Abstract
NRAGE, a neurotrophin receptor-interacting melanoma antigen-encoding gene homolog, is significantly increased in the nucleus of radioresistant esophageal tumor cell lines and is highly upregulated to promote cell proliferation in esophageal carcinomas (ECs). However, whether the overexpressed NRAGE promotes cell growth by participating in DNA-damage response (DDR) is still unclear. Here we show that NRAGE is required for efficient double-strand breaks (DSBs) repair via homologous recombination repair (HRR) and downregulation of NRAGE greatly sensitizes EC cells to DNA-damaging agents both in vitro and in vivo. Moreover, NRAGE not only regulates the stability of DDR factors, RNF8 and BARD1, in a ubiquitin-proteolytic pathway, but also chaperons the interaction between BARD1 and RNF8 via their RING domains to form a novel ternary complex. Additionally, the expression of NRAGE is closely correlated with RNF8 and BARD1 in esophageal tumor tissues. In summary, our findings reveal a novel function of NRAGE that will help to guide personalized esophageal cancer treatments by targeting NRAGE to increase cell sensitivity to DNA-damaging therapeutics in the long run.
In mammalian cells, DNA lesions are always endogenously and exogenously induced,1 among which double-strand breaks (DSBs) are the most lethal lesions.2 Therefore, efficient DNA-damage response (DDR) is essential to maintain genomic integrity.1, 3 However, DDR is a double-edged sword. In normal organisms, DDR protects cells from tumorigenesis, whereas in tumor cells, DDR enables cells to grow and resist DNA-damaging therapeutic agents.4 Thus, in order to correctly use DDR factors in cancer diagnosis and targeted therapy, it is important to study molecular pathways underlying DDR in cancer cells.5
NRAGE is a member of the melanoma-associated antigen (MAGE) family.6 Previous studies reveal that it is possibly a DDR factor: (1) upregulation of NRAGE in the nucleus is highly associated with the development of esophageal carcinoma (EC) by interacting with the proliferating cell nuclear antigen (PCNA);7 (2) the level of NRAGE is significantly increased in the nucleus of radioresistant EC cells;8 (3) NRAGE knockout (KO) mice exhibit depression-like behavior9 and disorder in circadian clock,10 both of which are analogous to DDR defective scenarios.11, 12 However, to our knowledge, there has been no direct report concerning the role of NRAGE in DDR.
In the present study, we aim to study the role and the underlying mechanisms of NRAGE in DDR. Our data support that NRAGE is a positive regulator in homologous recombination (HR) and regulates the chemoresistance of EC cells both in vivo and in vitro. From mechanism, it regulates the stability of RNF8 and BARD1 via the ubiquitin-proteasome pathway and interacts with the RING domains of the two proteins as a chaperon to form a novel ternary complex to participate in DDR. Furthermore, clinical characterization of NRAGE, RNF8, and BARD1 in squamous EC tissues shows that the expression of NRAGE protein is closely related with both RNF8 and BARD1. Therefore, we conclude that the nuclear localized NRAGE interacts with RNF8 and BARD1 to mediate the resistance of EC cells against DNA-damaging agents. And we think that NRAGE is likely to be a promising target for personalized DNA-damaging therapies in EC.
Results
NRAGE is specifically involved in HR repair (HRR)
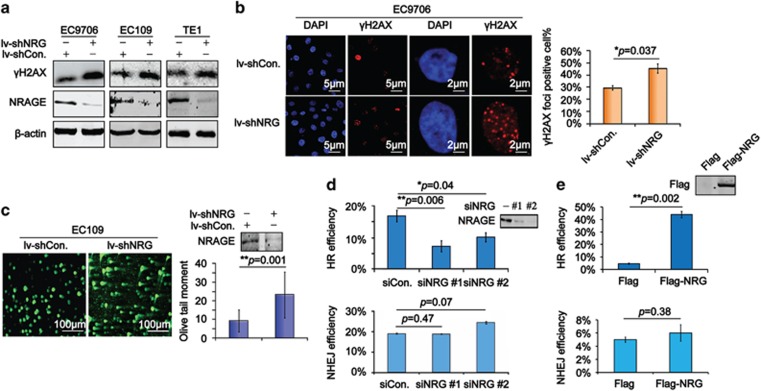
To investigate the direct role of NRAGE in DDR, we first detected the DSB biomarker, γH2AX,13 using immunoblotting assays in three different EC cells, which were stably knockdown of endogenous NRAGE or nonspecific sequence using lentivirus-mediated short hairpin RNA (lv-shNRG or lv-shCon., respectively). We found that NRAGE knockdown resulted in an obvious upregulation of γH2AX in all three EC cells (Figure 1a). Consistently, results from immunofluorescence (IF) showed that the percentage of cells with γH2AX foci was also greater in EC9706/lv-shNRG cells (Figure 1b). It is reported that the neutral comet assay is a sensitive method for visually detecting DSBs in cells.14 Therefore, we next conducted the assays to test the production of DSBs in EC109/lv-shCon. and EC109/lv-shNRG cells. As shown in Figure 1c, we found that DNA tails, illustrated by the olive tail moment (OTM), in EC109/lv-shNRG cells were apparently longer than those in EC109/lv-shCon. cells. Above all, we provide direct evidence that NRAGE guards the genomic stability by inhibiting the generation of endogenous DSBs in cancer cells.
Figure 1.
NRAGE is required for efficient homologous recombination repair (HRR) in esophageal cancer cells. (a) Total protein from EC9706, EC109, and TE1, stably knockdown of NRAGE using lentivirus mediated shRNA plasmids, were subjected to western blotting assays with the indicated antibodies. (b) The γH2AX foci formed in EC9706/lv-shCon. and EC9706/lv-shNRG cells were detected using IF, the scale bar is 5 and 2 μm, respectively. Cells with more than three γH2AX foci in the nucleus were defined as positive cells. The γH2AX foci-positive cells out of 300 cells (γH2AX foci positive%) were represented as means±S.D. and statistically analyzed (*P=0.037). (c) EC109/lv-shCon. and EC109/lv-shNRG cells were subjected to neutral comet assays to evaluate DSBs, the scale bar is 100 μm. OTM of 300 cells were statistically analyzed (**P=0.001) and graphically depicted. Error bar represents S.D. NRAGE interference efficiency was confirmed by western blottings. (d) 293T cells transfected with siCon. or siNRG, together with DR-GFP (−), GFP (+), NHEJ, or DR-GFP+Isce-I (HR), respectively, were subjected to flow cytometry to test GFP percentage (GFP%) in about 1 × 104 cells. The HR and NHEJ efficiency of siCon. and siNRG (#1, #2) cells were normalized to corresponding GFP (+). Data from three independent experiments were represented as means±S.D. and statistically analyzed (**P=0.006 for siNRG #1 and *P=0.04 for siNRG #2 in HR efficiency analysis). The efficiency of NRAGE knockdown was confirmed by western blottings. (e) 293T cells transfected with Flag or Flag-NRG, together with DR-GFP (−), GFP (+), or DR-GFP +Isce-I (HR) or NHEJ, respectively, were subjected to flow cytometry to test GFP%. The HR or NHEJ efficiency of Flag or Flag-NRG-transfected cells were normalized to the corresponding GFP (+), respectively. Data from three independent experiments are represented as means±S.D. and statistically analyzed (**P=0.002 for HR efficiency analysis). Overexpression of NRAGE was confirmed by western blottings
The lethal DSBs in cells are mainly repaired by HR and/or non-homologous end joining (NHEJ).15 Therefore, using the canonical HR and NHEJ reporter system,16 we evaluated the roles of NRAGE in HR and NHEJ. Interestingly, the HR efficiency was significantly reduced in siNRG-transfected cells (Figure 1d), whereas it was increased nearly 10-fold in NRAGE-overexpressed cells (Figure 1e). However, neither downregulation nor overexpression of NRAGE had any significant effects on NHEJ efficiency. Taken together, we conclude that NRAGE specifically participate in DSB repair via HR.
According to previous reports, NRAGE knockdown in EC cells results in G2/M phase arrest7 and HRR mainly functions in late S and G2 phases of the cell cycle.17 To further confirm whether the abnormal cell cycle distribution and the increase of γH2AX foci were an concomitant event in the absence of NRAGE, EC109 cells transfected with siCon. or siNRG (#1, #2) were coimmunostained with γH2AX and Cyclin A, a marker for S/G2 cells, using IF. As expected, the ratios of S/G2 cells with positive γH2AX foci were remarkably greater in siNRG-transfected cells (Supplementary Figures S1A and B). In line with the reduction of HR efficiency in NRAGE-deficient EC cells, the foci formation of BRCA1, the pivotal regulator in HR, was also obviously reduced (Supplementary Figure S1C). Thus our data support that NRAGE is a specific HR factor.
Knockdown of NRAGE sensitizes EC cells to DNA-damaging therapies both in vitro and in vivo
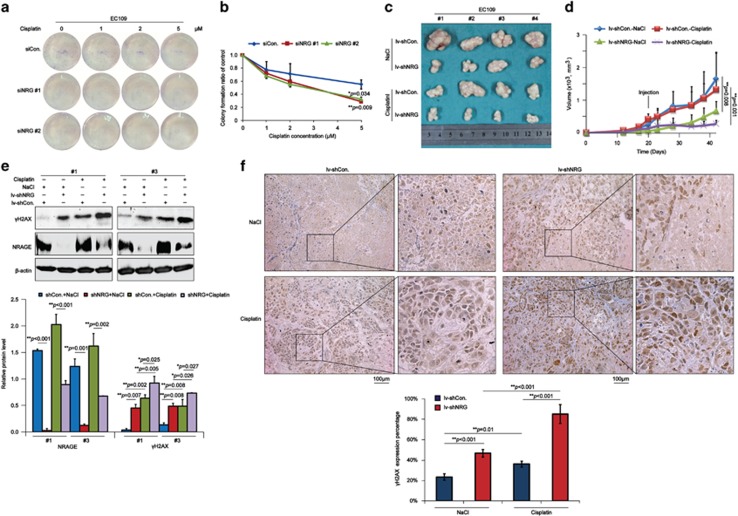
It is commonly known that cisplatin, a type of DNA-damaging agent, is basically a chemotherapeutic drug for EC; therefore, we further investigated the sensitivity of NRAGE-deficient EC cells to cisplatin. As indicated in Figures 2a and b, NRAGE knockdown significantly sensitized EC109 cells to cisplatin. Moreover, tumor volumes of EC109/lv-shNRG, especially when treated with cisplatin, reduced more obviously (Figures 2c and d, red and purple lines). To investigate whether the reduced tumor volumes were attributed to the increasing DSBs, we detected the γH2AX in the tumor tissues dissected from nude mice using immunoblotting and immunohistochemistry (IHC) assays. We found that in the absence of NRAGE, the DSBs were significantly increased, especially when treated with cisplatin (Figures 2e and f). Taken together, all the above data implicate that the novel HRR factor NRAGE regulates the chemoresistance of EC cells to cisplatin via DDR both in vitro and in vivo.
Figure 2.
NRAGE knockdown sensitizes EC cells to cisplatin both in vitro and in vivo. (a) EC109 cells transfected with siCon. or siNRG (#1, #2) were treated with the indicated concentrations of cisplatin for 2 h and left to form the colonies for 14 days. (b) The number of colonies with >50 cells in panel (a) was manually counted, followed by normalization to EC109 cells transfected with siCon. or siNRG (#1, #2) without cisplatin treatment, respectively. Data were represented as means±S.D. (*P=0.034 for siNRG #1, **P=0.009 for siNRG #2). (c) EC109/lv-shCon. and EC109/lv-shNRG cells were subcutaneously transplanted into male nude mice (N=4), and the mice were subsequently intraperitoneally injected with 0.9% NaCl or 10 mg/kg cisplatin when the bearing tumors were about 200 mm3. Finally, tumors were dissected and pictured. (d) Tumor volumes in panel (c) were measured and statistically analyzed (**P=0.008 for the NaCl group, **P=0.001 for the cisplatin group). Error bar represents S.D. (e) Tumor tissues from #1 and #3 in panel (c) were lysed and subjected to western blotting assays with the indicated antibodies. The gray values of western bands from three independent experiments were used for the ANOVA statistical analysis. (f) Tumors in panel (c) were detected of γH2AX using IHC assays (scale bar is 100 μm). The expression percentage of γH2AX was blindly evaluated by a professional doctor and finally used for the ANOVA statistical analysis
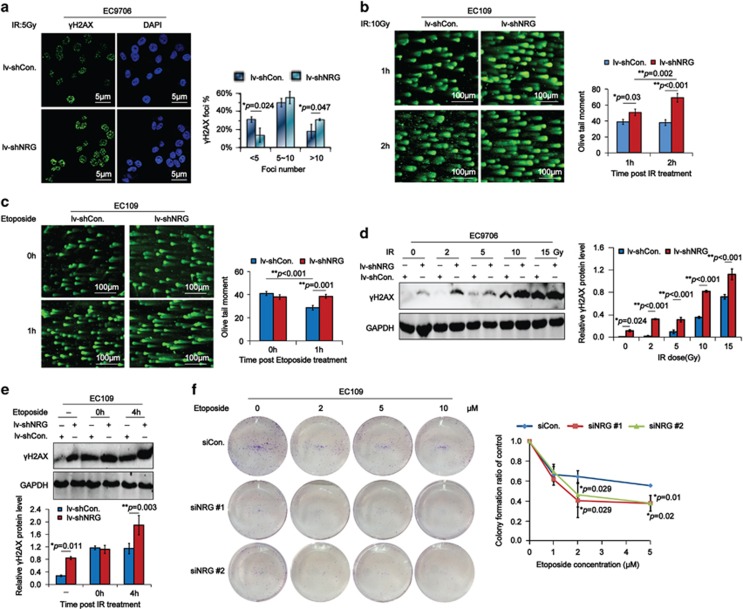
Furthermore, results from IF assays showed that the percentage of γH2AX foci in EC9706/lv-shNRG cells was more than that in EC9706/lv-shCon. cells in response to ionizing radiation (IR; Figure 3a). And results from neutral comet assays demonstrated that DSBs generated in IR irradiated EC109/lv-shNRG cells were much more serious (Figure 3b). Consistently, DSBs in etoposide-damaged EC109/lv-shNRG cells were more seriously generated at 1-h point after the treatment (Figure 3c). And the DSBs were more easily produced in NRAGE-deficient EC cells in response to different doses of IR (Figure 3d). Besides, DSBs in EC109/lv-shNRG cells were significantly increased at 4-h point after the etoposide treatment (Figure 3e). The sensitivity assays demonstrated that knockdown of NRAGE remarkably sensitized EC cells to etoposide as well (Figure 3f). Collectively, NRAGE protects EC cells from a wide range of DNA-damaging agents. Subsequently, we further detected the colocalization of γH2AX with NRAGE in EC cells using IF assays. We found that γH2AX colocalized with 53BP1, BRCA1, and MDC1in a foci–foci manner (Supplementary Figure S2A); however, it could only partially colocalize with BRCA2 and NRAGE (Supplementary Figure S2B).
Figure 3.
NRAGE knockdown sensitizes EC cells to a wide range of DNA-damaging agents. (a) EC9706/lv-shCon. and EC9706/lv-shNRG cells irradiated with 5 Gy IR were detected of the γH2AX foci using IF. The percentage of cells with γH2AX foci (termed γH2AX Foci%) from three independent experiments were statistically analyzed and graphically depicted (*P=0.024 for <5 foci, *P=0.047 for >10 foci). The scale bar is 5 μm. (b and c) EC109/lv-shCon. and EC109/lv-shNRG cells irradiated with 10 Gy IR or treated with 2 μM etoposide for 2 h were detected of the severity of DSBs using neutral comet assays at the indicated time points. Representative results were shown and the scale bar is 100 μm. OTM was used to statistically analyze the significance using the ANOVA tests. (d) EC9706/lv-shCon. and EC9706/lv-shNRG cells irradiated with different doses of IR were subjected to western blottings with the indicated antibodies. The gray values of bands were used for the ANOVA statistical analysis. (e) EC109/lv-shCon. and EC109/lv-shNRG untreated (−) or treated with 2 μM etoposide for 2 h were collected at the indicated time points and subjected to western blottings with the indicated antibodies. The gray values of bands were used for the ANOVA statistical analysis. (f) EC109 cells transfected with siCon. or siNRG (#1, #2) were treated with the indicated concentrations of etoposide for 2 h and left to grow for 14 days. The number of colonies with >50 cells were manually counted, followed by the normalization to EC109 cells transfected with siCon. or siNRG (#1, #2) without etoposide treatment, respectively. Data are represented as means±S.D. and statistically analyzed using Student's t-test
To exclude the possibility that DSBs repair capability of NRAGE are specific to EC cells, we examined the DDR capability of NRAGE in a panel of other cells. First of all, using IF assays, we found that NRAGE was significantly translocated into the nucleus of HaCat from the cytoplasm in response to ultraviolet (UV) damage and partially colocalized with γH2AX foci (Supplementary Figures S2C and S3A). Additionally, NRAGE in mouse embryonic fibroblast cells (MEFs) treated with UV was also obviously translocated into the nucleus from the cytoplasm (Supplementary Figure S3B). Besides, NRAGE in RKO cells treated with mitomycin C (MMC) was gradually translocated into the nucleus from the cytoplasm as well (Supplementary Figure S3C). Moreover, in response to hydroxyurea (HU) treatment, NRAGE was remarkably increased in both cytoplasm and nucleus (Supplementary Figure S3D). Above all, we concluded that NRAGE was ubiquitously involved in DDR. Afterwards, the HaCat/lv-shCon. and HaCat/lv-shNRG cells were exposed to UV and recovered for the indicated time points, followed by the detection of γH2AX foci. As shown in Supplementary Figure S3E, the γH2AX foci still exhibited a high level at 3-h point after the removal of UV in HaCat/lv-shNRG cells, whereas they were significantly reduced in HaCat/lv-shCon. cells, implicating that NRAGE depletion impaired the DSB repair capability. Consistently, the total protein extracted from HaCat cells irradiated with UV showed that γH2AX protein were remarkably increased in NRAGE-deficient cells (Supplementary Figure S3F). Furthermore, using NRAGE wild-type (WT) and KO mice, presented by professor Xiang Gao in Medical School of Nanjing University,10 we established the UV-irradiated skin tumor model to confirm the role of NRAGE in DDR. Excitingly, KO increased about five-fold susceptibility of the skin tumor in mice (Supplementary Figure S3G). And the IHC staining of γH2AX supported that DSBs were greatly increased in the skin tumor tissues in KO mice (Supplementary Figure S3H). Taken together, we consider that NRAGE is ubiquitously involved in DDR and acts as a two-edged sword similar to most of the DDR factors.
NRAGE regulates the stability of RNF8 and BARD1 to promote cell survival in EC cells
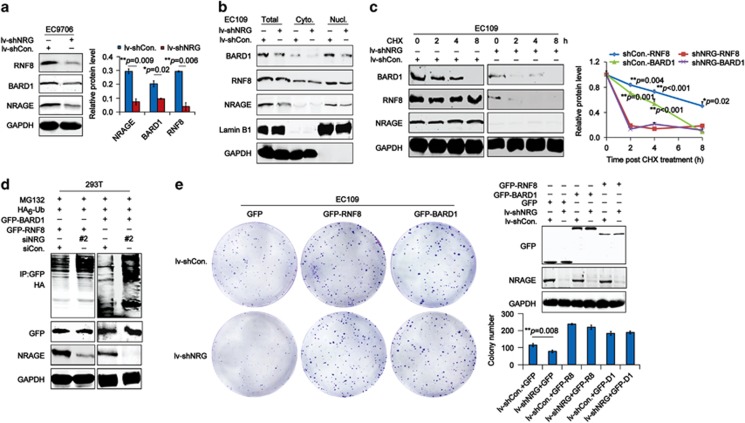
To explore the mechanisms of NRAGE in HR, we detected the expression of a band of DDR factors in NRAGE knockdown EC cells using the quantitative real-time PCR (qPCR) and immunoblotting assays. Surprisingly, downregulation of NRAGE resulted in no change in DDR genes at mRNA level (Supplementary Figure S4A); however, in EC9706 (Figure 4a) and EC109 (Supplementary Figure S4B) cells, it obviously led to a reduction of RNF8 and BARD1 proteins, without disrupting other DDR proteins, such as BRE, BRCC36, and PARP1. Furthermore, the subcellular protein separated from EC109/lv-shCon. and EC109/lv-shNRG cells confirmed that NRAGE knockdown obviously reduced the nuclear RNF8 and BARD1 (Figure 4b). Subsequently, the half-life of RNF8 and BARD1 in EC09/lv-shCon. and EC109/lv-shNRG cells treated with cycloheximide (CHX) were evaluated; we found that both RNF8 and BARD1 were apparently reduced at 2 h in EC109/lv-shNRG cells, whereas they maintained a relatively stable level in EC109/lv-shCon. cells (Figure 4c, blue and red lines for RNF8 expression; green and purple lines for BARD1 expression). These data suggested that NRAGE positively regulated the stability of RNF8 and BARD1 in EC cells. Furthermore, in vivo ubiquitination assays showed that two proteins were both highly polyubiquitinated in NRAGE-deficient cells (Figure 4d). Consequently, it is reasonable to consider that NRAGE functions in the DDR process by stabilizing RNF8 and BARD1 via the ubiquitin-proteasome pathway.
Figure 4.
NRAGE regulates the stability of both RNF8 and BARD1 proteins in a ubiquitin-proteolytic pathway to promote cell growth. (a) Total protein from EC9706/lv-shCon. and EC9706/lv-shNRG were subjected to western blotting assays with the indicated antibodies. Gray values of western bands from at least three independent experiments were represented as means±S.D. and statistically analyzed using Student's t-test (**P=0.009 for NRAGE, *P=0.02 for BARD1 and **P=0.006 for RNF8). (b) Subcellular protein extracted from EC109/lv-shCon. or EC109/lv-shNRG cells were subjected to western blottings with the indicated antibodies. Cyto., cytoplasmic protein; Nucl., nuclear protein. (c) E109/lv-shCon. and EC109/lv-shNRG cells were treated with 50 μg/ml CHX for the indicated time points, followed by the western blottings with the indicated antibodies. Gray values of western bands from at least three independent experiments were represented as means±S.D. and statistically analyzed using Student's t-test. (d) 293T cells cotransfected with siCon. or siNRG #2, together with GFP-RNF8 or GFP-BARD1 and HA6-Ub. plasmids and treated with 10 μM MG132 were lysed and subjected to IP with GFP antibody, followed by western blottings with HA antibodies. (e) EC109/lv-shCon. and EC109/lv-NRG cells transfected with GFP or GFP-RNF8 or GFP-BARD1 were subjected to colony-formation analysis. The transfection efficiency was confirmed by western blotting assays and the colony number was represented as means±S.D. and statistically analyzed using the ANOVA tests
It is well known that RNF8 regulates the histone H2AX ubiquitination to transduce DDR signals;18 therefore, we speculated that NRAGE knockdown impaired HRR by indirectly inhibiting downstream H2AX ubiquitination to prevent DDR signal transduction. As expected, the H2AX ubiquitination in EC109 cells transfected with siNRG (#1, #2) were significantly reduced, accompanied by the increase of γH2AX (Supplementary Figure S4C). Although RNF8 has also been reported to regulate the translocation of 53BP1 to the damaged foci,19, 20, 21 the recruitment of 53BP1 was not affected by downregulation of NRAGE (Supplementary Figure S4D). It is reported that RAP80 is the key factor to determine the role of RNF8 in assembling BRCA1 and 53BP1 to the damaged sites.1, 22 Consequently, we speculated that there might be a relationship between NRAGE and RAP80, which limited its roles in HR signaling. To confirm our hypothesis, we examined the interaction between NRAGE and RAP80 using immunoprecipitation (IP) assays. Excitingly, we found that NRAGE specifically interacted with RAP80 via its interspersed hexapeptide repeat domain (IRD) and MAGE homology domain (MHD) domains (Supplementary Figure S4E).
It is reported that DDR factors could promote cell survival by providing a safe survival environment in cancer cells. As a result, we overexpressed RNF8 and BARD1 in NRAGE-depleted EC cells, respectively, to examine whether the inhibition of cell growth could be reversed. Intriguingly, both RNF8 and BARD1 remarkably reversed the inhibition of cell growth in EC109/lv-shNRG cells (Figure 4e). Notably, both BARD1 (Supplementary Figure S5A) and GFP-RNF8 (Supplementary Figure S5B) foci formation in EC9706/lv-shNRG cells treated with cisplatin were also remarkably compromised, implying that NRAGE regulated the recruitment of these two proteins to the damaged sites as well. Moreover, overexpression of RNF8 and BARD1 obviously increased the resistance of EC cells transfected with siNRG #2 (Supplementary Figure S6). Above all, we conclude that NRAGE regulates the stability and recruitment of RNF8 and BARD1 to facilitate the chemoresistance of EC cells.
NRAGE chaperones the interaction between RNF8 and BARD1 via their RING domains
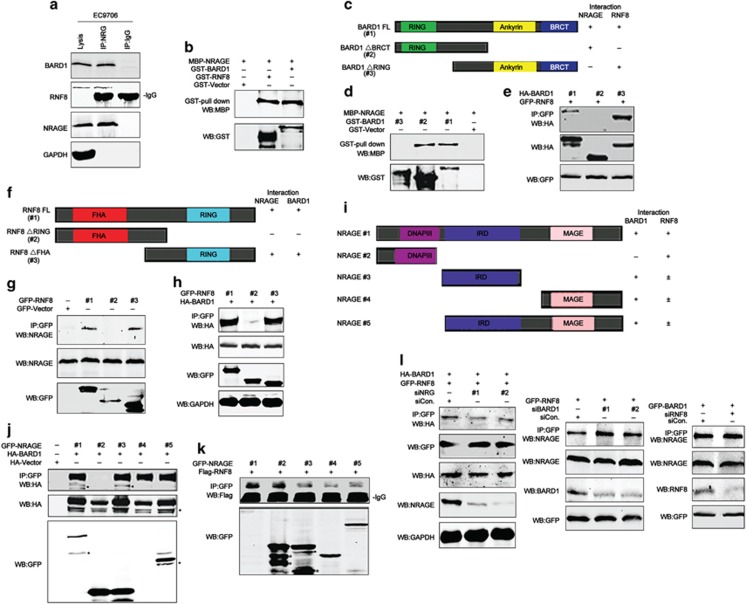
Next we explored the interaction between NRAGE and DDR factors. To our surprise, results from IP demonstrated that endogenous NRAGE could simultaneously interact with both RNF8 and BARD1 in EC9706 cells (Figure 5a). However, it could not bind with ATM, BRCA1, and ATR (Supplementary Figure S7). Further results from GST pull-down assays demonstrated that NRAGE directly interacted with RNF8 and BARD1 (Figure 5b), implicating that a novel complex comprised of NRAGE, RNF8, and BARD1 was formed. Subsequently, we constructed a series of BARD1 variants with different domains, as shown in Figure 5c. Results from GST pull-down showed that NRAGE directly interacted with the RING domain of BARD1 (Figure 5d). Moreover, IP results showed that RNF8 specifically interacted with the Ankyrin-BRCT domain of BARD1 (Figure 5e). In addition, data from IP assays demonstrated that NRAGE also specifically interacted with the RING domain of RNF8 (Figures 5f and g). And the deletion of RING domain of RNF8 also compromised its binding with BARD1 (Figure 5h), suggesting that BARD1 interacted with the RING domain of RNF8 as well. Further IP assays showed that BARD1 specifically interacted with the MHD–IRD of NRAGE (Figures 5i and 5j). As for RNF8, it interacted with the DNA polymerase III subunit (DNAPIII) domain of NRAGE (Figure 5k). Above all, in the ternary complex, NRAGE interacts with the RING domain of both RNF8 and BARD1 via its DNAPIII domain and MHD–IRD, respectively. Simultaneously, RNF8 interacts with the Ankyrin-BRCT domain of BARD1 via its RING domain.
Figure 5.
NRAGE simultaneously interacts with the RING domain of both RNF8 and BARD1 to form a novel ternary complex. (a) Protein from EC9706 cells were immunoprecipitated with NRAGE antibody, followed by western blottings with the indicated antibodies. (b) Prokaryotically expressed recombinant MBP-NRAGE protein and GST-RNF8 or GST-BARD1 protein were incubated, respectively, and then subjected to GST pull-down assays. (c) Graphical description of BARD1 domains, namely BARD1 (FL, #1), BARD1 (ΔBRCT, #2), and BARD1 (ΔRING, #3). (d) MBP-NRAGE protein was incubated with GST-tagged BARD1 domains and then subjected to GST pull-down assays. (e) 293T cells transfected with GFP-RNF8 and HA-BARD1 domain mutants were lysed and subjected to IP with GFP antibody, followed by western blottings with HA antibody. (f) Schematic description of RNF8 domains, RNF8 (FL, #1), RNF8 (ΔRING, #2), and RNF8 (ΔBRCT, #3). (g) 293T cells transfected with GFP-Vector or GFP-tagged RNF8 domains were subjected to IP with GFP antibody, followed by western blottings with NRAGE antibody. (h) 293T cells transfected with GFP-RNF8 domain mutants and HA-BARD1 were lysed and subjected to IP with GFP antibody, followed by western blottings with HA antibody. (i) Schematic presentation of NRAGE domains, #1~#5. (j) 293T cells cotransfected with HA-Vector or HA-BARD1 and GFP-tagged NRAGE domains were subjected to IP with GFP antibody, followed by western blottings with the indicated antibodies. (k) 293T cells cotransfected with GFP-NRAGE domain mutants and Flag-RNF8 were lysed and subjected to IP with GFP antibody, followed by western blottings with the indicated antibodies. (l) 293T cells cotransfected with siCon. or siRNAs against NRAGE, BARD1, or RNF8, respectively, together with GFP-RNF8 and HA-BARD1 or GFP-RNF8 or GFP-BARD1 were lysed and subjected to IP with GFP antibody, followed by western blottings with the indicated antibodies. *Indicates the unspecific bands
Afterwards, using siRNA knockdown skills, we found that NRAGE knockdown disrupted the binding between RNF8 and BARD1; on the contrary, the deletion of BARD1 slightly enhanced the interaction between RNF8 and NRAGE. As for the deletion of RNF8, there was no significant change in the binding between BARD1 and NRAGE (Figure 5l). Taken together, our studies demonstrate that NRAGE participates in the DDR not only by stabilizing and assembling RNF8 and BARD1 proteins but also by promoting the interaction between RNF8 and BARD1 to form a novel ternary complex.
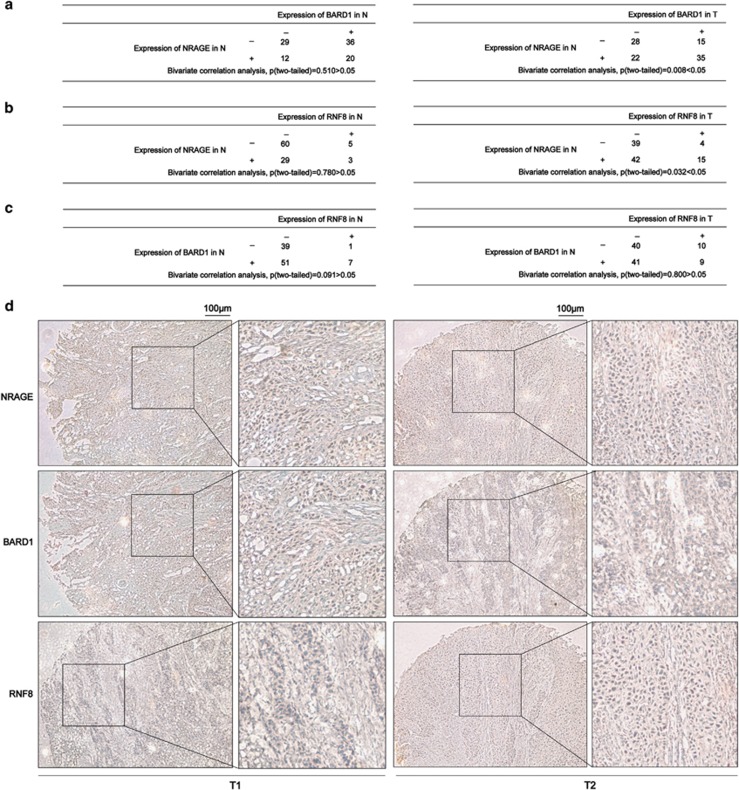
Finally, we characterized the expression of NRAGE, RNF8, and BARD1 in squamous esophageal tissue microarray, which we previously used.7 Bivariate correlation statistical analysis demonstrated that BARD1 and RNF8 were independent from the expression of NRAGE in the adjacent normal (N) tissues (Figures 6a and b, left panel); however, both RNF8 and BARD1 in esophageal tumor (T) tissues were closely correlated with NRAGE (Figures 6a, b, and d). Unfortunately, there was no expression correlation between RNF8 and BARD1 either in N or T (Figure 6c), implicating that the relationship between RNF8 and BARD1 might be more complicated than we had expected. Further studies on elucidating their relationship in EC tumorigenesis are required.
Figure 6.
Expression relationship among RNF8, BARD1, and NRAGE in clinical squamous esophageal tissues. (a) Bivariate correlation tests were used to analyze the expression dependence between NRAGE and BARD1 in esophageal tumor (T) tissues and adjacent normal (N) tissues (**P=0.008 in T tissues). (b) Bivariate correlation tests were used to analyze the expression dependence between NRAGE and RNF8 in esophageal T and N tissues (*P=0.032 in T tissues). (c) Bivariate correlation tests were used to analyze the expression correlation between BARD1 and RNF8 in esophageal T and N tissues. (d) Representative IHC results of NRAGE, BARD1, and RNF8 expression in the serial dissections of esophageal T tissues (T1, T2). The scale bar is 100 μm
Discussion
In the present study, we uncovered a novel biological function of NRAGE; it has a critical role in repairing DSBs via HR and regulates the chemoresistance of cells to DNA-damaging agents both in vitro and in vivo; it interacts with the RING domains of RNF8 and BARD1 to form a ternary complex and regulates their stability in a ubiquitin-proteolytic pathway. This discovery shows that NRAGE in squamous EC cells alternatively improves cell proliferation by employing a DDR mechanism, confirming our previous hypothesis that other proteins might synergically cooperate with PCNA to facilitate esophageal tumorigenesis.7
In the process of elaborating the mechanisms of NRAGE in the DDR process, we investigated the influence of NRAGE depletion on a majority of DDR genes and proteins using qPCR and immunoblotting assays. Interestingly, NRAGE knockdown in EC cells had no effects on the mRNA level of DDR genes. However, it strikingly reduced the expression of RNF8 and BARD1 proteins without influencing the expression of other DDR proteins, such as BRCA1, PARP1, BRE, and BRCC36, suggesting that NRAGE posttranslationally and selectively regulated the expression of RNF8 and BARD1. RNF8 has a critical role in the early DDR stage by facilitating the accumulation of checkpoint mediator proteins BRCA1 and 53BP1 to the damaged foci, on the one hand through the phospho-dependent FHA domain-mediated binding of RNF8 to MDC1, on the other hand via its role in ubiquitinating H2AX and possibly other substrates at damage sites.18 As for BARD1, it often interacts with BRCA1 to form a BRCA1-BARD1 heterodimer to transduce DDR signals in HR.23 Therefore, it is reasonable to think that NRAGE has a crucial role in HR by regulating the expression of RNF8 and BARD1. Notably, RNF8 is involved in both HR and NHEJ by regulating the accumulation of BRCA1 and 53BP1 to the damaging sites, respectively.20, 21, 24 However, although NRAGE regulated the stability of RNF8, it merely participated in HR signaling and affected the BRCA1 recruitment. It is reported that RAP80 is the key factor to determine the role of RNF8 in regulating the accumulation of BRCA1 and 53BP1 to the damaged sites.1, 22 Further IP assays demonstrated that the IRD and MHD domains of NRAGE specifically bound with RAP80, which helped to explain why it did not affect the translocation of 53BP1 to the damaged sites.
NRAGE and other MAGE family proteins have been reported to have a critical role in the ubiquitin-dependent protein degradation pathway.7, 9, 25 In the study, NRAGE negatively regulated the polyubiquitination of both RNF8 and BARD1. Notably, unlike PCNA, either RNF8 or BARD1 could dramatically reverse the cell survival of NRAGE-deficient EC cells, suggesting that NRAGE promotes cell survival of EC cells via RNF8 and/or BARD1. The IP or GST pull-down assays showed that NRAGE simultaneously and directly interacted with the RING domains of RNF8 and BARD1 via its DNAPIII and MHD–IRD, respectively. Additionally, RNF8 interacted with the Ankyrin-BRCT domain of BARD1 through its RING domain. The following siRNA transfection and IP assays revealed that NRAGE was required for the interaction between RNF8 and BARD1. However, owing to the same binding domain on the RNF8 protein of BARD1 and NRAGE, deletion of BARD1 could slightly increase the interaction between NRAGE and RNF8, suggesting that there was a competition between NRAGE and BARD1 when simultaneously interacted with RNF8. However, NRAGE is an indirect HR regulator via RING-containing complexes, which are more complicated than we have expected, and more studies are required to elucidate the relationship of NRAGE with different DDR complexes. Additional data from colocalization analysis demonstrated that NRAGE only partially colocalized with γH2AX. It has been reported that a lot of DDR proteins colocalized with γH2AX, such as ATM, BRCA1, 53BP1, and MDC1,3 contain the BRCT and FHA domains.1 In the study, NRAGE interacted with the RING domains of both RNF8 and BARD1, which have BRCT and/or FHA domains, suggesting that it has an indirect role in DDR via RNF8 and BARD1.
DDR is a double-edged sword in guarding genome integrity. A strong correlation among increased genomic instability, DNA repair defects, and cancer predisposition has been documented in cells isolated from individuals carrying germline mutations in BRCA1 or BRCA2 genes.26 On the contrary, cancers with nonfunctional BRCA1 and BRCA2 are particularly sensitive to combined treatment with DNA-damaging agents.4 Moreover, targeting DNA repair is widely recognized as a promising approach in cancer therapy.27 In our study, NRAGE deficiency is not embryonically lethal, and to date, there are no spontaneous tumors in our KO mice.7 However, by establishing UV-induced skin tumor model, we found that NRAGE deficiency greatly predisposed the tumorigenesis owing to the increase of DSBs. Clinical studies demonstrated that the expression of NRAGE was positively correlated with RNF8 and BARD1 in EC tissues, further supporting that NRAGE facilitated the DSB repair via RNF8 and BARD1 in vivo. It has been recently reported that ATM activates protein phosphatase 1 to antagonize Nek2-dependent phosphorylation of nucleophosmin (NPM), leading to the liberation of p14ARF from NPM to be degraded by the ULF E3-ubiquitin ligase and promoting tumor cell growth.28 Interestingly, NRAGE has also been suggested to act as an oncoprotein by transcriptionally inhibiting p14ARF expression via TBX2 in the nucleus.29 Therefore, we hypothesized that the functional interplay between nuclear NRAGE and DDR by regulating RNF8 and BARD1 stability was through ULF-dependent regulation of p14ARF protein, which will be further studied in our future studies.
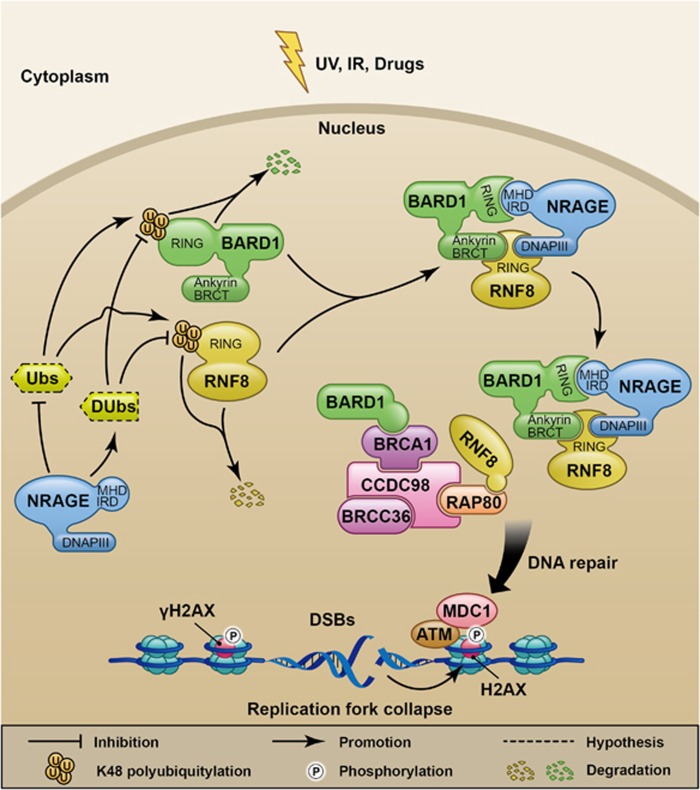
In conclusion, we propose a working model for NRAGE in the hierarchical DDR in EC tumorigenesis (Figure 7). Generally speaking, in response to DSBs, the upregulated NRAGE, by inhibiting some ubiquitin-related enzymes (Ubs) or promoting deubiquitination enzymes (DUbs) targeting K48-linked polyubiquitination of RNF8 and BARD1, stabilizes these two proteins to form a ternary complex to participate in the DDR signaling transduction. Finally, the DSBs are efficiently repaired, which facilitates cell survival and the chemotherapeutic resistance. However, more issues are raised and are needed to be explored in the future studies: what are the exact Ubs or DUbs that help NRAGE regulate the polyubiquitination of RNF8 and BARD1; how NRAGE coordinates cell cycle in HR process; how NRAGE facilitates the sequential accumulation of DDR factors to the damaged sites; and what are the relationships between NRAGE and the RAP80–RNF8–BRCA1–BARD1–BRCC36 complex in DDR process. However, our findings shed a bright future for the application of NRAGE in the personalized EC molecular targeted-therapy in the long run.
Figure 7.
Schematic illustration of NRAGE biological functions in DDR to facilitate cell survival in EC cells in response to DSBs
Materials and Methods
Cell culture and treatments
EC109, EC9706, TE1, HaCat, RKO, MEF, and 293T cells were all maintained in Dulbecco's modified Eagle's medium supplemented with 12% fetal bovine serum, 100 U/ml penicillin, and 100 U/ml streptomycin at 37 °C in a 5% CO2 incubator. For the DNA-damaging treatments, cells were treated with 2 μM etoposide for 2 h or 0.6 mJ/cm2 UV or 10 mM HU for 2 h or 5 μg/ml MMC for 2 h.
Plasmids, siRNAs, and transfection
cDNA fragments of BARD1 domains, including full length (FL, 1–777aa), Ankryrin-BRCT domain deletion (ΔBRCT) (1–550aa), and RING domain deletion (ΔRING) (125–777aa), were inserted between the endonuclease Xho I and Kpn I sites of pcDNA3.1(−) with an HA tag or pEGFPC3 vector (GFP-Vector) at the N-terminal. PCR products of RNF8 domains, including FL (1–480aa), ΔRING (1–320aa), ΔBRCT (200–480aa), and NRAGE domains, including #1 (1–778aa), #2 (1–150aa), #3 (450–650aa), #4 (650–778aa), and #5 (400–778aa), were all digested with XhoI and Kpn I and ligated into the GFP-Vector. GST-tagged BARD1 domains and RNF8 were made in pGEX-4 T-1 vectors (GST-Vector), and MBP-tagged NRAGE were made in MBP-NEB express vectors. Constructs of lv-shNRG and lv-shCon. were established according to our previous report.7 All the plasmids in the study were confirmed by sequencing. Small interferences targeting different sequences of NRAGE (siNRG #1, #2), BARD1 (siBARD1 #1, #2), and RNF8 (siRNF8) or nonspecific sequences (siCon.) were purchased from Sigma (St Louis, MO, USA). The primers of the siRNAs were summarized in Supplementary Table S1. For the transfection of plasmids, siRNAs, and the establishment of stable cell lines, we performed according to our previous report.7
HR and NHEJ reporter assays
HR and NHEJ assays were performed according to previous reports.30 In brief, 293T cells were co-transfected with siCon. or siNRG with a mutant GFP plasmid containing a specific Isce-I site (DR-GFP) and an Isce-I-expressing plasmid (Isce-I) or NHEJ plasmid, using the lipofectamine 2000 transfection agent (Invitrogen, Carlsbad, CA, USA). Notably, plamids of the GFP-Vector were transfected as a positive control to normalize the transfection discrepancy in 293T cells transfected with siCon. or siNRG, respectively. Forty-eight hours later, GFP percentage (GFP%) in 1 × 104 cells were analyzed using a FACS Canto II flow cytometer (BD Biosciences, San Jose, CA, USA). Finally, the HR and NHEJ efficiency were calculated using the formula: (GFP% of HR- or NHEJ-transfected cells)/(GFP% of corresponding GFP-Vector-transfected cells).
Cell sensitivity analysis
Cells transfected with siCon. or siNRG (#1, #2) (800 cells/well) were seeded in 12-well plates and treated with different concentrations of cisplatin or etoposide for 2 h. Afterwards, the cells were left to grow for 14 days in fresh medium, and the colonies were finally stained with 0.1% crystal violet (Beyotime, Nantong, Jiangsu, China) and manually counted the number of colonies with >50 cells.
Neutral comet assay
Briefly, in accordance with previous guidelines,31 cells after treatment were mixed with 0.6% low-melting point agarose and added to the frost slides embedded with 0.6% normal-melting point agarose, followed by solidification at 4 °C. Afterwards, the slides were submerged in precooling neutral lysis buffer (0.1 M Na2EDTA.2H2O, 2.5M NaCl, 1% TritonX-100, 10% DMSO, 10 mM Tris base, pH8.0) for 2 h and immersed in the neutral electrophoresis buffer (90 mM Tris buffer, 90 mM Boric acid, 2 mM Na2EDTA.2H2O, pH8.5) for 20 min. Subsequently, electrophoresis was performed at 4 °C for 20 min. Finally, the slides were stained with SYBR Green I Nucleic Acid Stain (Lonza, Rockland, ME, USA) for 10 min at room temperature away from light and visualized under a Leica inverted fluorescence microscope. OTM, a product of the tail length and percentage of tail DNA representing a measure of the relative fluorescent intensity in the head and tail,14 was automatically determined from 200 cells per sample from three independent analysis using the CASP_1.2.3 comet assay software (CaspLab, Wrocław, Poland).
Protein stability and ubiquitination analysis
To assess protein stability, cells were treated with 50 μg/ml CHX and collected at the indicated time points. As for the analysis of protein ubiquitination, 293T cells transfected with siCon. or siNRG, together with HA6-Ub, GFP-RNF8, or GFP-BARD1, respectively, were treated with 10 μM MG132 for 4 h, followed by IP with GFP antibody and western blotting with HA antibody.
Western blotting analysis and antibodies
After various treatments, cells were lysed with RIPA strong buffer (Beyotime) supplemented with protease inhibitors on ice. For tissue protein extraction, tumors from nude mice models were immersed in RIPA strong buffer with protease inhibitors and homogenated using an automatic sample grinding machine (Shanghai Jingxin Experimental Technology, Shanghai, China). After quantification, 80 μg protein were loaded on the SDS-PAGE gel for separation and then transferred to a nitrocellulose membrane (NC; GE Healthcare), followed by incubation with the indicated primary antibodies at 4 °C overnight. The next day, the membranes were incubated with appropriate fluorescence-conjugated secondary antibodies. Finally, the membranes were visualized using an Odyssey infrared imaging system (Licor, Lincoln, NE, USA). The gray scale values of western blotting bands were quantified using the WCIF ImageJ software (Toronto, Canada) and subjected to appropriate statistical analysis. Antibodies used in the study were as follows: β-actin (Sigma, A1978), GAPDH (Proteintech, 60004-1), NRAGE (Santa Cruze, Heidelberg, Germany, sc-28243 and sc-130434), γH2AX (Abcam, Cambridge, MA, USA, ab26350), RNF8 (Abcam, ab105362), BARD1 (Santa, sc-11438), BRCA1 (Proteintech, Wuhan, China, 22964-1), HA (Abcam, ab18181), Flag (Abcam, ab125243), GFP (Santa, sc-8334), GST (Earthox, Millbrae, CA, USA, E022040-01), MBP (Epitomics, Burlingame, CA, USA, 3380-1), H2AX (Abcam, ab11175), and RAP80 (Abcam, Ab124763).
Immunofluorescence
Cells on the slides were fixed with 4% PFA at room temperature for 20 min and perforated with 0.5%TritonX-100 at 37 °C for 10 min, followed by incubation with primary antibodies at 37 °C for 30 min. Finally, cells were stained and sealed with ProLong Gold Antifate Reagent plus 4',6-diamidino-2-phenylindole (Invitrogen). Slides were photographed under a Carl Zeiss (Oberkochen, Germany) fluorescence microscope. Antibodies used in IF assays in the study were as follows: γH2AX (abcam, 26350), NRAGE (Santa, sc-28243), BRCA1 (Santa, sc-642), Cyclin A (Santa, sc-751), 53BP1 (Santa, sc-22760), MDC1 (Abcam, Ab11171), and BRCA2 (Santa, sc-8326).
Immunoprecipitation
After treatments, cell pellets were lysed in NETN buffer (0.5% NP40, 1 M Tris HCl, 5 M NaCl, 0.5 M EDTA),32 supplemented with protease inhibitors. About 500 μg protein was incubated with 2 μg primary antibodies and 30 μl protein A/G plus-agarose (Santa Cruz) and precleared with cold NETN buffer, at 4 °C on the mute mixer (Suzhou Bing Lab Equipment CO., LTD, Suzhou, Jiangsu, China) overnight. Subsequently, the precipitated protein were washed with cold NETN buffer, denatured in boiling water, and subjected to western blotting assays with the indicated antibodies.
GST pull-down assay
According to previous reports,33 GST-tagged recombinant proteins were expressed in Rosetta bacteria in our study, induced by 0.5 mM isopropyl β-d-thiogalactoside (Takara, Dalian, Liaoning, China) for 10–16 h at 20 °C. MBP-tagged fused proteins were induced to express in NEB-express bacteria. Proteins extracted from GST-fused proteins or the GST-Vector were purified and incubated with glutathione sepharose 4 fast flow (GE Healthcare) at 4 °C overnight, followed by the addition of MBP-NRAGE. Finally, the mixture was washed and denatured in boiling water and subjected to western blottings with GST or MBP antibodies.
Immunohistochemistry
To assess the protein expression in tissues, IHC was used as previously described.7 Briefly, paraffin-embedded samples were generally deparaffinized and hydrated. The slides were then immersed in sodium citrate antigen repair solution at high pressure for 10 min, and the endogenous peroxidase was blocked with 3% H2O2 for 10 min at room temperature. Afterwards, the slides were incubated with appropriate primary antibodies overnight at 4 °C, followed by incubation with the correct secondary antibodies the next day. All the photographs were taken under a Leica inverted microscope.
In vivo esophageal tumor chemotherapy
Eight male nude mice aged 4 weeks were all subcutaneously injected with EC109/lv-shCon. or EC109/lv-shNRG cells to construct the esophageal tumor-bearing model as we previously described.7 When the tumor volumes reached 200 mm3, the mice were randomly divided into two groups and intraperitoneally injected with 0.9% NaCl or 10 mg/kg cisplatin three times per week. Twenty-eight days after the drug injection, tumors on nude mice were ethically dissected and photographed. All the above animal experiments were carried out in the animal center at Shanghai Tenth Hospital, which was approved by the Animal Experiment Management Committee of Shanghai.
Establishment of UV irradiated skin tumor model
The back hair of the WT (n=13) and KO (n=14) C57BL/6 mice were removed and periodically irradiated with 300 mJ/cm2 UV three times a week for 6 months. Then these mice were further cultured for about 5 months. Finally, the tumors on the back of these mice were pictured and the tumor tissues were stained with hematoxylin–erosin to confirm the pathology by a professional pathologist. It is worth noting that three of WT and five of KO mice died during the process.
Statistical analysis
The two-tailed Student's t-test was used to analyze the statistical significance between the two groups. The ANOVA analysis was applied for analyzing the statistical significance among more than three groups. Additionally, bivariate correlation tests were used to evaluate the relationship among NRAGE, RNF8, and BARD1 in the clinical esophageal tissues. The significance was defined and indicated as *P<0.05 or **P<0.01.
Acknowledgments
We thank Allan M Weissman at the University of Wisconsin for kindly providing us the HA6-Ub plasmid and also to Professor Lou Zhenkun at Mayo Clinic for generously giving us the HR and NHEJ reporter system. Moreover, we thank Professor Lou Zhenkun (Mayo Clinic), Professor Yuan Jian (East Hospital, Tongji University School of Medicine), and Professor Mao Zhiyong (Tongji University) for their valuable comments on the study. This research is supported by China National 973 Projects (Grant Nos.: 2012CB966904 and 20110402), National Natural Science Foundation of China (Grant Nos.: 81371913, 81472624, 81472124, 81502425), and the Shanghai young science and technology talents sailing program (Grant No. 15YF1409300).
Glossary
- EC
esophageal carcinoma
- DDR
DNA-damage response
- HRR
homologous recombination repair
- IR
ionizing radiation
- UV
ultraviolet
- DSB
double-strand break
- NHEJ
non-homologous end joining
- PCNA
proliferating cell nuclear antigen
- lv-shCon.
lv-shRNA targeting nonspecific sequence
- lv-shNRG
lentivirus-mediated short hairpin RNA targeting NRAGE
- IF
immunofluorescence
- OTM
olive tail moment
- WT
NRAGE wild-type
- KO
NRAGE knockout
- qPCR
quantitative real-time PCR
- CHX
cycloheximide
- IP
immunoprecipitation
- IHC
immunohistochemistry
- FL
full length
- ΔBRCT
Ankryrin-BRCT domain deletion
- ΔRING
RING domain deletion
- MHD
MAGE homology domain
- IRD
interspersed hexapeptide repeat domain
- DNAPIII
DNA polymerase III subunit
- Ub
ubiquitin-related enzyme
- DUb
deubiquitination enzyme
- DR-GFP
mutant GFP plasmid containing a specific Isce-I site
- Isce-I
Isce-I-expressing plasmid
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Oren
Supplementary Material
References
- Yan J, Jetten AM. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett 2008; 271: 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Mori E, Takahashi A. DNA double-strand breaks: their production, recognition, and repair in eukaryotes. Mutat Res 2009; 669: 8–12. [DOI] [PubMed] [Google Scholar]
- Lou Z, Chen J. Mammalian DNA damage response pathway. Adv Exp Med Biol 2005; 570: 425–455. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Traganos F, Wlodkowic D. Impaired DNA damage response—an Achilles' heel sensitizing cancer to chemotherapy and radiotherapy. Eur J Pharmacol 2009; 625: 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G, Chen J. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res 2013; 2: 107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi AH, Roux PP, Kubu CJ, Zeindler C, Bhakar A, Tannis LL et al. NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron 2000; 27: 279–288. [DOI] [PubMed] [Google Scholar]
- Yang Q, Ou C, Liu M, Xiao W, Wen C, Sun F. NRAGE promotes cell proliferation by stabilizing PCNA in a ubiquitin-proteasome pathway in esophageal carcinomas. Carcinogenesis 2014; 35: 1643–1651. [DOI] [PubMed] [Google Scholar]
- Xue XY, Liu ZH, Jing FM, Li YG, Liu HZ, Gao XS. Relationship between NRAGE and the radioresistance of esophageal carcinoma cell line TE13R120. Chin J Cancer 2010; 29: 900–906. [DOI] [PubMed] [Google Scholar]
- Mouri A, Sasaki A, Watanabe K, Sogawa C, Kitayama S, Mamiya T et al. MAGE-D1 regulates expression of depression-like behavior through serotonin transporter ubiquitylation. J Neurosci 2012; 32: 4562–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang J, Xing L, Shi G, Ruan H, Gu X et al. Interaction of MAGED1 with nuclear receptors affects circadian clock function. EMBO J 2010; 29: 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Langley B. Chromatin modifications associated with DNA double-strand breaks repair as potential targets for neurological diseases. Neurotherapeutics 2013; 10: 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis SJ, Boulton SJ. Emerging links between the biological clock and the DNA damage response. Chromosoma 2007; 116: 331–339. [DOI] [PubMed] [Google Scholar]
- Kuo LJ, Yang LX. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo 2008; 22: 305–309. [PubMed] [Google Scholar]
- Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol 2004; 26: 249–261. [DOI] [PubMed] [Google Scholar]
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair 2007; 6: 923–935. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Cavallo F, Brunet E, Jasin M. Homologous recombination assay for interstrand cross-link repair. Methods Mol Biol 2011; 745: 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol 2010; 17: 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007; 131: 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 2012; 19: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J 2012; 31: 1865–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson S, Rassoolzadeh H, Hedstrom E, Coucoravas C, Julner A, Goldstein M et al. The scaffold protein WRAP53beta orchestrates the ubiquitin response critical for DNA double-strand break repair. Genes Dev 2014; 28: 2726–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007; 316: 1198–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R, Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev 2002; 12: 86–91. [DOI] [PubMed] [Google Scholar]
- Lu CS, Truong LN, Aslanian A, Shi LZ, Li Y, Hwang PY et al. The RING finger protein RNF8 ubiquitinates Nbs1 to promote DNA double-strand break repair by homologous recombination. J Biol Chem 2012; 287: 43984–43994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell 2010; 39: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E, Magin S, Soni A, Iliakis G. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front Oncol 2013; 3: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damia G, D'Incalci M. Targeting DNA repair as a promising approach in cancer therapy. Eur J Cancer 2007; 43: 1791–1801. [DOI] [PubMed] [Google Scholar]
- Velimezi G, Liontos M, Vougas K, Roumeliotis T, Bartkova J, Sideridou M et al. Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat Cell Biol 2013; 15: 967–977. [DOI] [PubMed] [Google Scholar]
- Kumar S, Park SH, Cieply B, Schupp J, Killiam E, Zhang F et al. A pathway for the control of anoikis sensitivity by E-cadherin and epithelial-to-mesenchymal transition. Mol Cell Biol 2011; 31: 4036–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Zhang H, Wang L, Yuan J, Lou Z. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J 2012; 31: 3008–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 2006; 1: 23–29. [DOI] [PubMed] [Google Scholar]
- Yuan J, Luo K, Liu T, Lou Z. Regulation of SIRT1 activity by genotoxic stress. Genes Dev 2012; 26: 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci USA 2011; 108: 10448–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.