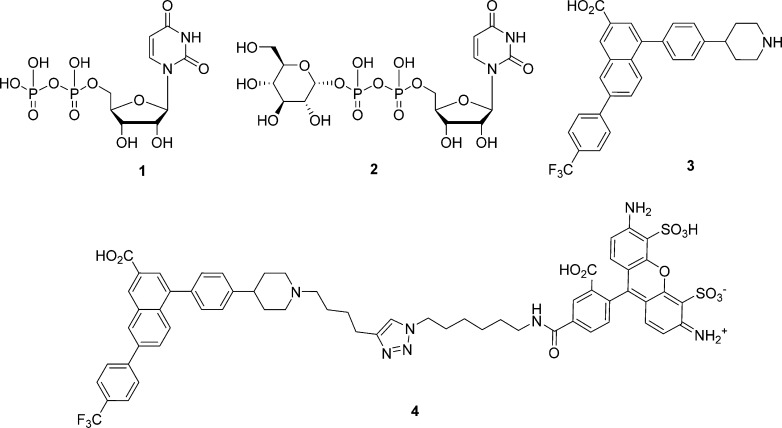
Abstract
UDP and UDP-glucose activate the P2Y14 receptor (P2Y14R) to modulate processes related to inflammation, diabetes, and asthma. A computational pipeline suggested alternatives to naphthalene of a previously reported P2Y14R antagonist (3, PPTN) using docking and molecular dynamics simulations on a hP2Y14R homology model based on P2Y12R structures. By reevaluating the binding of 3 to P2Y14R computationally, two alternatives, i.e., alkynyl and triazolyl derivatives, were identified. Improved synthesis of fluorescent antagonist 4 enabled affinity quantification (IC50s, nM) using flow cytometry of P2Y14R-expressing CHO cells. p-F3C-phenyl-triazole 65 (32) was more potent than a corresponding alkyne 11. Thus, additional triazolyl derivatives were prepared, as guided by docking simulations, with nonpolar aryl substituents favored. Although triazoles were less potent than 3 (6), simpler synthesis facilitated further structural optimization. Additionally, relative P2Y14R affinities agreed with predicted binding of alkynyl and triazole analogues. These triazoles, designed through a structure-based approach, can be assessed in disease models.
Introduction
Extracellular uridine-5′-diphosphate (1) and uridine-5′-diphosphoglucose (2, Chart 1) activate the P2Y14 receptor (P2Y14R), a G protein-coupled receptor (GPCR) belonging to the rhodopsin-like δ-branch, to modulate cell functions related to inflammation, diabetes, asthma, and other diseases.1,2 This receptor subtype is a member of the P2Y12R-like subfamily of nucleotide receptors, which inhibit the production of 3′,5′-cyclic adenosine monophosphate (cAMP) through Gi protein. The P2Y14R promotes hypersensitivity in microglial cells,3 the mobility of neutrophils,4 the release of mediators from mast cells,5 inflammation in renal intercalated cells,6 and mixed effects in insulin function.7,8 Thus, approaches to novel antagonists of nucleotide signaling at the P2Y14R would be desirable for exploration as novel therapeutics. The P2Y14R is also present in the CNS, where it suppresses release of matrix metalloproteinase-9 (MMP-9) and tumor necrosis factor (TNF) from astrocytes.9
Chart 1. Agonist and Antagonist Ligand Probes of the P2Y14Ra.
a Endogenous agonists 1 and 2 have functional EC50 values at the hP2Y14R of 160 and 261 nM, respectively.45 Antagonist 3 is highly potent at the hP2Y14R (KB from Schild analysis = 0.434 nM)14 but suffers from adverse physicochemical properties (highly hydrophobic/amphipathic) that make dissolution and purification difficult. Fluorescent antagonist 4 is highly potent at the hP2Y14R (functional Ki = 0.080 nM).15.
Only a limited set of P2Y14R antagonists are currently known. Several chemotypes based on naphthoic acid and pyrido[4,3-d]pyrimidine were reported originally in patents to provide potent P2Y14R antagonists, which however displayed low oral bioavailability.10−13 One of those naphthoic acid derivatives, 4-(4-(piperidin-4-yl)-phenyl)-7-(4-(trifluoromethyl)-phenyl)-2-naphthoic acid (PPTN, 3), was profiled pharmacologically at the entire family of eight P2YRs and found to display high affinity and selectivity (IC50 = 0.4 nM at P2Y14R and >10 μM at other P2YR subtypes).14 We demonstrated that the piperidine group of 3 was a suitable site for chemical derivatization and chain extension to prepare high affinity fluorescent probes of the P2Y14R. This conclusion was supported by molecular modeling and ligand docking, which showed the piperidine ring facing outward at the surface of the receptor.15 One such probe, 4, displayed exceptionally high affinity and low nonspecific binding when used as a tracer in a flow cytometric assay of the P2Y14R in whole Chinese hamster ovary (CHO) cells expressing the receptor.
Although previous approaches to modeling of P2YRs were subject to high uncertainty, we now have appropriate templates to obtain detailed docking predictions and structural explanations of previously determined structure–activity relationship (SAR) within the P2Y12R-like subfamily, e.g., uracil nucleotides binding to the P2Y14R.16 In the present study, a human (h) P2Y14R homology model based on recent hP2Y12R X-ray structures17,18 served as a template to conduct docking and molecular dynamics (MD) simulations. The immediate goal was to suggest bioisosteric alternatives to the hydrophobic and unwieldy naphthalene ring of 3 that would maintain a similar orientation of the piperidine and 4-(trifluoromethyl)-phenyl substituents when bound to the receptor and therefore preserve receptor affinity. We sought to simultaneously reduce the molecular weight and avoid the high lipophilicity of 3 that contributes to its low solubility and difficulty of purification.13
Results
The macromolecular starting point of our study is a previously obtained hP2Y14R MD-refined homology model based on the agonist-bound hP2Y12R X-ray structure (PDB ID: 4PXZ).16−18 The reference antagonist structure 3 (PPTN) has been docked into the model by using an Induced Fit Docking (IFD) protocol (see Methods section), and the complex has been refined by subjecting it to 10 ns of membrane MD simulations. With respect to the starting agonist-bound hP2Y14R homology model (Supporting Information, Figure S1A),16 the refined structure used in this study (Supporting Information, Figure S1B) featured a larger binding cavity extending toward the extracellular side. The adaptation of the binding site to the antagonist structure caused a rearrangement of the position of transmembrane domain (TM)2 and TM7 with respect to the TM bundle (Supporting Information, Figure S2A): in particular, TM7 was pushed outward, whereas the axis of TM2 became slightly bent toward TM3. Moreover, the extension of the binding site region toward the extracellular side reoriented the first and second extracellular loops (ECL1 and ECL2, respectively) away from the TM bundle (Supporting Information, Figure S2B,C). Notably, the differences described above in the MD refined agonist- and antagonist-bound hP2Y14R homology models mirror those experimentally observed for the hP2Y12R X-ray structures.17,18 Therefore, the final hP2Y14R MD-refined structure (Supporting Information) has been used as a template for all subsequent docking simulations.
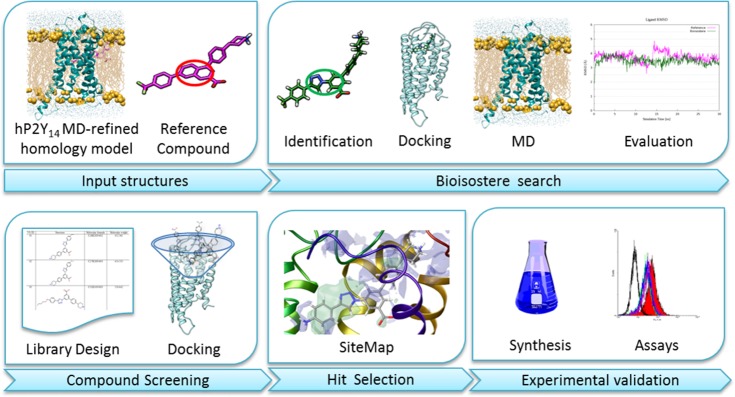
Starting from this refined template, we developed a computational pipeline comprising three subsequent phases (Figure 1): (i) bioisostere search, (ii) compound screening, and (iii) hit selection. The bioisostere search stage envisaged the use of docking runs followed by 30 ns of MD simulations to identify linking groups as suitable replacements of half of the naphthalene ring while preserving the ligand–receptor interactions observed for the reference compound. The selection of the linkers has been mainly guided by the knowledge gained in a previous study that led to the design of highly potent A3 adenosine receptor (A3AR) agonists.19 Once a new scaffold was found, a small library of hypothetical compounds was screened by means of docking simulations. Pose filtering and hit selection were based upon ligand–receptor complementarity and optimal overlap between ligand functional groups and computed protein interaction sites.
Figure 1.
Workflow of the computational pipeline. Selection of naphthalene bioisosteres was guided by a previous study focused on A3AR agonists. Between the two proposed alternatives, the triazole analogue resulted as being the most promising according to membrane MD simulations analysis of the ligand–protein complexes as compared with the reference compound. Consequently, a small library of 57 triazole analogues was designed and docked inside the hP2Y14R. Poses were selected by visual inspection. The synthesis and experimental validation of the compounds were prioritized according to the overlap between compounds functional groups and protein interaction sites.
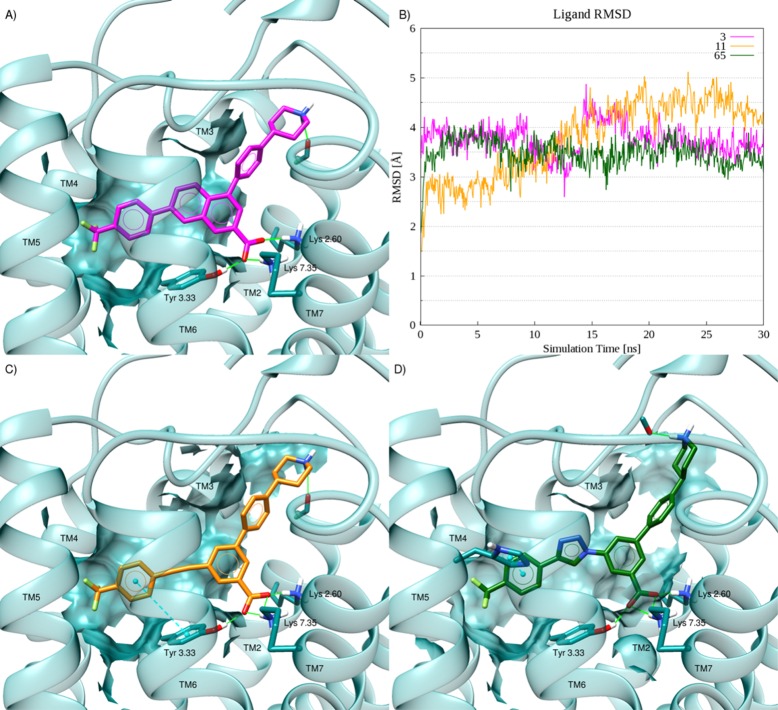
In the first instance, the parent structure 3 was redocked into the MD-refined model. The corresponding pose (Figure 2A) showed an overall root-mean-square deviation (RMSD) value with respect to the final MD snapshot of 0.42 Å (data not shown). In the predicted binding mode, the ligand resided in the orthosteric binding site in an orientation similar to that previously reported for hP2Y14R antagonist probes:15 the 2-naphthoic acid carboxylate bridged Lys772.60 (according to conventional TM numbering20), Lys2777.35, and Tyr1023.33, while the piperidine group pointed outward with the nitrogen atom establishing a H-bond interaction with the backbone of Gly802.63. The docking pose has been subjected to 30 ns of membrane MD simulation, and the RMSD with respect to the starting structure has been computed (Figure 2B, magenta line); this analysis served as a confirmation of the docking pose stability as well as a reference to compare newly proposed compounds containing bioisosteres of the naphthalene. During the MD simulation, compound 3 exhibited an average RMSD of 3.76 Å that can be ascribed mainly to motion of the solvent-exposed piperidine ring. As shown in the trajectory visualization (Supporting Information, Video S1), the naphthalene core with its carboxylate group is well anchored in the binding site during the 30 ns time frame by a tight H-bond network established with Lys772.60, Lys2777.35, and Tyr1023.33. However, the distal piperidine nitrogen suddenly moved apart from the pose predicted by docking and approached the backbone of Ile170 in the second extracellular loop (EL2) through the interplay of water molecules.
Figure 2.
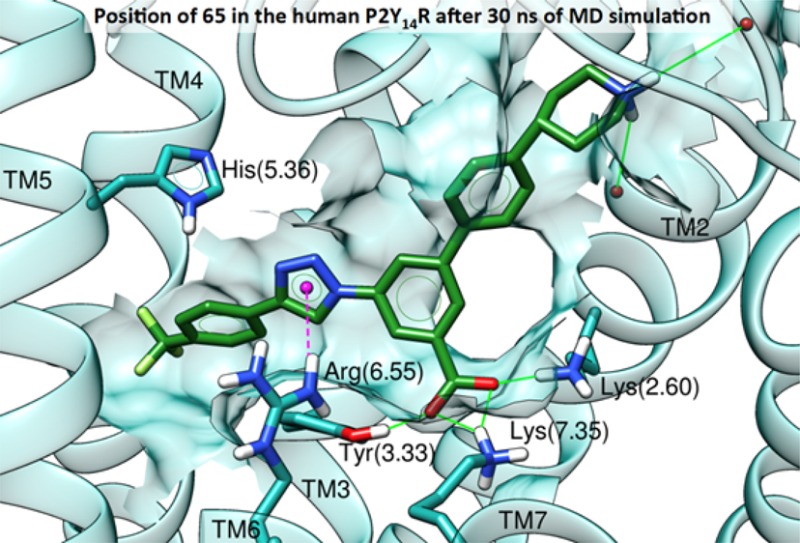
(A) Docking pose of reference compound 3 (magenta-colored carbons) at hP2Y14R. (B) RMSD plots for the considered compounds during 30 ns of membrane MD simulations. (C) Docking pose of the alkynyl derivative 11 (orange-colored carbons) at hP2Y14R. (D) Docking pose of the triazolyl derivative 65 (green-colored carbons) at the hP2Y14R. Side chains of residues important for ligand recognition are reported as sticks (dark-cyan carbon atoms). Side chains of residues establishing either van der Waals or hydrophobic contacts with the ligand are rendered as transparent surface. H-Bonds are pictured as green solid lines, whereas π–π stacking interactions as cyan dashed lines with the centroids of the aromatic rings displayed as cyan spheres. Nonpolar hydrogen atoms are omitted.
The first proposed alternative structure was an alkyne derivative (11, Scheme 1) containing a p-CF3-phenyl group, similar to 3. The compound was docked into the MD-refined hP2Y14R homology model and then subjected to 30 ns of MD simulations. In the resulting pose (Figure 2C), the trifluoromethylphenyl group was buried in a deep hydrophobic pocket, and the carboxylate group interacted with Lys772.60, Tyr1023.33, and Lys2777.35 as in the docking pose of 3 (Figure 2A). Although during the MD simulations of the hP2Y14–11 complex, the ligand atoms displayed an average RMSD value (3.79 Å) close to the one observed for the reference compound, the corresponding graph suggested that compound 11 was less stable than 3 in the binding pocket (Figure 2B, orange and magenta lines, respectively). In particular, in the first 15 ns of simulation, compound 11 experienced an increasing deviation from the starting pose, leading to a higher root-mean-square fluctuation (RMSF = 1.52 vs 1.16 Å for compounds 11 and 3, respectively). The trajectory analysis (Supporting Information, Video S2) revealed that, after a few ns, the piperidine ring of derivative 11 moved from the docking pose to establish an H-bond interaction with the backbone of Ile170 EL2 and Gln169EL2 in a way similar to that observed for the reference compound. On the other hand, after approximately 12 ns, the side chain of Tyr1023.33 moved to engage the trifluoromethylphenyl group in a T-shaped π–π stacking interaction. This movement caused a weakening of the H-bond network around the carboxylate moiety of 11 that increased the number of water molecules surrounding the group during the trajectory.
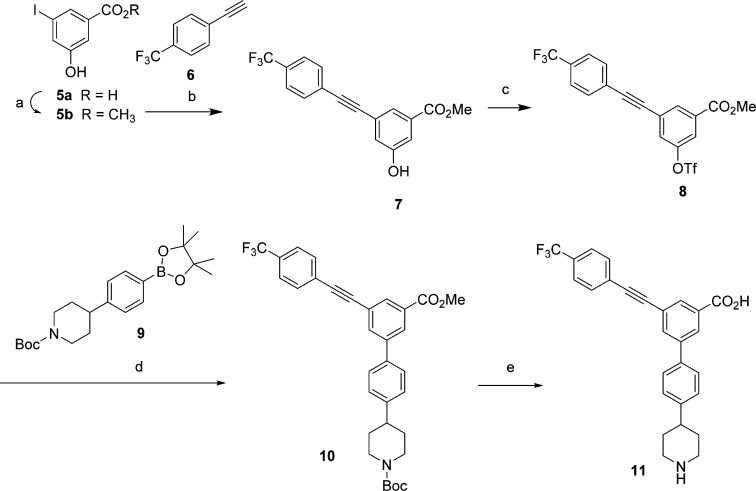
Scheme 1. Synthesis of an Alkynyl Derivative 11 as a P2Y14R Antagonist.
Reagents and conditions: (a) CH3OH, SOCl2, 0–23 °C (33%); (b) 1-ethynyl-4-(trifluoromethyl)benzene, CuI, PdCl2(PPh3)2, DMF, NEt3, 0–23 °C (81%); (c) (CF3SO2)2O, NEt3, CH2Cl2 (98%); (d) tert-butyl 4-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)piperidine-1-carboxylate, Pd(PPh3)4, K2CO3, DMF (67%); (e) LiOH (aqueous 0.5M), CH3OH reflux, then HCl (1 M aq), pH 1 (28%).
On the basis of the analogy observed for the docking poses and trajectory analyses of compound 11 with respect to the reference structure 3, we synthesized 11 using both Sonogashira21 and Suzuki22 cross-coupling reactions (Scheme 1). In particular, a Sonogashira coupling reaction of a iodo arene 5b with 4-(trifluoromethyl)phenylacetylene (6) in the presence of tetrakis(triphenylphosphine)palladium yielded derivative 7. Compound 5b was obtained by esterification of a commercial precursor 5a. The 3-hydroxy group of 7 was then converted to the corresponding aryl triflate 8 using triflic anhydride and pyridine. A Suzuki cross-coupling reaction between 8 and dioxaborolane derivative 9, prepared by conventional Suzuki–Miyaura reaction23 in the presence of tetrakis(triphenylphosphine)palladium catalyst, afforded compound 10. Finally, removal of the N-Boc protecting group followed by hydrolysis of the ester provided derivative 11. This route would allow a wide variety of substitutions to be introduced at a late stage in the synthetic sequence because of the commercial availability of numerous arylacetylene derivatives.
On the basis of the suggestion that the replacement of the naphthoic acid core of 3 with an alkynyl group might lead to the loss of an anchoring hydrogen bond with Tyr1023.33, other alternatives were considered as well. A similarly versatile synthetic approach could be used for introducing arylacetylene moieties in the form of 1,2,3-triazoles by copper-catalyzed [2 + 3] cycloaddition24 with an azido group present on the core of the molecule. The corresponding triazole derivative containing a p-CF3-phenyl group (65, Scheme 2) was subjected to the same computational protocol described for compound 11 (docking followed by 30 ns of MD simulations). The resulting docking pose (Figure 2D) suggested a placement of compound 65 in the binding site similar to that predicted for 3 and 11, which encompassed a tight H-bond network around the carboxylate group with the piperidine ring solvent-exposed. On the other side, a slightly higher placement within the binding site enabled compound 65 to establish an additional π–π stacking interaction with the side chain of His1845.36, while the piperidine ring was anchored by an H-bond with the Ile167EL2 backbone.
Scheme 2. Synthesis of Triazolyl Derivatives as P2Y14R Antagonists.
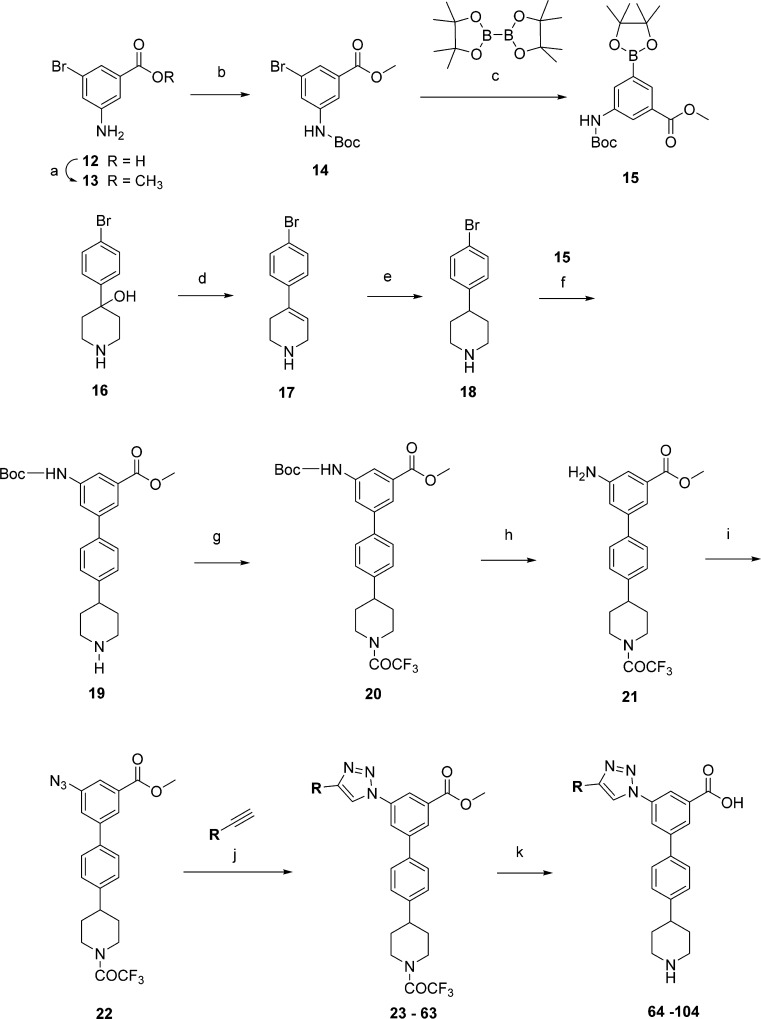
Reagents and conditions: (a) CH3OH, SOCl2, 0–23 °C (98%); (b) Boc2O, DMAP, CH2Cl2; (c) PdCl2(dppf), AcOK, DMF, 95 °C (74%); (d) F3CCO2H, 90 °C (97%); (e) H2, Rh/C, 100 psi (98%); (f) Pd(Ph3P)4, K2CO3, DME, 85 °C (71%); (g) (CF3CO)2O, NEt3, Et2O; (h) F3CCO2H, CH2Cl2 (70%); (i) (1) Ts-OH, NaNO2, H2O/acetonitrile, (2) NaN3, (83%); (j) CuSO4, sodium ascorbate (1 M aq); (k) KOH (1 M aq). R is defined in Tables 1 and 2.
MD simulations of the hP2Y14–65 complex resulted in lower average RMSD and RMSF values of 3.48 and 0.70 Å, respectively (Figure 2B, dark-green line). After a few ns (Supporting Information, Video S3), the ligand moved deeper in the binding site and was stabilized by persistent π–π stacking interactions established between the triazole group and Tyr1023.33 and between the trifluoromethylphenyl group and His1845.36. A π–cation interaction between the triazole ring and the side chain of Arg2536.55 further contributed to stabilizing the ligand position during the 30 ns time frame. Conversely to what was observed for compounds 3 and 11, the piperidine ring moved toward Gly802.63. The carboxylate group of 65 maintained a stable position, and the water molecules surrounding the carboxylate group of 11 were not observed during the simulation of the hP2Y14–65 complex.
On the basis of these favorable predictions, using a strategy similar to Scheme 1, triazolyl derivative 65 and its analogues were synthesized starting from the 3-amino-5-bromobenzoic acid (12) and 4-(4-bromophenyl)piperidin-4-ol (16, Scheme 2). The carboxylic group of 12 was first converted to the methyl ester 13, and then the amine function was protected to give Boc-derivative 14. The palladium-catalyzed condensation of aryl bromide 14 with bis(pinacolato)-diboron under basic conditions afforded dioxaborolane 15. The acid-catalyzed dehydration of 16 yielded alkene 17, which was reduced to provide compound 18. Derivative 19 was obtained by coupling 18 with compound 15 under Suzuki conditions.22 The conversion of the amino group of 19 to a trifluoroacetamide 20 was accomplished using trifluoroacetic anhydride in the presence of triethylamine. Removing the N-Boc protecting group of 20 gave the amine 21. Compound 21 was converted into aryl azide 22 from an arenediazonium tosylate that was generated in situ and subsequent addition of sodium azide.25 The protected 1,2,3-triazolyl derivatives 23–63 were synthesized via an azide–alkyne Huisgen cycloaddition (“click reaction”) involving aryl azide 22, various alkynes, Cu(II)sulfate salt, and sodium ascorbate.26 One-pot hydrolysis of the trifluoroacetamide and the ester in the presence of KOH yielded compounds 64–104, which were purified by semiprep HPLC and isolated either as acetate or triethylammonium salts.
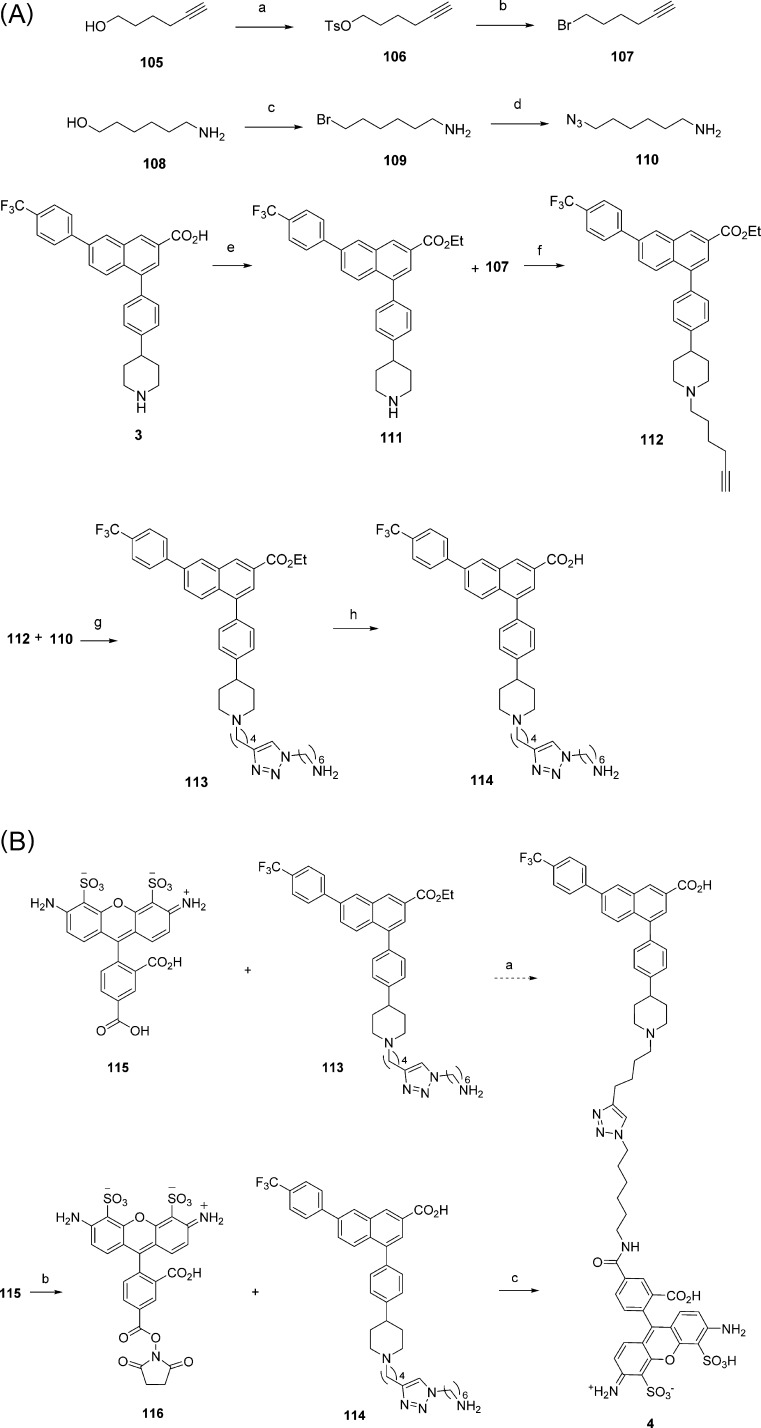
The synthesis of fluorescent antagonist 4 as previously reported15 suffered from a low yield in the final click cycloaddition step to link the azide-functionalized fluorophore and the alkyne-functionalized pharmacophore. Given the unusually high affinity of 4 and its low nonspecific character, we explored an alternate synthesis of 4. This was necessary to provide a sufficient supply of fluorescent probe 4 for use in routine assays, which we needed for the new putative antagonists 11 and 64–104. A more efficient route consisted of forming an amide as the ultimate or penultimate step. Thus, the pharmacophore was functionalized with an extended amine through a preformed triazole linker to provide intermediate 113 (Scheme 3A). The coupling of AlexaFluor488 fluorophore27 and pharmacophore was attempted by two methods, either: (1) condensation of the fluorophore as a 5-carboxylic acid 115 to the ethyl ester-protected derivative 113 of the pharmacophore followed by ester saponification, or (2) by reaction of the fluorophore that was activated in situ as a N-succinimidyl ester 116 with the amino derivative 114, having a deprotected carboxylic acid (Scheme 3B). However, only the second synthetic route provided compound 4 and at the same time improved the reaction yield compared to the previous synthetic method.15 Furthermore, we explored a different fluorescent antagonist analogue for possible use in screening, e.g., 130 containing a cyanine-5 (Cy5) fluorophore, but this compound was considerably less potent at the hP2Y14R in comparison to 4 (Supporting Information).
Scheme 3.
Reagents and conditions: (A) (a) TsCl, NEt3, DMAP, CH2Cl2, rt 15 h (88%); (b) LiBr, DMF, rt, 12 h (82%); (c) HBr 48% soln, 80 °C, 20 h (61%); (d) NaN3, H2O, reflux, 12 h (80%); (e) SOCl2, EtOH, 0 °C to rt (78%); (f) K2CO3, DMF (92%); (g) CuSO4 (15 mol %), sodium ascorbate (45 mol %), t-BuOH:H2O:CH2Cl2 (51%); (h) LiOH (aqueous 0.5 M), CH3OH reflux, then HCl (1 M aq), pH 1 (21%). (B) (a) (1) TSTU, N,N-diisopropylethylamine, DMF, (2) LiOH, 0.5 M, MeOH:H2O; (b) TSTU, N,N-diisopropylethylamine, DMF; (c) DMF, water, 0 °C.
Alkyne derivative 11 was tested in a functional assay of antagonism of the agonist-induced inhibition of cAMP production in the presence of 30 μM forskolin in Chinese hamster ovary (CHO-K1) cells stably expressing the hP2Y14R (P2Y14R-CHO cells, using an EC80 concentration of agonist 2 of 316 nM).14 Under these conditions, the IC50 values for 11 was 5690 ± 1440 (n = 3). Thus, this alkyne derivative was shown to be an antagonist with considerably less affinity than reference antagonist 3.
Although the alkyne 11 was not deemed a high priority lead compound because of its moderate potency, the triazoles achieved higher affinity. Compound 65 and the other newly synthesized triazole derivatives were assayed in a flow cytometry competition assay using whole cells (P2Y14R-CHO), and fluorescent antagonist ligand 4 (20 nM) as a tracer. Figure 3A shows typical flow cytometry traces in the presence of representative triazole inhibitors at a single concentration. The antagonist affinities of the various analogues were compared by this method, first by screening at a relatively high single concentration of inhibitor (3 μM) to identify the most potent analogues. The reference compound 65 inhibited fluorescent labeling by 92%; thus, it appeared to be a suitable highly potent lead compound for exploring the SAR in this series. Those IC50 values of triazole analogues inhibiting the fluorescent labeling by 80% or greater were determined in full concentration–response curves, which were sigmoidal, as shown for representative compounds in Figure 3B.
Figure 3.
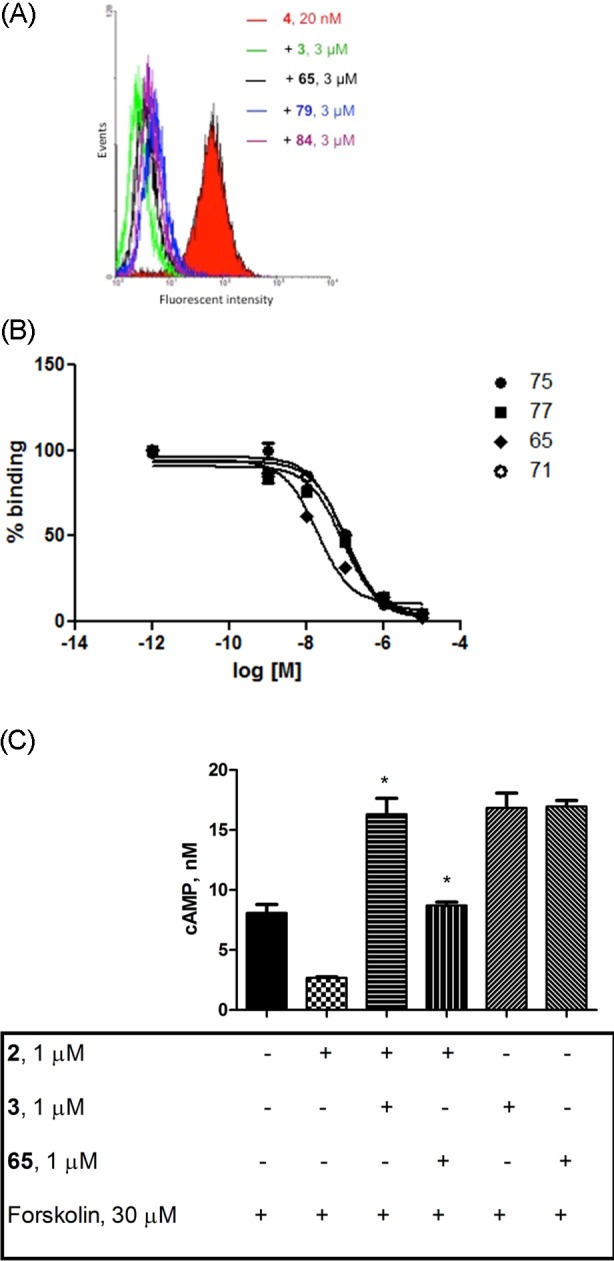
Biological characterization of triazole derivatives. Flow cytometric analysis (A) of the binding of selected triazolyl derivatives in comparison to reference naphthoic acid derivative 3 at the P2Y14R expressed in CHO cells, as detected through inhibition of fluorescent cell labeling with 4. Concentration–response curves for selected compounds (B) displayed a smooth concentration dependence of the inhibition. The IC50 values are given in Table 3. (C) Effects of P2Y14R antagonist 65 on cAMP levels in P2Y14R-expressing CHO cells.
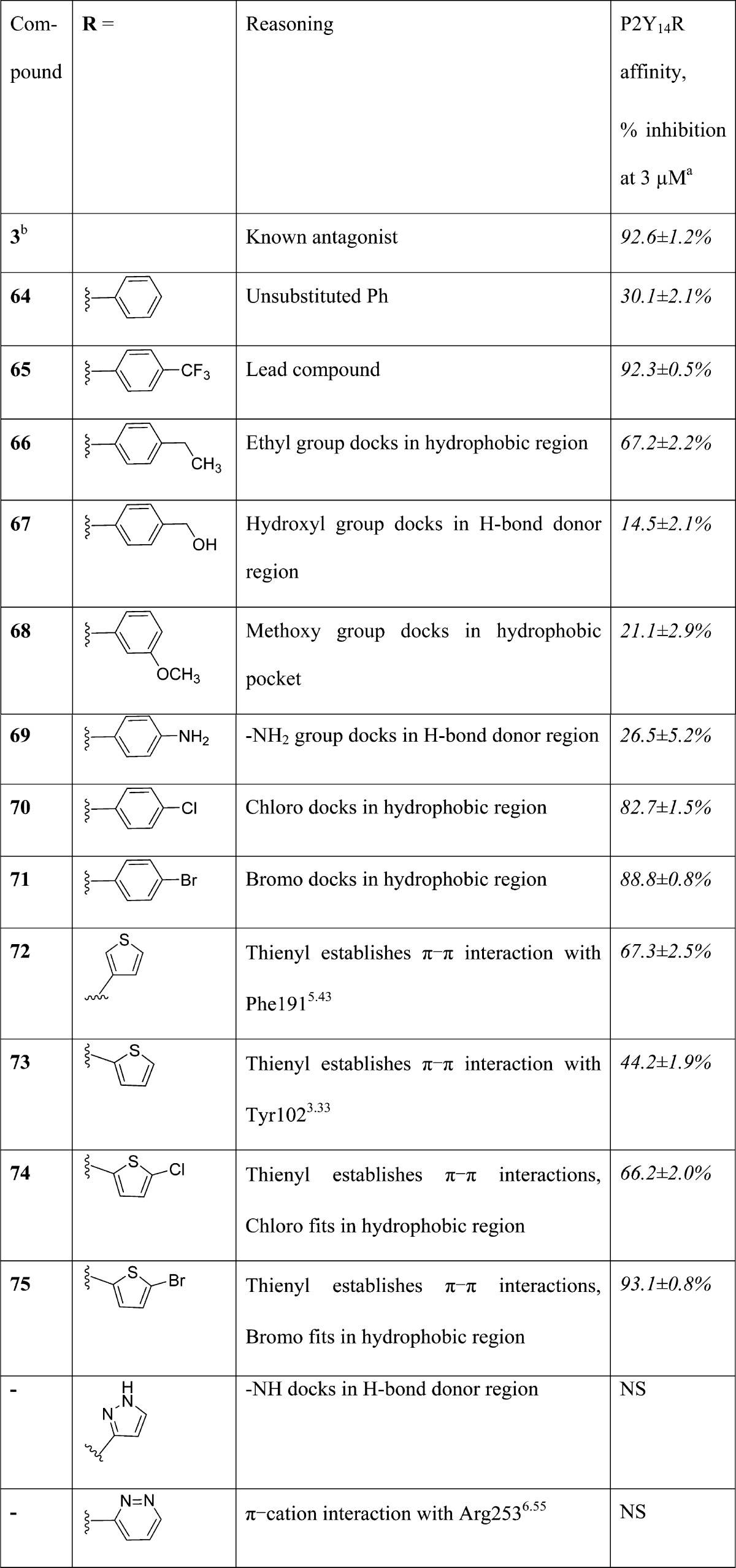
Selection of favored terminal aryl groups in the triazole series, other than 4-F3C-Ph (65), was based on predictions arising from the docking of a library of 57 hypothetical triazole derivatives (Supporting Information, Table S1), which could be easily synthesized by the same route as for 65. The criteria underlying the hit selection are reported in Table 1 and are based upon ligand–receptor complementarity and overlap of scaffold functional groups with computed receptor interaction sites (Supporting Information, Table S2). Initially, selected entries 66–75 were synthesized for testing using the fluorescent hP2Y14R assay. On the basis of the SAR determined for the initial set, a second group of P2Y14R antagonists 76–104 was prepared (Table 2) and evaluated similarly for the ability to inhibit fluorescent binding at the hP2Y14R.
Table 1. Selection of Terminal Aryl Group Based on Docking and Molecular Dynamics Simulation of Various Triazole Derivativesc.
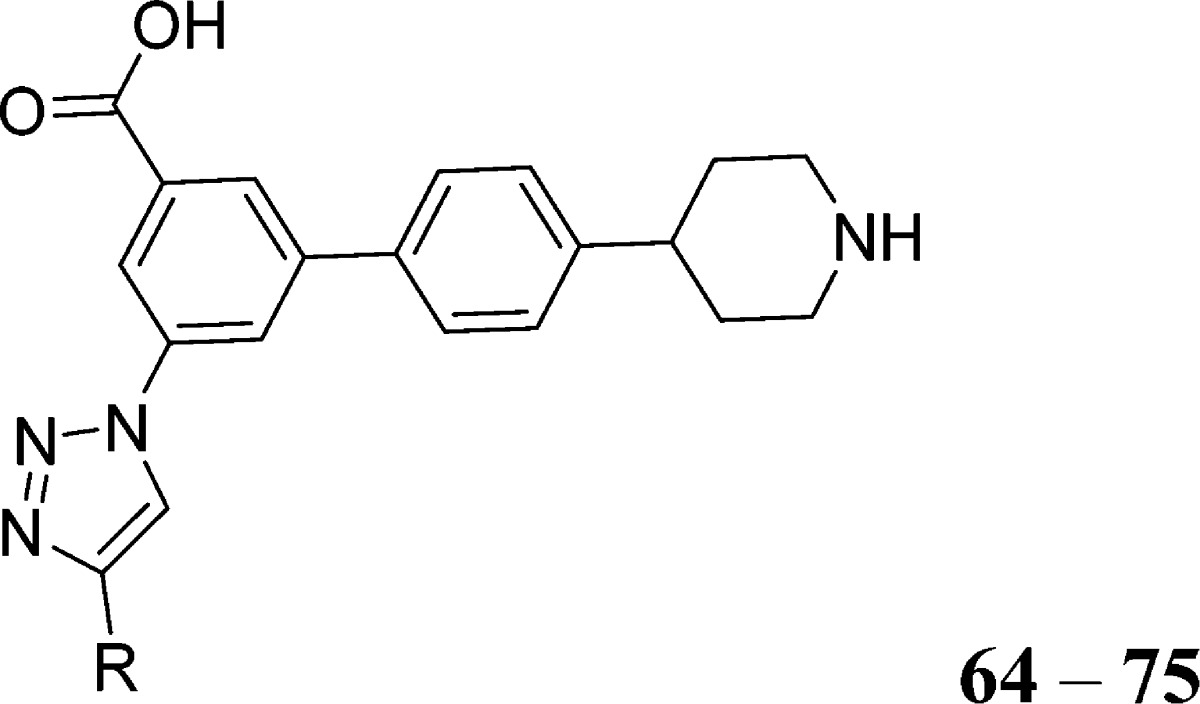
Table 2. Second Group of Triazolyl P2Y14R Antagonists That Was Prepared Based on Expanding the SAR Found in Table 1.
Percent inhibition at 3 μM of binding of fluorescent antagonist 4 (20 nM) in P2Y14R-CHO cells
The compounds that at 3 μM displayed inhibition of >80% of the fluorescent ligand (4, 20 nM) binding to the P2Y14R tended to be phenyl derivatives with 4-CF3 (65), 4-Cl (70), 4-Br (71), 4-n-Pr (77), 4-i-Pr (78), 4-t-Bu (79), 4-n-pentyl-O (82), 4-OCF3 (84), and 3,4-F2 (90) substituents. Also, a 5-bromothien-2-yl derivative (75) nearly completely inhibited P2Y14R fluorescent labeling. The compounds that displayed inhibition of 60%–80% contained a thien-3-yl (72) or 5-chlorothien-2-yl group (74) or were phenyl derivatives with 4-C2H5 (66), 4-CH3 (76), 4-t-Bu (79), 3-CF3 (80), 3-OCF3 (85), 4-F (87), 3,5-diF (91), 4-COCH3 (94), and benzofuran-2-yl (101) substituents. Among the weakest compounds, with <20% inhibition were 4-CH2OH (67), 2-OCH3 (83), 3-OH (96), and 3-NH2 (97) phenyl analogues and pyridyl (98), pyrazinyl (99), and thiazolyl (102) analogues. Both unsubstituted phenyl analogue 64 and the cyclohexyl analogue 103 inhibited P2Y14R binding in the intermediate range. Thus, recognizable patterns of SAR were: polar groups, H-bond donor groups, and heteroatom (especially N) substitution of an aryl ring were not well tolerated, while nonpolar phenyl substituents and especially para-substitution of the phenyl ring were favored.
The IC50 values of the compounds that were examined in the fluorescent assay with full concentration–response curves are given in Table 3. The IC50 values of the reference naphthoic acid 3 was 6.0 nM, and the values for 10 triazoles ranged from 31.7 nM (65) to 481 nM (78). The potencies were in the rank order of: 3 > 65 > 77 > 75 > 71, 82, 84 > 79, 90, 76, 70 > 78.
Table 3. Potencies of P2Y14R Antagonists in Flow Cytometry Assay Determined with Full Concentration–Response Curves.
| compd | P2Y14R potency, IC50 (nM)a |
|---|---|
| 3 | 6.0 ± 0.1 |
| 65 | 31.7 ± 8.0 |
| 70 | 290 ± 88 |
| 71 | 115 ± 15 |
| 75 | 91 ± 4 |
| 76 | 237 ± 90 |
| 77 | 72.4 ± 14.0 |
| 78 | 481 ± 81 |
| 79 | 224 ± 64 |
| 82 | 162 ± 25 |
| 84 | 131 ± 39 |
| 90 | 228 ± 63 |
Percent inhibition of binding of fluorescent antagonist 4 (20 nM) in P2Y14R-CHO cells (n = 3), over a concentration range of 10–9 to 10–5 M.
The most potently binding triazole derivative 65 was shown in measurements of cAMP to be an antagonist at the P2Y14R expressed in CHO cells, similar to known and potent antagonist 3 (Figure 3C). P2Y14R agonist 2 (1 μM) was applied in the absence or presence of antagonist 3 (1 μM) or compound 65 (1 μM) to cells stimulated with 30 μM forskolin. As expected, compound 2 significantly decreased forskolin-induced cAMP levels, but in the presence of either compound 3 or 65, P2Y14R agonist 2 did not inhibit cAMP accumulation. We also studied the effect of compound 65 on calcium mobilization induced by several other P2YRs. 65 (up to 10 μM) was inactive as either agonist or antagonist at the hP2Y1R and hP2Y6R expressed in 1321N1 astocytoma cells (Supporting Information, Figure S4). Compounds 11, 65, and 74 were separately evaluated by the Psychoactive Drug Screening Program (PDSP)28 at various P2YRs and found to be inactive (10 μM) as agonist or antagonist at human P2Y1, P2Y2, P2Y4, and P2Y11Rs (calcium transients) expressed in 1321N1 astrocytoma cells and protease-activated receptor (PAR)1 expressed in mouse KOLF cells.
Off-target activities for selected compounds were measured by the PDSP.28 Compounds 3 and 11 each showed only a few off-target interactions at <10 μM. At these sites, the measured Ki values (μM) of 3 were 6.79 (D3 dopamine receptor) and 2.75 (δ-opioid receptor), and the Ki values (μM) of 11 were 1.46 (σ1 receptor) and 3.60 (σ2 receptor). All other receptors, channels, and transporters in the standard diverse screen of the PDSP were not significant (i.e., <50% binding inhibition at 10 μM). A representative triazole derivative (74) showed no off-target interactions, but the trifluoromethyl analogue (65) bound weakly (Ki, μm) at H1 histamine (0.17) and α2A (1.56) and α2C (1.32) adrenergic receptors. Thus, only a few off-target interactions were detected in these chemical series.
Discussion
The aim of this project was the synthesis of a library of novel triazole-based structures as possible antagonists of the P2Y14R having improved physicochemical properties. A triazole moiety was proposed as an alternative bioisosteric replacement for the naphthoic acid core of the potent P2Y14R antagonist 3. On the basis of the results previously achieved with the same substitutions for A3AR agonists,19 alkyne derivatives and triazole derivatives were considered for components of the core to mimic the favorable interactions present in the naphthoic acid series. By docking to a homology model of the receptor, the envisaged structures were predicted to occupy the same binding site within the P2Y14R as 3, maintaining a similar orientation of the piperidine and 4-(trifluoromethyl)-phenyl substituents within the ligand binding pocket and preserving the affinity. Docking and MD simulation have suggested that the triazole scaffold can form additional interactions that stabilize the ligand within the receptor binding pocket. A p-CF3-phenyl group bearing triazole (65) proved to be of higher affinity than the corresponding alkyne (11); thus, the triazoles were explored in detail.
The synthetic route to the triazole series was versatile to allow the introduction of a wide range of functional groups on a terminal aryl substituent late in the synthesis. Both Suzuki and click cycloaddition reactions were applied sequentially to a benzoic ester moiety. The triazole derivatives were prepared by a late-stage diversification strategy, introducing the 1,2,3-triazole moiety at the end of the synthetic sequence by a copper-catalyzed [2 + 3] cycloaddition between an azide moiety and various arylacetylene derivatives. The choice of arylacetylene derivatives was initially guided by docking and MD studies. The overall yield from aniline 12 to protected azide 22 was 15%, with the last two steps (click reaction and deprotection) of variable, but usually high yield. One unexpected result was the hydrolysis of the 3-cyanophenyl group during the last deprotection step to a 3-carboxyphenyl group (92).
These new compounds were assayed in a convenient flow cytometric fluorescence competition assay with our previously reported antagonist probe 4 in P2Y14R-CHO cells, confirming the general docking predictions. Moreover, the expanded SAR exploration (Table 2) provided greater insight. The terminal aryl group attached to the triazole required hydrophobic substitution for high affinity, as the unsubstituted phenyl analogue (64) was weak in P2Y14R binding. The general preference for substitution of a phenyl ring at this position was p- > m- ≥ o-, as evidenced with the methoxy analogues (81 > 68 ≥ 83) and the fluoro analogues (87 > 88 ≈ 89). Similarly, there was a preference for p- over m- substitution in trifluoromethyl (65 > 80) and trifluoromethoxy (84 > 85) analogues. Thus, there is a hydrophobic pocket that tolerates considerable steric bulk, e.g., the 4-tert-butyl analogue (79), in this region of the receptor, as predicted in ligand docking to the P2Y14R homology model. However, introducing polar groups to form predicted H-bonding interactions with specific groups surrounding this aryl ring failed to enhance affinity. This trend could be explained by the observation that a residue side chain (Ser1875.39) that was predicted in the docking simulation to form H-bonds with the ligand polar groups was not available for ligand interaction when analyzed in the dynamic context of the membrane-embedded solvated receptor (data not shown). The most potent compounds, with IC50 values ranging from 32 to 131 nM, had substituents: 4-CF3-ϕ 65 > 4-n-propyl-ϕ 77 > 5-Br-thienyl 75 > 4-Br-ϕ 71, 4-n-pentyloxy-ϕ 82, and 4-CF3O-ϕ 84. Substitution of the 4-CF3 group of 65 with 4-CH3 in 76 considerably lowered the affinity 7-fold (IC50 237 nM). These affinities were not as potent as reference compound 4 (IC50 6.0 nM) in the same assay. The lead molecule 3 is a highly selective antagonist of the P2Y14R;14 although selectivity of these antagonists with respect to P2Y12R and P2Y13R remains to be determined, several derivatives were shown to be inactive at all P2Y1-like receptors.
The physicochemical properties of the triazole derivatives remain to be determined experimentally. However, an online tool for calculating small-molecule pharmacokinetic and toxicity properties predicted some advantage; 3 (2.7 μM solubility predicted) would be 100% bound to human plasma protein and 65 (6.9 μM solubility predicted) would be 1.5% unbound.29 The triazole ring benefited from increased polarity and additional H-bond accepting groups compared to the naphthalene core of 3. The cLogP of 3 is 5.65, which is more hydrophobic than the optimal range of ∼2–4, while the corresponding triazole derivative 65 had a cLogP of 4.59. The halogen substitution of a potent 3,4-difluorophenyl analogue 90 might impede potential oxidation by CYP450 enzymes in the liver.
Conclusion
In conclusion, we have used structural insights to discover a new scaffold 3-(4-aryl-1H-1,2,3-triazol-1-yl)-biphenyl) for P2Y14R antagonists. The high affinity among members of this chemical series of triazole derivatives provides new tools to aid in our understanding of P2Y14R pharmacology and potentially could lead to clinically useful drug candidates for inflammatory, endocrine, and other conditions.
Experimental Section
Chemical Synthesis
Reagents and Instrumentation
The proton and carbon nuclear magnetic resonance spectra were recorded using Bruker 400 MHz, Bruker 500, or Bruker 600 NMR spectrometer. Purification of final compounds was performed by preparative HPLC (column: Luna 5 μm C18(2) 100 Å, LC column 250 mm × 4.6 mm). Method A: eluent 0.1% TFA in water–CH3CN from 100:0 to 70:30 in 45 min with a flow rate of 5 mL/min. Method B: eluent 10 mM triethyammonium acetate buffer–CH3CN from 80:20 to 20:80 in 40 min, then 10 mM triethyammonium acetate buffer–CH3CN from 20:80 to 0:100 in 10 min with a flow rate of 5 mL/min. Purities of all tested compounds were ≥95%, as estimated by analytical HPLC (column: Zorbax SB-Aq 5 μm analytical column, 150 mm × 4.6 mm; Agilent Technologies, Inc.). Method: eluent 5 mM triethyammonium phosphate monobasic solution–CH3CN from 90:10 to 0:100 in 20 min, then triethyammonium phosphate monobasic solution–CH3CN from 0:100 to 90:10 in 5 min with a flow rate of 1 mL/min. Peaks were detected by UV absorption (254 nm) using a diode array detector. All derivatives tested for biological activity showed >95% purity in the HPLC system. Analytical thin-layer chromatography was carried out on Sigma-Aldrich TLC plates, and compounds were visualized with UV light at 254 nm. Silica gel flash chromatography was performed using 230–400 mesh silica gel. Unless noted otherwise, reagents and solvents were purchased from Sigma-Aldrich (St. Louis, MO). Compound 3 was synthesized as reported.14 Low-resolution mass spectrometry was performed with a JEOL SX102 spectrometer with 6 kV Xe atoms following desorption from a glycerol matrix or on an Agilent LC/MS 1100 MSD, with a Waters (Milford, MA) Atlantis C18 column. High-resolution mass spectroscopic (HRMS) measurements were performed on a proteomics optimized Q-TOF-2 (Micromass-Waters) using external calibration with polyalanine. cLogP was calculated using ChemDraw Professional (PerkinElmer, Boston, MA, v. 15.0).
6-Amino-9-(2-carboxy-4-((6-(4-(4-(4-(4-(3-carboxy-6-(4-(trifluoromethyl)phenyl)-naphthalen-1-yl)phenyl)piperidin-1-yl)butyl)-1H-1,2,3-triazol-1-yl)hexyl)carbamoyl)-phenyl)-3-iminio-5-sulfo-3H-xanthene-4-sulfonate (4)
To a solution of AlexaFluor 488 115 (4.44 mg, 7.08 μmol) and N,N-diisopropylethylamine (1.34 μL, 7.72 μmol) in dry DMF (400 μL), TSTU (2.42 mg, 7.72 μmol) was added at 0 °C. The resulting mixture was allowed to warm up at rt and stirred for 2–3 h. Then, a solution of 114 (4.5 mg, 6.44 μmol) and N,N-diisopropylethylamine (1.30 μL, 7.08 μmol) in dry DMF (300 μL) was added, and the reaction was stirred overnight at rt. After removal of the solvent, the residue was purified by preparative HPLC (method A, Rt = 24.9 min). The product 4 was obtained as an orange solid after lyophilization (0.8 mg, 10%). MS (ESI, m/z) 1212 [M – H]−. ESI-HRMS calcd m/z for C62H57F3N2O12S2 1212.3462, found 1212.3459 [M – H]−. HPLC purity 96.1% (Rt = 5.7 min).
4′-(Piperidin-4-yl)-5-((4-(trifluoromethyl)phenyl)ethynyl)-[1,1′-biphenyl]-3-carboxylic Acid Hydrochloride (11)
Lithium hydroxide (aqueous 0.5M, 70 μL, 25 μmol) was added to a solution of 10 (13 mg, 23 μmol) in methanol (0.2 mL), and the mixture was heated at reflux for 1.5 h. During this time, 10 was completely consumed. The mixture was allowed to cool to 23 °C and acidified with hydrochloric acid (1 M) until pH 1. The acidified mixture was stirred for additional 2 h before solvents were removed under reduced pressure. The residue was subjected to column chromatography (silica gel), eluting with chloroform/methanol/acetic acid 100/10/1 (v/v) mixture. Hydrochloric acid (1 M) was added to fractions containing the product, and the solvent was removed under reduced pressure to provide the desired product 11 as a hydrochloride salt (3.1 mg, 28%). MS (ESI, m/z) 450 [M + H]+. 1H NMR (400 MHz, DMSO-d6): δ (ppm) = 8.24 (s, 1H), 8.11 (t, J = 1.63 Hz, 1H), 8.08 (s, 1H), 7.85 (s, 1H), 7.79 (td, J = 1.51, 7.78 Hz, 1H), 7.73 (td, J = 1.38, 7.53 Hz, 1H), 7.69 (d, J = 8.03 Hz, 2H), 7.41 (t, J = 7.65 Hz, 1H), 7.35 (d, J = 8.28 Hz, 2H), 3.26 (br. s., 2H), 3.10–3.19 (m, 2H), 2.87 (d, J = 12.55 Hz, 3H), 2.42 (dt, J = 2.64, 7.22 Hz, 2H), 2.33 (td, J = 1.79, 3.70 Hz, 1H).
General Procedure A: Click Cycloaddition Reaction
To a solution of aryl azide (22, 1 equiv) and aryl alkyne (1.5 equiv) in 2 mL of THF:water (1:1), sodium ascorbate (freshly prepared 1 M aqueous solution) and CuSO4 (0.5 equiv) were sequentially added. The resulting reaction was vigorously stirred for 12 h at rt. The reaction mixture was then concentrated in vacuo and purified by flash chromatography (hexane:ethyl acetate = 6:4).
Methyl 5-(4-Phenyl-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (23)
Yellow solid. MS (ESI, m/z) 535 [M + H]+. ESI-HRMS calcd for C29H26F3N4O3 535.1952, found 535.1957 [M + H]+.
Methyl 5-(4-(4-(Trifluoromethyl)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (24)
Orange solid 6.9 mg (99%). MS (ESI, m/z) 603 [M + H]+. ESI-HRMS calcd for C30H25F6N4O3 603.1825, found 603.1831 [M + H]+.
Methyl 5-(4-(4-(Ethylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (25)
Yellow solid 2.8 mg (43%). MS (ESI, m/z) 563.2 [M + H]+. ESI-HRMS calcd for C31H30F3N4O3 563.2270, found 563.2274 [M + H]+.
Methyl 5-(4-(4-(Hydroxymethyl)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (26)
Yellow solid 1.6 mg (25%). MS (ESI, m/z) 565.2 [M + H]+. ESI-HRMS calcd for C30H28F3N4O4 565.2063, found 565.2068 [M + H]+.
Methyl 5-(4-(3-Methoxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (27)
Yellow solid 4 mg (80%). MS (ESI, m/z) 565.1 [M + H]+. ESI-HRMS calcd for C30H28F3N4O4 565.2063, found 565.2056 [M + H]+.
Methyl 5-(4-(4-Aminophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (28)
Yellow solid 1.7 mg (27%). MS (ESI, m/z) 550.2 [M + H]+. ESI-HRMS calcd for C29H27F3N5O3 550.2066, found 550.2075 [M + H]+.
Methyl 5-(4-(4-Chlorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (29)
Yellow solid 3.6 mg (55%). MS (ESI, m/z) 569.2 [M + H]+. ESI-HRMS calcd for C29H2535ClF3N4O3 569.1567, found 569.1561 [M + H]+.
Methyl 5-(4-(4-Bromophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl) piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (30)
Yellow solid 3.8 mg (54%). MS (ESI, m/z) 613.1 [M + H]+. ESI-HRMS calcd for C29H2579BrF3N4O3 613.1062, found 613.1057 [M + H]+.
Methyl 5-(4-(Thiophen-3-yl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (31)
Yellow solid 4.2 mg (67%). MS (ESI, m/z) 541.1 [M + H]+.
Methyl 5-(4-(Thiophen-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (32)
Yellow solid 1.6 mg (26%). MS (ESI, m/z) 541.2 [M + H]+. ESI-HRMS calcd for C27H24F3N4O332S 541.1521, found 541.1523 [M + H]+.
Methyl 5-(4-(5-Chlorothiophen-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)-piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (33)
Brown solid. MS (ESI, m/z) 575 [M + H]+. ESI-HRMS calcd for C27H23F3N4O3SCl 575.1131, found 575.1132 [M + H]+.
Methyl 5-(4-(5-Bromothiophen-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)-piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (34)
Orange solid 5.1 mg (72%). MS (ESI, m/z) 619 [M + H]+. ESI-HRMS calcd for calcd for C27H23F3N4O3S79Br 619.0626, found 619.0618 [M + H]+.
Methyl 5-(4-(4-Methylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (35)
Yellow solid 4 mg (80%). MS (ESI, m/z) 549.2 [M + H]+. ESI-HRMS calcd for C30H28F3N4O4 549.2114, found 549.2119 [M + H]+.
Methyl 5-(4-(4-Propylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (36)
Yellow solid 5 mg (76%). MS (ESI, m/z) 577.2 [M + H]+. ESI-HRMS calcd for C30H28F3N4O4 577.2427, found 577.2421 [M + H]+.
Methyl 5-(4-(4-Isopropylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (37)
Colorless oil 4 mg (60%). MS (ESI, m/z) 577.2 [M + H]+. ESI-HRMS calcd for C30H28F3N4O4 577.2427, found 577.2417 [M + H]+.
Methyl 5-(4-(4-(tert-Butyl)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (38)
Yellow solid 5.5 mg (81%). MS (ESI, m/z) 591.2 [M + H]+. ESI-HRMS calcd for C33H34F3N4O3 591.2583, found 591.2589 [M + H]+.
Methyl 5-(4-(3-(Trifluoromethyl)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (39)
Yellow solid 6.15 mg (89%). MS (ESI, m/z) 603.1 [M + H]+. ESI-HRMS calcd for C30H25F6N4O3 603.1831, found 603.1825 [M + H]+.
Methyl 5-(4-(4-Methoxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (40)
Yellow solid 3.2 mg (49%). MS (ESI, m/z) 565.2 [M + H]+. ESI-HRMS calcd for C30H28F3N4O4 565.2063, found 565.2062 [M + H]+.
Methyl 5-(4-(4-(Pentyloxy)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (41)
Pale-yellow oil 5.7 mg (81%). MS (ESI, m/z) 621.2 [M + H]+. ESI-HRMS calcd for C34H36F3N4O4 621.2689, found 621.2687 [M + H]+.
Methyl 5-(4-(2-Methoxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (42)
Yellow solid 6.5 mg (99%). MS (ESI, m/z) 565.1 [M + H]+. ESI-HRMS calcd for C30H28F3N4O4 565.2063, found 565.2061 [M + H]+.
Methyl 5-(4-(4-(Trifluoromethoxy)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (43)
Yellow solid 3.4 mg (48%). MS (ESI, m/z) 619.1 [M + H]+. ESI-HRMS calcd for C30H25F6N4O4 619.1780, found 619.1784 [M + H]+.
Methyl 5-(4-(3-(Trifluoromethoxy)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (44)
Yellow solid 6 mg (84%). MS (ESI, m/z) 619.2 [M + H]+. ESI-HRMS calcd for C30H25F6N4O4 619.1780, found 619.1778 [M + H]+.
Methyl 5-(4-(3-Chlorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl) piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (45)
Yellow solid 3.5 mg (54%). MS (ESI, m/z) 569.2 [M + H]+. ESI-HRMS calcd for C29H2535ClF3N4O3 569.1567, found 569.1570 [M + H]+.
Methyl 5-(4-(4-Fluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (46)
Yellow solid 3.9 mg (61%). MS (ESI, m/z) 553.1 [M + H]+. ESI-HRMS calcd for C29H25F4N4O3 553.1863, found 553.1855 [M + H]+.
Methyl 5-(4-(3-Fluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (47)
Yellow solid 4.5 mg (77%). MS (ESI, m/z) 553.2 [M + H]+. ESI-HRMS calcd for C29H25F4N4O3 553.1863, found 553.1872 [M + H]+.
Methyl 5-(4-(2-Fluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (48)
Yellow solid 4.9 mg (71%). MS (ESI, m/z) 553.2 [M + H]+. ESI-HRMS calcd for C29H25F4N4O3 553.1863, found 553.1866 [M + H]+.
Methyl 5-(4-(3,4-Difluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (49)
Pale-yellow solid 4.23 mg (65%). MS (ESI, m/z) 571.1 [M + H]+. ESI-HRMS calcd for C29H24F5N4O3 571.1769, found 571.1758 [M + H]+.
Methyl 5-(4-(3,5-Difluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (50)
Yellow solid 4.2 mg (63%). MS (ESI, m/z) 571.1 [M + H]+. ESI-HRMS calcd for C29H24F5N4O3 571.1769, found 571.1762 [M + H]+.
Methyl 5-(4-(4-Cyanophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (51)
Yellow solid 6 mg (94%). MS (ESI, m/z) 560.2 [M + H]+. ESI-HRMS calcd for C30H25F4N5O3 560.1909, found 560.1913 [M + H]+.
Methyl 5-(4-(4-Acetylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (53)
Yellow solid 4.45 mg (67%). MS (ESI, m/z) 577.2 [M + H]+. ESI-HRMS calcd for C31H28F3N4O4 577.2063, found 577.2056 [M + H]+.
Methyl 5-(4-(3,4-Dimethoxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (54)
Yellow solid 5.65 mg (83%). MS (ESI, m/z) 595.2 [M + H]+. ESI-HRMS calcd for C31H30F3N4O5 595.2168, found 595.2173 [M + H]+.
Methyl 5-(4-(3-Hydroxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (55)
Yellow solid 5.15 mg (81%). MS (ESI, m/z) 551.2 [M + H]+. ESI-HRMS calcd for C29H26F3N4O4 551.1906, found 551.1901 [M + H]+.
Methyl 5-(4-(3-Aminophenyl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (56)
Yellow solid 2.4 mg (38%). MS (ESI, m/z) 550.2 [M + H]+. ESI-HRMS calcd for C29H27F3N5O3 550.2066, found 550.2056 [M + H]+.
Methyl 5-(4-(Pyridin-4-yl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (57)
Yellow solid 2.3 mg (37%). MS (ESI, m/z) 536.2 [M + H]+. ESI-HRMS calcd for C28H25F3N5O3 536.1909, found 536.1913 [M + H]+.
Methyl 5-(4-(Pyrazin-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (58)
Yellow solid 3.5 mg (57%).
Methyl 5-(4-(Furan-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (59)
Yellow solid 3 mg (50%). MS (ESI, m/z) 525.1 [M + H]+. ESI-HRMS calcd for C27H24F3N4O4 525.1750, found 525.1744 [M + H]+.
Methyl 5-(4-(Benzofuran-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (60)
Brown solid 4 mg (60%). MS (ESI, m/z) 575.2 [M + H]+. ESI-HRMS calcd for C31H26F3N4O4 575.1906, found 575.1909 [M + H]+.
Methyl 5-(4-(Thiazol-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (61)
Yellow solid 4.6 mg (74%). MS (ESI, m/z) 542.1 [M + H]+. ESI-HRMS calcd for C26H23F3N5O332S 542.1474, found 542.1465 [M + H]+.
Methyl 5-(4-Cyclohexyl-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (62)
Yellow solid 5.4 mg (87%). MS (ESI, m/z) 541.2 [M + H]+. ESI-HRMS calcd for C29H32F3N4O3 541.2427, found 541.2419 [M + H]+.
Methyl 5-(4-Cyclopropyl-1H-1,2,3-triazol-1-yl)-4′-(1-(2,2,2-trifluoroacetyl)piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylate (63)
Yellow solid 5.7 mg (99%). MS (ESI, m/z) 499.2 [M + H]+. ESI-HRMS calcd for C26H26F3N4O3 499.1957, found 499.1959 [M + H]+.
General Procedure B: Deprotection of Piperidine N and Ester Hydrolysis
To a solution of protected ester (1 equiv) in 2 mL of MeOH:H2O (1:1), KOH (10 equiv) was added, and the resulting mixture was heated at 50 °C for 12 h. After removing the solvents under reduced pressure, the mixture was purified by semipreparative HPLC (method B).
5-(4-Phenyl-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (64)
Semipreparative HPLC: method B, Rt = 25.7 min. The product was obtained as a white solid by lyophilization (1.9 mg, 26%). MS (ESI, m/z) 425 [M + H]+. ESI-HRMS calcd for C26H25N4O2 425.1978, found 425.1980 [M + H]+. HPLC purity 97% (Rt = 8.9 min). 1H NMR (400 MHz, MeOD): δ (ppm) = 8.94 (s, 1H), 8.29 (d, J = 8 Hz, 2H), 7.88 (d, J = 8 Hz, 2H), 7.68 (d, J = 8 Hz, 2H), 7.39–7.43 (m, 2H), 7.31–7.35 (m, 3H), 2.90–2.96 (m, 5H), 1.94–1.97 (m, 2H), 1.77–1.80 (m, 2H).
5-(4-(4-(Trifluoromethyl)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (65)
Semipreparative HPLC: method B, Rt = 30.2 min. The product was obtained as a white salt by lyophilization (0.5 equiv CH3CO2H-salt, 6.01 mg, 99%). MS (ESI, m/z) 493.1 [M + H]+. ESI-HRMS calcd for C27H24F3N4O2 493.1851, found 493.1853 [M + H]+. HPLC purity 99% (Rt = 12.6 min), mp 280–288 °C d. 1H NMR (600 MHz, DMSO-d6): δ (ppm) = 9.66 (s, 1H), 8.39 (t, J = 1.6 Hz, 1H), 8.28 (t, J = 1.6 Hz, 1H), 8.22 (d, J = 8.1 Hz, 2H), 8.15 (t, J = 1.6 Hz, 1H), 7.88 (d, J = 8.1 Hz, 2H), 7.75 (d, J = 8.3 Hz, 2H), 7.41 (d, J = 8.3 Hz, 2H), 3.20–3.15 (m, 2H), 2.76 (td, J = 3.2, 12.1 Hz, 3H), 1.88 (s, 1.5 Hacetate), 1.77 (qd, J = 4.2, 12.9 Hz, 4H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 167.1, 146.5, 145.9, 143.9, 140.7, 137.1, 136.5, 134.4, 128.4, 128.2, 127.4 (2C), 127.2, 126.9 (2C), 126.0 (2C), 125.9 (2C), 125.2, 121.1, 119.2, 117.9, 55.0, 46.1 (2C), 41.7, 40.0, 33.4 (2C).
5-(4-(4-Ethylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (66)
Semipreparative HPLC: method B, Rt = 27.0 min. The product was obtained as a white solid by lyophilization (0.5 equiv CH3CO2H-salt, 3.4 mg, 93%). MS (ESI, m/z) 453.2 [M + H]+, ESI-HRMS calcd for C28H29N4O2 453.2291, found 453.2294 [M + H]+. HPLC purity 98% (Rt = 11.5 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.42 (s, 1H), 8.37 (s, 1H), 8.25 (s, 1H), 8.13 (s, 1H), 7.90 (d, J = 8.0 Hz, 2H), 7.75 (d, J = 7.9 Hz, 2H), 7.41 (d, J = 7.9 Hz, 2H), 7.35 (d, J = 8.0 Hz, 2H), 3.20 (d, J = 12.0 Hz, 2H), 2.81–2.71 (m, 3H), 2.66 (q, J = 7.6 Hz, 2H), 1.90 (s, 1.5Hacetate), 1.85–1.70 (m, 4H), 1.23 (t, J = 7.6 Hz, 3H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 167.1, 147.3, 143.8, 142.4, 137.8, 137.0, 128.8, 128.4 (2C), 127.9, 127.4 (2C), 126.9 (2C), 125.4 (2C), 125.2, 119.4, 119.0, 117.6, 28.0, 15.5.
5-(4-(4-(Hydroxymethyl)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (67)
Semipreparative HPLC: method B, Rt = 23.3 min. The product was obtained as a white solid by lyophilization (2 equiv CH3CO2H-salt, 1.4 mg, 87%). MS (ESI, m/z) 455.2 [M + H]+. ESI-HRMS calcd for C27H27N4O3 455.2083, found 455.2081 [M + H]+. HPLC purity 96% (Rt = 8.42 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.45 (s, 1H), 8.39 (s, 1H), 8.24 (s, 1H), 8.12 (s, 1H), 7.94 (d, J = 7.7 Hz, 1H), 7.73 (d, J = 7.8 Hz, 1H), 7.44 (d, J = 7.8 Hz, 2H), 7.37 (d, J = 7.7 Hz, 2H), 4.55 (s, 2H), 3.05 (d, J = 12.0 Hz, 2H), 2.72–2.53 (m, 3H), 1.77–1.51 (m, 10H (6Hacetate)). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 176.0 (2Cacetate), 167.8, 147.6, 147.1, 143.9, 142.9, 141, 137.4, 136.9, 129.2, 127.7 (2C), 127.3 (2C), 127.2, 127.2 (2C), 125.4 (2C), 119.9, 119.4, 118.1, 63.0, 46.9 (2C), 42.5, 39.4, 34.4 (2C), 24.7 (2Cacetate).
5-(4-(3-Methoxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (68)
Semipreparative HPLC: method B, Rt = 23.2 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 2.4 mg, 67%). MS (ESI, m/z) 455.2 [M + H]+. ESI-HRMS calcd for C27H27N4O3 455.2083, found 455.2083 [M + H]+. HPLC purity 99% (Rt = 10.1 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.52 (s, 1H), 8.42 (s, 1H), 8.33 (s, 1H), 8.19 (s, 1H), 7.78 (d, J = 7.8 Hz, 2H), 7.63–7.52 (m, 1H), 7.42 (m, 3H), 6.95 (d, J = 8.1 Hz, 1H), 6.81 (s, 1H), 3.84 (s, 3H, CH3), 2.90 (m, 3H), 1.91 (m, 4H). Proton signals for 2-CHpiperidine, 6-CHpiperidine and 3-CHpiperidine, 5-CHpiperidine are hidden under the H2O and DMSO-d6 signals. 13C NMR (500 MHz, DMSO-d6): δ (ppm) = 172.8 (1Cacetate), 168.7, 160.3, 147.7, 145.9, 141.3, 137.8, 137.3, 132.2, 130.7, 127.9 (2C), 127.5, 127.4 (2C), 120.5, 119.5, 119.1, 118.1, 114.5, 111.0, 55.7 (2C), 44.6 (2C), 30.8, 29.5 9 (1Cacetate).
5-(4-(4-Aminophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (69)
Semipreparative HPLC: method B, Rt = 17.4 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 2.84 mg, 95%). MS (ESI, m/z) 440.2 [M + H]+. ESI-HRMS calcd for C26H26N5O2 440.2087, found 440.2094 [M + H]+. HPLC purity 96% (Rt = 8.38 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.21 (s, 1H), 8.38 (s, 1H), 8.24 (s, 1H), 8.11 (s, 1H), 7.77 (d, J = 7.9 Hz, 2H), 7.69 (d, J = 8.2 Hz, 2H), 7.43 (d, J = 7.9 Hz, 2H), 6.95 (d, J = 8.2 Hz, 1H), 5.36 (s, 2HNH2), 3.10 (d, J = 12.0 Hz, 2H), 2.75–2.59 (m, 4H), 1.79 (d, J = 12.0 Hz, 2H), 1.65–1.51 (m, 2H).
5-(4-(4-Chlorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (70)
Semipreparative HPLC: method B, Rt = 22.7 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 3.3 mg, 99%). MS (ESI, m/z) 459.2 [M + H]+. ESI-HRMS calcd for C26H2435ClN4O2 459.1588, found 459.1588 [M + H]+. HPLC purity 98.4% (Rt = 11.4 min), mp 223–227 °C. 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.49 (s, 1H), 8.34 (s, 1H), 8.24 (s, 1H), 8.10 (s, 1H), 8.01 (d, J = 8.6 Hz, 2H), 7.72 (d, J = 8.4 Hz, 2H), 7.58 (d, J = 8.6 Hz, 2H), 7.38 (d, J = 8.4 Hz, 2H), 3.08 (d, J = 12.2 Hz, 2H), 2.69–2.59 (m, 3H), 1.81–1.70 (m, (3Hacetate), 5H), 1.60 (qd, J = 3.8, 12.4 Hz, 2H). 13C NMR (500 MHz, DMSO-d6): δ (ppm) = 173.6 (1Cacetate), 167.6, 145.0, 146.6, 144.2, 141.1 (2C), 137.5, 136.9, 133.0, 129.7, 129.5 (2C), 127.8 (2C), 127.5 (2C), 127.3 (2C), 120.6, 119.6, 118.3, 49.0, 46.5 (2C), 42.2, 33.9 (2C), 23.6 (1Cacetate).
5-(4-(4-Bromophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (71)
Semipreparative HPLC: method B, Rt = 24.2 min. The product was obtained as a white acetate salt by lyophilization (1 equiv CH3CO2H-salt, 3.5 mg, 99%). MS (ESI, m/z) 503.1 [M + H]+. ESI-HRMS calcd for C26H24N4O279Br 503.1083, found 503.1080 [M + H]+. HPLC purity 98.4% (Rt = 11.4 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.53 (s, 1H), 8.35 (s, 1H), 8.22 (s, 1H), 8.10 (s, 1H), 7.95 (d, J = 8.4 Hz, 2H), 7.72 (d, J = 7.9 Hz, 4H), 7.38 (d, J = 7.9 Hz, 2H), 3.08 (d, J = 12.0 Hz, 2H), 2.72–2.60 (m, 3H), 1.84 (s, 3Hacetate), 1.76 (d, J = 12.0 Hz, 2H), 1.60 (qd, J = 3.8, 12.4 Hz, 2H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.3 (1Cacetate), 167.6, 147.3, 146.9, 144.9, 141.3, 137.9, 137.2, 132.7 (2C), 130.4, 128.1 (2C), 128.0 (2C), 127.7, 127.5 (2C), 121.9, 120.9, 119.8, 118.4, 47.0 (2C), 42.6, 34.4 (2C), 22.9 (1Cacetate).
5-(4-(Thiophen-3-yl)-1H-1,2,3-triazol-1-yl)- 4′-(piperidin-4-yl)- [1,1′-biphenyl]-3-carboxylic Acid (72)
Semipreparative HPLC: method B, Rt = 22.3 min. The product was obtained as a white acetate salt by lyophilization (1 equiv CH3CO2H-salt, 3.77 mg, 99%). MS (ESI, m/z) 431.1 [M + H]+. ESI-HRMS calcd for C24H23N4O232S 431.1542, found 431.1548 [M + H]+. HPLC purity 99% (Rt = 9.7 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.40 (s, 1H), 8.39 (s, 1H), 8.27 (s, 1H), 8.13 (s, 1H), 8.05–8.01 (m, 1H), 7.77 (d, J = 7.8 Hz, 2H), 7.69 (d, J = 5.0 Hz, 1H), 7.43 (d, J = 7.8 Hz, 2H), 3.11 (d, J = 12.0 Hz, 2H), 2.76–2.63 (m, 3H), 1.87–1.76 (m, (3Hacetate), 6H), 1.63 (qd, J = 3.8, 12.0 Hz, 2H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.3 (1Cacetate), 166.9, 146.6, 144.3. 143.8, 140.6, 137.2, 136.5, 131.8, 127.4 (2C), 126.9, 126.8 (2C), 125.8, 121.3, 119.5, 119.0, 117.5, 46.5 (2C), 42.1, 33.9 (2C), 23.8 (1Cacetate), 11.8.
5-(4-(Thiophen-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (73)
Semipreparative HPLC: method B, Rt = 21.8 min. The product was obtained as a white acetate salt by lyophilization (0.5 equiv CH3CO2H-salt, 2.2 mg, 99%). MS (ESI, m/z) 431.2 [M + H]+. ESI-HRMS calcd for C24H23N4O232S 431.1542, found 431.1538 [M + H]+. HPLC purity 97% (Rt = 8.9 min). 1H NMR (400 MHz, DMSO-d6): δ (ppm) = 9.42 (s, 1H), 8.40 (s, 1H), 8.33 (s, 1H), 8.24 (d, J = 2.0 Hz, 1H), 7.81 (d, J = 7.9 Hz, 2H), 7.61 (d, J = 5.0 Hz, 1H), 7.56 (d, J = 3.6 Hz, 1H), 7.45 (d, J = 7.9 Hz, 2H), 7.19 (dd, J = 3.6, 5.1 Hz, 1H), 2.89–3.01 (m, 2H), 2.76–2.68 (m, 3H), 2.05–1.85 (m, (1.5Hacetate), 5H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 167.5, 145.5, 143.3, 141.8, 137.4, 137.3, 132.8, 128.5, 127.8 (2C), 127.7 (2C), 127.4, 126.4, 125.1, 119.6, 44.1 (2C), 40.5, 39.3, 29.9 (2C), 29.5.
5-(4-(5-Chlorothiophen-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (74)
Semipreparative HPLC: method B, Rt = 28.6 min. The product was obtained as a white solid by lyophilization (0.5 equiv CH3CO2H-salt, 2.8 mg, 38%). MS (ESI, m/z) 465 [M + H]+. ESI-HRMS calcd for C24H22N4O2SCl 465.1152, found 465.1151 [M + H]+. HPLC purity 98% (Rt = 10.6 min). 1H NMR (400 MHz, DMSO-d6): δ (ppm) = 9.34 (s, 1H), 8.25 (d, J = 2.4 Hz, 1H), 8.19 (s, 1H), 8.02 (d, J = 2.4 Hz, 1H), 7.65 (d, J = 8.1 Hz, 2H), 7.36–7.29 (m, 3H), 7.13 (d, J = 3.8 Hz, 1H), 3.12 (d, J = 12.0 Hz, 2H), 2.76–2.63 (m, 3H), 1.83–1.61 (m, (1Hacetate), 5H).
5-(4-(5-Bromothiophen-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (75)
Semipreparative HPLC: method B, Rt = 27.2 min. The product was obtained as a white solid by lyophilization (0.5 equiv CH3CO2H-salt, 2.31 mg, 52%). MS (ESI, m/z) 509.1 [M + H]+. ESI-HRMS calcd for C24H22N4O2SBr 509.0647, found 509.0648 [M + H]+. HPLC purity 98% (Rt = 11.8 min), mp 271–275 °C. 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.43 (s, 1H), 8.35 (s, 1H), 8.29 (s, 1H), 8.12 (s, 1H), 7.74 (d, J = 7.9 Hz, 2H), 7.48–7.35 (m, 3H), 7.33–7.27 (m, 1H), 3.23 (d, J = 12.0 Hz, 2H), 2.87–2.71 (m, 3H), 1.96–1.66 (m, (1.5Hacetate), 5H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.0, 167.8, 146.5, 142.4, 141.2, 137.6, 136.9, 134.9, 131.9, 127.9 (2C), 127.6, 127.4 (2C), 125.6, 119.8, 119.6, 119.3, 118.6, 111.5, 72.7, 46.0 (2C), 33.3 (2C), 29.2.
5-(4-(4-Methylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (76)
Semipreparative HPLC: method B, Rt = 24.4 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 3.31 mg, 94%). MS (ESI, m/z) 439.2 [M + H]+. ESI-HRMS calcd for C27H27N4O2 439.2134, found 439.2133 [M + H]+. HPLC purity 99% (Rt = 10.8 min), mp 280–286 °C. 1H NMR (600 MHz, DMSO-d6): δ (ppm) = 9.40 (s, 1H), 8.36 (s, 1H), 8.22 (s, 1H), 8.10 (s, 1H), 7.88 (d, J = 8.0 Hz, 2H), 7.72 (d, J = 8.2 Hz, 2H), 7.38 (d, J = 8.2 Hz, 2H), 7.31 (d, J = 8.0 Hz, 2H), 3.10 (d, J = 12.4 Hz, 2H), 2.72–2.62 (m, 3H), 2.36 (s, 3H), 1.84 (s, 3Hacetate), 1.81–1.73 (m, 2H), 1.63 (qd, J = 3.7, 12.4 Hz, 2H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 173.2, 167.5, 147.7, 146.9, 144.5, 141.0, 137.9, 137.6, 137.0, 129.9 (2C), 128.2, 127.8 (2C), 127.3 (2C), 125.7, 119.8, 119.5, 118.1, 46.7 (2C), 42.3, 40.5, 34.1 (2C), 22.8, 21.4.
5-(4-(4-Propylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (77)
Semipreparative HPLC: method B, Rt = 23.7 min. The product was obtained as a white solid by lyophilization (0.5 equiv CH3CO2H-salt, 4.5 mg, 98%). MS (ESI, m/z) 467.2 [M + H]+. ESI-HRMS calcd for C29H31N4O2 467.2447, found 467.2454 [M + H]+. HPLC purity 99.5% (Rt = 12.8 min). 1H NMR (600 MHz, DMSO-d6): δ (ppm) = 9.42 (s, 1H), 8.38 (s, 1H), 8.28 (s, 1H), 8.14 (s, 1H), 7.89 (d, J = 8.0 Hz, 2H), 7.76 (d, J = 7.2 Hz, 2H), 7.43 (d, J = 7.2 Hz, 2H), 7.32 (d, J = 8.0 Hz, 2H), 3.27–3.17 (m, 2H), 2.84–2.75 (m, 3H), 2.61 (t, J = 7.6 Hz, 2H), 1.90 (s, 1Hacetate), 1.87–1.77 (m, 4H), 1.64 (h, J = 7.4 Hz, 2H), 0.92 (t, J = 7.4 Hz, 3H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 149.5, 147.2, 142.2, 128.9 (2C), 127.9 (2C), 127.3 (2C), 125.3 (2C), 119.3, 40.0 (2C), 37.0 (2C), 24.0, 13.6.
5-(4-(4-Isopropylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (78)
Semipreparative HPLC: method B, Rt = 18.1 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 3.55 mg, 97%). MS (ESI, m/z) 467.2 [M + H]+. ESI-HRMS calcd for C29H31N4O2 467.2447, found 467.2440 [M + H]+. HPLC purity 98% (Rt = 12.5 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.48 (s, 1H), 8.42 (s, 1H), 8.29 (s, 1H), 8.17 (s, 1H), 7.96 (d, J = 7.8 Hz, 2H), 7.79 (d, J = 7.8 Hz, 2H), 7.48–7.40 (m, 4H), 3.18 (d, J = 12.0 Hz, 2H), 3.00 (hept, J = 7.0 Hz, 1H), 2.81–2.70 (m, 3H), 1.90 (s, 2Hacetate), 1.84 (d, J = 12.0 Hz, 2H), 1.71 (qd, J = 4.0, 12.0 Hz, 2H), 1.30 (d, J = 7.0 Hz, 4H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 172.5 (1Cacetate), 167.1, 148.4, 147.3, 146.4, 143.9, 140.6, 137.2, 136.6, 128.0, 127.4 (2C), 126.9 (4C), 125.4 (2C), 119.4, 119.0, 117.7, 45.9 (2C), 41.6, 39.4, 39.0, 33.3 (2C), 23.8 (1Cacetate), 22.0.
5-(4-(4-(tert-Butyl)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (79)
Semipreparative HPLC: method B, Rt = 31.1 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 5.03 mg, 99%). MS (ESI, m/z) 481.3 [M + H]+. ESI-HRMS calcd for C30H33N4O2 481.2604, found 481.2597 [M + H]+. HPLC purity 98% (Rt = 12.5 min), mp 298–303 °C. 0.3 Equiv Et3N and 1 equiv CH3CO2H-salt: 1H NMR (600 MHz, DMSO-d6): δ (ppm) = 9.41 (s, 1H), 8.37 (s, 1H), 8.25 (s, 1H), 8.13 (s, 1H), 7.91 (d, J = 8.5 Hz, 2H), 7.75 (d, J = 7.9 Hz, 2H), 7.52 (d, J = 8.4 Hz, 2H), 7.41 (d, J = 8.0 Hz, 2H), 3.19 (d, J = 12.1 Hz, 2H), 2.80–2.74 (m, 2H), 2.43 (q, J = 7.1 Hz, (0.3 equiv Et3N), 3H), 1.85 (s, 3Hacetate), 1.85–1.79 (m, 2H), 1.79–1.67 (m, 2H), 1.33 (s, 9H), 0.93 (t, J = 7.1 Hz, (0.3 equiv Et3N), 3H). 1 Equiv CH3CO2H-salt: 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 151.1, 147.7, 141.0, 137.8, 137.1, 128.4, 128.1, 127.8 (2C), 127.3, 127.4 (2C), 126.2 (2C), 125.6 (2C), 120.0, 119.8, 119.5, 40.5 (2C), 34.9 (2C), 31.6 (3C).
5-(4-(3-(Trifluoromethyl)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (80)
Semipreparative HPLC: method B, Rt = 27.6 min. The product was obtained as a white solid by lyophilization (0.5 equiv CH3CO2H-salt, 2.53 mg, 48%). MS (ESI, m/z) 493.1 [M + H]+. ESI-HRMS calcd for C27H24N4O2F3 493.1851, found 493.1851 [M + H]+. HPLC purity 99% (Rt = 11.8 min), mp 277–284 °C. 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.70 (s, 1H), 8.41 (s, 1H), 8.32 (s, 2H), 8.28 (s, 1H), 8.16 (s, 1H), 7.79–7.72 (m, 4H), 7.41 (d, J = 7.9 Hz, 2H), 3.20 (d, J = 12.0 Hz, 2H), 2.88–2.68 (m, 3H), 1.99–1.63 (m, (1.5Hacetate), 5H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 172.5, 167.6, 146.1, 145.8, 143.3, 140.8, 137.2, 136.6, 131.5, 130.3, 130.0, 129.8, 129.0, 127.4 (2C), 127.0, 126.9 (2C), 125.9 (q, JC–F = 274 Hz, 1C), 124.7, 121.8, 120.8, 119.0, 118.0, 45.7, 45.2 (2C), 40.8, 32.1 (2C), 29.0, 21.8, 11.7.
5-(4-(4-Methoxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (81)
Semipreparative HPLC: method B, Rt = 23.0 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 2.14 mg, 73%). MS (ESI, m/z) 455.2 [M + H]+. ESI-HRMS calcd for C27H27N4O3 455.2083, found 455.2086 [M + H]+. HPLC purity 97% (Rt = 10.1 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.35 (s, 1H), 8.35 (s, 1H), 8.21 (s, 1H), 7.92 (d, J = 8.7 Hz, 2H), 7.72 (d, J = 7.9 Hz, 2H), 7.38 (d, J = 7.9 Hz, 2H), 7.07 (d, J = 8.7 Hz, 2H), 3.82 (s, 3H), 3.07 (d, J = 11.8 Hz, 2H), 2.70–2.57 (m, 3H), 1.84–1.68 (m, (3Hacetate), 5H), 1.58 (qd, J = 4.0, 12.0 Hz, 2H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.8 (1Cacetate), 167.8, 159.9, 147.9, 147.3, 144.7, 141.3, 137.9, 137.3, 128.1 (2C), 127.6 (2C), 127.5, 127.4 (2C), 123.7, 119.7, 119.5, 118.3, 115.1 (2C), 55.9, 47.1 (2C), 42.7, 34.5 (2C), 29.8, 23.6 (1Cacetate).
5-(4-(4-(Pentyloxy)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (82)
Semipreparative HPLC: method B, Rt = 34.1 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 5.3 mg, 99%). MS (ESI, m/z) 511.3 [M + H]+. ESI-HRMS calcd for C31H35N4O3 511.2709, found 511.2703 [M + H]+. HPLC purity 99% (Rt = 14.4 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.40 (s, 1H), 8.40 (s, 1H), 8.28 (s, 1H), 8.16 (s, 1H), 7.95 (d, J = 8.4 Hz, 2H), 7.79 (d, J = 7.8 Hz, 2H), 7.45 (d, J = 7.9 Hz, 2H), 7.11 (d, J = 8.4 Hz, 2H), 4.08 (t, J = 6.5 Hz, 2H), 3.20 (d, J = 11.8 Hz, 2H), 2.83–2.68 (m, 3H), 1.94 (s, 3Hacetate), 1.89–1.78 (m, 4H), 1.77–1.65 (m, 3H), 1.52–1.38 (m, 4H), 0.97 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 137.0, 126.9 (2C), 126.3 (2C), 126.2 (2C), 120.6 (2C), 119.2, 118.8, 118.0, 114.4, 77.0, 72.3, 67.0, 46.7 (2C), 27.9, 27.2, 21.4, 13.5.
5-(4-(2-Methoxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (83)
Semipreparative HPLC: method B, Rt = 23.0 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 3.05 mg, 52%). MS (ESI, m/z) 455.2 [M + H]+. ESI-HRMS calcd for C27H27N4O3 455.2083, found 455.2086 [M + H]+. HPLC purity 98% (Rt = 10.44 min). 1H NMR (600 MHz, DMSO-d6): δ (ppm) = 9.02 (s, 1H), 8.33 (s, 1H), 8.24 (s, 1H), 8.22 (d, J = 7.5 Hz, 1H), 8.10 (s, 1H), 7.74 (d, J = 7.8 Hz, 2H), 7.43–7.34 (m, 3H), 7.18 (d, J = 8.3 Hz, 1H), 7.10 (t, J = 7.5 Hz, 1H), 3.98 (s, 3H), 3.14 (d, J = 12.0 Hz, 3H), 2.74–2.67 (m, 3H), 1.86 (s, 3Hacetate), 1.79 (d, J = 12.0 Hz, 2H), 1.73–1.62 (m, 2H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 173.1 (1Cacetate), 167.9, 156.0, 146.7, 143.7, 143.2, 141.1, 137.6, 137.1, 129.7, 127.7 (2C), 127.4 (2C), 127.3, 122.2, 121.0, 120.0, 119.1, 118.9, 111.9, 55.9, 46.3 (2C), 41.9, 40.4, 33.6 (2C), 22.4 (1Cacetate).
5-(4-(4-(Trifluoromethoxy)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (84)
Semipreparative HPLC: method B, Rt = 29.8 min. The product was obtained as a white acetate salt by lyophilization (1 equiv CH3CO2H-salt, 2.63 mg, 84%). MS (ESI, m/z) 509.2 [M + H]+. ESI-HRMS calcd for C27H24N4O3F3 509.1801, found 509.1817 [M + H]+. HPLC purity 96% (Rt = 12.3 min), mp 274–280 °C. 1H NMR (400 MHz, DMSO-d6): δ (ppm) = 9.52 (s, 1H), 8.35 (s, 1H), 8.23 (s, 1H), 8.15–8.05 (m, 3H), 7.72 (d, J = 8.0 Hz, 2H), 7.52 (d, J = 8.3 Hz, 2H), 7.38 (d, J = 8.0 Hz, 2H), 3.09 (d, J = 12.1 Hz, 2H), 2.71–2.64 (m, 3H), 1.85 (s, 3Hacetate), 1.77 (d, J = 12.1 Hz, 2H), 1.61 (qd, J = 4.0, 12.1 Hz, 2H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 167.6, 148.5, 146.9, 146.5, 141.2, 137.7, 137.0, 130.4, 127.9 (2C), 127.7 (2C), 127.6, 127.4 (2C), 122.3 (2C), 122.2 (q, JC–F = 260.4 Hz), 120.9, 119.7, 118.3, 46.5 (2C), 42.1, 40.7, 40.6, 33.7 (2C), 22.4.
5-(4-(3-(Trifluoromethoxy)phenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (85)
Semipreparative HPLC: method B, Rt = 29.2 min. The product was obtained as a white acetate salt by lyophilization (0.5 equiv CH3CO2H-salt, 3.63 mg, 66%). MS (ESI, m/z) 509.2 [M + H]+. ESI-HRMS calcd for C27H24N4O3F3 509.1801, found 509.1798 [M + H]+. HPLC purity 98% (Rt = 12.4 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.63 (s, 1H), 8.40 (s, 1H), 8.30 (s, 1H), 8.15 (t, J = 2.0 Hz, 1H), 8.04 (d, J = 7.8 Hz, 1H), 7.94 (s, 1H), 7.75 (d, J = 7.9 Hz, 2H), 7.65 (t, J = 8.0 Hz, 1H), 7.42 (d, J = 7.9 Hz, 2H), 7.38 (dd, J = 2.4, 8.1 Hz, 1H), 3.24 (d, J = 11.8 Hz, 2H), 2.88–2.75 (m, 3H), 1.88 (s, 1.5Hacetate), 1.86–1.79 (m, 4H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 171.9, 166.6, 148.5, 145.3, 140.2, 136.7, 135.9, 132.2, 130.7, 126.8 (2C), 126.4 (2C), 123.7, 120.2, 120.0, 119.5, 118.8 (q, JC–F = 257.4 Hz), 117.1, 45.1 (2C), 40.7, 32.1 (2C), 21.1.
5-(4-(3-Chlorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (86)
Semipreparative HPLC: method B, Rt = 21.2 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 2.2 mg, 68%). MS (ESI, m/z) 459.2 [M + H]+. ESI-HRMS calcd for C26H2435ClN4O2 459.1588, found 459.1583 [M + H]+. HPLC purity 98% (Rt = 11.1 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.59 (s, 1H), 8.36 (s, 1H), 8.22 (s, 1H), 8.10 (s, 1H), 8.05 (d, J = 1.9 Hz, 1H), 7.71 (d, J = 7.9 Hz, 2H), 7.55 (t, J = 7.9 Hz, 1H), 7.45 (dd, J = 2.1, 8.1 Hz, 1H), 7.38 (d, J = 7.9 Hz, 2H), 2.66 (s, 3H), 1.67 (m, (3Hacetate), 7H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 167.1, 146.9, 146.1, 140.8, 137.4, 136.6, 134.0, 132.9, 131.2, 128.1, 127.6 (2C), 127.3 (2C), 127.0, 125.3, 124.0, 120.8, 119.4, 117.7, 46.8, 42.4 (2C), 34.4 (2C), 29.2.
5-(4-(4-Fluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (87)
Semipreparative HPLC: method B, Rt = 20.6 min. The product was obtained as a salt by lyophilization (0.5 equiv CH3CO2H-salt, 3.06 mg, 99%). MS (ESI, m/z) 443.2 [M + H]+. ESI-HRMS calcd for C26H24FN4O2 443.1883, found 443.1890 [M + H]+. HPLC purity 96% (Rt = 10.8 min). 1H NMR (400 MHz, DMSO-d6): δ (ppm) = 9.46 (s, 1H), 8.36 (s, 1H), 8.23 (s, 1H), 8.10 (s, 1H), 8.06–7.95 (m, 2H), 7.71 (d, J = 7.7 Hz, 2H), 7.40–7.33 (m, 4H), 3.11–3.01 (m, 2H), 2.70–2.57 (m, 3H), 1.75 (m, 4H), 1.63–1.50 (m, 2H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 163.2, 161.2, 147.0, 146.8, 141.1, 137.6, 137.0, 127.8 (2C), 127.5 (2C), 127.3, 120.2, 119.9, 118.2, 116.5 (2C), 116.3 (2C), 72.7, 70.8, 60.7, 46.6, 42.3 (2C), 40.5, 34.2 (2C), 29.4, 20.7.
5-(4-(3-Fluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (88)
Semipreparative HPLC: method B, Rt = 21.5 min. The product was obtained as a white acetate salt by lyophilization (0.5 equiv CH3CO2H-salt, 3.4 mg, 84%). MS (ESI, m/z) 443.1 [M + H]+. ESI-HRMS calcd for C26H24FN4O2 443.1883, found 443.1884 [M + H]+. HPLC purity 98.2% (Rt = 10.5 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.57 (s, 1H), 8.37 (s, 1H), 8.27 (s, 1H), 8.13 (s, 1H), 7.85 (d, J = 7.9 Hz, 1H), 7.79 (dt, J = 2.6, 10.2 Hz, 1H), 7.75 (d, J = 7.9 Hz, 2H), 7.56 (td, J = 6.0, 8.0 Hz, 1H), 7.41 (d, J = 7.9 Hz, 2H), 7.22 (td, J = 2.6, 8.6 Hz, 1H), 3.19 (d, J = 11.8 Hz, 2H), 2.82–2.69 (m, 3H), 1.89 (s, 1.5Hacetate), 1.86–1.69 (m, 4H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.5, 167.5, 163.01 (d, JC–F = 243.6 Hz), 146.8, 146.5, 144.3, 141.0, 137.5, 136.8, 133.2, 133.2, 131.6, 131.5, 127.8 (2C), 127.4 (2C), 127.2, 121.7, 120.9, 119.4, 118.2, 115.3, 115.2, 112.4, 112.2, 46.4 (2C), 42.1, 33.8 (2C), 23.3.
5-(4-(2-Fluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (89)
Semipreparative HPLC: method B, Rt = 24.3 min. The product was obtained as a white salt by lyophilization (1 equiv CH3CO2H-salt, 2.64 mg, 60%). MS (ESI, m/z) 443.1 [M + H]+. ESI-HRMS calcd for C26H24FN4O2 443.1883, found 443.1887 [M + H]+. HPLC purity 99% (Rt = 10.4 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.28 (d, J = 3.0 Hz, 1H), 8.42 (s, 1H), 8.29 (s, 1H), 8.27 (td, J = 1.8, 7.6 Hz, 1H), 8.20 (t, J = 2.1 Hz, 1H), 7.81 (d, J = 8.0 Hz, 2H), 7.56–7.46 (m, 2H), 7.46–7.40 (m, 3H), 3.16 (d, J = 12.0 Hz, 2H), 2.81–2.69 (m, 3H), 1.91 (s, 3Hacetate), 1.83 (d, J = 12.0 Hz, 2H), 1.69 (qd, J = 6.4, 12.0 Hz, 2H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.0 (1Cacetate), 167.5, 159.1 (d, JC–F = 248.3 Hz), 146.9, 144.5, 141.3, 141.1, 137.7, 136.9, 130.5 (d, JC–C–F = 8.4 Hz), 128.3, 127.8 (2C), 127.6, 127.5 (2C), 125.5, 122.5 (d, JC–C–F = 10.1 Hz, H), 120.1, 118.7, 118.6, 116.7, 116.6, 46.6 (2C), 42.2, 33.9 (2C), 22.4 (1Cacetate).
5-(4-(3,4-Difluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (90)
Semipreparative HPLC: method B, Rt = 21.1 min. The product was obtained as a white solid by lyophilization (3.85 mg, 99%). MS (ESI, m/z) 461.2 [M + H]+. ESI-HRMS calcd for C26H24FN4O2 461.1789, found 461.1790 [M + H]+. HPLC purity 97% (Rt = 11.31 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.55 (s, 1H), 8.38 (s, 1H), 8.34 (s, 1H), 8.16 (s, 1H), 8.00 (ddd, J = 2.1, 7.8, 11.0 Hz, 1H), 7.87–7.81 (m, 1H), 7.78 (d, J = 7.9 Hz, 2H), 7.60 (dt, J = 8.5, 11.0 Hz, 1H), 7.45 (d, J = 7.8 Hz, 2H), 3.51–3.38 (m, 2H), 2.96–2.82 (m, 3H), 2.03–1.84 (m, 4H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 151.4 (d, JC–F = 245.3 Hz), 148.6, 146.2, 145.9, 143.9, 141.2, 140.6, 137.8, 136.7, 128.6, 127.9 (2C), 127.4 (2C), 122.6, 120.9, 119.5, 118.9 (d, JC–C–F = 17.7 Hz, H), 114.8 (d, JC–C–F = 18.6 Hz, H), 72.8, 60.7, 45.3, 32.7.
5-(4-(3,5-Difluorophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (91)
Semipreparative HPLC: method B, Rt = 22.2 min. The product was obtained as a white salt by lyophilization (1 equiv CH3CO2H-salt, 1.24 mg, 33%). MS (ESI, m/z) 461.2 [M + H]+. ESI-HRMS calcd for C26H24FN4O2 calcd for C26H23F2N4O2 461.1789, found 461.1782 [M + H]+. HPLC purity 99% (Rt = 11.40 min), mp 284–291 °C. 1H NMR (600 MHz, DMSO-d6): δ (ppm) = 9.61 (s, 1H), 8.34 (q, J = 1.4 Hz, 1H), 8.23 (t, J = 1.4 Hz, 1H), 8.08 (t, J = 1.4 Hz, 1H), 7.75–7.67 (m, 4H), 7.38 (d, J = 8.3 Hz, 2H), 7.27 (tt, J = 2.4, 9.3 Hz, 1H), 3.05 (d, J = 12.3 Hz, 2H), 2.69–2.56 (m, 2H), 1.78 (s, 3Hacetate), 1.75 (d, J = 12.4 Hz, 2H), 1.57 (qd, J = 3.8, 12.3 Hz, 2H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 173.4 (1Cacetate), 167.1, 164.2 (d, JC–C–F = 13.5 Hz), 162.6 (d, JC–C–F = 13.3 Hz), 147.2, 145.7, 145.0, 141.6, 141.0, 137.5, 136.7, 134.4 (t, JC–C–F = 10.7 Hz), 127.8 (2C), 127.6, 127.2 (2C), 121.6, 119.5, 117.9, 109.20–108.17 (m), 103.8 (t, JC–C–F = 25.8 Hz), 47.1, 42.6 (2C), 34.6 (2C).
5-(4-(4-Carboxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (92)
Semipreparative HPLC: method B, Rt = 9.4 min. The product was obtained as a white solid by lyophilization (4.6 mg, 86%). MS (ESI, m/z) 469.2 [M + H]+. ESI-HRMS calcd for C27H25F2N4O4 469.1876, found 469.1870 [M + H]+. HPLC purity 96% (Rt = 9.30 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.52 (s, 1H), 8.40 (s, 1H), 8.25 (s, 1H), 8.14 (s, 1H), 7.98 (q, J = 8.1 Hz, 4H), 7.77 (d, J = 7.9 Hz, 2H), 7.42 (d, J = 7.9 Hz, 2H), 3.23 (d, J = 12.2 Hz, 2H), 2.84–2.74 (m, 3H), 1.87–1.77 (m, 4H). Et3N-salt: 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.7 (3Ctriethylamine), 168.8, 167.4, 147.9, 145.0, 142.1, 140.9, 137.6, 136.9, 130.4, 129.9 (2C), 127.7 (2C), 127.2 (2C), 124.5 (2C), 119.9, 119.6, 117.9, 47.1 (2C), 42.7, 34.7 (2C), 26.5 (3Ctriethylamine).
5-(4-(3-Carboxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (93)
To a solution of aryl azide (22, 5 mg, 11.5 μmol, 1 equiv) and 3-ethynylbenzoic acid (6, R = 3-PhCO2H, 2.5 mg, 17 μmol, 1.5 equiv) in 2 mL of THF:water (1:1), sodium ascorbate (17 μmol, freshly prepared 1 M aqueous solution, 1.5 equiv), and CuSO4 (1.4 mg, 6 μmol, 0.5 equiv) were sequentially added. The resulting reaction was vigorously stirred for 12 h at rt. The reaction mixture was then concentrated in vacuo and added to a solution of 2 mL of MeOH:1.0 M KOH (1:1), and the mixture was heated at 50 °C for 12 h. After removing the solvents under reduced pressure, the mixture was purified by preparative HPLC (method B, Rt = 10.4 min). The product was obtained as a white salt by lyophilization (2 Et3N salt, 5.7 mg, 93%). MS (ESI, m/z) 469.2 [M + H]+. ESI-HRMS calcd for C27H25F2N4O4 469.1876, found 469.1884 [M + H]+. HPLC purity 97% (Rt = 9.32 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.69 (s, 1H), 8.55 (s, 1H), 8.49 (s, 1H), 8.36 (s, 1H), 8.30 (s, 1H), 8.20 (d, J = 7.8 Hz, 1H), 7.95 (d, J = 7.8 Hz, 1H), 7.83 (d, J = 7.9 Hz, 2H), 7.62 (t, J = 7.8 Hz, 1H), 7.44 (d, J = 7.9 Hz, 2H), 3.41 (d, J = 12.0 Hz, 2H), 3.07–3.00 (m, 3H), 2.95 (q, J = 7.4 Hz, 12Htriethylamine), 2.02–1.88 (m, 4H), 1.12 (t, J = 7.3 Hz, 18Htriethylamine). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 176.8, 173.6 (6Ctriethylamine), 168.8, 167.3, 148.2, 146.9, 144.0, 144.7, 142.5, 140.8, 137.5, 137.0, 129.6, 129.2, 127.9, 127.7 (2C), 127.2 (2C), 127.1, 126.6, 125.6, 119.8, 119.4, 117.8, 98.2, 46.9 (2C), 42.5, 40.5, 34.4 (2C), 24.6 (6Ctriethylamine).
5-(4-(4-Acetylphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (94)
Semipreparative HPLC: method B, Rt = 19.4 min. The product was obtained as a white salt by lyophilization (1 equiv CH3CO2H-salt, 3.88 mg, 95%). MS (ESI, m/z) 479.2 [M + H]+. ESI-HRMS calcd for C28H27N4O3 467.2083, found 467.2084 [M + H]+. HPLC purity 97% (Rt = 10.4 min). 1H NMR (400 MHz, DMSO-d6): δ (ppm) = 9.56 (s, 1H), 8.34 (s, 1H), 8.24 (s, 1H), 8.10 (m, 5H), 7.73 (d, J = 8.0 Hz, 2H), 7.37 (d, J = 8.0 Hz, 1H), 3.11 (d, J = 12.3 Hz, 2H), 2.74–2.64 (m, 3H), 2.61 (s, 3H), 1.79 (s, 3Hacetate), 1.63 (q, J = 10.9, 11.7 Hz, 4H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 198.2, 167.9, 146.8, 143.9, 141.3, 137.5, 136.9, 136.6, 135.2, 129.6 (2C), 129.3 (2C), 127.9, 127.7 (2C), 127.4 (2C), 125.8, 121.5, 119.9, 119.8, 118.9, 118.6, 72.7, 60.7, 46.4 (2C), 41.9, 33.6 (2C), 27.3.
5-(4-(3,4-Dimethoxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (95)
Semipreparative HPLC: method B, Rt = 13.5 min. The product was obtained as a white salt by lyophilization (1 equiv CH3CO2H-salt, 5.2 mg, 99%). MS (ESI, m/z) 485.2 [M + H]+. ESI-HRMS calcd for C28H29N4O4 485.2189, found 485.2195 [M + H]+. HPLC purity 97% (Rt = 9.8 min). 1H NMR (600 MHz, DMSO-d6): δ (ppm) = 9.37 (s, 1H), 8.34 (s, 1H), 8.20 (s, 1H), 8.07 (s, 1H), 7.71 (d, J = 7.8 Hz, 2H), 7.58 (d, J = 2.0 Hz, 1H), 7.55 (dd, J = 2.0, 8.2 Hz, 1H), 7.38 (d, J = 7.9 Hz, 2H), 7.08 (d, J = 8.3 Hz, 1H), 3.87 (s, 3H), 3.81 (s, 3H), 3.07 (s, 2H), 2.72–2.56 (m, 3H), 1.86–1.65 (m, (3Hacetate), 5H), 1.64–1.47 (m, 2H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 173.2 (1Cacetate), 167.0, 149.6, 149.3, 147.8, 147.1, 145.1, 140.9, 137.7, 136.9, 127.8 (2C), 127.3 (2C), 123.7, 119.4, 119.3, 118.2, 117.7, 112.6, 109.6, 56.0, 46.9 (2C), 42.5, 34.5 (2C), 23.7 (1Cacetate).
5-(4-(3-Hydroxyphenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (96)
Semipreparative HPLC: method B, Rt = 17.9 min. The product was obtained as a white salt by lyophilization (2 equiv CH3CO2H-salt, 4.7 mg, 99%). MS (ESI, m/z) 441.2 [M + H]+. ESI-HRMS calcd for C26H25N4O3 441.1927, found 441.1931 [M + H]+. HPLC purity 96.2% (Rt = 8.7 min). 1H NMR (600 MHz, DMSO-d6): δ (ppm) = 9.37 (s, 1H), 8.35 (t, J = 1.6 Hz, 1H), 8.21 (t, J = 1.5 Hz, 1H), 8.08 (t, J = 2.0 Hz, 1H), 7.72 (d, J = 8.2 Hz, 2H), 7.46 (t, J = 2.0 Hz, 1H), 7.40–7.33 (m, 3H), 7.27 (t, J = 7.8 Hz, 1H), 6.79 (dd, J = 2.4, 8.0 Hz, 1H), 3.05 (d, J = 11.7 Hz, 2H), 2.69–2.57 (m, 3H), 1.74 (d, J = 11.9 Hz, 2H), 1.68 (s, 6Hacetate), 1.57 (qd, J = 3.8, 12.4 Hz, 2H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 173.6, 167.2, 158.6, 147.8, 147.1, 144.9, 140.9, 137.6, 136.9, 132.0, 130.4, 127.8 (2C), 127.3 (2C), 127.3, 120.1, 119.5, 117.9, 116.5, 115.7, 112.7, 47.0 (2C), 42.6, 40.5, 34.6 (2C), 24.7.
5-(4-(3-Aminophenyl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (97)
Semipreparative HPLC: method B, Rt = 17.4 min. The product was obtained as a white solid by lyophilization (2 equiv CH3CO2H-salt, 2.2 mg, 99%). MS (ESI, m/z) 440.2 [M + H]+. ESI-HRMS calcd for C26H26N5O2 440.2087, found 440.2093 [M + H]+. HPLC purity 97% (Rt = 8.9 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.29 (s, 1H), 8.34 (s, 1H), 8.20 (s, 1H), 8.08 (s, 1H), 7.72 (d, J = 8.0 Hz, 2H), 7.37 (d, J = 7.9 Hz, 2H), 7.25 (s, 1H), 7.15–7.04 (m, 2H), 6.57 (d, J = 7.6 Hz, 1H), 5.21 (s, 2H), 3.04 (d, J = 12.0 Hz, 2H), 2.71–2.55 (m, 3H), 1.78–1.63 (m, (6Hacetate), 8H), 1.56 (qd, J = 3.8, 12.0 Hz, 2H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.9 (2Cacetate), 167.4, 149.6, 148.4, 147.2, 144.8, 140.9, 137.7, 137.1, 131.4, 129.9, 127.8 (2C), 127.3(2C), 119.8, 119.6, 118.0, 114.3, 113.8, 111.1, 47.1 (2C), 42.7, 34.6 (2C), 24.4 (2Cacetate).
5-(4-(Pyridin-4-yl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (98)
Semipreparative HPLC: method B, Rt = 15.7 min. The product was obtained as a white solid by lyophilization (2 equiv CH3CO2H-salt, 2 mg, 85%). MS (ESI, m/z) 426.1 [M + H]+. ESI-HRMS calcd for C25H24N5O2 426.1930, found 426.1933 [M + H]+. HPLC purity 97% (Rt = 9.2 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.71 (s, 1H), 8.74–8.66 (m, 2H), 8.36 (s, 1H), 8.23 (s, 1H), 8.11 (t, J = 2.0 Hz, 1H), 7.95 (d, J = 6.0 Hz, 1H), 7.71 (d, J = 8.2 Hz, 2H), 7.38 (d, J = 8.0 Hz, 2H), 3.05 (d, J = 13.0 Hz, 2H), 2.68–2.54 (m, 3H), 1.75–1.68 (m, (6Hacetate), 8H), 1.56 (qd, J = 3.9, 12.3 Hz, 2H). 13C NMR (1256 MHz, DMSO-d6): δ (ppm) = 174.4 (2Cacetate), 167.3, 150.9 (2C), 147.3, 145.4, 144.8, 141.1, 138.1, 137.5, 136.8, 127.9 (2C), 127.7, 127.3 (2C), 122.4, 120.1 (2C), 119.7, 118.3, 70.3, 47.0 (2C), 42.7, 40.3, 34.6 (2C), 24.4 (2Cacetate).
5-(4-(Pyrazin-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (99)
Semipreparative HPLC: method B, Rt = 16.8 min. The product was obtained as a white solid by lyophilization (4 equiv CH3CO2H-salt, 2.81 mg, 64%). MS (ESI, m/z) 427.2 [M + H]+. ESI-HRMS calcd for C24H23N6O2 427.1882, found 427.1881 [M + H]+. HPLC purity 98% (Rt = 8.43 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.70 (s, 1H), 9.41 (s, 1H), 8.81 (s, 1H), 8.73 (d, J = 2.7 Hz, 1H), 8.45 (s, 1H), 8.30 (s, 1H), 8.22 (d, J = 2.7 Hz, 2H), 7.82 (d, J = 7.9 Hz, 2H), 7.43 (d, J = 7.8 Hz, 2H), 3.10 (d, J = 12.1 Hz, 3H), 2.75–2.60 (m, 3H), 1.80 (d, J = 13.1 Hz, 2H), 1.71–1.57 (m, (12Hacetate), 14H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.9 (4Cacetate), 167.1, 147.1, 145.8, 145.8, 145.0, 144.8, 144.4, 141.9, 140.9, 137.4, 136.7, 127.7 (2C), 127.6, 127.3 (2C), 123.1, 119.9, 118.3, 46.9 (2C), 42.6, 34.5 (2C), 25.2 (4Cacetate).
5-(4-(Furan-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (100)
Semipreparative HPLC: method B, Rt = 19.8 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt 2.28 mg, 84%). MS (ESI, m/z) 415.2 [M + H]+. ESI-HRMS calcd for C24H23N4O3 415.1770, found 415.1767 [M + H]+. HPLC purity 98% (Rt = 9.2 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 9.30 (s, 1H), 8.38 (s, 1H), 8.29 (s, 1H), 8.15 (t, J = 1.9 Hz, 1H), 7.88 (d, J = 1.8 Hz, 1H), 7.79 (d, J = 8.0 Hz, 2H), 7.43 (d, J = 8.0 Hz, 2H), 6.97 (d, J = 3.3 Hz, 1H), 6.72 (dd, J = 3.3, 1.9 Hz, 1H), 3.11 (dt, J = 12.3, 2.9 Hz, 2H), 2.82–2.62 (m, 3H), 1.84–1.73 (m, 5H (3Hacetate)), 1.63 (qd, J = 12.6, 4.1 Hz, 2H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.6 (1Cacetate), 167.4, 147.1, 146.0, 144.5, 143.5, 141.1, 140.4, 137.5, 136.8, 127.8 (2C), 127.5, 127.4 (2C), 119.8, 119.6, 118.4, 114.9, 112.3, 107.5, 49.1, 46.9 (2C), 42.5, 34.3 (2C), 29.5, 24.0, 22.6 (1Cacetate), 14.5.
5-(4-(Benzofuran-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (101)
Semipreparative HPLC: method B, Rt = 21.9 min. The product was obtained as a white solid by lyophilization (1 equiv CH3CO2H-salt, 4.9 mg, 90%). MS (ESI, m/z) 465.2 [M + H]+. ESI-HRMS calcd for C28H25N4O3 465.1927, found 465.1936 [M + H]+. HPLC purity 97% (Rt = 11.4 min). 1H NMR (600 MHz, DMSO-d6): δ (ppm) = 9.54 (s, 1H), 8.40 (s, 1H), 8.26 (s, 1H), 8.16 (s, 1H), 7.76 (d, J = 7.9 Hz, 2H), 7.74 (d, J = 7.6 Hz, 1H), 7.66 (d, J = 8.1 Hz, 1H), 7.43–7.36 (m, 4H), 7.32 (t, J = 7.4 Hz, 1H), 3.10 (d, J = 12.5 Hz, 2H), 2.72–2.61 (m, 3H), 1.85 (s, 3Hacetate), 1.77 (d, J = 12.5 Hz, 2H), 1.63 (qd, J = 3.4, 12.0, 12.5 Hz, 2H). 13C NMR (150 MHz, DMSO-d6): δ (ppm) = 173.2 (1Cacetate), 167.5, 154.5, 148.3, 146.9, 144.5, 141.0, 139.9, 137.5, 136.7, 128.7, 127.8 (2C), 127.6, 127.4 (2C), 125.3, 123.9, 121.9, 121.3, 119.8, 118.4, 111.6, 103.4, 46.6 (2C), 42.3, 34.0 (2C), 29.5, 22.6 (1Cacetate).
5-(4-(Thiazol-2-yl)-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (102)
Semipreparative HPLC: method B, Rt = 18.7 min. The product was obtained as a white salt by lyophilization (2 equiv CH3CO2H-salt, 4.18 mg, 99%). MS (ESI, m/z) 431.2 [M + H]+. ESI-HRMS calcd for C23H22N5O232S 432.1494, found 432.1494 [M + H]+. HPLC purity 96.4% (Rt = 9.1 min). 1H NMR (400 MHz, DMSO-d6): δ (ppm) = 9.54 (s, 1H), 8.36 (s, 0H), 8.23 (s, 1H), 8.13 (s, 1H), 7.98 (d, J = 3.3 Hz, 1H), 7.83 (d, J = 3.3 Hz, 1H), 7.74 (d, J = 7.9 Hz, 2H), 7.35 (d, J = 7.9 Hz, 2H), 3.03 (d, J = 12.9 Hz, 2H), 2.72–2.56 (m, 3H), 1.73 (d, J = 12.9 Hz, 2H), 1.66 (s, 6Hacetate), 1.55 (qd, J = 3.1, 10.6, 11.4 Hz, 2H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 173.1 (2Cacetate), 167.5, 158.7, 146.8, 144.4, 144.2, 143.5, 141.1, 137.5, 136.8, 127.8 (2C), 127.7, 127.4 (2C), 121.3, 120.8, 120.1, 118.8, 46.4 (2C), 42.0, 33.6 (2C), 22.7 (2Cacetate).
5-(4-Cyclohexyl-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (103)
Semipreparative HPLC: method B, Rt = 21.2 min. The product was obtained as a white solid by lyophilization (2 equiv CH3CO2H-salt, 2.65 mg, 55%). MS (ESI, m/z) 431.3 [M + H]+. ESI-HRMS calcd for C26H31N4O2 431.2447, found 431.2444 [M + H]+. HPLC purity 97% (Rt = 10.4 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 8.77 (s, 1H), 8.35 (s, 1H), 8.25 (s, 1H), 8.09 (s, 1H), 7.76 (d, J = 7.8 Hz, 2H), 7.41 (d, J = 7.9 Hz, 2H), 3.14 (d, J = 11.9 Hz, 2H), 2.85–2.78 (m, 1H), 2.76–2.68 (m, 3H), 2.10 (d, J = 12.3 Hz, 2H), 2.00–1.72 (m, (6Hacetate), 10H), 1.72–1.59 (m, 2H), 1.57–1.26 (m, 6H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 168.2, 153.7, 147.0, 143.7, 141.0, 137.6, 137.3, 127.8 (2C), 127.3 (2C), 127.0, 119.7, 119.5, 118.3, 46.8 (2C), 42.4, 35.2, 34.2 (2C), 33.0 (2C), 26.2, 26.1 (2C), 23.37 (2Cacetate).
5-(4-Cyclopropyl-1H-1,2,3-triazol-1-yl)-4′-(piperidin-4-yl)-[1,1′-biphenyl]-3-carboxylic Acid (104)
Semipreparative HPLC: method B, Rt = 12.6 min. The product was obtained as a white solid by lyophilization (2 equiv CH3CO2H-salt, 5.1 mg, 87%). MS (ESI, m/z) 389.2 [M + H]+. ESI-HRMS calcd for C23H25N4O2 389.1978, found 389.1971 [M + H]+. HPLC purity 97% (Rt = 8.5 min). 1H NMR (500 MHz, DMSO-d6): δ (ppm) = 8.66 (s, 1H), 8.24 (s, 1H), 8.16 (s, 1H), 7.97 (s, 1H), 7.68 (d, J = 7.9 Hz, 1H), 7.35 (d, J = 7.9 Hz, 2H), 3.04 (d, J = 10.0 Hz, 2H), 2.69–2.54 (m, 3H), 2.02 (td, J = 4.4, 8.6 Hz, 1H), 1.75–1.65 (m, (6Hacetate), 8H), 1.60–1.51 (m, 2H), 0.96 (dt, J = 3.5, 8.9 Hz, 2H), 0.82 (dt, J = 3.5, 7.3 Hz, 3H). 13C NMR (125 MHz, DMSO-d6): δ (ppm) = 174.0 (2Cacetate), 167.5, 150.7, 147.0, 144.6, 140.9, 137.7, 137.2, 127.8 (2C), 127.3 (2C), 127.1, 119.4, 117.9, 46.9 (2C), 42.5, 34.4 (2C), 24.6 (2Cacetate), 8.3 (2C), 7.2.
Hex-5-yn-1-yl 4-Methylbenzenesulfonate (106)
To a solution of hex-5-yn-1-ol 105 (0.84 mL, 7.64 mmol), triethylamine (1.28 mL, 9.17 mmol), and 4-(dimethylamino)pyridine (20 mg, 0.15 mmol) in CH2Cl2 (25 mL) at 0 °C was added p-toluenesulfonyl chloride (1.53 g, 8.02 mmol) in three portions. The reaction mixture was brought to rt. and stirred for 15 h. Aqueous NaOH (1 N, 15 mL) was added, and the mixture was vigorously stirred for 15 min at rt. The aqueous phase was extracted with CH2Cl2. The combined organic fractions were washed with water, followed by brine, and then dried over Na2SO4, filtered, and concentrated in vacuo to give the title compound (1.68 g, 88%) as a yellowish oil. 1H NMR (400 MHz, CDCl3): δ (ppm) = 7.69 (2H, J = 8.0, d), 7.26 (d, J = 8.0 Hz, 2H), 3.96 (t, J = 6.0 Hz, 2H), 2.31 (s, 3H), 2.05–2.09 (m, 2H), 1.84 (t, J = 4.0 Hz, 1H), 1.65–1.68 (m, 2H), 1.42–1.48 (m, 2H). 13C NMR (100 MHz, CDCl3): δ 144.8, 133.0, 129.9, 127.0, 83.4, 69.9, 69.0, 27.7, 24.2, 21.6, 17.7.
6-Bromohex-1-yne (107)
LiBr (1.7 g, 19.6 mmol) was added to a stirred solution of 106 (1.64 g, 6.52 mmol) in dry DMF (20 mL). After the exothermic reaction, the mixture was stirred at room temperature for 24 h. Then water (25 mL) was added and the aqueous phase extracted with Et2O (3 × 25 mL). The collected organic fractions were dried over Na2SO4, filtered, and concentrated in vacuo. The residue was purified by flash chromatography using as eluent hexane:ethyl acetate (5:1) to afford a colorless oil (0.86 g, 82%). 1H NMR (400 MHz, CDCl3): δ (ppm) = 3.59 (t, J = 6.4, 2H), 2.24 (m, 2H), 1.90–1.98 (m, 3H), 1.66–1.70 (m, 2H). 13C NMR (100 MHz, CDCl3): δ (ppm) = 83.8, 69.2, 45.6, 29.7, 29.4, 19.7.
6-Bromohexan-1-amine Hydrochloride (109)
6-Aminohexanol 108 (0.5 g, 4.27 mmol) was slowly added to a stirring 48% HBr solution (5.1 mL) at 0 °C, and the resulting mixture was stirred at 80 °C for 20 h. The mixture was neutralized by adding 2N NaOH (20 mL) and extracted with ethyl acatate (3 × 20 mL). The combined organic fractions were washed with water (50 mL) followed by brine (50 mL) and then dried over Na2SO4, filtered, and concentrated in vacuo. The obtained viscous oil was dissolved in 4 M HCl solution in dioxane to give a sticky solid that was washed with Et2O and then filtered to afford a yellowish solid (0.55 g, 61%). 1H NMR (400 MHz, MeOD): δ (ppm) = 3.39 (t, J = 4.0 Hz, 2H), 2.86 (t, J = 8.0 Hz, 2H), 1.74–1.82 (m, 2H), 1.59–1.70 (m, 2H), 1.42–1.46 (m, 4H). 13C NMR (100 MHz, MeOD): δ (ppm) = 44.2, 32.8, 32.2, 27.2, 27.0, 25.2.
6-Azidohexan-1-amine (110)
To a solution of 109 (0.55 g, 2.54 mmol) in water (25 mL), NaN3 (0.49 g, 7.69 mmol) was added, and the resulting mixture was heated at 100 °C for 12 h. After cooling, 37% ammonia solution was added until a basic pH was reached, and the aqueous phase was extracted with Et2O (3 × 20 mL). The organic fractions were collected, dried over Na2SO4, and filtered, and the solvent removed under vacuum to give a yellow oil (0.29 g, 80%). 1H NMR (400 MHz, CDCl3): δ (ppm) = 3.18 (t, J = 8.0 Hz, 2H), 2.62 (t, J = 4.0 Hz, 2H), 2.15 (br s, 2H), 1.50–1.54 (m, 2H), 1.35–1.40 (m, 2H), 1.29–1.34 (m, 4H). 13C NMR (100 MHz, CDCl3): δ (ppm) = 51.4, 41.8, 33.1, 28.8, 26.6, 26.4.
Ethyl 4-(4-(Piperidin-4-yl)phenyl)-7-(4-(trifluoromethyl)phenyl)-2-naphthoate (111)
To a solution of 3 (0.50 g, 1.57 mmol) in EtOH (50 mL), thionyl chloride (1.37 mL, 18.84 mmol) was carefully added over 30 min at 0 °C. The reaction was allowed to warm up to rt and stirred overnight. The resulting mixture was quenched by adding 5% NaOH solution (25 mL). Then the solvent was evaporated under vacuum and the aqueous residue extracted with ethyl acetate (3 × 20 mL). The collected organic fractions were dried over Na2SO4, filtered, and the solvent removed under vacuum. The residue was purified by chromatography using as eluent CH2Cl2/MeOH/NH4OH (7:3:0.3). The title compound was obtained as a white solid (0.61 g, 78%). MS (ESI, m/z) 504 [M + H]+. ESI-HRMS calcd for C31H29F3NO2 504.2150, found 504.2150 [M + H]+. 1H NMR (400 MHz, MeOD): δ (ppm) = 8.58 (s, 1H), 8.26 (s, 1H), 7.83–7.90 (m, 4H), 7.76–7.79 (m, 1H), 7.69 (d, J = 8.0 Hz, 2H), 7.33–7.37 (m, 4H), 4.35 (q, J = 8.0 Hz, 2H), 3.15–3.19 (m, 2H), 2.73–2.79 (m, 3H), 1.81–1.84 (m, 2H), 1.65–1.69 (m, 2H), 1.35 (t, J = 4.0 Hz, 3H).
Ethyl 4-(4-(1-(Hex-5-yn-1-yl)piperidin-4-yl)phenyl)-7-(4-(trifluoromethyl)phenyl)-2-naphthoate (112)
K2CO3 (0.12 g, 0.9 mmol) was added to a solution of 111 (0.15 g, 0.3 mmol) in dry DMF (15 mL), and the resulting mixture was left stirring for 20 min. Compound 107 (0.06 g, 0.6 mmol) was subsequently added, and the reaction mixture was first stirred at rt for 2h and then at 50 °C for 2.5 h. After cooling down at rt, saturated NaHCO3 solution (15 mL) was added, and the aqueous phase extracted with ethyl acetate (3 × 20 mL). The collected organic fractions were dried over Na2SO4, filtered, and the solvent removed in vacuo. The residue was purified by chromatography using as eluent CH2Cl2/MeOH/NH4OH (9.5:0.5:0.05). The desired compound was obtained as a colorless oil (0.16 g, 92%). MS (ESI, m/z) 584 [M + H]+. ESI-HRMS calcd for C37H37F3NO2 584.2783, found 584.2776 [M + H]+. 1H NMR (400 MHz, CDCl3): δ (ppm) = 8.60 (s, 1H), 8.15 (s, 1H), 7.91–7.95 (m, 2H), 7.71–7.76 (m, 2H), 7.67–7.70 (m, 3H), 7.38 (d, J = 8.0 Hz, 2H), 7.33 (d, J = 8.0 Hz, 2H), 4.38 (q, J = 8.0 Hz, 2H), 3.10–3.13 (m, 2H), 2.56–2.59 (m, 1H), 2.40–2.44 (m, 2H), 2.06–2.17 (m, 5H), 1.90–1.99 (m, 4H), 1.65–1.69 (m, 2H), 1.51–1.55 (m, 2H), 1.38 (t, J = 4.0 Hz, 3H). 13C NMR (100 MHz, CDCl3): δ (ppm) = 166.6, 143.9, 140.5, 137.6, 133.3, 130.6, 130.1, 129.6, 128.0, 127.9, 127.4, 127.1, 127.0, 126.7, 125.9, 84.3, 68.6, 61.3, 58.3, 54.3, 42.3, 33.0, 29.7, 26.5, 25.8, 25.7, 18.4, 14.4.
Ethyl 4-(4-(1-(4-(1-(6-Aminohexyl)-1H-1,2,3-triazol-4-yl)butyl)piperidin-4-yl)phenyl)-7-(4-(trifluoromethyl)phenyl)-2-naphthoate (113)
To a solution of 112 (50 mg, 0.09 mmol) in CH2Cl2:t-BuOH:H2O (1:1:1) (2 mL), compound 110 was added, followed by copper(II) sulfate pentahydrate (15 mol %) and sodium ascorbate (45 mol %). The reaction mixture was stirred for 24 h at rt. The solvents were removed under vacuum and the residue rinsed with 37% ammonia solution (5 mL) and extracted with ethyl acetate (3 × 8 mL). The collected organic fractions were dried over Na2SO4, filtered, and the solvent removed under reduced pressure. The residue was purified by chromatography using as eluent a gradient of CH2Cl2/MeOH/NH4OH (from 9.5:0.5:0.05 to 7:3:0.3). The title product was obtained as a white solid (32 mg, 51%). MS (ESI, m/z) 726 [M + H]+. ESI-HRMS calcd for C43H51F3N5O2 726.3984, found 726.3995 [M + H]+. 1H NMR (400 MHz, CDCl3): δ (ppm) = 8.62 (s, 1H), 8.15 (s, 1H), 7.96–7.99 (m, 2H), 7.73–7.76 m, 2H), 7.66–7.70 (m, 3H), 7.38–7.40 (m, 2H), 7.30–7.33 (m, 2H), 7.21–7.23 (m, 1H), 4.38 (q, J = 8.0 Hz, 2H), 4.23–4.26 (m, 2H), 3.09–3.17 (m, 2H), 2.65–2.71 (m, 6H), 2.39–2.42 (m, 3H), 2.04–2.07 (m, 2H), 1.83–1.88 (m, 6H), 1.58–1.65 (m, 6H), 1.35–1.39 (m, 7H).
4-(4-(1-(4-(1-(6-Aminohexyl)-1H-1,2,3-triazol-4-yl)butyl)piperidin-4-yl)phenyl)-7-(4-(trifluoromethyl)phenyl)-2-naphthoic Acid (114)
To a solution of 113 (32 mg, 44 μmol) in MeOH (1 mL), a solution of 0.5 M LiOH (1 mL) was added and the resulting mixture was heated at 60 °C and stirred overnight. After cooling, the solvents were evaporated and the residue purified by preparative HPLC (Rt = 31.08 min). The product was obtained as a white solid by freeze-drying (6.5 mg, 21%). MS (ESI, m/z) 698 [M + H]+. ESI-HRMS calcd for C41H47F3N5O2 698.3690, found 698.3682 [M + H]+. 1H NMR (400 MHz, MeOD): δ (ppm) = 8.49 (s, 1H), 8.27 (s, 1H), 7.89–7.94 (m, 4H), 7.70–7.72 (m, 4H), 7.66–7.70 (m, 3H), 7.38 (d, J = 8.0 Hz, 2H), 7.32 (d, J = 8.0 Hz, 2H), 4.30 (t, J = 8.0 Hz, 2H), 3.09–3.12 (m, 2H), 2.79 (t, J = 8.0 Hz, 2H), 2.66–2.70 (m, 3H), 2.46–2.50 (m, 2H), 2.19–2.25 (m, 2H), 1.81–1.86 (m, 6H), 1.52–1.64 (m, 6H), 1.26–1.33 (m, 4H).
Pharmacological Assays
Procedures for functional assays at the P2Y14R reported previously were followed.14,15Ki values were calculated from IC50 values using the Cheng–Prusoff equation.30
Fluorescent Ligand Binding
Fluorescent ligand binding experiments were performed using FCM for all the final triazole derivatives. An initial experiment involved screening of the compounds at a single concentration (3 μM) to determine the percentage inhibition, followed by a full concentration–response curve for the more potent compounds.
For FCM binding experiments in hP2Y14R-CHO cells using 4 as fluorescent tracer,15 CHO cells expressing hP2Y14Rs were grown in 12-well plates and used for the assay when the cells were 80% confluent. CHO cells were incubated with a test compound (nonfluorescent antagonist) for 30 min, followed by continued incubation for 30 min after the addition of 20 nM of fluorescent antagonist 4. After the incubation, the cells were prepared for FCM analysis as described below. Briefly, cells were washed 3 times with ice-cold PBS and detached with 0.2% EDTA, which was neutralized with media after 5 min incubation at 37 °C. Cell suspensions were transferred to polystyrene round-bottom BD Falcon tubes (BD, Franklin Lakes, NJ), and centrifuged twice at 400g for 5 min. Cells were then suspended in PBS and proceeded to FCM using a FACSCalibur flow cytometer (BD, Franklin Lakes, NJ), with a 635 nm laser. MFIs were recorded in FL-1 channel in log mode. All data were analyzed by nonlinear least-squares analysis, and Ki values were calculated using Prism (GraphPad Software, San Diego, CA) for all assays.
To quantify the number of receptor-bound ligands, we used quantitative fluorescence calibration. To convert measured fluorescence intensity (MFI) values into molecules of equivalent soluble fluorochrome (MESF) values, we used Quantum Alexa fluor 488 MESF calibration beads and QuickCal program v. 2.3 (Bangs Laboratories, Inc., Fishers, IN) according to the instructions of the manufacturer. The detailed procedure is explained in the Supporting Information (Figure S3).
Compounds 79, 82, 95, and 101 displayed low solubility in DMSO, and therefore their 5 mM DMSO-solutions were heated to 50 °C for 5 min prior to the dilution step. To confirm the physicochemical stability of the derivatives, 1H NMR spectra were taken before and after heating, and no change in the 1H NMR was observed.
Quantification of cAMP Accumulation
cAMP accumulation in cells was quantified by competitive enzyme-linked immunoassay. Cells were plated in 12-well plates 24 h before the assay, and 30 min before the assay the cells were washed with PBS and the medium was changed to 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-buffered serum-free DMEM. The assays were performed in the presence of 10 μM rolipram and were initiated by the addition of 30 μM forskolin, agonist and/or antagonists.14 Incubation was for 15 min at 37 °C and were terminated by washing the cells twice with ice-cold PBS, and the cells were lysed using 100 μL of cell lysis buffer (Cell Signaling, Danvers, MA). Then 50 μL of the cell lysate were used for the quantification of cAMP using a Cyclic AMP XP assay kit (Cell Signaling, Danvers, MA) following manufacturer’s instructions.
Molecular Modeling
Homology Model Refinement
Compound 3 was built using Maestro and prepared with the Ligprep module31 using the OPLS-2005 force field.32 The ligand was docked to the previously obtained UDP-bound MD-refined hP2Y14R homology model16 by means of the Induced Fit Docking (IFD) procedure based on Glide search algorithm and SP scoring function.33 In particular, a Glide grid was positioned on the centroid of residues located within 5 Å from the previously identified cavity.16 The Glide grid was built using an inner box of 10 Å × 10 Å × 10 Å and an outer box that extended 20 Å in each direction around the inner box. The ligand was docked rigidly, and side chains within 5 Å of the ligand were refined. The top ranked docking pose was then subjected to 10 ns of MD simulations and the final trajectory snapshot of the receptor used as a template to redock compound 3 and to dock compounds 11 and 65 along with its structural analogues.
Docking of Compounds 3, 11, and 65
Ligands were built using Maestro and prepared using the Ligprep module31 and OPLS_2005 force field.32 Molecular docking of ligands to the 3-bound hP2Y14R MD-refined model was performed by means of the Glide software package34 from the Schrodinger suite, using the XP scoring function.35 The binding site was centered on the ligand barycenter and a Glide grid was computed (inner box side = 10 Å; outer box side = 30 Å). The top ranked docking poses for each ligand were subjected to 30 ns of membrane MD simulations.
Molecular Dynamics
The setup of the MD simulation was performed as previously described.16 In brief, the hP2Y14R homology model was uploaded to the “Orientations of Proteins in Membranes (OPM)” server36 and a suggested orientation was provided based on the 2MeSADP-bound hP2Y12R orientation (PDB: 4PXZ).18 The model was then positioned in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayer (70 Å × 70 Å) generated by a grid-based method using the VMD37 Membrane Plugin tool, and overlapping lipids within 0.6 Å were removed upon combining the protein and the membrane system. Each protein–membrane system was then solvated with TIP3P water using the Solvate 1.0 VMD37 Plugin tool and neutralized by 0.154 M Na+/Cl– counterions.
High-performance computational capabilities of the Biowulf Linux GPU cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov) were exploited to run MD simulations with periodic boundary conditions using the Nanoscale Molecular Dynamics (NAMD) software package38 and the CHARMM36 Force Field.39,40 The ligands were parametrized by analogy using the ParamChem service (1.0.0) and implementing the CHARMM General Force Field for organic molecules (3.0.1).41,42 A 10000-step conjugate gradient minimization was initially performed to minimize steric clashes. The protein and ligand atoms were kept fixed during an initial 8 ns equilibration of the lipid and water molecules. Atom constraints were then removed, and the entire system was allowed to equilibrate. The temperature was maintained at 300 K using a Langevin thermostat with a damping constant of 3 ps–1. The pressure was maintained at 1 atm using a Berendsen barostat. An integration time step of 1 fs was used, while hydrogen–oxygen bond lengths and hydrogen–hydrogen angles in water molecules were constrained using the SHAKE algorithm.43 VMD 1.937 was used for trajectory analysis and movie making. Each structure was simulated for 30 ns without constraints. RMSD plots for ligand atoms during the 30 ns trajectories were used to compare relative ligand stability in the binding pocket during the simulation.
Docking of Triazole Analogues
A library of 57 triazole compounds was designed and screened using the 3-refined P2Y14 model as receptor template. The binding pose of 3 was set as a constraint for the docking, including exposure of the positively charged piperidine ring toward the solvent and 2-naphthoic acid carbonyl interactions with Lys772.60 and Lys2777.35. Molecular docking of ligands to the hP2Y14R MD-refined model was performed by means of the Glide package,34 using the SP scoring function. The ligand binding site was defined as above-described. Protein interaction sites were calculated by means of the SiteMap package44 as implemented in the Schrodinger suite on the apo form of the hP2Y14R MD-refined model by generating at least 15 points per site with the OPLS_2005 force field.32 The top ranked site was used to select the compounds according to optimal overlap between hydrophobic and hydrogen bond donor/acceptor protein sites and ligand functional groups evaluated by visual inspection.
Acknowledgments
We acknowledge support from the NIDDK Intramural Research Program (ZIA DK031116-28) and technical assistance of Ryan Surujdin and Jinha Yu (NIDDK). Financial support of Anna Junker by the Deutsche Forschungsgemeinschaft (German Research Foundation) is gratefully acknowledged. We thank Bryan L. Roth (University of North Carolina at Chapel Hill) and National Institute of Mental Health’s Psychoactive Drug Screening Program (contract no. HHSN-271-2008-00025-C) for screening data. We thank Rolf Swenson and Zhen-Dan Shi (Imaging Probe Development Center, NHLBI) for reagents 115 and 116.
Glossary
Abbreviations Used
- AF488
Alexa Fluor 488
- CHO
Chinese hamster ovary
- DMEM
Dulbecco’s Modified Eagle’s Medium
- DMF
N,N-dimethylformamide
- DPPF
1,1′-bis(diphenylphosphino)ferrocene
- ECL
extracellular loop
- EDC
N-ethyl-N′-dimethylaminopropylcarbodiimide
- HATU
1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-bpyridinium 3-oxide hexafluorophosphate
- PLC
phospholipase C
- PPTN
4-(4-(piperidin-4-yl)-phenyl)-7-(4-(trifluoromethyl)-phenyl)-2-naphthoic acid
- RMSD
root-mean-square deviation
- SAR
structure–activity relationship
- UDPG
uridine-5′-diphosphoglucose
- THF
tetrahydrofuran
- TM
transmembrane helix
- TSTU
N,N,N′,N′-tetramethyl-O-(N-succinimidyl)-uronium tetrafluoroborate
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.6b00044.
3D coordinates of the hP2Y14R MD-refined model (PDB)
3D coordinates of hP2Y14R in complex with 3 (PDB)
3D coordinates of hP2Y14R in complex with 11 (PDB)
3D coordinates of hP2Y14R in complex with 65 (PDB)
Video of MD simulations of hP2Y14R in complex with 3 (AVI)
Video of MD simulations of hP2Y14R in complex with 11 (AVI)
Video of MD simulations of hP2Y14R in complex with 65 (AVI)
Supplementary modeling figures (comparison with agonist- and antagonist-bound hP2Y14R homology models) and tables (structures of 57 initially docked triazoles, docking poses of selected compounds, and overlap with protein interaction sites), Schemes S1 and S2 and synthetic procedures, and NMR and mass spectra of selected compounds (PDF) (PDF)
Molecular formula strings (CSV)
Author Contributions
A.J., E.U., and E.K. contributed equally
The authors declare no competing financial interest.
Supplementary Material
References
- Lazarowski E. R.; Harden T. K. Signalling and pharmacological properties of the P2Y14 receptor. Mol. Pharmacol. 2015, 88, 151–160. 10.1124/mol.115.098756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbracchio M. P.; Burnstock G.; Boeynaems J. M.; Barnard E. A.; Boyer J. L.; Kennedy C.; Fumagalli M.; King B. F.; Gachet C.; Jacobson K. A.; Weisman G. A. International Union of Pharmacology LVIII: update on the P2Y G protein-coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacol. Rev. 2006, 58, 281–341. 10.1124/pr.58.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K.; Yamanaka H.; Yanamoto F.; Okubo M.; Noguchi K. Multiple P2Y subtypes in spinal microglia are involved in neuropathic pain after peripheral nerve injury. Glia 2012, 60, 1529–1539. 10.1002/glia.22373. [DOI] [PubMed] [Google Scholar]
- Sesma J. I.; Kreda S. M.; Steinckwich-Besancon N.; Dang H.; García-Mata R.; Harden T. K.; Lazarowski E. R. The UDP-sugar-sensing P2Y14 receptor promotes Rho-mediated signaling and chemotaxis in human neutrophils. Am. J. Physiol. - Cell Physiol. 2012, 303, C490–C498. 10.1152/ajpcell.00138.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z.-G.; Ding Y.; Jacobson K. A. UDP-glucose acting at P2Y14 receptors is a mediator of mast cell degranulation. Biochem. Pharmacol. 2010, 79, 873–879. 10.1016/j.bcp.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azroyan A.; Cortez-Retamozo V.; Bouley R.; Liberman R.; Ruan Y. C.; Kiselev E.; Jacobson K. A.; Pittet M. J.; Brown D.; Breton S. Renal intercalated cells sense and mediate sterile inflammation via the P2Y14 receptor. PLoS One 2015, 10 (3), e0121419. 10.1371/journal.pone.0121419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Morinaga H.; Oh D.; Li P.; Chen A.; Talukdar S.; Lazarowski E.; Olefsky J. M.; Kim J. J. GPR105 Ablation prevents inflammation and improves insulin sensitivity in mice with diet-induced obesity. J. Immunol. 2012, 189, 1992–1999. 10.4049/jimmunol.1103207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister J.; Le Duc D.; Ricken A.; Burkhardt R.; Thiery J.; Pfannkuche H.; Polte T.; Grosse J.; Schöneberg T.; Schulz A. The G protein-coupled receptor P2Y14 influences insulin release and smooth muscle function in mice. J. Biol. Chem. 2014, 289, 23353–23366. 10.1074/jbc.M114.580803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M.; Nasu-Tada K.; Fujishita K.; Sato K.; Koizumi S. Secretion of matrix metalloproteinase-9 from astrocytes by inhibition of tonic P2Y14-receptor-mediated signal(s). Cell. Mol. Neurobiol. 2013, 33, 47–58. 10.1007/s10571-012-9869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belley M.; Deschenes D.; Fortin R.; Fournier J.-F.; Gagne S.; Gareau Y.; Gauthier J. Y.; Li L.; Robichaud J.; Therien M.; Tranmer G. K.; Wang Z.. Substituted 2-naphthoic acids as antagonists of GFPR105 activity. WO2009070873 A1, June 11, 2009.
- Gauthier J. Y.; Belley M.; Deschênes D.; Fournier J.-F.; Gagné S.; Gareau Y.; Hamel M.; Hénault M.; Hyjazie H.; Kargman S.; Lavallée G.; Levesque J. F.; Li L.; Mamane Y.; Mancini J.; Morin N.; Mulrooney E.; Robichaud J.; Thérien M.; Tranmer G.; Wang Z.; Wu J.; Black W. C. The identification of 4,7-disubstituted naphthoic acid derivatives as UDP-competitive antagonists of P2Y14. Bioorg. Med. Chem. Lett. 2011, 21, 2836–2839. 10.1016/j.bmcl.2011.03.081. [DOI] [PubMed] [Google Scholar]
- Guay D.; Beaulieu C.; Belley M.; Crane S. N.; DeLuca J.; Gareau Y.; Hamel M.; Henault M.; Hyjazie H.; Kargman S.; Chan C. C.; Xu L.; Gordon R.; Li L.; Mamane Y.; Morin N.; Mancini J.; Thérien M.; Tranmer G.; Truong V. L.; Wang Z.; Black W. C. Synthesis and SAR of pyrimidine-based, non-nucleotide P2Y14 receptor antagonists. Bioorg. Med. Chem. Lett. 2011, 21, 2832–2835. 10.1016/j.bmcl.2011.03.084. [DOI] [PubMed] [Google Scholar]
- Robichaud J.; Fournier J.-F.; Gagné S.; Gauthier J. Y.; Hamel M.; Han Y.; Hénault M.; Kargman S.; Levesque J.-F.; Mamane Y.; Mancini J.; Morin N.; Mulrooney E.; Wu J.; Black W. C. Applying the pro-drug approach to afford highly bioavailable antagonists of P2Y14. Bioorg. Med. Chem. Lett. 2011, 21, 4366–4368. 10.1016/j.bmcl.2010.12.113. [DOI] [PubMed] [Google Scholar]
- Barrett M. O.; Sesma J. I.; Ball C. B.; Jayasekara P. S.; Jacobson K. A.; Lazarowski E. R.; Harden T. K. A selective high-affinity antagonist of the P2Y14 receptor inhibits UDP-glucose–stimulated chemotaxis of human neutrophils. Mol. Pharmacol. 2013, 84, 41–49. 10.1124/mol.113.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev E.; Barrett M.; Katritch V.; Paoletta S.; Weitzer C. D.; Brown K. A.; Hammes E.; Yin A. L.; Zhao Q.; Stevens R. C.; Harden T. K.; Jacobson K. A. Exploring a 2-naphthoic acid template for the structure-based design of P2Y14 receptor antagonist molecular probes. ACS Chem. Biol. 2014, 9, 2833–2842. 10.1021/cb500614p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo K.; Paoletta S.; Kiselev E.; Jacobson K. A. Molecular modeling of the human P2Y14 receptor: A template for structure-based design of selective agonist ligands. Bioorg. Med. Chem. 2015, 23, 4056–4064. 10.1016/j.bmc.2015.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.; Zhang J.; Gao Z. G.; Zhang D.; Zhu L.; Han G. W.; Moss S. M.; Paoletta S.; Kiselev E.; Lu W.; Fenalti G.; Zhang W.; Müller C. E.; Yang H.; Jiang H.; Cherezov V.; Katritch V.; Jacobson K. A.; Stevens R. C.; Wu B.; Zhao Q. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 2014, 509, 115–118. 10.1038/nature13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.; Zhang J.; Gao Z. G.; Paoletta S.; Zhang D.; Han G. W.; Li T.; Ma L.; Zhang W.; Müller C. E.; Yang H.; Jiang H.; Cherezov V.; Katritch V.; Jacobson K. A.; Stevens R. C.; Wu B.; Zhao Q. Agonist-bound structure of the human P2Y12R receptor. Nature 2014, 509, 119–122. 10.1038/nature13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosh D. K.; Paoletta S.; Chen Z.; Crane S.; Lloyd J.; Gao Z. G.; Gizewski E. T.; Auchampach J. A.; Salvemini D.; Jacobson K. A. Structure-based design, synthesis by click chemistry and in vivo activity of highly selective A3 adenosine receptor agonists. MedChemComm 2015, 6, 555–563. 10.1039/C4MD00571F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros J. A.; Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- Chinchilla R.; Najera C. The Sonogashira reaction: A booming methodology in synthetic organic chemistry. Chem. Rev. 2007, 107 (3), 874–922. 10.1021/cr050992x. [DOI] [PubMed] [Google Scholar]
- Suzuki A. Cross-coupling reactions of organoboranes: An easy way to construct C-C bonds (Nobel Lecture). Angew. Chem., Int. Ed. 2011, 50 (30), 6722–6737. 10.1002/anie.201101379. [DOI] [PubMed] [Google Scholar]
- Ishiyama T.; Murata M.; Miyaura N. Palladium(0)-catalyzed cross-coupling reaction of alkoxydiboron with haloarenes: A direct procedure for arylboronic esters. J. Org. Chem. 1995, 60, 7508–7510. 10.1021/jo00128a024. [DOI] [Google Scholar]
- Kolb H. C.; Finn M. G.; Sharpless K. B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem., Int. Ed. 2001, 40, 2004–2021. . [DOI] [PubMed] [Google Scholar]
- Kutonova K. V.; Trusova M. E.; Postnikov P. S.; Filimonov V. D.; Parello J. A simple and effective synthesis of aryl azides via arenediazonium tosylates. Synthesis 2013, 45 (19), 2706–2710. 10.1055/s-0033-1339648. [DOI] [Google Scholar]
- Himo F.; Lovell T.; Hilgraf R.; Rostovtsev V. V.; Noodleman L.; Sharpless K. B.; Fokin V. V. Copper(I)-catalyzed synthesis of azoles. DFT study predicts unprecedented reactivity and intermediates. J. Am. Chem. Soc. 2005, 127 (1), 210–216. 10.1021/ja0471525. [DOI] [PubMed] [Google Scholar]
- a Brunet A.; Aslam T.; Bradley M. Separating the isomers—Efficient synthesis of the N-hydroxysuccinimide esters of 5 and 6-carboxyfluorescein diacetate and 5 and 6-carboxyrhodamine B. Bioorg. Med. Chem. Lett. 2014, 24, 3186–3188. 10.1016/j.bmcl.2014.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mitronova G. Y.; Belov V. N.; Bossi M. L.; Wurm C. A.; Meyer L.; Medda R.; Moneron G.; Bretschneider S.; Eggeling C.; Jakobs S.; Hell S. W. New fluorinated rhodamines for optical microscopy and nanoscopy. Chem. - Eur. J. 2010, 16, 4477–4488. 10.1002/chem.200903272. [DOI] [PubMed] [Google Scholar]
- Besnard J.; Ruda G. F.; Setola V.; Abecassis K.; Rodriguiz R. M.; Huang X. P.; Norval S.; Sassano M. F.; Shin A. I.; Webster L. A.; Simeons F. R.; Stojanovski L.; Prat A.; Seidah N. G.; Constam D. B.; Bickerton G. R.; Read K. D.; Wetsel W. C.; Gilbert I. H.; Roth B. L.; Hopkins A. L. Automated design of ligands to polypharmacological profiles. Nature 2012, 492, 215–220. 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. V. E.; Blundell T. L.; Ascher D. B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. C.; Prusoff W. H. Relationship between inhibition constant (K1) and concentration of inhibitor which causes 50% inhibition (I50) of an enzymatic-reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- LigPrep, version 3.1; Schrödinger, LLC: New York, 2014.
- Banks J. L.; Beard H. S.; Cao Y.; Cho A. E.; Damm W.; Farid R.; Felts A. K.; Halgren T. A.; Mainz D. T.; Maple J. R.; Murphy R.; Philipp D. M.; Repasky M. P.; Zhang L. Y.; Berne B. J.; Friesner R. A.; Gallicchio E.; Levy R. M. Integrated modeling program, applied chemical theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Induced Fit Docking Protocol; Glide version 6.4; Schrödinger, LLC: New York, 2014;; Prime version 3.7; Schrödinger, LLC: New York, 2014.
- Glide, version 6.4; Schrödinger, LLC: New York, 2014.
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Lomize M. A.; Lomize A. L.; Pogozheva I. D.; Mosberg H. I. OPM: orientations of proteins in membranes database. Bioinformatics 2006, 22, 623–625. 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD - visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R. B.; Zhu X.; Shim J.; Lopes P. E. M.; Mittal J.; Feig M.; MacKerell A. D. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; Hatcher E.; Acharya C.; Kundu S.; Zhong S.; Shim J.; Darian E.; Guvench O.; Lopes P.; Vorobyov I.; Mackerell A. D. Jr. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; MacKerell A. D. Jr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; Raman E. P.; MacKerell A. D. Jr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J. P.; Ciccotti G.; Berendsen H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- SiteMap, version 3.2; Schrödinger, LLC: New York, 2014.
- Trujillo K.; Balasubramanian R.; Uliassi E.; Brown K. A.; Trujillo K.; Katritch V.; Hammes E.; Stevens R. C.; Harden T. K.; Jacobson K. A. Design, synthesis and pharmacological characterization of a fluorescent agonist of the P2Y14 receptor. Bioorg. Med. Chem. 2015, 25, 4733–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.