Sequence Ontology definitions) of the top cis-eSNPs of associated transcripts (FDR<0.01). Interestingly, 79.6% of the top cis-eSNPs were located within or near gene regions (±5 kb), and only 20.4% were intergenic. Most of the top cis-eSNPs were intronic (43.1%) or in the 3’ or 5’ untranslated regions (UTR, 16.3%, Figure 3). Utilizing the TRANSFAC Database, SNPnexus (Dayem Ullah et al., 2012) annotation predicted the disruption of transcription factor binding sites by 80 and 77 top cis-eSNPs in adipose and muscle, respectively.
Figure 2. Physical location of cis-eSNPs with respect to transcription start sites of each gene.
Distance distribution of each transcript’s most strongly associated cis-eSNPs (FDR<0.01) to its TSS in adipose (A) and muscle (B). Each bar represents the count of top cis-eSNPs in 10kb bins. Plots C and D show distribution of significance level (−log10 p-values) of all genotyped cis-eSNPs (FDR<0.01) relative to TSS in adipose and muscle.
Figure 3. Genomic distribution and functional annotation of cis-eSNPs.
Bar graph shows functional annotation of each adipose and muscle tissue transcript’s most strongly associated cis-eSNPs (FDR<0.01) in the genome.
In silico analyses indicate functional significance of cis-eGenes
Gene-annotation enrichment analysis with DAVID (Huang et al., 2009) indicated an enrichment of mitochondrial genes (GO:0005739) among cis-eGenes in both adipose (p=4.67×10−11, 141 transcripts) and muscle (p=4.84×10−30, 235 transcripts). Although not strongly enriched, cis-eGenes in both adipose (34 genes, p=0.03, FDR=26.5%) and muscle (47 genes, p=0.0079, FDR= 7.8%) included genes involved in diabetes based on Reactome pathway annotation (Reactome pathway, REACT_15380: Diabetes pathways). IPA predicted HNF4A (a transcription factor) as the most common upstream regulatory factor for adipose (p-value of overlap=2.23×10−15, 254 transcripts) and muscle (p=3.82×10−22 , 288 genes) cis-eGenes identified. Expression of a subset of cis-eGenes (362 in adipose; 42 in muscle) was associated with SI in this non-diabetic African American cohort (Sharma et al., 2016).
Replication of cis-eQTLs using publically available data
Published studies for eQTLs in insulin-responsive tissues in African Americans are not available. Replication of eQTLs was assessed by mining published adipose and muscle eQTLs in Caucasians (populations of European ancestry). Adipose eQTL data from MuTHER (Grundberg et al., 2012) were assessed for replication of 190 top cis-eGenes identified in AAGMEx (included probes for 100 top cis-eGenes and 100 top SI-associated cis-eGenes). Information for 155 of the 190 selected probes was available in MuTHER. Comparison of data sets showed an association of 114 cis-eGenes (probes) with the same cis-eSNPs in both studies (Supplementary Table 5), supporting replication of 73.5% of top adipose cis-eQTLs in our AAGMEx cohort.
Muscle cis-eQTLs as reported by Keildson et al. (Keildson et al., 2014), were searched to assess replication of these cis-eGenes (n=287) in the AAGMEx cohort, considering all probes representing significant (up to FDR <0.04) cis-eGenes. Comparison of the two muscle-eQTL data sets showed an association of 144 cis-eGenes (represented by 191 probes, Supplementary Table 6). Despite the small sample and different design of the previous study by Keildson et al., 50.2% of their muscle-eGenes were replicated in our AAGMEx African American cohort. Despite different methods of transcript quantification (RNA-seq) and sample characteristics (N=298 and 361 cadaver donors for subcutaneous adipose and skeletal muscle, respectively), results from GTEx (GTEx_Analysis_v6 updated: 2015-06-18, dbGaP Accession phs000424.v6.p1) (GTEx consortium, 2015) supported replication of 965 adipose cis-eGenes and 1016 muscle cis-eGenes from AAGMEx (data not shown).
To determine the replication of adipose and muscle tissue cis-eGenes in surrogate tissues, publically available eQTL data for transformed lymphoblastoid cell lines (LCL) were searched. The LCL-eQTL data from a small African ancestry cohort (Geuvadis RNA sequencing project of 1000 Genomes YRI samples, N= 89; http://www.geuvadis.org/web/geuvadis/RNAseq-project), replicated 96 and 95 adipose and muscle tissue-identified cis-eGenes, respectively. However, replication was much higher when larger LCL-eQTL datasets, e.g. HapMap3 consensus cis-eQTL (SeeQTL; http://www.bios.unc.edu/research/genomic_software/seeQTL) were searched for comparison. Considering cis-eGenes at q<0.01 from SeeQTL data, ~24% of AAGMEx adipose and muscle cis-eGenes (448 in adipose; 443 in muscle) were replicated in LCLs. Thus, expression of a subset of transcripts is genetically regulated across tissues, and the eQTL data from LCLs may be used as a proxy for identifying a small subset of adipose and muscle tissue cis-eGenes.
Association of cis-eSNPs with T2D, obesity and related metabolic phenotypes
The NHGRI Catalog of Published GWAS (from UCSC table browser) was mined (Hindorff et al., 2009) for association of all significant cis-eSNPs (FDR<0.01) for adipose and muscle transcripts to identify potential roles in T2D susceptibility and related phenotypes. This catalog only includes phenotype-associated index SNPs (p-values <1.0×10−5), mostly from people not of recent African descent. Association of cis-eSNPs was detected for >50 genes with gluco-metabolic phenotypes including T2D (e.g. PIK3C2A, RBMS1, UFSP1, ACHE), fasting plasma glucose (e.g. NOSTRIN, RREB1), hemoglobin A1c (e.g. FN3KRP), and BMI and obesity-related traits (e.g. POMC, MARCH6, NINJ1, RBP1, HMBOX1, CHURC1, CPEB4; Supplementary Table 7).
GWAS data from Caucasians in the MAGIC cohort (Scott et al., 2012) were assessed for glucose homeostasis phenotypes; Supplementary Table 8 lists cis-eSNPs for 54 adipose and 63 muscle genes with evidence for association (p<0.01) with glucose homeostasis phenotypes. An eSNP (both adipose and muscle) for inositol polyphosphate-5-phosphatase (INPP5E) was strongly associated with fasting glucose (rs1128905, p=5.81×10−9), and an eSNP for ERAP2 was strongly associated with 2h-glucose (rs1019503, p=8.97×10−9 ). Interestingly, rs560887, located in the intron of the glucose-6-phosphatase catalytic subunit 2 gene (G6PC2) was strongly associated with fasting glucose (p=1.4×10−178) in Caucasians in the MAGIC cohort, as was a cis-eSNP for NOSTRIN (nitric oxide synthase trafficker gene) in adipose of people in the AAGMEx cohort. Muscle eSNPs rs2068834 (sorting nexin 17 gene, SNX17; p=9.78×10−20) and rs11715915 (macrophage stimulating 1 gene, MST1; p=4.90×10−8) associated with fasting glucose; eSNP rs6912327 (UHRF1 binding protein 1 gene, UHRF1BP1) associated with BMI-adjusted fasting insulin (p=2.26×10−8).
A search for association of eSNPs (FDR <0.01) with T2D and BMI was performed in GWAS from MEDIA (Ng et al., 2014) and AAAGC (Monda et al., 2013). Cis-eSNPs for 72 genes in adipose and 80 genes in muscle showed nominal evidence (p<0.01) of association with T2D in African Americans in the MEDIA cohort (Supplementary Table 9). Three cis-eSNPs show stronger association (p<1.0×10−4) with T2D (Table-2A). A cis-eSNP for the transcript of ATP synthase subunit s-like protein (ATP5SL) gene was most significantly associated with T2D (rs7259208, p=1.20×10−5). Cis-eSNPs for 65 genes in adipose and 91 genes in muscle showed nominal evidence of association (p<0.01) with BMI in African Americans in the AAAGC cohort (Supplementary Table 10). Four cis-eSNPs show stronger association (p<1.0×10−4) with BMI (Table-2B). Among the selected subset of cis-eSNPs, rs4074110 (methylcrotonoyl-CoA carboxylase 1, MCCC1) showed the most significant association with BMI (p=6.11×10−6) in meta-analyses from AAAGC data. Thus, cis-eSNPs may modulate the risk for T2D and obesity in African Americans.
Table 2.
cis-eSNPs for adipose and muscle tissue transcripts associated with T2D and BMI in GWAS meta-analysis in African American Subjects
| A) eSNPs (FDR 1%) from the AAGMEx cohort are associated with T2D (p≤ 0.0001) in the "Meta-analysis of Type 2 Diabetes in African Americans” Consortium cohorts. | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | SNP Chr |
SNP Pos |
A1 | MAF | Illuminaprobe_ID | Tissue | β | P Valuea |
Symbol | Entrez Gene _ID |
Allele1 | Freq1 | OR (95%CI) |
P Valueb |
N |
| rs1594 | 2 | 202025621 | C | 0.443 | ILMN_1770020 | Ad | −0.166 | 5.90E-09 | PPIL3 | 53938 | A | 0.527 | 0.912 (0.873–0.953) |
4.17E-05 | 23737 |
| ILMN_1789830 | Ad | 0.119 | 5.07E-07 | CFLAR | 8837 | ||||||||||
| ILMN_1770020 | Mu | −0.114 | 1.01E-05 | PPIL3 | 53938 | ||||||||||
| ILMN_1789830 | Mu | 0.086 | 2.45E-05 | CFLAR | 8837 | ||||||||||
| rs2930532 | 15 | 44133890 | C | 0.191 | ILMN_1725043 | Ad | −0.068 | 1.08E-05 | ADAL | 161823 | T | 0.813 | 1.137 (1.069–1.210) |
4.89E-05 | 21261 |
| ILMN_1725043 | Mu | −0.065 | 1.32E-07 | ||||||||||||
| rs7259208 | 19 | 41894678 | A | 0.094 | ILMN_1809027 | Ad | 0.163 | 5.64E-09 | ATP5SL | 55101 | A | 0.096 | 1.184 (1.098–1.277) |
1.20E-05 | 22755 |
| ILMN_1809027 | Mu | 0.156 | 1.18E-06 | ||||||||||||
| B) eSNPs (FDR 1%) from the AAGMEx cohort are associated with BMI (p≤ 0.0001) in the "African Ancestry Anthropometry Genetic Consortium" cohorts | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | SNP Chr |
SNP Pos |
A1 | MAF | Illumina probe_ID |
Tissue | β | P Valuea |
Symbol | Entrez Gene _ID |
Allele1 | Freq1 | Effect (SE) |
P Valueb |
N |
| rs4074110 | 3 | 182728669 | C | 0.140 | ILMN_1760174 | Mu | 0.109 | 6.65E-08 | MCCC1 | 56922 | A | 0.830 | −0.051 (0.011) |
6.11E-06 | 37033 |
| rs6005881 | 22 | 29183133 | A | 0.336 | ILMN_1809433 | Mu | 0.082 | 2.60E-06 | XBP1 | 7494 | A | 0.351 | −0.034 (0.008) |
3.62E-05 | 38497 |
| rs7018469 | 9 | 114195274 | C | 0.140 | ILMN_1704531 | Ad | 0.429 | 2.81E-18 | PTGR1 | 22949 | T | 0.870 | −0.058 (0.013) |
1.86E-05 | 29370 |
| ILMN_2225537 | Ad | 0.463 | 4.50E-22 | PTGR1 | |||||||||||
| ILMN_1704531 | Mu | 0.173 | 9.68E-17 | PTGR1 | |||||||||||
| ILMN_2225537 | Mu | 0.205 | 7.85E-20 | PTGR1 | |||||||||||
| rs7949567 | 11 | 85251005 | G | 0.346 | ILMN_1784847 | Ad | −0.071 | 2.17E-07 | CREBZF | 58487 | A | 0.633 | 0.036 (0.008) |
7.10E-06 | 39130 |
| ILMN_2336609 | Mu | −0.225 | 4.21E-10 | SYTL2 | 54843 | ||||||||||
| ILMN_2217809 | 0.211 | 6.42E-24 | TMEM126A | 84233 | |||||||||||
In eQTL analysis A1, Minor Allele; MAF, Minor Allele Frequency; β, effect size of minor allele (A1);
P-valuea, significance in additive model; Q-value, false discovery rate. In T2D and BMI GWAS meta-analysis Allele1, effect allele; Freq1, frequency of effect allele;
P-valueb, GWAS Meta-analysis p-values. Ad, subcutaneous adipose tissue; Mu, skeletal muscle tissue. Other T2D or BMI associated (p<0.01) eSNPs are shown in supplementary table 9 and 10.
Integration of AAGMEx eQTL results and GWAS of gluco-metabolic traits suggested putative target genes for GWAS-identified SNPs. A total of 216 and 249 target cis-eGenes in adipose and muscle, respectively, were identified. Among these target cis-eGenes, mRNA expression of 55 genes in adipose, and 20 genes in muscle were significantly associated (p<0.001) with the glucose homeostasis traits (SI and AIRG derived from FSIGT; HOMA-IR and Matsuda index derived from OGTT), or obesity (BMI) phenotypes of AAGMEx participants (Supplementary Table 11).
DISCUSSION
Despite successes in GWAS, the majority of loci accounting for T2D heritability remain unknown and the diversity of its pathophysiology, molecular mechanisms, and variants explaining enhanced susceptibility in African Americans are poorly understood. The present study combined gene expression in tissues important to insulin action, and genome-wide genotype data in African Americans to fill these gaps. Results provide a comprehensive map of genetically regulated transcripts in African Americans, which is critical for prioritizing GWAS-identified SNPs in replication studies and detecting functional roles of variants involved in T2D and related traits.
Integration of genome-wide expression and genotype data enabled mapping of loci involved in the regulation of gene expression. Association of SNPs with transcript levels of nearby (cis) or distal (trans) genes were identified. Compared to adipose, slightly more cis-eQTL-transcripts (cis-eGenes) were found in muscle. Overlap of 885 cis-eQTL transcripts (~45% of cis-eGenes) was seen in both tissues indicating tissue- independent expression regulatory elements. Significant cis-eGenes observed in both tissues included USMG5 and ERAP2. The USMG5 gene, also known as the diabetes-associated protein in insulin-sensitive tissue gene (DAPIT), is differentially modulated in insulin-responsive tissues of streptozotocin-treated diabetic rats (Kontro et al., 2012; Paivarinne and Kainulainen, 2001). ERAP2 is involved in maturation of many proteins in the endoplasmic reticulum (ER), and has been implicated in regulation of angiogenesis and blood pressure (Cifaldi et al., 2012). An eSNP for ERAP2 was strongly associated with 2h-glucose in the MAGIC cohort. Consistent with published eQTL studies (Stranger et al., 2005; Stranger et al., 2007; Veyrieras et al., 2008), top cis-eSNPs for 60% of transcripts in both tissues were within ±50 kb of the TSS. The genomic distribution of cis-eSNPs fits with existing knowledge on the genetic regulatory architecture of transcript expression. Further bioinformatic annotation of these loci indicated the disruption of transcription factor binding by eSNPs and provided evidence for regulatory motifs. Thus, the identified eQTLs support the concept that functional regulatory genomic regions exist in glucose homeostasis-regulating tissues. Many cis-eQTLs identified in this African American cohort were replicated in non-African cohorts. Thus, a subset of genetic regulatory mechanisms of transcript expression is common between African Americans and non-Africans. Further studies will be required to confirm, whether other subsets of genetic regulatory mechanisms of transcript expression predominately influence particular ancestral groups.
Enrichment (DAVID analysis) of mitochondrial genes was identified among adipose and muscle cis-eGenes, indicating a role for genetic factors in modulation of this pathway. IPA revealed enrichment of pathways involved in mitigating oxidative stress (including glutathione-mediated detoxification, p=8.32×10−3–3.72×10−5; NRF2-mediated oxidative stress response, p=0.02-4.47×10−4) among cis-eGenes. These biological pathways may play key roles in modulating insulin sensitivity in African Americans.
Compared to cis-eQTLs, the effect sizes of trans-eQTLs are generally small, requiring larger sample sizes for robust detection of trans-eQTLs (Grundberg et al., 2012). A recent eQTL analysis in adipose tissue from Caucasian female twins (MuTHER Project, N=856) identified 3,529 cis-eQTLs (at FDR 1%) and 639 trans-eQTLs (at FDR 10%) (Grundberg et al., 2012). A stringent threshold (FDR<1%, with corresponding uncorrected p-values <2.6×10−9 in adipose tissue) was used to account for the large number of tests performed for trans-eQTL analysis in the AAGMEx cohort, and it identified 322 and 591 trans-eGenes in adipose and muscle, respectively. Thus, the number of trans-eGenes identified in AAGMEx is consistent with expectations and comparable to published studies on adipose and other tissues.
Mining of the NHGRI catalogue of GWAS (Hindorff et al., 2009) and MAGIC GWAS meta-analysis (Scott et al., 2012) results revealed association of cis-eSNPs in this study with T2D and related phenotypes. Although these SNP-disease association results are primarily from cohorts of individuals not of African descent, integration of eQTL data from our African American participants suggests molecular mechanisms that are putatively regulated by these SNPs and sequentially modulating disease susceptibility.
Cis-eSNPs for many adipose and muscle transcripts showed association with T2D and BMI in MEDIA (Ng et al., 2014) and AAAGC (Monda et al., 2013) African Americans, supporting roles for these transcripts in T2D. Genes modulated by disease-associated cis-eSNPs (e.g., CD36, CAMK2A, IRS2, POMC, TLR4, XBP1) are involved in the pathophysiology of T2D, obesity and related traits; whereas the roles of other cis-eGenes (e.g. ADAL, ATP5SL, MCCC1) are unknown. Interestingly, among the target cis-eGenes for these GWAS-identified SNPs, mRNA expression of 55 genes in adipose, and 20 genes in muscle was significantly associated with glucose homeostasis or obesity phenotypes in AAGMEx African Americans. This observation suggests a putative role for these GWAS-identified SNPs and respective cis-eGenes in the pathophysiology of T2D and related metabolic diseases. Association summary statistics were available from the MEDIA and AAAGC cohorts for directly genotyped SNPs and HapMap reference panel imputed SNPs. Association results for a subset of cis-eSNPs or their proxies (those that were not among the HapMap SNPs) were not available from these GWAS. Thus, the role of cis-eSNPs of several transcripts identified in this study cannot be evaluated in the MEDIA and AAAGC cohorts.
Target cis-eGenes of T2D-associated SNPs from MEDIA African Americans overlapped with target cis-eGenes of glucose homeostasis trait-associated SNPs from MAGIC Caucasians. Cis-eSNPs for 18 genes (ACAD10, CUL3, G3BP2, GIN1, HLA-DPA1, HLA-DPB1, HSPA1B, KCTD10, NOTCH4, PFDN1, PPM1M, RNF41, SNX17, ST7L, STARD10, TIPARP, TMEM116, and WDR6) were associated with T2D in MEDIA African Americans and were also associated with glucose homeostasis phenotypes (e.g., fasting glucose, 2h-OGTT glucose and fasting insulin) in Caucasians from MAGIC. Similarly, target cis-eGenes of BMI-associated SNPs from AAAGC African Americans overlapped with target cis-eGenes of glucose homeostasis trait-associated SNPs from MAGIC Caucasians. Cis-eSNPs for 12 genes (APIP, ASAP3, BCS1L, GIN1, HSPA1B, PPP1CB, RABEP1, RPP40, SNX17, TMEM60, TOM1, and UHRF1BP1) were associated with BMI in AAAGC African Americans and were also associated with glucose homeostasis phenotypes in MAGIC Caucasians. Thus, regulatory SNP-mediated modulation of the transcript expression of some target genes may modulate susceptibility to T2D and related gluco-metabolic phenotypes in individuals with either African or European ancestry.
In conclusion, this study identified genetic loci influencing the expression of several genes in adipose and muscle of African Americans. Additionally, this study provides data on molecular mechanisms putatively regulated by eSNPs and sequentially modulating susceptibility for T2D and related metabolic phenotypes in African Americans.
Supplementary Material
Acknowledgments
We thank the dedicated staff of the Clinical Research Unit at WFSM and Kurt A. Langberg (WFSM-Endocrinology) for support of the clinical studies and assistance with data management. We thank Mrs. Joyce Byers for support in participant recruitment. We thank staff in the genomics core laboratory at Center for Genomics and Personalized Medicine Research, WFSM, especially Dr. Siqun Zheng, Shelly Smith, Tracey Young and Dr. Ge Li for their extensive support in genotyping, and gene expression analysis using the Illumina microarray platform. We acknowledge the support of the Center for Public Health Genomics, WFSM for computational resources. SKD and CDL are the guarantors of this work, and as such, had full access to all study data and take responsibility for integrity of the data and accuracy of data analysis.
Grants: This work was supported by National Institutes of Health Grant R01 DK090111 (SKD).
Footnotes
Duality of interest: No potential conflicts of interest relevant to this article were reported.
Author Contribution statement: S.P.S. performed quality control and statistical genetic analysis of data, and reviewed/edited the manuscript; N.K.S. performed biochemical and molecular genomic studies, analyzed data, and reviewed/edited the manuscript; J.W.C. performed quality control and analysis of genetics and genomic data, and reviewed/edited the manuscript; J. C-E., J.D., and S.R. performed studies including collection of tissue biopsy, and analyzed clinical data; L.Ma. and N.D.P. analyzed molecular genetic data, and reviewed/edited the manuscript; D.R.M. and J.B. contributed bioinformatic and computational tools; J.B. also developed the eQTL database; M.E.C. helped in experimental design and statistical analysis; L.M., E.K., and D.D. contributed to clinical studies and subject recruitment, and E.K. also reviewed/edited the manuscript; M.B., S.J.B., and L.E. performed physiological studies including OGTT and FSIVGT; the MEDIA consortium (M.N., N.M., S.P.,L.B., L.L., X.G., M.S., K.C.) and AAAG Consortium (K.M., G.C., K.T., C.P., T.E., K.N., C.H.) authors contributed meta-analysis data; D.W.B. contributed to study design, and reviewed/ edited the manuscript; B.I.F supervised participant recruitment and clinical studies, analyzed clinical data, and reviewed/edited manuscript; C.D.L. contributed to study design, supervised all statistical analysis of genetic/genomic data, and reviewed/edited the manuscript; S.K.D. designed the project, supervised physiological and molecular genomic studies, researched and analyzed data, led the interpretation of data, and wrote/ reviewed/ edited the manuscript.
REFERENCES
- Bergman RN, Stefanovski D, Kim SP. Systems analysis and the prediction and prevention of Type 2 diabetes mellitus. Curr Opin Biotechnol. 2014;28:165–170. doi: 10.1016/j.copbio.2014.05.007. Epub@2014 Jun 27.:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifaldi L, Romania P, Lorenzi S, Locatelli F, Fruci D. Role of endoplasmic reticulum aminopeptidases in health and disease: from infection to cancer. Int J Mol Sci. 2012;13:8338–8352. doi: 10.3390/ijms13078338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowie CC, Harris MI, Silverman RE, Johnson EW, Rust KF. Effect of multiple risk factors on differences between blacks and whites in the prevalence of non-insulin-dependent diabetes mellitus in the United States. Am J Epidemiol. 1993;137:719–732. doi: 10.1093/oxfordjournals.aje.a116732. [DOI] [PubMed] [Google Scholar]
- Cowie CC, Rust KF, Byrd-Holt DD, Gregg EW, Ford ES, Geiss LS, Bainbridge KE, Fradkin JE. Prevalence of diabetes and high risk for diabetes using A1C criteria in the U.S. population in 1988–2006. Diabetes Care. 2010;33:562–568. doi: 10.2337/dc09-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Sharma NK. Expression quantitative trait analyses to identify causal genetic variants for type 2 diabetes susceptibility. World J Diabetes. 2014;5:97–114. doi: 10.4239/wjd.v5.i2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40:W65–W70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groop L, Pociot F. Genetics of diabetes--are we missing the genes or the disease? Mol Cell Endocrinol. 2014;382:726–739. doi: 10.1016/j.mce.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, Bell JT, Yang TP, Meduri E, Barrett A, Nisbett J, Sekowska M, Wilk A, Shin SY, Glass D, Travers M, Min JL, Ring S, Ho K, Thorleifsson G, Kong A, Thorsteindottir U, Ainali C, Dimas AS, Hassanali N, Ingle C, Knowles D, Krestyaninova M, Lowe CE, Di MP, Montgomery SB, Parts L, Potter S, Surdulescu G, Tsaprouni L, Tsoka S, Bataille V, Durbin R, Nestle FO, O'Rahilly S, Soranzo N, Lindgren CM, Zondervan KT, Ahmadi KR, Schadt EE, Stefansson K, Smith GD, McCarthy MI, Deloukas P, Dermitzakis ET, Spector TD. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Keildson S, Fadista J, Ladenvall C, Hedman AK, Elgzyri T, Small KS, Grundberg E, Nica AC, Glass D, Richards JB, Barrett A, Nisbet J, Zheng HF, Ronn T, Strom K, Eriksson KF, Prokopenko I, Spector TD, Dermitzakis ET, Deloukas P, McCarthy MI, Rung J, Groop L, Franks PW, Lindgren CM, Hansson O. Expression of phosphofructokinase in skeletal muscle is influenced by genetic variation and associated with insulin sensitivity. Diabetes. 2014;63:1154–1165. doi: 10.2337/db13-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontro H, Hulmi JJ, Rahkila P, Kainulainen H. Cellular and tissue expression of DAPIT, a phylogenetically conserved peptide. Eur J Histochem. 2012;56:e18. doi: 10.4081/ejh.2012.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, Saleheen D, Wang X, Zeggini E, Abecasis GR, Adair LS, Almgren P, Atalay M, Aung T, Baldassarre D, Balkau B, Bao Y, Barnett AH, Barroso I, Basit A, Been LF, Beilby J, Bell GI, Benediktsson R, Bergman RN, Boehm BO, Boerwinkle E, Bonnycastle LL, Burtt N, Cai Q, Campbell H, Carey J, Cauchi S, Caulfield M, Chan JC, Chang LC, Chang TJ, Chang YC, Charpentier G, Chen CH, Chen H, Chen YT, Chia KS, Chidambaram M, Chines PS, Cho NH, Cho YM, Chuang LM, Collins FS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Danesh J, Das D, de FU, Dedoussis G, Deloukas P, Dimas AS, Dina C, Doney AS, Donnelly PJ, Dorkhan M, van DC, Dupuis J, Edkins S, Elliott P, Emilsson V, Erbel R, Eriksson JG, Escobedo J, Esko T, Eury E, Florez JC, Fontanillas P, Forouhi NG, Forsen T, Fox C, Fraser RM, Frayling TM, Froguel P, Frossard P, Gao Y, Gertow K, Gieger C, Gigante B, Grallert H, Grant GB, Grrop LC, Groves CJ, Grundberg E, Guiducci C, Hamsten A, Han BG, Hara K, Hassanali N, Hattersley AT, Hayward C, Hedman AK, Herder C, Hofman A, Holmen OL, Hovingh K, Hreidarsson AB, Hu C, Hu FB, Hui J, Humphries SE, Hunt SE, Hunter DJ, Hveem K, Hydrie ZI, Ikegami H, Illig T, Ingelsson E, Islam M, Isomaa B, Jackson AU, Jafar T, James A, Jia W, Jockel KH, Jonsson A, Jowett JB, Kadowaki T, Kang HM, Kanoni S, Kao WH, Kathiresan S, Kato N, Katulanda P, Keinanen-Kiukaanniemi KM, Kelly AM, Khan H, Khaw KT, Khor CC, Kim HL, Kim S, Kim YJ, Kinnunen L, Klopp N, Kong A, Korpi-Hyovalti E, Kowlessur S, Kraft P, Kravic J, Kristensen MM, Krithika S, Kumar A, Kumate J, Kuusisto J, Kwak SH, Laakso M, Lagou V, Lakka TA, Langenberg C, Langford C, Lawrence R, Leander K, Lee JM, Lee NR, Li M, Li X, Li Y, Liang J, Liju S, Lim WY, Lind L, Lindgren CM, Lindholm E, Liu CT, Liu JJ, Lobbens S, Long J, Loos RJ, Lu W, Luan J, Lyssenko V, Ma RC, Maeda S, Magi R, Mannisto S, Matthews DR, Meigs JB, Melander O, Metspalu A, Meyer J, Mirza G, Mihailov E, Moebus S, Mohan V, Mohlke KL, Morris AD, Muhleisen TW, Muller-Nurasyid M, Musk B, Nakamura J, Nakashima E, Navarro P, Ng PK, Nica AC, Nilsson PM, Njolstad I, Nothen MM, Ohnaka K, Ong TH, Owen KR, Palmer CN, Pankow JS, Park KS, Parkin M, Pechlivanis S, Pedersen NL, Peltonen L, Perry JR, Peters A, Pinidiyapathirage JM, Platou CG, Potter S, Price JF, Qi L, Radha V, Rallidis L, Rasheed A, Rathman W, Rauramaa R, Raychaudhuri S, Rayner NW, Rees SD, Rehnberg E, Ripatti S, Robertson N, Roden M, Rossin EJ, Rudan I, Rybin D, Saaristo TE, Salomaa V, Saltevo J, Samuel M, Sanghera DK, Saramies J, Scott J, Scott LJ, Scott RA, Segre AV, Sehmi J, Sennblad B, Shah N, Shah S, Shera AS. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Mohlke KL, Boehnke M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum Mol Genetddv. 2015;264 doi: 10.1093/hmg/ddv264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, Ng MC, Adeyemo AA, Allison MA, Bielak LF, Chen G, Graff M, Irvin MR, Rhie SK, Li G, Liu Y, Liu Y, Lu Y, Nalls MA, Sun YV, Wojczynski MK, Yanek LR, Aldrich MC, Ademola A, Amos CI, Bandera EV, Bock CH, Britton A, Broeckel U, Cai Q, Caporaso NE, Carlson CS, Carpten J, Casey G, Chen WM, Chen F, Chen YD, Chiang CW, Coetzee GA, Demerath E, Deming-Halverson SL, Driver RW, Dubbert P, Feitosa MF, Feng Y, Freedman BI, Gillanders EM, Gottesman O, Guo X, Haritunians T, Harris T, Harris CC, Hennis AJ, Hernandez DG, McNeill LH, Howard TD, Howard BV, Howard VJ, Johnson KC, Kang SJ, Keating BJ, Kolb S, Kuller LH, Kutlar A, Langefeld CD, Lettre G, Lohman K, Lotay V, Lyon H, Manson JE, Maixner W, Meng YA, Monroe KR, Morhason-Bello I, Murphy AB, Mychaleckyj JC, Nadukuru R, Nathanson KL, Nayak U, N'diaye A, Nemesure B, Wu SY, Leske MC, Neslund-Dudas C, Neuhouser M, Nyante S, Ochs-Balcom H, Ogunniyi A, Ogundiran TO, Ojengbede O, Olopade OI, Palmer JR, Ruiz-Narvaez EA, Palmer ND, Press MF, Rampersaud E, Rasmussen-Torvik LJ, Rodriguez-Gil JL, Salako B, Schadt EE, Schwartz AG, Shriner DA, Siscovick D, Smith SB, Wassertheil-Smoller S, Speliotes EK, Spitz MR, Sucheston L, Taylor H, Tayo BO, Tucker MA, Van Den Berg DJ, Edwards DR, Wang Z, Wiencke JK, Winkler TW, Witte JS, Wrensch M, Wu X, Yang JJ, Levin AM, Young TR, Zakai NA, Cushman M, Zanetti KA, Zhao JH, Zhao W, Zheng Y, Zhou J, Ziegler RG, Zmuda JM, Fernandes JK, Gilkeson GS, Kamen DL, Hunt KJ, Spruill IJ, Ambrosone CB, Ambs S, Arnett DK, Atwood L, Becker DM, Berndt SI, Bernstein L, Blot WJ, Borecki IB, Bottinger EP, Bowden DW, Burke G, Chanock SJ, Cooper RS, Ding J, Duggan D, Evans MK, Fox C, Garvey WT, Bradfield JP, Hakonarson H, Grant SF, Hsing A, Chu L, Hu JJ, Huo D, Ingles SA, John EM, Jordan JM, Kabagambe EK, Kardia SL, Kittles RA, Goodman PJ, Klein EA, Kolonel LN, Le ML, Liu S, McKnight B, Millikan RC, Mosley TH, Padhukasahasram B, Williams LK, Patel SR, Peters U, Pettaway CA, Peyser PA, Psaty BM, Redline S, Rotimi CN, Rybicki BA, Sale MM, Schreiner PJ, Signorello LB, Singleton AB, Stanford JL, Strom SS, Thun MJ, Vitolins M, Zheng W, Moore JH, Williams SM, Ketkar S, Zhu X, Zonderman AB, Kooperberg C, Papanicolaou GJ, Henderson BE, Reiner AP, Hirschhorn JN, Loos RJ, North KE, Haiman CA. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45:690–696. doi: 10.1038/ng.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MC, Shriner D, Chen BH, Li J, Chen WM, Guo X, Liu J, Bielinski SJ, Yanek LR, Nalls MA, Comeau ME, Rasmussen-Torvik LJ, Jensen RA, Evans DS, Sun YV, An P, Patel SR, Lu Y, Long J, Armstrong LL, Wagenknecht L, Yang L, Snively BM, Palmer ND, Mudgal P, Langefeld CD, Keene KL, Freedman BI, Mychaleckyj JC, Nayak U, Raffel LJ, Goodarzi MO, Chen YD, Taylor HA, Jr, Correa A, Sims M, Couper D, Pankow JS, Boerwinkle E, Adeyemo A, Doumatey A, Chen G, Mathias RA, Vaidya D, Singleton AB, Zonderman AB, Igo RP, Jr, Sedor JR, Kabagambe EK, Siscovick DS, McKnight B, Rice K, Liu Y, Hsueh WC, Zhao W, Bielak LF, Kraja A, Province MA, Bottinger EP, Gottesman O, Cai Q, Zheng W, Blot WJ, Lowe WL, Pacheco JA, Crawford DC, Grundberg E, Rich SS, Hayes MG, Shu XO, Loos RJ, Borecki IB, Peyser PA, Cummings SR, Psaty BM, Fornage M, Iyengar SK, Evans MK, Becker DM, Kao WH, Wilson JG, Rotter JI, Sale MM, Liu S, Rotimi CN, Bowden DW. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 2014;10:e1004517. doi: 10.1371/journal.pgen.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paivarinne H, Kainulainen H. DAPIT, a novel protein down-regulated in insulin-sensitive tissues in streptozotocin-induced diabetes. Acta Diabetol. 2001;38:83–86. doi: 10.1007/s005920170018. [DOI] [PubMed] [Google Scholar]
- Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, Magi R, Strawbridge RJ, Rehnberg E, Gustafsson S, Kanoni S, Rasmussen-Torvik LJ, Yengo L, Lecoeur C, Shungin D, Sanna S, Sidore C, Johnson PC, Jukema JW, Johnson T, Mahajan A, Verweij N, Thorleifsson G, Hottenga JJ, Shah S, Smith AV, Sennblad B, Gieger C, Salo P, Perola M, Timpson NJ, Evans DM, Pourcain BS, Wu Y, Andrews JS, Hui J, Bielak LF, Zhao W, Horikoshi M, Navarro P, Isaacs A, O'Connell JR, Stirrups K, Vitart V, Hayward C, Esko T, Mihailov E, Fraser RM, Fall T, Voight BF, Raychaudhuri S, Chen H, Lindgren CM, Morris AP, Rayner NW, Robertson N, Rybin D, Liu CT, Beckmann JS, Willems SM, Chines PS, Jackson AU, Kang HM, Stringham HM, Song K, Tanaka T, Peden JF, Goel A, Hicks AA, An P, Muller-Nurasyid M, Franco-Cereceda A, Folkersen L, Marullo L, Jansen H, Oldehinkel AJ, Bruinenberg M, Pankow JS, North KE, Forouhi NG, Loos RJ, Edkins S, Varga TV, Hallmans G, Oksa H, Antonella M, Nagaraja R, Trompet S, Ford I, Bakker SJ, Kong A, Kumari M, Gigante B, Herder C, Munroe PB, Caulfield M, Antti J, Mangino M, Small K, Miljkovic I, Liu Y, Atalay M, Kiess W, James AL, Rivadeneira F, Uitterlinden AG, Palmer CN, Doney AS, Willemsen G, Smit JH, Campbell S, Polasek O, Bonnycastle LL, Hercberg S, Dimitriou M, Bolton JL, Fowkes GR, Kovacs P, Lindstrom J, Zemunik T, Bandinelli S, Wild SH, Basart HV, Rathmann W, Grallert H, Maerz W, Kleber ME, Boehm BO, Peters A, Pramstaller PP, Province MA, Borecki IB, Hastie ND, Rudan I, Campbell H, Watkins H, Farrall M, Stumvoll M, Ferrucci L, Waterworth DM, Bergman RN, Collins FS, Tuomilehto J, Watanabe RM, de Geus EJ, Penninx BW, Hofman A, Oostra BA, Psaty BM, Vollenweider P, Wilson JF, Wright AF, Hovingh GK, Metspalu A, Uusitupa M, Magnusson PK, Kyvik KO, Kaprio J, Price JF, Dedoussis GV, Deloukas P, Meneton P, Lind L, Boehnke M, Shuldiner AR, van Duijn CM, Morris AD, Toenjes A, Peyser PA, Beilby JP, Korner A, Kuusisto J, Laakso M, Bornstein SR, Schwarz PE, Lakka TA, Rauramaa R, Adair LS, Smith GD, Spector TD, Illig T, de FU, Hamsten A, Gudnason V, Kivimaki M, Hingorani A, Keinanen-Kiukaanniemi SM, Saaristo TE, Boomsma DI, Stefansson K, van der Harst P, Dupuis J, Pedersen NL, Sattar N, Harris TB, Cucca F, Ripatti S, Salomaa V, Mohlke KL, Balkau B, Froguel P, Pouta A, Jarvelin MR, Wareham NJ, Bouatia-Naji N, McCarthy MI, Franks PW, Meigs JB, Teslovich TM, Florez JC, Langenberg C, Ingelsson E, Prokopenko I, Barroso I. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma NK, Sajuthi SP, Chou JW, Calles-Escandon J, Demons J, Rogers S, Ma L, Palmer ND, McWilliams R, Beal J, Comeau M, Cherry K, Hawkins GA, Menon L, Kouba E, Davis D, Burris M, Byerly SJ, Easter L, Bowden DW, Freedman BI, Langefeld CD, Das SK. Tissue-specific and Genetic Regulation of Insulin Sensitivity-Associated Transcripts in African Americans. J Clin Endocrinol Metab. 2016;101:1455–1468. doi: 10.1210/jc.2015-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM. Gene-expression variation within and among human populations. Am J Hum Genet. 2007;80:502–509. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavare S, Deloukas P, Dermitzakis ET. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, Sekowska M, Smith GD, Evans D, Gutierrez-Arcelus M, Price A, Raj T, Nisbett J, Nica AC, Beazley C, Durbin R, Deloukas P, Dermitzakis ET. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, Montgomery S, Tavare S, Deloukas P, Dermitzakis ET. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, Dolan ME. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.