Abstract
In spite of its prevalence in animals and plants, endogenous nitric oxide (NO) has been rarely reported in fungi. We present here our observations on production of intracellular NO and its possible roles during development of Neurospora crassa, a model filamentous fungus. Intracellular NO was detected in hypha 8–16 hours after incubation in Vogel’s minimal liquid media and conidiophores during conidiation using a fluorescent indicator (DAF-FM diacetate). Treatment with cPTIO, an NO scavenger, significantly reduced fluorescence levels and hindered hyphal growth in liquid media and conidiation, whereas exogenous NO enhanced hyphal extension on VM agar media and conidia formation. NO scavenging also dramatically diminished transcription of con-10 and con-13, genes preferentially expressed during conidiation. Our results suggest that intracellular NO is generated in young hypha growing in submerged culture and during conidia development and regulate mycelial development and conidia formation.
Microorganisms endogenously produce reactive oxygen and nitrogen species, although reactive species have anti-microbial effects1,2,3. Indeed, regulatory roles of reactive oxygen species such as superoxide (O2·−) and hydrogen peroxide (H2O2) in cell differentiation have been demonstrated in many microorganisms4,5,6. Comparably, reactive nitrogen species in microbial cells have been explored infrequently, but their involvement in a wide variety of cellular functions has been recently unveiled7,8,9. Studies show that nitric oxide (NO), known as a ubiquitous signaling molecule in mammalian and plant cells, can be endogenously synthesized and is involved in the regulation of many processes in microbial cells8,9,10,11,12,13,14,15. Although NO activities identified in microbial cells are somewhat similar to those found in animals and plants, differences in origin and functions are also known.
In many bacteria, NO is synthesized from L-arginine and oxygen by the catalytic activity of nitric oxide synthases (NOSs), which are structurally different from mammalian NOSs14,16. Mammalian NOS has three isoforms, neuronal NOS (nNOS), inducible NOS (iNOS), and endothelial NOS (eNOS), and all forms have N-terminal catalytic oxygenase domains and C-terminal reductase domains17. In contrast, most bacterial NOSs are structurally simple consisting of an oxygenase domain, with some exceptions possessing additional domains18. While endogenously produced NO in mammals and plants is involved in neurotransmission, vasodilation, and regulation of abiotic and biotic stresses, NO synthesized by bacterial NOS plays roles in alleviation of oxidative stress and UV damage, transcriptional regulation, and biosynthesis of nitrated compounds16,19,20.
Compared to bacteria, the biosynthesis and functions of endogenous NO in fungi have not been thoroughly explored. Using NO donors, mammalian NOS inhibitors, and NO scavengers, production and cellular functions of NO were indirectly demonstrated in fungi such as Blastocladiella emersonii, Candida tropicalis, Collectotrichum coccodes, Coniothyrium minitans, Flammulina velutipes, Neurospora crassa, Phycomyces blakesleeanus, Physarum polycephalum, and Saccharomyces cerevisiae7,9,10,12,21,22,23,24,25. Recently, intracellular NO has been directly detected using a fluorescent indicator in several fungi such as Aspergillus nidulans and Magnaporthe oryzae8,26,27. In these fungi, NO regulates conidiation, fruiting body formation, pseudomycelial formation, conidial germination, and infection. Compared to mammals, plants, and bacteria, NO in fungi shared similar roles during cell differentiation but also possess distinct functions in fungal life. Particularly, synthesis of NO in fungi does not seem to follow NOS dependent pathways, as shown in animals, plants, and bacteria. Despite the discovery of NOS like activities in several fungi, evidence for fungal NOS is still limited8,12,25,28.
As eukaryotic microorganisms, fungi share many common cellular processes with other eukaryotic organisms, but NO biology in fungi seems to be different in terms of mode of production and biological functions. In addition, evidence for the direct detection of NO in fungal cells is few, and the origin of intracellular NO and its functions are still unknown in many fungi. In this study, we analyzed intracellular production and functions of NO during fungal development using a model filamentous fungus, N. crassa. Recent work in N. crassa indirectly demonstrated the presence of NO and showed that NOS inhibitors enhanced conidiation and exogenous NO inhibited light induced conidiation21. However, the majority of NO biology in N. crassa is still unknown due to lack of direct detection of intracellular NO and lack of NOS like genes in the N. crassa genome. Therefore, our study was designed to elucidate endogenous generation and biological functions of NO in N. crassa.
Results
Intracellular NO is elevated during hyphal growth and conidia formation
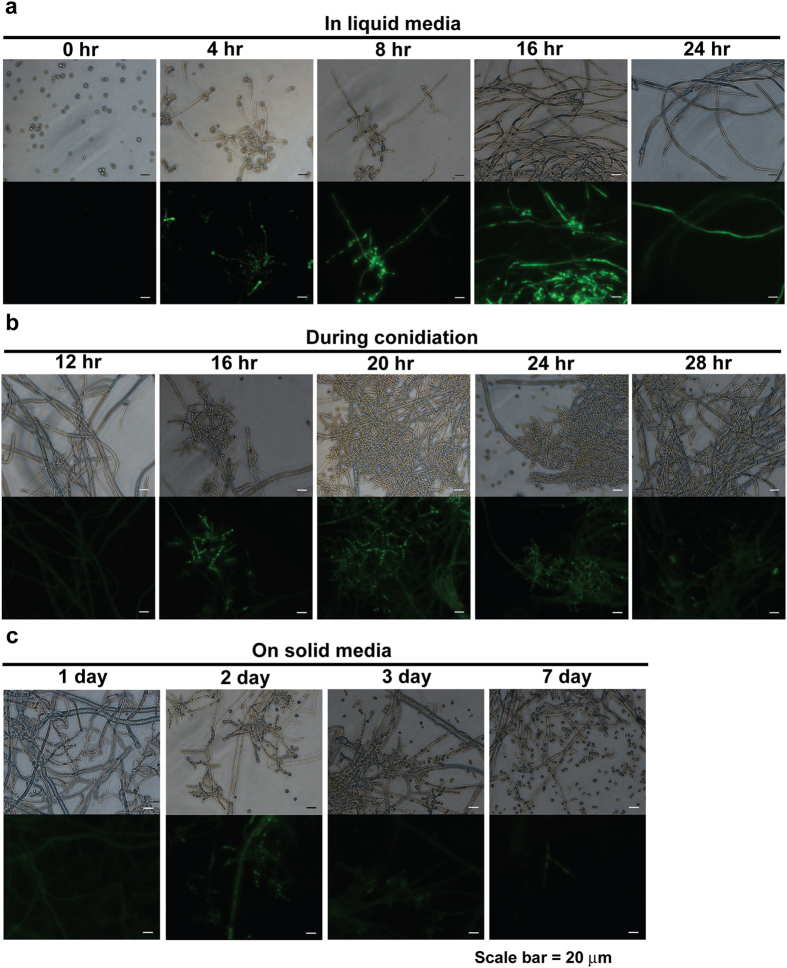
Intracellular NO levels were measured using a cell permeable fluorescent indicator, DAF-FM diacetate. After DAF-FM diacetate enters a cell, the acetyl group is hydrolyzed by intracellular esterase and DAF-FM, which is not fluorescent, becomes benzotriazole and fluoresces when it reacts with NO29. Fluorescent intensity therefore indicates the level of intracellular NO. Using this method, fluorescence detected by DAF-FM diacetate was increased in fungal hypha grown for 8–16 hours in VM (Vogel’s Minimal) liquid (Fig. 1a) and in conidiophores after 16–20 hours on VM agar plates, used to induce synchronized aerial hypha formation and conidiophore development (Fig. 1b). When spores were inoculated on VM agar plates and incubated, higher NO levels were observed in a 2 day-old culture (Fig. 1c). Development of conidiophores was more prominent after 2 days on VM agar plates, and the majority of fluorescence appeared in conidiophores (Fig. 1c).
Figure 1. NO production during fungal development.
Intracellular NO detected using DAF-FM diacetate during (a) vegetative growth in VM liquid, (b) conidiation on VM agar media, and (c) on VM agar media. Top and bottom of each picture indicate images captured in optical and fluorescent (465–495 nm excitation filter) mode, respectively.
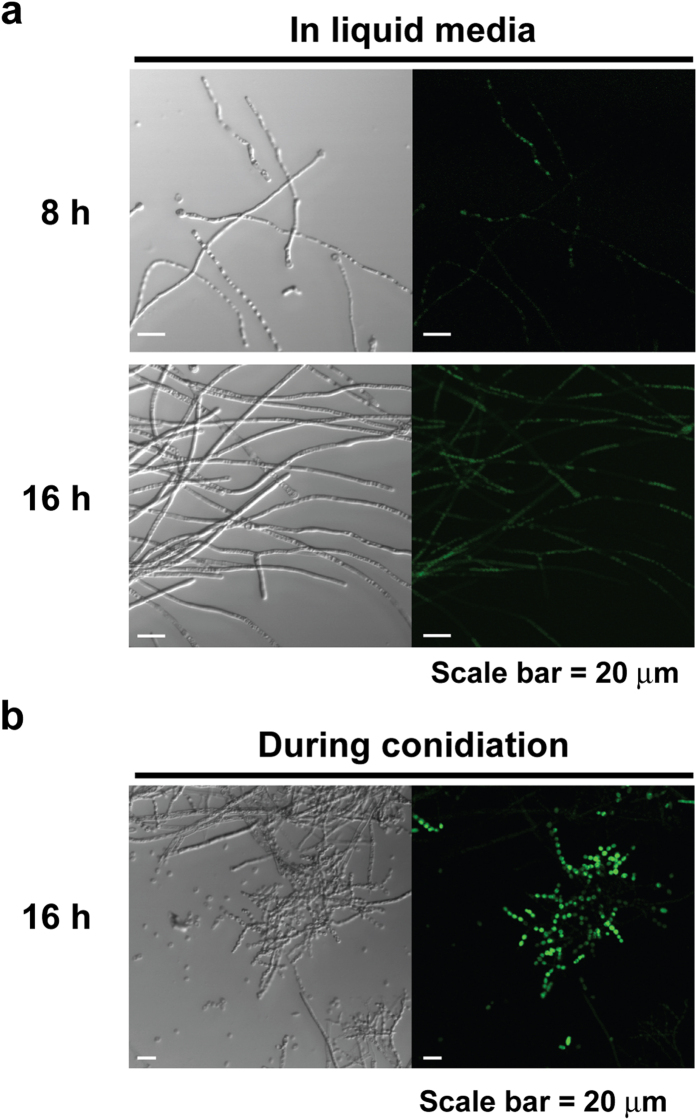
To identify the location of intracellular NO, fungal samples were further analyzed using confocal microscopy. This analysis showed that high fluorescence detected in 8 and 16 hour VM liquid cultures was mostly in vacuole like structures (Fig. 2a). In conidiation on VM agar media incubated for 16 hours, areas containing conidiophores were highly fluorescent compared to hypha (Fig. 2b).
Figure 2. Intracellular NO analyzed by confocal microscopy.
Intracellular NO detected by DAF-FM diacetate was observed in fungal hypha grown in VM liquid for (a) 8 and 16 hours and (b) in fungus during conidiation. Left and right of each picture indicate images captured in optical and fluorescent (465–495 nm excitation filter) mode, respectively.
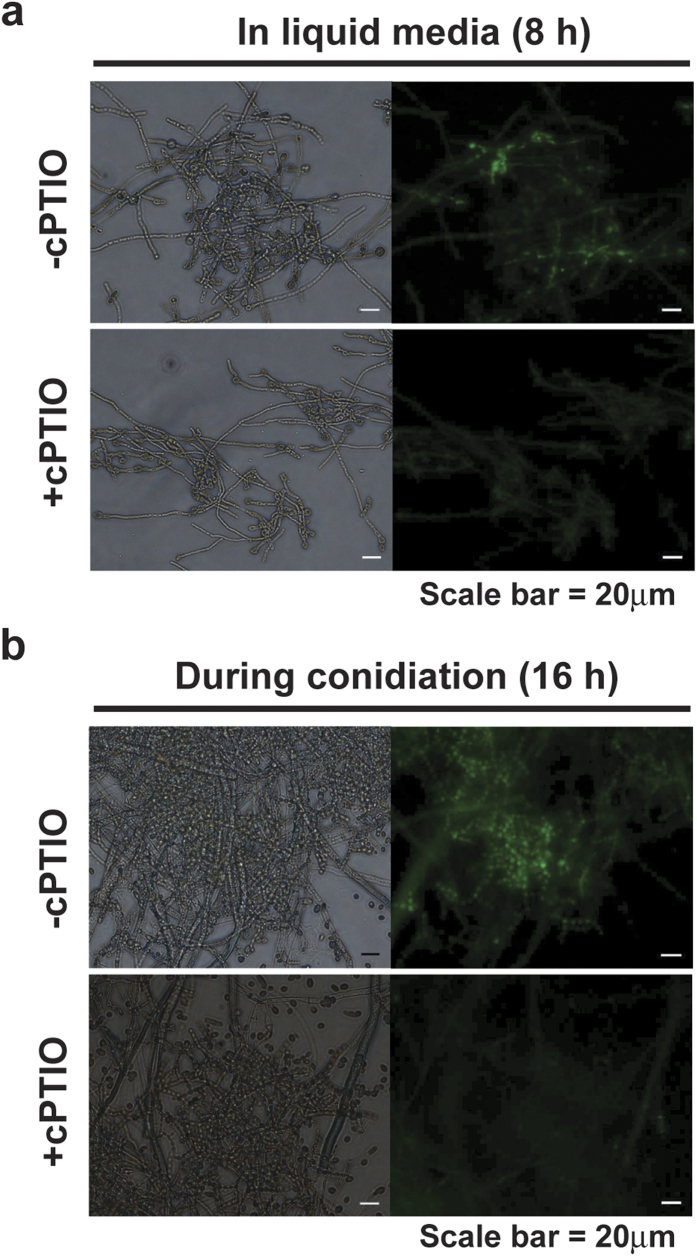
We treated fungi with an NO scavenger, cPTIO (2-(4-carboxyphenyl)-4,5-dihydro-4,4,5,5-tetramethy-1 H-imidazolyl-1-oxy-3-oxide), to confirm that fluorescence measured by DAF-FM was due to NO. As shown in Fig. 3a, bright fluorescence was exhibited in fungal hypha grown in VM liquid for 8 hours, whereas much lower levels of fluorescence were detected in fungal hypha after treatment with cPTIO (Fig. 3a). Fluorescence observed in conidiophores after 16 hours in conidiation induction culture was also significantly decreased after cPTIO treatment (Fig. 3b). These results show that removal of NO by cPTIO causes reduced fluorescence, therefore indicating the presence of NO.
Figure 3. Effects of cPTIO on intracellular NO levels.
(a) Intracellular NO detected by DAF-FM diacetate in hypha grown for 8 hours in VM liquid after treatment with or without 10 mM cPTIO for 20 minutes. (b) Intracellular NO in fungus incubated for 16 hours after conidiation was induced, with or without 10 mM cPTIO. Left and right of each picture indicate images captured in optical and fluorescent (465–495 nm excitation filter) mode, respectively.
NO scavenger treatment hinders hyphal growth and slows conidiation
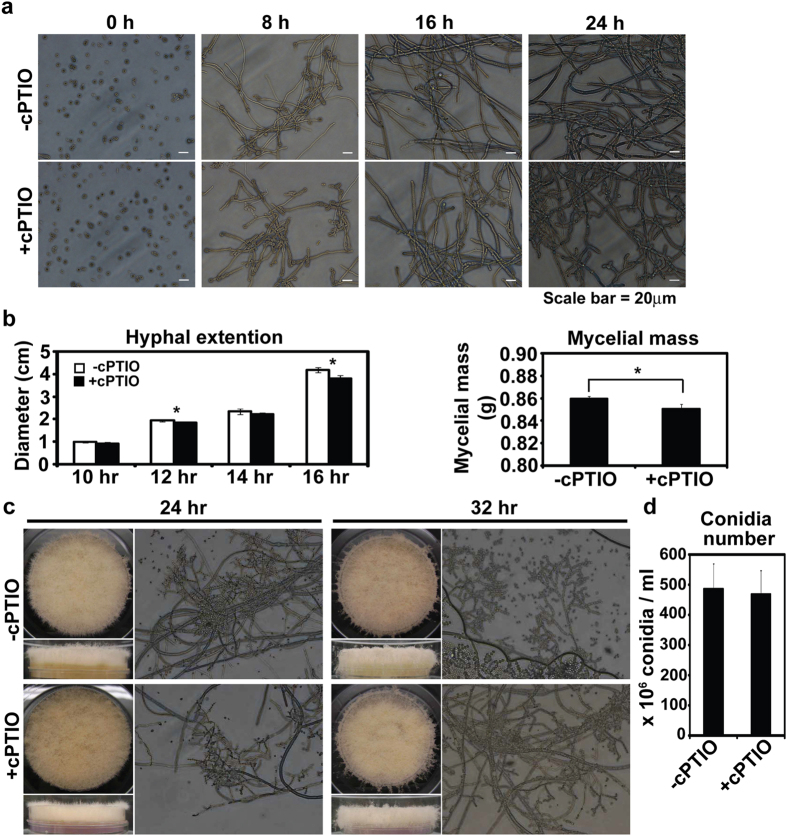
Because intracellular NO was increased in fungal hypha and conidiophores, we hypothesized that NO plays a role in regulating hyphal development and conidiation. To test this hypothesis, fungal development was monitored after NO was removed using an NO scavenger. In VM liquid culture, the presence of cPTIO modestly inhibited hyphal branching as observed under microscope (Fig. 4a). When cPTIO was added to VM agar plates (60 mm in diameter), basal hyphal extensions were significantly reduced compared to control after 16 hours (Fig. 4b). The dry weight of hyphal mass collected after 24 hours was also significantly smaller in cPTIO treated samples than control (Fig. 4b). On cPTIO treated VM agar, development of conidiophores was delayed compared to control (Fig. 4c), but aerial hyphal development of mycelia was not different. More conidiophores were observed on control than cPTIO treated VM agar plates after 24 hours (Fig. 4c), and, after 32 hours, conidia were released from conidiophores in control while cPTIO treated conidiophores were still holding conidia (Fig. 4c). Conidia were harvested after 5 days, revealing that the number of conidia per ml suspension was slightly decreased in cPTIO treated samples compared to control, although the difference was not significant (Fig. 4d).
Figure 4. Effects of cPTIO on fungal development.
(a) Hyphal growth in VM liquid with or without 10 mM cPTIO. (b) Hyphal extension on VM agar media (left panel) and weight of mycelia grown in VM liquid (right panel) in the presence of cPTIO. All values represent mean ± standard deviation of 3 replicate measurements. (c) Conidiation synchronously induced on VM agar media after treatment with or without cPTIO. (d) Number of conidia collected from 5 day-old cultures after cPTIO treatment. All values represent mean ± standard deviation of 3 replicate measurements.
NO scavenger treatment decreases transcription of conidiation related genes
During development of aerial hypha and conidiophores from fungal mycelia placed on VM agar, we observed that transcription of several con genes (known as preferentially expressed during conidiation) was increased (Fig. 5a). Among 9 putative genes involved in the regulation of conidiation (acon-2, acon-3, con-3, con-6, con-8, con-10, con-13, fluffy, ccg-2), the level of con-6, con-10, con-13, and ccg-2 transcripts was significantly increased compared to the other genes 16 hours after conidiation was induced (Fig. 5a). After 24 hours, transcript levels of these 4 genes decreased, although levels were still higher than those of other genes (Fig. 5a). When fungus was treated with the NO scavenger, cPTIO, a significant reduction in transcription of con-10, con-13, and ccg-2 was observed 16 hours after conidiation was induced compared to the control (Fig. 5b). Particularly, transcript level of con-13 after cPTIO treatment was much lower than that of control (Fig. 5b). These results indicate that removal of NO by cPTIO suppressed the expression of these 3 genes that are specifically associated with conidial development and maturation.
Figure 5. Transcription of con genes during conidiation after cPTIO treatment.
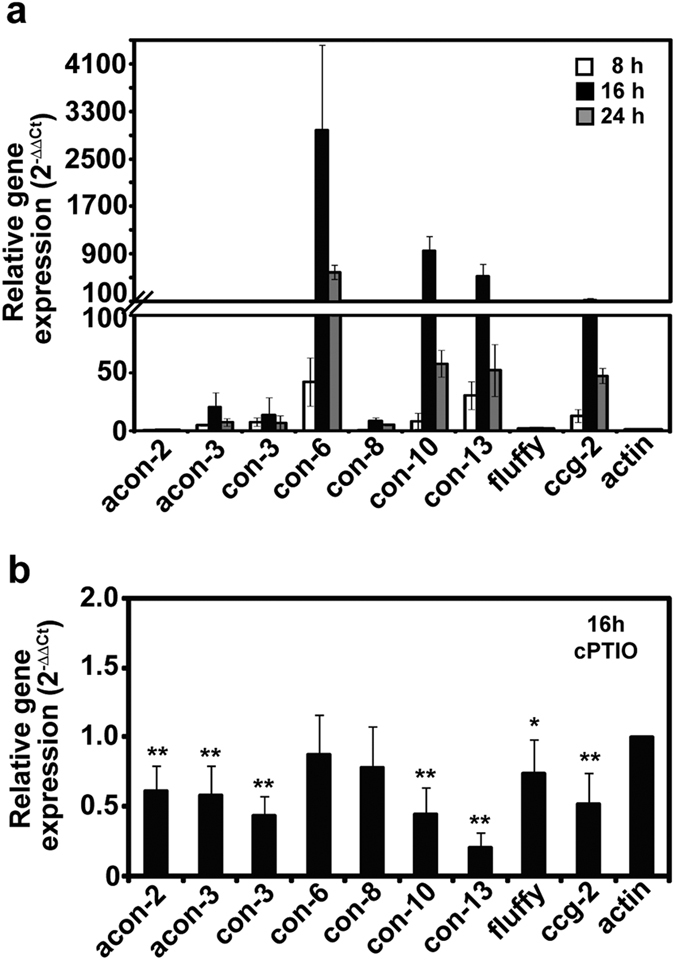
(a) Transcript levels of con genes in wild type N. crassa after synchronous conidiation was induced. All values are mean ± standard deviation of 6 replicate measurements from 2 independent experiments. (b) con gene transcripts 16 hours after conidiation was initiated in wild type treated with cPTIO. Student t-test was performed between control and each treatment; *p < 0.05, **p < 0.01.
Basal hyphal growth and conidia formation are increased by exogenous NO
Since scavenging intracellular NO by cPTIO caused retardation in vegetative hyphal growth and conidial development, addition of exogenous NO may also influence these developmental processes. We treated wild type N. crassa with exogenous NO using SNP (sodium nitroprusside), an NO releasing chemical, and AEMP3, an NO releasing nanoparticle. SNP is a well known NO producing chemical, and NO concentrations in SNP solution, measured as nitrite (NO2−) level, are shown in Fig. 6a. NO levels increased linearly as SNP concentration increased (Fig. 6a). NO reached a concentration of 20 μM in 1 mM SNP solution (Fig. 6a).
Figure 6. Vegetative growth and conidiation in N. crassa treated with SNP and AEMP3.
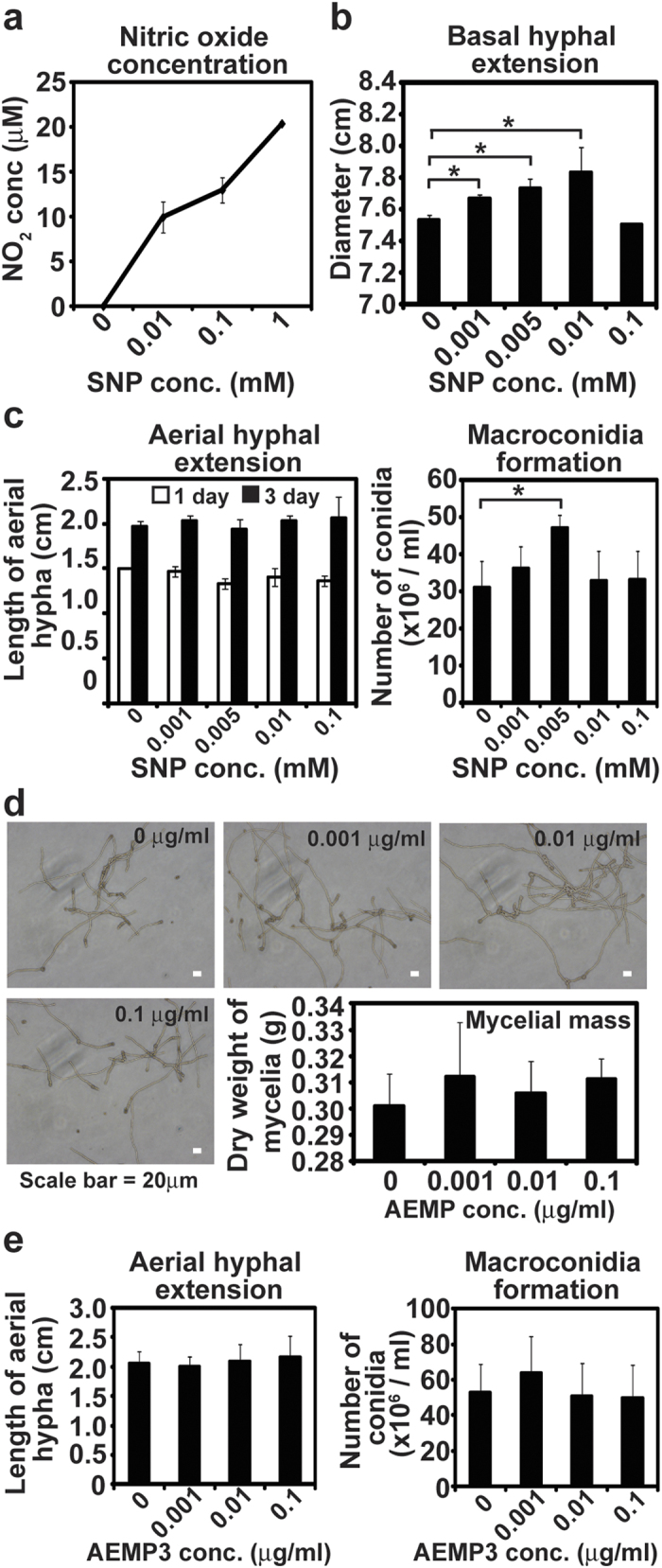
(a) NO released in different concentrations of SNP solution. All measurements were performed in 3 replicates. (b) Extension of basal hyphae on VM agar media supplemented with SNP. (c) Aerial hyphal extension (left panel) on the surface of VM liquid containing SNP and number of conidia (right panel) harvested from the culture after 5 days. All values are mean ± standard deviation of 3 replicate measurements and the experiments were repeated 3 times. Student t-test was performed between control and each treatment; *p < 0.05. (d) Hyphal development after 8 hours incubation in VM liquid supplemented with NO releasing nanoparticle AEMP3 (microscopic photographs). After 3 days, dry weight of mycelia mass harvested (graph). (e) Aerial hyphal extension (left panel) on the surface of VM liquid containing different concentrations of AEMP3 and number of conidia (right panel) harvested from cultures after 5 days. All values are mean ± standard deviation of 3 replicate measurements and the experiments were repeated 3 times.
After 24 hours of incubation, basal hyphal extension significantly increased on VM agar media when SNP was added (Fig. 6b). The diameter of hyphal extensions was also greatest in media with 0.01 mM SNP (about 10 μM NO), whereas there was no dramatic change in hyphal extension in the presence of 1 mM SNP, compared to control (Fig. 6b). During conidial development, addition of SNP did not affect aerial hyphal growth, although the number of conidia, after a 5 day incubation, was significantly higher in the presence of 0.005 mM SNP (about 5 μM NO) compared to control (Fig. 6c).
Nitric oxide-releasing AEMP3 silica nanoparticles were employed as another NO donor30. The NO release properties were characterized in phosphate buffered saline (PBS; 0.01 M, pH 7.4) at 37 °C as follows (see Supplementary Fig. S1). The total amount of NO released (t[NO]) and maximum NO flux ([NO]m) from the AEMP3 nanoparticle system were 0.59 μmol·mg−1 and 1180 ppb·mg−1, respectively. The NO release kinetics from the AEMP3 silica nanoparticles were relatively slow compared to other NO-releasing silica nanoparticle systems30 with a NO release half-life (t1/2) of 130 minutes. In addition, the time required to reach the maximum NO flux (tm) of 1180 ppb·mg−1 was determined by 5 min after immersion in buffer solution. As characterized by scanning electron microscopy (SEM), the size of the 40 mol% AEMP3 nanoparticles was 23 ± 2 nm. When NO releasing AEMP3 nanoparticles were added in VM liquid, fungal hyphal growth and asexual development (aerial hyphal development and conidia formation) were not significantly changed. However, slight enhancement was observed in hyphal mass and conidia number after treatment with AEMP3 in every repeated experiment (Fig. 6d,e). The dry weight of fungal mycelia grown for 3 days in the presence of AEMP3 was slightly higher than that of control (Fig. 6d). The height of aerial hypha developed on the surface of VM liquid was not different between control and AEMP3 treated samples, whereas the number of conidia collected after 5 days was slightly greater in the culture supplemented with 0.001 μg/ml AEMP3 (Fig. 6e).
NO level is not changed in knockout mutants of NOS like genes in N. crassa
To find the mechanism(s) for NO synthesis, we first searched candidate genes homologous to NOS (nitric oxide synthase) from other organisms in N. crassa genome. Several Neurospora genes showing high homology to NOSs of human17 and other fungi8,9,11 (M. oryzae, P. polycephalum, I. obliquus) were identified (Table 1). Homology of these genes to human NOSs was relatively lower than that to fungal NOSs (Table 1). Among candidate genes, NCU05006, NCU05185, and NCU09741 appeared to be highly homologous to human and other fungal NOSs (lowest expected value) (Table 1).
Table 1. N. crassa candidate genes homologous to NOS of other organisms.
| Organisms | Homologous genes in N. crassa | ||
|---|---|---|---|
| Homo sapiens | nNOS | eNOS | iNOS |
| aNCU09741 (b5.2E-43) | NCU09741 (3.9E-37) | NCU09741 (1.8E-42) | |
| NCU05185 (1.1E-35) | NCU05185 (9.1E-24) | NCU09727 (6.4E-30) | |
| NCU04077 (2.8E-23) | NCU04077 (6.4E-19) | NCU04077 (1.6E-29) | |
| NCU09727 (2.7E-19) | NCU09727 (2.9E-10) | NCU05185 (1.6E-29) | |
| Magnaporthe oryzae | NOL2 | NOL3 | |
| NCU05185 (0) | NCU05185 (0) | ||
| NCU05006 (0) | NCU05006 (0) | ||
| NCU08062 (2.4E-21) | NCU09741 (4.9E-27) | ||
| NCU09727 (2.6E-20) | NCU01086 (1.7E-21) | ||
| NCU09741 (8.6E-19) | NCU06327 (7.9E-19) | ||
| Physarum polycephalum | Physnosa | Physnosb | |
| NCU09741 (0) | NCU09741 (0) | ||
| NCU05185 (1.4E-45) | NCU05185 (2.8E-45) | ||
| NCU04077 (2.6E-35) | NCU04077 (1.8E-34) | ||
| NCU09727 (2.6E-22) | NCU09727 (1.9E-34) | ||
| Inonotus obliquus | KM519595 | KP052853 | |
| NCU09741 (0) | NCU05185 (0) | ||
| NCU05185 (0) | NCU05006 (0) | ||
| NCU04077 (3.5E-27) | NCU09741 (0) | ||
| NCU09727 (3.4E-25) | NCU01086 (1.1E-24) | ||
| NCU03697 (4.7E-17) | NCU02031 (2.3E-24) | ||
| NCU05238 (1.4E-10) | NCU04077 (1.5E-23) | ||
aHomologous gene ID number in N. crassa starts with NCU.
bNumber in parenthesis is expected value.
Intracellular NO was detected using DAF-FM diacetate in knockout mutants of these candidate genes. Since there were no available homokaryotic mutants for several genes including NCU09741 (probably essential gene for viability or sexual development), we examined NO in knockout mutants of only NCU01086, NCU04077, NCU05006, and NCU05185 grown for 16 hours after synchronous conidiation was induced. No dramatic difference in intracellular NO level was observed between wild type and mutants as fluorescence intensity in conidiophores was similar in all samples (Fig. 7). This indicates that these tested genes may not be the genes coding for NOS or NO synthesis in N. crassa does not follow NOS dependent way. Nevertheless, we still need to examine mutants of all candidate genes.
Figure 7. NO production in knockout mutants of NOS like genes in N. crassa.
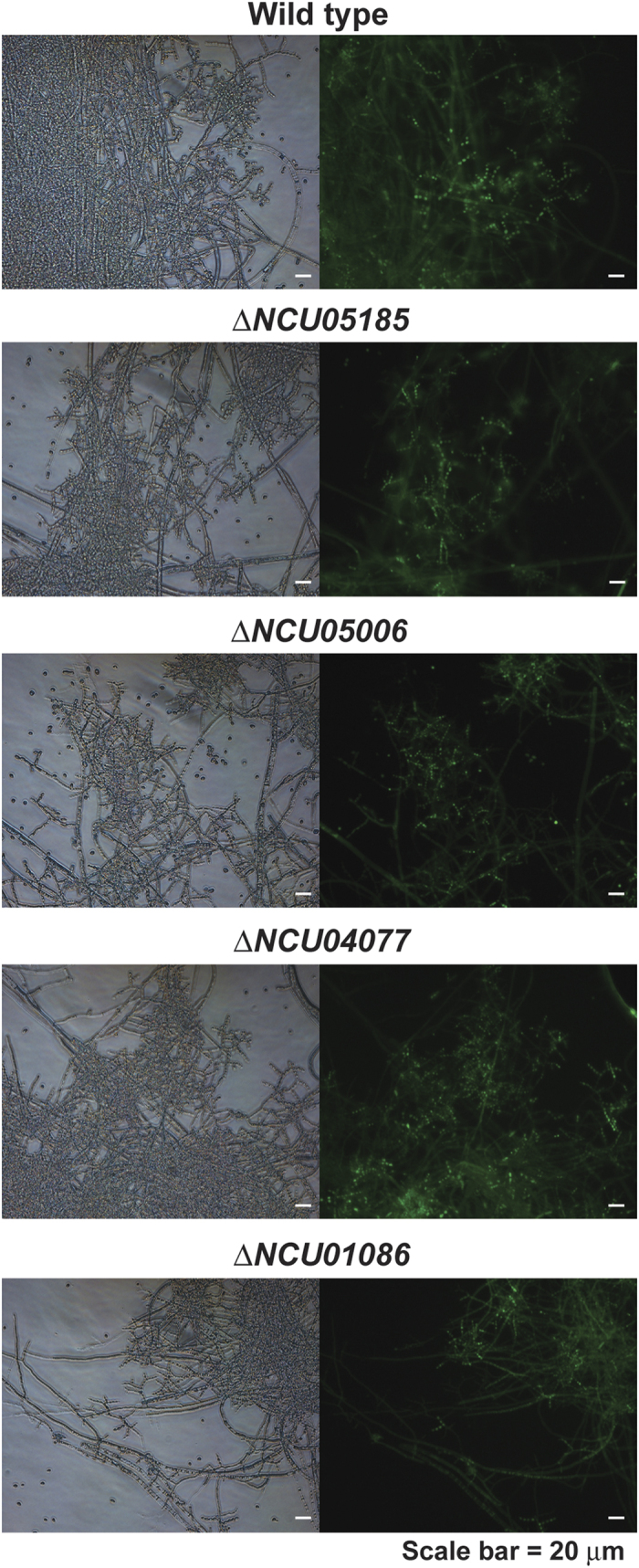
Intracellular NO was detected in wild type and knockout mutants of NCU05185, NCU05006, NCU04077, and NCU01086 homologous to NOS in human and other fungi. NO was detected using DAF-FM diacetate in fungal samples incubated for 16 hours after conidiation was induced. Left and right of each picture indicate images captured in optical and fluorescent (465–495 nm excitation filter) mode, respectively.
NCU01086, NCU04077, NCU05006, and NCU05185 seem to be dispensable to the phenotypes associated with intracellular NO level such as vegetative growth and conidiation in N. crassa. Knockout mutants of these genes did not show any significant changes in vegetative growth and conidiation, showing similar phenotypes to wild type (Supplementary Figs S2 and S3). It is likely that NCU01086, NCU04077, NCU05006, and NCU05185 are not much involved in NO synthesis and related pathways although further study is necessary.
Discussion
Endogenous generation of reactive species in microbial cells is an intriguing process, because reactive species have antimicrobial activities. However, increasing numbers of studies demonstrate that intracellular reactive species play critical roles in microbial vitality, including management of stresses and differentiation2,14. Among all reactive species, NO has been considered a universal signaling molecule in most organisms including microorganisms. However, very little is known about the synthesis and functions of NO in fungi. In most fungi, endogenous NO generation has been indirectly verified by identifying NOS-like activity or using NO scavengers7,10,12,21,22,23,24,25. Few studies have demonstrated direct detection of intracellular NO8,26,27.
In our study, endogenous NO in N. crassa was directly detected using the NO indicator, DAF-FM diacetate. Our data clearly show that NO is generated in cells during conidia formation on agar media and hyphal branching in liquid VM. Therefore, our data have provided direct evidence for endogenous NO production, which was not directly detected in Ninnemann and Maier’s study21. High fluorescence detected inside vacuole like structures in fungal hyphae may indicate the subcellular place of NO synthesis or be a result of accumulation of DAF-FM dye in vacuole. We also observed high DAF-FM fluorescence inside vacuolar structures in Fusarium oxysporum hyphae during sporulation in VM liquid (Supplementary Fig. S4). Intracellular NO level was increased during microconidia formation (16–24 h) in F. oxysporum but fluorescence was highly detected in vegetative hyphae not in phialides and microconidia (Supplementary Fig. S4). This result was different from that observed in N. crassa in which NO was detected highly in conidiophores during conidiation. In Arabidopsis thaliana, NO is detected inside the vacuole in immature growing root hairs and then the cytoplasm after maturation31. Vacuole is known as a storage reservoir for basic amino acids such as arginine in filamentous fungi32. It may be possible that NOS dependent NO synthesis can occur in vacuole but other possibilities should be also considered.
The origin of intracellular NO in N. crassa is still obscure. We identified several putative genes homologous to human and other fungal NOSs in the N. crassa genome (Table 1). None of the knockout mutants tested showed reduced NO levels and phenotypes during vegetative growth and conidiation. Since we did not examine mutants of all candidate genes, further analyses may be needed. Our results also suggest a possibility of more various routes for NO synthesis (NOS dependent and independent) in Neurospora, compared to animal. Neurospora NOSs may have a very different genome sequence or non-enzymatic synthesis can be possible. Both NOS dependent and independent pathways can be considered in Neurospora NO production as demonstrated in plants in which sequence of NOS gene is different from that of mammalian NOS33, and NO is also generated during nitrate reduction34, by the action of xanthine oxidase35, or by non-enzymatic ways36,37. It is recently suggested in A. nidulans that NO production can be catalyzed by nitrate reductase during development and more than one pathways can be involved in NO synthesis27. The implication here is that NO synthesis in fungi can be quite distinct from those found in other organisms. Our preliminary data show that deletion of a putative xanthine oxidase gene (NCU03350) does not impair NO synthesis during submerged hyphal development in N. crassa (Supplementary Fig. 5). We also did screening knockout mutants of transcription factors and serine/threonine protein kinases that exhibited defects in vegetative hyphal growth and conidiation for recovery of defects by exogenous NO (see Supplementary methods and Table S1). Aerial hyphal growth and conidia formation were partially recovered in knockout mutant of NCU05658 (stk-36) (Supplementary Table S2), a homolog of yeast SKY1 that is known to regulate polyamine transport and ion homeostasis38. Since exogenous polyamine is known to enable NO production in A. thaliana39, it may be possible that polyamine metabolism is related to NO synthesis in N. crassa. Further investigation will be needed on this.
Our data suggest several possible roles for NO in Neurospora development. First, because intracellular NO is detected in conidiophores and the NO scavenger cPTIO delays conidiophore development and reduces the number of conidia, NO may be involved in formation of macroconidia. Roles for NO in conidiation have been demonstrated in fungi, where production of NO is verified11,12,21,23. However, our results are somewhat contrary to the observation made in the Nimmemann and Maier’s study, in which inhibition of NOS activity enhanced conidiation and exogenous NO reduced conidiation21. This may be because homologous NOS genes in N. crassa do not seem to be related to NO generation, recall that use of a mammalian NOS inhibitor did not reduce NO levels. In addition, the concentration of SNP, an exogenous NO donor, used in their study (1 mM) was too high based on our data, which may have produced harmful effects21.
Interestingly, our study revealed that NO produced during conidiation is closely associated with con gene expression. In particular, NO scavenging significantly suppressed transcription of ccg-2, con-10 and con-13, which are highly expressed in conidiophores40. This suggests that NO may regulate conidiophore development by controlling expression of these genes. However, deletion of con-10 and con-13 did not exhibit any clear phenotypes during vegetative growth and conidiation in our study (Supplementary Figs S2 and S3). Same observation was already demonstrated in a previous study41. It is obscure how NO is involved in controlling conidiation with con-10 and con-13 although ccg-2 encodes an internal component of conidia42. A possibility is that NO may be involved in the regulation of circadian rhythm because ccg-2 and con-10 are known as clock controlled genes in Neurospora40,42,43. As a consequence, conidia development which is controlled by circadian rhythm can be affected by NO. If NO is associated with expression of clock controlled genes, it is possible that level of intracellular NO may be oscillated during a day. Endogenous ROS level is known to follow circadian rhythm in Neurospora44. However, there is no report for RNS level. In our study, NO was detected during conidiation in conidiophores 16 hours after conidiation induction, but not at 8 or 24 hours, indicating that intracellular NO may be rhythmically changed during a day. Further investigation should be needed for this.
NO may also regulate hyphal growth during submerged culture in VM. Intracellular NO levels were increased at 8–16 hours in culture and decreased after 24 hours, and NO was mostly detected in the cytosol or vacuoles of hyphae. NO production may occur early during hyphal development and is not associated with late stages of vegetative growth. A previous study demonstrated that ROS generated by NADPH oxidase Nox1 is involved in the regulation of hyphal growth in N. crassa45. However, ROS generated by Nox enzymes controls sexual development in many fungi46. Cooperation of ROS (reactive oxygen species) and RNS (reactive nitrogen species) should be considered in the regulation of fungal vegetative growth.
In conclusion, our results demonstrate that NO is endogenously generated during early vegetative growth and conidia formation on conidiophores in N. crassa. Generated NO may be involved in the regulation of submerged hyphal growth in liquid and conidia formation (maturation), and transcription of con-10 and con-13 is closely associated with intracellular NO levels.
Methods
Reagents and materials
Tetraethoxysilane (TEOS) and sodium methoxide (NaOMe) were purchased from Fluka (Buchs, Switzerland). (Aminoethylaminomethyl)phenethyltrimethoxysilane (AEMP3) was purchased from Gelest (Tullytown, PA). Ethanol (EtOH) and ammonia solution (NH4OH, 30 wt% in water) were purchased from Fisher Scientific (Fair Lawn, NJ). Nitric oxide (NO, 99.99%) and argon (Ar) gases were obtained from Dong-A Scientific (Seoul, South Korea) or Sung-Kang Special Gas (Seoul, South Korea). Other solvents and chemicals were analytical-reagent grade and used as received. A Millipore Milli-Q UV Gradient A10 System (Bedford, MA) was used to purify distilled water to a final resistivity of 18.2 MΩ·cm and a total organic content of ≤6 ppb.
Synthesis and characterization of NO-releasing silica nanoparticles
The synthesis and characterization of NO-releasing silica nanoparticles have been described previously30. Briefly, an aminoalkoxysilane solution was prepared by dissolving AEMP3 (1.86 mmol) in 20 mL of EtOH and 4 mL of MeOH in the presence of NaOCH3 (1.86 mmol). The solution was then placed into 10 mL vials equipped with stir bars. The vials were placed in a Parr bottle, connected to an in-house NO reactor, and flushed with Ar six times to remove O2 in the solution. The reaction bottle was pressurized to 10 atm NO for 3 days with continuous stirring of the silane solution. Prior to removing the N-diazeniumdiolate-modified AEMP3 silane sample (AEMP3/NO), unreacted NO was purged from the chamber with Ar. The silane solution was prepared by mixing TEOS (2.80 mmol) and AHAP3/NO (1.86 mmol; corresponding to 40 mol%, balance TEOS) in the EtOH/MeOH solution for 2. The silane solution was then added into 22 mL of EtOH and 6 mL of ammonia catalyst (30 wt % in water) and mixed vigorously for 30 min at 4 °C. The precipitated nanoparticles were collected by centrifugation (5000 rpm, 5 min), washed with EtOH several times, dried under ambient conditions for 1 h, and stored in a sealed container at –20 °C until used.
Nitric oxide release from the N-diazeniumdiolate-modified AEMP3 silica nanoparticles was measured in deoxygenated PBS (0.01 M, pH 7.4) at 37 °C using a Sievers NOA 280i chemiluminescence NO analyzer (Boulder, CO). Nitric oxide released from the donors was transported to the analyzer by a stream of N2 (70 mL·min−1) passed through the reaction cell. The instrument was calibrated with air passed through a NO zero filter (0 ppm NO) and a 24.1 ppm NO standard gas (balance N2).
The size of NO-releasing silica nanoparticles was characterized by scanning electron microscopy (SEM). To obtain high-quality images, samples were coated with a thin layer of platinum (~5 nm thickness) using a Cressington 108 auto sputter coater (Watford, England). SEM images of AEMP3 silica nanoparticles were collected on a Hitachi S4700 field-emission SEM (Tokyo, Japan) using an accelerating voltage of 10 keV (source working distance of 7.7 mm).
Fungus and culture condition
Wild type Neurospora crassa (ORS-SL6a, mat a) and knockout mutant strains were obtained from the Fungal Genetics Stock Center (FGSC, Manhattan, KS, USA) and conserved as conidial masses in silica gel at −20 °C. Culture of fungal strains from silica stocks was initiated by placing silica gels on VM agar media with (mutants) or without (wild type) hygromycin (200 μg/ml). Agar culture flasks and tubes were incubated at 30 °C in the dark for 2–3 days and then at 25 °C in the light for 11–12 days. For liquid culture, VM liquid inoculated with fungal spores (106/ml) and incubated at 30 °C in the dark with shaking (130 rpm).
Detection of intracellular nitric oxide (NO)
To detect intracellular NO during hyphal development in liquid media, VM liquid (15 ml) was inoculated with conidia (final concentration of 106 conidia/ml) and incubated at 30 °C with shaking. Fungal mycelia were harvested at each indicated time and washed with 1x phosphate buffered saline (PBS). Fungal mycelia were then resuspended in new PBS. To detect intracellular NO, DAF-FM diacetate (amino-5-methylamino-2′,7′-difluorescein diacetate; Life Technologies, Grand Island, NY, USA) was added to the mycelial suspension to make the final concentration of 20 μM and incubated at room temperature for 1 hour. The suspension was centrifuged at 10,000 rpm for 5 minutes and the supernatant was discarded. Fungal mycelia were washed with 1x PBS three times to remove residual DAF-FM diacetate and resuspended in new PBS. Suspended mycelia were mounted on a glass slide and observed under a fluorescence microscope using a 465–495 nm excitation filter (Nikon, Tokyo, Japan).
Production of intracellular NO was also examined during conidiation which was synchronously induced from the mycelial mat placed on VM agar media. Spores were inoculated into 15 ml VM liquid (106 spores/ml) and incubated at 30 °C for 20 hours with shaking. Fungal mycelia were collected on a filter paper disc (Whatman No. 1, 9 cm in diameter) and dried. The mycelial mat on filter paper was then placed on the surface of VM agar media and incubated at 30 °C in the dark for 3 days. At each indicated time, aliquots of fungal cultures were sampled and suspended in PBS. Intracellular NO was detected using DAF-FM diacetate (final concentration 20 μM) as described earlier.
Treatment with cPTIO, an NO scavenger
The NO scavenger, cPTIO (2-(4-carboxyphenyl)-4,5-dihydro-4,4,5,5-tetramethy-1 H-imidazolyl-1-oxy-3-oxide; Cayman, Ann Arbor, MI, USA), was used to confirm production of intracellular NO and investigate the effect of NO scavenging on hyphal development and conidiation. VM liquid (15 ml) was inoculated with conidia (final concentration of 106 conidia/ml) and incubated at 30 °C with shaking. Aliquots of fungal mycelia were harvested at each indicated time and washed with 1x phosphate buffered saline (PBS). Fungal mycelia were then resuspended in new PBS and treated with cPTIO (10 mM final concentration in PBS) for 20 minutes. After treatment with cPTIO, fungal mycelia were washed with PBS three times and intracellular NO was measured using DAF-FM diacetate (20 μM) as described above.
To treat fungi with cPTIO during conidiation, spores were inoculated into 15 ml of VM liquid and incubated at 30 °C for 20 hours with shaking. Fungal mycelia were harvested, dried on a filter paper disc (Whatman No. 1, 9 cm in diameter), and placed on the surface of VM agar media. VM agar plates were incubated at 30 °C. Fungal mycelia were harvested from VM agar plates and suspended in PBS at indicated times. cPTIO was added to mycelia suspensions (10 mM final concentration) and incubated for 20 minutes. After washing with PBS three times, fungal mycelia were resuspended in new PBS and DAF-FM diacetate was added (final concentration 20 μM). Detection of intracellular NO was performed as described previously.
To examine the effects of cPTIO on fungal development, cPTIO was added to VM liquid culture (final concentration 10 mM) or spread on VM agar media. Fungal spores were inoculated (106/ml) in 15 ml of VM liquid containing cPTIO (10 mM) and inoculated culture tubes were incubated at 30 °C with shaking. At indicated times, fungal cultures were sampled and observed under an optical microscope. cPTIO was also spread on VM agar in 60 mm petri dishes (100 μl of 10 mM cPTIO). To determine effects on basal hyphae extension, approximately 104 spores were inoculated in the center of a VM agar plate and the diameter of hyphal extension was measured. After 24 hours, hypha were harvested from the plate and dried, and the hyphal mass was measured. For the effects on conidiation, fungal mycelia collected from liquid culture were spread on the surface of VM agar media covered by cPTIO. Plates were incubated at 30 °C in the dark for 2 days and then at 25 °C in the light. Aerial hyphal development and conidia formation were analyzed using an optical microscope. After 1 week, plates were placed in larger petri-dishes (100 × 40 mm) and 50 ml of sterile water was added to cover culture plates. Petri-dishes were agitated and suspensions were filtered through 2 layers of miracloth (EMD Millipore, Billerica, MA, USA) to remove fungal hypha. Spores in the filtered spore suspensions were counted using a hemacytometer (Marienfeld, Germany).
Confocal microscopy
To further analyze the production of NO within fungal cells, we examined samples treated with DAF-FM diacetate using confocal microscopy (IX-83, Olympus, Tokyo, Japan). For liquid culture, fungal spores were inoculated in 15 ml of liquid VM (106/ml) and incubated with shaking at 30 °C. For conidiation culture, a fungal mycelial mat was placed on VM agar media and incubated at 30 °C as described previously. At each indicated time during culture, fungal mycelia were sampled and submerged in PBS. Treatment with DAF-FM diacetate was carried out and samples were observed and imaged. Sections in the Z-axis were acquired at 1.5 μm intervals.
Assay for the effect of exogenous NO on fungal development
Sodium nitroprusside (SNP; Sigma-Aldrich, St. Louis, MO, USA), a chemical that generates NO, and nitric oxide (NO)-releasing AEMP3 silica nanoparticles were used in the treatment with exogenous nitric oxide. NO released in SNP solution at different concentrations was quantitated by measuring the total concentration of nitrite (NO2−) and nitrate (NO3−), as NO is quickly oxidized to nitrite and nitrate. Measurements were performed using an NO colorimetric assay kit (Biovision, Milpitas, CA, USA) following the manufacturer’s protocol.
To determine the effects of exogeneous NO on fungal vegetative growth, SNP was added to VM agar media (0.001–0.1 mM) and wild type fungal spores (104 spores) were inoculated in the center of the plates. After incubation at 30 °C in the dark for 24 hours, the diameter of mycelial growth was measured. AEMP nanoparticles were added in VM liquid media at the concentration of 0–0.1 μg/ml and fungal spores were inoculated (104/ml). Liquid culture flasks were incubated at 30 °C with shaking for 3 days. After fungal mycelia were harvested and dried by filtering the culture through filter paper and vaccuum-drying overnight, the weight of dried mycelia was measured.
To examine the effects of SNP on conidiation, SNP or AEMP nanoparticles were added to 1 ml of VM liquid placed in a glass tube (13 × 10 mm) at 0.001–0.1 mM concentrations. Wild type fungal spores (104) were inoculated on the surface of VM liquid. Tubes were incubated at 30 °C in the dark for 3 days and then moved to 25 °C in the light. Growth of aerial hypha and conidiation were monitored during incubation. After 5 days, 3 ml of water was added into the tubes, which were vigorously shaken. The resulting mixtures were filtered through 2 layers of miracloth to produce conidial suspensions, and conidia were counted using a hemacytometer.
Quantitative PCR analysis
Expression of putative conidiation related genes (NCU08769, con-6; NCU09235, con-8; NCU07325, con-10; NCU07324, con-13; NCU08726, fluffy; NCU08457, ccg-2; NCU09873, con-3; NCU00478, acon-2; NCU07617, acon-3) was analyzed during formation of aerial hypha and conidia. Fungal mycelia collected from VM liquid culture (as described above) were placed on a filter paper disc (Whatman No. 1, 9 cm in diameter) and dried. The mycelial mat on filter paper was placed on the surface of VM agar media and incubated at 30 °C in the dark. After 8, 16, and 24 hours, aerial hypha and conidia were harvested and total RNA was extracted using the Takara RNAiso Plus kit according to the manufacturer’s protocol (Takara Bio Inc., Shiga, Japan). Total RNA was treated with RNase-free DNase (Toyobo Co., Ltd., Osaka, Japan) at 37 °C for 1 hour to remove genomic DNA, and RNA concentration was measured using a nanodrop (Biotek, Winooski, VT, USA). Equal amount of RNA (16 ng) from all samples was used to synthesize cDNA using the ReverTra Ace® qPCR RT mater mix with gDNA remover kit (Toyobo Co., Ltd., Osaka, Japan). Thermocycle conditions were as follows: 60 min at 37 °C and 5 min at 95 °C. Target genes (10 putative conidiation related genes) were amplified using gene specific primers (Table 2) and quantified at every thermal cycle using iQ SYBR Green Supermix (BioRad, Hercules, CA, USA) and the CFX96TM real time RT-PCR system (Bio-Rad, Hercules, CA, USA). Actin was amplified as a reference gene. The cycle threshold (Ct) was determined and the relative target gene expression level between the sample and the control was calculated as previously described47: ratio = 2−{(Ct target−Ct reference)sample−(Ct target−Ct reference)control}.
Table 2. Primers used in quantitative PCR analyses of conidiation related genes.
| No. | Sequence | Primer details |
|---|---|---|
| GP9F | CCAAGGAGCACTCCAAGAAG | NCU08769 (con-6) forward |
| GP9R | ACACTTTGGGGTTGTTGAGG | NCU08769 (con-6) reverse |
| GP11F | GGGCTTGATGGATCAAAA | NCU09235 (con-8) forward |
| GP11R | ATCCCACATCTTGGCAAT | NCU09235 (con-8) reverse |
| GP12F | AGGAAGAGGTTCAGGCCATC | NCU07325 (con-10) forward |
| GP12R | GGCTCAAAGCTGCCGCTGGA | NCU07325 (con-10) reverse |
| GP13F | AAGACTGGAAGGATACCGTT | NCU07324 (con-13) forward |
| GP13R | GATTGACCATACAGCCGACA | NCU07324 (con-13) reverse |
| GP14F | TCCAGCATCTCGTTGTCATC | NCU08726 (fluffy) forward |
| GP14R | GGTGAAAAACGGGAGGAAAT | NCU08726 (fluffy) reverse |
| GP15F | TTACTGCTGCCAGTCTATGT | NCU08457 (ccg-2) forward |
| GP15R | TAACATCGTCCTTGCAGCAC | NCU08457 (ccg-2) reverse |
| GP16F | CCATCGAGTACATCCCCAAC | NCU09873 (con-3) forward |
| GP16R | GCCGGAGATAAAGTGGAACA | NCU09873 (con-3) reverse |
| GP17F | AGAAGCCTTCACCCTTCTCC | NCU00478 (acon-2) forward |
| GP17R | CAGTAGCTGGTGCTGTGGAA | NCU00478 (acon-2) reverse |
| GP18F | GGATCAGGCGTTGAAGAAAA | NCU07617 (acon-3) forward |
| GP18R | TAGCCCGTTGAAGATTGGTC | NCU07617 (acon-3) reverse |
Statistical analysis
All data were obtained from at least 3 replicate measurements and 2 or 3 replicate experiments. Student’s t test was performed to determine significance between data points. Significant differences were established at p < 0.05 or p < 0.01 (*denotes p < 0.05 and **denotes p < 0.01).
Additional Information
How to cite this article: Pengkit, A. et al. Identification and functional analysis of endogenous nitric oxide in a filamentous fungus. Sci. Rep. 6, 30037; doi: 10.1038/srep30037 (2016).
Supplementary Material
Acknowledgments
Wild-type and mutant N. crassa strains were obtained from the Fungal Genetics Stock Center (Kansas City, Missouri USA). This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (2011-0014825, 2013R1A1A3011245, 2010-0027963) and partially supported by a Research Grant from Kwangwoon University in 2015.
Footnotes
Author Contributions A.P., S.S.J. and S.J.S. performed experiments, J.H.S. and G.P. did data analysis and wrote the main manuscript text, and E.H.C. and K.Y.B. helped data analysis. All authors reviewed the manuscript.
References
- Sudhamsu J. & Crane B. R. Bacterial nitric oxide synthases: what are they good for? Trends Microbiol. 17, 212–218 (2009). [DOI] [PubMed] [Google Scholar]
- Aguirre J. & Lambeth J. D. Nox enzymes from fungus to fly to fish and what they tell us about Nox function in mammals. Free Radic. Biol. Med. 49, 1342–1353 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J. & Tudzynski P. Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390 (2011). [DOI] [PubMed] [Google Scholar]
- Tudzynski P., Heller J. & Siegmund U. Reactive oxygen species generation in fungal development and pathogenesis. Curr. Opin. Microbiol. 15, 653–659 (2012). [DOI] [PubMed] [Google Scholar]
- Kim H. J. Exploitation of reactive oxygen species by fungi: roles in host-fungus interaction and fungal development. J. Microbiol. Biotechnol. 24, 1455–1463 (2014). [DOI] [PubMed] [Google Scholar]
- Dirschnabel D. E. et al. New insights into the roles of NADPH oxidases in sexual development and ascospore germination in Sordaria macrospora. Genetics 196, 729–744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. & Higgins V. J. Nitric oxide has a regulatory effect in the germination of conidia of Colletotrichum coccodes. Fungal. Genet. Biol. 42, 284–292 (2005). [DOI] [PubMed] [Google Scholar]
- Samalova M. et al. Nitric oxide generated by the rice blast fungus Magnaporthe oryzae drives plant infection. New Phytol. 197, 207–222 (2013). [DOI] [PubMed] [Google Scholar]
- Zhao Y. et al. Correlation of nitric oxide produced by an inducible nitric oxide synthase-like protein with enhanced expression of the phenylpropanoid pathway in Inonotus obliquus cocultured with Phellinus morii. Appl. Microbiol. Biotechnol. 99, 4361–4372 (2015). [DOI] [PubMed] [Google Scholar]
- Song N.-K., Jeong C.-S. & Choi H.-S. Identification of nitric oxide synthase in Flammulina velutipes. Mycologia 92, 1027–1032 (2000). [Google Scholar]
- Golderer G., Werner E. R., Leitner S., Grobner P. & Werner-Felmayer G. Nitric oxide synthase is induced in sporulation of Physarum polycephalum. Genes Dev. 15, 1299–1309 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X. et al. L-arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans. Fungal. Genet. Biol. 44, 1368–1379 (2007). [DOI] [PubMed] [Google Scholar]
- Zheng W. et al. Nitric oxide mediates the fungal-elicitor-enhanced biosynthesis of antioxidant polyphenols in submerged cultures of Inonotus obliquus. Microbiol. 155, 3440–3448 (2009). [DOI] [PubMed]
- Crane B. R., Sudhamsu J. & Patel B. A. Bacterial nitric oxide synthases. Annu. Rev. Biochem. 79, 445–470 (2010). [DOI] [PubMed] [Google Scholar]
- Kong W., Huang C., Chen Q., Zou Y. & Zhang J. Nitric oxide alleviates heat stress-induced oxidative damage in Pleurotus eryngii var. tuoliensis. Fungal Genet. Biol. 49, 15–20 (2012). [DOI] [PubMed] [Google Scholar]
- Rafferty S. Nitric oxide synthases of bacteria and other unicellular organisms Open Nitric Oxide J. 3, 25–32 (2011). [Google Scholar]
- Alderton W. K., Cooper C. E. & Knowles R. G. Nitric oxide synthases: structure, function and inhibition. Biochem. J. 357, 593–615 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane B. R. et al. Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science 279, 2121–2126 (1998). [DOI] [PubMed] [Google Scholar]
- Yun H. Y., Dawson V. L. & Dawson T. M. Neurobiology of nitric oxide. Crit. Rev. Neurobiol. 10, 291–316 (1996). [DOI] [PubMed] [Google Scholar]
- Lipton S. A. Physiology. Nitric oxide and respiration. Nature 413, 118–119 (2001). [DOI] [PubMed] [Google Scholar]
- Ninnemann H. & Maier J. Indications for the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem. Photobiol. 64, 393–398 (1996). [DOI] [PubMed] [Google Scholar]
- Wilken M. & Huchzermeyer B. Suppression of mycelia formation by NO produced endogenously in Candida tropicalis. Eur. J. Cell Biol. 78, 209–213 (1999). [DOI] [PubMed] [Google Scholar]
- Maier J., Hecker R., Rockel P. & Ninnemann H. Role of nitric oxide synthase in the light-induced development of sporangiophores in Phycomyces blakesleeanus. Plant Physiol. 126, 1323–1330 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida B. et al. NO-mediated apoptosis in yeast. J. Cell. Sci. 120, 3279–3288 (2007). [DOI] [PubMed] [Google Scholar]
- Vieira A. L., Linares E., Augusto O. & Gomes S. L. Evidence of a Ca(2+)-(*)NO-cGMP signaling pathway controlling zoospore biogenesis in the aquatic fungus Blastocladiella emersonii. Fungal Genet. Biol. 46, 575–584 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. MoARG1, MoARG5, 6 and MoARG7 involved in arginine biosynthesis are essential for growth, conidiogenesis, sexual reproduction, and pathogenicity in Magnaporthe oryzae. Microbiol. Res. 180, 11–22 (2015). [DOI] [PubMed] [Google Scholar]
- Marcos A. T. et al. Nitric oxide synthesis by nitrate reductase is regulated during development in Aspergillus. Mol. Microbiol. 99, 15–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats E., Carver T. L. & Mur L. A. Pathogen-derived nitric oxide influences formation of the appressorium infection structure in the phytopathogenic fungus Blumeria graminis. Res. Microbiol. 159, 476–480 (2008). [DOI] [PubMed] [Google Scholar]
- Kojima H. et al. Development of a fluorescent indicator for nitric oxide based on the fluorescein chromophore. Chem. Pharm. Bull. 46, 373–375 (1998). [DOI] [PubMed] [Google Scholar]
- Shin J. H. & Schoenfisch M. H. Inorganic/organic hybrid silica nanoparticles as a nitric oxide delivery scaffold. Chem. Mater. 20, 239–249 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo M. C. & Lamattina L. Nitric oxide is essential for vesicle formation and trafficking in Arabidopsis root hair growth. J. Exp. Bot. 63, 4875–4885 (2012). [DOI] [PubMed] [Google Scholar]
- Bowman E. J. & Bowman B. J. Vacuoles in filamentous fungi in Cellular and Molecular Biology of Filamentous Fungi (eds Borkovich K. A. & Ebbole D. J.) 179–190 (ASM Press, 2010). [Google Scholar]
- Guo F. Q., Okamoto M. & Crawford N. M. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302, 100–103 (2003). [DOI] [PubMed] [Google Scholar]
- Rockel P., Strube F., Rockel A., Wildt J. & Kaiser W. M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53, 103–110 (2002). [PubMed] [Google Scholar]
- Godber B. L. J. et al. Reduction of nitrite to nitric oxide catalyzed by xanthine oxidoreductase. J. Biol. Chem. 275, 7757–7763 (2000). [DOI] [PubMed] [Google Scholar]
- Bethke P. C., Badger M. R. & Jones R. L. Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell 16, 332–341 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier J. L., Samouilov A. & Kuppusamy P. Non-enzymatic nitric oxide synthesis in biological systems. Biochim. Biophys. Acta 1411, 250–262 (1999). [DOI] [PubMed] [Google Scholar]
- Erez O. & Kahana C. Screening for modulators of spermine tolerance identifies Sky1, the SR protein kinase of Saccharomyces cerevisiae, as a regulator of polyamine transport and ion homeostasis. Mol. Cell. Biol. 21, 175–184 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalasekera R., Tebartz F. & Scherer G. F. E. Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 181, 593–603 (2011). [DOI] [PubMed] [Google Scholar]
- Hager K. M. & Yanofsky C. Genes expressed during conidiation in Neurospora crassa: molecular characterization of con-13. Gene 96, 153–159 (1990). [DOI] [PubMed] [Google Scholar]
- Springer M. L., Hager K. M., Engele C. G. & Yanofsky C. Timing of synthesis and cellular localization of two conidiation-specific proteins of Neurospora crassa. Dev. Biol. 152, 255–262 (1992). [DOI] [PubMed] [Google Scholar]
- Bell-Pederson D., Dunlap J. C. & Loros J. J. The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 6, 2382–2394 (1992). [DOI] [PubMed] [Google Scholar]
- Lauter F. R. & Yanofsky C. Day/night and circadian rhythm control of con gene expression in Neurospora. Proc. Natl. Acad. Sci. USA 90, 8249–8253 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyöngyösi N. & Káldi K. Interconnections of reactive oxygen species homeostasis and circadian rhythm in Neurospora crassa. Antioxid. Redox Signal. 20, 3007–3023 (2014). [DOI] [PubMed] [Google Scholar]
- Cano-Dominguez N., Alvarez-Delfin K., Hansberg W. & Aguirre J. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot. Cell 7, 1352–1361 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott B. & Eaton C. J. Role of reactive oxygen species in fungal cellular differentiations. Curr. Opin. Microbiol. 11, 488–493 (2008). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.