Abstract
The origin and integration of novel traits are fundamental processes during the developmental evolution of complex organisms. Yet how novel traits integrate into pre-existing contexts remains poorly understood. Beetle horns represent a spectacular evolutionary novelty integrated within the context of the adult dorsal head, a highly conserved trait complex present since the origin of insects. We investigated whether otd1/2 and six3, members of a highly conserved gene network that instructs the formation of the anterior end of most bilaterians, also play roles in patterning more recently evolved traits. Using ablation-based fate-mapping, comparative larval RNA interference (RNAi) and transcript sequencing, we found that otd1/2, but not six3, play a fundamental role in the post-embryonic formation of the adult dorsal head and head horns of Onthophagus beetles. By contrast, neither gene appears to pattern the adult head of Tribolium flour beetles even though all are expressed in the dorsal head epidermis of both Onthophagus and Tribolium. We propose that, at least in beetles, the roles of otd genes during post-embryonic development are decoupled from their embryonic functions, and that potentially non-functional post-embryonic expression in the dorsal head facilitated their co-option into a novel horn-patterning network during Onthophagus evolution.
Keywords: evolutionary novelty, insect head, co-option, post-embryonic development, gene function
1. Introduction
Any complex organism can be seen as a mosaic of discrete traits. Each trait originated at some point along a lineage's evolutionary history, and had to become integrated within a pre-existing context without compromising critical, ancestral functions. The developmental mechanisms that enable the integration of new traits into pre-existing contexts are poorly understood, as is their role in biasing, facilitating, or hindering evolutionary innovation. A growing body of work proposes that at least one major route for the origin and integration of novel traits lies in the co-option and recombination of pre-existing gene network components into new developmental contexts. Redeployment of gene networks on tissues or stages different from the ones in which these networks play their ancestral role may relax pleiotropic constraints and facilitate phenotypic innovation. This decoupling of gene networks and ancestral functional contexts may thus allow the origin of novel gene functions without the evolution of novel genes or pathways, e.g. as in the co-option of Hox genes to pattern vertebrate limbs [1], or the use of arthropod appendage patterning genes during butterfly eye spot formation [2]. If such spatial/temporal decoupling of gene networks and ancestral functions captures a general feature of innovation and integration of novel traits in developmental evolution, then even genes with deeply conserved, core functions would be available for co-option to facilitate the developmental evolution of novel structures. Furthermore, developmental contexts that are especially conducive to decoupling would then be expected to become hotspots for the evolution of novelties.
One spectacular example of a hotspot for evolutionary innovation is the dorsal head of insects. Despite having originated as part of the basic and highly conserved hexapod body plan more than 420 million years ago, the insect head has nevertheless managed to accommodate an enormous diversity of novel structures [3], such as the eye stalks of stalk-eyed flies, the weevil rostrum, or the dramatically exaggerated head horns of scarab beetles. Beetle head horns lack obvious homologues in other insects [4] and as such qualify as evolutionarily novel even by the strictest definition [5]. Head horns carry out critical functions as weapons used during male head-to-head combat [4], and previous work showed that their developmental evolution was enabled through the redeployment of several components of the arthropod appendage patterning gene network into the novel, appendage-free developmental context of the dorsal adult head [6,7]. Horned beetles in the genus Onthophagus, a model system for studying the origin and diversification of horn development, stand out with over 2 000 extant species and an enormous diversity of head horns [8]. However, how Onthophagus horns, a recent innovation, became integrated into the formation of the adult dorsal head, a conserved region in existence since the origin of insects, is entirely unclear, as are the developmental means by which head horns are positioned at species- and sex-specific locations. Here, we address these and related questions by investigating the differential redeployment and neofunctionalization of embryonic head patterning genes in adult beetle head formation.
Most arthropod body regions are composed of serially homologous segments whose identities are determined by Hox genes. However, anterodorsal head tissues pose an exception to this rule and are believed to derive from an anterior pre-segmental domain ancestral to all bilaterians (figure 1a) [9]. This anterior domain is patterned by a gene network distinctly different from the one that directs regular segment development. In the red flour beetle Tribolium castaneum, experimental embryonic knockdown of members of this network, which includes orthologues of orthodenticle, six3/optix, cap'n’collar, and tailless, results in heads with severe developmental defects [9–12]. Interestingly, it is within this cephalic domain where the diversity of Onthophagus head horns has emerged.
Figure 1.
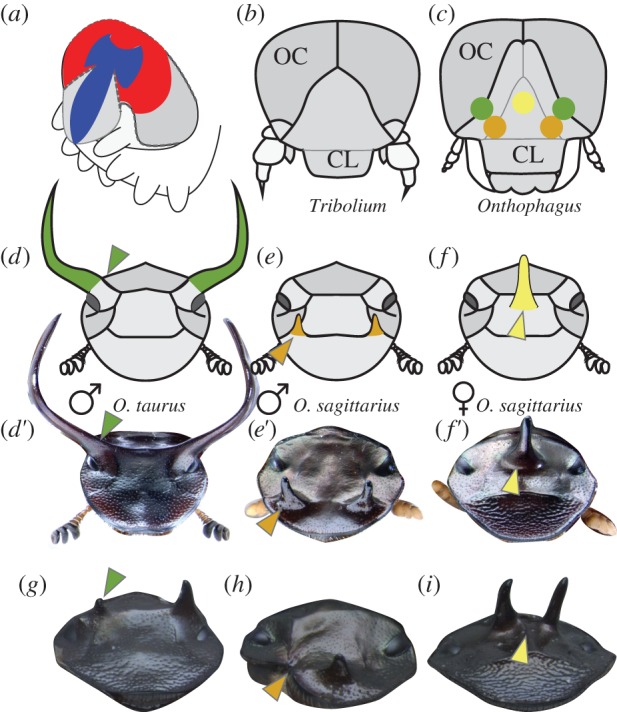
Fate maps of head horns in Onthophagus beetles. (a) Schematic of a generic late embryonic beetle head, showing the expression domains of six3 (red) and otd (blue) (after [9]). (b,c) Diagram of larval heads from Tribolium (b) and Onthophagus (c) beetles; colour dots in (c) represent regions ablated in fate-mapping experiments; OC: ocular region; CL: clypeolabral region. (d–f) Schematic and representative images of adult heads of Onthophagus taurus male (d,d′), O. sagittarius male (e,e′), and O. sagittarius female (f,f′); head horn colours represent mapping locations on the larval head shown in (c). (g–i) Examples of typical ablation phenotypes, including reduction of a posterior horn in an O. taurus male (g, arrowhead; n = 9), removal of an anterior horn in an O. sagittarius male (h, arrowhead; n = 19), and splitting of the medial posterior horn in an O. sagittarius female (i, arrowhead; n = 16). Sample size n represents the number of treated larvae surviving to adulthood; more than 90% of these showed an ablation phenotype.
In this study, we investigated the general hypothesis that genes instructing the embryonic patterning of this pre-segmental cephalic domain have been co-opted in post-embryonic Onthophagus development to pattern and position head horns. To do so, we took advantage of the fact that in beetles the adult head develops directly from the larval tissue (in contrast to dipterans like Drosophila, where all larval tissue dies and is replaced from imaginal discs and histoblasts), and developed a basic fate map to elucidate which embryonic/larval head regions are responsible for the formation and positioning of adult head horns in two closely related yet phenotypically highly divergent species of Onthophagus: O. taurus and O. sagittarius (figure 1d–f). We then measured and manipulated the gene expression of orthologues of otd and six3/optix, known core members of the embryonic anterior head gene network, in the same two species of horned beetles as well as in the red flour beetle Tribolium castaneum, which lacks horns and better reflects basal beetle head morphology. We find that, despite being expressed post-embryonically in the head epidermis of both genera, otd but not six3 dramatically affects dorsal head formation including horn development and positioning in Onthophagus, but has no effect in Tribolium.
2. Material and methods
(a). Animal husbandry and head fate mapping
Onthophagus horned beetles and Tribolium red flour beetles used in this study were obtained and reared as previously described [13,14]. The head fate map was achieved by ablating select epidermal cell groups underlying the larval head cuticle with a Hyfrecator 2000 electrosurgical unit (ConMed, Utica, NY, USA) equipped with an epilation needle electrode (714-S, ConMed). Details can be found in the electronic supplementary material.
(b). Candidate gene scan and gene expression analyses
Candidate gene sequence data for six3 and otd orthologues were retrieved from existing genomic and transcriptomic databases. Stage and tissue-specific expression levels of these genes were characterized using quantitative real-time polymerase chain reaction (qPCR) in Tribolium, and by in silico analyses of high-throughput RNAseq datasets. Details can be found in the electronic supplementary material.
(c). Cloning of six3 and otd fragments, dsRNA synthesis and injection
Using primers designed against Onthophagus six3 and otd orthologues, we amplified and cloned fragments of each gene from complementary DNA (cDNA) libraries. Cloned fragments were used as templates to synthesize dsRNA and inject it into beetle larvae as described previously [6]. We injected up to 3.0 µg of dsRNA into 417 O. taurus last instar larvae (Ot-six3: 70, Ot-otd1: 250, Ot-otd2: 53, Ot-otd1 + 2: 44) and 383 O. sagittarius larvae (Os-six3: 81, Os-otd1: 302). Sham control injections (47 O. taurus and 37 O. sagittarius) were made exactly as described above, except that larvae were injected with 1 µg of dsRNA from a 167 bp PCR product derived from a pBluescript SK vector. For Tribolium RNA interference (RNAi) we used previously described clones of Tc-otd1 and Tc-otd2 [15], and de novo synthesized 500 bp fragments (Integrated DNA Technologies (IDT) gBlock service, www.idtdna.com) based on the published Tc-six3 sequence [10]. We injected up to 0.7 µg of dsRNA; for each Tribolium gene, we made two sets of RNAi experiments targeting different transcript regions. Tribolium RNAi knockdown efficiency was validated by qPCR. We phenotypically scored 144 beetles that survived to adulthood (tc-six3: 29, tc-otd1: 41, tc-otd2: 40, tc-otd1 + 2: 34). Details can be found in the electronic supplementary material.
(d). Imaging and morphometric measurements
RNAi-treated and control Onthophagus pupae and adults were weighed on an analytical scale to the nearest milligram, and imaged through a Leica dissecting microscope (Leica, Buffalo Grove, IL, USA) mounted with a digital camera (Scion, Frederick, MD, USA) using ImageJ, and measurements recorded to the nearest 0.01 mm [16]. Thorax width was used as a measure of pupal and adult body size. Length of the pupal pronotum lateral profile was used to quantify pronotal horn size; head horn length was measured as described in [17]. For Tribolium, adults were fixed in 95% ethanol overnight. Images were captured by Zeiss Discovery V12 with an AxioCam MRc 5. The Zeiss AxioVision Extended Focus module was used to obtain images with increased depth of focus. Some pictures were enhanced only for brightness and contrast with Adobe Photoshop CS5. Details can be found in the electronic supplementary material
3. Results and discussion
(a). Onthophagus head horns map to the anterior pre-segmental region of the embryo
We sought to test the general hypothesis that genes used to pattern embryonic head development may have become redeployed during post-embryonic head differentiation. Embryonic head fate-mapping research in Tribolium has shown that the anterodorsal larval head, including the clypeolabral and ocular regions, derives from the terminal anterior region of the embryo, which undergoes a dorsal bending and subsequent dorsal closure, or zipping, of the subterminal region (a morphogenetic model known as bend and zipper [9,18], figure 1a). This region is considered pre-segmental and is characterized by a unique anterior gene network [9]. Because Tribolium and Onthophagus larvae exhibit very similar larval dorsal head morphologies (figure 1b,c), we reasoned that by relating larval to adult head regions in Onthophagus we would be able to use existing models on embryonic-to-larval transitions in Tribolium to inform our understanding of putative candidate pathways relevant to head horn development in Onthophagus. Thus, we first determined which regions of the larval head map to the adult head regions responsible for horn positioning in two closely related but morphologically highly divergent Onthophagus beetle species: O. taurus and O. sagittarius. Using selective ablation of small regions of the heads of last instar larvae, we established that posterior head horns as seen in male O. taurus are positioned in a head region homologous to the boundary between the ocular and clypeolabral head segments (figure 1d,d′,g). By contrast, the anterior paired horns of male O. sagittarius (figure 1e,e′,h) and posterior medial horn of the corresponding female (figure 1f,f′,i) are positioned in head regions more firmly contained within the clypeolabral segment. Even though our method only yields a very rough fate map and cannot discriminate whether loss of a feature is caused directly (by ablation of precursor tissue) or indirectly (by removal of inducers), our results strongly suggest that all three horn types appear to derive from the anterodorsal region of the larval head, itself derived from the embryonic anterior pre-segmental region.
(b). The embryonic anterior patterning genes sine oculis 3 and orthodenticle are expressed throughout metamorphosis in horned beetles
Since the anterior pre-segmental region is patterned by a unique gene network during embryogenesis [9], we tested whether components of this network have been co-opted to pattern horns during post-embryonic development. We focused our analysis on sine oculis 3/optix (six3) and orthodenticle (otd/otx), two genes coding for DNA-binding transcription factors, which in Tribolium exhibit complementary expression domains at the clypeolabral-ocular boundary of the presumptive embryonic head (figure 1a) [10,18,19] and, thus, the region which in Onthophagus is responsible for horn growth and integration, and which more generally play fundamental roles in patterning the anterior end across several metazoan phyla [20]. We used previously published sequences of Tribolium Tc-six3 [10], Tc-otd1, and Tc-otd2 [19] to search for orthologous genes in the O. taurus genome and an O. sagittarius transcriptome. We found a single sine oculis 3 orthologue, Ot-six3 (figure 2a) and two orthodenticle orthologues, Ot-otd1 and Ot-otd2 (figure 3a) in O. taurus. In our O. sagittarius transcriptome, we found one sine oculis 3 orthologue, Os-six3 (figure 2a), and only a single orthodenticle orthologue, Os-otd1 (figure 3a). However, we were able to use PCR to amplify a 660 bp fragment from O. sagittarius genomic DNA closely matching Ot-otd2, most likely part of Os-otd2. Alignment with Ot-otd2 shows a substitution rate of 0.05 (all synonymous) and 38.2% CG within exonic coding regions (477/660 bp), and a rate of 0.25 and 20.1% CG within intronic regions (183/660 bp). This suggests that, despite being absent from our transcript data, Os-otd2 is a functional gene, likely expressed earlier in development.
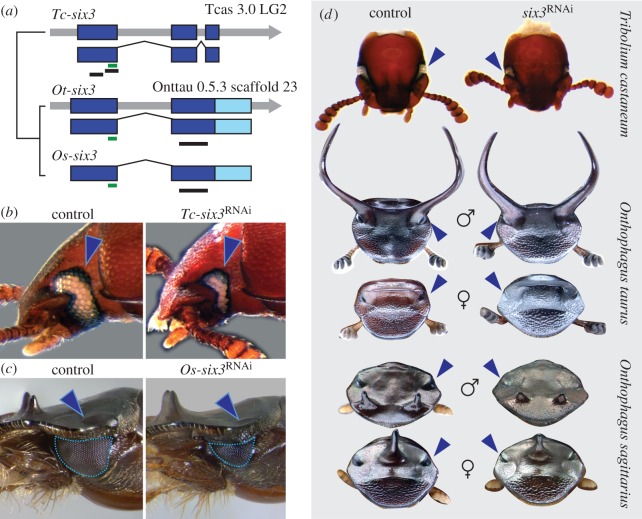
Figure 2.
Sine oculis 3/optix (six3) controls eye size but not adult dorsal head shape in beetles. (a) Gene structure of six3 orthologues from Tribolium castaneum (Tc-six3), Onthophagus taurus (Ot-six3), and O. sagittarius (Os-six3). Thick colour bars represent exons (dark blue for the coding region, light blue for the untranslated region (UTR), thin green bars highlight the homeodomain region and thin black bars show the fragments used for dsRNA interference. (b,c) Sham control (left) and six3 RNAi (right) phenotypes in Tribolium castaneum (b) and O. sagittarius (c); arrowheads point at eyes; outlines shows measured eye area. (d) Sham control (left) and six3 RNAi (right) dorsal head phenotypes in Tribolium castaneum (top), O. taurus (second and third row from top), and O. sagittarius (bottom two rows); arrowheads point at eyes.
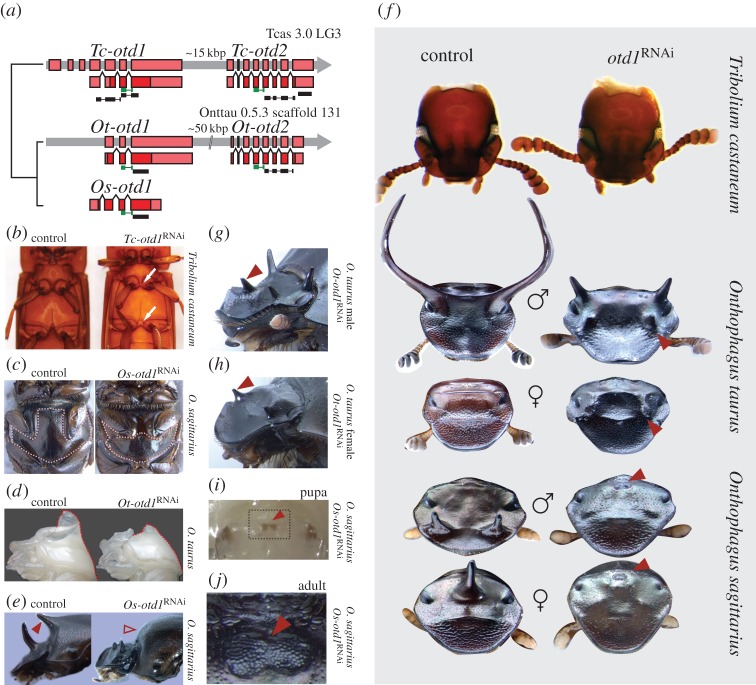
Figure 3.
Orthodenticle (otd) regulates adult body midline development in beetles, and adult dorsal head shape in Onthophagus, but not Tribolium. (a) Gene structure of otd orthologues from Tribolium castaneum (Tc-otd1 and Tc-otd2), Onthophagus taurus (Ot-otd1 and Ot-otd2), and O. sagittarius (Os-otd1). Thick colour bars represent exons (dark red for coding region, light red for UTR), thin green bars highlight the homeodomain region, and thin black bars show the fragments used for dsRNA interference. (b,c) Sham control (left) and otd1RNAi (right) ventral midline phenotypes in Tribolium castaneum (b) and O. sagittarius (c); the dotted outline demarcates the mesosternun. (d) Sham control (left) and otd1 RNAi (right) pupal horn phenotypes in O. taurus; dotted line shows pronotum profile. (e) Sham control (left) and otd1 RNAi (right) adult horn phenotypes (filled and empty arrowheads show, respectively, the presence and absence of a thoracic horn) in O. sagittarius females. (f) Sham control (left) and otd1 RNAi (right) dorsal head phenotypes in Tribolium castaneum (top), O. taurus (second and third row from top), and O. sagittarius (bottom two rows); arrowheads highlight ectopic structures. (g,h) Ectopic anterior horns (arrowhead) in adult male (g) and female (h) of O. taurus after otd1 RNAi. (i,j) Ectopic dorsal eyes (arrowhead) in pupal (i) and adult (j) of O. sagittarius after otd1 RNAi; arrowheads point at the intersection of the posterior edge of the ectopic eyes and the head midline; the boxed area in (i) corresponds approximately to the area shown in (j).
Sequence analysis and gene family phylogenetic reconstruction show that Ot-six3 and Os-six3 are indeed orthologous to Tribolium and other known insect sine oculis 3/optix genes (electronic supplementary material, figure S1). Similar analyses show that Ot-otd1 and Os-otd1 cluster with Tribolium Tc-otd1 and Drosophila otd, while Ot-otd2 groups with Tc-otd2 in a sister cluster missing in dipterans and lepidopterans (electronic supplementary material, figure S2).
Expression patterns and functions of six3, otd1, and otd2 during embryogenesis are well characterized in Tribolium [10,19], as is the function of otd genes during adult eye formation [21]. However, neither expression nor function of these genes are known in other post-embryonic contexts in Tribolium, nor during any stages of Onthophagus ontogeny. Because horns do not form until the final days of the last larval instar [13], we first sought to assess expression of six3 and otd just prior, during, and right after metamorphosis in both taxa. In Tribolium, we used quantitative PCR to confirm the expression of Tc-six3, Tc-otd1, and Tc-otd2 transcripts in late-last larval instars, pre-pupae, early pupae, as well as adults (electronic supplementary material, figure S3). In Onthophagus, we used high-throughput sequencing of total mRNA (RNAseq) across four stages of O. taurus (late-last instar larva to adult) and eight stages of O. sagittarius (mid-last instar larva to adult). We found that Onthophagus six3 is expressed during most stages we assayed, with the highest median expression found in late-last instar larvae (electronic supplementary material, figure S3). Similarly, we found that Ot-otd1 and Ot-otd2 are expressed throughout metamorphosis (electronic supplementary material, figure S3). Os-otd1 is also expressed throughout metamorphosis, albeit at very low levels; by contrast, Os-otd2 expression was not detected. We independently confirmed the presence of Ot-otd1, Ot-otd2, and Os-otd1 transcripts, and the absence of Os-otd2 transcripts, via PCR amplification of cDNA libraries from different metamorphic stages (data not shown). Our overall results show that orthologues of both six3 and otd genes, well known for their embryonic role in patterning the anterior end of metazoans [20], are expressed throughout post-embryonic beetle development.
(c). Beetle six3 is involved in post-embryonic eye development but not in horn formation
We next used larval RNAi to experimentally knockdown six3 expression and assess its potential role in adult head patterning and horn formation. We injected dsRNA fragments targeting Tc-six3, Ot-six3, and Os-six3 (figure 2a, black bars) into early-to-mid last instar larvae, followed the injected individuals through metamorphosis, and compared their phenotypes to those of control individuals injected with a non-sense dsRNA construct. In all three beetle species, six3 RNAi consistently reduced eye size compared with controls (Ot-six3: 48/50; Os-six3: 10/22; Tc-six3: 29/40+; figure 2b,c; electronic supplementary material, figure S4). However, we failed to detect any other obvious phenotypic differences between six3 RNAi and control beetles in the head (figure 2d; electronic supplementary material, figure S5) or the remainder of the body in any of the three species. In particular, we found no obvious differences in horn size or position between knockdown and control animals in either Onthophagus species. Taken together, these results suggest that six3 is involved in the regulation of post-embryonic eye development in both Onthophagus and Tribolium, but does not play a role in patterning major components of horn development in horned beetles.
(d). Otd regulates development of non-cephalic midline structures, including the thoracic horn
We used the same approach as above to investigate the potential function of Tc-otd1, Tc-otd2, Ot-otd1, Ot-otd2, and Os-otd1 (figure 3a, black bars). In all three beetle species, otd1RNAi resulted in the incomplete development of sternal structures located along the ventral midline (figure 3b,c). This phenotype was mild but evident in Tribolium (figure 3b): Tc-otd1RNAi resulted in a reduction of the T1 postcoxal bridge, T2 mesosternellum, T3 intercoxal process, and median groove, and A3 intercoxal process (Tc-otd1_F1R1: 31/36; Tc-otd1_F2R2: 10/25; see note on the electronic supplementary material, table S2). Tc-otd2RNAi individuals had either a more subtle version of the above defects (Tc-otd2_F2R2: 10/25; see note on the electronic supplementary material, table S2) or were indistinguishable from control animals (Tc-otd2_F1R1: 30/35). Similarly, in both Onthophagus species, otd1RNAi resulted in a strong reduction of the metasternal plate (Ot-otd1: 98/102; Os-otd1: 61/105), while otd2RNAi resulted in no visible phenotype (Ot-otd2: 0/33) (figure 3c; electronic supplementary material, figure S5a,b).
In Onthophagus beetles, otd1RNAi also affected thoracic horns, a dorsal midline structure. All Onthophagus pupae develop such thoracic horns on their pronotum (dorsal T1 segment) to aid in ecdysis of the larval head capsule during the larval-to-pupal moult [17,22], though only a subset of species then convert them to adult structures, while the remainder resorb them during the pupal stage via programmed cell death [23]. Among the two species studied here, only O. sagittarius females convert their pupal thoracic horn into an adult counterpart. otd1RNAi drastically reduced the size of pupal thoracic horns in males and females of both Onthophagus species (figure 3d; electronic supplementary material, figure S5c-d), and removed thoracic horns in adult female O. sagittarius (figure 3e). In Tribolium flour beetles, which lack thoracic horns in pupae or adults, neither Tc-otd1RNAi nor Tc-otd2RNAi caused any obvious change in the pronotum relative to controls. These results suggest that orthodenticle class genes play a critical role in post-embryonic patterning of epidermal midline structures in beetles. This finding is congruent with earlier work documenting midline expression of otd genes in the ventral nerve cord of late Tribolium embryos [19] and parallels results on otd function in the development of medial structures of larval and adult Drosophila [24], suggesting that the role of orthodenticle in instructing the development of midline structures may be an ancestral feature of post-embryonic insect development.
(e). otd1RNAi dramatically alters adult head formation in Onthophagus but not Tribolium
otd1RNAi resulted in strong cephalic phenotypes in both O. taurus and O. sagittarius but not in Tribolium (figure 3f). Specifically, otd1RNAi caused a reduction or deletion of the dorsal cephalic midline region, resulting in an apparent ‘lateralization’ of the head relative to sham controls. By contrast, no defects were observed on the ventral head. otd1RNAi also dramatically affected horn formation, resulting in complete loss or severe reduction of the paired posterior head horns in larger males of O. taurus (figure 3f; electronic supplementary material, figure S5e; 9/9 males large enough to be horned; note that smaller males are normally hornless in this species), the paired anterior horns of male O. sagittarius (25/30), and the single medial posterior horn of female O. sagittarius (20/21) (figure 3f; electronic supplementary material, figure S5f).
Strikingly, otd1 RNAi not only reduced or deleted head horns, but also consistently induced the formation of an ectopic pair of anterior horns in O. taurus (69/86, figure 3g,h). Ectopic horns were present in both males and females, and located at a position homologous to the lateral ends of an anterior cephalic ridge usually found in wild-type O. taurus females, thus resembling the paired anterior horns of wild-type male O. sagittarius in placement (figure 3f).
While ectopic anterior horns were consistently found in O. taurus Ot-otd1RNAi individuals, they were never observed in O. sagittarius. Instead, in O. sagittarius, Os-otd1 RNAi resulted in a fairly penetrant and completely unexpected phenotype: development of single or paired compound eyes at or near the midline of the posterodorsal head (43/61, figure 3f,i,j; electronic supplementary material, figure S6a–c). Ectopic dorsal eyes became evident during early pupal development and varied in size, but consistently presented all external features typical of regular compound eyes, including hexagonal ommatidia and lenses. By contrast, we never observed ectopic eyes in O. taurus Ot-otd1RNAi individuals or in sham controls in either species (but see below).
In Tribolium, in contrast, larval Tc-otd1 RNAi using constructs targeting two different regions of the gene had no discernible effect on pupal or adult head morphology (figure 3f), despite all individuals showing midline defects at other regions of the body (figure 3c). qPCR measurements detected significantly lower levels of Tc-otd1 transcripts in Tc-otd1RNAi pupae relative to controls, both for whole-body and tissue-specific samples (electronic supplementary material, figure S9), indicating that the lack of a head phenotype is not due to systemically or locally ineffective gene knockdown after dsRNA injections. These results stand in stark juxtaposition to the striking otd1RNAi phenotypes found in Onthophagus beetles, suggesting that the lineage including Tribolium either secondarily lost the ancestral role of otd1 in post-embryonic head development, or alternatively, that such a role may be a novel developmental feature unique to the lineage leading to Onthophagus.
(f). Nonlinear effects of Ot-otd1 and Ot-otd2 RNAi suggest partial redundancy of orthodenticle class transcription factors
In contrast to the striking phenotypes resulting from otd1 RNAi, otd2RNAi individuals exhibited no detectable morphological differences compared to controls in any obvious aspect of head or horn morphology (electronic supplementary material, figure S5). This may be due to otd2 simply being non-functional during post-embryonic development, or alternatively, to otd2 being partially redundant to otd1, so that it can be functionally replaced by otd1, but itself can only partially replace otd1 functions.
To discriminate between these alternate hypotheses, we performed simultaneous larval RNAi against otd1 and otd2 (otd1 + 2) in O. taurus and Tribolium. In O. taurus, Ot-otd1 + 2RNAi resulted in strong reduction of metasternal plates and thoracic pupal horns (electronic supplementary material, figure S5a, c), and complete deletion of paired posterior horns in males (13/14) and a reduced proportion of ectopic anterior horns compared with Ot-otd1RNAi individuals (Ot-otd1 + 2: 10/25; Ot-otd1: 69/86). Surprisingly, four out of 25 otd1 + 2RNAi animals surviving to adulthood also developed ectopic compound eyes at a medial posterodorsal region of the head, similar to Os-otd1RNAi individuals in O. sagittarius (electronic supplementary material, figure S6d-f). Similarly, double knockdown of Tc-otd1 and Tc-otd2 in Tribolium resulted in defects along the ventral midline more pronounced than those described for Tc-otd1 RNAi alone (electronic supplementary material, figure S7), but had no detectable effects on the dorsal head shape Together, these results support the hypothesis that orthodenticle class transcription factors show asymmetric functional redundancy [21]. If correct, this could also explain the difference in otd1 phenotypes observed between O. taurus and O. sagittarius: since we were not able to detect Os-otd2 transcripts in O. sagittarius, it is conceivable that this gene is not actively transcribed during post-embryonic development in this species and thus unable to partially rescue Os-otd1RNAi individuals.
(g). Beetle otd1 and otd2 are expressed in epidermal tissues at early pupal stages
Our otd RNAi experiments dramatically affected the formation of head and thoracic horns, i.e. evolutionary novel structures of strictly epidermal origin. However, current knowledge of otd embryonic expression in Tribolium emphasizes that in older embryos the expression domain of otd genes becomes restricted to the brain, ventral nerve cord, and eyes [19]. This raises the possibility that otd expression in Onthophagus either remains restricted to nervous tissues during post-embryonic development, and epidermal development is indirectly controlled by otd through signalling between tissues or, alternatively, that otd is post-embryonically redeployed in epidermal tissues and directly patterns local epidermal development. To distinguish between these scenarios, we used RNAseq of early O. taurus pupae to estimate tissue-specific gene expression for three epidermally derived tissues that vary in the degree to which they are affected by otd knockdown—dorsal head epidermis, T1 dorsal epidermis, and posterior ventral abdominal epidermis—as well as brain tissue. Our results indicate that Ot-otd1 expression levels relative to abdominal epidermis are approximately four times higher in the brain, approximately 24 times higher in the T1 dorsal epidermis, and approximately 42 times higher in the dorsal head epidermis (electronic supplementary material, figure S8, bottom row). By contrast, Ot-otd2 levels are only approximately two times higher in the brain, one half in the T1 dorsal epidermis, and approximately five times higher in the dorsal head epidermis (electronic supplementary material, figure S8). Thus, our results support a scenario whereby otd genes are redeployed in epidermal tissues during post-embryonic development.
Next, we sought to determine whether otd expression in the dorsal head epidermis is restricted to horned beetles. Using tissue-specific qPCR in early pupae of Tribolium, a taxon belonging to a family lacking any type of head and thoracic horns, we found that both Tc-otd1 and Tc-otd2 exhibit relatively high expression levels in the dorsal head epidermis (electronic supplementary material, figure S8, top row), suggesting that post-embryonic expression of otd class genes during head development may be a shared ancestral feature, at least among beetle families.
Importantly, independent work on Drosophila also documented late post-embryonic otd expression and function in the medial region of the eye-antennal, leg, and genital discs, as well as ventral epidermis [24]. Moreover, adult midline defects resulting from otd knockdown in Drosophila [24,25] are at least partly reminiscent of those described here for beetles, including defects to the dorsal head. However, it is worth emphasizing that head formation in cyclorrhaphan flies such as Drosophila is highly derived and non-representative compared with that of most other holometabolous insects, including beetles. Specifically, in complete contrast to head formation in Onthophagus and Tribolium, no larval head epidermis is used during metamorphosis to build the adult head of Drosophila, which instead derives anew from anterior imaginal discs and histoblasts [26]. It is therefore difficult to ascertain whether superficial similarities in otd function during dorsal adult head formation reflect a synapomorphy shared across holometabolous insects but lost in Tribolium, or independent functional co-options of this gene class in Onthophagus horned beetles and fruit flies. Furthermore, we cannot completely exclude the possibility that otd does have a function in Tribolium adult head formation, but defects caused by RNAi are too subtle to be detected by our methods. Even though a wider taxonomic sampling is clearly necessary to address this question, our results nevertheless highlight the conservation of post-embryonic otd expression in insects, and its independent co-option in the formation and integration of novel head structures in Onthophagus.
(h). Orthodenticle genes regulate post-embryonic patterning of evolutionary novel structures
Insects in general, and beetles in particular, present an enormous diversity of head morphologies. Moreover, from the stalks of stalk-eyed flies and the rostrum of weevils to the horns of scarab beetles, insects have managed to integrate numerous novel cephalic features into head formation, yet how such innovation and integration was enabled during developmental evolution remains largely unclear [9]. A growing body of evidence emphasizes the recruitment of modular developmental genetic components into new ontogenetic contexts as a central mechanism facilitating the integration of novel traits into ancestral contexts without necessitating the origin of novel genes or pathways [27,28]. Here, we examined six3 and otd, two members of an ancient gene network controlling embryonic head formation across diverse phyla [20]. In insects, these transcription factors tightly interact and cooperate to pattern the larval head from the anterior pre-segmental region of the embryo [9–12]. However, despite this deep phylogenetic and functional entrenchment, these genes appear to have become disconnected during post-embryonic development of the adult beetle head: six3 regulates adult compound eye size, but is not involved in shaping other adult structures. By contrast, otd regulates post-embryonic development of epidermal midline structures along the body in both horned and flour beetles and, most significantly, regulates the formation of cephalic and thoracic horns, true and rather recent evolutionary novelties in the genus Onthophagus. As such, the adult six3 and otd RNAi phenotypes we describe here are markedly different from those seen after embryonic knockdown [10–12]. Furthermore, we found no evidence that the tight embryonic interaction between these genes is present during post-embryonic development, showing that gene coupling is strongly context dependent. If this is true for other genes that pattern the anterior pre-segmental region, this suggests that the insect dorsal head may be decoupled from, and less constrained than, its appendage-bearing ventral counterpart, thus explaining why this region is such a hotspot of morphological diversity.
(i). Neofunctionalization of orthodenticle may have been facilitated by latent yet non-adaptive tissue-specific expression in beetle heads
Recruitment of ancestral transcription factors into new developmental contexts requires the evolution of novel spatio-temporal expression domains, developmental genetic interactions or both. Here, we document a novel function for orthodenticle class genes in patterning apomorphic structures in the dorsal heads of adult Onthophagus beetles. Notice that our approach was not predicated on the implicit expectation of gene sequence or structure change upon acquisition of a novel function. Function emerges through context and interactions, and changes in functionality during development may therefore result from changes in the associated genes themselves, their interactions, or the context within which they occur. Thus, just as a hammer (whose original function is hammering nails) can be used for a novel function as a nutcracker without any fundamental changes in the hammer itself, a gene can acquire a novel function by virtue of being redeployed in a novel context or by changes in its regulatory interactions with other genes, without a single structural change within its coding region. A scenario of evolution by contextual novelty is also supported by our results showing that both otd orthologues are expressed at relatively high levels in the dorsal head epidermis of early pupae, suggesting that post-embryonic development of dorsal head structures including horns is directly regulated by tissue-specific expression of orthodenticle class genes, rather than inductive interactions with the nervous system. Strikingly, we also found evidence for the expression of otd in the dorsal head epidermis of Tribolium flour beetle pupae, even though otdRNAi individuals showing knockdown phenotypes in the ventral midline did not show any defect on their dorsal heads. Although a wider taxonomic sampling is needed to ascertain dorsal head specific post-embryonic otd expression and function, it is tempting to speculate that if latent yet non-functional expression of otd is common to most beetles, then such pre-existing, function-less, yet already tissue- and body region-specific expression patterns could have provided a critical developmental genetic substrate for the evolution of novel head morphologies. More generally, the strong spatio-temporal phylogenetic conservation of otd expression on one side, coupled with the frequent acquisition of diverse functions observed across Metazoan phyla on the other [11–13,25,29–32], suggests that orthodenticle class transcription factors may be particularly suitable to provide positional information facilitating the patterning of evolutionarily novel structures.
Supplementary Material
Acknowledgements
We would like to thank Erik Parker, Adam Moore, Alexander Neufeld, Justin Song, Keeley Newsom, Justine Christian, Hailey Riggs, Peyton Joachim, and Bonnie Young for expert help with beetle collecting and care. We also thank Melissa Pespeni for generating the developmental RNAseq dataset, Cris Ledon-Rettig, James Ford, and the Indiana University Center for Genomics and Bioinformatics for help with tissue-specific RNAseq. Finally, we would like to thank our two anonymous reviewers who provided valuable, detailed, and insightful feedback that greatly helped improve this manuscript.
Data accessibility
The datasets supporting the conclusions of this article are available from the Dryad repository (https://datadryad.org/resource/doi:10.5061/dryad.nd1js) [33].
Authors' contributions
E.E.Z. and A.P.M. designed the experiments; H.A.B. performed ablation experiments in Onthophagus spp.; E.E.Z. performed all other experiments in Onthophagus spp.; D.M.L. and Y.T. performed all experiments in Tribolium castaneum; E.E.Z. and A.P.M. analysed the data and wrote the manuscript. All authors read and approved the manuscript for publication.
Competing interests
All authors declare that they have no competing interests.
Funding
Funding for this study was provided by National Science Foundation grants IOS 1256689 and IOS 1120209 to A.P.M., and IOS 0950964 and IOS 1557936 to Y.T.
References
- 1.Zakany J, Duboule D. 2007. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 17, 359–366. ( 10.1016/j.gde.2007.05.011) [DOI] [PubMed] [Google Scholar]
- 2.Carroll SB, Gates J, Keys DN, Paddock SW, Panganiban GE, Selegue JE, Williams JA. 1994. Pattern formation and eyespot determination in butterfly wings. Science 265, 109–114. ( 10.1126/science.7912449) [DOI] [PubMed] [Google Scholar]
- 3.Grimaldi D, Engel MS. 2005. Evolution of the insects. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 4.Moczek AP. 2005. The evolution and development of novel traits, or how beetles got their horns. BioScience 55, 937–951. ( 10.1641/0006-3568(2005)055%5B0937:TEADON%5D2.0.CO;2) [DOI] [Google Scholar]
- 5.Müller GB, Wagner GP. 1996. Homology, hox genes, and developmental integration. Am. Zool. 36, 4–13. ( 10.1093/icb/36.1.4) [DOI] [Google Scholar]
- 6.Moczek AP, Rose DJ. 2009. Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc. Natl Acad. Sci. USA 106, 8992–8997. ( 10.1073/pnas.0809668106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kijimoto T, Pespeni M, Beckers O, Moczek AP. 2013. Beetle horns and horned beetles: emerging models in developmental evolution and ecology. Wiley Interdiscip. Rev. Dev. Biol. 2, 405–418. ( 10.1002/wdev.81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emlen DJ, Marangelo J, Ball B, Cunningham CW. 2005. Diversity in the weapons of sexual selection: horn evolution in the beetle genus Onthophagus (coleoptera: Scarabaeidae). Evolution 59, 1060–1084. ( 10.1111/j.0014-3820.2005.tb01044.x) [DOI] [PubMed] [Google Scholar]
- 9.Posnien N, Schinko JB, Kittelmann S, Bucher G. 2010. Genetics, development and composition of the insect head—a beetle's view. Arthropod. Struct. Dev. 39, 399–410. ( 10.1016/j.asd.2010.08.002) [DOI] [PubMed] [Google Scholar]
- 10.Posnien N, Koniszewski NDB, Hein HJ, Bucher G. 2011. Candidate gene screen in the red flour beetle Tribolium reveals Six3 as ancient regulator of anterior median head and central complex development. PLoS Genet. 7, e1002416 ( 10.1371/journal.pgen.1002416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schinko JB, Kreuzer N, Offen N, Posnien N, Wimmer EA, Bucher G. 2008. Divergent functions of orthodenticle, empty spiracles and buttonhead in early head patterning of the beetle Tribolium castaneum (Coleoptera). Dev. Biol. 317, 600–613. ( 10.1016/j.ydbio.2008.03.005) [DOI] [PubMed] [Google Scholar]
- 12.Kotkamp K, Klingler M, Schoppmeier M. 2010. Apparent role of Tribolium orthodenticle in anteroposterior blastoderm patterning largely reflects novel functions in dorsoventral axis formation and cell survival. Development 137, 1853–1862. ( 10.1242/dev.047043) [DOI] [PubMed] [Google Scholar]
- 13.Moczek AP, Nagy LM. 2005. Diverse developmental mechanisms contribute to different levels of diversity in horned beetles. Evol. Dev. 7, 175–185. ( 10.1111/j.1525-142X.2005.05020.x) [DOI] [PubMed] [Google Scholar]
- 14.Philip B, Tomoyasu Y. 2011. Gene knockdown analysis by double-stranded RNA injection. In Molecular methods for evolutionary genetics (eds Orgogozo V, Rockman MV), pp. 471–497. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- 15.Linz DM, Tomoyasu Y. 2015. RNAi screening of developmental toolkit genes: a search for novel wing genes in the red flour beetle, Tribolium castaneum. Dev. Genes Evol. 225, 11–22. ( 10.1007/s00427-015-0488-1) [DOI] [PubMed] [Google Scholar]
- 16.Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11, 36–42. [Google Scholar]
- 17.Moczek AP. 2006. Pupal remodeling and the development and evolution of sexual dimorphism in horned beetles. Am. Nat. 168, 711–729. ( 10.1086/509051) [DOI] [PubMed] [Google Scholar]
- 18.Posnien N, Bucher G. 2010. Formation of the insect head involves lateral contribution of the intercalary segment, which depends on Tc-labial function. Dev. Biol. 338, 107–116. ( 10.1016/j.ydbio.2009.11.010) [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Brown SJ, Hausdorf B, Tautz D, Denell RE, Finkelstein R. 1996. Two orthodenticle-related genes in the short-germ beetle Tribolium castaneum. Dev. Genes Evol. 206, 35–45. ( 10.1007/s004270050028) [DOI] [PubMed] [Google Scholar]
- 20.Steinmetz PR, et al. 2010. Six3 demarcates the anterior-most developing brain region in bilaterian animals. EvoDevo 1, 14 ( 10.1186/2041-9139-1-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahato S, Morita S, Tucker AE, Liang X, Jackowska M, Friedrich M, Shiga Y, Zelhof AC. 2014. Common transcriptional mechanisms for visual photoreceptor cell differentiation among pancrustaceans. PLoS Genet. 10, e1004484 ( 10.1371/journal.pgen.1004484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moczek AP, Cruickshank TE, Shelby A.. 2006. When ontogeny reveals what phylogeny hides: gain and loss of horns during development and evolution of horned beetles. Evolution 60, 2329–2341. ( 10.1111/j.0014-3820.2006.tb01868.x) [DOI] [PubMed] [Google Scholar]
- 23.Kijimoto T, Andrews J, Moczek AP. 2010. Programed cell death shapes the expression of horns within and between species of horned beetles. Evol. Dev. 12, 449–458. ( 10.1111/j.1525-142X.2010.00431.x) [DOI] [PubMed] [Google Scholar]
- 24.Wieschaus E, Perrimon N, Finkelstein R. 1992. orthodenticle activity is required for the development of medial structures in the larval and adult epidermis of Drosophila. Development 115, 801–811. [DOI] [PubMed] [Google Scholar]
- 25.Royet J, Finkelstein R. 1996. hedgehog, wingless and orthodenticle specify adult head development in Drosophila. Development 122, 1849–1858. [DOI] [PubMed] [Google Scholar]
- 26.Haynie JL, Bryant PJ. 1986. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J. Exp. Zool. 237, 293–308. ( 10.1002/jez.1402370302) [DOI] [PubMed] [Google Scholar]
- 27.Shubin N, Tabin C, Carroll S. 2009. Deep homology and the origins of evolutionary novelty. Nature 457, 818–823. ( 10.1038/nature07891) [DOI] [PubMed] [Google Scholar]
- 28.Monteiro A, Podlaha O. 2009. Wings, horns, and butterfly eyespots: how do complex traits evolve? PLoS Biol. 7, e1000037 ( 10.1371/journal.pbio.1000037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen S, Reichert H, VijayRaghavan K. 2013. Conserved roles of ems/Emx and otd/Otx genes in olfactory and visual system development in Drosophila and mouse. Open Biol. 3, 120177 ( 10.1098/rsob.120177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nederbragt AJ, te Welscher P, van den Driesche S, van Loon AE, Dictus WJ. 2002. Novel and conserved roles for orthodenticle/otx and orthopedia/otp orthologs in the gastropod mollusc Patella vulgata. Dev. Genes Evol. 212, 330–337. ( 10.1007/s00427-002-0246-z) [DOI] [PubMed] [Google Scholar]
- 31.Blanco J, Pandey R, Wasser M, Udolph G. 2011. Orthodenticle is necessary for survival of a cluster of clonally related dopaminergic neurons in the Drosophila larval and adult brain. Neural Dev. 6, 34 ( 10.1186/1749-8104-6-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. 2003. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev. Cell 5, 621–633. ( 10.1016/S1534-5807(03)00293-4) [DOI] [PubMed] [Google Scholar]
- 33.Zattara E, Busey H, Linz D, Tomoyasu Y, Moczek A. 2016. Data from: Neofunctionalization of embryonic head patterning genes facilitates the positioning of novel traits on the dorsal head of adult beetles. ( 10.5061/dryad.nd1js) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are available from the Dryad repository (https://datadryad.org/resource/doi:10.5061/dryad.nd1js) [33].