Abstract
Heat shock transcription factor (Hsf) family is one of the most important regulators in the plant kingdom. Hsf has been demonstrated to be involved in various processes associated with plant growth, development as well as in response to hormone and abiotic stresses. In this study, we carried out a comprehensive analysis of Hsf family in desert poplar, Populus euphratica. Total of 32 genes encoding Hsf were identified and they were classified into three main classes (A, B, and C). Gene structure and conserved motif analyses indicated that the members in each class were relatively conserved. Total of 10 paralogous pairs were identified in PeuHsf family, in which nine pairs were generated by whole genome duplication events. Ka/Ks analysis showed that PeuHsfs underwent purifying selection pressure. In addition, various cis-acting elements involved in hormone and stress responses located in the promoter regions of PeuHsfs. Gene expression analysis indicated that several PeuHsfs were tissue-specific expression. Compared to Arabidopsis, more PeuHsf genes were significantly induced by heat, drought, and salt stresses (21, 19, and 22 PeuHsfs, respectively). Our findings are helpful in understanding the distinguished adaptability of P. euphratica to extreme environment and providing a basis for functional analysis of PeuHsfs in the future.
As the terminal components of stress signal transduction chain, heat shock transcription factors (Hsfs) play an important role in mediating the expression of genes responsive to various abiotic stresses, especially heat stress (HS)1. Hsfs specifically recognize the binding motifs ‘AGAAnnTTCT’, called heat stress elements (HSEs) conserved in promoters of HS-inducible genes2.
The structure of Hsfs was similar to other transcription factors, a classical Hsf was composed by N-terminal highly conserved DNA-binding domain (DBD), oligomerization domain (OD), nuclear localization signal (NLS), nuclear export signal (NES), repressor domain (RD), and C-terminal activator peptide motif (AHA)2. The DBD is characterized by a central helix-turn-helix motif that specifically binds to the HSEs in promoters of target genes, this domain is the best preserved domain during the evolution3. The OD with a pattern of hydrophobic heptad repeats, referred to as the HR-A/B region, form a coiled-coil structure mediate oligomerization of Hsfs2. According to the length of the linker between DBD and HR-A/B regions and the amino acid length inserted into the HR-A/B region, plant Hsfs are grouped into three main classes (A, B, and C)4. The NLS and NES mediate the intracellular distribution, while activity of Hsfs depends on the shutting balance between nuclei and cytoplasm5. The RD are characterized by the tetrapeptide LFGV in the C-terminal of class B Hsfs, except HsfB5, which is function as repressor motif through interaction with the corepressor6. The C-terminal AHA motifs confer the transcriptional activator function of Hsfs and are class A Hsf specific and not found in class B or C2,4.
Numerous studies have indicated that Hsfs play important roles in plant responses to various environmental stresses, including heat, cold, drought, salinity, and oxidative stresses3. Within class A Hsf, the members of HsfA1 subclass are considered as master regulators of HS response. The tomato HsfA1a has been reported as the single master regulator in thermotolerance7. And HsfA1a is required for the nuclear retention and transcriptional activation of HsfA28. However, the situation might be inconsistent in different plant species. For example, single or multiple mutants of four HsfA1s (a, b, d, and e) revealed no such master HsfA1 regulating the thermotolerance in Arabidopsis9. In many plant species, such as tomato, Arabidopsis, rice, Populus trichocarpa, and Salix suchowensis, HsfA2 is the most strongly induced Hsf under heat stress10,11,12,13. In heat stressed cells, HsfA2 becomes the dominant Hsf after long-term HS or repeated cycles of HS and recovery14. Although HsfA2 can function to improve thermotolerance in the absence of HsfA1s when overexpressed in a quadruple knockout mutant Arabidopsis (athsfA1a,b,d,e)15. The hetero-oligomerization of HsfA1 and HsfA2 leads to form a superactivator complex, which activates downstream HS-inducible genes in a stronger manner than individuals10. In contrast, the HsfA4 and HsfA5 can also form oligomers, but their activities are inhibited. HsfA4 controls the ROS levels and is considered as an antiapoptotic factor, while HsfA5 specifically represses HsfA4 by inhibiting its DNA binding activity and acting as a proapoptotic factor16. HsfA3 is involved in drought and salt stress responses and is located in downstream of the DREB2A transcriptional cascade system17. In comparison to class A Hsfs, class B Hsfs have no typical transcriptional activity as they lack of an activator domain2. In tomato, the HS-induced HsfB1 act as a coactivator of HsfA1a by assembling into an enhanceosome-like complex14. Tomato HsfA1a, HsfA2, and HsfB1 form a triad to activate the expression of HS-inducible genes during HS response and recovery2. But HsfB1 from Arabidopsis is described as a repressor of HS-responsive Hsfs18. This indicates the Hsf members, even the orthologous genes, play divergent roles in different species. Recently, genome-wide expression profiles analyses of Hsfs were performed in many plant species, including two woody species P. trichocarpa and S. suchowensis13,19. However, little is known about Hsfs in the desert poplar, P. euphratica.
P. euphratica, along with the two other genome sequenced woody species, P. trichocarpa and S. suchowensis, belong to the Salicaceae family. Their morphological characteristics and stress tolerance performances were significantly different. As a native species in desert regions, P. euphratica shows distinguished adaptability to various abiotic stresses20.
To explore the potential roles of P. euphratica Hsfs in abiotic stress responses, the current study identified 32 PeuHsf genes and analyzed their evolutionary relationships, gene structures, conserved domains, cis-acting elements, and expression patterns across different tissues and under various abiotic stresses. In addition, the expression profiles of PeuHsfs under different abiotic stresses were compared to the well-studied Hsfs in Arabidopsis. In conclusion, the present study is helpful in understanding the distinguished stress tolerance of P. euphratica and providing the basis for further functional studies of PeuHsfs.
Results
Genome-wide identification of the Hsf genes in P. euphratica
To identify the Hsf genes in P. euphratica, the conserved Hsf domain (PF00447) from the Pfam database was used to search against the P. euphratica genome. In addition, the amino acid sequences of 27 AtHsfs and 31 PtHsfs were used as query to perform BLASTP search in the P. euphratica genome. After detection of the conserved DBD domain and the coiled-coil structure from the SMART database, a total of 32 PeuHsfs were identified (Table 1). The subfamily classification of PeuHsf was based on the length of the linker between DBD and HR-A/B regions and the amino acid length inserted into the HR-A/B regions. Moreover, the results were also confirmed in the Heatster database (http://www.cibiv.at/services/hsf/)2. The identified PeuHsfs encode proteins ranging from 207 to 737 amino acids (aa) in length (average of 384 aa), with molecular weight (MW) of 23.89 to 81.18 kDa (average of 42.91 kDa) and isoelectric point (pI) of 4.72 to 9.28. Among the 32 PeuHsf proteins, the percentage of negatively charged residues (Asp + Glu) ranged from 8.8% to 17.7% and the percentage of positively charged residues (Arg + Lys) ranged from 7.8% to 16.7%. Based on the instability index analysis, all the PeuHsf proteins were unstable. In addition, the aliphatic index had a range of 57.58 to 79.06 and the grand average of hydropathicity ranged from −0.903 to −0.415 (Table 1).
Table 1. The Hsf genes identified from the P. euphratica.
| Gene Name | ID | Map position (bp) | Length (aa) | MW (kDa) | pI | n.c.r. (%) | p.c.r. (%) | I.I. | Stability | A.I. | GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PeuHsf-A1a | CCG006597.1 | scaffold1541.1:1132–3828(+) | 491 | 53.7 | 4.95 | 63 (12.8%) | 41 (8.4%) | 61.54 | unstable | 72.75 | −0.553 |
| PeuHsf-A1b | CCG033924.1 | scaffold96.1:588160–589431(−) | 423 | 46.6 | 5.53 | 50 (11.8%) | 38 (9%) | 65.03 | unstable | 65.93 | −0.626 |
| PeuHsf-A1c | CCG014424.1 | scaffold25.1:1905477–1910713(−) | 510 | 56 | 4.79 | 67 (13.1%) | 45 (8.8%) | 64.06 | unstable | 67.73 | −0.647 |
| PeuHsf-A1d | CCG024768.1 | scaffold49.1:1143998–1146694(−) | 523 | 57.2 | 4.95 | 64 (12.2%) | 41 (7.8%) | 62.77 | unstable | 76.33 | −0.486 |
| PeuHsf-A2 | CCG009416.1 | scaffold19.1:1223913–1226045(−) | 425 | 48.1 | 5.04 | 65 (15.3%) | 47 (11.1%) | 53.2 | unstable | 71.29 | −0.573 |
| PeuHsf-A3 | CCG023633.1 | scaffold4512.1:713–3120(+) | 522 | 58.2 | 4.78 | 82 (15.7%) | 51 (9.8%) | 66.9 | unstable | 65.17 | −0.613 |
| PeuHsf-A4a | CCG026694.1 | scaffold56.1:184555–187033(+) | 404 | 45.9 | 5.22 | 57 (14.1%) | 43 (10.6%) | 56.17 | unstable | 65.17 | −0.801 |
| PeuHsf-A4b | CCG015090.1 | scaffold26.1:1129045–1132041(−) | 444 | 51 | 5.62 | 64 (14.4%) | 49 (11%) | 62.36 | unstable | 65.43 | −0.794 |
| PeuHsf-A4c | CCG030319.1 | scaffold739.1:61353–63562(−) | 427 | — | — | 61 (14.3%) | 45 (10.5%) | 55.87 | unstable | 63 | −0.772 |
| PeuHsf-A5a | CCG001724.1 | scaffold1039.1:58159–61226(−) | 485 | 54.1 | 5.96 | 63 (13%) | 56 (11.5%) | 56.93 | unstable | 67.2 | −0.783 |
| PeuHsf-A5b | CCG019200.1 | scaffold323.1:258192–261325(−) | 489 | 54.6 | 5.92 | 65 (13.3%) | 57 (11.7%) | 59.16 | unstable | 70.43 | −0.785 |
| PeuHsf-A5c | CCG019201.1 | scaffold323.1:281255–285809(−) | 737 | 81.2 | 7.02 | 82 (11.1%) | 81 (11%) | 54.67 | unstable | 77.84 | −0.577 |
| PeuHsf-A6a | CCG007863.1 | scaffold17.1:1250083–1251797(−) | 358 | 41.4 | 5.14 | 58 (16.2%) | 43 (12%) | 54.42 | unstable | 67.23 | −0.788 |
| PeuHsf-A6b | CCG000940.1 | scaffold1.1:8456623–8458194(+) | 331 | 38.2 | 5.08 | 56 (15.8%) | 41 (11.5%) | 59.91 | unstable | 72.15 | −0.736 |
| PeuHsf-A7a | CCG013195.1 | scaffold23.1:595273–598538(−) | 355 | 40.2 | 5.33 | 55 (15.5%) | 45 (12.7%) | 66.56 | unstable | 64.31 | −0.882 |
| PeuHsf-A7b | CCG017804.1 | scaffold30.1:760950–762628(−) | 359 | 41.2 | 5.44 | 54 (15%) | 47 (13.1%) | 55.04 | unstable | 68.75 | −0.788 |
| PeuHsf-A7c | CCG034159.1 | scaffold988.1:37988–39666(−) | 359 | 41.2 | 5.44 | 54 (15%) | 47 (13.1%) | 55.25 | unstable | 68.75 | −0.782 |
| PeuHsf-A8 | CCG033049.1 | scaffold9.1:2064027–2067752(−) | 392 | 44.7 | 4.72 | 66 (16.8%) | 37 (9.4%) | 43.46 | unstable | 72.6 | −0.698 |
| PeuHsf-A9 | CCG013896.1 | scaffold240.1:34097–35820(+) | 501 | 55.8 | 5.28 | 76 (15.2%) | 58 (11.6%) | 57.98 | unstable | 71.86 | −0.586 |
| PeuHsf-B1 | CCG009216.1 | scaffold188.1:576141–580111(+) | 288 | 31.4 | 4.73 | 51 (17.7%) | 35 (12.2%) | 38.99 | unstable | 58.26 | −0.903 |
| PeuHsf-B2a | CCG001341.1 | scaffold1000.1:34084–35186(+) | 294 | 32.4 | 5.02 | 44 (15%) | 34 (11.6%) | 49.29 | unstable | 68.54 | −0.669 |
| PeuHsf-B2b | CCG024466.1 | scaffold48.1:934135–936766(−) | 340 | 36.6 | 4.95 | 47 (13.8%) | 35 (10.3%) | 53.42 | unstable | 69.76 | −0.552 |
| PeuHsf-B2c | CCG005948.1 | scaffold148.1:57661–59281(+) | 287 | 31.9 | 5.02 | 47 (16.4%) | 37 (12.9%) | 52.39 | unstable | 79.06 | −0.507 |
| PeuHsf-B2d | CCG007214.1 | scaffold16.1:1230405–1231507(+) | 294 | 32.4 | 5.02 | 44 (15%) | 34 (11.6%) | 49.29 | unstable | 68.54 | −0.669 |
| PeuHsf-B3 | CCG001458.1 | scaffold101.1:1374018–1376473(−) | 228 | 26.5 | 7.62 | 37 (16.2%) | 38 (16.7%) | 55.59 | unstable | 72.28 | −0.736 |
| PeuHs -B4a | CCG027759.1 | scaffold6.1:2805693–2807677(+) | 364 | 40.4 | 8.15 | 32 (8.8%) | 34 (9.3%) | 51.71 | unstable | 73.9 | −0.466 |
| PeuHsf-B4b | CCG020996.1 | scaffold38.1:136369–138015(+) | 271 | 31.5 | 6.49 | 34 (12.5%) | 31 (11.4%) | 56.94 | unstable | 71.51 | −0.585 |
| PeuHsf-B4c | CCG010253.1 | scaffold2.1:1006270–1007951(+) | 368 | 41 | 7.72 | 33 (9%) | 34 (9.2%) | 55.68 | unstable | 69.16 | −0.552 |
| PeuHsf-B4d | CCG023235.1 | scaffold44.1:1597707–1599573(−) | 270 | 31.3 | 7.18 | 32 (11.9%) | 32 (11.9%) | 58.82 | unstable | 68.93 | −0.651 |
| PeuHsf-B5a | CCG006244.1 | scaffold15.1:1276216–1278351(+) | 209 | 24.1 | 9.24 | 26 (12.4%) | 33 (15.8%) | 55.35 | unstable | 71 | −0.649 |
| PeuHsf-B5b | CCG032077.1 | scaffold84.1:65593–67546(−) | 207 | 23.9 | 9.28 | 22 (10.6%) | 30 (14.5%) | 52.67 | unstable | 57.58 | −0.744 |
| PeuHsf-C1 | CCG004861.1 | scaffold135.1:519792–520919(+) | 338 | 37.8 | 5.92 | 40 (11.8%) | 37 (10.9%) | 49.6 | unstable | 75.86 | −0.415 |
Notes: pI, isoelectric point; n.c.r., total number of negatively charged residues (Asp + Glu); p.c.r., total number of positively charged residues (Arg + Lys); I.I., instability index; A.I., aliphatic index; GRAVY, grand average of hydropathicity.
Phylogenetic relationship of PeuHsf genes
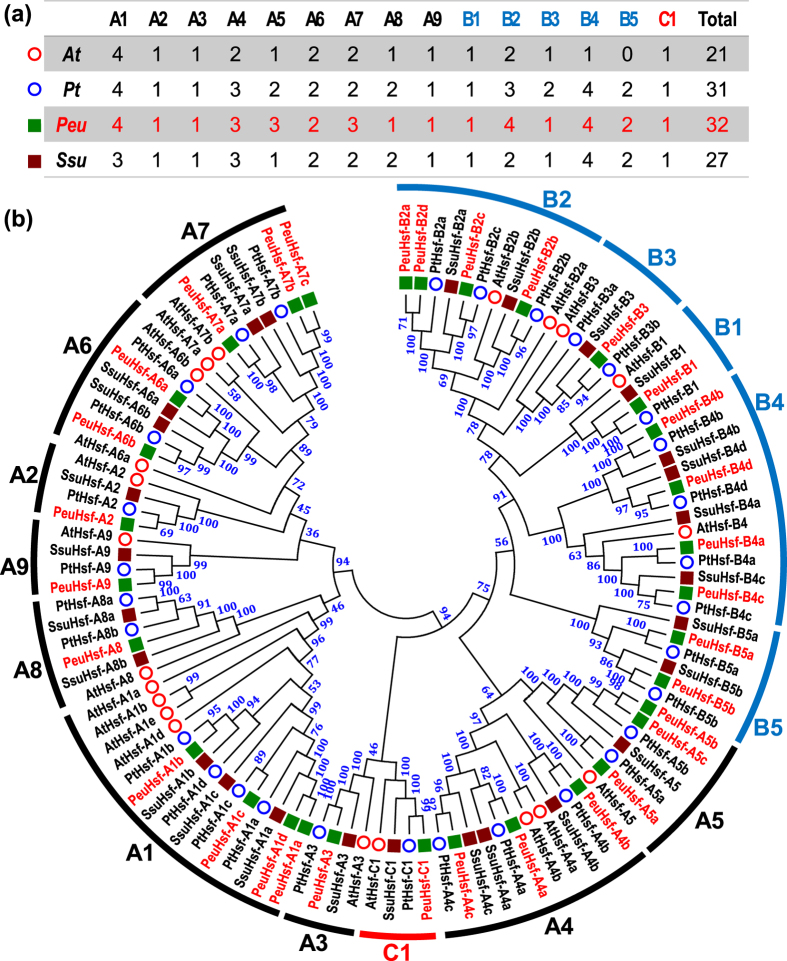
To explore the evolutionary characteristics of the PeuHsf genes, an unrooted phylogenetic tree was generated using the Hsf protein sequences from P. euphratica, P. trichocarpa, S. suchowensis, and A. thaliana. Compared to the two species in Salicaceae (P. trichocarpa and S. suchowensis) and model plant Arabidopsis, the desert poplar P. euphratica is the largest in size of the Hsf family (Fig. 1a). According to the phylogenetic tree, the PeuHsfs could be grouped into three main classes (A, B, and C) (Fig. 1b). Among the three main classes, class A was the largest class consisting of 19 members from nine subclasses (A1–A9), class B consisted of 12 members from five subclasses (B1–B5), while class C only contained one gene (Fig. 1). The length of class A PeuHsfs ranged from 331 to 737 aa (average of 449 aa), which was relatively longer than class B (207 to 368 aa with average of 285 aa) and class C (338 aa) (Table 1). All the three analyzed Salicaceae species contained Hsf members in subclass B5, while Arabidopsis did not include any member in subclass B5. Compared to the Hsfs in P. trichocarpa, P. euphratica containing one more in subclasses A5, A7, and B2, but one less in subclasses A8 and B3 (Fig. 1 and Supplementary Table S1).
Figure 1. Hsf gene family and their phylogenetic relationships in P. euphratica (Peu), P. trichocarpa (Pt), S. suchowensis (Ssu), and A. thaliana (At).
(a) The members of Hsf from four species were classified into three mainly subfamilies (class A, B, and C). (b) The phylogenetic tree was constructed using full-length of amino acid sequences of Hsf proteins in the four species by the neighbor-joining (NJ) method with 1,000 bootstrap replicates. Bootstrap support values are indicated on each node.
Structural analysis of PeuHsfs
Gene structures
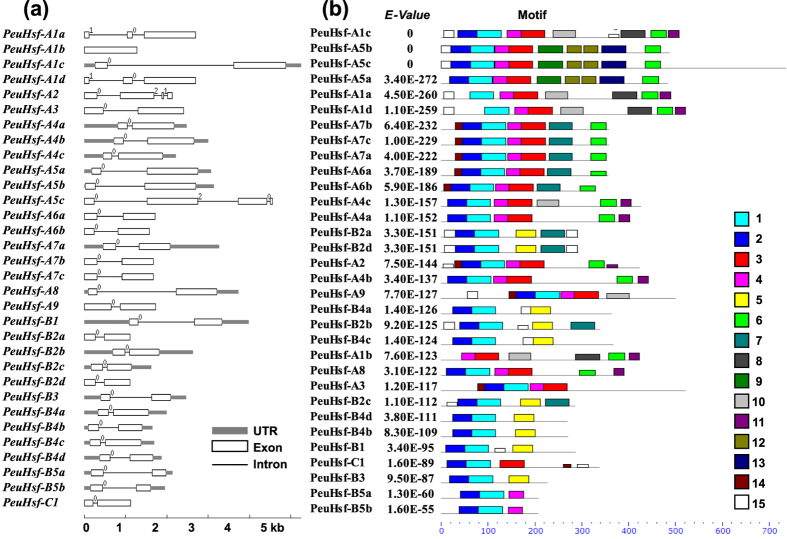
The gene structures of PeuHsfs were analyzed by comparing the cDNA sequence and genomic DNA sequence. As shown in Fig. 2a, two PeuHsfs (-A2 and -A5c) were comprised of four exons, two PeuHsfs (-A1a and -A1d) were comprised of three exons, while PeuHsf-A1b was only comprised of one exon without splice. Except for the aforementioned five PeuHsfs, all the other 27 PeuHsfs were comprised of two exons. Introns can be classified into phase 0, phase 1, and phase 2 depending on their position relative to the reading frame. All the two-exon containing PeuHsfs showed the same intron phase (phase 0). Over all, the gene structure and the intron phases were significantly conserved among the members in each subclass of PeuHsf family.
Figure 2.
Gene structure (a) and conserved protein motifs (b) of members in PeuHsf family. (a) Grey box, blank box, and black line were represented UTR, exon, and intron, respectively. The number 0, 1, and 2 on the black line were intron phase. (b) Total of 15 conserved motifs were identified using MEME. The detail were listed in Supplementary Table S2.
Conserved motifs
Based on the known information of Hsfs in Arabidopsis, six conserved domains including DBD, HR-A/B, NLS, NES, AHA, and RD were identified in PeuHsf proteins (Table 2). As the binding domain to the downstream functional genes, the DBD existed in all the PeuHsfs. The DBD was comprised of three α-helices and four β-sheets in the form of α1-β1-β2-α2-α3-β3-β4 (Supplementary Fig. S1). All the members of PeuHsf included NLS, which consists of nucleus localization of the transcription factor. With the exception of the members in subclasses A3 and A9, all the other class A PeuHsfs had NES domains. AHA motifs were only detected in class A PeuHsfs (except for PeuHsf-A9), four proteins in subclasses A2 and A4 had two AHA motifs, while PeuHsf-A3 had a longer AHA motif (W-X17-W-X20-W-X15-W) than the other members (Table 2).
Table 2. Functional domains of PeuHsfs.
| Gene Name | DBD | HR-A/B | NLS | NES | AHA | RD |
|---|---|---|---|---|---|---|
| PeuHsf-A1a | 33–110 | 143–190 | (223) NKKRRLKQ | (475) VDQLTEQME | (432) SSFWDDLLVQ | N.D. |
| PeuHsf-A1b | 1–35 | 61–108 | (141) SKKRRLPR | (409) MNRLAEQMG | (361) DVFWEQFLTA | N.D. |
| PeuHsf-A1c | 33–126 | 159–206 | (239) NKKRRLKQ | (493) MDQLTEQMG | (450) SSFWDDLLAQ | N.D. |
| PeuHsf-A1d | 78–142 | 175–222 | (255) NKKRRLKQ | (507) VDQLTEQME | (464) SSFWDDLLVQ | N.D. |
| PeuHsf-A2 | 40–133 | 157–201 | (229) RR-X8-RKRR | (411) LDSTALYVGFL | (318) ETIWEEFLTD; (357) DWSDDFQE | N.D. |
| PeuHsf-A3 | 90–183 | 209–253 | (270) ARLKQKKEQ | N.D. | (446) W-X17-W-X20-W-X15-W | N.D. |
| PeuHsf-A4a | 10–103 | 129–171 | (204) DRKRRL | (391) LTEQMGHL | (257) LTFWENMVHD; (340) DVFWEQFLTE | N.D. |
| PeuHsf-A4b | 11–104 | 125–178 | (203) NKKRKL | (431) LAKNMGHI | (253) LKFLEDFLYA; (378) DLFWQHFLTE | N.D. |
| PeuHsf-A4c | 10–103 | 129–174 | (204) DRKRRL | (394) LTEQMGHL | (258) LTFWENMVND; (343) DVFWEQFLTE | N.D. |
| PeuHsf-A5a | 14–107 | 126–177 | (196) SK-X10-KKRR | (480) MEQLSL | (433) DGFWEQFLTE | N.D. |
| PeuHsf-A5b | 18–111 | 133–181 | (200) RK-X10-KKRR | (484) MEQLSL | (438) DVFWEQFLTE | N.D. |
| PeuHsf-A5c | 18–111 | 133–181 | (200) RK-X10-KKRR | (484) MEQLSL | (438) DVFWEQFLTE | N.D. |
| PeuHsf-A6a | 40–133 | 159–197 | (238) KKKRR | (346) LVEQLGYL | (322) EAFWEDLLNE | N.D. |
| PeuHsf-A6b | 17–110 | 138–180 | (215) KKRRR | (317) LGSEGED | (299) EGFWEDLLNE | N.D. |
| PeuHsf-A7a | 42–135 | 163–234 | (231) KKKELEEAMTKKRRR | (343) LAERMGYL | (322) EGFWEELLNE | N.D. |
| PeuHsf-A7b | 42–135 | 163–208 | (231) KRKELEEAMTKKRR | (344) LAERLGYL | (322) EGFWEELLNE | N.D. |
| PeuHsf-A7c | 42–135 | 163–208 | (231) KRKELEEAMTKKRR | (344) LAERLGYL | (322) EGFWEELLNE | N.D. |
| PeuHsf-A8 | 8–101 | 135–175 | (161) NKLLLLRDR | (380) TEQMGLL | (298) DGSWEQLLLA | N.D. |
| PeuHsf-A9 | 157–250 | 272–313 | (341) KR-X12-KKRR | N.D. | N.D. | N.D. |
| PeuHsf-B1 | 6–99 | 151–179 | (255) LFGV-X6-KKKR | N.D. | N.D. | (255) LFGV |
| PeuHsf-B2a | 27–120 | 161–189 | (168) RLRK | N.D. | N.D. | (225) IFGV |
| PeuHsf-B2b | 36–129 | 196–222 | (282) LFGV-X4-KRVR | N.D. | N.D. | (282) LFGV |
| PeuHsf-B2c | 32–125 | 173–214 | (177) RLRK | N.D. | N.D. | (235) IFGV |
| PeuHsf-B2d | 27–120 | 161–189 | (168) RLRK | N.D. | N.D. | (225) IFGV |
| PeuHsf-B3 | 15–108 | 146–192 | (195) LFGV-X9-RKRK | N.D. | N.D. | (195) LFGV |
| PeuHsf-B4a | 21–114 | 190–218 | (318) LFGV-X4-KKR | N.D. | N.D. | (318) LFGV |
| PeuHsf-B4b | 21–114 | 156–188 | (254) LFGV-X4-NKR | N.D. | N.D. | (254) LFGV |
| PeuHsf-B4c | 21–114 | 198–221 | (322) LFGV-X4-KKR | N.D. | N.D. | (322) LFGV |
| PeuHsf-B4d | 21–114 | 152–186 | (253) LFGV-X4-NKR | N.D. | N.D. | (253) LFGV |
| PeuHsf-B5a | 29–131 | 164–182 | (119) RGRR | N.D. | N.D. | N.D. |
| PeuHsf-B5b | 28–128 | 158–182 | (164) NKNLRR | N.D. | N.D. | N.D. |
| PeuHsf-C1 | 9–102 | 131–157 | (193) KKQR | N.D. | N.D. | N.D. |
N.D.: no motifs detectable by sequence similarity search.
In addition, the conserved motifs of PeuHsfs were also analyzed using MEME. Among the 15 detected motifs, motifs 1 and 2 existed in almost all of the PeuHsfs; the two motifs including the highly conserved DBD. Motif 3 represented coiled-coil structure in classes A and C PeuHsfs, while which was replaced by motif 5 in class B PeuHsfs. Moreover, motif 6 corresponding to the AHA motif nearby the C-terminus of class A PeuHsfs (Fig. 2b).
Expansion of Hsf family in P. euphratica
Duplication events
To analyze the expansion of Hsf genes in P. euphratica, the PeuHsfs were mapped onto each scaffold based on publicly available information provided in the P. euphratica genome database. A total of 32 PeuHsfs were mapped onto 31 scaffolds (Supplementary Fig. S2). The synteny analysis was performed to identify the duplicated blocks. Based on the chromosomal location, duplicated blocks, phylogenetic relationships, gene structures, and conserved motifs of the PeuHsfs, total of 10 paralogous pairs were identified in PeuHsf family (Table 3). With the exception of one paralogous pair (PeuHsf-A5b/PeuHsf-A5c) that was generated by tandem duplication event, the other nine pairs were generated by whole genome duplication (WGD) (Supplementary Fig. S2). Two pairs (PeuHsf-A1a/PeuHsf-A1d and PeuHsf-A5b/PeuHsf-A5c) were generated in the latest stage (~0.49 MYA), three pairs (PeuHsf-A4a/PeuHsf-A4c, PeuHsf-A6a/PeuHsf-A6b, and PeuHsf-A7a/PeuHsf-A7b) were generated in ~13 MYA, and the other five pairs were generated between 17–20 MYA (Table 3). Comparatively, the generation date of P. trichocarpa Hsf paralogous pairs was relatively concentrated, one pair was generated in ~7.87 MYA, five pairs were generated in ~13 MYA, four pairs were generated in 17–20 MYA19.
Table 3. Divergence between paralogous PeuHsf gene pairs.
| Subfamily | Gene 1 | Gene 2 | Type | Ka | Ks | Ka/Ks | Date (MYA) |
|---|---|---|---|---|---|---|---|
| A1 | PeuHsf-A1a | PeuHsf-A1d | WGD | 0.004 | 0.009 | 0.389 | 0.49 |
| A1 | PeuHsf-A1c | PeuHsf-A1d | WGD | 0.173 | 0.343 | 0.503 | 18.85 |
| A4 | PeuHsf-A4a | PeuHsf-A4c | WGD | 0.069 | 0.253 | 0.274 | 13.90 |
| A5 | PeuHsf-A5b | PeuHsf-A5c | Tandem | 0.003 | 0.009 | 0.303 | 0.49 |
| A6 | PeuHsf-A6a | PeuHsf-A6b | WGD | 0.116 | 0.204 | 0.568 | 11.20 |
| A7 | PeuHsf-A7a | PeuHsf-A7b | WGD | 0.077 | 0.279 | 0.275 | 15.31 |
| B2 | PeuHsf-B2c | PeuHsf-B2d | WGD | 0.141 | 0.362 | 0.388 | 19.91 |
| B4 | PeuHsf-B4a | PeuHsf-B4c | WGD | 0.022 | 0.325 | 0.068 | 17.86 |
| B4 | PeuHsf-B4b | PeuHsf-B4d | WGD | 0.061 | 0.316 | 0.194 | 17.38 |
| B5 | PeuHsf-B5a | PeuHsf-B5b | WGD | 0.096 | 0.335 | 0.286 | 18.38 |
Among the four analyzed species, 10, 10, 5, and 4 Hsf paralogous pairs were detected in P. euphratica, P. trichocarpa, S. suchowensis, and A. thaliana, respectively (Supplementary Table S3). With respect to the total number of Hsfs in each species, the proportions of duplicated Hsfs were relatively high in two Populus species (10 pairs in 31 PtHsfs or 32 PeuHsfs). Although the same number of Hsf paralogous pairs were identified in the allied species P. euphratica and P. trichocarpa, the composition of paralogous pairs was significantly different - two subclass A1 Hsf paralogous pairs in P. euphratica but only one pair in P. trichocarpa, Hsf paralogous pair in subclass B5 was specifically in P. euphratica while Hsf paralogous pairs in subclasses A8 and B3 were specifically in P. trichocarpa (Supplementary Table S3).
Ka/Ks values
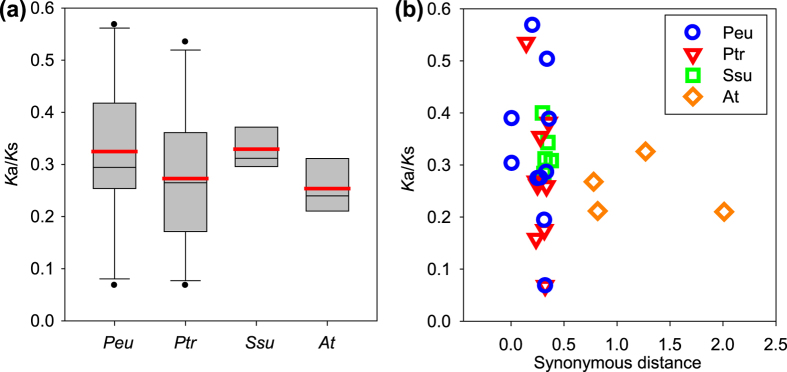
Subsequently, the substitution rate ratio of non-synonymous (Ka) vs. synonymous (Ks) substitutions was analyzed to verify the Darwinian position selection. The Ka/Ks ratios of all the 10 PeuHsf paralogous pairs were less than 0.6 (Table 3), indicating that the PeuHsf family had undergone purifying selection pressure. Compared to S. suchowensis and A. thaliana, the two Populus species had wide range of Ka/Ks, where the average Ka/Ks in P. euphratica was larger than that in P. trichocarpa (Fig. 3a). When combined, looking at the synonymous distance, we found that the synonymous distance of all the Hsf paralogous pairs were less than 0.5 in Salicaceae (P. euphratica, P. trichocarpa, and S. suchowensis) but more than 0.7 in A. thaliana, which resulted in the distribution of AtHsf paralogous pairs departing from PeuHsf, PtHsf, and SsuHsf paralogous pairs (Fig. 3b).
Figure 3. Estimates of Ka/Ks ratios in pairwise comparisons in four species Hsf families.
(a) Average molecular evolutionary rate (Ka/Ks) for Hsf genes in P. euphratica (Peu), P. trichocarpa (Ptr), S. suchowensis (Ssu), and A. thaliana (At). Red line in each box indicates average value of Ka/Ks in each species. (b) The Ka/Ks ratios of the duplicated Hsf genes in four species are shown in the scatter plots, the y and x axes denote the Ka/Ks ratio and synonymous distance for each pair. The detail of the Ka and Ks in the four species was listed in Supplementary Table S3.
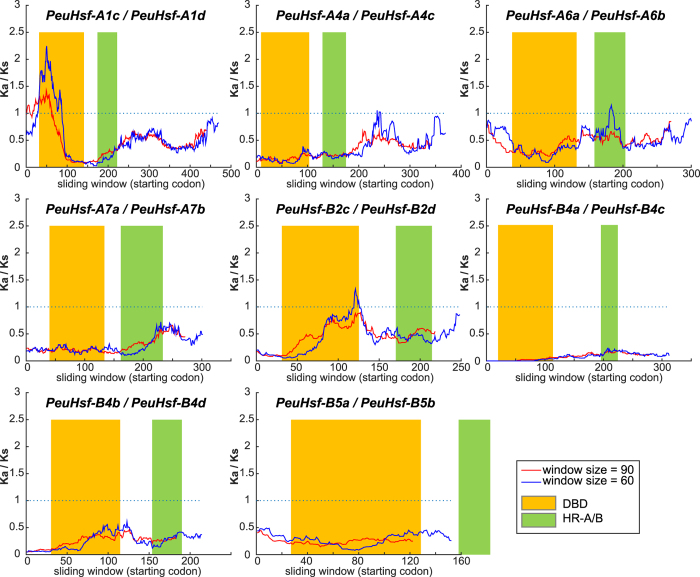
Furthermore, a sliding window analysis of the Ka/Ks ratios was carried out for pairwise comparison. Two pairs (PeuHsf-A1a/PeuHsf-A1d and PeuHsf-A5b/PeuHsf-A5c) did not obtain the results because of their extremely similar sequences. Among the other eight PeuHsf paralogous pairs, the N-terminus of DBD in PeuHsf-A1c/PeuHsf-A1d pair had a significant peak higher than 1 that was under positive selection, while all the other regions were under purifying selection (Fig. 4). However, all the DBD and HR-A/B regions in Arabidopsis Hsf paralogous pairs were conserved with very low Ka/Ks ratio, except for the region behind HR-A/B in AtHsf-A1a/AtHsf-A1c pair had a peak more than 1 (Supplementary Fig. S3).
Figure 4. Sliding window plots of representative duplicated Hsf genes in P. euphratica.
The window sizes were 90 bp and 60 bp separately.
Variety of cis-acting elements in the promoter regions of PeuHsf genes
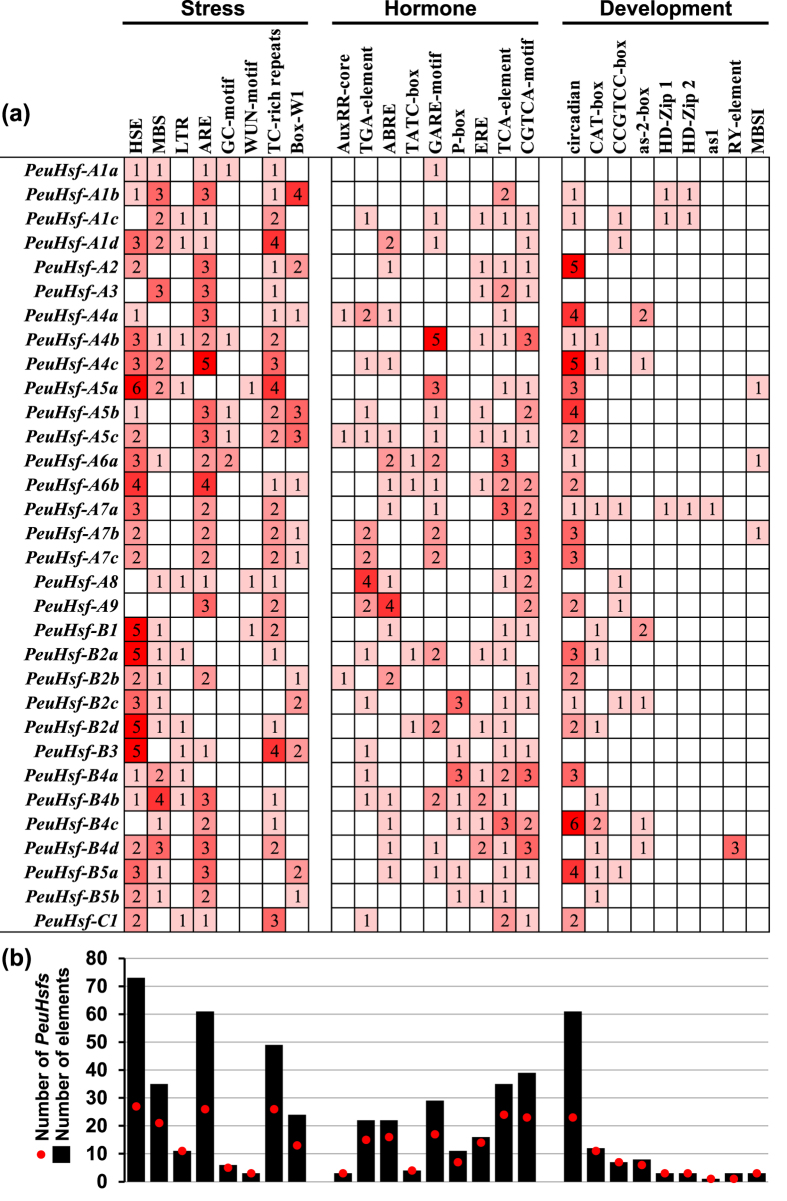
The 2 kb upstream sequences of the translation initiating site of the PeuHsfs were searched in the PlantCARE database to identify the potential cis-acting elements. As shown in Fig. 5, the identified cis-acting elements were classified into three mainly functional classes: stress, hormone, and development. In stress-related cis-acting elements, HSE, anaerobic induction element (ARE), TC-rich repeats, and MYB binding site involved in drought inducibility (MBS) were detected in the promoters of 27, 26, 26, and 21 PeuHsfs, respectively. Among these stress-related cis-acting elements, HSE was the most enriched element with enrichment level 2.7 (total of 73 HSE located in 27 PeuHsfs promoters, i.e. 73/27), followed by ARE (enrichment level 2.3), TC-rich repeats (enrichment level 1.9), and MBS (enrichment level 1.7). In hormone-related cis-acting elements, a total of 39 MeJA responsive elements (CGTCA-motif), 35 salicylic acid responsive elements (TCA-element), 29 gibberellin responsive elements (GARE-motif), 22 abscisic acid responsive elements (ABRE), and 22 auxin responsive elements (TGA-element) were detected in the promoters of 23, 24, 17, 16, and 15 PeuHsfs, respectively. Among the development-related cis-acting elements, 23 PeuHsfs had circadian related elements (enrichment level 2.7) and 16 PeuHsfs contained meristem related cis-acting elements (CAT-box or CCGTCC-box). Three PeuHsfs (-A1b, -A1c, and -A7a) had leaf development related elements (HD-Zip1 or HD-Zip2). In addition, three PeuHsfs (-A5a, -A6a, -A7b) contained MYB binding site involved in flavonoid biosynthetic gene regulation (MBSI).
Figure 5. cis-acting elements in the promoter region of PeuHsf genes.
(a) Number of each cis-acting element in the promoter region (2 kb upstream of translation initiation site) of PeuHsf genes. (b) The statistics of total number of PeuHsf genes including corresponding cis-acting elements (red dot) and total number of cis-acting elements in PeuHsf gene family (black box). Based on the functional annotation, the cis-acting elements were classified into three major classes: stress-, hormone-, or development-related cis-acting elements. (HSE, heat stress responsive element; MBS, MYB binding site involved in drought inducibility; LTR, low temperature responsive element; ARE, essential for the anaerobic induction; GC-motif, enhancer-like element involved in anoxic specific inducibility; WUN-motif, wound responsive element; TC-rich repeats, defense responsive element; Box-W1, fungal elicitor responsive element; AuxRR-core and TGA-element, auxin responsive element; ABRE, abscisic acid responsive element; TATC-box, GARE-motif and P-box, gibberellin responsive element; ERE, ethylene responsive element; TCA-element, salicylic acid responsive element; CGTCA-motif, MeJA responsive element; circadian, circadian control; CAT-box, meristem expression; CCGTCC-box, meristem specific activation; as-2-box, shoot-specific expression and light responsiveness; HD-Zip1, differentiation of the palisade mesophyll cells; HD-Zip2, control of leaf morphology development; as1, root-specific expression; RY-element, seed-specific regulation; MBSI, MYB binding site involved in flavonoid biosynthetic genes regulation.)
Expression profiles of PeuHsfs across different tissues and response to various abiotic stresses
Tissue-specific expression
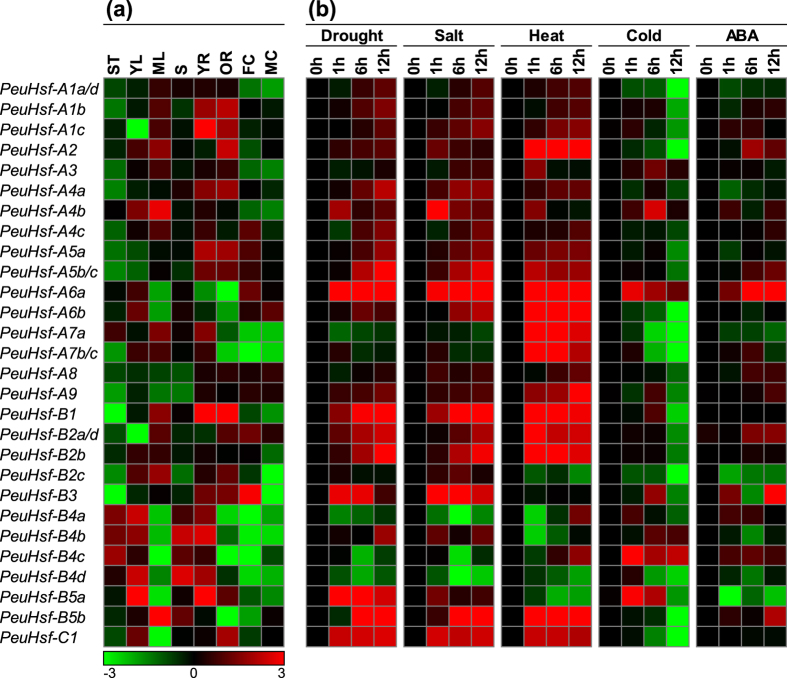
We then analyzed the spatial and temporal expression profiles of PeuHsfs in eight tissues including shoot tip (ST), young leaf (YL), mature leaf (ML), stem (S), young root (YR), old root (OR), female catkin (FC), and male catkin (MC). Because of the highly similarity of four pairs (A1a/A1d, A5b/A5c, A7b/A7c, B2a/B2d), no gene-specific primers could be designed to distinguish their expression patterns in each of the pairs. Among the 28 analyzed PeuHsfs, three of four members in subclass B4 PeuHsfs were highly expressed in shoot tip. Eight PeuHsfs (-A1b, -A1c, -A2, -A4b, -A7a, -B1, -B2c, and -B5b) were highly expressed in mature leaf compared with those in young leaf. Four members in subclass B4 PeuHsf were highly expressed in stem. Eight PeuHsfs (-A1b, -A1c, -A4a, -A5a, -A5b/c, -B1, -B3, and -B5a) were highly expressed in both young root and old root. Noticeably, PeuHsf-B3 was highly expressed in female catkin but scarcely expressed in male catkin (Fig. 6a).
Figure 6.
Expression patterns of PeuHsf genes across different tissues (a) and under various abiotic stresses (b) using qRT-PCR. (a) The expression of PeuHsfs in shoot tip (ST), young leaf (YL), mature leaf (ML), stem (S), young root (YR), old root (OR), female catkin (FC), and male catkin (MC) from P. euphratica. (b) The expression patterns of PeuHsfs after treated for 0, 1, 6, or 12 h under drought (25% PEG w/v), salt (300 mM NaCl), heat (42 °C), cold (4 °C), or 100 μM abscisic acid (ABA). To illustrate, the expression of PeuHsfs in different tissues were compared with the median value among these tissues. For different treatment, the expression of PeuHsfs in 1, 6, or 12 h were compared with the control in 0 h. The different colors correspond to log2 transformed value, green indicates down-regulation and red represents up-regulation. The expression patterns of orthologous Hsf genes in Arabidopsis across various tissues and under abiotic stresses were shown in Supplementary Fig. S4 and Supplementary Fig. S5. Primers used for qRT-PCR are listed in Supplementary Table S3.
Stress responses
To identify the roles of PeuHsfs in stress responses, the expression patterns of PeuHsfs were analyzed in the leaves of P. euphratica seedlings treated with drought (25% PEG w/v), salt (300 mM NaCl), heat (42 °C), cold (4 °C), or ABA (Fig. 6b). As the name declares, PeuHsfs were dramatically responsive to heat stresses, as a total of 10 PeuHsfs (-A2, -A5b/c, -A6a, -A6b, -A7a, -A9, -B1, -B2b, -B5b, and -C1) showed prompt induction by heat stress at 1 h. Aside from heat stress, most of the PeuHsfs (e.g. PeuHsf-A4b, -A6a, -B1, -B3, -B5a, -B5b, -C1) were responsive to both drought and salinity stresses. In contrast to the strong responsiveness of PeuHsfs to heat, drought or salt stresses, only three PeuHsfs (-A6a, -B4c, and -B5a) were promptly induced after 1 h under cold stress. It is important to note that PeuHsf-A6a was promptly and significantly induced by all the tested treatments in this study (Fig. 6b).
Expression divergence between paralogous pairs
The expression patterns of duplicated PeuHsf genes were divergent during the evolution. As indicated in Figs 6 and 7, most PeuHsfs in paralogous pairs showed different expression patterns. Among the 10 PeuHsf paralogous pairs, genes in two paralogous pairs (PeuHsf-A1a/PeuHsf-A1d and PeuHsf-A5b/PeuHsf-A5c) could not be effectively distinguished by qRT-PCR in our study because of their highly sequence similarity in each pair. The genes in three pairs (PeuHsf-A6a/PeuHsf-A6b, PeuHsf-B4a/PeuHsf-B4c, and PeuHsf-B4b/PeuHsf-B4d) showed similar expression patterns across different tissues (Fig. 6a), but had significant differences under various stresses (Fig. 6b). In contrast, the genes in the remaining five PeuHsf paralogous pairs presented different patterns both in tissues and under stress conditions (Fig. 6).
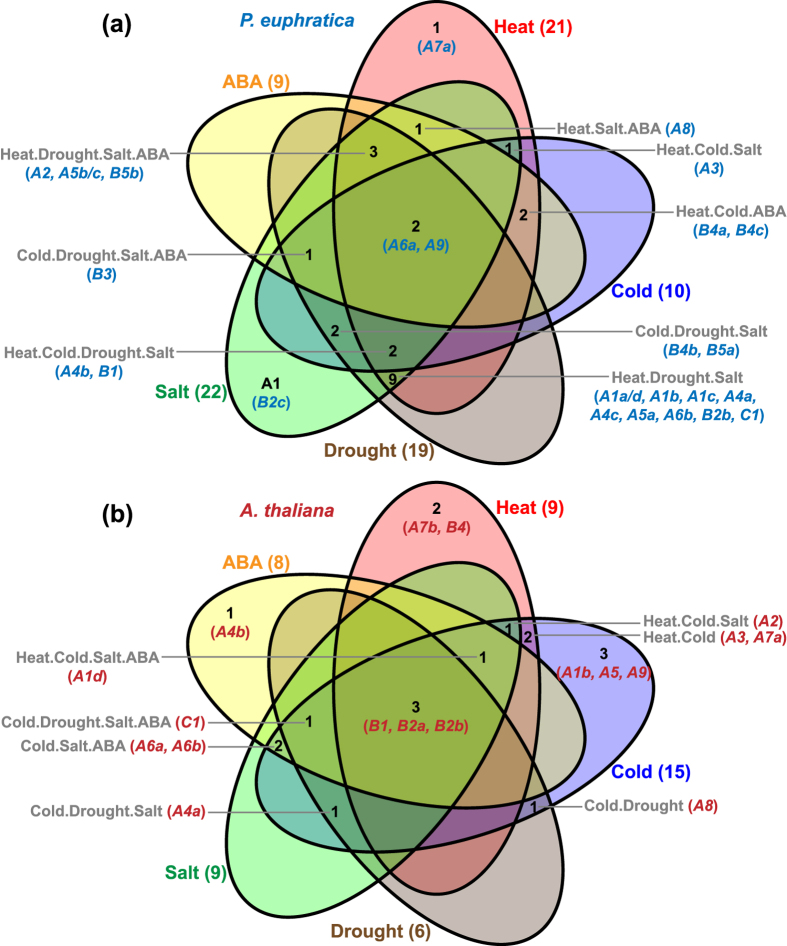
Figure 7.
Different stress response model of Hsf family in P. euphratica (a) and A. thaliana (b). (a) The results were based on our qRT-PCR in Fig. 6b. (b) The results were based on the public available microarray data (see Supplementary Fig. S5).
Divergence of expression patterns between P. euphratica and A. thaliana
To reveal the potential mechanism of distinguished stress tolerance of P. euphratica, the stress response profiles of PeuHsfs were also compared to AtHsfs (see Supplementary Fig. S5). As shown in Fig. 7, the responses of Hsf genes in the two species were significantly different. In Arabidopsis, with the exception of the 9 Hsfs that responded to heat stress, there were 15, 6, 9, and 8 Hsfs that were highly induced by cold, drought, salt, or ABA treatment, respectively (Fig. 7b). While in P. euphratica, more Hsfs were induced by these stresses. Total of 21 PeuHsfs were induced by heat stress. In addition, 22 and 19 PeuHsfs were induced by salt and drought stresses. Total of 10 and 9 PeuHsfs were induced by cold and ABA treatments. The orthologous genes in the two species showed significantly divergent expression patterns. For example, AtHsfA2 was induced by heat, cold, and salt stresses but PeuHsf-A2 was induced by heat, drought, salt, and ABA treatments. In Arabidopsis, three class B AtHsfs (-B1, -B2a, and -B2b) were widely induced by heat, cold, drought, salt, and ABA treatments; while in P. euphratica, only two class A PeuHsfs (-A6a and -A9) were induced by all the five treatments (Fig. 7).
Discussion
As a class of ubiquitous transcription factors, the Hsf gene family plays an important role in various biological processes including plant development and stress tolerance. In this study, the evolution and divergence of the Hsf genes in desert poplar, P. euphratica, were studied to identify specific and subtle changes in these genes resulting in subfunctionalization or possible neofunctionalization.
After a comprehensive analysis, a total of 32 Hsf genes were identified in P. euphratica (Table 1). The size of the Hsf family in P. euphratica was larger than that of other species in Salicaceae such as P. trichocarpa and S. suchowensis13,19. Phylogenetic analysis revealed that Hsf genes in P. euphratica followed a similar distribution pattern as in other plant species (Fig. 1). Based on the previous classification, the PeuHsfs was classified as three main classes (A, B, and C). Classes A and B were further divided into A1-A9 and B1-B5 subclasses. Differing from P. trichocarpa, P. euphratica is a native species in desert area and has great tolerance to drought and salinity20. Compared with the stress sensitive poplar P. trichocarpa, the size of Hsf subclasses A5, A7, and B2 was larger in P. euphratica (Fig. 1). Meanwhile, the members in these subclasses had abundant stress-related cis-acting elements (Fig. 5) and showed significant stress responses (Fig. 6), which might contribute to the distinguished stress tolerance in P. euphratica.
It has been reported that the intron densities were significantly lower in the genes with rapid expression induction in response to stresses. Introns affect the expression efficiency through at least three possible aspects: delay transcript production by 1) splicing, 2) the added length of nascent transcript, or 3) the added energetic cost from increased transcript length21. Our analysis suggests that most of PeuHsfs contain one or two introns, which is helpful to their prompt induction under stress conditions. Interestingly, HsfA2 in P. euphratica has three introns, while its orthologous genes have two and one intron(s) in P. trichocarpa and S. suchowensis, respectively13,19.
Gene duplication plays a critical role in the generation of new genes, increasing the number of these genes, and dispersing them in the genome22. In this study, the expansion of PeuHsf family was primarily due to the WGD events. The duplication events in PeuHsf gene family might introduce neofunctionalization, subfunctionalization, or nonfunctionalization. Most duplicated PeuHsfs display different spatial expression patterns or stress responses (Fig. 6), indicating that PeuHsfs are going through subfunctionalization or neofunctionalization. As dioecious and cross-pollinated plants, the gene flows in Salicaceae plants were stronger than self-pollinated plants such as Arabidopsis, which provided chance for subfunctionalization or neofunctionalization of duplicated genes. Despite P. euphratica, P. trichocarpa, and S. suchowensis belonging to the Salicaceae family, the genome sizes of P. euphratica and P. trichocarpa are significantly larger than S. suchowensis, which might be the reason for the broad range of Ka/Ks value in P. euphratica and P. trichocarpa (Fig. 3).
During the long stage of evolution, selection pressure plays an important role in shaping gene families, resulting in different evolutionary patterns among gene families or even in one gene family23. Our study demonstrates that the Ka/Ks ratios in PeuHsf HR-A/B regions were smaller than those in DBD domains (especially in paralogous pairs PeuHsf-A1c/PeuHsf-A1d and PeuHsf-B2c/PeuHsf-B2d) (Fig. 4), indicating that the HR-A/B region is relatively conserved in PeuHsf family. As the recognition and binding region, DBD domains play crucial roles in determining the specific downstream HS-inducible genes. The relatively high variation in DBD domains between PeuHsf paralogous pairs is helpful in enhancing the binding specificity and diversity of the P. euphratica Hsf family.
Among various cis-acting elements in promoter regions of PeuHsfs, HSE was the most abundant cis-acting element (Fig. 5). In the 32 members of PeuHsf family, total of 27 PeuHsf including 73 HSE in their promoter regions, and with high enrichment level (2.7). Based on our expression analysis (Fig. 6), 12 PeuHsfs (-A2, -A6a, -A6b, -A7a, -A7b, -A7c, -B1, -B2a, -B2b, -B2d, -B5b, and -C1) were highly induced by heat stress, which including 37 HSE in their promoter regions with a relatively higher enrichment level (3.1) than that in all HSE-including genes (2.7). These results suggest that there is a positive correlation between the cis-acting elements and gene expression patterns in PeuHsf gene family.
In P. euphratica, more than 60% Hsf genes were induced by heat, drought, or salt stresses (21, 19, and 22 PeuHsfs, respectively), which were significantly more than that in Arabidopsis (9, 6, and 9 AtHsfs, respectively) (Fig. 7). The expression patterns of PeuHsfs might be the evolved adaptive mechanism of P. euphratica to face the frequent heat, drought, and salt stresses in desert areas. In tomato and Arabidopsis, HsfA2 has a high activator potential to activate the expression of Hsp genes. It has been identified as the dominant Hsf and strongly accumulated under heat stress7. In P. euphratica, the PeuHsfA2 was also highly induced by heat stress, reaching ~500 folds at 1 h after heat stress (Fig. 6). The result implied that HsfA2 might also be the dominant Hsf in P. euphratica similar to the tomato and Arabidopsis.
The members in Hsf family showed different stress responses across various plant species. The P. euphratica HsfA2 was responsive to heat, drought, salt, and ABA treatments (Figs 6 and 7), whileArabidopsis HsfA2 was responsive to heat, cold, and salt stresses (Fig. 7, Supplementary Fig. S5), indicating that HsfA2 might be involved in ABA-independent pathway in Arabidopsis but ABA-dependent pathway in P. euphratica. Noticeably, some heat-insensitive Hsfs were involved in other stress tolerances. For instance, overexpression of wheat HsfA4a enhanced Cd tolerance but did not improve thermotolerance in transgenic rice24. In Arabidopsis, HsfA4a did not response to heat, but could be induced by cold, drought, and salt stresses (Fig. 7b). Consistently, overexpression of a dominant negative mutant form of AtHsfA4a increased oxidative stress sensitivity in transgenic Arabidopsis25. In contrast, HsfA4a in P. euphratica was not only induced by drought and salt stresses, but also induced by heat stress. Moreover, the other members in A4 subclass (PeuHsfA4b and PeuHsfA4c) were also induced by both heat and other stresses (Fig. 7a). It implies that these Hsf genes showed wide stress response patterns in the desert poplar.
It has been revealed that HsfA1 and HsfA2 can form a superactivator complex to activate downstream genes in a stronger manner than individual factors10, although overexpression of AtHsfA2 also improved thermotolerance in quadruple knock-out Arabidopsis mutant (athsfA1a, b, d, e)15. In Arabidopsis, only one member (A1d) in A1 subclass showed multiple stress response to heat, cold, salt, and ABA (Fig. 7b), it could be cooperated with HsfA2 in these stress conditions. In contrast, many more A1 members (A1a/d, A1b, and A1c) in P. euphratica showed multiple stress responses to heat, drought, and salt stresses, and is similar to the expression patterns of PeuHsfA2 (response to heat, drought, salt, and ABA treatments) (Fig. 7a). The high similarity of expression patterns of PeuHsfA1s and PeuHsfA2 can form more superactivator complexes under heat, drought, and salt stresses in P. euphratica, which might be one reason for high stress tolerances of P. euphratica in arid and saline environments.
In conclusion, identification and detailed analysis of the Hsf gene family has been carried out in desert poplar, P. euphratica. Analysis suggests that Hsf gene family in P. euphratica has diverged during evolution and is widely responsive to various abiotic stresses. These findings are helpful in understanding the distinguished adaptability of P. euphratica to severe desert environments and provide the basis for functional analysis of PeuHsfs in the future.
Methods
Identification of Hsfs in P. euphratica and other plant species
For Hsf identification, the Hsf domain (PF00447) from the Pfam database (http://pfam.xfam.org/) was used to search against the P. euphratica genome (http://me.lzu.edu.cn/stpd/#main_tabs=0)20. In addition, the Hsf protein sequences of A. thaliana26 and P. trichocarpa19 were used as queries to perform a BLASTP search against the P. euphratica genome. The Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/) was used to analyze the DBD domain and the coiled-coil structure.
Sequence analysis
Full-length protein sequences of Hsf from four species, including model species A. thaliana and three Salicaceae species - P. euphratica (this study), P. trichocarpa19, and Salix suchowensis13, were aligned using the Clustal X2.127 and the phylogenetic tree was constructed using MEGA 528 with the neighbor-joining (NJ) method with 1,000 bootstrap replicates. The P. euphratica Hsfs (PeuHsfs) were named according to their subfamily classification and their phylogenetic relationships with the AtHsfs and PtHsfs (see Supplementary Table S1). The gene structures including exon and intron were displayed using Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/index.php). The conserved motifs of PeuHsfs were defined by Multiple Em for Motif Elicitation (MEME, http://meme-suite.org/). The parameters (e.g. molecular weight, isoelectric point, instability index, aliphatic index, GRAVY, and so on) of PeuHsf proteins were obtained from the ExPASy database (https://www.expasy.org/).The instability index provides an estimate of the stability of the protein in a test tube. A protein whose instability index is smaller than 40 is predicted as stable, a value above 40 predicts that the protein may be unstable. The aliphatic index of a protein is described as the relative volume occupies by the amino acids such as alanine, valine, isoleucine and leucine, which have an aliphatic side chain in their structure. The GRAVY value for a protein or a peptide is calculated by adding the hydropathy value of each amino acid residues and dividing by the number of residues in the sequence or length of the sequence. Increasing positive score indicates a greater hydrophobicity.
Chromosome location and duplication analysis
The PeuHsf genes were mapped onto P. euphratica scaffolds based on the publicly available information provided in the P. euphratica genome database. For duplication of Hsf genes in A. thaliana and P. trichocarpa, the duplication events were obtained from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/). For duplication of Hsf genes in P. euphratica and S. suchowensis, the duplicated events were identified based on the duplicated block from synteny analysis using MicroSyn29. A duplicated block was defined as a region where three or more conserved homologs were located within 15 genes up- and down-stream between scaffolds30.
In silico analysis of cis-acting elements of PeuHsfs
The cis-acting elements in promoter region (2 kb upstream of translation initiation site) of the PeuHsf genes were identified using PlantCARE31.
Calculation of Ka/Ks values
The paralogous pairs were aligned using Clustal X2.1 and analyzed using PAL2NAL (http://www.bork.embl.de/pal2nal/) to calculation the Ka and Ks substitution rates. The divergence time (T) was calculated according to T = Ks/(2 × 9.1 × 10−9) MYA for Populus22.
Plant growth conditions and treatments
P. euphratica were planted in the greenhouse at Chinese Academy of Forestry (Beijing, China). For different tissues, shoot tip (ST), young leaf (YL), mature leaf (ML), stem (S), young root (YR), old root (OR), female catkin (FC), and male catkin (MC) from P. euphratica were collected. For various abiotic stresses, 4-month-old P. euphratica seedlings were water-cultured using Hoagland solution32. The seedlings were treated with 25% (w/v) polyethylene glycol 6000 (PEG6000, for drought stress), 300 mM NaCl (for salt stress), 42 °C (for heat stress), 4 °C (for cold stress), or 100 μM abscisic acid (ABA). The dosages of the abiotic stresses and hormone treatment were determined based on treatments in poplar19,33. Considering the outstanding salt tolerance of P. euphratica, the concentration of NaCl was increased to 300 mM from 150 mM. The leaves mixed from eight individuals at four time points (0, 1, 6, and 12 h) during the treatments were collected and frozen immediately in liquid nitrogen until to use. Three biological replicates were performed for each sample.
The expression data of Arabidopsis Hsf genes response to various abiotic stresses and hormone was download from AtGenExpress database (http://jsp.weigelworld.org/expviz/expviz.jsp). The Arabidopsis seedlings were treated under 38 °C (for heat stress, and recovery at 25 °C), 4 °C (for cold stress), 150 mM NaCl (for salt stress), 300 mM mannitol (for osmotic stress), 1.5 μg/ml bleomycin + 22 μg/ml mitomycin (for genotoxic stress), 10 μM methyl viologen (for oxidative stress), punctured with pins (for wounding stress), or 10 μM ABA. For drought stress, the Arabidopsis seedlings were stressed by 15 min dry air stream (clean bench) until 10% loss of fresh weight, then incubation in closed vessels in the climate chamber34. To compare the responses of Hsf genes to various abiotic stresses and hormone treatment between P. euphratica and A. thaliana, the significantly induced genes were selected by the following criteria: P < 0.05 and the value of Log2 (fold change) ≥1.
RNA extraction and qRT-PCR analysis
Total RNA was isolated from P. euphratica materials using the RNeasy Plant Mini Kit (Qiagen) according to the manufacture’s protocol. Approximately 4 μg of total RNA were reverse transcribed using the SuperScript III reverse transcription kit (Invitrogen) to generate cDNA. qRT-PCR was performed on the LightCycler® 480 Real Time PCR System (Roche) using SYBR Premix Ex TaqTM Kit (Takara) according to the manufacturer’s procedure. The PeuActin gene was used as an internal control. The final threshold cycle (Ct) values were the mean of four values for each sample and four technical replicates. All primers used in this study are listed in Supplementary Table S3.
Statistical analyses
Statistical analyses of qRT-PCR were carried out using SPSS 16.0 software (SPSS Inc, Chicago, IL, USA). Data was compared using Student’s t test. Differences were considered to be significant if P < 0.05.
Additional Information
How to cite this article: Zhang, J. et al. Molecular evolution and expression divergence of the Populus euphratica Hsf genes provide insight into the stress acclimation of desert poplar. Sci. Rep. 6, 30050; doi: 10.1038/srep30050 (2016).
Supplementary Material
Acknowledgments
This work was supported by Forestry Industry Research Special Funds for Public Welfare Projects [201404101] and National Nonprofit Institute Research Grant of CAF [CAFYBB2014ZX001-4].
Footnotes
Author Contributions J.Z. carried out all the experiments, data analysis and manuscript preparation. H.J. performed most of the qRT-PCR experiments. J.L. and Y.L. helped in data collection, sample preparation and RNA extraction. J.Z. and J.H. conceived the project and designed the experiments. M.L. and J.H. supervised the analysis and critically revised the manuscript. All authors read and approved the final manuscript.
References
- von Koskull-Doring P., Scharf K. D. & Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci 12, 452–457 (2007). [DOI] [PubMed] [Google Scholar]
- Scharf K.-D., Berberich T., Ebersberger I. & Nover L. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. BBA-Gene Regul Mech 1819, 104–119 (2012). [DOI] [PubMed] [Google Scholar]
- Guo M. et al. The plant heat stress transcription factors (HSFs): structure, regulation and function in response to abiotic stresses. Front Plant Sci 7, 114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover N. et al. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperon 177–189 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Port M., Ganguli A., Bicker F. & Koskull-Döring V. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39, 98–112 (2004). [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E., Pan S., Salem T. & Gurley W. B. Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol Biol 56, 57–75 (2004). [DOI] [PubMed] [Google Scholar]
- Mishra S. K. et al. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16, 1555–1567 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz D., Döring P., Bonzelius F., Winkelhaus S. & Nover L. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol Cell Biol 21, 1759–1768 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. C., Liao H. T. & Charng Y. Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34, 738–751 (2011). [DOI] [PubMed] [Google Scholar]
- Chan-Schaminet K. Y., Baniwal S. K., Bublak D., Nover L. & Scharf K.-D. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J Biol Chem 284, 20848–20857 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F. et al. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol 60, 759–772 (2006). [DOI] [PubMed] [Google Scholar]
- Mittal D., Chakrabarti S., Sarkar A., Singh A. & Grover A. Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem 47, 785–795 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. The heat shock factor gene family in Salix suchowensis: a genome-wide survey and expression profiling during development and abiotic stresses. Front Plant Sci 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkostefanakis S., Roeth S., Schleiff E. & SCHARF K. D. Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ 38, 1881–1895 (2015). [DOI] [PubMed] [Google Scholar]
- Liu H. C. & Charng Y. Y. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol 163, 276–290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal S. K., Chan K. Y., Scharf K.-D. & Nover L. Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J Biol Chem 282, 3605–3613 (2007). [DOI] [PubMed] [Google Scholar]
- Schramm F. et al. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 53, 264–274 (2008). [DOI] [PubMed] [Google Scholar]
- Ikeda M. & Ohme-Takagi M. A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol 50, 970–975 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genomics 16, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T. et al. Genomic insights into salt adaptation in a desert poplar. Nat Commun 4 (2013). [DOI] [PubMed] [Google Scholar]
- Jeffares D. C., Penkett C. J. & Bähler J. Rapidly regulated genes are intron poor. Trends Genet 24, 375–378 (2008). [DOI] [PubMed] [Google Scholar]
- Lynch M. & Conery J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000). [DOI] [PubMed] [Google Scholar]
- Yang Z. L., Liu H. J., Wang X. R. & Zeng Q. Y. Molecular evolution and expression divergence of the Populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol 197, 1353–1365 (2013). [DOI] [PubMed] [Google Scholar]
- Shim D. et al. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant cell 21, 4031–4043 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S. et al. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant cell 17, 268–281 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübel A. & Schöffl F. Arabidopsis heat shock factor: isolation and characterization of the gene and the recombinant protein. Plant Mol Biol 26, 353–362 (1994). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B., Yang X., Tuskan G. A. & Cheng Z.-M. MicroSyn: a user friendly tool for detection of microsynteny in a gene family. BMC Bioinformatics 12, 1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Comparative genomic analysis of the WRKY III gene family in populus, grape, arabidopsis and rice. Biol Direct 10, 1–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M. et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30, 325–327 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D. R. & Arnon D. I. The water-culture method for growing plants without soil. Circular. California Agricultural Experiment Station 347 (1950). [Google Scholar]
- Shao Y. et al. Genome-Wide Analysis of BURP Domain-Containing Genes in Populus trichocarpa. J Integr Plant Biol 53, 743–755 (2011). [DOI] [PubMed] [Google Scholar]
- Kilian J. et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50, 347–363 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.