Abstract
Background:
Copy-number gain of the oncostatin-M receptor (OSMR) occurs frequently in cervical squamous cell carcinoma (SCC) and is associated with adverse clinical outcome. We previously showed that OSMR overexpression renders cervical SCC cells more sensitive to the major ligand oncostatin-M (OSM), which increases migration and invasion in vitro. We hypothesised that a major contribution to this phenotype would come from epithelial–mesenchymal transition (EMT).
Methods:
We performed a comprehensive integrated study, involving in vitro cell line studies, in vivo animal models and numerous clinical samples from a variety of anatomical sites.
Results:
In independent sets of cervical, head/neck and lung SCC tissues, OSMR expression levels correlated with multiple EMT-associated phenotypic markers and transcription factors. OSM treatment of OSMR overexpressing cervical SCC cells produced consistent EMT changes and increased tumour sphere formation in suspension culture. In a mouse model, OSMR overexpressing SCC cells treated with OSM showed significant increases in lung colonisation. The biological effects of exogenous OSM were mirrored by highly significant adverse overall survival in cervical SCCs with OSMR overexpression (N=251).
Conclusions:
OSM:OSMR interactions are able to induce EMT, increased cancer stem cell-like properties and enhanced lung colonisation in SCC cells. These changes are likely to contribute to the highly significant adverse outcome associated with OSMR overexpression in cervical SCCs.
Keywords: Cervix, squamous cell carcinoma, oncostatin-M receptor, epithelial-mesenchymal transition, metastasis, STAT3
There are ∼300 000 deaths from cervical carcinoma worldwide each year (Forman et al, 2012). Most cases are squamous cell carcinomas (SCC), which develop from precursor squamous intraepithelial lesions (SILs; Groves and Coleman, 2015). Vaccination against high-risk human papillomavirus (HPV), the necessary cause of cervical SCC, is of no benefit to women who are already persistently infected with the virus (Stanley et al, 2014). Improved understanding of cancer biology can be translated into new treatments for cervical SCC, where novel targeted therapies would be an important advance.
Early studies identified oncogenes of potential significance in cervical SCC, by screening for genes located in regions of frequent copy-number gain (CNG) that showed significant associations between expression levels and copy number (Ng et al, 2007). One of the most common regions of CNG in advanced cervical SCC is on chromosome 5p, where a candidate driver gene is the oncostatin-M receptor (OSMR), a cell surface cytokine receptor that is suitable for antibody-mediated therapeutic inhibition (Caffarel and Coleman, 2014). In our previous work, we observed gain of the genomic locus containing OSMR in 61% of 36 cervical SCC samples using array CGH and also found OSMR gain in 61% of 110 independent cervical SCCs using fluorescence in situ hybridisation (Ng et al, 2007). SCCs with OSMR CNG showed significantly higher OSMR expression than those without CNG (P<0.001; Ng et al, 2007). OSMR overexpression (which we define as levels at least twofold greater than the mean of normal cervix samples) was seen in 50% of 18 non-selected cervical SCCs (Ng et al, 2007). Other groups independently showed chromosome 5p CNG in >60% of advanced cervical carcinomas (Narayan et al, 2007; Scotto et al, 2008), while OSMR was overexpressed at 3.1-fold greater levels in 30 cervical carcinomas compared with 20 cases of normal cervical squamous epithelium (Scotto et al, 2008). Importantly, we found that OSMR CNG was associated with significantly adverse clinical outcome in cervical SCC, with a relative risk of death vs cases without CNG of 3.6 (Ng et al, 2007).
In cervical SCC cells in vitro, OSMR overexpression conferred greater sensitivity to the major ligand oncostatin-M (OSM), which induced increased cell migration and invasion (without affecting cell proliferation), plus an ability to induce angiogenesis (Winder et al, 2011). Expression profiling of OSM-treated cell lines and cervical SCC tissue identified consistent patterns of gene induction following OSM:OSMR interactions, which led to functional analyses of OSM-induced promalignant effects in cervical SCC cells (Winder et al, 2011). We identified that one mechanism by which OSMR confers increased motility and invasiveness to cervical SCC cells is by inducing transglutaminase-2 (tissue transglutaminase), which, among other functions, interacts with cell surface integrin-α5β1 and fibronectin (Caffarel et al, 2013). In the present study, we hypothesised that a further mechanism mediating the promalignant effects of OSMR overexpression in cervical SCC cells is induction of epithelial–mesenchymal transition (EMT), a process linked with invasion and metastasis (Polyak and Weinberg, 2009).
EMT is a dynamic and plastic mechanism that involves complex transcriptional and post-transcriptional cellular changes (Craene and Berx, 2013). These lead to downregulation of epithelial genes and upregulation of mesenchymal markers, orchestrated through EMT-associated transcription factors (EMT-TFs), including Snail, Slug (Snail2), Zeb1 and Zeb2 (Lamouille et al, 2014). EMT is characterised by changes in cell–cell contact, shape, motility and invasiveness and may also be associated with the acquisition of stem cell-like properties (Mani et al, 2008), including the ability to grow as cell spheres in suspension culture. In carcinomas, EMT and cancer stem cell (CSC)-like features are thought to arise through common mechanisms of induction, including those originating in the tumour microenvironment (TME; Korkaya et al, 2011; Jung et al, 2015). We previously identified that OSMR levels in SCCs correlated positively with those of fibronectin and integrin-α5 (Caffarel et al, 2013), both regarded as mesenchymal markers (Ocaña et al, 2012).
We investigated the functional associations between OSMR overexpression and EMT by examining cervical SCC cells in vitro and in preclinical metastasis models, and by analysing multiple clinical sample data sets. Our evidence further supports therapeutic targeting of OSMR in SCCs of the cervix and potentially other anatomical sites.
Materials and Methods
Tissue samples and gene expression profiles
All clinical tissue sample sets studied are summarised in Supplementary Table S1. We measured OSMR expression levels by qRT–PCR in: 13 cervical SCCs and 3 samples of normal ectocervical squamous epithelium (set 1; Scottish HPV Reference Laboratory, Edinburgh); 27 SCCs of the larynx and 9 samples of normal larynx (set 2; CNIO Tumor Bank Core Unit, Madrid, Spain); 18 skin SCCs and 7 samples of normal skin (set 3; CNIO Tumor Bank Core Unit, Madrid, Spain). All three tissue sample sets were studied with Local Research Ethics Committee approval. In addition, we reanalysed our previous microarray gene expression profiling of 32 samples of lesional epithelium microdissected from cervical high-grade SILs (HSILs) and low-grade SILs (LSILs) and 12 samples of normal ectocervical squamous epithelium (set 4; Gene Expression Ommibus Accession Number GSE27678; Ng et al, 2007; Winder et al, 2011; Caffarel et al, 2013).
We also reanalysed published gene expression profiles from: 33 cervical SCCs plus 21 normal cervix (GSE9750, set 5; Scotto et al, 2008); 35 oral SCCs plus 6 normal oral mucosa samples (GSE10121, set 6; Sticht et al, 2008); 31 tongue SCCs plus 26 normal tongue, including 20 matched sample pairs (GSE13601, set 7; Estilo et al, 2009); 13 vulvar SCCs plus 5 normal vulva (GSE38230, set 8; Micci et al, 2013); 15 skin SCCs, 14 actinic keratosis and 13 normal skin, including 13 matched sample pairs (GSE32628, set 9; Hameetman et al, 2013); 5 skin SCCs, 4 actinic keratosis and 6 normal skin (GSE2503, set 10; Nindl et al, 2006); 12 matched sample pairs of skin SCC and normal skin (set 11; Hudson et al, 2010).
Analysis of next-generation sequencing data
We analysed expression levels of OSMR and other EMT genes in the publically available RNA sequencing results of The Cancer Genome Atlas (TCGA). Computational analysis and statistical testing of the next-generation sequencing (NGS) data were conducted using the R statistical programming language. Filtered and log 2 normalised RNA expression data, along with all available clinical data, were downloaded from the GDAC firehose database (run: stddata__2015_06_01) for each gene of interest from the CESC (cervical cancer), HNSCC (head and neck squamous cell carcinoma) and LUSC (lung squamous cell carcinoma) collections (sets 12-14; Supplementary Table S1). Data downloads were performed using the FirebrowseR database access R package.
Correlation testing for associations between expressed genes was performed using the cor.test function in R, to calculate the coefficient of variation (r) and test for significant deviation from no correlation. A list of 16 EMT-related genes (Supplementary Table S2) was derived from published literature (Polyak and Weinberg, 2009; Nieto, 2011; Craene and Berx, 2013; Lamouille et al, 2014). Plotting of the correlation data was performed using the ggplot2 R package. Survival analysis was performed using the survival R package. Kaplan–Meier estimated survival curves were constructed using the clinical data. Statistical testing of differences between survival curves used the G-rho family of tests, as implemented in the survdiff function of the survival package.
In addition, we analysed published NGS data from 24 cervical SCC samples, in which the carcinoma and tumour-infiltrating cells had been separated by flow sorting, followed by RNA sequencing of the two cell populations (set 15; Supplementary Table S1; Punt et al, 2015).
Cells and cell culture
We used the representative OSMR overexpressing cervical SCC cell lines CaSki (HPV16-positive) and SW756 (HPV18-positive), in which OSMR was expressed at 3.4- and 4.5-fold greater levels than in a pool of four independent cultures of basal-type normal cervical squamous cells (Ng et al, 2007). There was no overexpression of OSM in either cell. The cells were obtained from the American Type Culture Collection (ATCC-LGC, Middlesex, UK) and verified by short tandem repeat profiling (Muralidhar et al, 2011). Cell culture and OSM treatment were performed as described (Winder et al, 2011). OSM (R&D Systems, Abingdon, UK) was added at 10 ng ml−1, in accordance with previous studies examining long-term effects of OSM in tissue culture (Winder et al, 2011). OSM secretion into culture medium was measured by sandwich ELISA (human OSM DuoSet ELISA, R&D Systems), according to the manufacturer's instructions.
Sphere forming assay
Cells in monolayer cultures were trypsinised, filtered through a 30-μm filter (Sysmex, Milton Keynes, UK) and resuspended in DMEM medium containing 1% methylcellulose (R&D systems), 20 ng ml−1 EGF and 20 ng ml−1 b-FGF (both PeproTech, London, UK). Cells were then added to non-adhesive 96-well plates (Greiner bio-one Cellstar, Monroe, NC, USA), at 1000 cells per well, and treated every three days with medium containing 10 ng ml−1 OSM, or control medium. The total number of spheres in each well was counted after 10 days and numbers in OSM-treated wells compared with those of control wells. Spheres were then isolated by centrifugation and the RNA extracted.
Real time quantitative PCR and western blotting
Relative mRNA levels were measured by QuantiTect One-Step SYBR-Green qRT–PCR (Qiagen, Crawley, UK) as described (Caffarel et al, 2013), using the primers and conditions listed in Supplementary Table S3. Expression ratios were calculated using the comparative threshold cycle (Ct) method (Pfaffl, 2001) and normalised using three housekeeping genes; ACTB, TBP and HMBS (Vandesompele et al, 2002). Western blotting and densitometry were performed as described (Caffarel et al, 2013), using the primary antibodies listed in Supplementary Table S4. In keeping with our previous work (Muralidhar et al, 2007; Muralidhar et al, 2011; Winder et al, 2011), we defined OSMR overexpression in SCCs as levels twofold or more greater than the mean for corresponding normal cervical squamous epithelium samples. The latter represented primary cultures for cell lines and microdissected epithelium for tissues.
Generation of OSM overexpressing SW756 cells and bioluminescent SW756 cells
SW756 cells were stably transfected with 2 μg of pUNO1-hOSM expression construct (InvivoGen, Toulouse, France), using Lipofectamine LTX with PLUS reagent (Thermo Fisher Scientific, Loughborough, UK) followed by Blasticidin (InvivoGen) selection at 5 μg ml−1. Control transfections were performed simultaneously using 2 μg of empty vector. For in vivo experiments, bioluminescent SW756 cells were generated by stable transfection of pGL4.51 luciferase reporter vector (Promega, Southampton, UK), using Lipofectamine LTX with PLUS reagent followed by G418 (Thermo Fisher Scientific) selection at 1.2 mg ml−1.
Small-molecule inhibition and RNA interference
Cells were treated for 2 h with medium supplemented with TG101348 (Axon Medchem BV, Groningen, The Netherlands), a small-molecule inhibitor of JAK2. This was used at 1 μM, which was the lowest concentration that showed inhibition of STAT3 phosphorylation on western blotting. Negative control cells were treated with equivalent volumes of DMSO vehicle (vol/vol) only.
STAT3 or OSMR were each depleted using four pooled siRNAs (ON-TARGETplus SMARTpool L-003544 or L-008050 respectively, both Dharmacon, Lafayette, CO, USA), compared with cells treated with non-targeting control (NTC) pooled siRNAs (siCONTROL non-targeting siRNA-1, Dharmacon). Individual siRNAs were used at 10 nM, producing 40 nM pools. Cells were transfected at 20–30% confluence using Lipofectamine RNAiMAX (Invitrogen, Paisley, UK), as described (Hanning et al, 2013a; Hanning et al, 2013b). OSM or PBS was added 24 h after transfection and cells treated for 48 h.
Tail vein injection and subcutaneous xenografts
For experimental metastasis experiments, bioluminescent SW756 cells (1 × 106 in 100 μl) were injected into the lateral tail vein of 5–6-week-old female NOD SCID (NOD.CB17-Prkdcscid/NcrCrl) mice (Charles River, Oxford, UK), using 29-gauge insulin needles. Growth of lung tumours was monitored non-invasively using optical whole-body imaging at weekly intervals. Mice were injected with D-luciferin intraperitoneally and imaged 15 min later using the IVIS Spectrum preclinical imaging system (Perkin Elmer, Waltham, MA, USA). Mice were treated three times a week with intraperitoneal 100 μl injections containing 1 μg OSM or PBS only. Animals were killed 11 weeks after tail vein injection and the lungs examined histologically. The tissue samples were fixed in 4% formaldehyde in PBS and embedded in paraffin. Five-micron sections were stained with H&E, observed with a DM LB light microscope (Leica, Wetzlar, Germany) and photographed using a DC500 digital camera (Leica).
To establish subcutaneous xenografts, 5–6-week old female NOD SCID (NOD.CB17-Prkdcscid/NcrCrl) mice were implanted subcutaneously in the flank with 5 × 106 bioluminescent SW756 cells. Mice were treated three times a week with peri-tumoural injections of 100 μl containing 1 μg OSM or PBS only. All mice were palpated three times a week until tumours were detected and thereafter tumours were measured with callipers. Tumour volumes were calculated as (4π/3) × (width/2)2 × (length/2). Animals were killed nine weeks after cell transplantation and the subcutaneous tumours collected. Metastases in visceral organs and brain were evaluated by quantification of bioluminescent signal and by histopathology. To quantify skeletal metastases, visceral organs were removed and the mice assessed by optical imaging.
All mice were maintained in conventional cages within a specific pathogen free animal facility. They were sacrificed by neck dislocation. All animals were treated in strict accordance with the local ethical committee (University of Cambridge Licence Review Committee) and the UK Home Office guidelines.
Statistics
For comparisons between groups we used Student's T test and ANOVA, with post-hoc analysis by the Student–Newman–Keuls' test. Unless otherwise stated, results are expressed as mean±s.e.m. Timecourses for the development of lung metastases in vivo were compared using linear mixed effects modelling, implemented in R via the lme4 package.
Results
OSMR expression associates with EMT markers in cervical and other SCCs
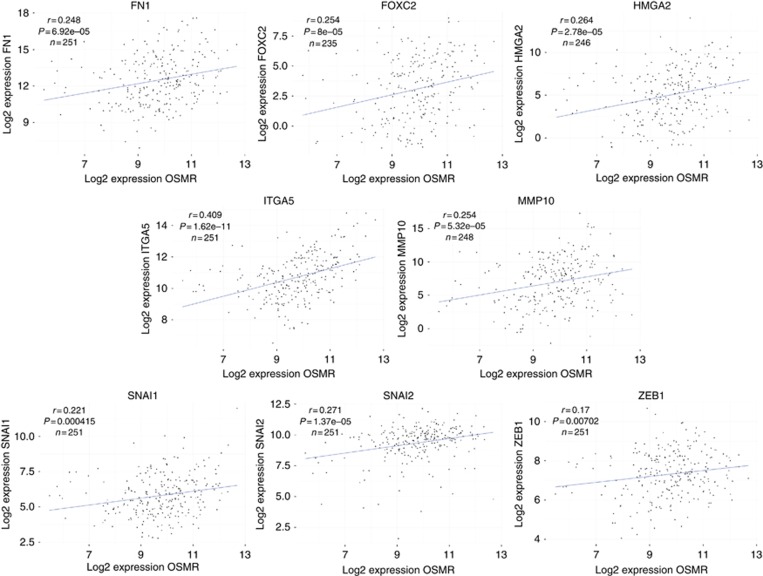
We used TCGA gene expression data for multiple SCC sample sets to test for correlations between OSMR levels and those of 16 genes associated with EMT (Supplementary Table S2). In 251 cervical SCCs (sample set 12), we observed strong and significant (P<0.05) correlations between OSMR and eight of the 16 genes, namely fibronectin (FN1), FOXC2, HMGA2, integrin-α5 (ITGA5), MMP10, Snail (SNAI1), Slug (SNAI2) and ZEB1 (Figure 1). Importantly, similar correlations were also seen in SCCs from other anatomical sites. OSMR levels correlated with 14 of the 16 EMT genes in 514 head/neck SCCs (sample set 13) and with all 16 genes in 492 lung SCCs (sample set 14) (Supplementary Table S2).
Figure 1.
Higher OSMR expression in cervical SCC tissue samples is associated with increased expression of mesenchymal markers. Linear regression analysis of expression levels of EMT-associated genes versus OSMR in cervical SCC samples from TCGA (N=251 maximum).
OSM induces EMT and CSC-like features in OSMR-overexpressing cervical SCC cells in vitro
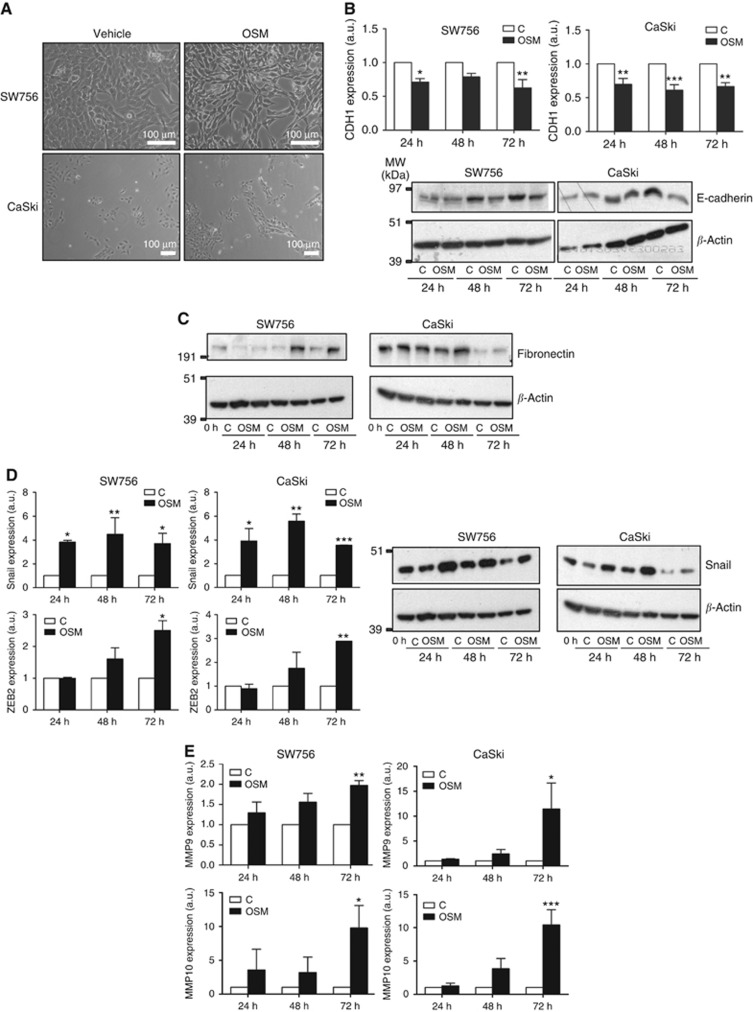
As SCC tissue samples include both epithelial and stromal components, we used cervical SCC cells in vitro (Ng et al, 2007) to investigate the direct associations between OSMR overexpression and EMT. In both CaSki and SW756, treatment with exogenous OSM at 10 ng ml−1 for 72 h induced a more mesenchymal morphology, with cell elongation, reduced cohesion and a more refractile appearance on bright-field microscopy (Figure 2A). Consistent with these changes, the OSM-treated cells showed reduced mRNA and protein expression of the epithelial marker E-cadherin (Figure 2B), together with increased expression of fibronectin (Figure 2C), the EMT-TFs Snail and ZEB2 (Figure 2D), and the matrix metalloproteinases MMP9 and MMP10 (Figure 2E). The mesenchymal marker vimentin was also induced in CaSki cells (Supplementary Figure S1). The effects of OSM could be reduced or abrogated following depletion of OSMR using pooled siRNAs (Supplementary Figure S1). SW756 cells that constitutively overexpressed OSM following stable transfection also showed a more mesenchymal morphology, identical to that of cells treated with exogenous OSM (Supplementary Figure S2A), and also showed increased levels of fibronectin, Snail, ZEB2, MMP9 and MMP10, compared with control-transfected cells (Supplementary Figure S2B).
Figure 2.
OSM induces EMT in OSMR overexpressing cervical SCC cells. (A) Effects of OSM treatment (72 h) on the morphology of OSMR overexpressing cells in monolayer culture. (B–E) Levels of EMT-associated markers in OSMR overexpressing cervical SCC cells treated with OSM for the indicated time points, compared with control treated cells (C). Graphs show transcript levels and blots show protein levels. B, CDH1 (E-cadherin), C, fibronectin, D, EMT-TFs Snail and ZEB2, E, MMP9 and 10. Transcript quantification values are from three independent experiments, whereas the western blots show the data from one representative experiment. The protein loading control was β-actin (lower row of the blots). As the blots in C and D were done on the same membrane, the same loading controls are shown for each. *P<0.05, **P<0.01, ***P<0.001.
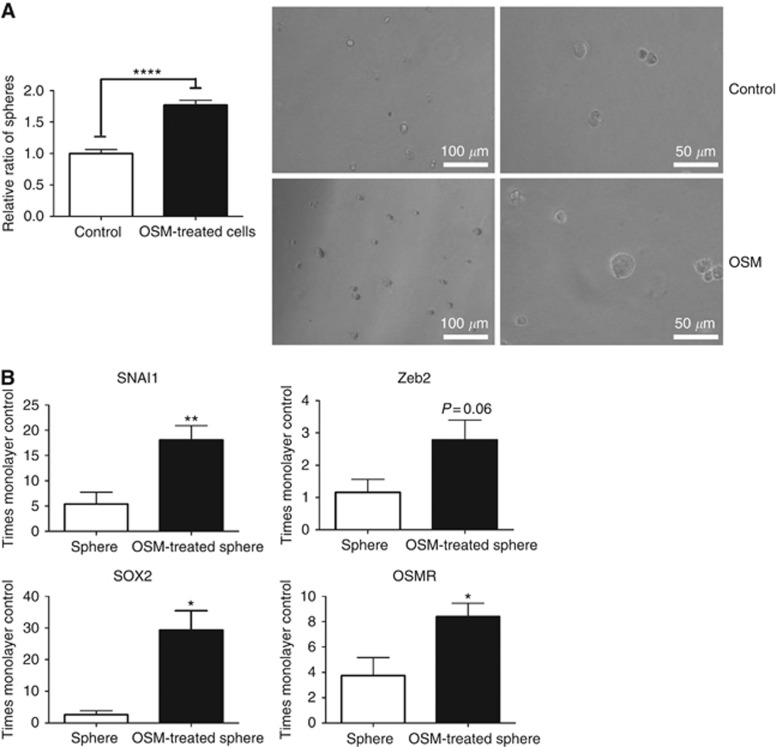
In addition, treatment with exogenous OSM increased the formation of tumour spheres by SW756 cells grown in suspension culture for 10 days (Figure 3A), consistent with increased CSC-like properties (Wang et al, 2013). When compared with untreated control spheres, the OSM-treated spheres showed increased levels of Snail and ZEB2, as well as the pluripotency marker SOX2 and OSMR itself (Figure 3B). The untreated spheres showed increased levels of Snail and OSMR compared with SW756 cells in monolayer culture. Untreated CaSki cells formed spheres very efficiently and there were no additional effects of OSM treatment.
Figure 3.
OSM induction of EMT is associated with increased CSC-like properties. (A) The graph shows the number of SW756-derived spheres in the presence of OSM or vehicle only (control). Representative images of the spheres are shown in the right-hand panel. Data were derived from two independent experiments. (B) Relative gene expression in OSM-treated versus control-treated spheres. All expression values were normalised to those in untreated SW756 cells in monolayer culture. Data were derived from three independent experiments. Key for all panels: *P<0.05, **P<0.01, ****P<0.0001.
EMT induction requires JAK2/STAT3 signalling
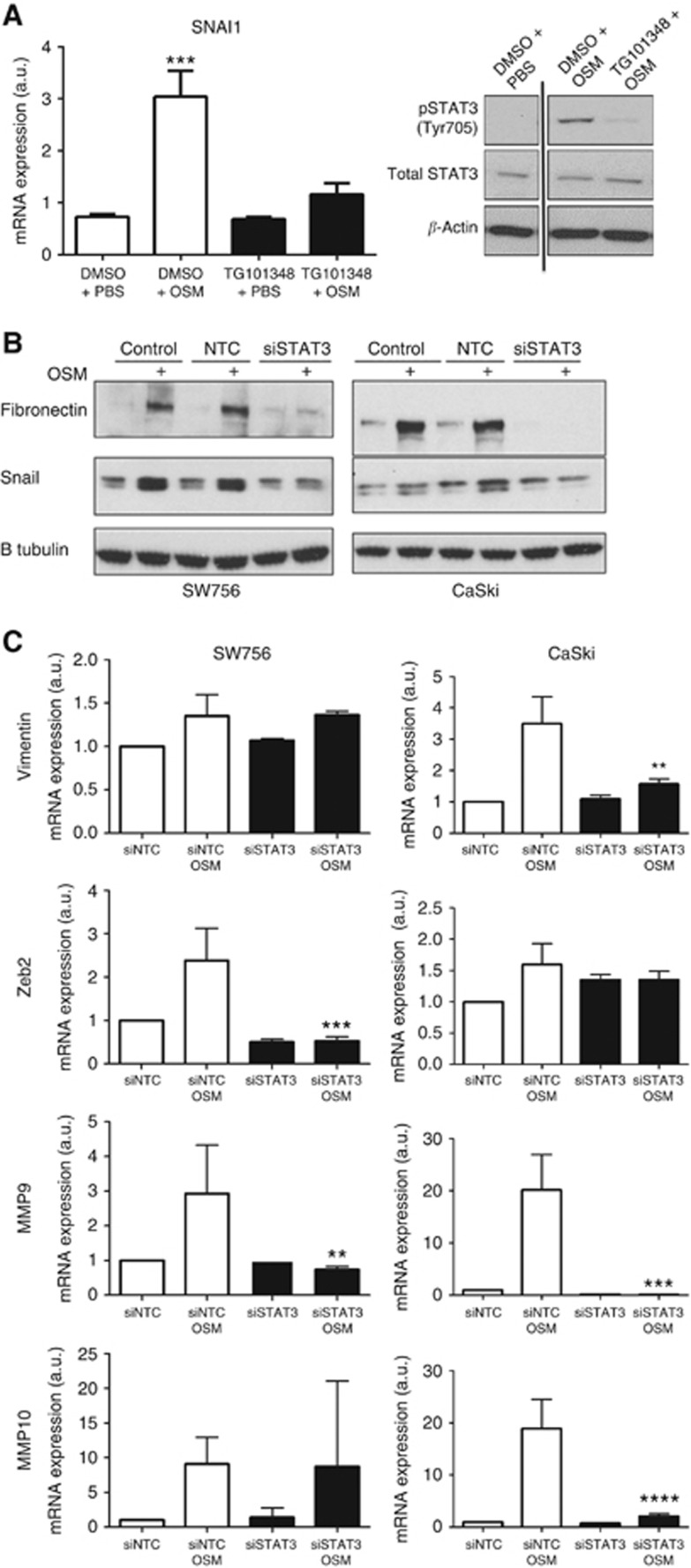
As OSM:OSMR interactions are known to induce STAT3 (Ng et al, 2007), we tested the importance of JAK/STAT3 signalling in the induction of EMT-related genes. To investigate the role of JAK2, cells were treated with OSM for 15 min, followed by assessment of STAT3 phosphorylation by western blotting after 30 min and measurement of Snail transcript levels after 24 h. In this experiment, cells pretreated with the small-molecule JAK2 inhibitor TG101348 for 2 h prior to the addition of OSM showed reduced STAT3 phosphorylation and Snail induction (Figure 4A). In addition, STAT3 depletion using pooled siRNAs (Supplementary Figure S3) inhibited the induction of fibronectin and Snail proteins following OSM treatment for 48 h (both Figure 4B) and also inhibited expression of vimentin, ZEB2, MMP9 and MMP10 in one or both of the OSMR overexpressing cell lines (Figure 4C).
Figure 4.
OSM induction of EMT is JAK2 and STAT3 dependent. (A) Effects of pre-treatment for 2 h with JAK2 inhibitor TG101348 or DMSO only on OSM induction of Snail transcripts (left panel) and STAT3 phosphorylation (right panel) in SW756 cells. The OSM was added for 15 min, followed by assessment of STAT3 phosphorylation after 30 min and Snail transcript levels after 24 h. In the left panel, transcript levels were referenced to those in untreated cells. The data were derived from four independent experiments. In the right panel, β-actin (lowest row) was used as the loading control. (B and C) Effects of STAT3 depletion on OSM induction of fibronectin and Snail proteins (B) and vimentin, ZEB2, MMP9 and MMP10 transcripts (C). In B, β-tubulin (lowest row) was used as the loading control. In C, asterisks refer to the comparison between siSTAT3 OSM cells and siNTC OSM cells. Data were derived from two or three independent experiments. Key for all panels: **P<0.01, ***P<0.001, ****P<0.0001.
OSM increases metastasis of OSMR-overexpressing SW756 cervical SCC cells in vivo
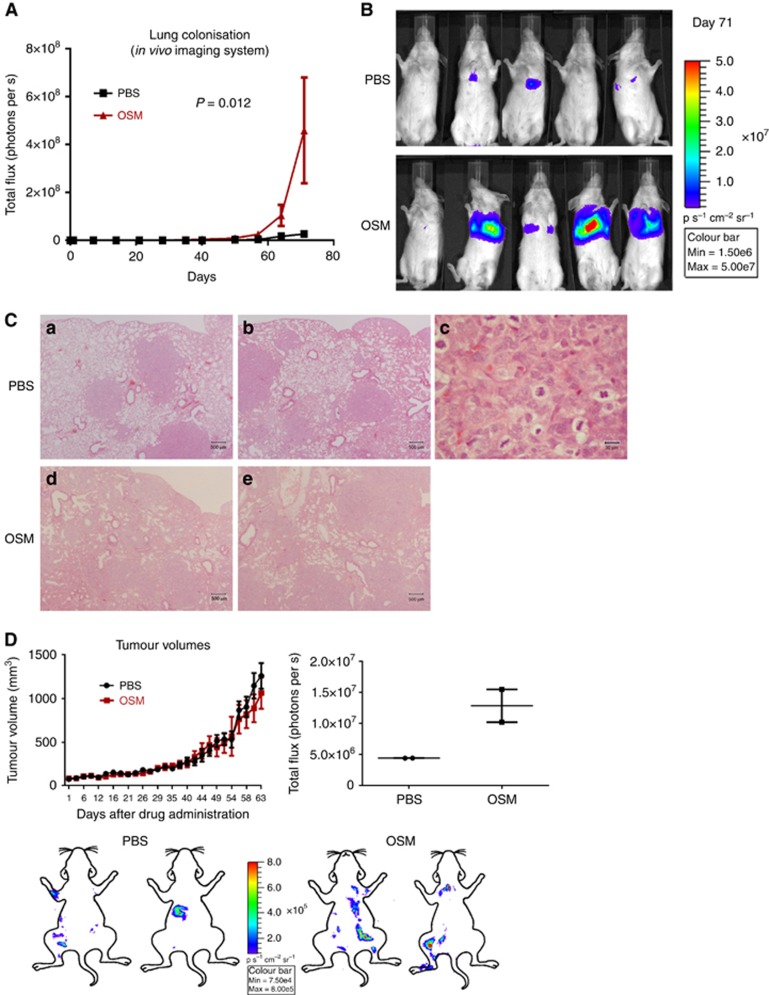
Mice injected intravenously with luciferase-expressing SW756 cells and treated three times per week with intraperitoneal OSM injections showed significantly increased lung metastasis, compared with PBS-treated control animals (Figure 5A and B). There were no histological differences between the lung metastases seen in the two experimental groups (n=5 mice per group; Figure 5C).
Figure 5.
OSM increases lung colonisation by OSMR overexpressing SW756 cervical SCC cells. (A) Time course quantification of the thoracic bioluminescent signal from mice following tail vein injection of SW756 cells and intraperitoneal injection of OSM or PBS (n=5 per group). (B) Luminescence images from the end point of the experiment shown in A. (C) Representative images of lung tissue at the end point of the experiment shown in A. Images a–c are from PBS-treated animals, while images d and e are from OSM-treated animals. Image c is a high-magnification view of a region of image b. (D) The line graph (upper row, left) shows growth of primary subcutaneous xenografts of SW756 cells treated with peri-tumoral injections of OSM or PBS (n=4 per group). The images (lower row) were generated from bioluminescence analysis of mice at the end of the experiment, after removal of the xenografts and visceral organs. The images were overlapped with cartoon outlines to indicate the approximate locations of the skeletal luminescence signals. However, they are not precisely localised, due to variations in the orientation of the mice during optical imaging. The dot plot (upper row, right) shows the relatively greater levels of skeletal luminescence in mice bearing xenografts treated with OSM versus those treated with PBS (n=2 per group).
Subcutaneous xenografts of SW756 cells treated three times per week with peri-tumoural OSM injections showed no difference in growth rates compared with PBS-treated control xenografts (n=4 per group; Figure 5D). There were no macroscopic or histological differences between the primary xenografts from the two experimental groups. However, the OSM-treated group showed more skeletal metastases (n=2 per group; Figure 5D). There was no evidence of metastasis to other sites, including liver and lung.
OSMR is overexpressed in multiple SCCs and associated with worse overall survival
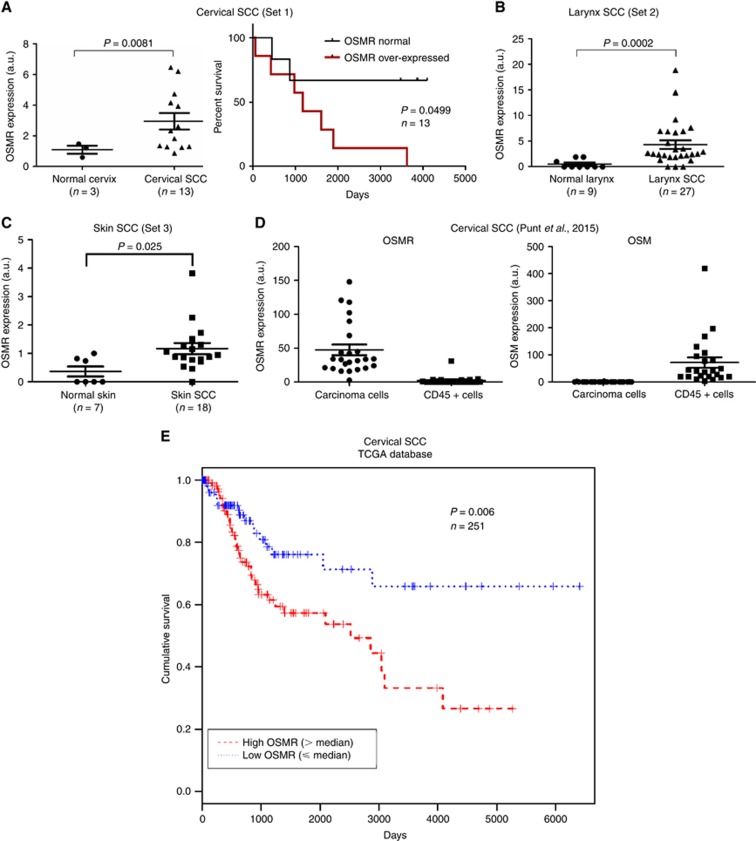
In view of the effects of OSM:OSMR interactions on metastasis in vivo, we investigated the frequency of OSMR overexpression in SCCs, as well as the associations with overall survival. When compared with equivalent normal tissues, OSMR was overexpressed in SCCs from numerous sites, including cervix, head/neck, vulva and skin (sets 1–3, 5–11) (Figure 6A–C; Supplementary Figure S4). In cells flow-sorted from cervical SCCs (set 15; Punt et al, 2015), OSMR was overexpressed by the carcinoma cell fraction only, with very little expression in the CD45+ tumour-infiltrating immune cells (Figure 6D). In contrast, OSM was expressed strongly by the immune cells, with minimal expression in the carcinoma cells (Figure 6D).
Figure 6.
OSMR is overexpressed in SCCs from multiple sites and is associated with adverse overall survival in cervical SCC. (A–C) Levels of OSMR expression in SCC versus normal tissue from cervix (A), larynx (B) and skin (C). The line graphs in A show overall survival in the 13 cervical SCCs in sample set 1, according to OSMR expression levels. (D) Levels of OSMR and OSM in flow-sorted carcinoma and CD45+ infiltrating immune cells from cervical SCCs (sample set 15). (E) Kaplan–Meier curves showing overall survival for cervical SCCs with high vs low expression of OSMR, as determined from TCGA data (sample set 12).
Lower-level increases in OSMR expression were also seen in premalignant squamous lesions. In the cervix, there were gradual increases in expression levels from normal cervix, through LSILs to HSILs (set 4; Supplementary Figure S4B). OSMR overexpression (levels at least twofold greater than the mean of the 12 normal cervix samples) was seen in 3/21 HSILs, 0/11 LSILs and 0/12 normal cervix. In skin tissue, OSMR levels in actinic keratosis, a potentially premalignant condition (Fu and Cockerell, 2003), were intermediate between those of normal skin and cutaneous SCC (sets 9 and 10; Supplementary Figure S4F and S4G).
We next examined the associations between OSMR expression levels and clinical outcome, using cervical SCCs from TCGA. In this analysis, we were unable to define OSMR overexpression by reference to normal cervix samples, which were not included in the data set. Accordingly, we divided the SCCs into two groups, those with OSMR expression levels above the median for the data set (n=126) and those with expression levels at or below the median (n=125). Importantly, the SCCs with high OSMR expression showed a highly significant adverse overall survival, compared with those with low expression (P=0.006; Figure 6E).
This finding was confirmed in a small independent set of 13 cervical SCCs (set 1). Of these, seven (54%) overexpressed OSMR, defined as levels at least twofold greater than the mean of the three normal cervix samples in the data set. These seven SCCs showed significantly worse overall survival than the six SCCs without OSMR overexpression (Figure 6A). None of the three normal cervix samples in this data set showed OSMR overexpression.
Discussion
This is the first study to show that OSMR overexpression is associated with EMT features and adverse clinical outcome in SCC samples and that OSM can enhance EMT, CSC-like properties and metastasis in SCC cells. We identified that OSMR is frequently overexpressed in SCCs from multiple anatomical sites and provides the malignant cells with increased responsiveness to the major ligand OSM. The frequency of OSMR overexpression observed in the cervical SCCs of sample set 1 (54%) was similar to our previous observations in an independent data set (50%) (Ng et al, 2007). In total, OSMR overexpression was present in 52% of the cervical SCCs we examined (16/31), consistent with the observed frequency of OSMR CNG (61%) and our evidence of significant associations between gene copy number and expression (Ng et al, 2007; Winder et al, 2011).
In cervical SCC samples, OSMR was almost exclusively expressed by the carcinoma cells, while OSM was almost exclusively expressed by the infiltrating immune cells. The latter observation is consistent with evidence that OSM may be produced by a wide range of cells in the TME, including macrophages, dendritic cells and T-lymphocytes (Tanaka and Miyajima, 2003). We previously demonstrated that OSM stimulation of OSMR overexpressing cervical SCC cells leads to increased invasion and migration in vitro (Winder et al, 2011). We now show that such changes are accompanied by increased metastatic ability in vivo and that EMT is an important association of these malignant features.
Our present and previous (Caffarel et al, 2013) in vitro data demonstrate that exogenous OSM rapidly induces a consistent pattern of EMT-like changes in OSMR overexpressing cervical SCC cells, with downregulation of E-cadherin and upregulation of several mesenchymal markers. The latter include MMP9 and MMP10, which are key mediators of ECM destruction during carcinoma cell invasion (Nistico et al, 2012). All seven of the genes that we tested and showed to be upregulated in the SCC cells (fibronectin, MMP9, MMP10, Snail, vimentin, Zeb2 and integrin-α5 (Caffarel et al, 2013)), also showed significant positive associations with OSMR levels in one or more of the TCGA SCC tissue data sets (Figure 1; Supplementary Table S2). We concluded that the correlations between OSMR and multiple mesenchymal markers in clinical samples are likely to include direct effects on the carcinoma cells. However, contributions to these observations from cells of the TME cannot be excluded.
The effects of OSM treatment of OSMR overexpressing SCC cells also included increased CSC-like features, as measured using suspension cultures. The spheres formed by OSM-treated cells showed increased expression of mesenchymal genes, as well as the pluripotency marker Sox2, similar to observations in breast adenocarcinoma cells (West et al, 2014). Interestingly, OSMR was overexpressed in untreated cell spheres, compared with cells in monolayer culture, and was further induced by OSM. Altogether, these data indicate that OSMR is directly linked with CSC-like properties in cervical SCC cells. As EMT is a dynamic, reversible process (Thiery et al, 2009), reversion of OSM-exposed SCC cells to a more stem-like state would support clonal expansion and the development of tumour heterogeneity, within both primary lesions and metastatic deposits. It might also be expected that high levels of OSMR and/or OSM in SCCs would confer resistance to chemotherapy and radiotherapy (Nieto, 2011). Indeed, this effect may have contributed to the association between OSMR levels and adverse clinical outcome that we observed in multiple cervical SCC data sets, including TCGA samples.
The enhanced responsiveness to OSM in OSMR overexpressing cervical SCC cells is associated with induction of multiple signalling pathways, including phosphorylation of STAT3 (Ng et al, 2007). Our present data indicate that STAT3 is important in inducing mesenchymal markers and the major EMT-TF Snail. One mediator of STAT3 phosphorylation is JAK2, as indicated by specific small-molecule inhibition. It is likely that other signalling pathways induced by OSM in OSMR overexpressing SCC cells are also of functional significance. In breast adenocarcinoma, MAPK signalling is associated with downregulation of oestrogen receptor (an epithelial marker; West et al, 2012) and PI3K signalling with increased CSC-like properties (West et al, 2014). Detailed dissection of the roles of signalling pathways in the multiple promalignant effects of OSM in cervical SCC cells is now required.
There are likely to be multiple effectors of the transcriptional changes characteristic of EMT. Our current data, from in vitro experiments and clinical samples, indicate a role for the EMT-TFs Snail, Slug, ZEB1 and ZEB2. It will now be important to investigate the mechanisms responsible for the induction of these EMT-TFs, including similarities and differences with other cell types. For example, the OSMR target transglutaminase-2 (Caffarel et al, 2013) has been linked to EMT in ovarian malignancy (Shao et al, 2009) and may also have a role in SCCs. In breast adenocarcinoma cells OSM induces Lin28, which represses Let7 microRNAs and so upregulates protein-coding gene targets including HMGA2 (Guo et al, 2013). The latter is a non-histone chromatin protein that acts as an architectural transcription factor and is known to bind the SNAI1 promoter (Thuault et al, 2008). Whether similar mechanisms apply in tumours of squamous histogenesis remains to be determined, particularly given the lineage specificity of microRNA profiles (Lu et al, 2005). However, we note the significant correlations between levels of OSMR and HMGA2 across the TCGA SCCs (Figure 1 and Supplementary Table S2). There may also be differences in mechanisms of EMT induction in SCCs at different sites. For example, OSMR levels correlated with some EMT markers (e.g., Twist2 and MMP2) in head/neck and lung SCCs but not cervical SCCs, while the high-risk HPV oncogenes E6 and E7 that drive cervical and some head/neck SCCs have been linked with EMT induction (Qureshi et al, 2015).
Our in vivo data indicate that the metastatic capacity of OSMR overexpressing cervical SCC cells increases following exposure to OSM, as assessed by a lung colonisation assay. This demonstrates an important role for OSMR early in the metastatic process, for example via increased extravasation and infiltration of secondary sites (Nguyen et al, 2009). We observed no histological differences between the lung metastases at the end of the experiment, consistent with the reversibility of EMT, via the reciprocal process of mesenchymal–epithelial transition (Brabletz, 2012). In mice bearing cervical SCC xenografts, the increased skeletal metastases may have been due to several consequences of OSM:OSMR interactions, including enhanced cell motility/detachment, invasion, blood vessel intravasation and extravasation/colonization (Nguyen et al, 2009; Caffarel and Coleman, 2014). Our observations support the clinical evidence that OSMR overexpression in cervical SCC is associated with significantly worse overall survival, as well as with gene expression profiles indicating EMT. These data resonate with those in breast adenocarcinoma, where high OSMR expression correlates with shorter overall survival and recurrence free survival, together with features of EMT (West et al, 2014). Recently, evidence for an association between OSMR overexpression and adverse outcome has also been identified in a non-epithelial malignancy, glioblastoma multiforme (GBM), again with evidence of an association with EMT (Natesh et al, 2015).
Our data have indicated an important mechanism by which OSMR overexpression provides a selective advantage in cervical SCCs. Cells capable of greater migration/invasion are more likely to access conditions of improved oxygenation and nutrient availability and would therefore be provided with a competitive advantage in mixed cell populations. Induction of EMT/CSC-like properties increases metastasis and therapeutic resistance (Polyak and Weinberg, 2009), both of which are likely to contribute to the adverse clinical outcome associated with OSMR overexpression in cervical SCC. Together, our findings further strengthen the case for inhibiting OSM:OSMR interactions as a targeted anti-cancer therapy (Caffarel and Coleman, 2014). The observation that OSMR is commonly overexpressed in a range of SCCs, as well as other carcinomas and GBMs, indicate that effective targeting of this promalignant pathway may produce clinical benefits in a wide range of common cancers.
Acknowledgments
This work was supported by Cancer Research UK (Programme Grant A13080). We thank Professor Tony Green, Department of Haematology, Cambridge University for providing TG101348; Dr Alison Taylor for help with tail vein injections and Mrs Helen Skelton for histology assistance.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Brabletz T (2012) To differentiate or not—routes towards metastasis. Nat Rev Cancer 12: 425–436. [DOI] [PubMed] [Google Scholar]
- Caffarel MM, Chattopadhyay A, Araujo AM, Bauer J, Scarpini CG, Coleman N (2013) Tissue transglutaminase mediates the pro-malignant effects of oncostatin M receptor over-expression in cervical squamous cell carcinoma. J Pathol 231: 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarel MM, Coleman N (2014) Oncostatin M receptor is a novel therapeutic target in cervical squamous cell carcinoma. J Pathol 232: 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craene BD, Berx G (2013) Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 13: 97–110. [DOI] [PubMed] [Google Scholar]
- Estilo CL, Oc P, Talbot S, Socci ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y, Boyle JO, Kraus DH, Patel S, Shaha AR, Wong RJ, Huryn JM, Shah JP, Singh B (2009) Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer 9: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, Vignat J, Ferlay J, Bray F, Plummer M, Franceschi S (2012) Global burden of human papillomavirus and related diseases. Vaccine 30(Suppl 5): F12–F23. [DOI] [PubMed] [Google Scholar]
- Fu W, Cockerell CJ (2003) The actinic (solar) keratosis: a 21st-century perspective. Arch Dermatol 139: 66–70. [DOI] [PubMed] [Google Scholar]
- Groves IJ, Coleman N (2015) Pathogenesis of human papillomavirus-associated mucosal disease. J Pathol 235: 527–538. [DOI] [PubMed] [Google Scholar]
- Guo L, Chen C, Shi M, Wang F, Chen X, Diao D, Hu M, Yu M, Qian L, Guo N (2013) Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene 32: 5272–5282. [DOI] [PubMed] [Google Scholar]
- Hameetman L, Commandeur S, Bavinck JN, Wisgerhof HC, de Gruijl FR, Willemze R, Mullenders L, Tensen CP, Vrieling H (2013) Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer 13: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanning JE, Groves IJ, Pett MR, Coleman N (2013. a) Depletion of polycistronic transcripts using short interfering RNAs: cDNA synthesis method affects levels of non-targeted genes determined by quantitative PCR. Virol J 10: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanning JE, Saini HK, Murray MJ, van Dongen S, Davis MP, Barker EM, Ward DM, Scarpini CG, Enright AJ, Pett MR, Coleman N (2013. b) Lack of correlation between predicted and actual off-target effects of short-interfering RNAs targeting the human papillomavirus type 16 E7 oncogene. Br J Cancer 108: 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson LG, Gale JM, Padilla RS, Pickett G, Alexander BE, Wang J, Kusewitt DF (2010) Microarray analysis of cutaneous squamous cell carcinomas reveals enhanced expression of epidermal differentiation complex genes. Mol Carcinog 49: 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HY, Fattet L, Yang J (2015) Molecular pathways: linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin Cancer Res 21: 962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H, Liu S, Wicha MS (2011) Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest 121: 3804–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R (2014) Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micci F, Panagopoulos I, Haugom L, Dahlback HS, Pretorius ME, Davidson B, Abeler VM, Trope CG, Danielsen HE, Heim S (2013) Genomic aberration patterns and expression profiles of squamous cell carcinomas of the vulva. Genes Chromosomes Cancer 52: 551–563. [DOI] [PubMed] [Google Scholar]
- Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, Barbosa-Morais NL, Mukherjee G, Thorne NP, Roberts I, Pett MR, Coleman N (2007) Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol 212: 368–377. [DOI] [PubMed] [Google Scholar]
- Muralidhar B, Winder D, Murray M, Palmer R, Barbosa-Morais N, Saini H, Roberts I, Pett M, Coleman N (2011) Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J Pathol 224: 496–507. [DOI] [PubMed] [Google Scholar]
- Narayan G, Bourdon V, Chaganti S, Arias-Pulido H, Nandula SV, Rao PH, Gissmann L, Durst M, Schneider A, Pothuri B, Mansukhani M, Basso K, Chaganti RS, Murty VV (2007) Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer 46: 373–384. [DOI] [PubMed] [Google Scholar]
- Natesh K, Bhosale D, Desai A, Chandrika G, Pujari R, Jagtap J, Chugh A, Ranade D, Shastry P (2015) Oncostatin-M differentially regulates mesenchymal and proneural signature genes in gliomas via STAT3 signaling. Neoplasia 17: 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng G, Winder D, Muralidhar B, Gooding E, Roberts I, Pett M, Mukherjee G, Huang J, Coleman N (2007) Gain and overexpression of the oncostatin M receptor occur frequently in cervical squamous cell carcinoma and are associated with adverse clinical outcome. J Pathol 212: 325–334. [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, Massague J (2009) Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 9: 274–284. [DOI] [PubMed] [Google Scholar]
- Nieto MA (2011) The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol 27: 347–376. [DOI] [PubMed] [Google Scholar]
- Nindl I, Dang C, Forschner T, Kuban RJ, Meyer T, Sterry W, Stockfleth E (2006) Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol Cancer 5: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistico P, Bissell MJ, Radisky DC (2012) Epithelial-mesenchymal transition: general principles and pathological relevance with special emphasis on the role of matrix metalloproteinases. Cold Spring Harb Perspect Biol 4: a011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocaña OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA (2012) Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22: 709–724. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9: 265–273. [DOI] [PubMed] [Google Scholar]
- Punt S, Corver WE, van der Zeeuw SA, Kielbasa SM, Osse EM, Buermans HP, de Kroon CD, Jordanova ES, Gorter A (2015) Whole-transcriptome analysis of flow-sorted cervical cancer samples reveals that B cell expressed TCL1A is correlated with improved survival. Oncotarget 6: 38681–38694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi R, Arora H, Rizvi MA (2015) EMT in cervical cancer: its role in tumour progression and response to therapy. Cancer Lett 356: 321–331. [DOI] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Subramaniyam S, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Schneider A, Arias-Pulido H, Murty VV (2008) Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer 7: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Cao L, Shen C, Satpathy M, Chelladurai B, Bigsby RM, Nakshatri H, Matei D (2009) Epithelial-to-mesenchymal transition and ovarian tumor progression induced by tissue transglutaminase. Cancer Res 69: 9192–9201. [DOI] [PubMed] [Google Scholar]
- Stanley MA, Sudenga SL, Giuliano AR (2014) Alternative dosage schedules with HPV virus-like particle vaccines. Expert Rev Vaccines 13: 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticht C, Freier K, Knopfle K, Flechtenmacher C, Pungs S, Hofele C, Hahn M, Joos S, Lichter P (2008) Activation of MAP kinase signaling through ERK5 but not ERK1 expression is associated with lymph node metastases in oral squamous cell carcinoma (OSCC). Neoplasia 10: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Miyajima A (2003) Oncostatin M, a multifunctional cytokine. Rev Physiol Biochem Pharmacol 149: 39–52. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890. [DOI] [PubMed] [Google Scholar]
- Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A (2008) HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem 283: 33437–33446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.0031–research0034.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Bailey JM, Rovira M, Leach SD (2013) Sphere-forming assays for assessment of benign and malignant pancreatic stem cells. Methods Mol Biol 980: 281–290. [DOI] [PubMed] [Google Scholar]
- West NR, Murphy LC, Watson PH (2012) Oncostatin M suppresses oestrogen receptor-alpha expression and is associated with poor outcome in human breast cancer. Endocr Relat Cancer 19: 181–195. [DOI] [PubMed] [Google Scholar]
- West NR, Murray JI, Watson PH (2014) Oncostatin-M promotes phenotypic changes associated with mesenchymal and stem cell-like differentiation in breast cancer. Oncogene 33: 1485–1494. [DOI] [PubMed] [Google Scholar]
- Winder DM, Chattopadhyay A, Muralidhar B, Bauer J, English WR, Zhang X, Karagavriilidou K, Roberts I, Pett MR, Murphy G, Coleman N (2011) Overexpression of the oncostatin M receptor in cervical squamous cell carcinoma cells is associated with a pro-angiogenic phenotype and increased cell motility and invasiveness. J Pathol 225: 448–462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.