Abstract
Objectives
Various terms and definitions are used to describe personalised approaches to medicine and healthcare, but in ambiguous and inconsistent ways. They mostly have been defined in a top-down manner. However, actual practices might take different paths. Here, we aimed to provide a ‘practice-based’ perspective on the debate by analysing the content of ‘personalised’ practices published in the literature.
Methods
The search in PubMed and EMBASE (April 2014) using the terms frequently used for personalised approaches resulted in 5333 records. 2 independent researchers used different strategies for screening, resulting in 157 articles describing 88 ‘personalised’ practices that were implemented/presented on at least 1 individual/patient case. The content analysis was grounded on these data and did not have a priori analytical frameworks.
Results
‘Personalised medicine/healthcare’ can be a commodity in the healthcare market, a way how health services are provided, or a keyword for emerging applications. It can help individuals/patients to gain control of their health, health professionals to provide better services, healthcare organisations to increase effectiveness and efficiency, or national health systems to increase performance. Country examples indicated that for integration of practices into health services, attitude towards innovations and health system and policy context is important. Categorisation based on the terms or the technologies used, if any, was not possible.
Conclusions
This study is the first to provide a comprehensive content analysis of the ‘personalised’ practices in the literature. Unlike the top-down definitions, our findings highlighted not the technologies but real-life issues faced by the practices. ‘Personalised medicine’ and ‘personalised healthcare’ can be differentiated by using the former for specific tools available and the latter for health services with a holistic approach, implemented in certain contexts. To realise integration of ‘personalised medicine/healthcare’ into real life, science, technology, health policy and practice, and society domains must work together.
Keywords: QUALITATIVE RESEARCH, BIOTECHNOLOGY & BIOINFORMATICS, integration of innovations into health care, personalized medicine / personalized health care
Strengths and limitations of this study.
This is the first report that investigates personalised medicine and the personalised healthcare field from a bottom–up view, as opposed to the conventional top-down approach where opinion leaders and committees describe what the field is about and in which direction it should proceed.
The practices were identified with a systematic search and inclusion strategy within the scientific literature, where two researchers worked first independently, then reached a consensus.
A special combination of methods, which included triangulation among others, was designed and used at every step of the analysis due to the uniqueness of the research aim and heterogeneity of the practices presented in the literature as ‘personalised’ approaches.
A limitation is that the research was carried out in the scientific literature alone without inclusion of information from websites of the ‘personalised’ practices.
Owing to limitation of data on integration of the identified practices into health services, the issue of integration could not be elaborated on all practices; however, only three prominent and contrasting country examples were used to demonstrate some of the integration issues.
Introduction
A trend towards ‘personalisation’ in medicine and healthcare has become increasingly prominent in the past 10–15 years. It has been perceived and described from different angles by various opinion leaders, institutions and stakeholder organisations. The terms used include ‘personalized medicine’, ‘personalized health care (healthcare)’, ‘P4 Medicine—Predictive, Personalized, Preventive/Preventive, and Participatory Medicine’, ‘individualized medicine’, ‘precision medicine’ and ‘systems (bio)medicine’.
These terms are heavily used in the scientific and grey literature. However, until now, there has been no consensus on their precise meaning and how they are to be distinguished. The situation is complicated by the ambiguous and inconsistent use of terminology. For example, in some cases, terms are being used interchangeably (eg, ‘individualized medicine’ and ‘personalized medicine’ at Medical Subject Headings of PubMed),1 whereas in others each is used to describe different concepts.2 Different individuals, research groups or manuscripts seem to have a preferred term and working definition for it. Moreover, many publications seem to use ‘personalised medicine’ as a buzzword and/or a ‘catch-all’ term, resulting in a steep increase in its appearance in the literature.3 4 These create chaos within terminology in the literature. Within the present article, the abbreviation PM/PHC will be used to address the wide scope of personalised medicine, personalised healthcare and others.
Problems with the terminology have been the subject of several publications,2–10 the most recent of them being a comprehensive overview by Pokorska-Bocci et al.3 So far, influential definitions and descriptions of the field have mainly been launched by institutions,11–14 organisations representing major stakeholder groups such as industry and academia,15–17 and respected opinion leaders.18–23 The body of literature that addressed or discussed the terminology has mostly referred to these influential ‘top-down’ definitions. There is an implicit assumption that there is such a thing as a best definition or classification of PM/PHC and that the practices in this field will tend to develop accordingly. However, as the studies in other advanced technological domains have shown,24 in reality, actual practices in healthcare might take their own path, thus ‘enacting’ in their practices implicit definitions of what is at stake in their field. These definitions then determine what direction the field is heading towards.
The problem with the definitions and terminology is not just a linguistic one. In the broader context, it is very likely to be connected with the issue of unexpectedly slow integration of personalised practices in real life. PM/PHC has been on the agenda of scientific fora for over a decade. Personalised approaches were proposed to be solutions to the main problems of today's health systems, including prevention and treatment of chronic complex diseases, responsible for the main burden of disease globally.19 23 25 26 However, PM/PHC has not fulfilled the expectations so far. With the exception of cancer-related programmes in some countries, diffusion of ‘personalised’ practices in regular health services has been limited. Can the underlying assumptions and origins of the top-down definitions be contributing to this problem? If so, can the practice side of the coin give us clues about what matters for integration of personalised approaches in healthcare?
Inspired by these ideas, we decided to look at the literature for the PM/PHC practices that use the various terms listed above. We assumed that these practices actually reflect their implicit views on PM/PHC and their analysis may provide better insight into what is happening on the practice side of the field. Thus, this study aims to start developing an understanding of how the field of PM/PHC can be defined from a ‘practice-based’ perspective. This can make a valuable contribution to the debate about the terms and definitions, and help see how the field is actually evolving. We will then look at what the findings mean for integration of PM/PHC into real life. In the discussion, we will compare the implicit notions on PM/PHC, as enacted in actual practices, with notions underlying the ‘top-down’ definitions, and propose a ‘practice-based’ look at the healthcare integration problem in the field and possible solutions.
Methods
Approach
In order to approach the field from a ‘bottom-up’ or ‘practice-based’ perspective and avoid the influence of the top-down definitions, we needed to be careful about common assumptions seen in ‘top-down’ definitions. We identified two of them as important.
The first one is linked to the drivers of the personalised vision. Two drivers seem to have a prominent place in the background of top-down definitions: (1) the general societal trend towards personalisation and (2) the scientific discoveries and technological developments in various fields, mainly genome-based and information/communication technologies (ICT).2 The top-down definitions seem to be influenced particularly by the second driver. Most of them tend to define the field from the perspective of a certain sector or discipline, that is, via the use of certain interventions (drugs, etc) or technologies (genomics, ICT, etc).1 11 12 15 18 19 In this work, we took a sector-independent, discipline-independent and technology-independent stand. This can help us identify new emerging innovative approaches that use tools and disciplines that are not traditionally covered in top-down descriptions of the personalised approaches.
The second assumption we avoided was thinking in silos of the ‘pipeline’ approach to innovations in healthcare. In the top-down approaches, innovative practices or interventions are often perceived to be an end result of the research and (technological) development process. The whole process is seen as stages, for instance basic research, applied research, development, pilot and implementation, which are assumed to follow one another in a linear way. However, in real life, neither scientific advancements nor technologies get into health practice in a predictable pattern or ‘pipeline’.27 Innovations stemming from actual practices might not necessarily follow those in a structured way and the borders between these stages are not always clear.24
Systematic search and selection strategy
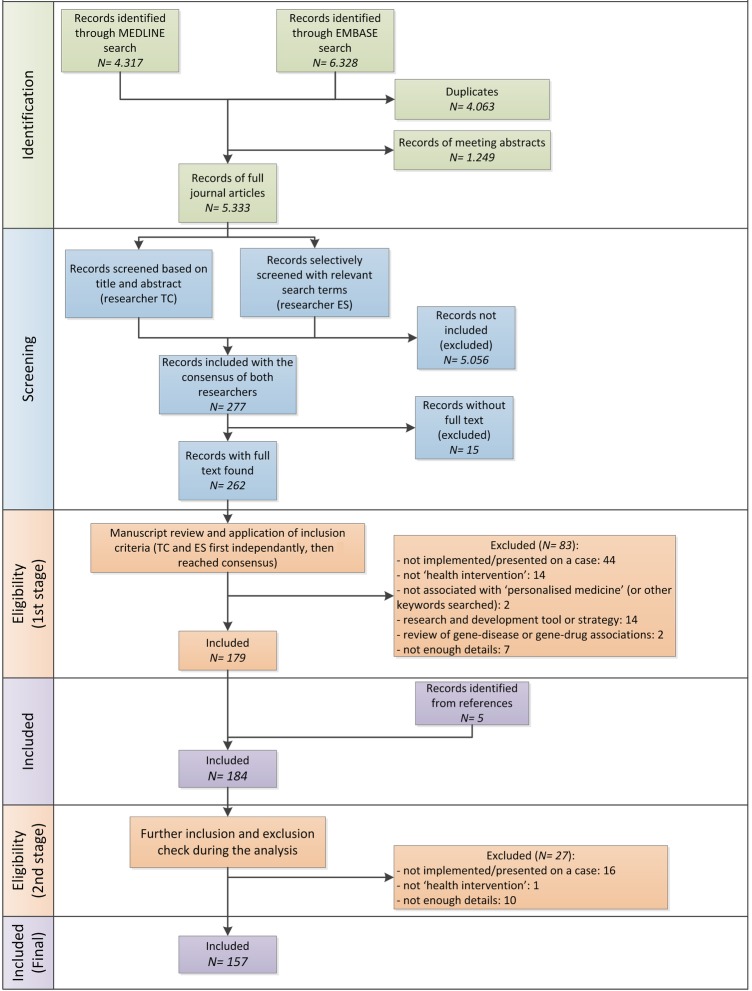
In order to identify the practices, we used a systematic search and selection strategy summarised in figure 1. It is based on the PRISMA flow diagram,28 modified according to the purpose of our study. For full details of the process, refer to online supplementary material 1.
Figure 1.
Flow diagram of data collection process.
bmjopen-2015-010243supp1.pdf (60.1KB, pdf)
Information sources and search strategy
PubMed (via PubMed.gov on 1 April 2014) and EMBASE (via OVID on 14 April 2014) were chosen for the search as they cover the highest number of journals related to ‘personalised medicine’.
The following terms were searched in the title or abstract of the manuscripts: personalized health care (healthcare), personalized medicine, P4 medicine, precision medicine, stratified medicine, systems (bio)medicine, individualized medicine and their UK English versions (personalised and individualised). The search was limited to English articles, with no time limitations.
The search within PubMed resulted in 4.317 records and 6.328 in EMBASE, all of which were imported to EndNote. Duplicates and meeting/conference abstracts were removed. This resulted in 5.333 records.
The search was restricted to scientific publications only and did not include websites, reports and other grey literature sources, since it is not always possible to assess the validity of claims made about the practices in those media.
Screening and selection process
The focus of this study is practices presented in relation to PM/PHC. Since ‘health practice’ or ‘medical practice’ do not have a formal and/or operational definition, we used the concept of ‘health intervention’, as “…an activity performed for, with or on behalf of a person or a population whose purpose is to improve, assess or modify health, functioning or health conditions.”29 Thus, our inclusion criteria identified the articles describing a health intervention that is developed (designed) and/or implemented/presented in association with one of the terms used to describe PM/PHC (see the search terms).
More precisely, we searched for health intervention/practices that (1) were implemented on at least one real-life case, applied to the patient/individual or used her/his data (ie, a case, series of cases, feasibility studies, pilots, trials, reports on products/services on the market, etc) and (2) were provided with enough details in the article(s). The stage of research, development and implementation, the scientific evidence on the practice, how well the practice is established or the technological components were not included in the analysis.
The selection was done independently by two researchers (TC and ES) on two occasions. First, during the initial screening, resulting in 277 articles, and second, during the eligibility screening. Within the screening phase, independent researchers used two different strategies for triangulation. The results of both researchers were compared and differences were discussed until consensus was reached. At this stage, 179 articles were included. On inclusion of some of the articles identified from the references and exclusion of those not providing enough information, the final list of 157 articles was formed. The reasons for exclusion are summarised in figure 1.
Identification of practices from the articles
Practices were extracted from the articles; different articles describing the same practice were grouped and a list of the practices was formed. From this stage onwards, practices (not the articles) became the unit of analysis in this study. Some further group clustering was performed, based on the similarities of practices. For example, various drug-genetic test pairs could be grouped under ‘pharmacogenetics’, since no apparent differences were expected during the analysis and the available data in the articles were also presented to support grouping. Eventually, nine groups of practices were formed (treated as one practice in the rest of this article) in addition to 79 individual ones, summing up to 88 practices (see table S1 in online supplementary material 2). The ‘P-numbers’ in online supplementary table S1 are used as the identifier of each practice in the rest of the article and referred with a ‘P-’ in front of it in brackets. For example, (P−1) refers to ‘group: pharmacogenetics (drug—gene)’. Details of the group of practices are presented in online supplementary table S2.
bmjopen-2015-010243supp2.pdf (491.2KB, pdf)
Content analysis
The author (TC) read the articles, investigated the practices from different perspectives, identified their distinctive properties and discussed various categorisation possibilities with the coauthors (ES, AK, FF). We decided to start with the axis of ‘application form and context’, since many articles provided ample information on that topic (axis-1). While comparing similarities and contrasting differences among the practices within and across categories, the categorisation was refined and redefined. This resulted in reorganisation of the practices under the categories. This took several rounds until the description of the categories matched the content of the practices included and the categories became mutually exclusive.
The second area we decided to investigate focused on ‘who is served by this practice?’. This involves ‘users’ or ‘clients’ (axis-2). Using a method similar to axis-1, we identified various groups who are eventually served by this practice and clustered them in four groups. Many of the practices served more than one group and some of these were minor or optional clients, or implied to be served in the articles. We preferred to note these in the tables as well (with notes such as ‘minor’, ‘optional’, ‘indirect’) because many of the practices are in the conception or infancy phases now and in the future may evolve to different directions. Eventually, the groups that emerged in this axis were not mutually exclusive. Therefore, we called them ‘groups’ rather than ‘categories’.
During several discussions, we identified the issues such as for whom the practice is available, who controls access to it and who benefits from it are important topics for integration of practices into (health) services. However, these themes are closely related to the health system, including the health financing, of the country where the practice is implemented. Since we collected our data from the scientific literature, where information on revenue models, benefits, availability, access, etc, are not reported regularly, we chose to present three country examples to compare how the practices are integrated.
The analysis was performed by the author, and coauthors challenged it, providing critical feedback in several rounds. The discussions among the team continued until an agreement on the axes and their components was reached. MS Excel was used for the analysis.
Before and after the analysis process, we explored if there was an identifiable pattern in the use of different terms referring to the personalised approaches. However, we did not observe a consistent pattern in their use in terms of technologies and/or disciplines involved, the purpose (prevention, therapy, disease management, etc), target groups, etc. Neither did we observe a pattern in terminology according to the categories we identified.
After the analysis, the results were summarised into tables, where short descriptions based on the articles were added. Since we based the analysis on only the articles, changes that happened after they were published, such as stopping serving a practice or changing its form, were not taken into account. For practices that were reported to have a website, the websites were checked to see if the provider has apparently changed or stopped services after the publication and this was put as a brief note in the relevant tables as necessary.
Results
The basic properties of the 88 practices identified from the articles are presented in table S1 in online supplementary material 2. Figure 2 presents the general view of the axes and their components, which are summarised in the below text with examples from the practices.
Figure 2.
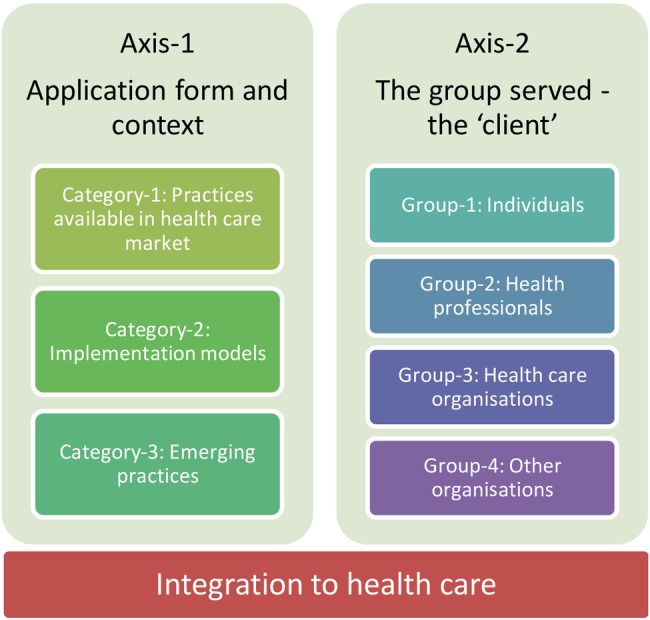
The axes and their components that emerged from the analysed practices.
Axis-1: application form and context
Data analysis revealed three main categories in terms of the application form and context: (1) practices available for use in health services, implying that PM/PHC is a commodity that is provided on the healthcare market; (2) implementation models where the product or service is developed and/or implemented in a wider context (ie, in an institution), implying that PM/PHC is a way how health services are organised and provided and (3) emerging practices which have not evolved to become one of the first two categories yet, implying PM/PHC as a keyword which indicates a ‘future’ or ‘novel’ potential.
All practices were categorised in only one of the three categories, except for Gentest (P-20)26 and GeneInsight (P-32)30 as these two practices had two service facets. Practices are summarised in separate tables for each category in online supplementary material 2 (tables S3–S5 for categories 1–3, respectively).
Category-1: practices available in the healthcare market
This category includes products/services that are available on the healthcare sector/market. Their uptake and use depend on the decision of the health professional and/or individuals/patients. Tests, test-drug pairs, drugs, (bio)informatics services, devices, online tools and websites fall into this group. Most of these are commodities available on the healthcare market and have a cost for the payer (whoever the payer is, ie, third party payer, the patient or others). An example is pharmacogenetic tests and related drugs (P-1), which are ordered by the physician (see box 1).
Box 1. Examples to practices available in the healthcare market (category-1).
Pharmacogenetic tests and related drugs (P-1)
▸ In pharmacogenetic testing, the sample of the patient is tested for a certain (genetic or genomic) biomarker in the laboratory to predict the response of the individual to a certain drug. The physician can use this information to choose the most appropriate drug or decide its dose. For example, individuals respond differently to warfarin, an anticoagulant drug used for prevention and treatment of thrombosis and thromboembolism. Therefore, the warfarin dose must be titrated according to the response of the individuals, as measured by the international normalised ratio (INR) value in the blood. The therapeutic index for INR is rather narrow, and higher and lower than desired levels increase the risk of serious complications (haemorrhage) and lack of desired effect (thrombosis). Genetic polymorphisms in CYP2C9 and VKORC1 genes are responsible for 40–60% of the variability in the therapeutic warfarin dose. Testing individuals for these polymorphisms before the initiation of warfarin therapy can help to optimise the starting dose of the warfarin and might reduce the risk of haemorrhagic complications.31–36
Coronary risk scores (P-12)
▸ The risk of cardiovascular diseases can be calculated using classical scoring systems such as the Framingham risk score (USA).37 The scoring is available in the literature or can be calculated automatically in the internet when the personal clinical variables of the individuals, such as age, gender, total and high-density lipoprotein cholesterol levels, smoking status and hypertension/blood pressure information, are entered (http://cvdrisk.nhlbi.nih.gov/).
A few interventions are available publicly, not commercially, for example, the tools for predicting the risk or prognosis of a disease, such as coronary risk scores, which are published in the literature and available freely on the internet (P-12; see box 1).
Category-2: implementation models
This category includes products or services that are developed for and/or implemented in a certain context, that is, a healthcare institution, a country and worldwide. Practices can be a part of a project, a programme or a service framework that aims to implement, pilot or assess the feasibility of the implementation of products or services, as well as related tools in a certain setting. The implemented products or services can already be available on the market (category-1) or developed specifically for that setting. These PM/PHC practices imply that PM/PHC is not just a tool, product or service available (category-1), but also the way how health services are organised and delivered.
We labelled the practices in this category as ‘implementation models’ because they do not solely focus on the product/service or its technical use, but take into account its implementation in the organisational setting. By definition, they are expected to involve a health professional for their administration, except Athlete Biological Passport (explained below). Examples of three implementation contexts are presented below (see box 2).
Institutional context: projects, programmes or service frameworks developed for and/or implemented at a specific healthcare institution are included under the ‘institutional context’. They can be as small as a physician practice, or as large as a chain or group of major academic medical centres, including home health services. A variety of practices are implemented in an institutional context. For example, a number of programmes explore, develop and pilot the ways of implementing pharmacogenomics in clinical practice in major medical centres (P-25, P-57, P-62, P-66, P-68, P-82). Some of the practices are service frameworks that are developed at specific institutions, focusing on other aspects of personalising the health services, such as psychosocial needs (P-86).
National context: the only example of implementation on a national scale is the French National Cancer Institute’s (French NCI) nationwide programme for tumour genomics (P-44).
International context: currently, the only practice in this subcategory is Athlete Biological Passport (P-42), a programme offered by the World Anti-Doping Agency and implemented by antidoping agencies around the globe.
Box 2. Examples to implementation models (category-2).
Pharmacogenetics-related programmes (P-25, P-57, P-62, P-66, P-68, P-82)
▸ A number of programmes involve genotyping a group of patients for identified genotypes before the relevant drug and testing is indicated (pre-emption), and aim to investigate the results of this practice in various aspects, such as outputs, patient outcomes and costs, and others: CLIPMERGE PGx Program at Mount Sinai Medical Center (P-25),38 PREDICT project at Vanderbilt (P-57),39 Personalized Medicine Program at University of Florida Health (P-62),40 Clinical Pharmacogenomics Implementation Program at University of Chicago (P-66),41 42 Coriell Personalized Medicine Collaborative (Pharmacogenomics; P-68)43 44 and Preemptive PGx protocol at Mayo Clinic (P-82),45 all in the USA.
Youth area for adolescent and young adult patients with cancer (P-86)
▸ ‘Youth area’ at the Centro di Riferimento Oncologico (CRO) National Cancer Institute, Italy, is a special unit for adolescents and young adults at a cancer research hospital to provide assistance to them, in particular for their psychosocial needs.46
French National Cancer Institute’s (NCI) nationwide programme for tumour genomics (P-44)
▸ The programme aims to provide available genomic tests to all relevant patients with cancer in France. French NCI runs it with a network of 28 laboratories in France. Each laboratory is located in a specific geographic area to provide tumour genomics services to all cancer patients (public and private), thus covering the whole of France. Funded by the French NCI and Ministry of Health, all patients have free access to the identified biomarkers that predict response to certain cancer treatments.47
Athlete Biological Passport (P-42)
▸ Athletes are monitored longitudinally with tests on haematological and steroidal profiles. Recent changes and disparities in the parameters can be a result of performance-enhancing drugs or a medical condition that needs to be investigated. The programme is developed by the World Anti-Doping Agency. Antidoping agencies around the globe can implement it using the relevant guidelines.48
Category-3: emerging practices
Practices in this category include the ones not fitting in the previous categories. The practices achieved ‘contact with the patient (data)’; however, information on their current use is not (yet) available in the literature. These, unlike practices in category-1, are not yet available on the market (at the time of the data collection) and are not developed for a specific (institutional) context like the ones in category-2. They may evolve into these two, or to other directions (see, eg, box 3).
Box 3. Examples to emerging practices (category-3).
IT-based Diagnostic Instrument System (P-13)
▸ It is a set of diagnostic medical instruments which have been integrated into a personal wearable device (wrist-worn health monitoring device), which measure a variety of parameters in the body, a blood glucometer integrated to a cell phone and a home telehealth system including biosignal measurement systems that can measure, for example, ECG, body temperature and body fat ratio on a toilet seat. It is developed at Seul National University (Korea).49
GliomaPredict (P-28)
▸ GliomaPredict is an algorithm for prediction of the outcome of gliomas (a brain tumour) based on their transcriptomic profile. It is validated on data sets and available for use in one institution, the National Cancer Institute, USA.50
Computer-Assisted Brief Intervention for Tobacco (CABIT) Program (P-47)
▸ CABIT is an opportunistic computer-based tool to assist quitting smoking in clinical settings (Cooper University Hospital and Polaris Health Directions, USA). It is a self-administered computerised assessment to produce reports for healthcare providers and patients. On the basis of the assessment, the tool provides a video intervention to the individual (based on the stage of change for the individual’s smoking habit) and a referral for smoking cessation.51
A prototype for automated interpretation of a pharmacogenetic test (CYP3A5—tacrolimus dose; P-85)
▸ Tacrolimus is a drug used to prevent rejection in organ transplantation. The needed dose for this drug may change according to the CYP3A5 genotype. An algorithm and reporting system was developed for interpretation of the CYP3A5 test and recommending tacrolimus dose at Oslo University Hospital, Norway.52
Integrative Personal Omics Profile (iPOP’ project; P-50)
▸ Extensive omics profiling of blood components from a generally healthy individual was performed over a 14-month period at Stanford University, USA. The whole genome sequencing data were analysed with the RiskOGram algorithm, which integrates information from multiple alleles associated with disease risk. It indicated an increased risk of type 2 diabetes, which actually developed during the follow-up period and was diagnosed by a physician.53 The primary aim of the project was not to provide a ‘health practice’. However, the analysis made on the whole genome data, that is, risk assessment of various diseases, had implications as a health practice.
Quantified-self (P-59)
▸ An individual followed various parameters related to his body longitudinally using digital biomedical measurement devices and blood and stool biomarkers (USA). He observed that episodically elevated stool protein markers were accompanied by increased blood C reactive protein levels, indicating inflammation. He got colon imaging studies and biopsies, and was diagnosed with late-onset Crohn's disease, which is an inflammatory bowel disease.54
We did not observe a pattern according to which the practices per se can be grouped, but the ways they are reported in the articles seem to have three prominent ways:
Demonstration of applicability or proof of concept: these practices were applied or presented on cases (one or more) or data sets. A good example is the ‘IT-based Diagnostic Instrument System’ (P-13), a personal wearable device and home telehealth system for measurement of a variety of parameters and biosignals of the individuals. The applicability of algorithms or bioinformatics tools are demonstrated by applying them on data sets, such as the algorithm that predicts the outcome of gliomas based on their transcriptomic profile, GliomaPredict (P-28).
-
Trial implementation: these practices are implemented specifically for research purposes in one or more settings. This can be a pilot to assess feasibility, usability, clinical validity and utility (efficacy and effectiveness). Unlike the pilots reported in category-2 of axis-1, which were programmes developed for specific institutions; here, the product or service is not developed for the setting where it is tested.
An example is the Computer-Assisted Brief Intervention for Tobacco (CABIT) Program (P-47), a web-based multimedia tobacco intervention for use in opportunistic settings. For its initial evaluation, including patient and provider satisfaction and treatment initiation, it was piloted in a number of healthcare settings. It is unknown at this point if it became a long-term programme for those settings or became available for different settings. Another example explored physicians' reactions to the prototype of an algorithm and reporting system for a pharmacogenetic test (CYP3A5—tacrolimus dose; P-85).
Exploratory/investigational cases: these cases initially had an investigational or exploratory purpose, without knowing if it would have specific health practice implications. A specific health intervention was not (fully) designed prior to the start of the case, but it eventually led to a health practice. ‘integrative Personal Omics Profile (iPOP)’ project (P-50) carried out longitudinal extensive omics profiling on blood components from a generally healthy individual. Another individual followed various parameters related to his body longitudinally using digital biomedical measurement devices and blood and stool biomarkers (P-59). In both examples, the follow-up led to the clinical diagnosis of diseases.
Axis-2: the group served by the practice, that is, the ‘user/client’
This axis involves the question ‘who is the main group or entity served by this practice?’. In other words, ‘who turns out to be the main “user” or “client” of this practice, and what definition of PM/PHC does this imply?’.
Data analysis revealed four main groups: (1) individual (including the patient), implying that (PM/PHC) means tools that involve and/or empower the individuals to gain control over matters related to their health; (2) health professionals, implying that PM/PHC tools help them provide better tailored and therefore better quality health services; (3) healthcare organisations, implying that PM/PHC means ways to increase effectiveness and efficiency of healthcare delivery; (4) other organisations, such as a payer organisation, a national health system and international system.
In tables S3–S5 of the online supplementary material 2, axis-2 is summarised in a separate column.
Group-1: individuals
If the articles indicate that the practice is used by the individual for management of their health or disease (risk), it is considered to serve the individual. The term individual covers both ‘patients’, that is, people with disease, and healthy individuals (see, eg, box 4).
Box 4. Examples to practices which serve to individuals (group-1).
Direct-to-consumer genomic tests (‘personal’ or ‘consumer’ genomics; P-6)
▸ These tests are also called ‘consumer genomics’ or ‘personal genomics’ services, commonly exemplified by 23andMe.55–59 23andMe, a US company, provides a genome-wide DNA test (Personal Genome Scan) over the internet directly to consumers. A saliva sample of the individual is used to detect variations in the DNA, with the purpose of providing information on two areas: ancestry/genealogy and health. Under the ‘health’ section, disease risks based on common polymorphisms as well as highly penetrant variations, carrier status of monogenic disorders, some physical traits and response to drugs are provided online as results of the test. Individuals are free to take these to their doctors for discussion or medical follow-up. (This description is based on the articles included, with the latest from 2012. It is of note that the content of 23andMe changed in December 2013 due to the warning of the Food and Drug Administration of USA60 and the company no longer offers the ‘health’ section to new customers).61
Smartphone-based personal, pervasive health informatics services (P-9)
▸ Smartphone-based personal health informatics aim to improve diagnosis, acute and chronic treatment, and rehabilitation of diseases, as well as supporting self-care to stay healthy/prevention of diseases.62 They include applications of sensor, signal and imaging informatics. Many of these services are available for the individuals, for example, for fitness monitoring using GPS and accelerometers, sensors for heart rate monitors, and devices that combine these to a wrist-worn device such as FitBit.
A prominent example here is direct-to-consumer genetic tests that identify predisposition to or predict risk of certain diseases (P-6). The individual can order the test and receive the results without the involvement of a health professional. Another example includes various smartphone-based personal, pervasive health informatics services that support individuals' self-care to stay healthy and prevent diseases (P-9).
The practices serving primarily the individual place him/her in the centre of the service and help them be more involved with their health. Here, personalisation implies empowering persons to take care of themselves or to gain control of their own health.
This group also includes practices that serve the people surrounding the individuals, such as families of the adolescents and young adults in cancer wards (P-86; see box 2). This is considered as a continuum of the ‘individual centred’ approach; taking the individual with people who can support or help them.
Group-2: health professionals
If the health professionals use the practice for medical management of the disease (risk) or the patient, it is considered to serve this group. Laboratory tests ordered by the physician and the drugs prescribed are included here because they mainly assist the medical management of the disease, carried out by the physician and other health professionals. A prominent example here is tumour genetics/genomics tests and related drugs (P-2; see, eg, box 5).
Box 5. Examples to practices which serve to health professionals (group-2).
Tumour genetics/genomics tests and related drugs (P-2)
▸ Here, the laboratory test is carried out on the tumour tissue, to predict its response to a given cancer drug.63–68 The technology used for the test varies: genetic tests (specific mutation or sequencing), gene expression analysis, immunohistochemistry, in situ hybridisation, etc. In recent years, there have been an increasing number of cases of pairs of cancer drugs and accompanying laboratory tests (companion diagnostics), which were developed, received regulatory approval and presented to the healthcare market together.33 66 67 69–73
ALGA-C (questionnaire that measures and collects psychocognitive information about patients; P-84)
▸ ALGA-C is an electronic questionnaire focusing on cognitive, physical-related and psychological aspects of the patients. It is filled by the patients before meeting the doctor. The results provide a psychocognitive picture of the patient with whom the physician is about to meet. The physician can use this information to optimise the communication style and interaction with the patient. It is developed by the P-medicine project (Greece, Italy and the UK).74
Gentest (P-20)
▸ Gentest is an integrative preventive model that uses an individual’s personal health information, family history, detailed lifestyle analysis (nutritional habits, food consumption using a food portion size atlas, physical activity and exercise, and smoking), anthropometric and bioimpedance measurements, genotype, and other biomarkers to stratify individuals into risk groups to prevent complex diseases in a targeted way. It is provided to individuals via authorised physicians. On the basis of the results of the aforementioned components, risk assessments are made for the major complex diseases including heart attack, stroke, type 2 diabetes, cancers (lung, breast, prostate, colon and gastric cancers) and osteoporosis and an optimum lifestyle plan, including personal menu plans and food exchange lists, exercise plans, smoking cessation recommendations and a medical follow-up plan, are drawn. It is developed by GENAR Institute for Public Health and Genomics Research in Turkey.26
An electronic questionnaire that identifies the psychocognitive status of the patients before the physician meets them (ALGA-C, P-84) provides feedback to the physician to optimise his/her communication style and interaction with the patient. Thus, the questionnaire is filled in by the patient but it eventually is a tool that serves the physician.
A few practices serve primarily both the individuals and the health professionals. One of them is Gentest, which is a preventive practice model provided to individuals via authorised physicians (P-20). The output of the service is a report which includes, among others, the summary of assessments made for the individual, an optimum lifestyle programme and a medical follow-up programme, thus serving both the individual and the physician.
The practices that primarily serve the health professional are PM/PHC tools that help the health professional (mostly a physician) deliver even better or more accurate health services to the individuals, potentially leading to more successful prevention, diagnosis and treatment. They can be perceived as ‘high-profile’ or elaborate products or services used by professionals.
Group-3: healthcare organisations
Practices which had implications on the organization of the health services are included here. They had components such as involvement of different stakeholders (disciplines/departments) within an organisation or across organisations for development and implementation of the practice and clinical and/or laboratory workflows. They eventually targeted, to increase effectiveness and efficiency in a healthcare institution. These are actually issues that are related to the goals of a healthcare provider institution, that is, effectiveness, efficiency (costs) and patient and employee satisfaction. Thus, PM/PHC becomes tools or ways for healthcare organisations to perform better in their services, which may help to improve their businesses in the competitive environment (see, eg, box 6).
Box 6. Examples to practices which serve to healthcare organisations (group-3).
Mayo Clinic's Individualized Medicine Clinic (P-71)
▸ Two lines of services were introduced in this clinic: (1) tumour genomics services for patients with advanced cancer who fail standard therapy; (2) whole exome sequencing and interpretation services for patients with a suspected genetic condition for whom previous genetic testing did not reveal an aetiology. A common infrastructure for these was built and processes were designed with involvement of several disciplines and functions. These included IT, bioinformatics, bioethics, patient access management, education, revenue cycle, operations and the research sequencing facilities under the Individualized Medicine Clinic Working Group (USA).75
Geisinger blood component usage programme (P-33)
▸ Geisinger Medical Center (USA) initiated a programme with the overall goal of reducing unnecessary blood component usage for patients undergoing surgery. Within this programme, algorithms were developed to clarify the conditions for using blood components and a system was set for communication between the pathologist and the patient’s clinical team 7/24. When too much blood components were used for a given patient in the given time frame, the system automatically involved the clinical pathologist. The programme was developed by the Transfusion Committee of the hospital.76
The Mayo Clinic's Individualized Medicine Clinic (P-71) is an example of an initiative that opened a new service line within an organisation. This initiative set up and organized an infrastructure which involved with the laboratory, coordination, systems to handle samples and report results, education materials and others. All relevant disciplines within the organisation were involved for the clinical and administrative workflows, which included, among others, coverage of test costs by the insurance.
Geisinger Medical Center introduced algorithms and warning systems in their service lines with the aim of reducing unnecessary blood component usage for patients undergoing surgery (P-33). This involved several stakeholders at the hospital and served to increase the effectiveness and efficiency of patient care.
This group is strongly related to the second category in axis-1, that is, implementation models. All the practices in category-2 had a direct or indirect component of healthcare organisations, as summarised above.
Group-4: other organisations
Three practices go beyond the borders of healthcare organisations. The first one eventually serves a national health system: the French NCI's nationwide programme for tumour genomics (P-44; see box 2). The programme aims increasing the effectiveness and efficiency of cancer treatment, as well as to providing equitable services to the population. These correspond to the main goals of a health system (effectiveness, efficiency, patient and employee satisfaction and equity). Thus, PM/PHC becomes a part of tools/activities for health systems to perform better.
The second example is of a healthcare payer organisation: a social health insurance fund, Techniker Krankenkasse, in Germany, which implies that PM/PHC is both for healthcare providers and for payers. Within the ‘Telemedicine for the Heart’ programme, telemedicine services for patients with heart failure within their insured population were provided (P-60; see box 7). In the case of the payer organisation, PM/PHC becomes a tool in the service package of the payer organisation to provide more efficient and effective services.
Box 7. Examples to practices which serve to other organisations (group-4).
Telemedicine for the Heart (P-60)
▸ A telemedicine programme which consisted of nurse calls to motivate patients to perform regular self-measurements (blood pressure, pulse, weight) and their recording was offered to the members of Techniker Krankenkasse, which is a sickness fund (social health insurance fund) in Germany. In case the measured values were outside the set limits, calls to physicians were placed for therapy adjustments.77
The third practice is of an international system of national and local antidoping agencies, as well as the World Anti-Doping Agency (P-42; see box 2).
Integration of practices into real life
The two axes summarised above can give new perspectives on PM/PHC. However, they are not enough to provide a wider picture of issues around integration of these practices into real-life (health) services. It is widely assumed that one of the main barriers towards integration is the lack of clinical utility of ‘personalised’ practices. Nevertheless, when we look at the practices identified, we see various other issues surrounding them in their context. Are these practices available to everyone in their respective countries as commodities in the healthcare market (category-1)? Are the implementation models (category-2) available in all health institutions in a given country? Even if they are available, can the individuals access them without geographical, financial or other barriers? What about the situation globally?
For most of the practices, it was not possible to answer these questions because issues around availability and accessibility are not regularly reported in articles describing practices in the scientific literature. Moreover, many of these issues are inherent in the health system in which the provider is placed. That is why we chose three country examples among the practices in category-2: the USA, France and Taiwan, which included some information on these dimensions.
Cases: USA, France and Taiwan
In category-2 (implementation models), we see that several medical centres (centres that combine healthcare service provision, research and medical education) and other major healthcare providers in the USA have initiated some sort of a programme or project on ‘personalised’, ‘individualised’ or ‘precision’ medicine in the past 5 years. These programmes often involve a genetic component, such as tumour and cancer genomics and pharmacogenomics (P-25, P-30,78 P-32, P-38,79 P-41,80 P-56,81 P-57, P-61,82 P-62, P-71, P-75,83 P-77,84 P-80,85 P-82; see box 2 and box 6). Many of the institutions initialising these projects are world-renowned major medical centres, which are known to pioneer introduction of innovations in mainstream medicine in the world. Many of their ‘personalised’ programmes in category-2 are probably among the first of their kind in the USA or worldwide.
However, from the ‘availability’ point of view, these ‘personalised’ programmes are actually ‘prestigious’ services that are available to a very limited number of people with geographical and financial access to these major medical centres. For the rest of the US population, the availability is unknown.
Moreover, the US health system has serious problems with accessibility of health services. The uninsured/underinsured population and ineffectiveness of the insurance-based health finance are major problems that lead to disparities in the system.86
In France, the NCI's nationwide initiative provides tumour genetics/genomics services to all patients with cancer and is funded by the NCI and Ministry of Health (P-44, see box 2). Thus, tumour genomics services are not limited to certain hospitals or geographical regions and are available and accessible (geographically and financially) to all patients with cancer who need it. This indicates ‘equity’.
The third example is from Taiwan. A programme called Ubiquitous Healthcare was developed and introduced at the National Taiwan University Hospital (P-24, see box 8). It is a set of telehealth/telecare solutions, which aim to provide services to patients living in remote areas. In a sense, it is an attempt to overcome geographic accessibility problems, in the context of a country that widely embraced mobile technologies. However, the fact that this service is being promoted among the target group of patients by a marketing team suggests that it is to be paid out of pocket. The costs do not seem to be extremely high (eg, monthly cost of a cardiovascular patient is around €150). Nevertheless, for people from lower income groups, it can still be unaffordable. Thus, at a stage where the service is not yet covered by the National Health Insurance of Taiwan, it is available only to people with access to the university hospital (at least once), and possibilities to afford the service.
Box 8. Example of a practice provided with out-of-pocket payment.
Ubiquitous Healthcare (P-24)
▸ National Taiwan University Hospital (Taipei, Taiwan) combined the home healthcare service network and sensors network into a new service known as the Ubiquitous Healthcare (U-Health) service system in order to continually provide personalised healthcare services to patients who live in remote areas. The system included six service components that are introduced in an integrated manner: teleeducation, telediagnosis, telemonitoring, teleconsultation, teletherapy and lifetime health. The services were targeted towards the following patient segments: postdischarge patients, patients suffering from cardiovascular diseases, patients suffering from chronic diseases and patients in hospice care. It reduced rehospitalisation rate and emergency room visits during the postdischarge period.87
Contextual factors: attitude towards innovations and health systems and policies
These cases indicate that two factors, among others, may be important for integration of personalised practices in services: attitude of healthcare providers and society towards innovations and the health system and policy context.
The USA has long been regarded as a global leader in medical technology innovation88 and, as a matter of fact, PM/PHC has been closely linked to technological advancements. Apart from the publication tradition of these high-profile medical centres in scientific journals, the high number of publications from the USA may give the impression that uptake of innovation there is higher than in many other countries. However, we do not know much about the availability, that is, whether it is limited to high-profile centres or widely available. There is a strikingly different example from the other side of the Atlantic: French NCI's initiative, making tumour genomics services available and accessible for everyone who needs it.
These contrasting cases illustrate that uptake and implementation of personalised practices is influenced by the attitude of the country towards health innovations, as well as by the health system and its policy values. The French health system has ‘equity’ as a central policy value, reflected by the universal health coverage for all its citizens. In line with this, France provides all services in an equitable way to all citizens. Contrastingly, a prominent policy value in the US health system is ‘liberty’, supporting the free market economy largely even in the health system, at the cost of equity.86 89 Thus, prestigious high-profile medical centres compete for newer and more innovative applications. Lack of equity as a policy value also results in a huge proportion of the uninsured and underinsured population who cannot access health services. This means that even if the individuals can geographically access the high-profile medical centres, they may not be able to access the ‘prestigious’ ‘personalised’ services due to their inability to pay for them.
We also see the effect of components of health systems on these examples. Such contextual issues are not explicitly addressed, but implied in various articles. For example, some of the healthcare institutions from the USA refer to ‘insurance coverage’ for the practices they offer.75 82 90 91 In the country context, this implies several issues within the US health system, which create a barrier to operationalisation of these services. Among them is the difficulty of dealing with multiple payers (insurances) which decide independently on coverage of these new, not yet standardised, ‘personalised’ services. On the other hand, in a single payer health system, that is, France, it was possible to cover the whole population for tumour genomics services with a special programme and dedicated funds for it.
In the case of Taiwan, where out-of-pocket services account for almost 40% of the healthcare expenditure,92 we see that a telecare practice enters the market as an out-of-pocket service. For some countries, it is customary for new technologies to enter the healthcare market as an out-of-pocket service and gradually become covered by third party payers. This means that the availability and accessibility of a practice in a given country can evolve over time.
Discussion
Summary of the main findings
In this study, a systematic search and inclusion strategy was used to identify ‘personalised’ practices within the scientific literature. Neither the terminology used to describe the practices (personalised, individualised, systems, medicine, healthcare, etc) nor the technologies used showed a consistent pattern among the included practices. Thus, a different angle grounded on the practices was taken to analyse them to reveal notions of PM/PHC.
The analysis of ‘the application form and context’ indicated that PM/PHC can be a commodity for the health market, a way in which health services are organised/provided, or ‘future’ or ‘novel’ emerging applications. When reviewing whom these practices were aiming to serve, we identified that PM/PHC can serve individuals (or patients) to empower them to gain control of their health, health professionals to help them provide ‘even’ better health services, healthcare organisations to help increase the effectiveness/efficiency of healthcare delivery in the competitive environment or national health systems to help increase their performance in terms of effectiveness/efficiency and equity. During the analysis, it was noted that for successful integration of the practices into health services, both the attitude of the country towards innovations and the health system and policies context are crucially important.
Strengths and limitations
This article aimed to investigate the field of ‘personalised’ care from a practice-based perspective, not restricted by current (top-down) theoretical frameworks. Owing to the uniqueness of the research aim and heterogeneity of the included practices, a combination of methods was used at every step of the analysis. Among these were: triangulation with different strategies applied by two independent researchers during the screening phase, usage of the ‘health intervention’ definition of ‘International Classification of Health Interventions (ICHI)’ to assist the selection process, and content analysis grounded on the data (practices/articles) for identification of relevant patterns. This combination of methods and tools allowed us to include a broad range of sectors/technologies and research/development stages, which enabled us to demonstrate the spectrum of ‘personalised’ practices reported in the literature. To the best of our knowledge, this makes the paper unique in the field.
The main limitation of our study is that the search was carried out using the scientific literature alone without inclusion of information from websites. As a consequence, some well-established existing practices may not have been included in the analysis. This restriction was deliberately made in order to ensure the quality and credibility of the practices analysed.
Data on integration of practices into health services were very limited. Therefore, three prominent and contrasting country examples were identified to discuss some of the important factors that affect availability and accessibility. Since only the identified articles were analysed, we were not able to cross-check or elaborate on all aspects. Further research with a targeted approach using various other data sources can yield a more comprehensive analysis of the issues raised.
The scientific evidence base of the ‘personalised’ practices, or its lack thereof, is a major issue discussed in scientific fora for implementation of personalised practices in health services. This study took a different perspective on PM/PHC and aimed to develop an understanding of how the field can be defined from a ‘practice-based’ perspective. Since the review of the evidence on these practices was not a part of the research question, it was not taken as a dimension in the analysis. Nevertheless, this article may inspire others to identify the ‘personalised’ practices in the literature with a practice-based approach and/or use the inventory provided in online supplementary tables S1–S5 to investigate them from other perspectives, such as patient and practitioner acceptance and clinical utility.
Implications of the findings for the integration of personalised approaches
Since the early 2000s, there has been a growing expectation that ‘personalised medicine’ would enter the health services within a decade and gradually become common practice. In the early stages, it was mostly genomics that fuelled these expectations, but later ‘personalisation’ became a broader theme. However, the genome-based applications did not enter routine practices as was initially anticipated.
There have been several discussions in the scientific fora on the slow progress. It is widely recognised that the lack of robust evidence demonstrating clinical utility is a major problem in the field. Nevertheless, as seen in the results, the personalised ‘practices’ are very diverse in terms of the purposes and groups they serve. Therefore, it is not possible to talk about the clinical utility of all of them at once. Even in pharmacogenomics, which is considered to be ‘one area’, we may see different results for different drugs and genetic factors tested. For example, interventional and retrospective studies demonstrated the clinical utility of screening for the HLA-B*5701 genotype to predict the hypersensitivity reaction to treatment with abacavir, an antiviral drug used in treatment of HIV infections.93 94 However, clinical trials failed to demonstrate the clinical utility of adding genetic testing to warfarin dose prediction algorithms that use clinical variables.95 96
Although genome-based applications are not a single entity, their slower than expected progression to enter routine clinical practice led to questioning whether personalisation of medicine is a ‘myth’ rather than reality.97–99 It is now expected that clear examples of application of genome-based information in medicine and healthcare may start to flourish in this decade, but will probably become more prominent in the period beyond the year 2020.100 The below discussion on the implications of our ‘practice-based’ findings for integration of PM/PHC may provide insights on some of the possible factors contributing to these unfulfilled claims. Among them are: excessive technological focus, issues with definitions, issues with the transformation of practices and discrepancy between initial vision and practice.
Excessive technological focus
There is a striking contrast in terms of the technological orientation, in particular genomics, between the top-down descriptions and the findings of this study. The axes that emerged in our study did not point technologies or methods to define PM/PHC. Instead, they reflected on some of the real-life issues that practices face.
Many of the ‘top-down’ descriptions build the definition of the personalised approaches on the use of genome-based technologies and molecular markers.12 14 15 18 However, many of the practices identified in this study did not have a genomic or molecular component. This is illustrated in the Results section by giving at least two examples to each category and group: one that includes and one that does not include genomic/molecular technologies. Besides, there are some practices that have no technological component at all (eg, P-12, P-81,101 P-86, P-18).102 Thus, our findings indicate that it is not the use of certain technologies or methods, including genomic or molecular markers, that makes PM/PHC ‘personalised’.
The problems around equating PM/PHC to genomics have already been raised. First of all, this causes attribution of currently unfulfilled expectations of genomic medicine to the broader application of PM/PHC.5 Furthermore, genomic markers can be one of the components but it is not the only factor that makes individuals unique or medicine personalised.2 There are various other factors that determine health, including the lifestyle we lead, the environment we are exposed to (also termed as ‘exposome’),103 the microbiota we host on our bodies,104 our socioeconomic context and the health services we receive.
An example is the ‘personalisation’ of warfarin dosing. The standard practice has been to start warfarin therapy with a fixed dose and to modify it based on the international normalised ratio (INR) response (see box 1). However, this has been a clinically challenging task since the values above or below the narrow therapeutic window of INR can lead to serious clinical conditions. To minimise them, using genetic tests for guiding warfarin dosing was considered as one of the most promising pharmacogenomics practices. To assess the clinical utility of such tests, different randomised clinical trials were conducted comparing pharmacogenomics-guided warfarin dosing with standard fixed and/or clinical algorithm-guided dosing. The clinical algorithms included various variables of individuals, such as age, gender and body surface area. The results of the clinical trials indicated that a genotype-guided strategy for warfarin dosing is more accurate than the standard fixed dosing, but not more accurate than the clinical algorithm-guided dosing.105 106
This indicates that moving towards ‘personalisation’ of warfarin therapy using clinical variables, which is something not done routinely in clinical practice, is likely to be valuable, but adding the genetic bit on top of these algorithms did not show additional value. When ‘personalisation’ is equated with genetic testing, this seems to be a ‘failure’ of personalising warfarin therapy. However, if ‘personalisation’ is conceptualised as using all relevant clinical variables of the individual, the results indicate which variables can be, with the available evidence, useful in the algorithms for personalising warfarin therapy.
The definitions
The axes that emerged in this study were very different from the dimensions highlighted by the more common top-down definitions. However, the practices still did not yield a single common theme shared among them, which was also the case for top-down definitions as addressed in the Introduction section. Our findings indicated that PM/PHC practices are present in several forms and contexts, serving a variety of groups, having different connotations in terms of availability and accessibility around the world. It is not a realistic expectation to have one definition to cover all of these. Is PM/PHC a commodity that can be bought or sold? Is it a tool to empower individuals to gain more control of their health? Is it a tool for health professionals to provide even better healthcare? Is it the way how health services are organised and provided? Is it something or anything novel (with or without genomics) that is emerging? Is it ‘prestigious’ clinical applications in ‘high-profile’ medical centers? All or none of these?
Thus, similar to the case of top-down definitions, a consistent and unified definition of the personalised approaches cannot be made from the practice-based perspective either. This means that people may presume that they talk about the same concepts when they are using the same terms, while they actually have very different perceptions and ideas. This scene can be recognised by many people who try to follow the field via the literature or by participating in related meetings, in particular the multidisciplinary ones. Lack of coherence in the use of concepts and terminology can be a significant factor contributing to the unfulfilled claims.
Looking at some specific aspects of existing definitions, the distinction made earlier between ‘personalised medicine’ and ‘personalised healthcare’5 seems to apply to the practices as well. The findings of the axis-1 indicated that having a product or service available for use in the market is one thing (category-1), and actually implementing it in a certain context is another (category-2).
The ‘personalised healthcare’ description5 23 107 108 seems to match with the category-2, which includes practices that are implemented in a certain context, mostly a healthcare provider setting. Typical examples of ‘personalised healthcare’ are My PREVENT Plan (P-49)109 and Gentest (P-20), which use personal health profiles and ‘personalised medicine tools’ in a broader context to prepare personalised health plans (the former focus more on chronic disease management, whereas the latter focus on their prevention).
In the framework of Simmons et al,5 personalised healthcare is differentiated from personalised medicine. While the former is focused on a strategic approach to patient care, that is, health services, the latter deals with the use of specific ‘tools’ in medicine. Further, the ‘personalised medicine’ definition by Personalized Medicine Coalition, USA,15 has some sort of a match with category-1 in the sense that the Coalition also positions personalised medicine as a commodity in the healthcare market, but with particular focus on diagnostic tests and related drugs which involve genomic and molecular markers.110 Most practices in category-1 are some sort of tests or drugs that are, however, not restricted to genomics or molecular markers.
On a map of the various concepts defining the personalised approaches, Pokorska-Bocci et al3 depicted personalised healthcare as a superset of all of them. This gives the impression that the difference is in their scope, but it is probably beyond that. There are two dimensions that distinguish them. The first is the application form and context, as supported by our findings and explained above (commodities/tools in the market vs implementation model in a healthcare context). The second is that personalised medicine is used to refer to specific tools, whereas personalised healthcare is used for a broader, holistic approach to health.
In brief, our findings are in line with Simmons et al5 and indicate that ‘personalised medicine’ should be used for specific tools that are available for practice in medicine (such as tests, test-drug pairs, predictive algorithms, etc), as in category-1, whereas personalised healthcare should be used for health services that have a holistic approach to the individual to be implemented in certain contexts.
Transformation of practices
PM/PHC is a new and dynamic field; thus, it is possible that the available approaches transform or converge from one category to another. For example, an implementation model designed by one institution (category-2) may become a solution available to other stakeholders (category-1), that is, GeneInsight Suite (P-32), or a generic practice available within the healthcare market (category-1), that is, Gentest (P-20). Practices in category-3 (emerging practices) might converge to those available on the market (category-1) or institutional models (category-2). It is possible that some will evolve in different ways that have not been identified in this study. Targeted research using different data sources can explore how the practices evolve and transform over time.
For example, iPOP (P-50) and an example of ‘quantified-self’ movement by Smarr (P-59) are categorised as emerging practices (category-3) in axis-1. They are single cases that use longitudinal health data, in line with the P4 medicine vision of Hood and Price.18 19 25 A new initiative called the 100 K Wellness Project takes the P4 medicine vision to a larger scale longitudinal research project with a practice component, where health coaches are trained to guide the participating individuals to change their lifestyle based on available ‘actionable’ data.25 If we apply the distinction between personalised medicine (tools/commodities) and personalised healthcare (implementation models) to this case, the 100 K Wellness Project is essentially an example of personalised healthcare, indicating transition from P4 medicine towards P4 healthcare.
Within the dynamism of the field, there are various promising scientific and technological fields that are emerging but could not be captured within the scope of this study which focused on published ‘practices’ (see the ‘Systematic search and selection strategy’ section, as well as supplementary material 1). For example, ‘metabonomics’ aims to analyse the metabolic response of living systems to stressors or interventions.111 ‘Pharmacometabonomics’ takes this concept further and uses metabolic profiling for prediction of an individual's response to a drug.112 While pharmacogenomics focuses on genomic factors, pharmacometabonomics is sensitive to genetic and environmental factors such as diet, drug intake and the individual's microbiome.113 This makes pharmacometabonomics a promising technological area for personalisation of drug therapy.
Another example to emerging fields that were not identified as a ‘practice’ at the time we carried out our literature search is ‘metagenomics’. In the context of human health, it involves the study of the genetic material of the microbiota living on humans, in particular in the gut. Since an increasing number of studies indicate the significance of gut microbiota in human health and disease, metagenomics continues to be a promising technology that can be used in health practice.114
The transformations in practices and the variety of new technologies that are emerging at a rapid pace indicate the highly dynamic nature of the field, which is making the definitions more difficult. Moreover, by the time one definition framework is agreed on, it runs a risk of becoming outdated as new advances emerge.
Discrepancy between initial descriptions and practice
One of the factors that contribute to why the field of PM/PHC has not fulfilled the expectations might be how PM/PHC was envisaged and defined in the first place. The top-down descriptions focus on the theoretical promises of available and forthcoming technologies and try to guide the field in that direction. They were designed mostly by highly influential individuals and committees on the science and technology side and assume that if the technology alone is good enough, it can solve the problems of today's healthcare. For example, P4 medicine was proposed to eventually lead to a universal democratisation of healthcare by increasing the effectiveness of preventive and curative interventions, and reducing the costs, thus lessening the social and financial burden of diseases.19 25
There is clearly a difference between how the field is depicted in these theoretical models and real life, as reflected in the practices we identified and analysed. In real life, innovations do not diffuse and enter services just because they are technically possible.115 Policymakers, practitioners and/or individuals do not (decide to) use tools just because they exist. In line with the authors such as Bijker116 and Berg and Mol,117 the development and integration of new practices is much more complex than science and technology alone. It involves people, functions and bodies in health services in the whole context of the health system, in interaction with the society.
The health practice side of the coin has been widely neglected. Practitioners are not ‘empty vessels’, who only absorb all the literature, guidance and visions set forth by the commissions and opinion leaders. As Green27 states, they are “…full of prior knowledge, attitudes, beliefs, values and, above all, contextual constraints at any given point in practice time”. Practitioners adjust methods and technologies they use in order to successfully adapt them to the context, which inevitably changes and transforms them.117 At the end of the day, it will be those healthcare practices that will actually construct the field and shape how personalised approaches will actually take place in medicine and healthcare.
The health systems and policies context has also been under-represented. For example, can inequity in a health system be solved by personalised practices alone? As seen in the case examples of the USA, France and Taiwan, strengths and weaknesses of health systems have direct implications on the personalised practices. The practice cannot be isolated from the context. Moreover, issues related to health systems cannot be solved in isolation either. Various publications have discussed and reported the barriers, challenges or agendas for/of personalised medicine. They list a number of items that need to be solved, such as reimbursement, data issues, physician education and others, and try to tackle each individually, one at a time, independent from each other and the general context. This isolated and piecemeal approach is unlikely to solve real-life problems efficiently because they are all interlinked with each other and the context. The health system must be addressed in its entirety.
Proposed solution: towards a true multidisciplinary approach in the field
Realisation of the PM/PHC depends on science, technology, healthcare and society domains.118 The healthcare domain includes the health professionals who are in the front line of health services, as well as other professionals, functions and bodies that make healthcare services available within the local, regional and national context, thus covering both health policy and practice. When defining and working towards PM/PHC, predominance of specific domains hinders the progress by creating unrealistic expectations. Instead, the field must be created with a collaborative effort of all domains.
These efforts should not be limited to higher level meetings, where academicians assemble and discuss the topics. To bring PM/PHC into practice, these domains must come together effectively in teams working on concrete projects. An inspiring example relevant to this field is the Institute for Systems Biology (ISB) in the USA. Established in 2000, cofounded by Hood, ISB has been pioneering the field of systems biology. To study complex biological systems in a holistic way, a cross-disciplinary culture of research which uses technology to solve biological questions was required. For that, ISB created a special and unusual institutional platform, where scientists from different disciplines learn one another's languages and work effectively in teams, instead of disciplinary departments.119
To bring PM/PHC to life, we also need an integrative ‘systems’ approach. The system here is much broader than the organism. It includes science, technology, health and society domains. Just like the interaction of genes, lifestyle and environmental factors, socioeconomic factors and health services, which determine health and disease, individuals from different disciplines and sectors need to come together, interact with each other and form real collaborations to realise PM/PHC. Various mechanisms can be used to facilitate this, such as double appointments in science, policy and practice, financial incentives to make different parties work together, and events across domains to trigger cross-fertilisation. In terms of facilitating structures, ISB can be an inspiring example. In addition, education of health professionals (undergraduate and postgraduate), in particular physicians, is crucially important for them to be able to adopt innovations in practice.
PM/PHC can fulfil its promises only if it is tackled in a collective way. If the field focuses only on science and technology, it can make significant discoveries and perhaps bring out tools for the healthcare market, but their real-life use will be questionable.
Conclusion
This work presents an important endeavour of looking at the practice side of the coin of PM/PHC. For the first time, such an analysis was based on empirical data in such an extensive way. The perspective presented in this article, methods used and the findings may trigger discussions on new ways to describe personalised approaches. The practices identified and earlier publications show that personalised medicine and personalised healthcare are different entities, one involving commodities and tools available in the healthcare market and the other involving holistic approaches to individuals and their implementation in actual settings. Science or technology alone is not enough to have an impact on the health of individuals. The health policy and practice side of the coin must also be taken into account to address innovation for health in its entirety. For that, science, technology, healthcare (including policy and practice) and society domains should work collectively towards realisation of PM/PHC.
Acknowledgments
We thank Dr Serdar Savas for his feedback on the manuscript, in particular on health systems-related issues.
Footnotes
Contributors: The study was initiated by TC, and conceived and designed by all authors collectively. The steps of the literature search, that is, identification, screening, eligibility and inclusion, were carried out mainly by TC, with significant contribution from ES, as described under the Methods section. The initial analysis was carried out by TC, enhanced with regular discussions and critical feedback of ES, AK and FF. TC drafted the manuscript. All authors commented on, revised sequential drafts and approved the final version of the manuscript.
Funding: This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: All relevant data are within the paper and its online supplementary files.
References
- 1.Individualized Medicine at MeSH (Medical Subject Headings). http://www.ncbi.nlm.nih.gov/mesh/?term=personalized+medicine (accessed 20 Jun 2014).
- 2.Cesuroglu T, van Ommen B, Malats N et al. . Public health perspective: from personalized medicine to personal health. Personalized Med 2012;9:115–19. 10.2217/pme.12.16 [DOI] [PubMed] [Google Scholar]
- 3.Pokorska-Bocci A, Stewart A, Sagoo GS et al. . ‘Personalized medicine’: what's in a name? Personalized Med 2014;11:197–210. 10.2217/pme.13.107 [DOI] [PubMed] [Google Scholar]
- 4.Schleidgen S, Klingler C, Bertram T et al. . What is personalized medicine: sharpening a vague term based on a systematic literature review. BMC Med Ethics 2013;14:55 10.1186/1472-6939-14-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simmons LA, Dinan MA, Robinson TJ et al. . Personalized medicine is more than genomic medicine: confusion over terminology impedes progress towards personalized healthcare. Personalized Med 2012;9:85–91. 10.2217/pme.11.86 [DOI] [PubMed] [Google Scholar]
- 6.European Science Foundation. ESF Forward Look—personalised medicine for the European citizen. Strasbourg, France: European Science Foundation, 2012. [Google Scholar]
- 7.Redekop WK, Mladsi D. The faces of personalized medicine: a framework for understanding its meaning and scope. Value Health 2013;16(Suppl 6):S4–9. 10.1016/j.jval.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 8.Roden DM, Tyndale RF. Genomic medicine, precision medicine, personalized medicine: what's in a name? Clin Pharmacol Ther 2013;94:169–72. 10.1038/clpt.2013.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siest G. Systems medicine, stratified medicine, personalized medicine but not precision medicine. Drug Metabol Drug Interact 2014;29:1–2. 10.1515/dmdi-2013-0068 [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Campos GH, Lopez-Alonso V, Martin-Sanchez F. Is precision medicine different from personalised medicine? A biomedical informatics perspective. Stud Health Technol Inform 2014;202:20–3. [PubMed] [Google Scholar]
- 11.National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease. Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. Washington DC, USA: The National Academies Press (US), 2011. [PubMed] [Google Scholar]
- 12.President's Council of Advisors on Science and Technology (US). Priorities for personalized medicine. Washington DC: Office of Science and Technology Policy, Executive Office of the President (US), 2008. [Google Scholar]
- 13.United States Department of Health and Human Services. Personalized Health Care: Opportunities, Pathways, Resources, 2007. https://www.genome.gov/pages/policy/personalizedmedicine/dhhsreportpersonalizedhealth.pdf (accessed 01 Jul 2014).
- 14.Stratification biomarkers in personalised medicine (Summary report of the “workshop to clarify the scope for stratification biomarkers and to identify bottlenecks in the discovery and the use of such biomarkers”. European Commission, Brussels, 10–11 June 2010). Brussels: European Commission, DG Research, 2010. http://ec.europa.eu/research/health/pdf/biomarkers-for-patient-stratification_en.pdf (accessed 03 Jul 2014). [Google Scholar]
- 15.Abrahams E, Ginsburg GS, Silver M. The Personalized Medicine Coalition: goals and strategies. Am J Pharmacogenomics 2005;5:345–55. 10.2165/00129785-200505060-00002 [DOI] [PubMed] [Google Scholar]
- 16. CASyM Europe—Coordinating Action Systems Medicine: Implementation of Systems Medicine across Europe. CASyM Report: 1st CASyM open stakeholder consultation conference, Lyon, March 2013. https://www.casym.eu/index.php?index=90 (accessed 03 Jul 2014).
- 17.Bousquet J, Anto JM, Sterk PJ et al. . Systems medicine and integrated care to combat chronic noncommunicable diseases. Genome Med 2011;3:43 10.1186/gm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hood L. A personal journey of discovery: developing technology and changing biology. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:1–43. 10.1146/annurev.anchem.1.031207.113113 [DOI] [PubMed] [Google Scholar]
- 19.Hood L. A Doctor's Vision of the Future of Medicine. Newsweek, 13 Jul 2009. http://www.newsweek.com/id/204227 (accessed 4 Jul 2014).
- 20.Topol EJ. Transforming medicine via digital innovation. Sci Transl Med 2010;2:16cm4 10.1126/scitranslmed.3000484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topol E. The creative destruction of medicine: how the digital revolution will create better health care. New York, NY: Basic Books, 2012. [Google Scholar]
- 22.Snyderman R, Williams RS. Prospective medicine: the next health care transformation. Acad Med 2003;78:1079–84. 10.1097/00001888-200311000-00002 [DOI] [PubMed] [Google Scholar]
- 23.Snyderman R. Personalized health care: from theory to practice. Biotechnol J 2012;7:973–9. 10.1002/biot.201100297 [DOI] [PubMed] [Google Scholar]
- 24.Bijker WE, Hughes TP, Pinch T, eds. The social construction of technological systems. Anniversary edition. Boston, MA: The MIT Press, 2012. [Google Scholar]
- 25.Hood L, Price ND. Demystifying disease, democratizing health care. Sci Transl Med 2014;6:225ed5 10.1126/scitranslmed.3008665 [DOI] [PubMed] [Google Scholar]
- 26.Cesuroglu T, Karaca S, Erge S. A practice model for personalized healthcare with a public health genomics perspective. Personalized Med 2009;6:567–77. 10.2217/pme.09.37 [DOI] [PubMed] [Google Scholar]
- 27.Green LW. Making research relevant: if it is an evidence-based practice, where's the practice-based evidence? Fam Pract 2008;25(Suppl 1):i20–4. 10.1093/fampra/cmn055 [DOI] [PubMed] [Google Scholar]
- 28.Moher D, Liberati A, Tetzlaff J et al. . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WHO-FIC Family Development Committee. International Classification of Health Interventions (ICHI)—alpha version. World Health Organization, 2012:409. [Google Scholar]
- 30.Aronson SJ, Clark EH, Babb LJ et al. . The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat 2011;32:532–6. 10.1002/humu.21470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donnan JR, Ungar WJ, Mathews M et al. . A cost effectiveness analysis of thiopurine methyltransferase testing for guiding 6-mercaptopurine dosing in children with acute lymphoblastic leukemia. Pediatr Blood Cancer 2011;57:231–9. 10.1002/pbc.22936 [DOI] [PubMed] [Google Scholar]
- 32.Babic N. Clinical pharmacogenomics and concept of personalized medicine. J Med Biochem 2012;31:281–6. 10.2478/v10011-012-0021-2 [DOI] [Google Scholar]
- 33.Cohen J, Wilson A, Manzolillo K. Clinical and economic challenges facing pharmacogenomics. Pharmacogenomics J 2013;13:378–88. 10.1038/tpj.2011.63 [DOI] [PubMed] [Google Scholar]
- 34.Faruki H, Lai-Goldman M. Application of a pharmacogenetic test adoption model to six oncology biomarkers. Personalized Med 2010;7:441–50 10.2217/pme.10.37. [DOI] [PubMed] [Google Scholar]
- 35.Mini E, Nobili S. Pharmacogenetics: implementing personalized medicine. Clin Cases Miner Bone Metab 2009;6:17–24. [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie MD. The success of pharmacogenomics in moving genetic association studies from bench to bedside: study design and implementation of precision medicine in the post-GWAS era. Hum Genet 2012;131:1615–26. 10.1007/s00439-012-1221-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Topol EJ, Lauer MS. The rudimentary phase of personalised medicine: coronary risk scores. Lancet 2003;362:1776–7. 10.1016/S0140-6736(03)14941-0 [DOI] [PubMed] [Google Scholar]
- 38.Gottesman O, Scott SA, Ellis SB et al. . The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther 2013;94:214–17. 10.1038/clpt.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulley JM, Denny JC, Peterson JF et al. . Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther 2012;92:87–95. 10.1038/clpt.2011.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weitzel KW, Elsey AR, Langaee TY et al. . Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet 2014;166:56–67. 10.1002/ajmg.c.31390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Donnell PH, Danahey K, Jacobs M et al. . Adoption of a clinical pharmacogenomics implementation program during outpatient care-initial results of the University of Chicago “1,200 Patients Project”. Am J Med Genet C Semin Med Genet 2014;166:68–75. 10.1002/ajmg.c.31385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolan ME, Maitland ML, O'Donnell PH et al. . Institutional profile: University of Chicago Center for Personalized Therapeutics: research, education and implementation science. Pharmacogenomics 2013;14:1383–7. 10.2217/pgs.13.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gharani N, Keller MA, Stack CB et al. . The Coriell personalized medicine collaborative pharmacogenomics appraisal, evidence scoring and interpretation system. Genome Med 2013;5:93 10.1186/gm499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang RS, Gamazon ER. Translating pharmacogenomics discoveries into the clinic: an implementation framework. Genome Med 2013;5:94 10.1186/gm497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bielinski SJ, Olson JE, Pathak J et al. . Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 2014;89:25–33. 10.1016/j.mayocp.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mascarin M, Truccolo I, Byther E et al. . Cancer, adolescence, and their peers: “They'll give you a story”. J Cancer Educ 2014;29:434–40. 10.1007/s13187-014-0635-9 [DOI] [PubMed] [Google Scholar]
- 47.Andre F, Nowak F, Arnedos M et al. . Biomarker discovery, development, and implementation in France: a report from the French National Cancer Institute and cooperative groups. Clin Cancer Res 2012;18:1555–60. 10.1158/1078-0432.CCR-11-2201 [DOI] [PubMed] [Google Scholar]
- 48.Sottas PE, Robinson N, Rabin O et al. . The athlete biological passport. Clin Chem 2011;57:969–76. 10.1373/clinchem.2011.162271 [DOI] [PubMed] [Google Scholar]
- 49.Chun H, Kang J, Kim KJ et al. . IT-based diagnostic instrumentation systems for personalized healthcare services. Stud Health Technol Inform 2005;117:180–90. [PubMed] [Google Scholar]
- 50.Li A, Bozdag S, Kotliarov Y et al. . GliomaPredict: a clinically useful tool for assigning glioma patients to specific molecular subtypes. BMC Med Inform Decis Mak 2010;10:38 10.1186/1472-6947-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boudreaux ED, Bedek KL, Byrne NJ et al. . The Computer-Assisted Brief Intervention for Tobacco (CABIT) program: a pilot study. J Med Internet Res 2012;14:e163 10.2196/jmir.2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lærum H, Bremer S, Bergan S et al. . A taste of individualized medicine: physicians’ reactions to automated genetic interpretations. J Am Med Inform Assoc 2014;21:e143–6. 10.1136/amiajnl-2012-001587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen R, Mias GI, Li-Pook-Than J et al. . Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012;148:1293–307. 10.1016/j.cell.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smarr L. Quantifying your body: a how-to guide from a systems biology perspective. Biotechnol J 2012;7:980–91. 10.1002/biot.201100495 [DOI] [PubMed] [Google Scholar]
- 55.Arribas-Ayllon M, Sarangi S, Clarke A. Promissory accounts of personalisation in the commercialisation of genomic knowledge. Commun Med 2011;8:53–66. 10.1558/cam.v8i1.53 [DOI] [PubMed] [Google Scholar]
- 56.Palmer JE. Genetic gatekeepers: regulating direct-to-consumer genomic services in an era of participatory medicine. Food Drug Law J 2012;67:475–524, iii. [PubMed] [Google Scholar]
- 57.Platt D. When consumers get their genomes. Personalized Med 2009;6:669–79 10.2217/pme.09.50. [DOI] [PubMed] [Google Scholar]
- 58.Richards M. Reading the runes of my genome: a personal exploration of retail genetics. New Genet Soc 2010;29:291–310 10.1080/14636778.2010.507486. [DOI] [Google Scholar]
- 59.Tenenbaum JD, James A, Paulyson-Nunez K. An altered treatment plan based on direct to consumer (DTC) genetic testing: personalized medicine from the patient/pin-cushion perspective. J Personalized Med 2012;2:192–200 10.3390/jpm2040192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inspections, Compliance, Enforcement, and Criminal Investigations—23andMe, Inc. 11/22/13. 2013. http://www.fda.gov/iceci/enforcementactions/warningletters/2013/ucm376296.htm (accessed 21 Dec 2013).
- 61.23ANDME, INC. PROVIDES UPDATE ON FDA REGULATORY REVIEW. http://mediacenter.23andme.com/press-releases/23andme-inc-provides-update-on-fda-regulatory-review/ (accessed 21 Dec 2013).
- 62.Wac K. Smartphone as a personal, pervasive health informatics services platform: literature review. Yearb Med Inform 2012;7:83–93 [published Online First: 2012/08/15]. [PubMed] [Google Scholar]
- 63.André F, Ciccolini J, Spano JP et al. . Personalized medicine in oncology: where have we come from and where are we going? Pharmacogenomics 2013;14:931–9. 10.2217/pgs.13.79 [DOI] [PubMed] [Google Scholar]
- 64.Arnedos M, Vielh P, Soria JC et al. . The genetic complexity of common cancers and the promise of personalized medicine: is there any hope? J Pathol 2014;232:274–82. 10.1002/path.4276 [DOI] [PubMed] [Google Scholar]
- 65.Bailey AM, Mao Y, Zeng J et al. . Implementation of biomarker-driven cancer therapy: existing tools and remaining gaps. Discov Med 2014;17:101–14. [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng S, Koch WH, Wu L. Co-development of a companion diagnostic for targeted cancer therapy. N Biotechnol 2012;29:682–8. 10.1016/j.nbt.2012.02.002 [DOI] [PubMed] [Google Scholar]
- 67.Donnelly E. XalKori: A new paradigm of therapeutic and companion diagnostic co-development. Regul Rapporteur 2013;10:9–11. [Google Scholar]
- 68.Van Allen EM, Foye A, Wagle N et al. . Successful whole-exome sequencing from a prostate cancer bone metastasis biopsy. Prostate Cancer Prostatic Dis 2014;17:23–7 10.1038/pcan.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waring PM. Matching patients with drugs: triumphs and challenges. Personalized Med 2006;3:335–44. 10.2217/17410541.3.3.335 [DOI] [PubMed] [Google Scholar]
- 70.Ong FS, Das K, Wang J et al. . Personalized medicine and pharmacogenetic biomarkers: progress in molecular oncology testing. Expert Rev Mol Diagn 2012;12:593–602. 10.1586/erm.12.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love D, Stratton E, Stocum M. Best practices for companion diagnostic and therapeutic development: translating between the stakeholders. New Biotechnol 2012;29:689–94 10.1016/j.nbt.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 72.Leopold C, Vogler S, Habl C et al. . Personalised medicine as a challenge for public pricing and reimbursement authorities—a survey among 27 European countries on the example of trastuzumab. Health Policy 2013;113:313–22. 10.1016/j.healthpol.2013.09.018 [DOI] [PubMed] [Google Scholar]
- 73.Ou SH, Bartlett CH, Mino-Kenudson M et al. . Crizotinib for the treatment of ALK-rearranged non-small cell lung cancer: a success story to usher in the second decade of molecular targeted therapy in oncology. Oncologist 2012;17:1351–75. 10.1634/theoncologist.2012-0311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kondylakis H, Kazantzaki E, Koumakis L et al. . Development of interactive empowerment services in support of personalised medicine. Ecancermedicalscience 2014;8:400 http://ecancer.org/journal/8/400-development-of-interactive-empowerment-services-in-support-of-personalised-medicine.php 10.3332/ecancer.2014.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lazaridis KN, McAllister TM, Babovic-Vuksanovic D et al. . Implementing individualized medicine into the medical practice. Am J Med Genet C Semin Med Genet 2014;166:15–23. 10.1002/ajmg.c.31387 [DOI] [PubMed] [Google Scholar]
- 76.Brown RE, Dorion RP, Trowbridge C et al. . Algorithmic and consultative integration of transfusion medicine and coagulation: a personalized medicine approach with reduced blood component utilization. Ann Clin Lab Sci 2011;41:211–16. [PubMed] [Google Scholar]
- 77.Sohn S, Helms TM, Pelleter JT et al. . Costs and benefits of personalized healthcare for patients with chronic heart failure in the care and education program “Telemedicine for the Heart”. Telemed J Health 2012;18:198–204 10.1089/tmj.2011.0134. [DOI] [PubMed] [Google Scholar]
- 78.MacDonald DJ, Blazer KR, Weitzel JN. Extending comprehensive cancer center expertise in clinical cancer genetics and genomics to diverse communities: the power of partnership. J Natl Compr Canc Netw 2010;8:615–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khan NA, Peterson JF. A surveillance tool to support quality assurance and research in personalized medicine. AMIA Annu Symp Proc 2011;2011:701–8. [PMC free article] [PubMed] [Google Scholar]
- 80.Roychowdhury S, Iyer MK, Robinson DR et al. . Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med 2011;3:111ra21 10.1126/scitranslmed.3003161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Printz C. Universities bring personalized medicine to the forefront of patient care: two programs aim to make customized treatments easily accessible. Cancer 2012;118:2563–4. 10.1002/cncr.27600 [DOI] [PubMed] [Google Scholar]
- 82.Teng K, Eng C, Hess CA et al. . Building an innovative model for personalized healthcare. Cleve Clin J Med 2012;79(Suppl 1):S1–9. 10.3949/ccjm.79.s1.01 [DOI] [PubMed] [Google Scholar]
- 83.Meric-Bernstam F, Farhangfar C, Mendelsohn J et al. . Building a personalized medicine infrastructure at a major cancer center. J Clin Oncol 2013;31:1849–57. 10.1200/JCO.2012.45.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patel CJ, Sivadas A, Tabassum R et al. . Whole genome sequencing in support of wellness and health maintenance. Genome Med 2013;5:58 10.1186/gm462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vorderstrasse AA, Ginsburg GS, Kraus WE et al. . Health coaching and genomics-potential avenues to elicit behavior change in those at risk for chronic disease: protocol for personalized medicine effectiveness study in air force primary care. Glob Adv Health Med 2013;2:26–38. 10.7453/gahmj.2013.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Savas BS, Cesuroglu T. Health for America: European arguments for a paradigm shift. In: Shah A, Colombano J, eds. Learning from the world: new ideas to redevelop. America Palgrave Macmillan, 2013:167–84. [Google Scholar]
- 87.Chang HL, Shaw MJ, Lai F et al. . U-Health: an example of a high-quality individualized healthcare service. Personalized Med 2010;7:677–87. 10.2217/pme.10.64 [DOI] [PubMed] [Google Scholar]
- 88.PricewaterhouseCoopers. PwC Medical Technology Innovation Scorecard Highlights 2013. http://www.pwc.com/InnovationScorecard (accessed 14 Nov 2014).
- 89.Jennings B. Liberty: Free and Equal. In: Crowley M, ed. Connecting American Values with Health Reform. New York The Hastings Center, 2009:1–3. [PubMed] [Google Scholar]
- 90.Farrugia G, Weinshilboum RM. Challenges in implementing genomic medicine: the Mayo clinic center for individualized medicine. Clin Pharmacol Ther 2013;94:204–6. 10.1038/clpt.2013.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McClure JB, Swan GE, St John J et al. . Pharmacogenetic smoking cessation intervention in a health care setting: a pilot feasibility study. Nicotine Tob Res 2013;15:518–26. 10.1093/ntr/nts173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Checking up on Taiwan healthcare—Market challenges and opportunities 2012. https://www.pwc.tw/en_TW/tw/industries/publications/assets/healthcare-en.pdf (accessed 18 Dec 2014).
- 93.Mallal S, Phillips E, Carosi G et al. . HLA-B*5701 screening for hypersensitivity to abacavir. New Engl J Med 2008;358: 568–79. 10.1056/NEJMoa0706135 [DOI] [PubMed] [Google Scholar]
- 94.Saag M, Balu R, Phillips E et al. . High sensitivity of human leukocyte antigen-B*5701 as a marker for immunologically confirmed abacavir hypersensitivity in white and black patients. Clin Infect Dis 2008;46:1111–18. 10.1086/529382 [DOI] [PubMed] [Google Scholar]
- 95.Kimmel SE, French B, Kasner SE et al. . A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med 2013;369:2283–93. 10.1056/NEJMoa1310669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pirmohamed M, Burnside G, Eriksson N et al. . A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 2013;369:2294–303. 10.1056/NEJMoa1311386 [DOI] [PubMed] [Google Scholar]
- 97.Dabrock P, Braun M, Ried J. Individualized medicine between hype and hope: exploring ethical and societal challenges for healthcare. Münster: LIT Verlag Munster, 2012. [Google Scholar]
- 98.Chabner BA, Ellisen LW, Iafrate AJ. Personalized medicine: hype or reality. Oncologist 2013;18:640–3. 10.1634/theoncologist.2013-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Greenberg T. Conference scene: personalized medicine comes to Harvard. Personalized Med 2012;9:17–18. 10.2217/pme.11.92 [DOI] [PubMed] [Google Scholar]
- 100.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature 2011;470:204–13. 10.1038/nature09764 [DOI] [PubMed] [Google Scholar]
- 101.Barnard KD, Lloyd CE, Dyson PA et al. . Kaleidoscope model of diabetes care: time for a rethink? Diabet Med 2014;31:522–30. 10.1111/dme.12400 [DOI] [PubMed] [Google Scholar]
- 102.Kazak AE. Pediatric Psychosocial Preventative Health Model (PPPHM): research, practice, and collaboration in pediatric family systems medicine. Fam Syst Health 2006;24:381–95. 10.1037/1091-7527.24.4.381 [DOI] [Google Scholar]
- 103.Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science 2010;330:460–1. 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sekirov I, Russell SL, Antunes LCM et al. . Gut microbiota in health and disease. Physiol Rev 2010;90:859–904. 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- 105.Stergiopoulos K, Brown DL. Genotype-guided vs clinical dosing of warfarin and its analogues: meta-analysis of randomized clinical trials. JAMA Intern Med 2014;174:1330–8. 10.1001/jamainternmed.2014.2368 [DOI] [PubMed] [Google Scholar]
- 106.Scott SA, Lubitz SA. Warfarin pharmacogenetic trials: is there a future for pharmacogenetic-guided dosing? Pharmacogenomics 2014;15:719–22. 10.2217/pgs.14.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Simmons LA, Wolever RQ, Bechard EM et al. . Patient engagement as a risk factor in personalized health care: a systematic review of the literature on chronic disease. Genome Med 2014;6:16 10.1186/gm533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Snyderman R, Yoediono Z. Perspective: prospective health care and the role of academic medicine: lead, follow, or get out of the way. Acad Med 2008;83:707–14. 10.1097/ACM.0b013e31817ec800 [DOI] [PubMed] [Google Scholar]
- 109.Burnette R, Simmons LA, Snyderman R. Personalized health care as a pathway for the adoption of genomic medicine. J Pers Med 2012;2:232–40. 10.3390/jpm2040232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Personalized Medicine Coalition. The case for personalized medicine (4th Edition). 4th edn Washington DC: USA: PMC, 2014:66. [Google Scholar]
- 111.Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Mol Syst Biol 2006;2:52 10.1038/msb4100095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Clayton TA, Baker D, Lindon JC et al. . Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA 2009;106:14728–33. 10.1073/pnas.0904489106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Everett JR, Loo RL, Pullen FS. Pharmacometabonomics and personalized medicine. Ann Clin Biochem 2013;50:523–45. 10.1177/0004563213497929 [DOI] [PubMed] [Google Scholar]
- 114.Klymiuk I, Högenauer C, Halwachs B et al. . A physicians’ wish list for the clinical application of intestinal metagenomics. PLoS Med 2014;11:e1001627 10.1371/journal.pmed.1001627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rogers EM. Diffusion of innovations. 3rd ed NY, USA: The Free Press, 1983. [Google Scholar]
- 116.Bijker WE. How is technology made?—That is the question! Camb J Econ 2009;34:63–76. 10.1093/cje/bep068 [DOI] [Google Scholar]
- 117.Berg M, Mol A, eds. Differences in medicine: unraveling practices, techniques, and bodies. Durham, London, NC: Duke University Press, 1998. [Google Scholar]
- 118. Periodic Report of P4-MEDICINE@EU project—Evaluation of a Best Practice Model for Personalized Health Care within Public Health Genomics (Project No.:272563). Submitted to the European Commission, Research Executive Agency, Marie Curie Actions (Intra-European Fellowships for Career Development) on 05.06.2014. http://cordis.europa.eu/result/rcn/150015_en.html (accessed 30 Jun 2016).
- 119.Hood L. Systems biology and p4 medicine: past, present, and future. Rambam Maimonides Med J 2013;4:e0012 10.5041/RMMJ.10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2015-010243supp1.pdf (60.1KB, pdf)
bmjopen-2015-010243supp2.pdf (491.2KB, pdf)