Abstract
Glaucoma related proteomic changes have been documented in cell and animal models. However, proteomic studies investigating on human retina samples are still rare. In the present work, retina samples of glaucoma and non-glaucoma control donors have been examined by a state-of-the-art mass spectrometry (MS) workflow to uncover glaucoma related proteomic changes. More than 600 proteins could be identified with high confidence (FDR < 1%) in human retina samples. Distinct proteomic changes have been observed in 10% of proteins encircling mitochondrial and nucleus species. Numerous proteins showed a significant glaucoma related level change (p < 0.05) or distinct tendency of alteration (p < 0.1). Candidates were documented to be involved in cellular development, stress and cell death. Increase of stress related proteins and decrease of new glaucoma related candidates, ADP/ATP translocase 3 (ANT3), PC4 and SRFS1-interacting protein 1 (DFS70) and methyl-CpG-binding protein 2 (MeCp2) could be documented by MS. Moreover, candidates could be validated by Accurate Inclusion Mass Screening (AIMS) and immunostaining and supported for the retinal ganglion cell layer (GCL) by laser capture microdissection (LCM) in porcine and human eye cryosections. The workflow allowed a detailed view into the human retina proteome highlighting new molecular players ANT3, DFS70 and MeCp2 associated to glaucoma.
Glaucoma is a neurodegenerative ocular disease complex with multifactorial etiology displaying a characteristic damage pattern in the optic nerve head manifested by a progressive loss of retinal ganglion cells (RGCs), their axons and visual field1,2. In 2010, worldwide 60.5 million people suffered from glaucoma and over 70 million people are expected to develop glaucoma by 2020 making glaucoma to one of the leading causes of blindness affecting people of all ages, however with increasing prevalence with age3. Inflammatory4 and autoimmune processes5,6 implementing oxidative stress7 and mitochondrial dysfunction8,9 have been documented for glaucoma so far and underlying molecular mechanisms are shifting in focus of research. Accordingly, numerous studies have demonstrated proteomic changes referring to experimental in-vivo and in-vitro models, regarding rodents10,11,12 and primates13. Glaucoma related proteomic alterations have also been reported for human sample material, e.g. aqueous humor14,15,16,17,18,19, trabecular meshwork20,21 and tears22 proposing key proteins and biomarker candidates. Focusing on human glaucomatous retina samples, Tezel et al. documented hemoglobin upregulation using immunohistochemistry23, whereas the research groups of Yang and Luo revealed proteomic changes linked to TNF-α/TNFR124 and Toll-like receptors (TLRs)25 using mass spectrometry. However, proteomic investigations on human retinal sample species are rare due to limitation of donors and human sample material. Therefore, the purpose of the present study was to analyze human retinal samples of glaucoma (N = 5) and non-glaucoma subjects (N = 5) by use of a state-of-the-art “bottom-up” high performance liquid chromatography electro spray ionization mass spectrometry (BU LC ESI MS) workflow. Retinal sample exploration should provide an in-depth view to the human retina proteome, reveal glaucomatous proteomic alterations and should contribute to a better understanding of the molecular pathomechanism of glaucoma. Moreover, new molecular candidates linked to glaucomatous neurodegeneration should be proposed giving direction for future glaucoma research projects.
Results
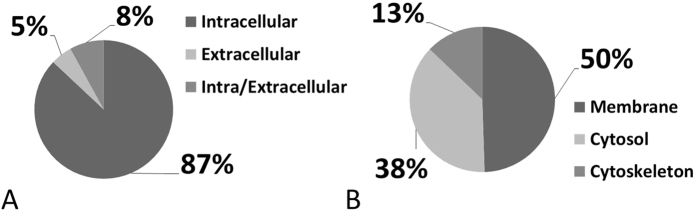
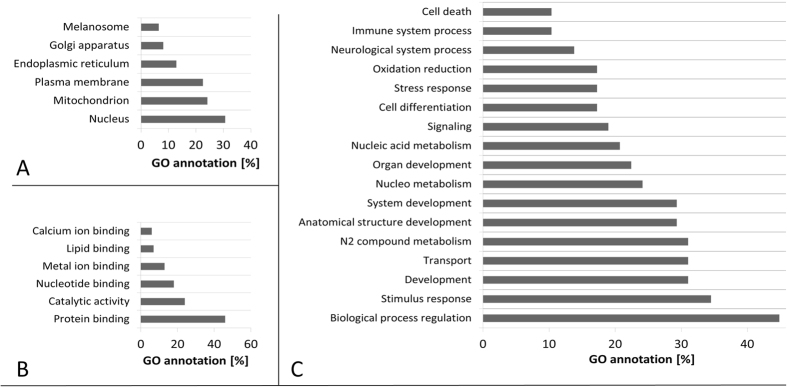
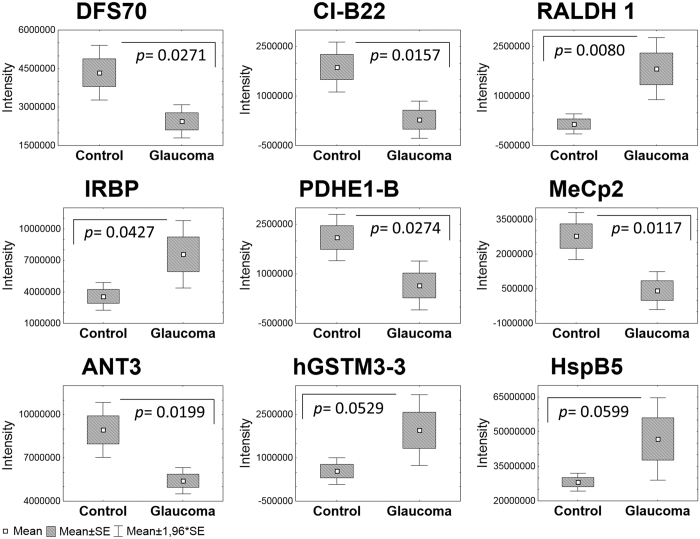
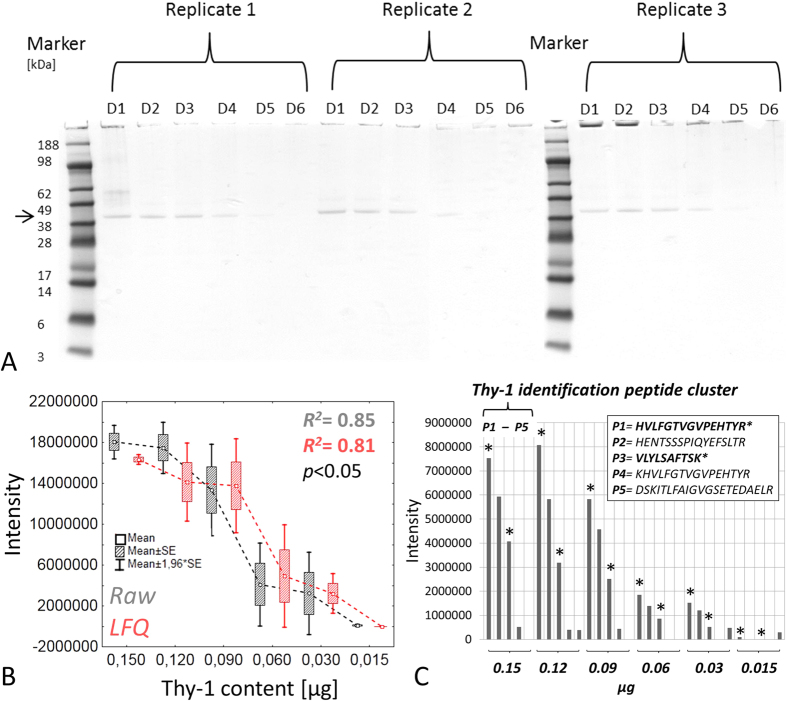
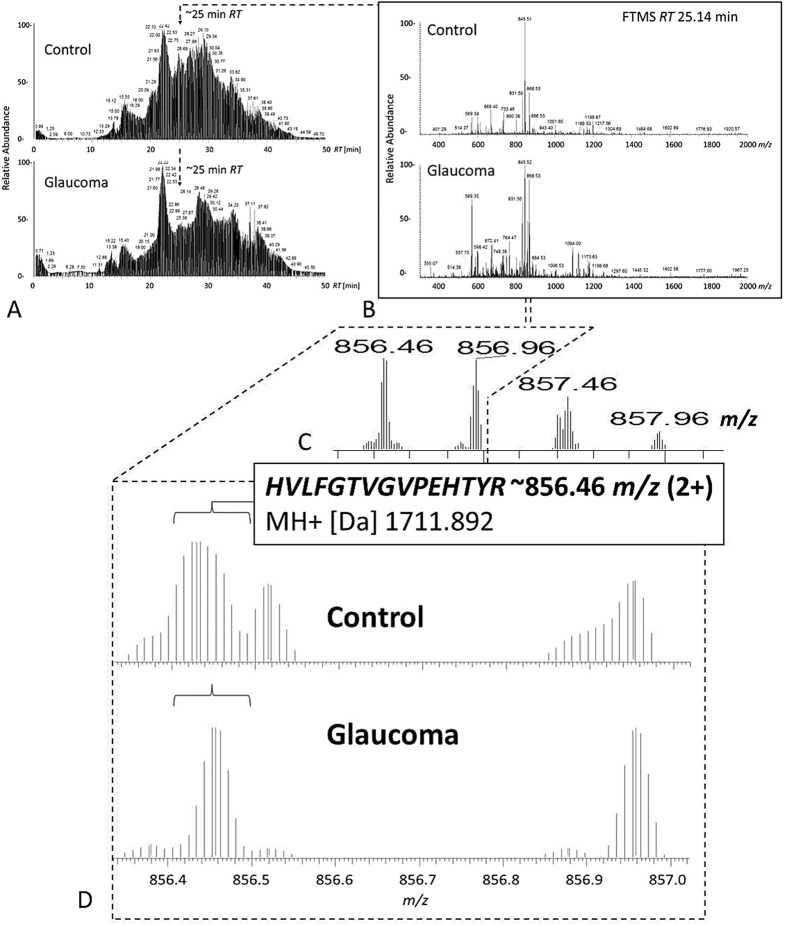
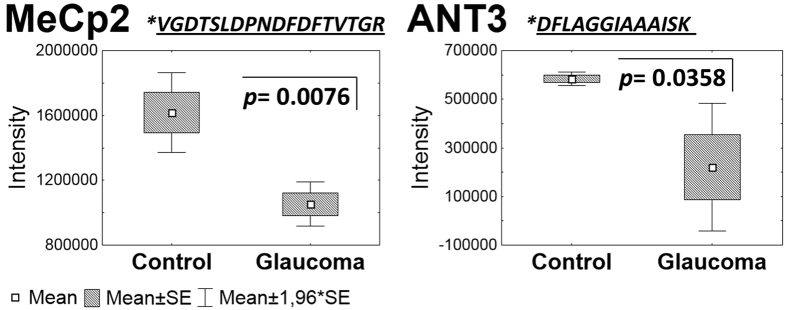
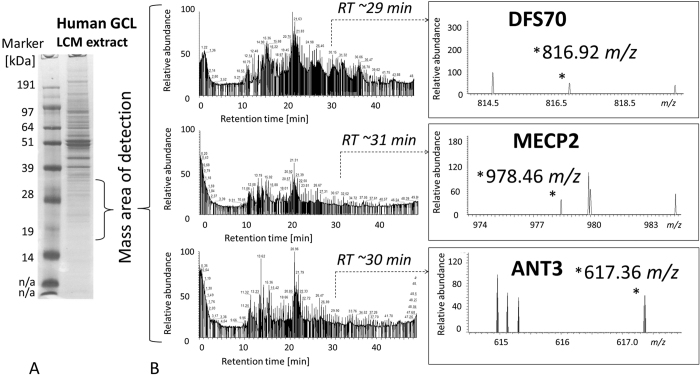
Individual human retina samples (Glaucoma, N = 5; Control, N = 5) showed a high pattern congruency regarding 1D-SDS-PAGE with the exception of glaucoma donor #G3, which displayed an exclusively distinct protein cluster between ~15 and 20 kDa (Fig. 1), primarily referring to exorbitant crystalline upregulation. Due to the risk of masking of low abundant effects glaucoma donor #G3 was excluded from quantitative statistical analysis. However, some samples generated more background than others without affecting the proteomic pattern. BU LC ESI MS analysis of human retina extracts resulted in highly qualitative total ion current (TIC) chromatograms displaying proper peptide elution within 50 min HPLC gradient period leading to accurate protein identification (Fig. 2). Thereby, a sensitive view to the human retina proteome could be obtained. In summary, more than 600 retinal proteins were identified with stringent false discovery rate (FDR < 1%; Supplementary information). For qualitative description of the human retina proteome, exemplary photoreceptor specific proteins like interphotoreceptor matrix proteoglycan 126, rod outer segment membrane protein 127 or rhodopsin28, but also numerous synaptic neuroproteins, e.g. neuroplastin29, synaptogyrin-130, synaptophysin31, synaptosomal-associated protein 2532 or synaptotagmin-133 could be mapped. Additional retina and optic nerve resident proteins, e.g. astrocytic phosphoprotein PEA-1534, neuronal cell adhesion molecule35, glial abundant proteins neurocalcin-Σ36 and vimentin37 could be detected. Beside reported RGC abundant proteins, Thy-1 membrane glycoprotein, tubulin-ß 3 chain and neurofilament light polypeptide, the RGC selective marker protein RNA binding protein with multiple splicing (RBPMS)38,39 could not be confirmed in human retinal samples in this work. In contrast, neuronal relevant retinal proteins, non-POU domain-containing octamer-binding protein40 and glial fibrillary acidic protein (GFAP)41, could be recovered. Therefore, analyzed proteins qualitatively represent the functional human retina proteome featuring the ganglion cell layer (GCL). Interestingly, MUC-9 and proline-rich protein 4 representing characteristic ocular surface proteins could be detected in the samples, most likely artificially derived from the ocular surface during sample preparation. Regarding functional analysis, 95% of mapped retinal proteins could be GO annotated and have been primarily revealed as intracellular proteins. Inferred from label-free quantification (LFQ) statistics, approximately 10% of the whole retina proteome showed distinct alterations between glaucoma and healthy samples emphasized by “unique” LFQ attendance, significant group-specific abundance alterations (p < 0.05) or distinct changes by trend (p < 0.1) (Table 1). In particular, altered candidate proteins predominantly represent intracellular, membrane-resided species (Fig. 3), within the nucleus and/or mitochondria (Fig. 4A). Moreover, candidates were dominated by binding and catalytic features and were found to be involved in cellular key processes like development, cellular transport and cell death (Fig. 4B,C). Proteins like V-type proton ATPase 116 kDa subunit A isoform 1 (VPP1) or arrestin-C (cArr) were revealed below the LFQ abundance threshold in glaucoma samples. Three nucleus proteins, methyl-CpG-binding protein 2 (MeCp2), PC4 and SRFS1-interacting protein 1 (DFS70) and 40S ribosomal protein S7, were revealed to be significantly diminished (p < 0.05) in glaucoma retina samples (Table 1). In contrast, nucleus proteins, guanine nucleotide-binding protein G(i) subunit α-2 and histone H1.0, showed strong tendencies of glaucoma associated level decrease (p < 0.1) (Table 1). Beside nucleus proteins, numerous mitochondrial proteins displayed glaucoma associated level decrement, e.g. ADP/ATP translocase 3 (ANT3), pyruvate dehydrogenase component subunit ß (PDHE1-B) and cytochrome C oxidase subunit 7A2 (COX7A2) (Table 1, Fig. 5). In contrast, retinal dehydrogenase 1 (RALDH 1), retinol-binding protein 3 (IRBP) (Fig. 5) and vesicle-associated membrane protein-associated protein B/C, showed significant level enhancement referring to glaucoma (p < 0.05) (Table 1). Also, stress related proteins including crystallins, serotransferrin and glutathione metabolic proteins indicated level elevation in samples of glaucoma subjects (Table 1, Fig. 5). Since Thy-1, an important RGC marker protein, Thy-1 membrane glycoprotein42, could exclusively been LFQ substantiated in glaucoma samples (Table 1), Thy-1 recovery was further inspected. Thereby, BU LC MS determination of the dynamic range of Thy-1 using recombinant protein showed a linear correlation between MS abundance and Thy-1 content between 0.15 and 0.015 μg before and after quantification (R2 > 0.8; p < 0.05). In the range <0.03 μg Thy-1 raw intensities approximate a value of 1*E6 generating a LFQ zero value during quantification. Moreover, in this low abundant range only one of initial five Thy-1 specific peptides is detectable, which indicate critical detection of Thy-1 <0.03 μg (Fig. 6). Reanalysis of the study samples supported the presence of Thy-1 in both groups; glaucoma and control, based on raw intensities. Thereby, the protein detection corresponded to two Thy-1 characteristic peptides, which abundances could be recorded close to the detection threshold. BU LC MS analysis of a glaucoma and a control pool of remaining sample material supported identification of Thy-1 in glaucoma samples based on one characteristic peptide by MS/MS. Nevertheless, manual inspection of corresponding chromatograms also supported existence of Thy-1 in the control pool indicated by the identical diagnostic reporter peptide, however not leading to MS/MS based identification (Fig. 7). Using Laser capture microdissection (LCM) several proteins proposed to be particularly glaucoma-associated, e.g. MeCp2, VPP1 or ANT3, could be significantly recovered (p < 0.05) from the GCL of porcine retinae utilizing the BU LC ESI MS platform (Fig. 8, Table 1). Representative validation of glaucoma related proteomic changes was realized in pooled in-solution digests of retina lysates by targeted Accurate Inclusion Mass Screening (AIMS) strategy considering limited sample material (Table 1, Fig. 9). Thereby, selection of a five candidate subset including ANT3, MeCp2, DFS70, cArr and HspB5 was based on functional representation (mitochondria, nucleus, stress indication) and/or GCL support as well as on possibility of redetection of diagnostic reporter peptides in in-solution digested samples. Reporter peptides, one for each candidate protein, were preselected based on their unique amino acid sequence (Table 2). Whereas HspB5 and cArr failed for validation, level diminishment of ANT3, DFS70 and MeCp2 could be supported by targeted AIMS analysis in pooled samples (N = 4/group) (Fig. 9). Despite material limitation for sufficient MS/MS fragmentation, these unique reporter peptides corresponding to MeCp2, DFS70 and ANT3 could be detected in a human non-glaucoma GCL LCM preparation also providing evidence of their presence in the human GCL (Fig. 10). Regarding immunohistological validation of selected candidates; ANT3, DFS70 and MeCp2; all three candidates were detectable in all examined human retinae. Thereby, significant changes were observed in the overall intensity between control and glaucomatous tissue samples. ANT3 was found to be distributed in large deposits in the GCL, the inner nuclear layer (INL) and in the outer plexiform layer (OPL) and showed levels of 38.94 ± 8.2 I in control samples compared to 24.15 ± 7.8 I in glaucomatous tissue (−38%, p < 0.04) (Fig. 11A). DFS70 occurred throughout the retina with GCL accumulations and was degraded in glaucomatous retina with 21.23 ± 1.13 I compared to 31.31 ± 1.8 I in control retina (−32%, p < 0.001) (Fig. 11B). MeCp2 displaying accumulations in the GCL and INL showed levels of 51.85 ± 8.39 compared to 31.85 ± 9.31 in control tissue (−39%, p < 0.02) (Fig. 11C). Moreover, STRING analysis indicated evidence for strong interaction between MeCp2 and DFS70. Also, their interaction with nucleus proteins, e.g. histone 1, high mobility group proteins (HMGs) and further proteins, e.g. endoplasmin (HSP90B1) and opticin (Fig. 12) could be documented. In summary, a detailed view to the human retina proteome could be achieved providing a subset of candidate proteins showing level alterations in glaucomatous retina samples. Thereby, special evidence for glaucoma correlation could be provided for ANT3, DFS70 and MeCp2 representative for glaucoma related proteomic alterations in human retinae.
Figure 1. 1D SDS PAGE pattern of individual retina lysate samples of glaucoma and control donors (N = 5/group).
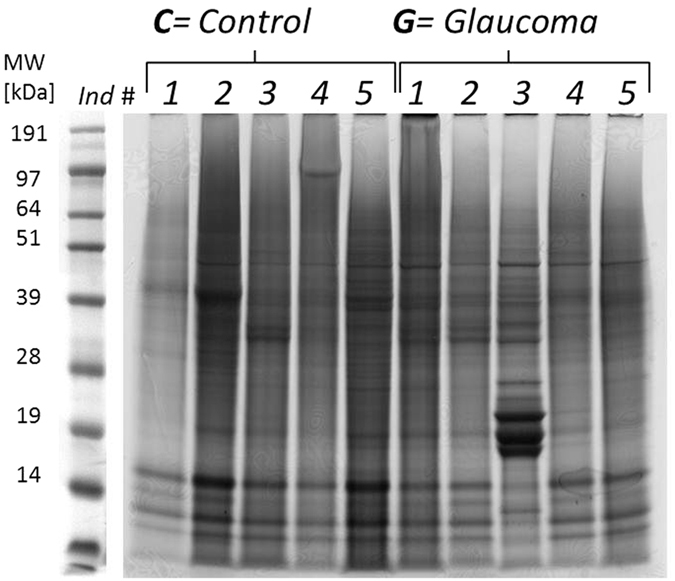
Sample lanes, containing each 80 μg of total protein, have been equally divided into 17 slices following tryptic in-gel digestion. Sample #G3 was excluded from later statistical analysis due to a unique protein cluster between ~20 and ~15 kDa.
Figure 2.
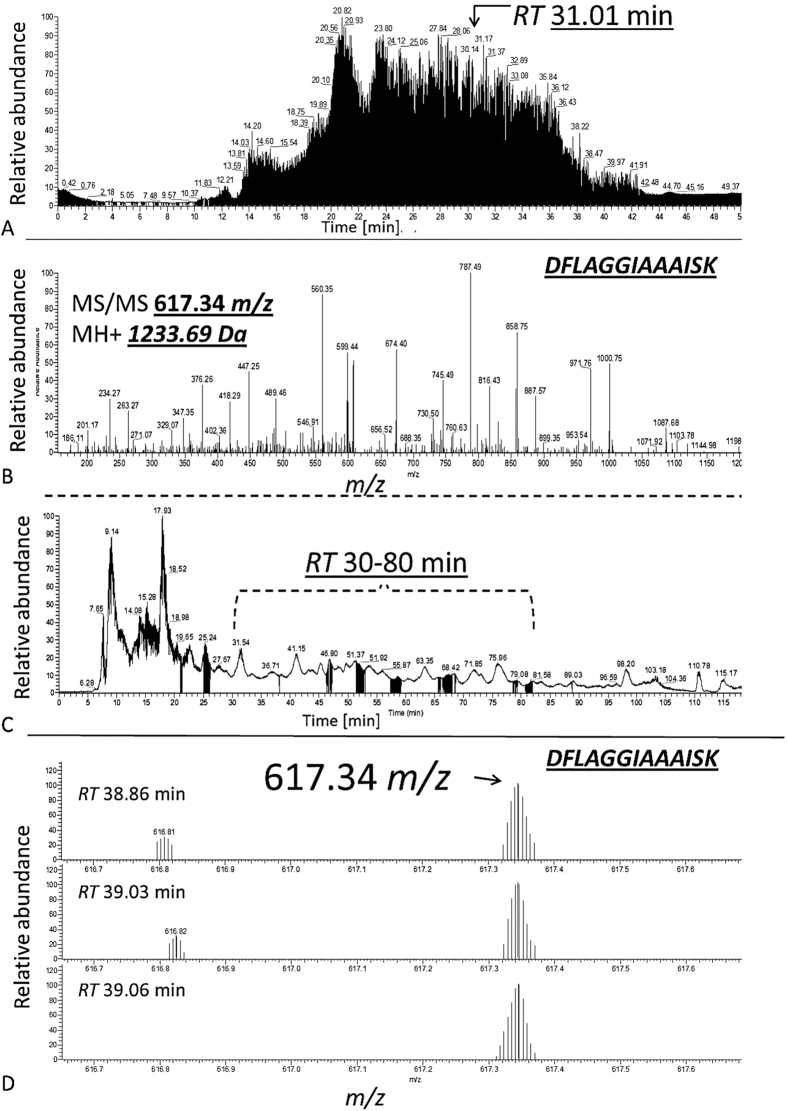
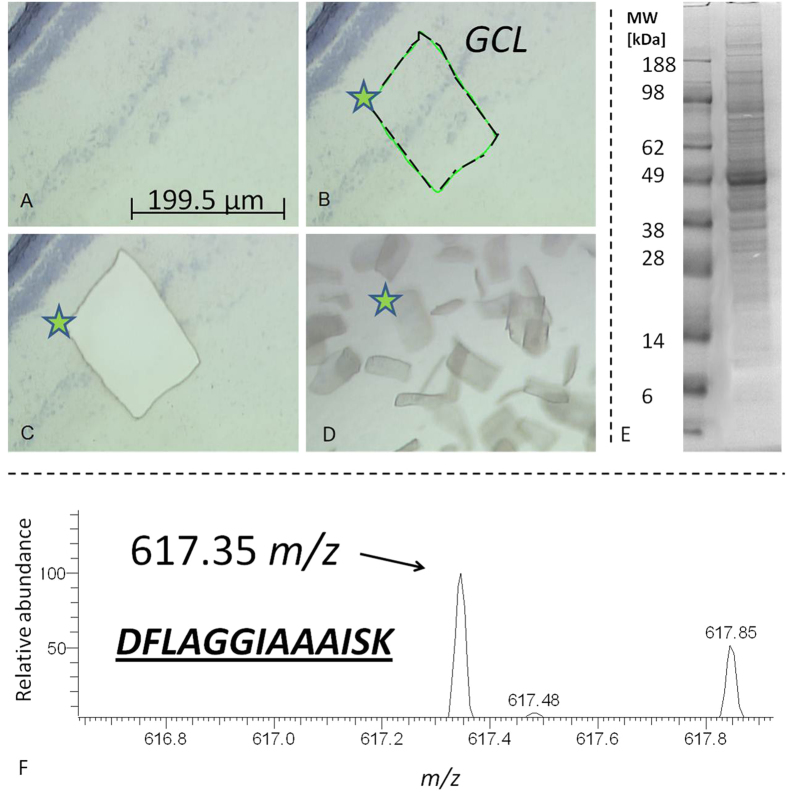
(A) Exemplary LC ESI MS TIC chromatogram of a single gel slice peptide extract corresponding to a control retina lysate highlighting the exemplary identification of ANT3 at RT 31.01 min. within the 50 min gradient (B). MS/MS fragment spectrum of 617.34 m/z (2+; MH+ 1233.69 Da) contributing to the identification of ANT3 (C). Elution range of the unique ANT3 reporter peak at 617.34 m/z between RT 30–80 min corresponding to an in-solution digest of a control retina lysate pool (D). Exemplary MS spectra at three RTs showing the ANT3 reporter peak 617.34 m/z corresponding to the amino acid sequence: DFLAGGIAAAISK.
Table 1. Human candidate retinal proteins (FDR <1%) that were A1. documented exclusively in the control group based on zero LFQ values in the glaucoma group and therefore named “unique”; A2. significantly (p < 0.05) or distinctly (p < 0.1) decreased in the glaucoma group; B1. documented exclusively in the glaucoma group based on zero LFQ values in the control group and therefore named “unique”; B2. significantly (p < 0.05) or distinctly (p < 0.1) increased in the glaucoma group.
| Protein name | Gene name | Entry | Entry name | LCM | ||
|---|---|---|---|---|---|---|
| A1: “Unique” control retinal proteins | ||||||
| V-type proton ATPase 116 kDa subunit a isoform 1 (VPP1) | ATP6V0A1 | B7Z641 | B7Z641_HUMAN | √ | ||
| Arrestin-C (cArr) | ARR3 | P36575 | ARRC_HUMAN | |||
| Protein NipSnap homolog 3A | NIPSNAP3A | Q9UFN0 | NPS3A_HUMAN | |||
| Heterogeneous nuclear ribonucleoprotein Q (hnRPQ) | SYNCRIP | F6UXX1 | F6UXX1_HUMAN | |||
| 6-phosphofructokinase, liver type | PFKL | P17858 | K6PL_HUMAN | |||
| Elongation factor Tu, mitochondrial | TUFM | P49411 | EFTU_HUMAN | |||
| 40S ribosomal protein S19 | RPS19 | P39019 | RS19_HUMAN | √ | ||
| Peripherin-2 | PRPH2 | P23942 | PRPH2_HUMAN | |||
| Protein SET | SET | Q01105 | SET_HUMAN | |||
| Reticulocalbin-2 | RCN2 | Q14257 | RCN2_HUMAN | |||
| A2: Glaucoma decreased retinal proteins | Glaucoma/Control | p-value | ||||
| Calcium-binding mitochondrial carrier protein Aralar2 | SLC25A13 | Q9UJS0 | CMC2_HUMAN | 0,15 | 0,0060 | |
| Methyl-CpG-binding protein 2 (MeCp2) | MECP2 | P51608 | MECP2_HUMAN | 0,15 | 0,0117 | √ |
| NADH dehydrogenase [ubiquinone] 1 ß subcomplex sub. 9 (CI-B22) | NDUFB9 | Q9Y6M9 | NDUB9_HUMAN | 0,15 | 0,0157 | |
| 40S ribosomal protein S7 | RPS7 | P62081 | RS7_HUMAN | 0,17 | 0,0748 | |
| 3-hydroxyacyl-CoA dehydrogenase type-2 | HSD17B10 | Q99714 | HCD2_HUMAN | 0,18 | 0,0357 | |
| Trifunctional enzyme subunit α, mitochondrial | HADHA | P40939 | ECHA_HUMAN | 0,19 | 0,0498 | |
| Myosin-11/9 | MYH11 | P35749 | MYH11_HUMAN | 0,21 | 0,0968 | |
| 60S acidic ribosomal protein P0; P0-like | RPLP0 | P05388 | RLA0_HUMAN | 0,27 | 0,0238 | √ |
| Pyruvate dehydrogenase E1 comp. sub. ß, mitochondrial (PDHE1-B) | PDHB | P11177 | ODPB_HUMAN | 0,31 | 0,0274 | √ |
| High mobility group protein HMG-I/HMG-Y | HMGA1 | P17096 | HMGA1_HUMAN | 0,40 | 0,0239 | |
| Small nuclear ribonucleoprotein Sm D3 | SNRPD3 | P62318 | SMD3_HUMAN | 0,43 | 0,0109 | |
| (ATP-dependent) 6-phosphofructokinase, muscle type | PFKM | P08237 | PFKAM_HUMAN | 0,47 | 0,0743 | √ |
| Vesicle-fusing ATPase | NSF | P46459 | NSF_HUMAN | 0,49 | 0,0657 | √ |
| Heterogeneous nuclear ribonucleoprotein L | HNRNPL | P14866 | HNRPL_HUMAN | 0,51 | 0,0777 | |
| PC4 and SFRS1-interacting protein (DFS70) | PSIP1 | O75475 | PSIP1_HUMAN | 0,56 | 0,0271 | |
| Cytochrome c oxidase subunit 7A2, mitochondrial (COX7A2) | COX7A2 | P14406 | CX7A2_HUMAN | 0,58 | 0,0893 | |
| ADP/ATP translocase 3 (ANT3) | SLC25A6 | P12236 | ADT3_HUMAN | 0,61 | 0,0199 | √ |
| Ras-related protein Rab-11A; B | RAB11A | Q15907 | RB11B_HUMAN | 0,62 | 0,0174 | |
| Heterogeneous nuclear ribonucleoprotein U | HNRNPU | Q00839 | HNRPU_HUMAN | 0,62 | 0,0494 | |
| Histone H1.0 | H1F0 | P07305 | H10_HUMAN | 0,67 | 0,0729 | |
| Putative elongation factor 1-α-2;1; like 3 | EEF1A1P5 | Q5VTE0 | EF1A3_HUMAN | 0,69 | 0,0148 | √ |
| Guanine nucleotide-binding protein G(i) subunit α-2 | GNAI2 | P04899 | GNAI2_HUMAN | 0,69 | 0,0930 | √ |
| B1: “Unique” glaucoma retinal proteins | ||||||
| Protein name | Gene name | Entry | Entry name | LCM | ||
| Glutathione synthetase | GSS | P48637 | GSHB_HUMAN | |||
| Septin-8 | SEPT8 | Q92599 | SEPT8_HUMAN | |||
| Opticin | OPTC | Q9UBM4 | OPT_HUMAN | |||
| Calcium-binding mitochondrial carrier protein Aralar1 | SLC25A12 | O75746 | CMC1_HUMAN | √ | ||
| Serpin B6 | SERPINB6 | P35237 | SPB6_HUMAN | |||
| Neuroplastin | NPTN | Q9Y639 | NPTN_HUMAN | |||
| Carbonyl reductase [NADPH] 1 | CBR1 | P16152 | CBR1_HUMAN | |||
| 4-trimethylaminobutyraldehyde dehydrogenase | ALDH9A1 | B4DXY7 | B4DXY7_HUMAN | |||
| ß-crystallin A3;isoform A1/Σ4/Σ7/Σ8 | CRYBA1 | P05813 | CRBA1_HUMAN | |||
| Ig γ-2 chain C region | IGHG2 | P01859 | IGHG2_HUMAN | |||
| ß-crystallin B1 | CRYBB1 | P53674 | CRBB1_HUMAN | |||
| Phakinin | BFSP2 | Q13515 | BFSP2_HUMAN | |||
| α-1-antitrypsin;Short peptide from AAT | SERPINA1 | P01009 | A1AT_HUMAN | |||
| Sideroflexin-1 | SFXN1 | Q9H9B4 | SFXN1_HUMAN | √ | ||
| X-ray repair cross-complementing protein 5 | XRCC5 | P13010 | XRCC5_HUMAN | |||
| N(G), N(G)-dimethylarginine dimethylaminohydrolase 1 | DDAH1 | B4DGT0 | B4DGT0_HUMAN | |||
| Hippocalcin-like protein 1 | HPCAL1 | P37235 | HPCL1_HUMAN | |||
| Thy-1 membrane glycoprotein | THY1 | J3QRJ3 | J3QRJ3_HUMAN | |||
| V-type proton ATPase subunit G 1/G2 | ATP6V1G1 | O75348 | VATG1_HUMAN | |||
| Cytosolic non-specific dipeptidase | CNDP2 | Q96KP4 | CNDP2_HUMAN | |||
| B2: Glaucoma increased retinal proteins | Glaucoma/Control | p-value | ||||
| Retinal dehydrogenase 1 (RALDH 1) | ALDH1A1 | P00352 | AL1A1_HUMAN | 11,73 | 0,0080 | |
| High mobility group protein B1; Putative B1-like 1 | HMGB1 | Q5T7C4 | Q5T7C4_HUMAN | 7,39 | 0,0745 | |
| Vesicle-associated membrane protein-associated protein B/C | VAPB | O95292 | VAPB_HUMAN | 6,51 | 0,0023 | √ |
| Annexin A2; Putative A2-like | ANXA2 | P07355; | ANXA2_HUMAN | 4,81 | 0,0962 | |
| Reticulon-3 | RTN3 | O95197-3 | RTN3_HUMAN | 3,69 | 0,0922 | √ |
| Glutathione S-transferase Mu 3 (hGSTM3-3) | GSTM3 | P21266 | GSTM3_HUMAN | 3,61 | 0,0529 | |
| Dihydropteridine reductase | QDPR | P09417 | DHPR_HUMAN | 2,37 | 0,0950 | |
| Retinol-binding protein 3 (IRBP) | RBP3 | P10745 | RET3_HUMAN | 2,12 | 0,0427 | |
| α-crystallin B chain (HspB5) | CRYAB | P02511 | CRYAB_HUMAN | 1,66 | 0,0599 | √ |
| Endoplasmin | HSP90B1 | P14625 | ENPL_HUMAN | 1,58 | 0,0575 | √ |
| Serotransferrin | TF | P02787 | ENPL_HUMAN | 1,46 | 0,0872 | √ |
| Rab GDP dissociation inhibitor α | GDI1 | P31150 | ENPL_HUMAN | 1,40 | 0,0541 | |
In case of multiple isoforms, only the first gene, entry accession and entry name are given. In case of candidate recovery in the porcine retinal GCL using LCM, candidates are highlighted by check mark in the LCM column. ANT3, MECP2 and DFS70 presence was supported by MS detection of diagnostic m/z peaks in human LCM preparation (illustrated in Fig. 10, not highlighted in the table).
Figure 3. Cellular component distribution of human retinal proteins displaying glaucoma related level alterations.
(A) The majority of affected proteins are intracellular species, representing predominantely (B) membrane proteins.
Figure 4. Localization and functional GO analysis of human retinal proteins showing glaucoma related level changes.
(A) Predominantly, nucleus and mitochondrial proteins showed level changes associated to glaucoma. (B) Candidates display binding and catalytic properties. (C) A high portion of candidates are involved in regulatory biological processes encircling developmental, transport processes and cell death.
Figure 5. Exemplary human retinal proteins showing significant differences (p < 0.05) to distinct alteration tendencies (p < 0.1) in protein levels revealed by BU LC ESI MS following label-free quantification and t-test.
Figure 6. Dynamic MS detection range of recombinant Thy-1.
(A) Recombinant Thy-1 1D SDS PAGE dilution experiment (43.34 kDa, mass migration area highlighted by arrow; D1 = 0.3, D2 = 0.24, D3 = 0.18, D4 = 0.12, D5 = 0.06, D6 = 0.03 μg; N = 3 replicate runs). Thy-1 content <0.12 μg is not visible by CBB staining. (B) Dynamic MS range of recombinant Thy-1 (50% of gel content is introduced to the LC MS system for technical reasons; therefore the X-axis displays corrected values) generated by MaxQuant software LC MS detection threshold can be determined <0.03 μg describing intensity values < 1*E6, whereby the LFQ value is calculated zero. (C). Separate evaluation of the Thy-1 specific peptides (P1–P5) indicating that for Thy-1 content <0.03 μg only two Thy-1 specific peptides can be detected (P1 & 3; highlighted by asterisk); which represent MS/MS identification peptides in study sample runs. Since in sample runs only two of five characteristic peptides (P1 & P3) could be detected showing raw output values <1*E6, Thy-1 can be attributed to the low detection range and is consequently underrepresented in the retina samples leading to critical quantification output.
Figure 7. MS detection of Thy-1 in pooled retina samples (glaucoma vs. control).
(A,B) Thy-1 could be identified in pooled glaucoma samples by MS/MS of 856.46 m/z. The corresponding peak is also detectable in the identical RT range at ~25 min in the control pool, however not leading to identification. (C) Isotopic pattern of Thy-1 specific peptide showing monoisotopic peak at ~856.46 m/z. (D). Zoom view to Thy-1 specific peptide isotopic pattern corresponding to MS spectrum of control and glaucoma pool sample supporting Thy-1 presence for both groups.
Figure 8.
(A–C) Laser capture microdissection (LCM) of ganglion cell layer (GCL) tissue areas (indicated by asterisk) of porcine retina cryosections. (D) Exemplary GCL exudates. (E) 1D-SDS-PAGE Protein pattern of a GCL exudate extract pool destinated for BU LC ESI MS analysis. (F) Exemplary recovery of ANT3 in GCL exudates indicated by ANT3 unique reporter peak 617.35 m/z corresponding to the amino acid sequence DFLAGGIAAAISK.
Figure 9. AIMS validation of selected retinal protein candidates: MeCp2 and ANT3 are decreased in pooled glaucoma samples (N = 4 individuals/group) supported by technical replicate runs (N = 4).
Unique reporter peptide sequences are highlighted by asterisk.
Table 2. Subset of representative human retinal candidate proteins selected for AIMS validation in retina sample pools.
| Protein | Gene names | Mass [Da] | m/z | Amino acid sequence | Charges | Glaucoma/Control | Valid |
|---|---|---|---|---|---|---|---|
| ADP/ATP translocase 3 (ANT3) | SLC25A6 | 1232,68 | 617,34 | DFLAGGIAAAISK | 2 | 0,38 | √ |
| PC4 and SFRS1-interacting protein (DFS70) | PSIP1 | 1631,76 | 816,88 | GFNEGLWEIDNNPK | 2 | 0,80 | √ |
| Methyl-CpG-binding protein 2 (MeCp2) | MECP2 | 1954,89 | 978,45 | VGDTSLDPNDFDFTVTGR | 2 | 0,65 | √ |
| α-crystallin B chain (HspB5) | CRYAB | 1164,65 | 583,33 | VLGDVIEVHGK | 2 | 1,08 | × |
| Arrestin-C (cArr) | ARR3 | 1821,94 | 911,97 | VQFAPPEAGPGPSAQTIR | 2 | 0,99 | × |
Analysis of technical replicate runs (N = 4; N = 4 individuals/pool sample) supported glaucoma related significant level diminishment of ANT3, MeCp2 and distinct decline of DFS70 highlighted by check mark in the column.
Figure 10.
Detection of unique reporter peaks (indicated by asterisks in the MS spectra in B) corresponding to the three marker candidates DFS70, MECP2 and ANT3 in the retinal ganglion cell layer (GCL) of a human ophthalmic LCM preparation following (A). 1D SDS PAGE and (B). LC MS analysis. The preparation was based on 30 μm cryosections of a female non-glaucoma donor bulbus of a 69 year old donor.
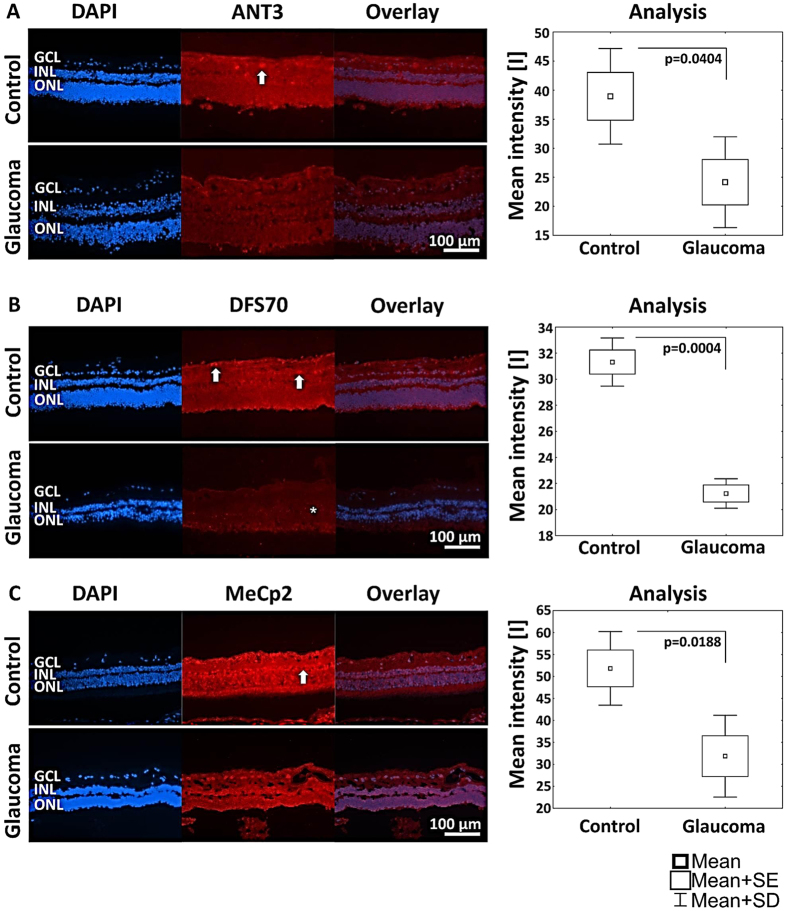
Figure 11. Immunofluorescence was detected and analyzed against candidates ANT3, DFS70 and MeCp2 in control and glaucomatous retinal tissue supporting significantly lower abundance of candidates in glaucomatous retinae compared to control retinae.
(A) ANT3 shows depositions in the ganglion cell layer (GCL; indicated by arrow). (B) DFS70 is distributed in the entire retina with accumulations in the GCL. The distribution of DFS70 in glaucomatous retinal tissue seems to be accumulated in the inner and outer nuclear layer (INL and ONL; highlighted by asterisk). (C) MeCp2 shows a strong immunofluorescence in the entire retina with accumulations in the GCL and the INL (indicated by arrow).
Figure 12. STRING analysis leads to isolation of an interaction network between glaucoma associated candidates.
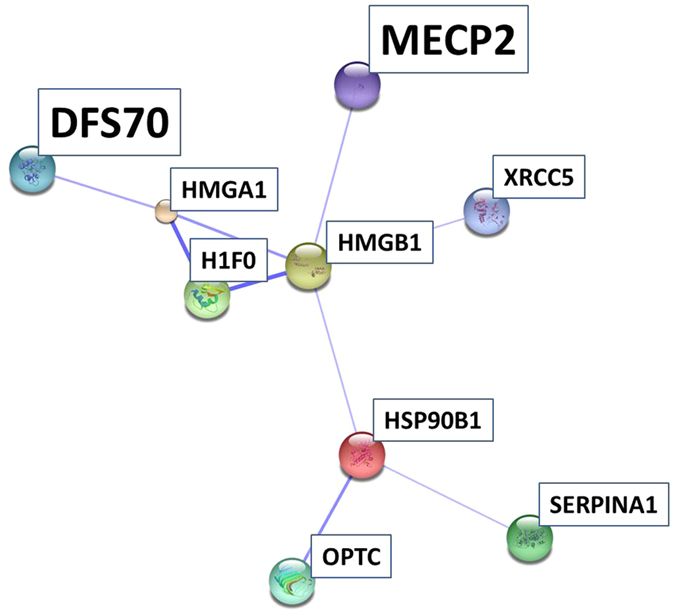
MECP2 and DFS70 are two highlighted members of the interaction network emphasizing their potential role in the disease.
Discussion
By use of a state-of-the-art BU LC ESI approach43 we could identify more than 600 proteins with FDR < 1%, incorporating numerous neuronal relevant protein species. Moreover, the proteomic output fits well to prior mammalian retina studies, e.g. reporting 400 proteins to be visualized from rat retina10, more than 900 2D gel protein spots44,45 and more than 800 proteins identified from mouse retina samples46,47. Recently, Zhang and colleagues catalogued over 3000 retinal proteins from healthy human donors48. Nevertheless, proteomic investigations on human retina, especially diseased human retina samples are still rare. Ethen and colleagues identified over 500 proteins in human AMD donor retinae49, 72 secretory proteins were reported from human RPE cells50 and Yang and collegues reported several hundred proteins from human retina samples24. Qualitatively, many of the identified retinal proteins could be supported by proteomic mammalian retina maps of other working groups focusing on rodents51,52 or primates53. Regarding the human retina proteome examined in the present work, most of the revealed proteins could be classified as intracellular species indicating a high recovery of cellular proteins in contrast to secretory or extracellular proteins. The large amount of membrane proteins probably reflects ionization priority of hydrophobic peptides in ESI experiments54,55. Moreover, since retina samples in the present study were based on processed retina tissue after removing blood vessels and vitreous residuals the proteomic output can be suggested to refer to the “core proteome” of the human retina explaining a number of >600 proteins and limited representation of secretory proteins. Regarding glaucoma affected retinal proteins, numerous candidates could be recovered from the GCL of porcine retinae by LCM analysis. “Unique” LFQ documentation of RGC specific Thy-1 in glaucoma samples was found contrary to reported reduced levels indicative for RGC loss56,57,58. Thy-1 related reanalysis showed that the protein is present in samples of both groups, control and glaucoma in low abundance and could be identified by two specific peptides, which correspond to the detection threshold <0.03 μg. In a further experiment with pooled samples of remaining tissue, Thy-1 could be identified in glaucoma samples by MS/MS analysis. Nevertheless, Thy-1 could be indirectly affirmed for the control group by reporter peak detection. In consequence of the low abundant recovery of Thy-1 in the study samples, Thy-1 quantification has to be viewed critical. This is a clear study limitation and has to be considered for future experimental designs. However, the detection of Thy-1, even in low abundance supports RGC representation in retinal samples. Moreover, regarding the majority of altered proteins, in first instance nucleus and mitochondrial species were found affected and could be verified to be involved in cellular key processes, e.g. transport, system development and cell death. Mitochondrial dysfunction, involvement in cell death and associated protein alteration has been intensively studied in neurodegeneration59 including glaucoma60,61. In accordance, diminishment of retinal mitochondrial proteins has been documented for neurodegenerative processes, e.g. in a Parkinson monkey model62. Confidently, ocular levels of COX7A2, decreased in glaucoma retina samples in the present study, were found to be reduced in glaucomatous primates63 and are involved in mitochondrial stress in POAG64. In this work, levels of mitochondrial ANT3 were clearly found diminished in glaucomatous retinae demonstrated by MS and immunohistology results. Moreover, ANT3 localization could be shown for the GCL of porcine and human retina fitting well with its documented high abundance in CNS neurons (www.proteinatlas.org). Lately, ANT was found to be part of the mitochondrial ATP synthasome65 emphasizing its role for cellular energy supply. In fact, level elevation of ANT3 was reported to enhance cell survival66 supporting its key function in glaucomatous neurodegeneration. Level declines of mitochondrial key enzymes ANT3, COX7A2, PDHE1-B and VPP1 support the entanglement of the cellular energy supply system in glaucoma60. Confidently, increase of ANT expression and activity has been shown to reduce cellular oxidative stress and apoptosis67. Furthermore, ANT3 interactions have been documented for secretogranin-1 (BiomarkerDigger: www.biomarkerdigger.org; Hit Predict: hintdb.hgc.jp), which was demonstrated to play a key role in Parkinson disease68. Also, interactions were reported between ANT3 and members of the tumor necrosis factor (TNF) receptor superfamily (Hit Predict: hintdb.hgc.jp) involved in glaucomatous neurodegeneration69. Beside mitochondrial protein alterations, levels of certain cytoplasmic proteins were found to be altered in glaucoma, e.g. cArr, which has been documented to be involved in glaucoma photoreceptor degeneration70 and found to be diminished in AMD macula71. Interestingly, anti-cArr auto-antibodies have been detected in multiple sclerosis72 supporting cArr association to neurodegenerative processes. However, even if cArr could not been validated in this work, it needs to be considered for future prospects. Additionally, further proteomic alterations can be associated to neurodegenerative processes in the retina. Glaucoma related level decrease of protein NipSnap homolog 3A is in confidence with downregulation of NipSnap proteins in Alzheimer disease73 and hyperoxia/erythropoietin stressed brain tissue74. NipSnap 3 was reported to be involved in apoptosis via antagonizing x-linked inhibitor of apoptosis protein (XIAP) caspase 3 inhibition75. A comparable role was proposed for nucleus proteins, heterogeneous nuclear ribonucleoproteins (hnRPs) C1 and C276. In accordance, a key role of hnRNP A1 in neurodegeneration was highlighted77. Confidently, the observed decline of hnRNP Q and U most likely refer to neuronal damages in glaucoma retinae. Diminishment of MeCp2, a retinal48 and neuron expressed nucleus key trigger protein in Rett syndrome78,79 fits well with its high CNS expression levels, turnover, histone modification, DNA and protein binding features as well as its level declines observed in neurodegenerative diseases80. Beside the neuronal key regulator function of MeCp2 in gene expression81, MeCp2 deficiency was suggested to affect directly the neuronal cytoskeleton82 triggering nucleus stability, proliferation and cell viability83. Since there is growing evidence of MeCp2 participation in neurodegenerative pathologies84, Bogdanovic & Veenstra proposed a “well-balanced requirement of MeCp2 levels for proper function of the mammalian CNS”85, most likely encircling retinal health. Despite its high abundance in neuronal cells, which is in confidence with the supported MeCp2 retinal GCL localization in the present work, it is not known why MeCp2 level diversification exclusively affect neuronal cells86. Also, its high expression level and its associated role in the retina remains controversial87. From a functional view MeCp2 has been demonstrated to display multiple binding features to several proteins80, e.g. to methyl-cytosine binding domain protein 2 (MBD2)88 or neurological relevant proteins like α-thalassemia/mental retardation, X-linked protein (ATRX)89 and γ-synuclein90, which underlines its potential to modulate molecular pathways in the course of neurodegeneration. Especially, MeCp2 DNA binding and neuronal gene expression regulating features91 give new perspective to neurodegenerative processes in the glaucomatous retina. Interestingly, DFS70 displaying glaucoma related decline, shares functional similarities to MeCp2, e.g. nucleus residence and DNA- and protein binding properties (UniProt: www.uniprot.org), suggestible for neurodegenerative involvement. Moreover, STRING interaction analysis supported strong association of MeCp2 and DFS70, both detected in the retina GCL in the present work, in confidence with their high abundance in CNS neurons (www.proteinatlas.org). Consistently, Leoh and colleagues documented direct binding of MECP2 to DFS7092. Misregulation of MeCp2, histone H1 and HMGs were suggested indicative for chromatin composition changes in mouse myoblasts93 and could reflect neurodegenerative related chromatin alterations94. Regarding MeCp2/HMGB1 interaction95, the existence of a chromatin-binding protein interaction complex affected in the glaucomatous retina is thinkable. However, the exact role of these chromatin-binding proteins has not been uncovered so far and needs to be addressed in future studies, e.g. in glaucoma animal models. Also, an indirect interaction between chromatin-binding proteins to glaucoma related proteins, opticin96 and endoplasmin (HSP90B1)97, could be observed. Beside elevated endoplasmin levels, increased levels of further characteristic stress marker proteins could be detected in glaucomatous retinae, e.g. glutathione synthetase associated to the glutathione system involved in neurodegeneration98,99. Furthermore, glutathione S-transferase (GST) has been described as a glaucoma associated stress marker100 and increased serum GST immunoreactivity has been documented in glaucoma101. In fact, various GST species were found to be implemented to degenerative stress related pathologies102,103,104,105. Increase of serotransferrin as an acute phase protein106 fits well with differential expression of the protein observed in serum of glaucoma patients107. Also, indicative alterations of crystallins51,108 have been documented for diabetic retinopathy109,110 and glaucoma111 and especially upregulation in retinal disorders have been reviewed112. Retina associated protective potential has been discussed for crystallins113 and retinol dehydrogenase114, probably explaining the observed enhancements of these proteins. Exorbitant increase of crystallins was observed in the excluded glaucoma subject #G3 and consequently other stress factors beyond neurodegenerative processes cannot be precluded for crystalline increment. Decrease of 6-phosphofructokinase levels observed in glaucoma retinae is in confidence with downregulation in neurodegenerative Alzheimer115 and Down syndrome brains116 and supports energy metabolism implementation. Increase of α-1-antitrypsin could be indicative for elevated autoreactivity for this protein observed in serum of glaucoma patients demonstrated by our group117. Moreover, increase of the protein is in confidence with overexpression in serum of newly recruited glaucoma patients107 and α-1-antitrypsin is suggestive to play a role in Alzheimer´s disease118. In conclusion, based on proteomic data three functional classes could be supported to be involved in glaucomatous processes: mitochondrial, stress and nucleus proteins indicating an impairment of energy metabolism, stress response and gene expression alterations in the course of retinal neurodegenerative processes. Numerous proteins could be supported to be GCL expressed revealed by LCM, underlining their involvement at the site of damage. However, the present study is limited by the number of donor retinae and sample material. Another limitation is the focus on progressed glaucoma and use of postmortem retinal material. Accordingly, the role of mitochondrial proteins like ANT3 and the function of nucleus proteins, especially hnRNPs, DFS70 and MeCp2 in the course of neurodegeneration need to be in the focus of future projects determining accurate candidate protein dynamics during glaucoma progression. In conclusion, the present work provides a sensitive image of the human “retina core proteome” supporting mitochondrial and nucleus proteome entanglement of glaucomatous neurodegeneration and highlights new molecular players, e.g. ANT3, DFS70 and MeCp2 to be considered in future glaucoma studies.
Methods
Patients and samples
Donor eyes were provided by the Cornea Bank of Rhineland-Palatine, Department of Ophthalmology, University Medical Center Mainz, Germany. Organ donations followed the guidelines for corneal donations regulated by the German Transplantation Law in agreement with the requirements of the Declaration of Helsinki. The use of remaining ocular donor tissue not required for transplantation for study purposes has been explicitly approved by the relatives of the deceased and the local ethics committee. Retina tissue samples were prepared from post-mortem donor eyes (right or left eye). Glaucoma donors (N = 5; 3 females, 2 males) were 86 +/−9 years old, control donors (N = 5; 3 females, 2 males) were 82 +/−7 years old. For LCM experiments an additional eye bulb of a 69-year old female non-glaucoma donor was used. Donor exclusion criteria were defined as the following: any kind of ocular diseases including infection and tumors, systemic infections (viral, fungal, bacterial) incl. hepatitis, HIV, malignat neoplasia, autoimmune diseases including multiple sclerosis, Creutzfeldt-Jakob, Parkinson, Alzheimer, amyotrophic lateral sclerosis as well as need of dialysis. Ophthalmologic background and clinical history were requested from closest relatives, general practitioner and last attending doctor. Clinical phenotype of glaucoma could not be defined in donors, which is a distinct limitation of the present study. Retina tissue was extracted from eye cups (sagittal half of bulbs) and isolated from pigment epithelium, ciliar body, blood vessels and vitreous residuals in phosphate buffered saline under a binocular microscope. Liquid nitrogen snap frozen retina tissue samples were grinded by mortar and pestle, added 0.1% SDS, mixed for 20 min at room temperature and centrifuged at 14000 rpm at 4 °C. Supernatants were used for total protein measurement using the Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, USA) following the manual of the manufacturer.
BU LC ESI MS
Retina samples (Glaucoma, N = 5; Control, N = 5; 40 μg/sample) were run on 12% Bis-Tris 10-well prepared SDS PAGE minigels under reducing condition at 150 V followed by Coomassie Brilliant Blue staining (Invitrogen, Carlsbad, CA, USA). Sample lanes (individual and pools) were divided into 17 slices/lane followed by trypsin digestion using a modified protocol of Shevchenko et al.119. Dehydrated gel pieces were soaked with 100 μl of sequencing grade modified trypsin (Promega Corporation, Madison, WI, USA) solution [15 ng/μl] and incubated overnight at 37 °C. Lyophilized peptides were double purified by ZIPTIP C18 solid phase extraction (Millipore, Billerica, MA, USA) prior to MS analysis according to the protocol of the manufactor following lyophilization and storage −20 °C prior to MS analysis. Resolubilized peptides (20 μl 0.1% TFA/slice digest) were analyzed by use of LC ESI MS. The LC system consisted of a Rheos Allegro pump (Thermo Scientific, Rockford, IL, USA) downscaled to a capillary system (6.7+/− 0.03 μl/min flow rate) as already described with weak modifications (21, 22). The column system consisting of a 30 × 0.5 mm BioBasic C18 coupled to a 150 × 0.5 mm BioBasic C18 analytical column (Thermo Scientific, Rockford, IL, USA) and protected by an A316 online precolumn filter (Upchurch Scientific, Washington, DC, USA) was run under gradient elution (buffers: A = 98% H2O, 1.94% ACN, 0.06% methanol, 0.05% formic acid; B = 95% ACN, 3% methanol, 2% H2O, 0.05% formic acid). The 50 min gradient was programmed as follows: 15–20% B (0–2 min), 20–60% B (2–35 min), 60–100% B (35–40 min), 100–0% B (40–45 min), 0% B (45–50 min). The LC system was directly connected to the ESI source of a hybrid linear ion trap (LTQ) Orbitrap (FT) XL instrument (Thermo Fisher Scientific, Rockford, IL, USA) introducing ions by use of a low flow metal needle (Thermo Fisher Scientific, Rockford, IL, USA). 5 μl of peptides were injected for each run by use of a PAL HTS robot (CTC Analytics, Zwingen, Switzerland). To avoid sample carryover effects 30 min washing runs injecting 80% ACN to the system were realized. The whole workflow was administered by XCalibur 2.0.7 SP1 (Thermo Fisher Scientific Rockford, IL, USA). For FT MS measurements a range of 300–2000 m/z in the instruments positive mode was selected and the following parameters were adjusted: max. injection time =50 ms (LTQ), 500 ms (FT), activation = collision induced decay (CID), normalized collision energy = 35, activation time = 30 ms, activation Q = 0.25. The top five centroid detected monoisotopic peaks (charge state = 1+, 2+, 3+, 4+; intensity > 500) were selected within an isolation width of 2 m/z for fragmentation in each scan event considering dynamic exclusion of 90 s, repeat duration of 30 s and resolution of 30000.
Protein identification and quantification
Combined LC MS raw files were transferred to MAXQuant version 1.4.1.2 (Max Planck Institute of Biochemistry, Martinsried, Germany; www.maxquant.org) and its built-in Andromeda search engine for protein identification and quantification. MS/MS fragment spectra were search against the SwissProt human database (SwissProt_111101, 533049 sequences, 189064225 residues). Thereby, settings were set as follows: peptide mass tolerance =±30 ppm, fragment mass tolerance =±0.5 Da, fixed modification = carbamidomethylation (C), variable modifications: oxidation (M), acetylation (protein N-term), enzyme = trypsin, allowed missed cleavages: 2. Proteins were identified with a false discovery rate (FDR) <1% with ≥6 amino acid residues and only ‘unique plus razor peptides’ that belong to a protein, which was also included for quantification. LFQ normalized peak intensities120 were transferred to Statistica (Version 10, Statsoft, Tulsa, OK, USA) for group specific protein level comparison. Protein LFQ normalized peak intensities of glaucoma and control subjects were analyzed using t-test except donor sample #G3, which was excluded from analysis due to its deviated gel pattern. Proteins displaying significant group-specific level alterations (p < 0.05) or distinct tendencies (p < 0.1) were assigned candidates. For LCM analysis human as well as porcine RGC based MS raw files were combined and matched against the SwissProt human database using Proteome Discoverer version 1.1 (Thermo Scientific, Rockford, IL, USA) and MASCOT version 1.4.1.2 using a more relaxed strategy based on a 95% probability identification threshold.
Functional analysis (GO and interaction analysis)
Candidate proteins were inspected by gene ontology (GO) analysis using Cytoscape version 2.8.3 with integrated BINGO 2.44 plugin (www.cytoscape.org). Interaction analysis was realized by use of STRING version 10 (http://string-db.org)121 considering a medium confidence score of 0.4 for interactions. Moreover particular candidates were manually searched for localization and interaction using online databases (www.proteinatlas.org, www.biomarkerdigger.org, Hit Predict: hintdb.hgc.jp).
Targeted MS
Validation of a selected candidate protein subset was realized by an already established in-solution tryptic digestion/LC ESI MS protocol122 focusing on pooled samples for proper protein amount (10 μg/subject; 40 μg/group, N = 4/group, N = 4 technical replicates) following Accurate Inclusion Mass Screening (AIMS) strategy based on quantification of unique reporter peptides123, each representing one particular candidate protein. Selection of these peptides for the inclusion list was carried out manually based on the msms.txt file resulting from MaxQuant analysis. For inclusion list-dependent acquisition the use of global parent list was enabled, the dynamic exclusion segment was disabled and the m/z tolerance around targeted precursors was set ±10 ppm. The acquired MS spectra were analyzed by MaxQuant for peptide and protein identification. Extracted spectra were searched against Uniprot/Swissprot human database using settings with peptide mass tolerance of ±10 ppm, fragment mass tolerance of ±0.5, peptide charge state of +2 with FDR < 1% for identification. The output of the generated ‘peptides.txt’ from MaxQuant were exported in an excel workbook format (Microsoft Office Excel 2010) and the sum absolute intensity of the signature peptides for each proteins were transferred to Statistica for unpaired t-test analysis (p < 0.05).
Laser-capture microdissection (LCM)
Due to their retina model comparability124 freshly enucleated porcine (Sus scrofa) eye bulbs (N = 3) obtained from the local slaughterhouse (Robert-Bosch-Str. 23–25, 55232 Alzey, Germany) were embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, Tokyo, Japan), deep frozen in liquid nitrogen and sliced at −27 °C to 30 μm transversal sections using a CM 1850 cryotome (Leica Biosystems Nussloch GmbH, Nussloch, Germany). Slices were collected on UV-pretreated PEN membrane slides and transferred to cool 70% ethanol, stained with hematoxylin for 1 min and dehydrated with 70%, 96% and 100% ethanol for 2 min. RGC tissue LCM was realized by use of a PALM MicroBeam Laser system (Zeiss, Oberkochen, Germany) using 5x magnification. For each LCM extract a RGC layer tissue area on average of 5.3 mm2 has been dissected approximately representing 19950 cells. Single cutouts had a size of 0.024 mm2 and were captured in AdhesiveCap 500 opaque caps (Carl Zeiss Jena GmbH, Jena, Germany) under RoboLPC laser control. Tissue preparations were extracted by shaking using a KS250 Basic Intellimixer (Kika Labortechnik, Cologne, Germany) at 25 rpm and 4 °C for 2.5 h. Suspensions were transferred to BU LC ESI MS as already described. Finally, to recover candidates in human GCL, the above described LCM BU LC ESI MS workflow was applied on 30 μm transversal sections of a single non-glaucoma donor eye bulb approximately recovering 15500 cells. Corresponding LC MS TIC chromatograms were screened for the occurrence of selected candidate (DFS70, ANT3 and MeCp2) unique reporter peptides to indicate their existence in the human retina GCL.
Immunodetection of DFS70, ANT3 and MeCp2 in retinal cross sections of healthy and glaucomatous humans
Retina specimen from sagittally split bulbi of age and gender matched donor eyes (N = 4 glaucoma, N = 4 control) corresponding to the proteomic analysis were fixed in 4% formaldehyde (Carl Roth, Karlsruhe, Germany) for 10 days before paraffin embedding by use of an automated Tissue-Tek Auto TEC embedder (Sakura Finetek, Tokyo, Japan) applying the following steps: 1. fixation, 2. dehydration, 3. clearing and 4. paraffination. For each patient, four serial cross sections with 5 μm thickness were sliced on a 2030 Biocutrotation microtome (Reichert &Jung), transferred onto Superfrost plus slides (Thermo Scientific, Schwerte, Germany), dried and stored at 37 °C. For staining slices were dewaxed in xylene three times and rehydrated in a sequential ethanol gradient (100%, 100%, 96%, 96%, 70%, each 5 min) to Aqua dest. Antigen retrieval was performed in preheated Target Retrieval Solution (DAKO, Hamburg, Germany) at 87 °C for 30 min, followed by 20 min at room temperature before short washes in PBS (Dulbecco’s PBS, Sigma Aldrich, St. Louis, MO, USA). Slices were blocked with 1% normal donkey serum/0.5% bovine serum albumin/PBS for DFS70 staining or with 3% normal goat serum/0.1%TritonX/PBS for ANT3 and MeCp2 staining for 30 min. Primary antibodies were diluted 1:200 and incubated at 4 °C overnight. Spare antibodies were washed off using PBS twice for 15 min on a rotating platform. All secondary antibodies (ANT3: Ref. orb101369, Biorbyt Ltd., Cambridge, UK; DFS70: Ref. sc-33371, Santa Cruz Biotechnology, Dallas, TX, USA; MeCp2: Ref. ChIP Grade ab2828, Abcam, Cambridge, UK) were diluted 1:200 in PBS and incubated on tissue for 60 min. Again, spare antibodies were washed off before tissue was mounted using diamidin-2-phenylindol (DAPI) supplemented Vectashield (Vector, Burlingame, CA, USA). Negative controls were conducted through incubation of equally treated slides with secondary antibodies exclusively. For quantification mounted tissue was photographed immediately after staining using Eclipse TS 100 microscope with a DS-Fi1-U2 digital microscope camera and an ELWD 20x/0.45 S Plan Flour Ph1 ADM objective and recorded by the imaging software NIS Elements version 4.10 64 bit (microscope, camera, equipment and software were purchased from Nikon, Tokyo, Japan). Exposure time settings were the same for samples from healthy and glaucomatous patients. At least 30 pictures were taken per patient for analysis. Quantification of intensity differences was performed using Image J (rsb.info.nih.gov/ij, NIH, Bethesda, MD, USA) and statistical differences in the overall intensity (I) were calculated by using Breakdown ANOVA Test and unpaired t-test in Statistica.
Focused Thy-1 analysis
Dynamic range of recombinant human Thy-1 (P01, Ref. H00007070-P01, Abnova, Heidelberg, Germany) was determined by use of the established BU LC MS workflow encircling 1D-SDS PAGE and LC MS. At first, three identical sample dilution subsets of Thy-1 were run on three gels allowing triplicate analysis of each Thy-1 dilution in the LC MS system. In-gel digestion and ZIP TIP purification of Thy-1 dilution samples were carried out as already described. Accordingly, triplicate samples of 0.15, 0.12, 0.09, 0.03, 0.015 μg Thy-1 were analyzed by the LC MS system following MaxQuant analysis considering MaxQuant Raw and LFQ data acquisition. Mean MaxQuant Raw and LFQ intensity values of Thy-1 triplicate LC MS measurements were transferred to Statistica (version 10, Statsoft, Tulsa, OK, USA) for dynamic range determination by linear regression analysis. Furthermore, for validation of Thy-1 regulation, LC MS TIC raw files corresponding to Thy-1 SDS PAGE mass migration area of the control and the glaucoma sample pool (40 μg/lane) were analyzed by ProteomeDiscoverer and manually inspected for occurrence of Thy-1 specific unique peptide corresponding to 856.46 m/z.
Additional Information
How to cite this article: Funke, S. et al. Glaucoma related Proteomic Alterations in Human Retina Samples. Sci. Rep. 6, 29759; doi: 10.1038/srep29759 (2016).
Supplementary Material
Acknowledgments
We would like to thank Dr. Arno Schad and colleagues of the Pathological Institute of the University Medical Center Mainz, Germany for support with the LCM system. Furthermore, we would like to thank Lukas Weiler for his assistance during LCM analysis. Stiftung Rheinland-Pfalz für Innovation.
Footnotes
Author Contributions S.F. developed study design, realized proteomic mass spectrometric experiments and statistical analysis, contributed important intellectual content and drafted the manuscript. N.P. (N. Perumal) participated in study design, proteomic mass spectrometric experiments and contributed important intellectual content. S.B. participated in sample recruitment, sample preparation and contributed important intellectual content. S.G. participated in sample preparation and proteomic mass spectrometric experiments and contributed important intellectual content. C.S. participated in validation experiments and contributed important intellectual content. J.T. performed immunohistology and contributed important intellectual content. C.G. performed LCM analysis and contributed important intellectual content O.G. participated in sample recruitment, sample preparation and contributed important intellectual content. N.P. (N. Pfeiffer) critically reviewed the manuscript and contributed important intellectual content. F.G. performed study coordination, study design, participated in proteomic mass spectrometric experiments, contributed important intellectual content and participated in manuscript drafting. All authors read and approved the final manuscript.
References
- Foster P. J., Buhrmann R., Quigley H. A. & Johnson G. J. The definition and classification of glaucoma in prevalence surveys. Br J Ophthalmol 86, 238–242 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson R. J., Chidlow G., Wood J. P. M., Crowston J. G. & Goldberg I. Definition of glaucoma: clinical and experimental concepts. Clin Exp Ophthalmol 40, 341–349, 10.1111/j.1442-9071.2012.02773.x (2012). [DOI] [PubMed] [Google Scholar]
- Quigley H. A. & Broman A. T. The number of people with glaucoma worldwide in 2010 and 2020. Brit J Ophthalmol 90, 262–267, 10.1136/bjo.2005.081224 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohra R., Mai J. C. & Kolko M. The Role of Inflammation in the Pathogenesis of Glaucoma. Surv Ophthalmol 58, 311–320, 10.1016/j.survophthal.2012.08.010 (2013). [DOI] [PubMed] [Google Scholar]
- Grus F. & Sun D. Immunological mechanisms in glaucoma. Semin Immunopathol 30, 121–126, 10.1007/s00281-008-0105-8 (2008). [DOI] [PubMed] [Google Scholar]
- Bell K. et al. Does autoimmunity play a part in the pathogenesis of glaucoma? Prog Retin Eye Res 36, 199–216, 10.1016/j.preteyeres.2013.02.003 (2013). [DOI] [PubMed] [Google Scholar]
- Tezel G. Oxidative stress in glaucomatous neurodegeneration: Mechanisms and consequences. Prog Retin Eye Res 25, 490–513, 10.1016/j.preteyeres.2006.07.003 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong G. Y. X., Van Bergen N. J. & Trounce I. A. & Crowston, J. G. Mitochondrial Dysfunction and Glaucoma. Journal of Glaucoma 18, 93–100 (2009). [DOI] [PubMed] [Google Scholar]
- Chrysostomou V., Rezania F., Trounce I. A. & Crowston J. G. Oxidative stress and mitochondrial dysfunction in glaucoma. Curr Opin Pharmacol 13, 12–15, 10.1016/j.coph.2012.09.008 (2013). [DOI] [PubMed] [Google Scholar]
- Tezel G., Yang X. & Cai J. Proteomic identification of oxidatively modified retinal proteins in a chronic pressure-induced rat model of glaucoma. Invest Ophth Vis Sci 46 (2005). [DOI] [PubMed] [Google Scholar]
- Bhattacharya S. K. et al. Proteomics implicates peptidyl arginine deiminase 2 and optic nerve citrullination in glaucoma pathogenesis. Invest Ophthalmol Vis Sci 47, 2508–2514, 10.1167/iovs.05-1499 (2006). [DOI] [PubMed] [Google Scholar]
- Crabb J. W. et al. Preliminary quantitative proteomic characterization of glaucomatous rat retinal ganglion cells. Exp Eye Res 91, 107–110, 10.1016/j.exer.2010.04.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell C., Arbogast B., Cioffi G., Burgoyne C. & Zhou A. Retinal proteomic changes following unilateral optic nerve transection and early experimental glaucoma in non-human primate eyes. Exp Eye Res 93, 13–28, 10.1016/j.exer.2011.03.020 (2011). [DOI] [PubMed] [Google Scholar]
- Grus F. H. et al. Transthyretin and complex protein pattern in aqueous humor of patients with primary open-angle glaucoma. Mol Vis 14, 1437–1445 (2008). [PMC free article] [PubMed] [Google Scholar]
- Duan X. M. et al. Proteomic analysis of aqueous humor from patients with primary open angle glaucoma. Mol Vis 16, 2839–2846 (2010). [PMC free article] [PubMed] [Google Scholar]
- Izzotti A., Longobardi M., Cartiglia C. & Sacca S. C. Proteome Alterations in Primary Open Angle Glaucoma Aqueous Humor. J Proteome Res 9, 4831–4838, 10.1021/Pr1005372 (2010). [DOI] [PubMed] [Google Scholar]
- Anshu A. et al. Alterations in the aqueous humor proteome in patients with a glaucoma shunt device. Mol Vis 17, 1891–1900 (2011). [PMC free article] [PubMed] [Google Scholar]
- Bouhenni R. A. et al. Identification of differentially expressed proteins in the aqueous humor of primary congenital glaucoma. Exp Eye Res 92, 67–75, 10.1016/j.exer.2010.11.004 (2011). [DOI] [PubMed] [Google Scholar]
- Bagnis A., Izzotti A., Centofanti M. & Sacca S. C. Aqueous humor oxidative stress proteomic levels in primary open angle glaucoma. Exp Eye Res 103, 55–62, 10.1016/j.exer.2012.07.011 (2012). [DOI] [PubMed] [Google Scholar]
- Bhattacharya S. K. et al. Proteomics reveal cochlin deposits associated with glaucomatous trabecular meshwork. J Biol Chem 280, 6080–6084, 10.1074/jbc.M411233200 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. et al. Comparative genomic and proteomic analysis of cytoskeletal changes in dexamethasone-treated trabecular meshwork cells. Mol Cell Proteomics 12, 194–206, 10.1074/mcp.M112.019745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieragostino D. et al. Differential protein expression in tears of patients with primary open angle and pseudoexfoliative glaucoma. Mol Biosyst 8, 1017–1028, 10.1039/c1mb05357d (2012). [DOI] [PubMed] [Google Scholar]
- Tezel G. et al. Hemoglobin Expression and Regulation in Glaucoma: Insights into Retinal Ganglion Cell Oxygenation. Invest Ophth Vis Sci 51, 907–919, 10.1167/Iovs.09-4014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. et al. Neurodegenerative and inflammatory pathway components linked to TNF-alpha/TNFR1 signaling in the glaucomatous human retina. Invest Ophthalmol Vis Sci 52, 8442–8454, 10.1167/iovs.11-8152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C. et al. Glaucomatous tissue stress and the regulation of immune response through glial Toll-like receptor signaling. Invest Ophthalmol Vis Sci 51, 5697–5707, 10.1167/iovs.10-5407 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner H. H., Rayborn M. E., Osterfeld A. M. & Hollyfield J. G. Localization of Proteoglycan within the Extracellular-Matrix Sheath of Cone Photoreceptors. Exp Eye Res 44, 633–642, 10.1016/S0014-4835(87)80135-5 (1987). [DOI] [PubMed] [Google Scholar]
- Clarke G. et al. Rom-1 is required for rod photoreceptor viability and the regulation of disk morphogenesis. Nat Genet 25, 67–73 (2000). [DOI] [PubMed] [Google Scholar]
- Khorana H. G. Rhodopsin, Photoreceptor of the Rod Cell - an Emerging Pattern for Structure and Function. J Biol Chem 267, 1–4 (1992). [PubMed] [Google Scholar]
- Smalla K. H. et al. The synaptic glycoprotein neuroplastin is involved in long-term potentiation at hippocampal CA1 synapses. P Natl Acad Sci USA 97, 4327–4332, 10.1073/pnas.080389297 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenius K., Janz R., Sudhof T. C. & Jahn R. Structure of synaptogyrin (p29) defines novel synaptic vesicle protein. J Cell Bio 131, 1801–1809, 10.1083/jcb.131.6.1801 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L. et al. Identification of Synaptophysin as a Hexameric Channel Protein of the Synaptic Vesicle Membrane. Science 242, 1050–1053, 10.1126/science.2461586 (1988). [DOI] [PubMed] [Google Scholar]
- Oyler G. A. et al. The Identification of a Novel Synaptosomal-Associated Protein, Snap-25, Differentially Expressed by Neuronal Subpopulations. J Cell Biol 109, 3039–3052, 10.1083/jcb.109.6.3039 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit H. et al. Synaptotagmin-1 Docks Secretory Vesicles to Syntaxin-1/SNAP-25 Acceptor Complexes. Cell 138, 935–946, 10.1016/j.cell.2009.07.027 (2009). [DOI] [PubMed] [Google Scholar]
- Araujo H., Danziger N., Cordier J., Glowinski J. & Chneiweiss H. Characterization of Pea-15, a Major Substrate for Protein-Kinase-C in Astrocytes. J Biol Chem 268, 5911–5920 (1993). [PubMed] [Google Scholar]
- Bartsch U., Kirchhoff F. & Schachner M. Highly Sialylated N-Cam Is Expressed in Adult-Mouse Optic-Nerve and Retina. J Neurocytol 19, 550–565, 10.1007/Bf01257243 (1990). [DOI] [PubMed] [Google Scholar]
- Okazaki K., Obata N. H., Inoue S. & Hidaka H. S100-Beta Is a Target Protein of Neurocalcin-Delta, an Abundant Isoform in Glial-Cells. Biochem J 306, 551–555 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl D., Rueger D. C., Bignami A., Weber K. & Osborn M. Vimentin, the 57 000 molecular weight protein of fibroblast filaments, is the major cytoskeletal component in immature glia. Eur J Cell Biol 24, 191–196 (1981). [PubMed] [Google Scholar]
- Kwong J. M. K., Caprioli J. & Piri N. RNA Binding Protein with Multiple Splicing: A New Marker for Retinal Ganglion Cells. Invest Ophth Vis Sci 51, 1052–1058, 10.1167/Iovs.09-4098 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A. R., Muller L. P. D. & Brecha N. C. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J Comp Neurol 522, 1411–1443, 10.1002/Cne.23521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. G. et al. Change of gene expression profiles in the retina following optic nerve injury. Mol Brain Res 101, 82–92, 10.1016/S0169-328x(02)00171-7 (2002). [DOI] [PubMed] [Google Scholar]
- Gallego B. I. et al. IOP induces upregulation of GFAP and MHC-II and microglia reactivity in mice retina contralateral to experimental glaucoma. J Neuroinflamm 9, 10.1186/1742-2094-9-92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnstable C. J. & Drager U. C. Thy-1 antigen: a ganglion cell specific marker in rodent retina. Neuroscience 11, 847–855, 0306-4522 (1984). [DOI] [PubMed] [Google Scholar]
- Bell K., Funke S., Pfeiffer N. & Grus F. H. Serum and antibodies of glaucoma patients lead to changes in the proteome, especially cell regulatory proteins, in retinal cells. PLoS One 7, e46910, 10.1371/journal.pone (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T. et al. Circadian proteomics of the mouse retina. Proteomics 7, 3500–3508, 10.1002/pmic.200700272 (2007). [DOI] [PubMed] [Google Scholar]
- Kanamoto T., Ue T., Yokoyama T., Souchelnytskyi N. & Kiuchi Y. Proteomic study of DBA/2J mice retina: Down-regulation of Integrin beta 7 correlated with retinal ganglion cell death. Proteomics 9, 4962–4969, 10.1002/pmic.200800978 (2009). [DOI] [PubMed] [Google Scholar]
- Reidel B. et al. Proteomic profiling of a layered tissue reveals unique glycolytic specializations of photoreceptor cells. Mol Cell Proteomics 10, 10.1074/mcp (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A. et al. Retinal proteome alterations in a mouse model of type 2 diabetes. Diabetologia 57, 192–203, 10.1007/s00125-013-3070-2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. et al. The proteome of human retina. Proteomics 15, 836–840 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethen C. M., Reilly C., Feng X., Olsen T. W. & Ferrington D. A. The proteome of central and peripheral retina with progression of age-related macular degeneration. Invest Ophthalmol Vis Sci 47, 2280–2290, 10.1167/iovs.05-1395 (2006). [DOI] [PubMed] [Google Scholar]
- An E. et al. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. J Proteome Res 5, 2599–2610, 10.1021/pr060121j (2006). [DOI] [PubMed] [Google Scholar]
- Cavusoglu N. et al. Differential proteomic analysis of the mouse retina - The induction of crystallin proteins by retinal degeneration in the rd1 mouse. Mol Cell Proteomics 2, 494–505, 10.1074/mcp.M300029-MCP (2003). [DOI] [PubMed] [Google Scholar]
- Quin G., Len A. C., Billson F. A. & Gillies M. C. Proteome map of normal rat retina and comparison with the proteome of diabetic rat retina: new insight in the pathogenesis of diabetic retinopathy. Proteomics 7, 2636–2650, 10.1002/pmic.200600486 (2007). [DOI] [PubMed] [Google Scholar]
- Okamoto H. et al. Comparative proteomic analyses of macular and peripheral retina of cynomolgus monkeys (Macaca fascicularis). Exp Anim 59, 171–182 (2010). [DOI] [PubMed] [Google Scholar]
- Stapels M. D. & Barofsky D. F. Complementary use of MALDI and ESI for the HPLC-MS/MS analysis of DNA-binding proteins. Anal Chem 76, 5423–5430, 10.1021/Ac030427z (2004). [DOI] [PubMed] [Google Scholar]
- Stapels M. D., Cho J. C., Giovannoni S. J. & Barofsky D. F. Proteomic analysis of novel marine bacteria using MALDI and ESI mass spectrometry. J Biomol Tech 15, 191–198 (2004). [PMC free article] [PubMed] [Google Scholar]
- Nash M. S. & Osborne N. N. Assessment of Thy-1 mRNA levels as an index of retinal ganglion cell damage. Invest Ophth Vis Sci 40, 1293–1298 (1999). [PubMed] [Google Scholar]
- Chidlow G. & Osborne N. N. Rat retinal ganglion cell loss caused by kainate, NMDA and ischemia correlates with a reduction in mRNA and protein of Thy-1 and neurofilament light. Brain Res 963, 298–306, 10.1016/S0006-8993(02)04052-0 (2003). [DOI] [PubMed] [Google Scholar]
- Huang W., Fileta J., Guo Y. & Grosskreutz C. L. Downregulation of Thy1 in retinal ganglion cells in experimental glaucoma. Curr Eye Res 31, 265–271, 10.1080/02713680500545671 (2006). [DOI] [PubMed] [Google Scholar]
- Bauer M. F. & Neupert W. Import of proteins into mitochondria: A novel pathomechanism for progressive neurodegeneration. J Inherit Metab Dis 24, 166–180, 10.1023/A:1010314900814 (2001). [DOI] [PubMed] [Google Scholar]
- Schober M. S., Chidlow G., Wood J. P. & Casson R. J. Bioenergetic-based neuroprotection and glaucoma. Clin Experiment Ophthalmol 36, 377–385 (2008). [DOI] [PubMed] [Google Scholar]
- Tezel G. & Res F. A. P. O. The Role of Glia, Mitochondria, and the Immune System in Glaucoma. Invest Ophth Vis Sci 50, 1001–1012, 10.1167/Iovs.08-2717 (2009). [DOI] [PubMed] [Google Scholar]
- Campello L. et al. Alterations in energy metabolism, neuroprotection and visual signal transduction in the retina of Parkinsonian, MPTP-treated monkeys. PLoS One 8, e74439, 10.1371/journal.pone (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers J. C. et al. Magnocellular and parvocellular visual pathways are both affected in a macaque monkey model of glaucoma. Aust Nz J Ophthalmol 25, 239–243 (1997). [DOI] [PubMed] [Google Scholar]
- Osborne N. N. Mitochondria: Their role in ganglion cell death and survival in primary open angle glaucoma. Expl Eye Res 90, 750–757, 10.1016/j.exer (2010). [DOI] [PubMed] [Google Scholar]
- Nuskova H. et al. Mitochondrial ATP synthasome: Expression and structural interaction of its components. Biochem Biophys Res Commun 464, 787–793, 10.1016/j.bbrc (2015). [DOI] [PubMed] [Google Scholar]
- Won J. C. et al. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha overexpression prevents endothelial apoptosis by increasing ATP/ADP translocase activity. Arterioscler Thromb Vasc Biol 30, 290–297, 10.1161/ATVBAHA (2010). [DOI] [PubMed] [Google Scholar]
- Kim E. H., Koh E. H., Park J. Y. & Lee K. U. Adenine nucleotide translocator as a regulator of mitochondrial function: implication in the pathogenesis of metabolic syndrome. Korean Diabetes J 34, 146–153, 10.4093/kdj (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A. et al. Striatal Alterations of Secretogranin-1, Somatostatin, Prodynorphin, and Cholecystokinin Peptides in an Experimental Mouse Model of Parkinson Disease. Mol Cell Proteomics 8, 1094–1104, 10.1074/mcp.M800454-MCP200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G., Li L. Y., Patil R. V. & Wax M. B. TNF-alpha and TNF-alpha receptor-1 in the retina of normal and glaucomatous eyes. Invest Ophthalmol Vis Sci 42, 1787–1794 (2001). [PubMed] [Google Scholar]
- Loeffler K. U. & Mangini N. J. Anti-arrestin immunoreactivity in the human retina: Difference between light- and dark-adaptation. Curr Eye Res 14, 1165–1168 (1995). [DOI] [PubMed] [Google Scholar]
- Ethen C. M., Feng X., Olsen T. W. & Ferrington D. A. Declines in arrestin and rhodopsin in the macula with progression of age-related macular degeneration. Invest Ophth Vis Sci 46, 769–775, 10.1167/Iovs.04-0810 (2005). [DOI] [PubMed] [Google Scholar]
- Forooghian F., Cheung R. K., Smith W. C., O’Connor P. & Dosch H. M. Enolase and arrestin are novel nonmyelin autoantigens in multiple sclerosis. J Clin Immunol 27, 388–396, 10.1007/s10875-007-9091-1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahid S., Oellerich M., Asif A. R. & Ahmed N. Differential expression of proteins in brain regions of Alzheimer’s disease patients. Neurochem Res 39, 208–215, 10.1007/s11064-013-1210-1 (2014). [DOI] [PubMed] [Google Scholar]
- Kaindl A. M. et al. Erythropoietin protects the developing brain from hyperoxia-induced cell death and proteome changes. Ann Neurol 64, 523–534, 10.1002/ana.21471 (2008). [DOI] [PubMed] [Google Scholar]
- Verhagen A. M. et al. Identification of mammalian mitochondrial proteins that interact with IAPs via N-terminal IAP binding motifs. Cell Death Differ 14, 348–357, 10.1038/sj.cdd.4402001 (2007). [DOI] [PubMed] [Google Scholar]
- Holcik M., Gordon B. W. & Korneluk R. G. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol Cell Biol 23, 280–288, 10.1128/Mcb.23.1.280-288.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekenstein U. & Soreq H. Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: From structural insights to post-transcriptional regulatory roles. Mol Cell Neurosci 56, 436–446, 10.1016/j.mcn.2012.12.002 (2013). [DOI] [PubMed] [Google Scholar]
- Chen R. Z., Akbarian S., Tudor M. & Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet 27, 327–331, 10.1038/85906 (2001). [DOI] [PubMed] [Google Scholar]
- Amir R. E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23, 185–188 (1999). [DOI] [PubMed] [Google Scholar]
- Ausio J., de Paz A. M. & Esteller M. MeCP2: the long trip from a chromatin protein to neurological disorders Trends Mol Med 20, 487–498 (2014). [DOI] [PubMed] [Google Scholar]
- Chahrour M. et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229, 10.1126/science (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold W. A., Lacina T. A., Cantrill L. C. & Christodoulou J. MeCP2 deficiency is associated with reduced levels of tubulin acetylation and can be restored using HDAC6 inhibitors. J Mol Med , 10.1007/s00109-014-1202-x (2014). [DOI] [PubMed] [Google Scholar]
- Babbio F. et al. Knock-down of methyl CpG-binding protein 2 (MeCP2) causes alterations in cell proliferation and nuclear lamins expression in mammalian cells. BMC Cell Biol 13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales M. L. & LaSalle J. M. The role of MeCP2 in brain development and neurodevelopmental disorders. Curr Psychiatry Rep 12, 127–134, 10.1007/s11920-010-0097-7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O. & Veenstra G. J. C. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 118, 549–565 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausio J., Levin D. B., de Amorim G. V., Bakker S. & Macleod P. M. Syndromes of disordered chromatin remodeling. Clin Genet 64, 83–95 (2003). [DOI] [PubMed] [Google Scholar]
- Song C. et al. DNA methylation reader MECP2: cell type-and differentiation stage-specific protein distribution. Epigenetics & Chromatin 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A. et al. Direct homo- and hetero-interactions of MeCP2 and MBD2. PLoS One 8, e53730, 10.1371/journal.pone (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X. S. et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. P Natl Acad Sci USA 104, 2709–2714, 10.1073/pnas.0608056104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surgucheva I. & Surguchov A. Gamma-synuclein: cell-type-specific promoter activity and binding to transcription factors. J Mol Neurosci 35, 267–271, 10.1007/s12031-008-9074-6 (2008). [DOI] [PubMed] [Google Scholar]
- Mellen M., Ayata P., Dewell S., Kriaucionis S. & Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoh L. S. et al. The stress oncoprotein LEDGF/p75 interacts with the methyl CpG binding protein and influences its transcriptional activity. Mol Cancer Res 10, 378–391 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocher J., Vogel B. & Hock R. HMGA1 down-regulation is crucial for chromatin composition and a gene expression profile permitting myogenic differentiation. BMC Cell Biol 11, Artn 64 10.1186/1471-2121-11-64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski M. & Akbarian S. Epigenetic mechanisms in neurodevelopmental and neurodegenerative disease. Nat. Med. 18, 1194–1204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dintilhac A. & Bernues J. HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J Biol Chem 277, 7021–7028, 10.1074/jbc (2002). [DOI] [PubMed] [Google Scholar]
- Acharya M. et al. Evaluation of the OPTC gene in primary open angle glaucoma: functional significance of a silent change. BMC Mol Biol 8, 21, 10.1186/1471-2199-8-21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borras T. The effects of myocilin expression on functionally relevant trabecular meshwork genes: a mini-review. J Ocul Pharmacol Ther 30, 202–212, 10.1089/jop.2013.0218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg M. E. et al. Differential effect of nitric oxide on glutathione metabolism and mitochondrial function in astrocytes and neurones: implications for neuroprotection/neurodegeneration? J Neurochem 86, 228–237 (2003). [DOI] [PubMed] [Google Scholar]
- Sacca S. C., Izzotti A., Rossi P. & Traverso C. Glaucomatous outflow pathway and oxidative stress. Exp Eye Res 84, 389–399, 10.1016/j.exer.2006.10.008 (2007). [DOI] [PubMed] [Google Scholar]
- Aslan M., Cort A. & Yucel I. Oxidative and nitrative stress markers in glaucoma. Free Radic Biol Med 45, 367–376, 10.1016/j.freeradbiomed.2008.04.026 (2008). [DOI] [PubMed] [Google Scholar]
- Yang J., Tezel G., Patil R. V., Romano C. & Wax M. B. Serum autoantibody against glutathione S-transferase in patients with glaucoma. Invest Ophthalmol Vis Sci 42, 1273–1276 (2001). [PubMed] [Google Scholar]
- Singhal S. S. et al. Induction of glutathione S-transferase hGST 5.8 is an early response to oxidative stress in RPE cells. Invest Ophth Vis Sci 40, 2652–2659 (1999). [PubMed] [Google Scholar]
- Juronen E. et al. Polymorphic glutathione S-transferase M1 is a risk factor of primary open-angle glaucoma among Estonians. Exp Eye Res 71, 447–452, 10.1006/exer.2000.0899 (2000). [DOI] [PubMed] [Google Scholar]
- Yildirim O. et al. May glutathione S-transferase M1 positive genotype afford protection against primary open-angle glaucoma? Graef Arch Clin Exp 243, 327–333, 10.1007/s00417-004-1013-9 (2005). [DOI] [PubMed] [Google Scholar]
- Pae C. U. et al. Glutathione S-transferase M1 polymorphism may contribute to schizophrenia in the Korean population. Psychiat Genet 14, 147–150, 10.1097/00041444-200409000-00005 (2004). [DOI] [PubMed] [Google Scholar]
- Gruys E., Toussaint M. J., Niewold T. A. & Koopmans S. J. Acute phase reaction and acute phase proteins. J Zhejiang Univ Sci B 6, 1045–1056, 10.1631/jzus.2005.B1045 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-Inglesias H. et al. Comparative proteomic study in serum of patients with primary open-angle glaucoma and pseudoexfoliation glaucoma. J Proteomics 98, 65–78 (2014). [DOI] [PubMed] [Google Scholar]
- Kannan R., Sreekumar P. G. & Hinton D. R. Novel roles for alpha-crystallins in retinal function and disease. Prog Retin Eye Res 31, 576–604, 10.1016/j.preteyeres.2012.06.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D. S., Boskovska O. B., Knopf H. L., Lampi K. J. & Shiels A. A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. Am J Hum Genet 71, 1216–1221, 10.1086/344212 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P. E., Freeman W. M., Losiewicz M. K., Singh R. S. & Gardner T. W. The retinal proteome in experimental diabetic retinopathy: up-regulation of crystallins and reversal by systemic and periocular insulin. Mol Cell Proteomics 8, 767–779, 10.1074/mcp (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutjen-Drecoll E. et al. Localization of the stress proteins alpha B-crystallin and trabecular meshwork inducible glucocorticoid response protein in normal and glaucomatous trabecular meshwork. Invest Ophthalmol Vis Sci 39, 517–525 (1998). [PubMed] [Google Scholar]
- Thanos S. et al. Role of crystallins in ocular neuroprotection and axonal regeneration. Prog Retin Eye Res 42C, 145–161, 10.1016/j.preteyeres.2014.06.004 (2014). [DOI] [PubMed] [Google Scholar]
- Piri N., Kwong J. & Caprioli J. Crystallins in Retinal Ganglion Cell Survival and Regeneration. Mol Neurobiol 48, 819–828, 10.1007/s12035-013-8470-2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Y. et al. Retinol dehydrogenase 13 protects the mouse retina from acute light damage. Mol Vis 18, 1021–1030 (2012). [PMC free article] [PubMed] [Google Scholar]
- Bigl M., Bruckner M. K., Arendt T., Bigl V. & Eschrich K. Activities of key glycolytic enzymes in the brains of patients with Alzheimer’s disease. J Neural Transm 106, 499–511 (1999). [DOI] [PubMed] [Google Scholar]
- Perluigi M., Di Domenico F. & Buttterfield D. A. Unraveling the complexity of neurodegeneration in brains of subjects with Down syndrome: insights from proteomics. Proteomics Clin Appl 8, 73–85, 10.1002/prca.201300066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S. et al. Comparison of Anti-Ocular IgG and IgM Autoantibody Patterns in Glaucoma Patients. Invest Ophthalmol Vis Sci 51, E-Abstract 4817 (2010). [Google Scholar]
- Janciauskiene S. & Krakau T. Alzheimer’s peptide and serine proteinase inhibitors in glaucoma and exfoliation syndrome. Doc Ophthalmol 106, 215–223 (2003). [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J. V. & Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1, 2856–2860, 10.1038/nprot.2006.468 (2006). [DOI] [PubMed] [Google Scholar]
- Cox J. et al. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol Cell Proteomics in press (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal N., Funke S., Pfeiffer N. & Grus F. H. Characterization of lacrimal proline-rich protein 4 (PRR4) in human tear proteome. Proteomics 00, 1–12 (2014). [DOI] [PubMed] [Google Scholar]
- Jaffe J. D. et al. Accurate Inclusion Mass Screening A bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics 7, 1952–1962, 10.1074/mcp.M800218-MCP200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N., Rettenmeier A. W. & Schmitz-Spanke S. Recent advances in the use of Sus scrofa (pig) as a model system for proteomic studies. Proteomics 11, 776–793, 10.1002/pmic.201000320 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.