Abstract
Long noncoding RNAs (lncRNAs) play important regulatory roles in a variety of diseases, including many tumors. However, the functional roles of these transcripts and mechanisms responsible for their deregulation in pancreatic ductal adenocarcinoma (PDAC) are not thoroughly understood. In this study, we discovered that lncRNA MIR31HG is markedly upregulated in PDAC. Knockdown of MIR31HG significantly suppressed PDAC cell growth, induced apoptosis and G1/S arrest, and inhibited invasion, whereas enhanced expression of MIR31HG had the opposite effects. Online database analysis tools showed that miR-193b could target MIR31HG and we found an inverse correlation between MIR31HG and miR-193b in PDAC specimens. Inhibition of miR-193b expression significantly upregulated the MIR31HG level, while overexpression of miR-193b suppressed MIR31HG's expression and function, suggesting that MIR31HG is negatively regulated by miR-193b. Moreover, using luciferase reporter and RIP assays, we provide evidence that miR-193b directly targeted MIR31HG by binding to two microRNA binding sites in the MIR31HG sequence. On the other hand, MIR31HG may act as an endogenous ‘sponge' by competing for miR-193b binding to regulate the miRNA targets. Collectively, these results demonstrate that MIR31HG functions as an oncogenic lncRNA that promotes tumor progression, and miR-193b targets not only protein-coding genes but also the lncRNA, MIR31HG.
Introduction
Pancreatic ductal adenocarcinoma (PDAC), accounting for over 90% of all pancreatic tumor cases, is the fourth leading cause of cancer-related deaths in the United States and remains one of the most lethal malignancies worldwide. Although notable improvements in survival have been made for most cancers over the past three decades, the prognosis for pancreatic cancer patients has shown the least improvement.1 Against this backdrop, the search for novel, biologically relevant genes involved in tumorigenesis, especially those with prognostic and therapeutic significance in PDAC, takes on added importance.
Accumulating data from whole genome and transcriptome studies have revealed that only a very small percentage (1–2%) of the genome encodes proteins,2, 3 but the majority of the mammalian genome encodes vast numbers of noncoding RNAs.4, 5 Long noncoding RNAs (lncRNAs) are a new class of regulatory RNAs and do not have protein-coding capacity.6 Recent studies have demonstrated that lncRNAs play essential roles in a wide range of biological processes,7, 8 and appear to be involved in the progression of a variety of human diseases.5, 9, 10 Furthermore, it has been shown that several lncRNAs are deregulated in many tumors and may be involved in carcinogenesis or cancer progression.11, 12, 13 In the case of PDAC, cDNA microarrays using probes for lncRNAs have revealed that sets of intronic lncRNAs are differentially expressed in primary and metastatic pancreatic cancer.14 Kim et al.15 demonstrated that one such lncRNA, HOTAIR, exhibits pro-oncogenic activity and is a negative prognostic factor in pancreatic cancer. MIR31HG (NCBI no: NR_027054) is a recently identified 2166-nt lncRNA. Interestingly, miR-31 maps to the intronic region of MIR31HG, and the transcriptional activities of both are regulated by promoter hypermethylation.16 Recent study reported that MIR31HG expression levels were significantly elevated in lung and breast cancer,17, 18 and knockdown of MIR31HG reduces cell growth and induces a senescence phenotype in p16INK4A-dependent manner.19 However, the functional role of MIR31HG in PDAC is still unknown.
Although large numbers of lncRNAs have been found to be associated with human diseases, less is known about their regulatory mechanisms. MicroRNAs (miRNAs), which are ~22 nucleotides in length, play important roles in the regulation of target gene expression by base pairing to complementary sites in 3′-untranslated regions (3′UTRs), 5′UTRs, and/or coding regions of the target mRNA.20, 21, 22 According to the recently proposed competing endogenous RNA hypothesis, which has been validated in a number of studies,23, 24 multiple lncRNA transcripts can act as endogenous decoys for miRNAs through their miRNA-binding sites25 and thus may regulate the biological activity of one another. This raises the question of whether lncRNA MIR31HG could also interact with miRNAs in a manner similar to that of protein-coding genes.
In the present study, we investigated the expression and role of MIR31HG in human PDAC. First, we demonstrated that MIR31HG levels are remarkably upregulated in PDAC tumor tissues. MIR31HG promotes PDAC cell growth and invasion, implying a possible role of tumor oncogenic lncRNA in the PDAC progression. More importantly, we found that miR-193b negatively regulates MIR31HG by directly targeting the miRNA-binding site in the body of MIR31HG, and MIR31HG also acts as a molecular sponge for miR-193b and regulates its downstream miRNA-target genes.
Results
MIR31HG is upregulated in PDAC
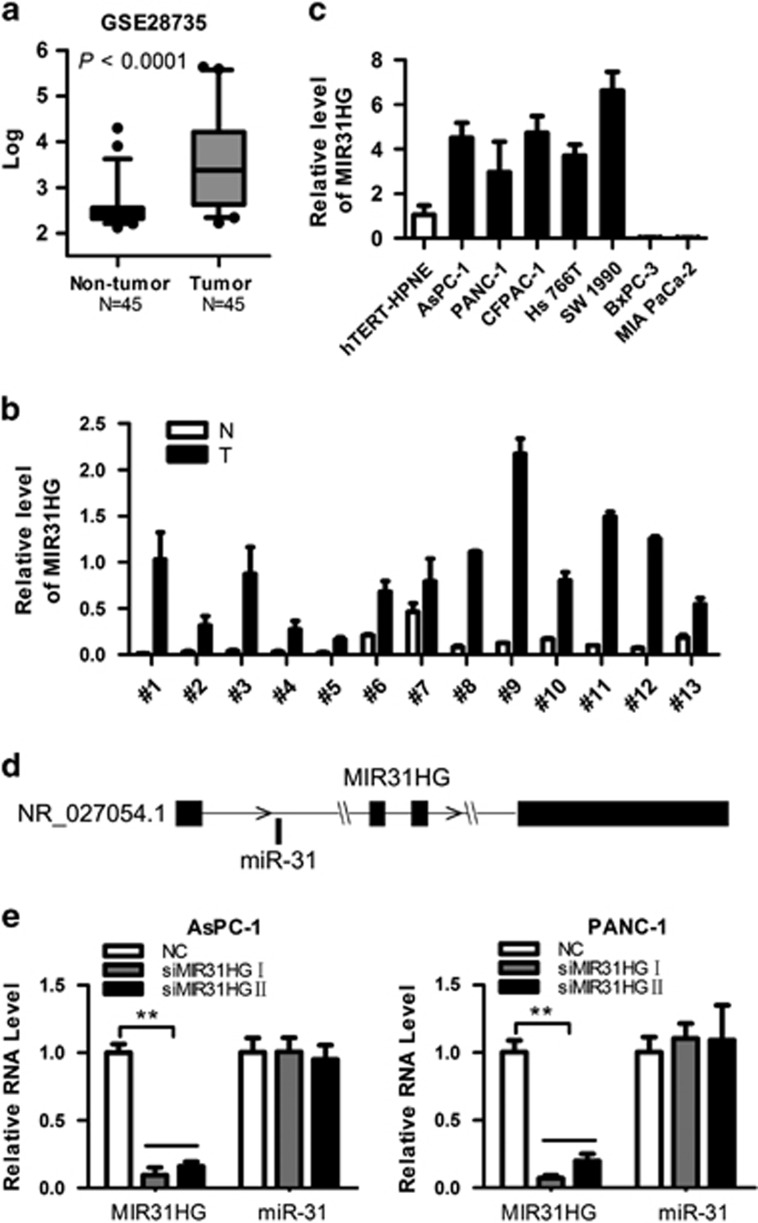
An analysis of pancreatic cancer patient gene profiling results (GSE28735)26 showed that, among 45 matched pairs of PDAC tumor and adjacent non-tumor tissues, MIR31HG was more highly expressed in pancreatic tumors (Figure 1a). To further validate this result, we evaluated the MIR31HG expression level in 13 paired PDAC and adjacent pancreatic tissue samples using reverse transcription and quantitative real-time PCR (RT–qPCR). The results showed that MIR31HG levels were significantly higher in tumor tissues (Figure 1b). We then examined the expression level of MIR31HG in PDAC cell lines (AsPC-1, PANC-1, CFPAC-1, Hs 766 T, SW 1990, MIA PaCa-2, and BxPC-3) and hTERT-HPNE cells, an immortalized, normal human pancreatic ductal cell line, using RT–qPCR. Compared with hTERT-HPNE cells, PDAC cells exhibited significantly higher levels of MIR31HG expression; strangely, MIR31HG expression was not detected in MIA PaCa-2 or BxPC-3 cells (Figure 1c; see Discussion). Collectively, these results suggest that MIR31HG is upregulated in PDAC. Moreover, Coding-Potential Assessment Tool (CPAT)27 was used to calculate the coding potential of MIR31HG, and the results showed that the possible ORF of MIR31HG is very short, and the coding probability of MIR31HG is very low (P<0.05; Supplementary Figure S1A). Then the possible ORF (termination codon removed) of MIR31HG was cloned into the eukaryotic expression vector pcDNA3.1 with C-terminal Flag tag. ACTB with C-terminal Flag tag severs as positive control.28 Western blot analysis showed that the ACTB-Flag vector transfection group could detect the expression of Flag-tagged protein, while both of MIR31HG-Flag-1 and MIR31HG-Flag-2 vectors transfection groups could not detected this protein (Supplementary Figure S1B). Together, these observations validated that MIR31HG does not have the coding capacity (Supplementary Figure S1B).
Figure 1.
MIR31HG expression is highly upregulated in PDAC tissues and cell lines. (a) Data mining of MIR31HG expression in the PDAC patients gene profiling results (n=45; NCBI/GEO/GSE28735). Values are median with 95% CI. (b) RT–qPCR analysis of MIR31HG expression in 13 pairs of PDAC tissues (T) and corresponding non-tumor pancreatic tissues (N). Transcript levels were normalized to GAPDH expression. (c) RT–qPCR analysis of MIR31HG expression in hTERT-HPNE cells and eight PDAC cell lines. Transcript levels were normalized to GAPDH expression. (d) Schematic representation of the genomic organization of MIR31HG. miR-31, indicated with a horizontal bar, is located within intron 1 of MIR3HG. (e) RT–qPCR analysis of the effect of MIR31HG downregulation on the expression level of miR-31. Transcript levels of MIR31HG and miR-31 were normalized to GAPDH and U6 expression, respectively. **P<0.01.
The full sequence of miR-31 is located within the intron of MIR31HG (Figure 1d), and the transcriptional activity of both is under the control of a promoter-associated CpG island.16 Our study found a significant positive correlation between MIR31HG and miR-31 in PDAC specimens and cell lines (Supplementary Figure 2A and B). However, siRNA-mediated knockdown of MIR31HG did not alter the levels of miR-31 (Figure 1e), and inhibition or overexpression of miR-31 had no effect on MIR31HG expression (Supplementary Figure 2C). Despite the interrelated location and co-transcription of MIR31HG and miR-31, they did not regulate each other's expression, suggesting that the role of MIR31HG in biological processes is independent of miR-31.
Knockdown of MIR31HG inhibits PDAC cell growth and invasion
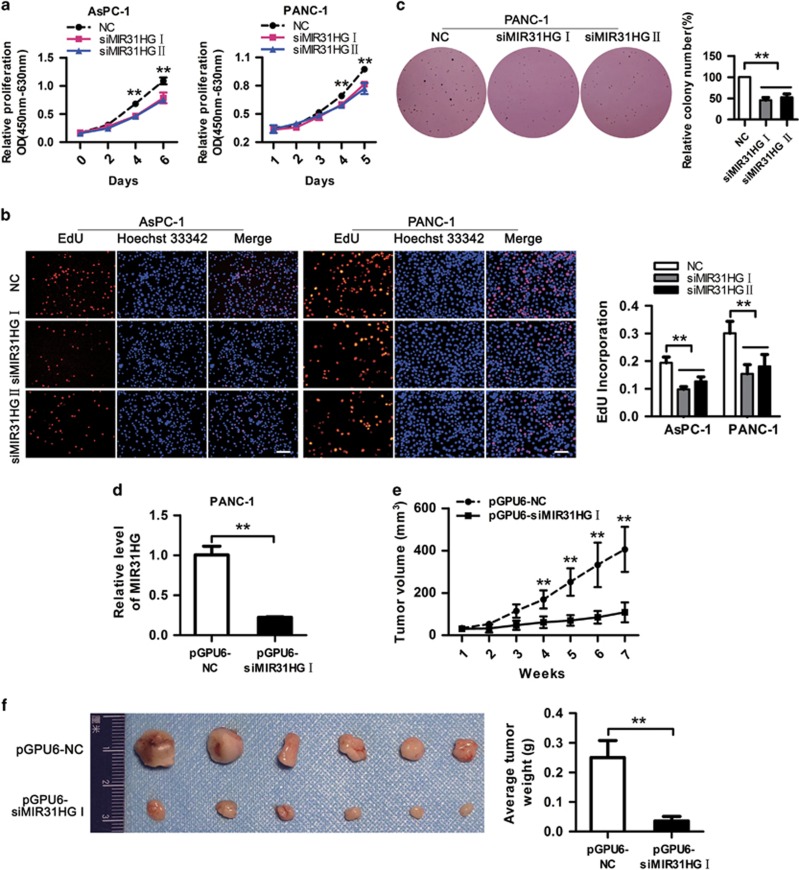
To evaluate the possible role of MIR31HG in PDAC, we transfected PDAC cell lines with two different siRNAs against MIR31HG (designated siMIR31HG I and II), both of which could efficiently knock down the endogenous MIR31HG level (Figure 1e and Supplementary Figure 3A). The growth curves detected by CCK8 showed that MIR31HG knockdown significantly decreased PDAC cell growth (Figure 2a and Supplementary Figure 3B). Consistent with the results of CCK8 assays, EdU incorporation was drastically reduced following MIR31HG downregulation (Figure 2b). Moreover, colony-formation assay revealed that MIR31HG downregulation significantly inhibited anchorage-independent growth of PANC-1 cells, as indicated by the formation of fewer and smaller colonies in soft agar (Figure 2c). These data provide evidence of the growth-promoting role of MIR31HG in vitro. To further verify the findings above, we constructed PANC-1 cell lines stably expressing siMIR31HG I or negative control using pGPU6/GFP/Neo vector (Figure 2d and Supplementary Figure 4). Then we injected nude mice subcutaneously with PANC-1 cells stably downexpressed or not for MIR31HG. It is obvious that tumors formed by MIR31HG-silencing cells grew much slower than those formed by control cells (Figure 2e) and the tumor weight from the MIR31HG knockdown group was reduced significantly when compared with the control group (Figure 2f). These in vivo data complement the above in vitro studies of MIR31HG and confirm MIR31HG is essential for regulating PDAC cell proliferation.
Figure 2.
Downregulation of MIR31HG inhibits PDAC cell growth in vitro and in vivo. (a) Growth curves of AsPC-1 and PANC-1 cells after transfection with siMIR31HGs or NC were determined by CCK8 assays. **P<0.01. (b) Cell proliferation was evaluated 48 h after transfection with siMIR31HGs or NC using EdU-incorporation assays. Proliferating cells label with EdU. Scale bars, 100 μm. **P<0.01. (c) Soft agar colony-formation assays were performed to determine the effect of MIR31HG downregulation on the anchorage-independent growth of PANC-1 cells. **P<0.01. (d) PANC-1 cells stably expressing siRNA-1 targeting MIR31HG or negative control were generated using pGPU6/GFP/Neo vector. **P<0.01. (e) Growth curves of xenograft tumors after the subcutaneous injection of PANC-1 cells stably knockdown of MIR31HG or negative control. The tumor volumes were measured every week after inoculation. **P<0.01, n=6. (f) Left: Representative images of tumors excised from mice 7 weeks after inoculation. Right: Tumor weights. **P<0.01.
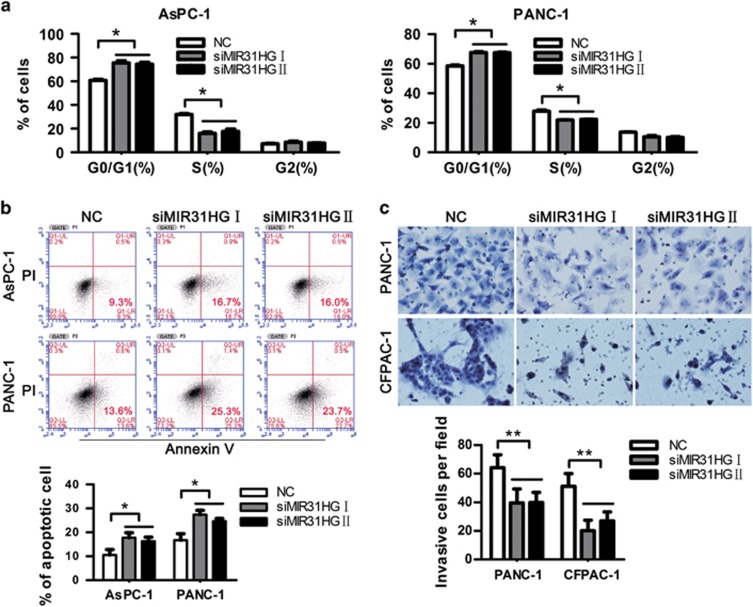
In order to explore the potential mechanisms underlying growth suppression after MIR31HG knockdown, we assessed its effect on cell cycle and apoptosis in AsPC-1 and PANC-1 cells. An analysis of cell-cycle distribution revealed that siMIR31HGs significantly decreased the percentage of S-phase cells and increased the percentage of G0/G1-phase cells, indicating that MIR31HG downregulation caused a G0/G1 to S-phase arrest in AsPC-1 and PANC-1 cells (Figure 3a and Supplementary Figure 5). Annexin V staining showed that the percentage of early apoptotic cells following siMIR31HGs treatment was drastically increased relative to that in control groups (Figure 3b), indicating that MIR31HG downregulation induced pancreatic cancer cell apoptosis. These data imply that G0/G1 to S-phase arrest and increased apoptosis may contribute to siMIR31HG-mediated growth inhibition.
Figure 3.
Downregulation of MIR31HG represses PDAC cell-cycle progression, induces apoptosis, and inhibits invasion in vitro. (a) The cell-cycle distribution after knockdown of MIR31HG was determined by PI staining and flow cytometer analysis in AsPC-1 and PANC-1 cells. *P<0.05. (b) The effect of MIR31HG knockdown on apoptosis in AsPC-1 and PANC-1 cells was determined by measuring the percentage of Annexin V-stained cells using flow cytometry. *P<0.05. (c) The effect of MIR31HG downregulation on the invasion of PANC-1 and CFPAC-1 cells was assessed using transwell assays. **P<0.01.
To further determine whether MIR31HG is associated with the progression of PDAC, we analyzed the effect of MIR31HG knockdown on the invasive behavior of PANC-1 and CFPAC-1 cells. Using a transwell system, we found that the invasive ability of PDAC cells was significantly decreased following siRNA-mediated downregulation of MIR31HG (Figure 3c). AsPC-1 cells were excluded from this analysis owing to their limited basal invasion properties. Taken together, these results suggest that MIR31HG has important roles in pancreatic cancer development and progression.
Overexpression of MIR31HG promotes PDAC cell proliferation and invasion
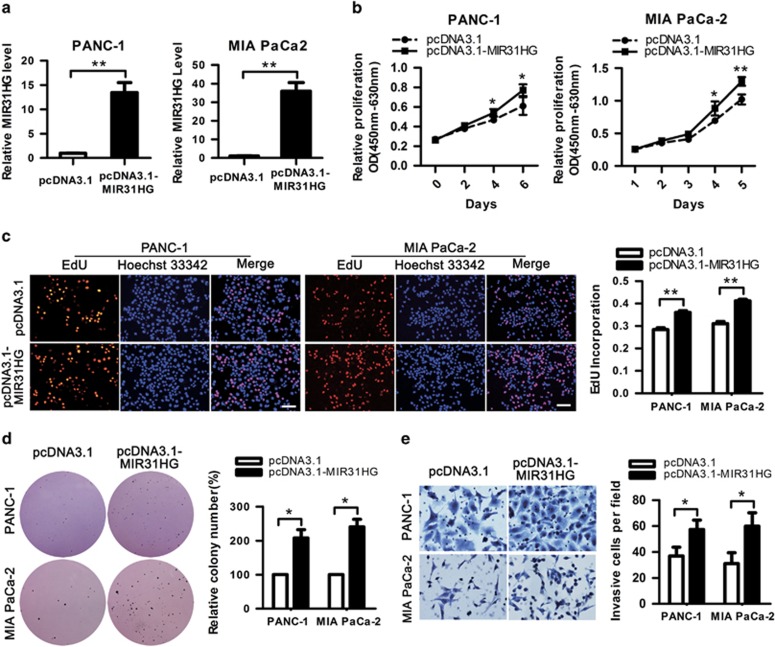
We further examined the role of MIR31HG by assessing the effect of its overexpression in PDAC cells using the pcDNA3.1-MIR31HG expression plasmid, focusing on cell lines with moderate (PANC-1) or very low (MIA PaCa-2) endogenous MIR31HG levels. MIR31HG expression in both PANC-1 and MIA PaCa-2 cells was drastically increased 48 h after transfection with pcDNA3.1-MIR31HG vector compared with cells transfected with empty pcDNA3.1 vector (Figure 4a). CCK8 assays showed that MIR31HG upregulation promoted cell proliferation in both cell types (Figure 4b). EdU-incorporation assay showed similar results that the percentage of EdU-incorporating cells was markedly increased following MIR31HG overexpression (Figure 4c). Moreover, in soft agar colony-formation assays, the number of colonies formed in pcDNA3.1-MIR31HG-transfected groups was significantly increased than that in control groups after culturing for 3 weeks (Figure 4d), indicating that overexpression of MIR31HG also enhanced the anchorage-independent growth of PDAC cells. In addition, enhanced expression of MIR31HG promoted cell invasion in PANC-1 and MIA PaCa-2 cells (Figure 4e). Taken together, these results indicate that overexpression of MIR31HG increases the tumorigenicity of PDAC cells.
Figure 4.
Upregulation of MIR31HG increases PDAC cell proliferation and invasion. (a) RT–qPCR analysis of MIR31HG expression in PANC-1 and MIA PaCa-2 cells transfected with pcDNA3.1-MIR31HG or empty pcDNA3.1 vector. Transcript levels were normalized to GAPDH expression. **P<0.01. (b–d) Effects of ectopic expression of MIR31HG on the proliferation of PDAC cells. (b) CCK8 assays were performed to determine the proliferation of pcDNA3.1-MIR31HG-transfected PDAC cells. *P<0.05, **P<0.01. (c) Representative profiles (left) and quantification of EdU-incorporating cells transfected with pcDNA3.1-MIR31HG or empty pcDNA3.1 vector (right) are shown. Scale bars, 100 μm. **P<0.01. (d) The effects of MIR31HG overexpression on the anchorage-independent growth of PDAC cells were analyzed by soft agar colony-formation assays. *P<0.05. (e) The invasive ability after upregulation of MIR31HG was assessed using transwell invasion assays. *P<0.05.
lncRNA MIR31HG is negatively regulated by miR-193b
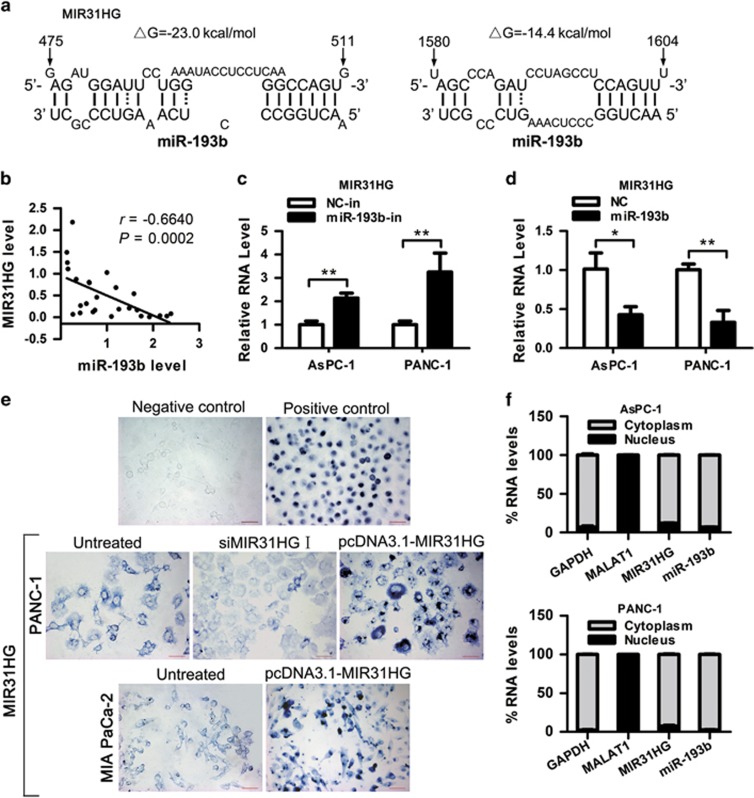
Using DIANA-LncBase29 and miRcode30 software, we selected several miRNAs predicted to be bound to MIR31HG with high scores. We transfected PANC-1 cell with mimics of these miRNAs and miR-193b had the most significant effect on MIR31HG expression (Supplementary Figure 6A). Thus, it was pursued for studies in detail. The predicted binding sites of miR-193b to the MIR31HG sequence are illustrated in Figure 5a. RT–qPCR showed that, whereas MIR31HG was upregulated in PDAC tissues (Figure 1b), miR-193b was downregulated in the same tumor specimens, resulting in a significant inverse correlation between MIR31HG and miR-193b (Figure 5b and Supplementary Figure 6B). Compared with hTERT-HPNE cells, PDAC cell lines displayed lower expression of miR-193b. In contrast, the MIR31HG level was in opposite trend in these cell lines except for SW 1990 (Figure 1c and Supplementary Figure 6C). To further assess the potential relationship between miR-193b and MIR31HG, we transfected AsPC-1 and PANC-1 cells with miR-193b inhibitor or mimic to decrease or increase miR-193b expression, respectively. Inhibition of miR-193b expression resulted in a significant upregulation of MIR31HG (>2-fold; Figure 5c and Supplementary Figure 6D), while overexpression of miR-193b significantly reduced MIR31HG expression (>50%) in both cell lines (Figure 5d and Supplementary Figure 6E). These results suggest that miR-193b negatively regulates lncRNA MIR31HG either directly or indirectly.
Figure 5.
Negative regulation of MIR31HG by miR-193b. (a) Predicted binding sites for miR-193b in MIR31HG sequences. Numbers show the nucleotides relative to the transcriptional start site of MIR31HG. (b) The expression of MIR31HG was negatively correlated with that of miR-193b (r=−0.6640, P=0.0002) in clinical specimens. (c, d) AsPC-1 and PANC-1 cells were transfected with miR-193b inhibitor (c) or mimic (d), and 48 h later total RNA was isolated and the expression of MIR31HG was analyzed by RT–qPCR. *P<0.05, **P<0.01. (e) RNA ISH assay of MIR31HG in PDAC cells. The blue signal is from the digoxigenin-labeled RNA probe. Negative control (Sense MIR31HG probe) shows no staining and positive control (MALAT1 probe) shows nuclear blue staining in PANC-1 cells. Scale bars, 50 μm. (f) RT–qPCR detection of the percentage of MIR31HG, miR-193b, GAPDH and MALAT1 in the cytoplasmic and nuclear fractions of AsPC-1 and PANC-1 cells. GAPDH and MALAT1 serve as a cytoplasmic and nuclear localization marker, respectively. Abbreviation: ISH, In situ hybridization.
In situ hybridization assay was performed on PDAC cells and the results showed that MIR31HG-specific staining was observed in the cytoplasm of PANC-1 cells of the untreated group, whereas nearly no staining was observed in the nucleus (Figure 5e), suggesting that MIR31HG was located in the cytoplasm. We transfected PANC-1 cells with siMIR31HGs or MIR31HG expression vectors, and observed a substantial decrease or increase of MIR31HG staining signals, and similar results were obtained in MIA PaCa-2 cells (Figure 5e). This result validated the specificity of MIR31HG probe and the efficiency of MIR31HG knockdown or overexpression. RT–qPCR of nuclear and cytoplasmic fractions of AsPC-1 and PANC-1 cells further validated that both MIR31HG and miR-193b were located in the cytoplasm (Figure 5f), providing prerequisite for reciprocal interaction between MIR31HG and miR-193b. As in SW 1990 cells, MIR31HG was almost cytoplasmic, whereas miR-193b located both in the cytoplasm and the nucleus (Supplementary Figure 7). The enrichment of miR-193b in the nucleus would diminish the interaction between MIR31HG and miR-193b, which may contribute to the non inverse correlation between MIR31HG and miR-193b in this cell line. However, the detailed mechanism by which miR-193b imported to the nucleus requires further investigation.
miR-193b suppresses MIR31HG function
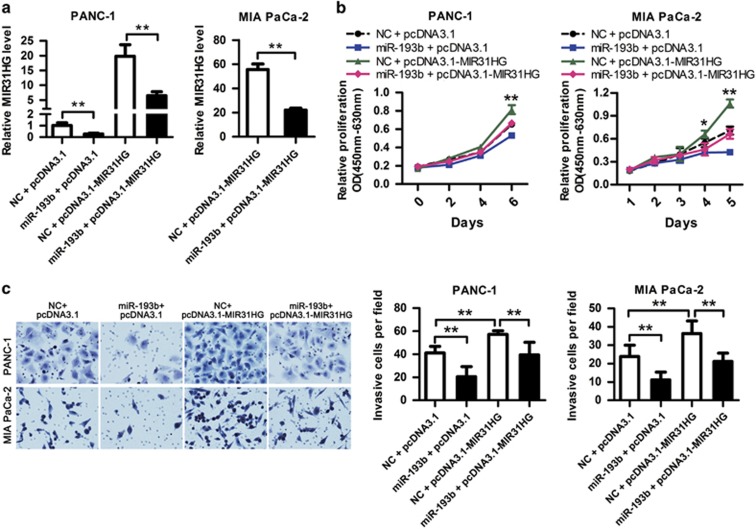
We transfected PANC-1 and MIA PaCa-2 cells with miR-193b mimic and MIR31HG expression vector to study the effects of miR-193b on cell proliferation and invasion mediated by MIR31HG. First, RT–qPCR showed that miR-193b could significantly decrease the ectopic wild-type MIR31HG expression in both cell lines (Figure 6a), but nearly have no effect on the expression of mutant one that was introduced with miR-193b-binding site mutation (Supplementary Figure 8). Then, CCK8 proliferation assays revealed that miR-193b suppressed cell proliferation and MIR31HG promoted cell proliferation, while co-transfection of miR-193b mimic and MIR31HG expression plasmid showed that miR-193b could abrogate the effect of MIR31HG in inducing cell proliferation (Figure 6b and Supplementary Figure 9). Moreover, transwell invasion assay demonstrated that miR-193b suppressed and MIR31HG promoted PDAC cell invasion, while co-transfection of miR-193b mimic and MIR31HG expression vector showed that miR-193b decreased cell invasion promoted by MIR31HG (Figure 6c). Those observations suggest that the effects of MIR31HG overexpression on the promotion of PDAC cell proliferation and invasion could be diminished by miR-193b, in accordance with the suppression of the ectopic MIR31HG expression by miR-193b (Figure 6a).
Figure 6.
miR-193b inhibits MIR31HG function. (a) PANC-1 and MIA PaCa-2 cells were co-transfected with miR-193b mimic and MIR31HG expression plasmid and the effect of miR-193b on ectopically expressed MIR31HG was analyzed by RT–qPCR. **P<0.01. (b) PDAC cells were co-transfected with negative control or miR-193b mimic and control plasmid (pcDNA3.1) or MIR31HG expression plasmid (pcDNA3.1-MIR31HG) and cell viability was determined using CCK8 assays. *P<0.05, **P<0.01. (c) Transwell invasion assays of PDAC cells after co-transfected with negative control or miR-193b mimic and control plasmid (pcDNA3.1) or MIR31HG expression plasmid (pcDNA3.1-MIR31HG). **P<0.01.
MIR31HG is directly targeted by miR-193b, further regulating miR-193b target genes
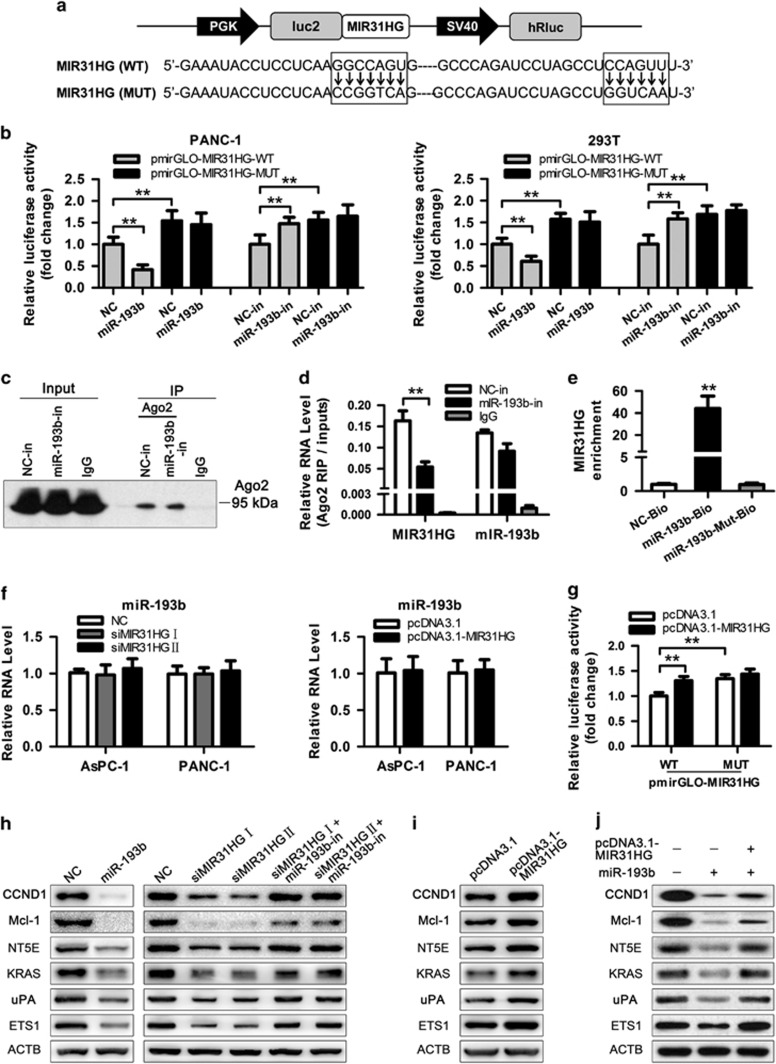
We next sought to examine whether miR-193b-mediated MIR31HG regulation occurs through direct targeting of miRNA-binding sites in the MIR31HG sequence. We subcloned full-length of MIR31HG into the pmirGLO dual luciferase reporter vector (Figure 7a) and performed luciferase assays in PANC-1 and 293 T cells, both of which express endogenous miR-193b (Supplementary Figures 6C and 10). As shown in Figure 7b, co-transfection of PANC-1 and 293 T cells with pmirGLO-MIR31HG-WT vector and miR-193b mimic significantly reduced luciferase reporter activity compared with the negative control (P<0.01). This repressive effect was abrogated by directed mutagenesis of the miR-193b-binding seed region in MIR31HG. Conversely, inhibition of miR-193b induced a remarkable increase in luciferase activity in both cell types, which was also abolished by binding sites mutation. In addition, cells co-transfected with pmirGLO-MIR31HG-MUT vector and negative control showed a higher level of luciferase activity with respect to the group of co-transfected with WT vector and negative control, which may due to the disrupt of miR-193b binding in the MUT construct (Figure 7b). Previous studies have demonstrated that miRNAs are present in the form of miRNA ribonucleoprotein complexes (miRNPs) that contains Ago2, the key component of the RNA-induced silencing complex.31, 32 To test whether MIR31HG associates with miR-193b, RNA-binding protein immunoprecipitation (RIP) experiments were performed on PANC-1 cells treated with miR-193b inhibitor or negative control (Supplementary Figure 11). Ago2 protein was efficiently immunoprecipitated from cell extracts by Ago2 antibody (Figure 7c). RIP assays revealed that while MIR31HG was detected in Ago2 immunoprecipitates from the control group, its levels were drastically reduced in Ago2 complexes purified from cells treated with miR-193b inhibitor (Figure 7d), indicating that MIR31HG is likely in the miR-193b–RISC complex. Further, we applied a biotin-avidin pulldown system to assay whether miR-193b could pull down MIR31HG. MIR31HG was pulled down as analyzed by RT-qPCR, but the introduction of mutations that disrupt base pairing between MIR31HG and miR-193b led to the inability of miR-193b to pull down MIR31HG (Figure 7e and Supplementary Figure 12), suggesting that miR-193b interacts with MIR31HG in a sequence-specific manner. Collectively, these results revealed that miR-193b exerts inhibitory effects on MIR31HG expression by directly targeting MIR31HG.
Figure 7.
MIR31HG is a direct target of miR-193b. (a) Schematic of wild-type and mutant pmirGLO-MIR31HG constructs. (b) Luciferase assays of the indicated cells transfected with pmirGLO-MIR31HG-WT or pmirGLO-MIR31HG-MUT reporter and NC or miR-193b mimic. **P<0.01. (c) Ago2 protein immunoprecipitated from cell extracts by Ago2 antibody or IgG was detected by western blot analysis. (d) The amount of MIR31HG and miR-193b bound to Ago2 or IgG was measured by RT–qPCR in the presence of miR-193b inhibitor or negative control. **P<0.01. (e) PANC-1 cells were transfected with biotinylated WT miR-193b (miR-193b-Bio) or biotinylated mutant miR-193b (miR-193b-Mut-Bio) or biotinylated NC (NC-Bio). Forty-eight hours after transfection, cells were collected for biotin-based pulldown assay. MIR31HG expression levels were analyzed by RT–qPCR. **P<0.01. (f) The expression level of miR-193b was detected by RT–qPCR following MIR31HG knockdown or overexpression. (g) Luciferase assays of 293 T cells transfected with pmirGLO-MIR31HG-WT or pmirGLO-MIR31HG-MUT reporter and pcDNA3.1-MIR31HG vector. **P<0.01. (h) Effect of miR-193b and MIR31HG-siRNAs on the expression of miR-193b targets: CCND1, Mcl-1, NT5E, KRAS, uPA and ETS1. PANC-1 cells were transfected with miR-193b mimic or MIR31HG-siRNAs or co-transfected with miR-193b inhibitor and MIR31HG-siRNAs. Cellular protein was isolated from the transfected cells 48 h later and used for western blot analysis. (i) Effect of transfected PANC-1 cells with MIR31HG expression plasmid on the expression of the miR-193b targets: CCND1, Mcl-1, NT5E, KRAS, uPA, and ETS1. (j) Effect of co-transfected PANC-1 cells with MIR31HG expression plasmid and miR-193b mimic on the expression of the miR-193b targets: CCND1, Mcl-1, NT5E, KRAS, uPA, and ETS1.
To further determine the effect of MIR31HG on miR-193b, we transfected siMIR31HGs or MIR31HG expression vector into PDAC cells. However, we did not observe obvious changes in miR-193b levels following MIR31HG knockdown or overexpression (Figure 7f). Luciferase assay of 293 T cells co-transfected with pmirGLO-MIR31HG vector and pcDNA3.1-MIR31HG vector showed that MIR31HG overexpression induced a significant increase in luciferase activity, which was abolished by miR-193b-binding sites mutation, suggesting that ectopically expressed MIR31HG specifically sequestered endogenous miR-193b, thereby preventing it from inhibiting luc2 expression (Figure 7g). miR-193b has been reported to target and repress CCND1, Mcl-1, NT5E, KRAS, uPA, and ETS133–36 expression. Western blot assays revealed that the expression level of these genes was remarkably lower in hTERT-HPNE cells than in PANC-1 cells (Supplementary Figure 13) and forced expression of miR-193b triggered a significant silencing effect on these genes expression in PANC-1 cells, confirming that they are targets of miR-193b (Figure 7h). We transfected PANC-1 cells with MIR31HG-siRNAs or co-transfected with miR-193b inhibitor and MIR31HG-siRNAs, and found that knockdown of MIR31HG significantly suppressed miR-193b targets expression, whereas inhibition of miR-193b partly abolish the silencing effect of MIR31HG knockdown on these genes (Figure 7h). Then we transfected PANC-1 cells with MIR31HG expression plasmid and observed that the expression of miR-193b targets was highly elevated following MIR31HG upregulation (Figure 7i). Furthermore, we co-transfected PANC-1 cells with MIR31HG expression plasmid and miR-193b mimic to investigate the effect of MIR31HG overexpression on miR-193b targets suppressed by miR-193b. In the presence of MIR31HG, the repression of these genes was partly restored compared with the miR-193b group (Figure 7j). In conclusion, these results suggest that MIR31HG could regulate the target genes of miR-193b through sequestered endogenous miR-193b. We provide evidence that MIR31HG may act as an endogenous ‘sponge' by binding miR-193b, thus abolishing the miRNA-induced repression on these genes.
Discussion
In the current study, we identified that the lncRNA, MIR31HG, is upregulated in PDAC and may serve as an oncogene in tumorigenesis. In agreement with microarray gene-expression profile (GSE28735) results, RT–qPCR showed that MIR31HG levels were significantly higher in PDAC tissues than in non-tumor pancreatic tissues. Similarly, compared with the normal hTERT-HPNE cells, MIR31HG expression was strikingly higher in six kinds of PDAC cell lines except BxPC-3 and MIA Paca-2 cell lines. CpG island methylation plays an important role in silencing both MIR31HG and miR-31 genes in breast cancer cells.16 Accordingly, we suspect that the low expression of MIR31HG in these two PDAC cell lines and the normal pancreatic cell line may be attributable, at least in part, to promoter methylation.
Although miR-31 is located within the intron of MIR31HG, and the transcriptional activity of miR-31 is under the control of MIR31HG,16 they did not regulate each other's expression. Using online software, we identified MIR31HG as a possible target of miR-193b. miR-193b, a putative tumor suppressor, is downregulated in a variety of tumors, including epidermal squamous cell carcinoma, melanoma, pancreatic cancer, hepatocellular carcinoma, and others.33, 34, 35, 36 The recent study found that overexpression of miR-193b significantly inhibits the proliferation of pancreatic cancer cells,33 and identified CCND1, NT5E, PLAU, STARD7, STMN1, and YWHAZ as target genes of miR-193b.33, 34, 35, 36 Restoration of miR-193b expression results in decreased cell growth, clonogenic potential, and migration through direct inhibition of KRAS and MAX in epidermal squamous cell carcinoma.34 miR-193b has also been found to inhibit melanoma cell growth by targeting Mcl-1, and function as a putative suppressor in melanoma pathogenesis and progression.35 Although miR-193b has been experimentally shown to target a large number of protein-coding genes, our study found that miR-193b also targets lncRNA MIR31HG. First, we found a negative correlation between MIR31HG and miR-193b in pancreatic clinical specimens. Moreover, overexpression of miR-193b reduced MIR31HG expression, whereas inhibition of miR-193b expression resulted in a significant upregulation of MIR31HG, suggesting that MIR31HG is negatively regulated by miR-193b. In addition, we provide evidence that miR-193b targets MIR31HG by directly binding to two miRNA-binding sites in MIR31HG sequence. Thus, our study establishes that increases in MIR31HG levels are due, in part, to a reduction in the tumor suppressor miR-193b, providing a possible mechanism for upregulation of MIR31HG in PDAC.
The mechanisms responsible for lncRNAs deregulation in cancer have not been thoroughly elucidated. lncRNAs have many features in common with mRNAs; they are polyadenylated, have complex splicing patterns, and are often transcribed by RNA polymerase II. Recently, accumulating evidence indicates that lncRNAs may be regulated in a similar manner to those of protein-coding genes. For instance, epigenetic regulation modulates the expression of lncRNA-LET11 and MEG337 through promoter-associated histone acetylation and DNA methylation, respectively, and the transcription factor c-Myc activates lncRNA CCAT1 expression by directly binding to an E-box element in the promoter region.38 Our study showed that miRNAs also negatively regulate lncRNA through binding to target sequence, a mechanism similar to that of miRNA-mediated silencing of target mRNAs. The data presented here are consistent with recent reports supporting direct miRNA regulation of lncRNAs.39, 40, 41 For example, miR-671 directs cleavage of a circular, noncoding antisense transcript in an Ago2-mediated manner,39 and miR-9 targets the lncRNA, MALAT1, and induces degradation of MALAT1 in the nucleus.40 Direct modulation of the expression of lncRNAs by miRNAs greatly expands the potential gene-regulatory network in the process of tumorigenesis.
Applying loss-of-function and gain-of-function approaches, we demonstrated that the lncRNA, MIR31HG, plays a key role in PDAC cell proliferation and invasion. Downregulation of MIR31HG significantly decreased PDAC cell growth, induced apoptosis and G1/S arrest, and inhibited invasion, whereas overexpression of this lncRNA had the opposite effects. These findings indicate that MIR31HG may function as a tumor oncogene, and its overexpression could contribute to pancreatic cancer development. However, the potential downstream pathways of MIR31HG remain unknown. Recently, a number of lncRNAs have been suggested to act as ‘sponges' to bind specific miRNAs and regulate their function.42, 43, 44 Here we provide evidence that MIR31HG may also function as endogenous decoy for miR-193b to affect its distribution on specific targets without inducing miR-193b destabilization, as the level of miR-193b was not affected following MIR31HG knockdown or overexpression. We found that similar to miR-193b mimic, downregulation of MIR31HG is able to suppress miR-193b targets, while ectopically expressed MIR31HG inhibits miR-193b function, leading to derepression of these genes. Therefore, effect of MIR31HG on PDAC cells proliferation and invasion could be explained, although in part, by its function as a molecular sponge of miR-193b, which provides a possible mechanism for MIR31HG functioning as a tumor oncogene.
In summary, we show that the lncRNA MIR31HG is upregulated in PDAC. Its effects on cell proliferation and invasion suggest that it exhibits oncogenic property in PDAC tumorigenesis. miR-193b directly targeted MIR31HG and suppressed MIR31HG's expression and function, while MIR31HG acts as a molecular sponge for miR-193b and regulates its targets. This reciprocal repression of miR-193b and MIR31HG may highlight the significance of RNA–RNA interaction and provide new insight into mechanisms underlying various aspects of tumorigenesis including tumor growth, invasion, and metastasis.
Materials and methods
Cell culture and human tissue samples
The PDAC cell lines AsPC-1, PANC-1, CFPAC-1, Hs 766 T, SW 1990, MIA PaCa-2, and BxPC-3; human pancreatic nestin-expressing cell line (hTERT-HPNE); and the embryonic kidney cell line 293 T were obtained from American Type Culture Collection (Rockville, MD, USA). These cell lines were tested 1 month before the study by methods of morphology check, growth curve assay, and mycoplasma detection according to the ATCC cell line verification test recommendations. All cells were maintained at 37 °C in a humidified 5% CO2 atmosphere. Thirteen pairs of primary PDAC and adjacent non-tumor tissues were obtained from patients undergoing surgery at Peking Union Medical College Hospital. All patients in this study met the following inclusion criteria: Resected samples were identified as PDAC by pathological examination; no radiotherapy or chemotherapy were given before surgery. Clinical characteristics of all patients enrolled in the study is summarized in Supplementary Table 1. All samples were collected immediately from resected PDAC cases, snap frozen in liquid nitrogen, and stored at −80 °C until RNA extraction. The study was approved by the Ethics Committee of Peking Union Medical College Hospital and all participants provided written informed consent.
RNA isolation, cDNA synthesis, RT–qPCR, siRNA and miRNA transfection, plasmid construction and transfection, and western blot analysis
RNA isolation, cDNA synthesis, RT–qPCR, siRNA and miRNA transfection, plasmid construction and transfection, and western blot analysis are described in the Supplementary Materials and Methods.
In situ hybridization
In situ hybridization was used to detect MIR31HG in PDAC clinical specimens and cell lines. A digoxigenin-UTP labeled antisense RNA probe was derived from 816 to 1614 nt of MIR31HG by in vitro transcription using DIG RNA Labeling Kit (Roche, Indianapolis, IN, USA). The digoxigenin-UTP labeled sense RNA probe derived from 816 to 1614 nt of MIR31HG was employed as negative control.
Cell cytoplasm/nucleus fraction isolation
NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, MA, USA) was employed to prepare cytoplasmic and nuclear extracts from PDAC cells. RNAs extracted from each of the fractions were subjected to following RT–qPCR analysis to demonstrate the levels of nuclear control transcript (MALAT1), cytoplasmic control transcript (GAPDH), lncRNA MIR31HG, and miR-193b.
CCK8 assay, EDU assay, colony-formation assay, flow cytometry analysis of cell cycle and apoptosis, and invasion assay
Detailed descriptions of CCK8 assay, EDU assay, colony-formation assay, flow cytometry analysis of cell cycle and apoptosis, and invasion assay can be found in the Supplementary Materials and Methods section.
Pancreatic tumor xenograft model
Female nude (BALB/c-nude) mice (4 weeks old) were purchased to examine tumorigenicity. Twelve mice were divided into two groups (6 for stably knockdown of MIR31HG and 6 for negative control) according to the completely randomized method. PANC-1 cells stably expressing siMIR31HG I or negative control were propagated and 1 × 107 cells were inoculated subcutaneously into the dorsal flanks of mice. Tumor growth was examined at the indicated time points and tumor volumes were measured by using the equation V (mm3)=A × B2/2, where A is the largest diameter and B is the perpendicular diameter. After 7 weeks, the mice were killed and tumors were removed and weighed. All animal procedures were performed in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the Peking Union Medical College Hospital.
Dual luciferase reporter assay
Cells (PANC-1 and 293 T) were seeded at 3 × 104 cells/well in 24-well plates and allowed to settle overnight. The next day, cells were co-transfected with pmirGLO-MIR31HG-WT or -MUT reporter plasmids and miR-193b mimic or inhibitor. Twenty-four hours after transfection, the relative luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and normalized against Renilla luciferase activity.
RNA-binding protein immunoprecipitation
RIP experiments were performed using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) and the Ago2 antibody (Abcam, Cambridge, MA, USA) following the manufacturer's protocol. Co-precipitated RNAs were subjected to RT–qPCR analysis.
Pulldown assay with biotinylated miRNA
PANC-1 cells were transfected with biotinylated miRNA, collected 48 h after transfection. The cell lysates were incubated with M-280 streptaviden magnetic beads (Invitrogen, San Diego, CA, USA) as described previously.45 The bound RNAs were purified using TRIzol reagent (Invitrogen) for further RT–qPCR analysis.
Statistical analysis
Statistical analyses were performed using SPSS 17.0 software (SPSS, Chicago, IL, USA). Differences between two groups were assessed using Student's t-test (two-tailed). Correlations between MIR31HG and miR-193b or miR-31 were analyzed by Spearman rank correlation. Each experiment was performed at least three times. Results of experiments are displayed as mean±standard deviation. A P-value<0.05 was considered to indicate statistical significance.
Acknowledgments
We sincerely thank Dr Yamei Niu for technical support and critical comments on the manuscript, Weilong Zhang for detailed bioinformatics analysis of MIR31HG, and Yongsong Yue for technical support of ISH. This work was supported by the National Nature Science Foundations of China (81472326 and 81172334 to JC, 31021091 to W-MT) and the China 973 (2011CBA01104 to W-MT).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63: 11–30. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J et al. Initial sequencing and analysis of the human genome. Nature 2001; 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N et al. The transcriptional landscape of the mammalian genome. Science (New York, NY) 2005; 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muers M. RNA: genome-wide views of long non-coding RNAs. Nat Rev Genet 2011; 12: 742. [DOI] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, Tung CS, Sanbonmatsu KY. Rise of the RNA machines: exploring the structure of long non-coding RNAs. J Mol Biol 2013; 425: 3731–3746. [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010; 39: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011; 472: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yan GY. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics (Oxford, England) 2013; 29: 2617–2624. [DOI] [PubMed] [Google Scholar]
- Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans 2012; 40: 902–906. [DOI] [PubMed] [Google Scholar]
- Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell 2013; 49: 1083–1096. [DOI] [PubMed] [Google Scholar]
- Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer 2013; 108: 2419–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut 2014; 63: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V et al. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer 2011; 10: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013; 32: 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer 2012; 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi S, Yang M, Tao Y, Xu H, Shan J, Inchauste S et al. Cigarette smoke induces C/EBP-beta-mediated activation of miR-31 in normal human respiratory epithelia and lung cancer cells. PloS One 2010; 5: e13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lu J, Zhou J, Tan X, He Y, Ding J et al. Long non-coding RNA Loc554202 regulates proliferation and migration in breast cancer cells. Biochem Biophys Res Commun 2014; 446: 448–453. [DOI] [PubMed] [Google Scholar]
- Montes M, Nielsen MM, Maglieri G, Jacobsen A, Hojfeldt J, Agrawal-Singh S et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun 2015; 6: 6967. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Sacco L, Masotti A. Recent insights and novel bioinformatics tools to understand the role of microRNAs binding to 5' untranslated region. Int J Mol Sci 2012; 14: 480–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. BioEssays 2014; 36: 617–626. [DOI] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011; 147: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011; 147: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, Ghadimi BM et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PloS One 2012; 7: e31507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W et al. Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 2013; 41: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science (New York, NY) 2014; 344: 310–313. [DOI] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM et al. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res 2013; 41: D239–D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics (Oxford, England) 2012; 28: 2062–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E. Elucidating the temporal order of silencing. EMBO Rep 2012; 13: 662–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9: 102–114. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Tanji E, Makino N, Kawata S, Furukawa T. MicroRNAs associated with mitogen-activated protein kinase in human pancreatic cancer. Mol Cancer Res 2012; 10: 259–269. [DOI] [PubMed] [Google Scholar]
- Gastaldi C, Bertero T, Xu N, Bourget-Ponzio I, Lebrigand K, Fourre S et al. miR-193b/365a cluster controls progression of epidermal squamous cell carcinoma. Carcinogenesis 2014; 35: 1110–1120. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang X, Lentz C, Abi-Daoud M, Pare GC, Yang X et al. miR-193b regulates Mcl-1 in melanoma. Am J Pathol 2011; 179: 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J et al. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer 2010; 46: 2828–2836. [DOI] [PubMed] [Google Scholar]
- Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011; 30: 4750–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y et al. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol 2013; 139: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011; 30: 4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci E, Patella F, Waage J, Holmstrom K, Lindow M, Porse B et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep 2013; 3: 2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ST, Huang Y, Zhang L, Heng MY, Ptacek LJ, Fu YH. MicroRNA-23a promotes myelination in the central nervous system. Proc Natl Acad Sci USA 2013; 110: 17468–17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen Y et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene 2015; 34: 1768–1779. [DOI] [PubMed] [Google Scholar]
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 2013; 52: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014; 13: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 2014; 5: 3596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.