Abstract
Histone deacetylase (HDAC) inhibition leads to dynamic changes in the epigenetic landscape that is postulated to alter the expression of critical mediators of cellular proliferation and death. While current HDAC inhibitors have shown to be efficacious in the treatment of specific hematologic malignancies, their therapeutic utility in epithelial-based cancers warrants further evaluation. Moreover, the mechanisms of HDAC inhibition-induced cancer cell death are not completely understood. Therefore, elucidation of the underlying pathways engaged by HDAC inhibition may enable the development of more effective therapeutic strategies. Here, we report that HDAC inhibition in human breast and lung carcinoma cells activates an apoptotic mechanism mediated by microRNA (miRNA) and induced by the oncogene MYC. Specifically, following HDAC inhibition, MYC, which normally represses miR-15 and let-7 families, transcriptionally activated their expression and MYC was required for this miRNA upregulation. As a result, transcript levels of the tumor-suppressive miR-15 and let-7 families increased, which targeted and decreased the expression of the crucial prosurvival genes BCL-2 and BCL-XL, respectively. MYC was also required for the downregulation of BCL-2 and BCL-XL following HDAC inhibition. Blocking the binding sites of the miR-15 and let-7 families in the 3′-untranslated regions of BCL-2 and BCL-XL protected against HDAC inhibition-induced apoptosis. These results provide important insight into the molecular underpinnings of HDAC inhibition-induced cell death in breast and lung cancer and reveal a tumor-suppressive role for MYC-regulated miRNA that is activated with HDAC inhibition.
Aberrant gene transcription is a defining feature of cancer, and alterations in transcription regulation often lead to cellular transformation.1 Complex mechanisms regulate transcription, including the addition or removal of chemical modifications, such as acetyl groups, to histone tails.2 Deregulation in the expression and/or activity of histone deacetylase (HDAC) enzymes, which remove acetyl groups, leads to alterations in gene expression and has been linked to the development of cancer.2 The predominant biological outcome following exposure of cells to inhibitors of HDACs has been the selective death of malignant cells.3 Although HDAC inhibitors have provided clinical benefit to the treatment of specific hematological malignancies, its impact on solid organ cancer treatment is less clear and the underlying mechanisms behind HDAC inhibition (HDACi)-induced tumor cell apoptosis remain unresolved.
Although the mechanism of action of HDAC inhibitors should favor chromatin decondensation and a global increase in gene transcription, only a small percentage of genes appears to be affected.4 This suggests that post-transcriptional mechanisms of gene regulation are likely involved in the molecular events following HDACi. One such mechanism that has been linked to HDAC regulation includes microRNA (miRNA).5, 6, 7, 8 miRNA comprise a class of noncoding RNA that post-transcriptionally regulate the expression of target mRNA, typically resulting in decreased translation.9 The potential for miRNA-guided regulation of gene expression is significant, as it is predicted that the majority of all mRNAs are under miRNA control and that a single miRNA can target many mRNA.9 Therefore, HDACi-induced changes in one or more miRNA are capable of eliciting a significant downstream biological response.
Cancers often present with reduced levels of mature miRNA as compared with normal tissue of the same origin.10, 11 In B-cell lymphomas, downregulation of miRNA expression was reported to be the result of widespread transcriptional repression by the oncogenic transcription factor MYC.12 Moreover, well-known tumor-suppressive miRNA, including the miR-15 and let-7 families, are repressed by MYC in human B-cell lymphoma.12 These miRNA have also been reported to be downregulated in breast and lung cancers,10, 11, 13 but the involvement of MYC in their repression in these malignancies is unknown. Recently, MYC was shown to repress miR-29 in B-cell lymphomas through the recruitment of HDAC3 to the miR-29 promoter.5 Here, we demonstrate that MYC repressed the miR-15 and let-7 families in breast and lung cancer, and that upon HDACi, these miRNA were transcriptionally activated by MYC. Blocking the ability of miR-15 and let-7 families from targeting BCL-2 and BCL-XL, respectively, resulted in carcinoma cell survival. Our data reveal a previously unknown mechanism of MYC-induced apoptosis mediated by miRNA that is activated in breast and lung cancer cells by HDACi.
Results
HDACi induces transcription of the miR-15 and let-7 families in breast and lung cancer cells
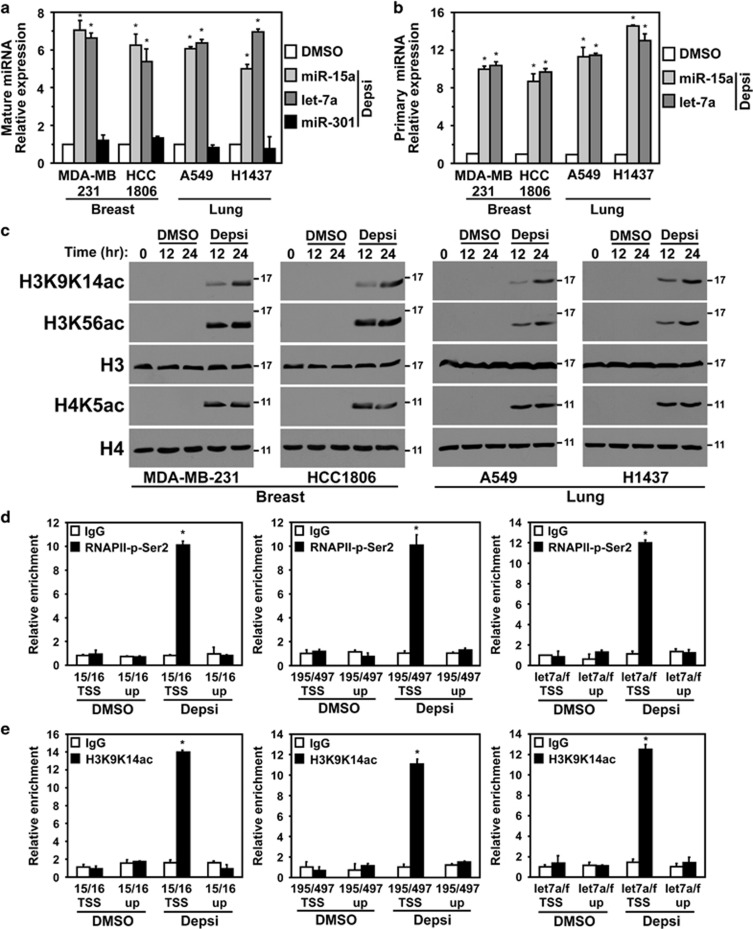
Cancer cells frequently select for downregulation of miRNA that inhibit growth and mediate apoptosis.14 For example, the miR-15 and let-7 families, both of which have been shown to induce apoptosis and inhibit tumorigenesis,15 are significantly downregulated in most human cancers, including breast and lung carcinoma.10, 11, 13, 14 A recent mechanism of miRNA repression was reported to be mediated, in part, by the recruitment of HDACs to miRNA promoters.5 As such, we sought to determine whether HDACs contributed to the transcriptional repression of the miR-15 and let-7 families in breast and lung cancers. Following HDACi with Depsipeptide (Depsi, pan-class I HDAC inhibitor), an increase in levels of representative members of the miR-15 and let-7 families (miR-15a and let-7a, respectively) was observed in all four breast and lung cancer cell lines evaluated compared to vehicle control (dimethyl sulfoxide (DMSO)) (Figure 1a). Expression of other miRNA (e.g., miR-301) did not change upon HDACi, indicating that HDACi did not lead to the upregulation of all miRNA. Analogous results were obtained with suberoylanilide hydroxamic acid (SAHA, a pan-HDAC inhibitor) and RGFP966 (966, an HDAC3-selective inhibitor) (Supplementary Figure 1A). The increase in expression of the miRNA was specific for transformed cells, as peripheral blood mononuclear cells (PBMCs) did not show an induction of miR-15a and let-7a following HDACi (Supplementary Figure 1B).
Figure 1.
miR-15 and let-7 families are transcriptionally upregulated following HDACi. Human breast (MDA-MB-231, HCC1806) and lung (A549, H1437) carcinoma cell lines were treated with vehicle control (DMSO) or Depsi. (a and b) Following 12 h with Depsi or DMSO, levels of the indicated mature miRNA (a) and primary miRNA (b) were determined by quantitative real-time-PCR (qRT-PCR) (triplicates) and normalized to small RNA RNU6b levels. Values for each miRNA are plotted relative to their respective DMSO sample, which was set at 1 (only 1 bar for the DMSO-treated samples is displayed.) (c) Western blot analysis of the indicated proteins at intervals following Depsi or DMSO vehicle control. Molecular weight (kilodalton) is indicated. (d and e) MDA-MB-231 breast cancer cells were treated with Depsi or DMSO for 4 h. ChIP with anti-RNAPII-phosphorylated on serine 2 (RNAPII-p-Ser2; d), H3K9K14ac (e) or isotype controls (immunoglobulin G (IgG)) was performed followed by qRT-PCR (triplicates) for the indicated promoter regions (transcriptional start site (TSS)) or the upstream regions (up; negative controls). Values are relative to input DNA and their respective IgG controls and plotted relative to the first DMSO sample, which was set at 1. Error bars for (a, b, d and e) are S.E.M.; (a and b) *P<0.002 and (d and e) *P<0.0015 were determined by comparison with DMSO control (t-tests)
We postulated that the increased miRNA levels we observed were likely due to increased transcription. To determine whether the miR-15 and let-7 families were actively being transcribed following HDACi, we first assessed the expression of the primary miRNA transcripts for miR-15a and let-7a, representative members of both families. Following HDACi, we detected an increase in levels of pri-miR-15a and pri-let-7a in the breast and lung cancer cell lines (Figure 1b). Additionally, western blotting revealed an increase in global histone acetylation marks associated with active transcription following HDACi with Depsi, SAHA or 966 (Figure 1c and Supplementary Figure 1C).
Global acetylation of histones is typically associated with open and active chromatin; therefore, we next evaluated the transcriptional status of the promoters of the miR-15 and let-7 families. We performed chromatin immunoprecipitation (ChIP) with the MDA-MB-231 breast cancer cell line following administration of HDACi or vehicle control. Specifically, we assessed the enrichment of a phosphorylated form of RNA polymerase II (serine 2 phosphorylated; RNAPII-p-Ser2), which is indicative of RNAPII engaged in transcriptional elongation. Notably, HDACi resulted in a significant enrichment of RNAPII-p-Ser2 at the promoter regions of the miR-15 family (miR-15a/16-1, miR-195/497) and the let-7a/f cluster (Figure 1d). RNAPII enrichment was not detected at regions upstream of the miRNA promoters or in cells that received DMSO vehicle control. Furthermore, H3K9K14ac (histone 3 with lysine 9 and 14 acetylation), an acetylation mark associated with transcriptionally active and open chromatin, was also enriched at the promoter regions of miR-15a/16-1, miR-195/497 and let-7a/f following HDACi (Figure 1e). These results demonstrate that HDACi alleviates the repression of the miR-15 and let-7 families observed in malignant breast and lung cells, inducing their transcriptional upregulation.
MYC is required to mediate the HDACi-induced increase in the miR-15 family and let-7a
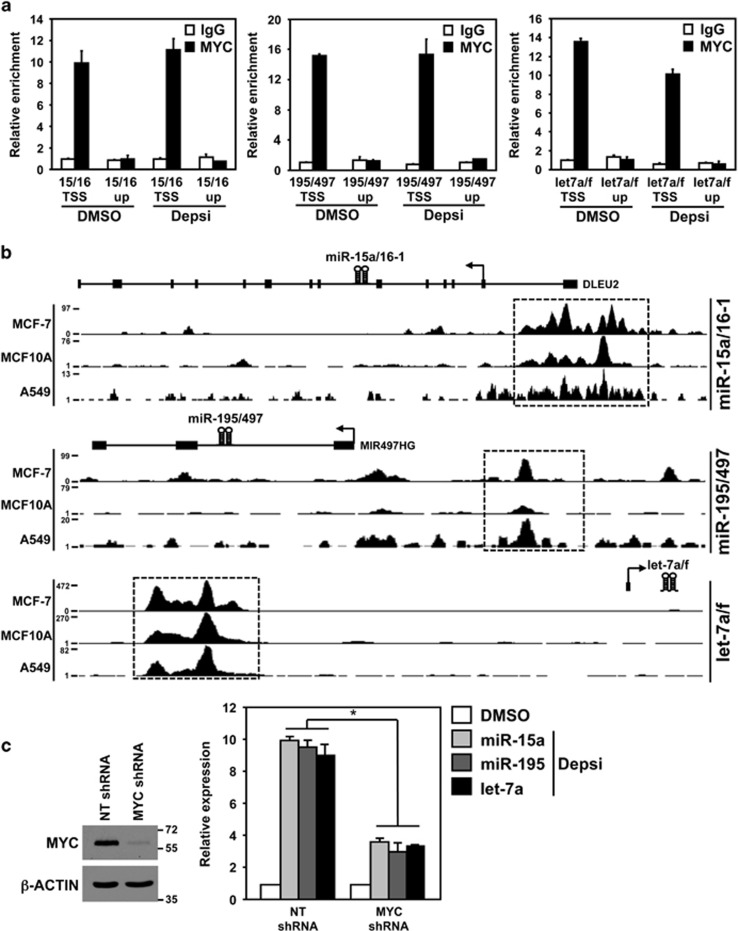
The oncogenic transcription factor MYC has been shown to transcriptionally activate or repress the expression of protein-coding genes and noncoding RNA, including miRNA.16 Previously, reduced expression of the miR-15 and let-7 families in human B-cell lymphoma was reportedly due to MYC-mediated transcriptional repression at their promoter regions.12 Therefore, we first investigated whether MYC was enriched at the promoter regions of the miR-15a/16-1, miR-195/497 and let-7a/f clusters in the presence and absence of HDACi using ChIP. MYC enrichment was detected in MDA-MB-231 cells that had received either DMSO vehicle control or the HDACi Depsi (Figure 2a). However, no enrichment was detected at regions upstream of the miRNA promoters (Figure 2a). In addition, ENCODE (encyclopedia of DNA elements) ChIP-seq data17 from human breast and lung cell lines was evaluated and also showed that MYC was enriched at these same miRNA promoter regions (Figure 2b). These results suggest that MYC is bound to the promoters of the miR-15 and let-7 families regardless of their transcriptional status and HDACi.
Figure 2.
MYC is required to transcriptionally upregulate the miR-15 and let-7 families upon HDACi. (a) MDA-MB-231 breast carcinoma cells were treated for 4 h with Depsi or vehicle control (DMSO). Following ChIP with antibodies against MYC or isotype control (immunoglobulin G (IgG)), quantitative real-time-PCR (qRT-PCR) for the indicated promoter regions (transcriptional start site (TSS)) or the upstream (up) regions MYC does not bind (negative controls) was performed (triplicates). Values are relative to input DNA and their respective IgG controls and plotted relative to the first DMSO sample, which was set at 1. (b) Evaluation of ENCODE MYC ChIP-sequencing data.17 Peaks demonstrate MYC enrichment at the promoter of the miR-15a/16-1, miR-195/497 and let-7a/f clusters (boxed) in breast adenocarcinoma (MCF-7), immortalized mammary epithelial cells (MCF10A) and lung adenocarcinoma (A549) cells. (c) MDA-MB-231 cells were transfected with a vector encoding either a MYC-specific shRNA or an NT shRNA control. MYC protein levels were evaluated by western blot 48 h after shRNA transfection (left). Molecular weight (kilodalton) is indicated. Relative expression of the indicated miRNA was determined by qRT-PCR (triplicates) following 12 h of HDACi with Depsi or DMSO (right). miRNA levels were normalized to the expression of small RNA RNU6b. Values for each miRNA are plotted relative to their respective DMSO sample, which was set at 1 (only 1 bar for the DMSO-treated samples is displayed.) Error bars are S.E.M.; *P<0.01 (t-tests)
To determine whether MYC was necessary to transcriptionally activate the expression of the miR-15 and let-7 families following HDACi, MDA-MB-231 breast cancer cells were transfected with a vector encoding a MYC-specific short hairpin RNA (shRNA) or a non-targeting (NT) shRNA (Figure 2c). Upon knockdown of MYC, the HDACi-induced increase in miR-15a, miR-195 and let-7a levels were significantly blunted (Figure 2c). These data demonstrate that MYC is required to mediate the increase in miR-15 and let-7 families induced by HDACi.
BCL-2 and BCL-XL expression is reduced by HDACi and requires MYC
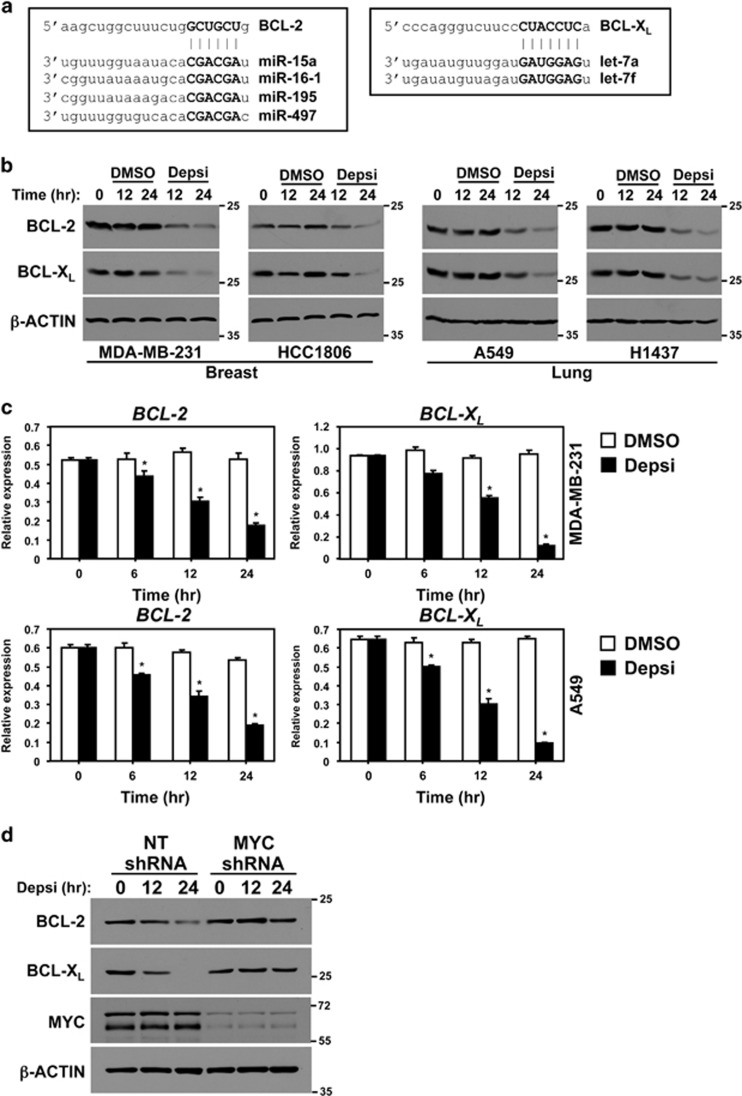
Post-transcriptional mRNA regulation is mediated, in part, by miRNA.9, 14 Given the significant increase in the levels of the miR-15 and let-7 families following HDACi, we questioned whether this would impact the expression of known targets of these two miRNA families. Specifically, we evaluated the expression of crucial prosurvival proteins BCL-2 and BCL-XL, as their 3′-UTRs are well-characterized targets of the miR-15 and let-7 families, respectively (Figure 3a).14, 15 To test this, breast and lung cancer cell lines were administered Depsi or vehicle control (DMSO). Compared with cells exposed to vehicle control, HDACi resulted in significantly decreased BCL-2 and BCL-XL protein levels in all four carcinoma lines (Figure 3b). Analogous results were obtained upon treatment with SAHA or 966 (Supplementary Figure 2a). Analysis of BCL-2 and BCL-XL mRNA in breast and lung cancer cells showed that both were significantly decreased upon HDACi, indicating that the decrease in protein expression was likely due to a reduction in BCL-2 and BCL-XL mRNA (Figure 3c). The decreased BCL-2 and BCL-XL mRNA in response to HDACi was specific for transformed cells, as BCL-2 and BCL-XL mRNA levels did not change in PBMCs following HDACi (Supplementary Figure 2b).
Figure 3.
MYC mediates HDACi-induced decrease of BCL-2 and BCL-XL protein expression. (a) Diagram of the miR-15 family and let-7 family binding site within the 3′-UTR of BCL-2 and BCL-XL, respectively. miRNA seed sequence is given in bold. (b) Western blots for the indicated proteins were performed with whole-cell protein lysates of the indicated human breast and lung carcinoma cell lines following the addition of vehicle control (DMSO) or Depsi for the indicated time. (c) BCL-2 and BCL-XL mRNA levels were evaluated by quantitative real-time-PCR (qRT-PCR) (normalized to β-ACTIN levels) in MDA-MB-231 and A549 cells at intervals following the addition of DMSO or Depsi. Error bars are S.E.M.; *P<0.03 was determined by comparison with DMSO (t-tests). (d) MDA-MB-231 cells were transfected with a vector encoding a MYC-specific shRNA or an NT shRNA control. Forty-eight hours after transfection, cells were treated with Depsi for the indicated time and western blots for the indicated proteins were performed. Molecular weight (kilodalton) is indicated
Because knockdown of MYC blunted the HDACi-induced increase of the miR-15 and let-7 families (Figure 2c), we tested the effects of MYC knockdown on their targets, BCL-2 and BCL-XL, following HDACi. MDA-MB-231 breast cancer cells were transfected with a vector encoding a MYC shRNA or an NT shRNA and then treated with Depsi. Levels of both BCL-2 and BCL-XL proteins decreased within 12 h following HDACi in the cells that had received the NT shRNA (Figure 3d). However, MYC knockdown greatly inhibited this decrease with only slightly reduced levels of BCL-2 and BCL-XL observed after 24 h of HDACi (Figure 3d). Taken together, the data indicate that HDACi results in reduced expression of the prosurvival proteins, BCL-2 and BCL-XL, and that MYC is necessary to mediate the HDACi-induced decrease of BCL-2 and BCL-XL.
HDACi induces apoptosis in breast and lung cancer cells
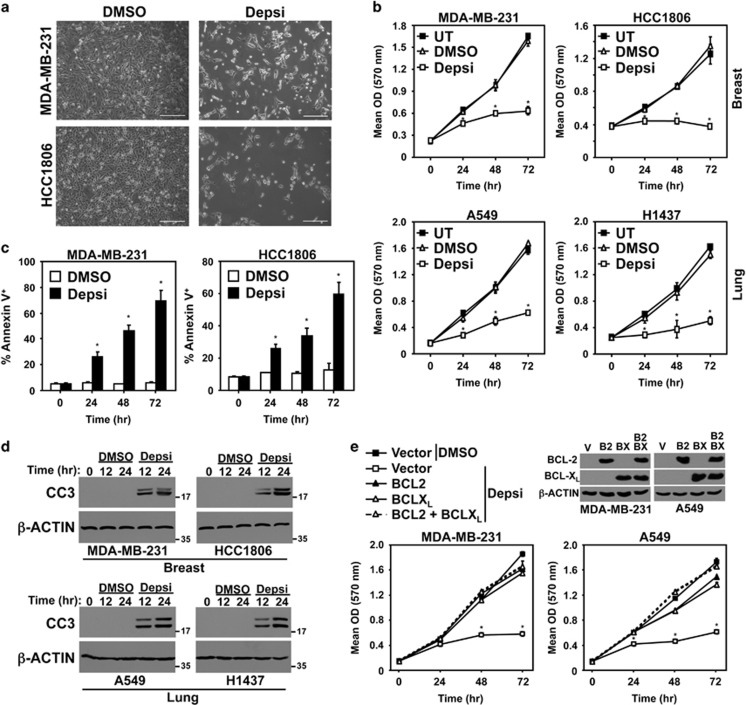
To test whether the decrease in the expression of prosurvival proteins BCL-2 and BCL-XL results in apoptosis, human breast and lung cancer cell lines were treated with Depsi or vehicle control (DMSO) and several assays were performed. There was an obvious reduction in cell number following HDACi (Figure 4a). Moreover, in MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays, all four breast and lung carcinoma cell lines showed decreased cell expansion compared with cells administered vehicle control or that were left untreated (Figure 4b). Assessment of Annexin-V positivity at intervals following HDACi showed a significant increase in the number of Annexin-V positive cells (Figure 4c). Furthermore, western blotting revealed an increase in caspase-3 cleavage after HDACi in all four carcinoma cell lines (Figure 4d). Similar results were obtained with SAHA or 966 treatment (Supplementary Figure 3). These data demonstrate that as BCL-2 and BCL-XL levels decrease in breast and lung cancer cells following HDACi, apoptosis occurs.
Figure 4.
HDACi induces apoptosis of breast and lung cancer cells. (a) Following addition of Depsi or vehicle control (DMSO) to MDA-MB-231 and HCC1806 cells, representative microscopic images were taken 72 h later (× 10 objective; line denotes 200 microns). (b–d) The indicated human breast and lung carcinoma cells remained untreated (UT) or were treated with DMSO or Depsi. MTT assays were performed (quadruplicates) every 24 h (b). Annexin-V positivity (triplicates) was measured at intervals by flow cytometry (c). At intervals, cleaved caspase-3 (CC3) was assessed by western blot (d); molecular weight is indicated in kilodaltons. (e) MDA-MB-231 and A549 cells were transfected with empty vector (V) or vectors encoding BCL-2 (B2) and/or BCL-XL (BX). Western blots were performed for the indicated proteins. Cells were treated with DMSO or Depsi and MTT assays performed (quadruplicates) every 24 h. Error bars are S.D.; *P<0.01 for (b) and *P<0.003 for (c) were determined by comparison with DMSO; *P<0.01 for (e) was determined by comparison with vector control cells treated with DMSO (t-tests)
To determine whether ectopic expression of BCL-2 and/or BCL-XL could protect human carcinoma cells from HDACi-induced cell death, BCL-2 and/or BCL-XL were overexpressed in MDA-MB-231 (breast) and A549 (lung) cancer cells. In the presence of Depsi, both cell lines overexpressing BCL-2 and/or BCL-XL continued to proliferate at a rate similar to those cells that received DMSO vehicle control (Figure 4e), demonstrating that they were resistant to Depsi-induced cell death. Collectively, the data show that overexpressing the antiapoptotic proteins BCL-2 and BCL-XL (targets of the miR-15 family and let-7a, respectively) significantly attenuated the induction of HDACi-mediated human breast and lung carcinoma cell death.
miR-15 and let-7 families mediate HDACi-induced apoptosis
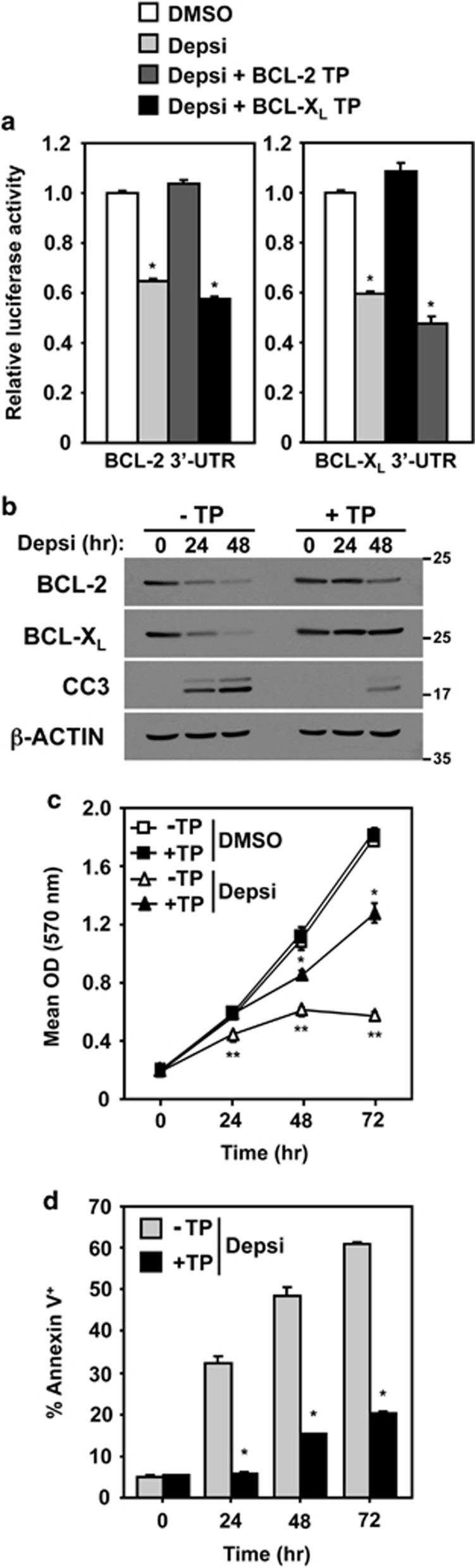
Our data suggest that the decrease observed in BCL-2 and BCL-XL protein following HDACi, resulting in apoptosis, was the direct consequence of the upregulation of the miR-15 and let-7 families. To test this directly, we transiently transfected MDA-MB-231 cells with modified RNA molecules termed Target Protectors that are designed to bind specific sites in the BCL-2 or BCL-XL 3′-untranslated region (UTR) and block the binding of the miR-15 and let-7 families, respectively. In addition, these cells were transfected with luciferase reporters harboring the BCL-2 or BCL-XL 3′-UTR that contain the miR-15 and let-7 family binding sites, respectively. Following HDACi with Depsi, there was a significant decrease in luciferase activity in cells that contained either the BCL-2 or the BCL-XL 3′-UTR, indicating that the miR-15 family and let-7a were upregulated and bound their respective sites in the BCL-2 and BCL-XL 3′-UTR, respectively (Figure 5a). In contrast, in the presence of the BCL-2 or BCL-XL Target Protector, a decrease in luciferase activity after HDACi was not observed (Figure 5a). These effects were specific for the individual miRNA and the 3′-UTR, as luciferase activity was not decreased following HDACi in cells containing the BCL-2 3′-UTR and the BCL-XL Target Protector or in cells with the BCL-XL 3′-UTR and the BCL-2 Target Protector (Figure 5a). These data provide direct evidence that the decrease in BCL-2 or BCL-XL expression following HDACi is due to an upregulation of the miR-15 and let-7 families, respectively, that bind and promote their downregulation.
Figure 5.
Blocking miR-15 and let-7 families from binding BCL-2 and BCL-XL protects from HDACi-induced apoptosis. MDA-MB-231 breast cancer cells were transiently transfected with Target Protectors (TP) that block the miR-15 family and let-7 family binding sites in the BCL-2 and BCL-XL 3′-UTR, respectively. (a) A luciferase expression vector containing the 3′-UTR of BCL-2 or BCL-XL was also transfected into MDA-MB-231 cells. Luciferase activity was measured (triplicates) after 24 h with vehicle control (DMSO) or Depsi. (b) Following the addition of Depsi for the indicated intervals, total cell protein lysates from MDA-MB-231 with (+TP) or without (−TP) TPs were western blotted for the indicated proteins; cleaved caspase-3 (CC3). Molecular weight (kilodalton) is indicated. (c and d) Cells with (+TP) or without (−TP) the BCL-2 and BCL-XL TPs were subjected to MTT assay (quadruplicates; c) and Annexin-V analysis (triplicates; d) by flow cytometry at intervals following the addition of Depsi or DMSO. Error bars are S.D. For (a) *P<0.003 and for (c) *P<0.01 and **P<0.001 were determined by comparison with DMSO; for (d), *P<0.02 was determined by comparison with cells without TPs (t-tests)
We also assessed levels of BCL-2 and BCL-XL protein in cells with Target Protectors, following HDACi. When both BCL-2 and BCL-XL Target Protectors were present, the levels of BCL-2 and particularly BCL-XL were maintained even after 24 and 48 h of HDACi (Figure 5b). Individually, the Target Protectors prevented HDACi-induced downregulation of either BCL-2 or BCL-XL (Supplementary Figure 4). Evaluation of the effects on cell expansion showed a rapid decrease in the MDA-MB-231 cells treated with Depsi that did not contain any Target Protectors, whereas cells that had received the BCL-2 and BCL-XL Target Protectors continued to proliferate with HDACi (Figure 5c). MDA-MB-231 cells with Target Protectors also showed a significant reduction (~40%) in the percentage of Annexin-V-positive apoptotic cells following HDACi (Figure 5d). In addition, there was less caspase-3 cleavage following HDACi in cells with Target Protectors (Figure 5b and Supplementary Figure 4). Therefore, blocking the miR-15 and let-7 families from binding BCL-2 or BCL-XL in breast cancer cells prevented the downregulation of BCL-2 and BCL-XL and blunted the apoptotic response resulting from HDACi. Collectively, the data reveal a novel miR-15 and let-7 family-mediated mechanism of apoptosis that underlies HDACi-induced breast and lung carcinoma cell death.
Discussion
In addition to acquiring genetic mutations, cancers can also arise from aberrant epigenetic alterations that modify chromatin accessibility, and thus gene expression.2 Development of small-molecule inhibitors of HDACs, which function to compact DNA, making it inaccessible, had immediate anticancer application because of their potential to reactivate aberrantly silenced tumor suppressors.3 Although a number of HDAC inhibitors are being evaluated in preclinical cancer models and clinical trials, much remains unknown about the mechanism of cell death they induce and the molecular determinants of cancer cell sensitivity. In our current study, data demonstrate that inhibition of HDACs in human breast and lung carcinoma cells activates a miRNA-mediated mechanism of apoptosis that is induced by MYC. Our results show that, following HDACi, the MYC-regulated miR-15 and let-7 families are transcriptionally upregulated and they target and downregulate the expression of BCL-2 or BCL-XL, triggering tumor cell apoptosis.
Recently, it has been appreciated that, in addition to protein-coding genes, deregulated expression of noncoding RNA, including miRNA, is a hallmark of tumorigenesis.1 Although miRNA expression profiling has revealed several miRNA to be upregulated in cancers relative to their normal tissue counterparts, the majority of miRNA are downregulated.10, 11 These alterations in miRNA expression are beginning to be understood and some have proved to be advantageous in tumorigenesis. For example, miRNA families shown to possess tumor- suppressive properties,15 such as the miR-15 and let-7 families, are commonly downregulated in human cancers, including breast and lung carcinoma.10, 11, 13, 14 Altered expression and/or recruitment of HDACs have been shown to contribute to dysregulation of specific miRNA (e.g., miR-29, miR-200b, miR-31, miR-125a, miR-125b and miR-205) in human breast and lung cancers.5, 6, 7, 8 Our data show that treatment with Depsi (class I HDAC inhibitor), SAHA (pan-HDAC inhibitor) or 966 (HDAC3-selective inhibitor) resulted in elevated levels of mature miRNA of the miR-15 and let-7 families. This increase in miRNA expression was selective as not all miRNA assessed were induced by HDACi. Moreover, this effect was not observed in normal, untransformed PBMCs, indicating tumor cell specificity for HDACi. Notably, the HDAC3-selective inhibitor induced miR-15a and let-7a to an equal extent as Depsi and SAHA, suggesting that HDAC3 is involved in mediating the repression of the miR-15 and let-7 families in carcinoma cells. HDAC3 was reported to repress miRNA expression in hematopoietic malignancies.5 Evaluation of primary miRNA transcript levels for the miR-15 family and let-7a indicated that the increase in mature miRNA following HDACi was a result of increased transcription of these miRNA. These data were supported by the observed enrichment of phosphorylated RNA polymerase II and the H3K9K14ac histone acetylation mark, which are both indicative of open and transcriptionally active chromatin. These results demonstrate that HDACs, and specifically HDAC3, contribute to the downregulation of the miR-15 and let-7 families in breast and lung cancer, which can be reversed by HDACi.
Dysregulation of the oncogenic transcription factor MYC is known to markedly alter the expression of genes to facilitate cellular proliferation and tumorigenesis.16 To elicit these effects, MYC coordinates the activation and repression of an extensive network of genes, including miRNA.18 MYC can repress protein-coding genes and miRNA by recruiting HDACs, including HDAC3.5, 19 Previously, MYC was reported to repress the miR-15 and let-7 families in B-cell lymphoma, as it was enriched at the promoter regions of these miRNA.12 Our results, in combination with genome-wide MYC ChIP-sequencing data,17 indicate that MYC was also present at the promoter regions of the miR-15 and let-7 families in human breast and lung cancer cells. Notably, by knocking down MYC, we determined that MYC was necessary for the upregulation of the miR-15 and let-7 families following HDACi. These data provide mechanistic evidence that MYC uses HDACs, and likely HDAC3 specifically, to transcriptionally silence the miR-15 and let-7 families in breast and lung cancer cells, and that HDACi converts MYC from a repressor to a transcriptional activator at these miRNA promoters.
While induction of tumor cell death has been the biological outcome most often reported following HDACi,3 and we observed this as well, the molecular determinants responsible for the apoptosis remained incompletely resolved. As such, we investigated known mRNA targets of the miR-15 and let-7 families that, when targeted by these miRNA, lead to cell death.15 The decrease in BCL-2 and BCL-XL protein we observed following HDACi indicated that the miR-15 and let-7 families were derepressed, and thus, transcriptionally upregulated, targeted BCL-2 and BCL-XL mRNA, respectively. This is supported by our data demonstrating that BCL-2 and BCL-XL mRNA is reduced in human breast and lung cancer cell lines following HDACi. Direct evidence that the cell death observed in the tumor cells following HDACi was due to the downregulation of BCL-2 and BCL-XL by the miR-15 and let-7 families, respectively, was obtained by blocking the binding sites of these miRNA families in the 3′-UTR of BCL-2 and BCL-XL. By inhibiting the miR-15 and let-7 families from binding the 3′-UTR of BCL-2 and BCL-XL, respectively, there was a significant decrease in cell death caused by HDACi. Similarly, overexpression of BCL-2 and/or BCL-XL protected breast and lung cancer cells from HDACi-induced cell death. Other groups have also reported that the forced expression of prosurvival BCL-2 family of proteins can protect multiple malignant cell types from the deleterious effects of HDACi.3 Additionally, when MYC was knocked down, HDACi had very little effect on BCL-2 and BCL-XL protein expression. This suggested that MYC was required to mediate the HDACi-induced increase in the miR-15 and let-7 families that subsequently targeted BCL-2 and BCL-XL. These results reveal a novel MYC-regulated, miRNA-mediated mechanism of apoptosis that is inactivated in carcinomas, but that can be reactivated by HDACi.
We have demonstrated that inhibition of HDACs in human breast and lung carcinoma cells induces a previously unappreciated tumor suppressor mechanism that uses miRNA to decrease the expression of critical prosurvival proteins, triggering tumor cell death. Activation of this miRNA-mediated apoptotic mechanism was dependent on MYC, revealing that HDACi redirected MYC towards apoptosis, which is inactivated in transformed cells. Given that MYC is dysregulated in the majority of human cancers,16 our results reveal a new MYC-mediated mechanism to induce cell death that could have therapeutic potential in breast and lung cancers. Furthermore, our data suggest that upregulation of the miR-15 and let-7 families and downregulation of prosurvival proteins BCL-2 and BCL-XL could offer a predictive measure of clinical response to HDACi.
Materials and Methods
Cell culture and transfection
Human breast (MDA-MD-231 and HCC1806) and lung (A549 and H1437) carcinoma cell lines were provided by Drs. Jennifer Pietenpol, Pierre Massion and William Pao. All cells were cultured as described by the American Type Culture Collection (Manassas, VA, USA). MDA-MB-231 and A549 cells were transfected using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). For the knockdown experiments, MDA-MB-231 cells were transfected with a SureSilencing vector encoding a MYC shRNA or NT control shRNA (Qiagen, Valencia, CA, USA) and cells were analyzed 48 h later. For the BCL-2 and BCL-XL overexpression experiments, MDA-MB-231 and A549 cells were transfected with pMIG vector control or pSFFV-BCL-2 and/or pMIG-BCL-XL (vectors obtained from Dr. Sandra Zinkel, Vanderbilt University, Nashville, TN, USA). For the 3′-UTR-blocking experiments, MDA-MB-231 cells were transfected with 200 nM miScript Target Protectors (Qiagen) designed to block miR-15 family and let-7 family binding sites in the 3′-UTR of BCL-2 and BCL-XL, respectively, and cells were evaluated 24 h later.
HDAC inhibition and cell survival assays
Cells were treated with 10 nM Depsi (Celgene, Summit, NJ, USA), 1 μM SAHA (Selleck Chemicals, Houston, TX, USA), 10 μM 966 (Repligen, Waltham, MA, USA) or vehicle control (DMSO). SAHA and 966 were provided by Dr. Scott Hiebert (Vanderbilt University). To assess proliferation, cells were plated in quadruplicate and MTT assays were performed according to the manufacturer's protocol (Sigma, St Louis, MO, USA; 570 nm). Apoptosis was evaluated by assessing caspase-3 cleavage by western blotting (see below) and by Annexin-V positivity using flow cytometry, as we reported previously.20 For the BCL-2 or BCL-XL overexpression experiments, 24 h after transfecting MDA-MB-231 and A549 cells with empty vector or vectors encoding BCL-2 and/or BCL-XL (described above), cells were administered Depsi or DMSO vehicle control and MTT assays were performed at intervals.
Luciferase assays
Luciferase reporter plasmids were constructed by cloning a 60-mer of the 3′-UTR of BCL-2 or BCL-XL that contained the predicted miR-15 family or let-7 family target sequence, respectively, into the multiple cloning site of pMIR-REPORT (Invitrogen, Carlsbad, CA, USA). MDA-MB-231 cells were transfected with a luciferase reporter, β-galactosidase control plasmid (Invitrogen) and 200 nM miScript Target Protectors (Qiagen). Luciferase and β-galactosidase activity was measured as described previously.21
Western blotting
Cells were harvested 48 h after transfection or at the indicated times following HDACi and were lysed as reported previously.22 Equal amounts of protein were resolved by SDS-PAGE and western blotted as described.22 Antibodies against BCL-2 and BCL-XL (BD Biosciences, Franklin Lakes, NJ, USA), cleaved caspase-3 (Cell Signaling, Danvers, MA, USA), MYC (Millipore, Billerica, MA, USA); β-ACTIN (Sigma), and H3K9K14ac, H3K56ac (histone 3 with lysine 56 acetylation), H4K5ac (histone 4 with lysine 5 acetylation), histone H3 and histone H4 (Abcam, Cambridge, MA, USA) were used.
RNA isolation and qRT-PCR
RNA was isolated, cDNA was generated and TaqMan MicroRNA Assays (Applied Biosciences, Grand Island, NY, USA) and SYBR Green (SA Biosciences, Valencia, CA, USA) were used to perform qRT-PCR, in triplicate, as described previously.21, 23 miRNA and mRNA expression were normalized to small RNA RNU6b levels and β-ACTIN, respectively, and presented as 2−ΔΔCt. Sequences of primers used to detect primary miRNA transcripts of the miR-15a/16-1, miR-195/497 and let-7a/f clusters are were designed such that ~100 bp of flanking sequence extended on either side of the stem loop. Stem-loop sequences of each miRNA were derived from the Sanger miRBase data repository and mapped to the human genome. miR-15a/16-1 – forward, 5′-CAATTACAGTATTTTAAGAG and reverse, 5′-GCATATTACATCAATGTTAT; miR-195/497 – forward, 5′-CCTCTCAGCTTCGTGCTGTC and reverse, 5′-CCTGCTCAGCCCGTCCCTGG; let-7a/f – forward, 5′-CTTATCACTCACACAGGAAAC and reverse, 5′-GACAAGAAGCAAAAGGTTTC. Primer sequences for BCL-2, BCL-XL and β-ACTIN mRNA expression were obtained from the Harvard Medical School Primer Bank (Boston, MA, USA).
Chromatin immunoprecipitation
ChIP was performed as described previously.24 The antibodies used for MYC (N-262) and the isotype controls were from Santa Cruz (Santa Cruz, CA, USA), and antibodies against H3K9K14ac, histone H3 and RNA polymerase II (Ser2 phosphorylated form) were from Abcam. Primer sequences for ChIP of the promoter or upstream regions (negative controls) for the miR-15a/16-1, miR-195/497 and let-7a/f clusters were described previously.12
ENCODE data
Data from the ENCODE project was used.17 The GEO accession numbers of the data sets evaluated are GSM1003607, GSM935441 and GSM822301.
Statistics
Student's t-tests were used to determine statistical significance.
Acknowledgments
We thank the members of the Eischen lab for helpful discussions. These studies were supported by F31CA165728 (CMA), R01CA177786 (CME) and the NCI Cancer Center Support Grant P30CA068485 using Flow Cytometry.
Glossary
- HDAC
histone deacetylase
- miRNA
microRNA
- 3′-UTR
3′-untranslated region
- HDACi
histone deacetylase inhibition
- Depsi
Depsipeptide
- SAHA
suberoylanilide hydroxamic acid
- 966
RGFP966
- DMSO
dimethyl sulfoxide
- PBMC
peripheral blood mononuclear cell
- ChIP
chromatin immunoprecipitation
- RNAPII-p-Ser2
RNA polymerase II Serine 2 phosphorylated
- H3K9K14ac
histone 3 with lysine 9 and 14 acetylation
- H3
histone 3
- H4
histone 4
- IgG
immunoglobulin G
- TSS
transcriptional start site
- ENCODE
encyclopedia of DNA elements
- GEO
gene expression omnibus
- shRNA
short hairpin RNA
- NT
non-targeting
- mRNA
messenger RNA
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- TP
Target Protector
- S.D.
standard deviation
- S.E.M.
standard error of the mean
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by Y Shi
Supplementary Material
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012; 150: 12–27. [DOI] [PubMed] [Google Scholar]
- West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014; 124: 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 2007; 26: 5541–5552. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhao X, Fiskus W, Lin J, Lwin T, Rao R et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-cell lymphomas. Cancer Cell 2012; 22: 506–523. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cho JH, Dimri M, Dimri GP. MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cellular senescence. J Biol Chem 2015; 290: 10555–10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DQ, Pan BZ, Huang JY, Zhang K, Cui SY, De W et al. HDAC 1/4-mediated silencing of microRNA-200b promotes chemoresistance in human lung adenocarcinoma cells. Oncotarget 2014; 5: 3333–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Huang J, Lyu H, Lee CK, Tan J, Wang J et al. Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis 2013; 4: e556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell 2012; 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838. [DOI] [PubMed] [Google Scholar]
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 2007; 67: 2456–2468. [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 2008; 40: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014; 9: 287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima RT, Busacca S, Almeida GM, Gaudino G, Fennell DA, Vasconcelos MH. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer 2011; 47: 163–174. [DOI] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell 2012; 149: 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui TV, Mendell JT. Myc: maestro of microRNAs. Genes Cancer 2010; 1: 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland JF, Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res 2008; 68: 3624–3629. [DOI] [PubMed] [Google Scholar]
- Adams CM, Eischen CM. Inactivation of p53 is insufficient to allow B cells and B-cell lymphomas to survive without Dicer. Cancer Res 2014; 74: 3923–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGirt LY, Adams CM, Baerenwald DA, Zwerner JP, Zic JA, Eischen CM. miR-223 regulates cell growth and targets proto-oncogenes in mycosis fungoides/cutaneous T-cell lymphoma. J Invest Dermatol 2014; 134: 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 1998; 12: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene 2008; 27: 1590–1598. [DOI] [PubMed] [Google Scholar]
- Grieb BC, Gramling MW, Arrate MP, Chen X, Beauparlant SL, Haines DS et al. Oncogenic protein MTBP interacts with MYC to promote tumorigenesis. Cancer Res 2014; 74: 3591–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.