Abstract
The rapidly expanding interest in, and availability of, digital tomography data to visualize casts of the vertebrate endocranial cavity housing the brain (endocasts) presents new opportunities and challenges to the field of comparative neuroanatomy. The opportunities are many, ranging from the relatively rapid acquisition of data to the unprecedented ability to integrate critically important fossil taxa. The challenges consist of navigating the logistical barriers that often separate a researcher from high‐quality data and minimizing the amount of non‐biological variation expressed in endocasts – variation that may confound meaningful and synthetic results. Our purpose here is to outline preferred approaches for acquiring digital tomographic data, converting those data to an endocast, and making those endocasts as meaningful as possible when considered in a comparative context. This review is intended to benefit those just getting started in the field but also serves to initiate further discussion between active endocast researchers regarding the best practices for advancing the discipline. Congruent with the theme of this volume, we draw our examples from birds and the highly encephalized non‐avian dinosaurs that comprise closely related outgroups along their phylogenetic stem lineage.
Keywords: Aves, brain, comparative neuroanatomy, computed tomography, endocast
Introduction
The last 25 years of evolutionary morphology have been witness to an explosion of digital techniques for observing, analyzing, and interpreting anatomical information. The impact of these innovations has been transformative in the field of comparative neuroanatomy where the various forms of computed tomography (CT) [i.e. viewing a three‐dimensional (3D) structure based on a parallel series of digitally acquired two‐dimensional (2D) images known as tomograms] are now widely used across vertebrate clades to visualize the endocranial space and to assess morphological details of this space as a proxy for brain morphology through the construction of digital endocasts (Fig. 1) (e.g. Maisey, 2004; Colbert et al. 2005; Zollikofer et al. 2005; Macrini et al. 2007a; Snitting, 2008; Witmer et al. 2008; Olori, 2010; Smith & Clarke, 2012; George & Holliday, 2013; Kawabe et al. 2013; Pradel et al. 2013; Racicot & Colbert, 2013; Racicot & Rowe, 2014). These advances have facilitated broad‐scale comparisons of 3D neuroanatomy across the vertebrate tree, and the non‐destructive nature of these methods has been a significant advance for vertebrate paleontologists, giving new energy to the discipline of paleoneurology (Rowe et al. 1995, 1997; Ketcham & Carlson, 2001; Carlson et al. 2003; Sutton, 2008; Cunningham et al. 2014; Rahman & Smith, 2014). Fossils are the only direct empirical windows into the deep history of morphological evolution and a study of their endocasts can help establish the timing and nature of critical neuroanatomical transformations (Conroy & Vannier, 1985; Marino et al. 2003; Witmer et al. 2003; Macrini et al. 2007a; Pradel et al. 2009, 2011; Silcox et al. 2010; Carlson et al. 2011; Rowe et al. 2011; Zelenitsky et al. 2011; Knoll et al. 2012; Dupret et al. 2014; Giles & Friedman, 2014; Kirk et al. 2014).
Figure 1.
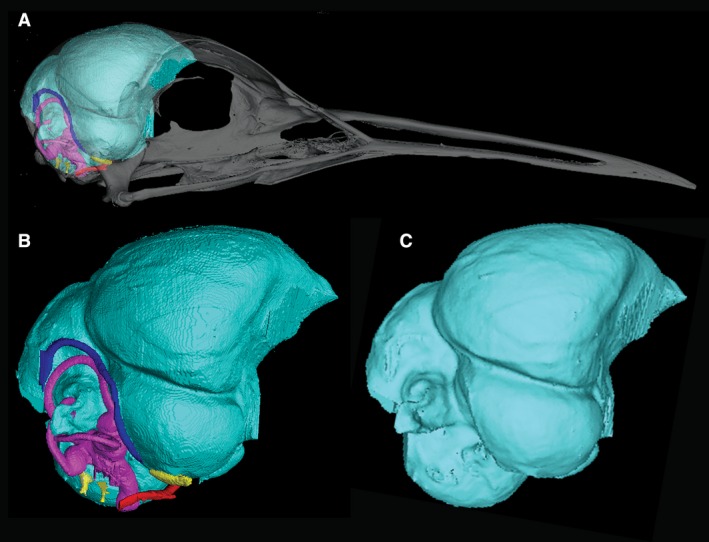
Three‐dimensional (3D) renderings of the cranial endocast of Eurypyga helias constructed from CT data. (A) Cranial endocast of E. helias with transparent skull. (B) Cranial endocast of E. helias with cranial nerves (yellow), arteries (red), veins (blue), and inner ear (pink). (C) Cranial endocast of E. helias with inner ear, nerves, and vasculature digitally removed. A smoothing algorithm (Laplacian Smooth) has been applied to the rendering.
As CT imaging has become standard practice in paleontology and as rapidly emerging techniques for differentially staining neural tissues promise to greatly expand the number and functional implications of endocast studies for modern taxa (Fig. 2) (Gignac & Kley, 2014), we think it prudent to articulate some best‐practice guidelines for constructing digital endocasts. One aim in making these suggestions is to maximize the efficiency and efficacy of what can be a time‐consuming and challenging process, especially for researchers just entering the field. Given that the ultimate goal of the field is to gain a synthetic understanding of neuroanatomical structure, function, and evolution across vertebrate history (and beyond), we expect that these guidelines will contribute to the general comparability of endocast data from diverse researchers. Recognizing and minimizing the biologically meaningless variation expressed in our data will likely become increasingly important as we refine the scope and sophistication of the questions that our field pursues.
Figure 2.
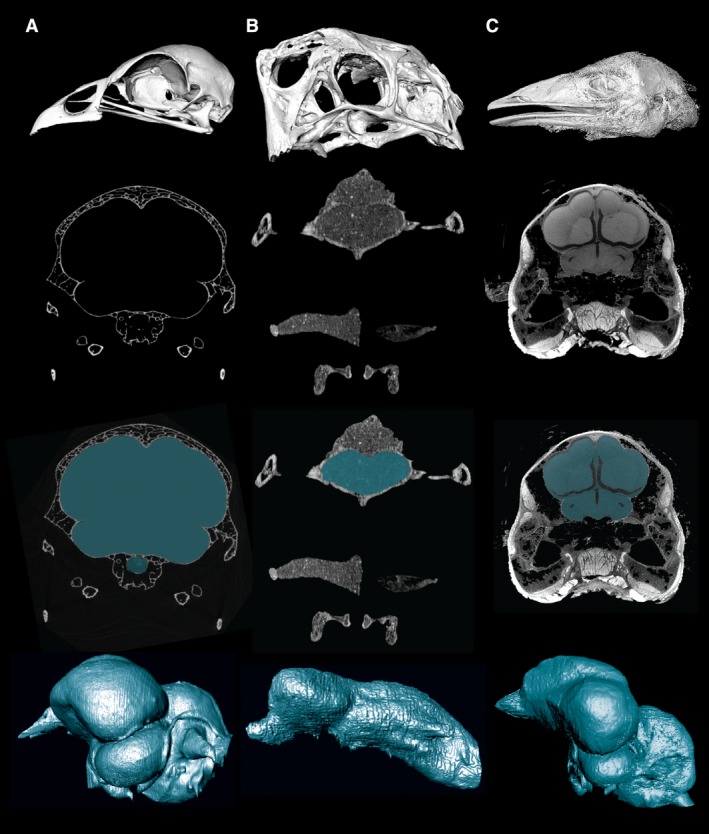
CT images and cranial endocasts from different specimen types and preparations. Top row, 3D reconstruction of skull/head from CT data. Second row, representative CT slice through the cranial cavity. Third row, CT slice with the cranial cavity segmented. Fourth row, digitally prepared cranial endocast. (A) A skeletal preparation of the extant galliform bird Alectura lathami. (B) A fossil oviraptorosaur dinosaur, Citipati osmolskae. (C) A contrast‐enhanced iodine‐stained preparation of the extant paleognathous bird Dromaius novaehollandiae. These staining techniques provide a relatively direct bridge between an endocast and the soft tissues it represents, thus expanding the future role of endocasts in comparative neuroscience.
The approach taken here is to propose and address a series of questions that someone interested in constructing and studying an endocast might ask. Following the general theme of this volume, we draw our examples from the avian crown and its stem lineage – a group that, along with the mammalian total group, has long been at the forefront of endocast research (Marsh, 1880; Osborn, 1912; Edinger, 1926, 1941, 1951; Janensch, 1935; Jerison, 1968, 1969, 1973; Hopson, 1979; Larsson et al. 2000; Witmer et al. 2003; Franzosa, 2004; Domínguez Alonso et al. 2004; Evans, 2005; Franzosa & Rowe, 2005; Kundrát, 2007; Sampson & Witmer, 2007; Sereno et al. 2007; Witmer et al. 2008; Witmer & Ridgely, 2008a, 2009; Balanoff et al. 2009, 2010, 2013, 2014; Evans et al. 2009; Norell et al. 2009; Milner & Walsh, 2009; Picasso et al. 2009, 2010; Bever et al. 2011, 2013; Miyashita et al. 2011; Walsh & Milner, 2011a; Smith & Clarke, 2012; Knoll et al. 2012, 2013; Ksepka et al. 2012; Lautenschlager et al. 2012; Kawabe et al. 2013; Walsh et al. 2013; Degrange et al. 2015; Sues et al. 2015).
What is an endocast, and what information can an endocast convey?
In the context employed here, a digital endocast is the 3D cast of any internal space that is digitally constructed from serially sectioned data (Fig. 1), usually generated by an X‐ray CT scanner. The cranial cavity housing the brain and associated tissues is most commonly targeted for casting, but the inner ear cavities, neurovascular canals, tympanic and paranasal pneumatic sinuses, and nasal cavities are just a few of the other intracranial spaces that are studied using this approach (Fig. 1B) (Spoor & Zonneveld, 1998; Bever et al. 2005; Rowe et al. 2005; Evans, 2006; Kundrát & Janáček, 2007; Georgi & Sipla, 2008; Witmer & Ridgely, 2008b, 2010; Evans et al. 2009; Walsh et al. 2009; Ekdale, 2010, 2011, 2013; Farke, 2010; Luo et al. 2010; Macrini et al. 2010; Ekdale & Rowe, 2011; Tahara & Larsson, 2011; Georgi et al. 2013; Racicot & Berta, 2013; Bourke et al. 2014; Lautenschlager et al. 2014; Ekdale & Racicot, 2015.
A standard endocast is generated at the interface between the skeleton (typically bone or cartilage) and the soft tissue (or fluid) lying immediately deep to it. In prepared skeletal material from extant taxa, this space is typically filled with air, whereas in fossils this space is commonly filled, at least in part, with sedimentary matrix. The endocast itself thus has a shape and a volume but no internal structure. In the cranial cavity, the soft tissue forming the interface with the surrounding skeleton is not the brain but the superficial surface of the dural meninges and vasculature enveloping the brain. The degree to which this dural surface reflects the morphology of the underlying neural tissue can vary widely between lineages and over ontogeny (Jerison, 1973; Hopson, 1979; Hurlburt, 1996; Evans, 2005; Macrini et al. 2007b; Witmer & Ridgely, 2008a; George & Holliday, 2013; Hurlburt et al. 2013). This variance largely reflects the degree to which the brain fills the cranial cavity. In general, highly encephalized taxa (those with large brains relative to body size) tend to have brains that nearly fill the cranial cavity, resulting in a strong correlation between the volume/morphology of the endocast and that of the brain. The crown radiations of mammals and birds are among the best examples of taxa with a high brain‐to‐endocranial cavity correlation index (BEC index) (Jerison, 1969, 1977; Hopson, 1979; Northcutt, 2002; Rowe et al. 2011; Balanoff et al. 2013), but the brain also nearly fills the endocranial space in some extant chondrichthyan and teleost lineages (Northcutt, 1977, 2002; Van Dongen, 1998; Kotrschal et al. 1998; Coates, 1999) and seemingly in some extinct lineages (e.g. pterosaurs; Witmer et al. 2003). The thickness of the dura and extent of dural sinuses ensure that even those taxa with a high BEC index express some disparity between the shape and volume of the endocast and that of the brain (Jerison, 1973).
There is a legitimate concern regarding the value of constructing an endocast when it is unclear whether the time and effort will produce a meaningful approximation of the brain. This is especially the case for fossils where poor contrast between matrix and bone and/or postmortem crushing or distortion are often limiting factors. The answer depends heavily on the hypothesis being tested, but virtually all endocasts convey some useful morphological information (even if it is only that the brain does not significantly fill the endocranial space). In cases of uncertainty, a phylogenetic perspective can be informative. If the taxon in question is nested within a phylogenetic bracket comprising lineages that express a reasonably uniform BEC index, it follows that a similar index can be inferred for the fossil (Farris, 1983; de Queiroz & Gauthier, 1992; Witmer, 1995). When one of the crown clades comprising the bracket differs in its BEC index, then the stem of that lineage should be witness to the apomorphic transformation, though the nature and timing of the transformation remains unknown until it is established empirically with endocasts from fossils. A good example of this phenomenon is found in crown‐group Archosauria. Adult crown‐group crocodylians express the plesiomorphic condition in which the brain fails to fill the cranial cavity. Thus, the braincase‐dural interface does not accurately reflect neural shape and volume (Hurlburt, 1996; George & Holliday, 2013; Hurlburt et al. 2013). The extant sister taxon to crown‐group crocodylians is crown‐group Aves (Gauthier, 1986; Gauthier et al. 1988), whose high BEC index endocasts closely approximate the morphology of the brain (Fig. 1). Much of the known diversity along the avian stem is plesiomorphic in this regard, with high indices being a relatively crown‐ward apomorphy (Jerison, 1969; Hopson, 1979; Larsson et al. 2000; Northcutt, 2002; Domínguez Alonso et al. 2004; Witmer & Ridgely, 2009; Balanoff et al. 2013). Endocasts from the earlier stem divergences are thus less likely to produce a meaningful brain approximation, but pinpointing the origin of the avian condition will require new fossil discoveries and endocast studies.
A common dilemma in paleoneurology is the challenge posed by incomplete or distorted specimens, which can affect morphology as well as volume (Walsh et al. 2014). Here, the focus of the study is critical, as some questions require pristinely preserved material to address whereas others can be addressed from rendering the deep surface of even a single bony element. For example, the deep surface of the avian frontal preserves the external signature of the cerebrum, including the dorsal pallium (Wulst) – a sensory processing center that appears to have had an important influence on modern avian taxonomic and ecological diversity (Emery & Clayton, 2005; Iwaniuk & Hurd, 2005; Reiner et al. 2005; Iwaniuk & Wylie, 2006; Iwaniuk et al. 2007; Jarvis et al. 2013). When damage or distortion is restricted to one side of the braincase, an endocast from the opposing side can be constructed and then mirrored to estimate the complete structure (e.g. Domínguez Alonso et al. 2004; Balanoff & Rowe, 2007; Lautenschlager et al. 2012, 2014; Balanoff et al. 2014). This approach is generally justified given the symmetry of vertebrate neuroanatomy but will clearly misrepresent the shape of asymmetrical features and may be subject to error if the medial extent of the undistorted side is not clear. When an endocast is constructed using right–left reflection, the method should be explicitly stated not only in the methods section but also in the appropriate figure captions to avoid any subsequent confusion or over‐interpretation of the data.
In cases where brittle deformation has left a break in the braincase it is best to follow the contour of the endocranium across the break with the realization that the volume of the cavity will likely be affected. Each case of brittle deformation is different and should be addressed in a way that minimally affects the shape of the endocast. In most instances this amounts to drawing straight lines across any openings. Any type of physical repair of this nature should be made explicit within the methods. Some types of postmortem distortion may not significantly affect the endocranial volume even when the shape relationships are altered.
The retrodeformation of both plastic and brittle distortion is theoretically possible using algorithmic corrections. These have been most actively pursued in the anthropological literature (Boyd & Motani, 2008; Tallman et al. 2014) but are beginning to be used on bird‐line archosaurs (see Witmer & Ridgely, 2008a).
What data are needed to construct an endocast?
Digital endocast construction requires procuring digital serial sections from the target specimen. These slices are generally obtained using either a medical, micro‐ (μ‐) or industrial CT scanner, although magnetic resonance imaging (MRI) and synchrotron scanning have also been employed (Tafforeau et al. 2006; Sutton, 2008; Cunningham et al. 2014; Rahman & Smith, 2014). MRI has the benefit of showing greater differentiation of tissues in specimens that are not alcohol‐preserved (e.g. formalin‐preserved, recently dead or live specimens) but offers only a slight improvement over CT in differentiating tissues in alcohol‐saturated preserved specimens (Van der Linden et al. 1998; Corfield et al. 2008). Synchrotron scanning, which uses a particle accelerator as an X‐ray source, can produce exceptional images for extinct taxa; it has even been able to distinguish organic soft tissue in some fossil specimens (Tafforeau et al. 2006; Pradel et al. 2009). However, MRI and synchrotron scans are generally costly or logistically complicated to acquire, and only need to be pursued when their unique capabilities are required.
Endocasts can also be constructed from physical sections made using a microtome or serial grinding (Sutton, 2008; Cunningham et al. 2014). Once digitized, these histological series can be aligned using one of the many available software programs designed to deal with serial data and then studied like any other digital dataset (Sutton, 2008; Corfield et al. 2012). Digitizing and aligning existing collections of histological series is time‐consuming, but may provide new information (Sutton, 2008; Cunningham et al. 2014).
If a fossil or extant specimen is broken such that the internal surface of the cranial cavity (or any space of interest) is exposed, a partial ‘endocast’ can also be constructed from surface scans of the internal surface of the endocranial region. Surface scans produce data that are accurate and can be easily manipulated and rendered in a range of polygon mesh or point cloud file types (e.g. STL and PLY models). The use of this technique essentially removes the outer surface from the hollow model of the braincase, so that only the internal surface is visible. This process can be accomplished in freeware programs such as meshlab (http://meshlab.sourceforge.net). Additionally, surface scanning can be used to digitize existing physical endocasts that are catalogued in museums. As with endocasts generated from CT data, the resulting visualization presents the interface of bone and dura, which is an approximation of the brain during life. Although physical tomography and surface scans are useful methods, the discussion here focuses on the use of X‐ray CT data, as those are the most commonly available and widely used data for constructing endocasts.
Are high‐resolution CT scans required or are medical‐grade scans adequate?
Although X‐ray CT scanners are available with a wide variety of options such as different beam shapes, they come in two general grades: medical and high‐resolution (μCT or industrial grade) (Fig. 3). The great benefit of medical‐grade scanners is that they are widely available, being present in essentially every community that has advanced medical care facilities. Medical‐grade scanners are also (typically) extremely economical. In many countries, gaining access to a medical CT facility for research purposes is not problematic if the scientific need for the scans is stated clearly. Cost and access will vary (especially internationally) depending upon a community's medical need for these scanners. Medical scanners, however, are designed for living subjects, which may limit their application to neuroanatomical comparative studies. Moreover, radiation technologists operating the scanners are trained to optimize patient safety and maintain X‐ray dosage within safe levels, and therefore are generally unfamiliar with parameters that are optimal for non‐human cadavers or fossils. Although medical scanners often have capabilities that make them suitable for many fossils, technologists may not be familiar with these capabilities. These scanners typically use relatively low energies (voltage) and modest power (amperage), which may be insufficient to penetrate dense fossils and/or surrounding matrix. Although medical scanners are now capable of approaching the slice thickness of high‐resolution systems (~ 0.3 mm in the z‐axis and no less than 0.4 mm2 in x‐ and y‐axes, respectively), such resolutions are generally inadequate for very small specimens. These effects can sometimes be mitigated if the technician operating the machine is made aware of requirements for higher X‐ray energies and image resolution prior to scanning. Technicians may require assurances that these settings will not damage their scanners or facilities. The following references provide examples of a variety of common medical scanner models and settings used to acquire endocasts of fossil and extant material (e.g. Chapla et al. 2007; Witmer & Ridgely, 2009; Kawabe et al. 2013; Sales & Schultz, 2014; Carril et al. 2015).
Figure 3.
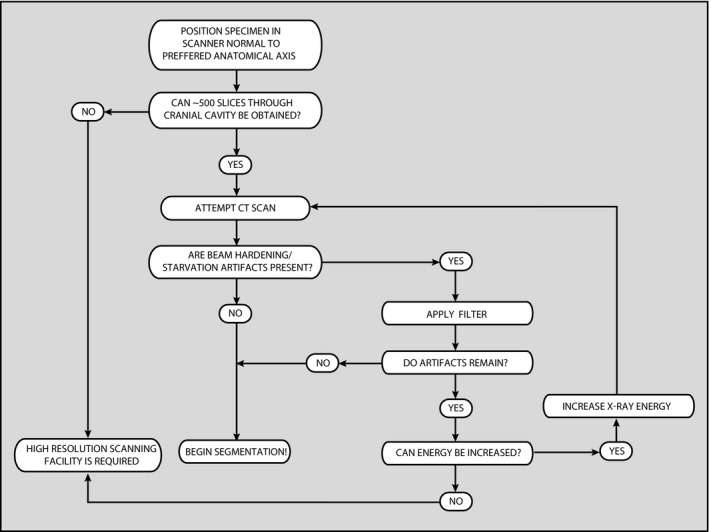
Flow chart illustrating procedures to help determine if high‐resolution CT scanning is the preferred method to deal with the specimen of interest.
Dataset resolution is an outcome of specimen size relative to the size of the voxels (voxels, or ‘volume elements’, are the 3D equivalents of pixels). Endocasts are constructed by segmenting (i.e. digitally isolating features of interest) the cranial cavity from a series of slices and then interpolating between those slices to create a 3D product (Fig. 2) (see below). Morphological descriptions and measurements using CT data require sufficient voxel coverage of the targeted features (i.e. the voxel size must be small enough to resolve the region of interest). If the voxels are too large, the interface between bone and air/matrix may at points appear blurred because it falls within a voxel (Walsh et al. 2014). This phenomenon, known as voxel partial volume averaging (see below), is best avoided by scanning with a resolution high enough to detect the boundary between materials of contrasting attenuation.
Note that most μCT scanners will typically deliver scan data in which the voxels are isotropic (i.e. x, y, and z axes are equal in length), but many older scanners, medical scanners, and linear detector industrial scanners produce data with anisotropic voxels (i.e. x and y lengths differ from z; with z corresponding to the slice spacing). The required resolution for cranial cavity endocasts will vary with the complexity of the space, but 500 slices probably lies on the low end of this distribution, with most endocasts now being constructed from more than 1000 slices. To put this in context, to obtain 500 medical‐grade slices (at an interslice spacing of 0.3 mm) through a braincase would require that the cranial cavity be 15 cm long – a length exceeding that of most vertebrates. That stated, medical scanning often produces good results (and is often the only option) for larger specimens.
The X‐rays produced by a medical scanner range up to ~ 140 kV, an energy optimized to minimize dosage while providing adequate X‐ray penetration of typical human subjects. Often medical scanners have preset parameters designed to highlight various tissue types (e.g. bone, heart, lungs), with a concomitant reduced emphasis on other tissues. For example, when scanning skeletonized specimens, the contrast in density between the skull and the air that fills the cranial cavity is typically sufficient to obtain good resolution with common medical presets. In fact, these scanners are equipped with settings (typically for dental amalgam, titanium pins, etc.) that can ameliorate ‘starburst’ artifacts caused by minerals such as pyrites and manganese that are commonly seen in fossils. However, when scanning fossils in which the cranial cavity is filled with hardened sediment, we advise the acquisition of a series of short (i.e. ~ 10 slices) preliminary scans to determine the optimal scanning parameters (i.e. highest resolution balanced with minimal artifacts; see below). Beam energies produced by medical scanners are sometimes inadequate to fully penetrate a densely permineralized fossil or one that is embedded in matrix that has not been fully prepared away physically. This generally leads to beam‐hardening or beam‐starvation artifacts (see Discussion). The shape of the specimen can further exacerbate these artifacts, particularly if the fossil is preserved on a slab (e.g. known specimens of Archaeopteryx lithographica), although problems of specimen geometry are not unique to medical scanners.
To mitigate artifacts related to inadequate X‐ray penetration, the scan protocol should take advantage of the highest energy X‐rays permitted by the system. Higher energy X‐rays will better penetrate dense materials than lower energy X‐rays. The X‐ray dose should also be maximized, scanning for the greatest amount of time possible, and with the highest permissible amperages. That being said, a consequence of using higher energies is a reduction of contrast and the ability to resolve materials of similar density (Carlson et al. 2003), which can be especially problematic in fossils that have bone and matrix of similar density.
Finally, the specimen should be mounted for scanning in a position that presents its smallest possible diameter to the X‐rays in order to minimize X‐ray filtration by the sample. For fossils, it is advisable to reduce the size of the surrounding matrix block prior to scanning, as this will reduce filtration of the X‐ray beam through the specimen. This does not necessarily mean removing the entirety of the matrix. Physical preparation is inherently dangerous because even with great care, structures can be damaged and additional artifacts can be introduced into the CT slices at the air–fossil interface (see below). Unfortunately, in many cases medical scanners will simply be inadequate for scanning some fossil specimens.
Assuming that there are no concerns about the adequacy of the employed X‐ray source to penetrate the sample, it is also advisable that the specimen be scanned in a plane normal to standard anatomical axes. This will facilitate interpretation and segmentation of the data by taking full advantage of the natural symmetries of the specimen. Non‐orthogonal scans can certainly be used, and most of the upper‐end image‐processing software facilitates realignment of a specimen relative to standard anatomical planes, although this type of realignment can result in loss of data through partial volume averaging. Regardless, proper alignment at scanning will save considerable time and effort.
What information should be gathered prior to scanning a specimen at a high‐resolution CT (HRCT) facility?
If logistics and costs permit, high‐resolution scanners, either μCT or industrial CT, are preferable to medical scanners because they are engineered for research. These scanners are typically restricted to research institutions (e.g. universities and museums) or industrial facilities and thus are generally less accessible than medical scanners, often requiring either an affiliation/collaboration with the institution and/or a fee that is based on the amount of required beam time and the volume of data generated.
High‐resolution scanners provide not only the benefit of much higher‐resolution (with many scanners attaining sub‐micron scale voxels), but also the capacity to employ higher‐energy X‐rays that can penetrate dense materials and/or large samples that cannot be imaged well with medical scanners. Such scanners also have flexible geometries that permit optimization of a wide range of object sizes and shapes. Some nano‐scanners even allow in vivo scanning. HRCT facilities are usually staffed with research scientists who understand how to maximize the quality of scan data; however, receiving high quality HRCT data that can be converted into a meaningful endocast may require clear communication of your research needs.
As described above, image resolution is a key parameter that is largely dictated by voxel size relative to specimen size. In general we recommend capturing approximately 1000 slices through the length of the braincase. Thus, if the braincase is 5 cm in length, we recommend ~ 50 μm voxel size (or interslice spacing for non‐cubic voxels). All data should be collected at as high a resolution as possible, but this can lead to extremely large, memory intensive datasets if one is scanning the entire skull, particularly for birds with long beaks (e.g. scanning a skull with a 5 cm braincase and a 30 cm total length would yield 6000 total slices). In these instances, the cost and size of the dataset can be reduced by scanning the braincase only. However, the skull is an integrated system and, if possible, it is best to procure a scan of the complete skull, even if this is not immediately needed for the project at hand. Another option is to scan the entire skull at a low resolution and the braincase at a higher resolution. The endocranial structures can then be registered with the full skull for illustrations and animations. As with medical scans, proper orientation of the specimen and, if possible, trimming excess matrix from fossil specimens will generally improve scan quality due to improved X‐ray penetration and reduced specimen size.
Finally, with both medical and high‐resolution CT data, it is important that the grayscale values in the reconstructed CT data are appropriately optimized to best showcase meaningful contrast in the specimen. The grayscale values in CT data are correlated to X‐ray attenuation through the scanned materials (attenuation coefficient), which is a function of density and elemental composition of the scanned object. By convention, darker grays are mapped to less attenuating materials, with air generally being the least dense material in the scan. To facilitate accurate thresholding (see below) and to improve 3D renderings, air should be assigned a range of values above the minimum (i.e. pure black) grayscale value. Similarly, the most attenuating materials should not be mapped to the maximum gray value (i.e. pure white), unless it is clear that the material is not of biological significance.
How are CT artifacts recognized and corrected?
A number of characteristic CT artifacts can potentially negatively affect segmentation results. There are many techniques that a skilled scan operator can employ to improve the quality of the scan (see Ketcham & Carlson, 2001). In general, it is important to mitigate CT artifacts as well as possible during the scanning and reconstruction stage, as they can complicate subsequent processing of the data.
Among the most common CT artifacts are those related to ‘beam hardening’. Beam hardening is a phenomenon occurring in all CT data employing polychromatic X‐ray sources (i.e. all scans apart from those collected with synchrotron sources). Polychromatic X‐ray sources, such as those used in medical and HRCT scanners, generate a spectrum of energies, of which the reported energy value generally corresponds to the peak energy. This necessarily results in differential filtration of X‐rays as they pass through the scanned specimen, with less‐energetic X‐rays being more highly attenuated than higher‐energy X‐rays. This results in an increase in the average energy recorded by the detectors (hence the ‘beam‐hardening’ moniker). In the data, this is manifested as an artificial brightening of the edge of objects compared with the interior. Accordingly, the gray values associated with a particular material may differ depending on their location in the scan, and a threshold value may not be valid to define the boundary between materials (e.g. bone vs. matrix in a fossil specimen) at all locations in the object of interest.
Beam‐hardening artifacts are best minimized as the scan is acquired. Preventative measures include pre‐filtering the X‐ray beam to remove the lower energy X‐rays, and the use of higher‐energy (filtered) X‐rays to provide more of the signal that is able to penetrate the specimen. Copper filters are commonly employed, but the CT technician typically will be aware of which filter is most appropriate. Note that the use of higher‐energy X‐rays will lower the contrast between materials, and the use of filters will reduce the overall signal, leading to noisier image data. This phenomenon can be compensated for by increasing the amperage, or X‐ray flux, which unfortunately will itself lead to a defocusing of the X‐rays, causing slightly blurrier data. The ultimate choice of parameters is thus almost always a compromise between competing variables.
Beam hardening can generally be partially corrected by application of a software filter to the raw data as they are being reconstructed into CT slices, typically using proprietary software packages that are bundled with the CT scanner. Such software filters generally involve warping the gray values in the CT slices to minimize the grayscale variability. When examining data to look for the occurrence of beam‐hardening artifacts it is important to be aware that overcorrected data can manifest the opposite pattern, with the specimen becoming artificially brightened in the interior.
Beam starvation is another class of artifacts related to beam hardening, occurring when there is an inadequate X‐ray signal through the specimen. This often manifests as wavy lines running through the data, or as a marked increase of noise towards the interior of specimens. Apart from rescanning with higher energy (if possible), there is little that can be done to correct beam‐starvation artifacts.
Rings are another common class of CT artifact and are the manifestation of detectors being out of calibration relative to their neighbors. Rings are generally exacerbated in dense, uniform materials. Different scanners have various strategies for minimizing rings including use of filters while scanning, dithering, and post‐reconstruction ring correction. Minor rings tend to be more of a visual distraction than an artifact resulting in loss of data, and when they are encountered in the data, one is advised to try to look past them.
What software applications are available for endocast construction and visualization?
A plethora of programs are available for viewing and segmenting CT data in order to digitally construct 3D endocasts (Zollikofer & Ponce de León, 2005; Martin et al. 2013; Cunningham et al. 2014; Atala & Yoo, 2015; http://biocomp.stanford.edu/3dreconstruction/software.html). Most offer the same basic features, differing primarily in available types of segmentation tools, smoothing algorithms, allowable size of datasets, quality of rendering, and price. Several software programs are available free for download. A few popular examples of this freeware include 3d slicer (Fedorov et al. 2012), the non‐medical version of osiriX (Rosset et al. 2004), drishti (http://sf.anu.edu.au/Vizlab/drishti/index.shtml), spiers (http://spiers-software.org/), seg3D (http://www.sci.utah.edu/cibc-software/seg3d.html), and imageJ (Abràmoff et al. 2004). Although some of the aforementioned packages are written for specific operating systems (e.g. osiriX is available only for mac os x 10.8 or later), most can be used on multiple platforms. In these cases, the best operating system for processing these data is a matter of user preference. Most of these programs offer some variation of a free‐hand and threshold tool for segmentation, can visualize the 3D object either as a volumetric or isosurface rendering, and can export volumetric data as a polygon model (e.g. STL or PLY). Those functions encompass most of the needed features for endocast construction, but some of these programs cannot handle a dataset that is over 1 gigabyte (GB) in size. For datasets larger than 1 GB (which includes almost all those generated by μCT scanners), the free programs available may be insufficient. The proprietary programs, not surprisingly, are able to import larger datasets. The most popular of these for segmenting the endocranial space include mimics, vgstudio max, avizo, and amira, to name only a few.
What file format should be used to visualize data?
Different types of scanners output their data in different file formats. The medical scanning community has almost universally adopted the DICOM (.dcm) format. DICOM files are useful because they include many scanning parameters and volume dimensions as part of the header information, and that information generally is incorporated automatically by the wide variety of software packages available for visualizing and processing serial image data (see below). Loading other, non‐proprietary forms of slice data (e.g. TIFF, BMP, PNG) into these programs minimally requires manually entering the voxel dimensions. We discourage the use of JPEGs or any type of compressed images. Providing the correct dimensions is imperative, lest the size and aspect ratio of the specimen be distorted and any subsequent linear or shape measurements (see below) become inaccurate. In all cases, it is vital to obtain information regarding voxel size before generating endocasts. Voxels from most recent μCT scanners models are cubic (isometric), and are often reported as ‘voxel size’. Older μCT scanners and some high‐energy scanners with linear detectors generate non‐cubic voxels (anisometric). In either case, voxel metadata from such scanners are provided as either x, y, and z spacing or as the field of reconstruction and the interslice spacing. In the latter case, the x and y spacing must be calculated by dividing the 2D field of reconstruction (mm) by the number of pixels that make up that field (usually 512, 1024 or, more recently, 2048).
HRCT scanners can output data in a variety of image file types, although standard TIFFs are commonly used. TIFF images usually are saved as either 8‐bit or 16‐bit grayscale formats. The bit depths refer to the number of gray values in each voxel (28, or 256 grays for 8‐bit data; and 216, or 65536 grays for 16‐bit data). The 16‐bit data are much larger than the 8‐bit data but can be useful, particularly for many fossils, when there is minimal X‐ray contrast between bone and matrix. Similarly, visualization of soft tissues in differentially stained modern specimens may be facilitated by use of 16‐bit data (Fig. 2C). Generally speaking, 8‐bit data are adequate for resolving modern osteological specimens, where there is inherently greater X‐ray contrast between air and bone (Fig. 2A). The main reason to prefer 8‐bit data over 16‐bit data is because the large size of 16‐bit CT data sets may be computationally intractable on many desktop computers.
Because of the relatively large size of CT datasets, it is advisable to use a computer equipped with a large amount of RAM, fast processors, and a high‐end graphics card that has ample RAM. The amount of computer power required, of course, depends on the size of the CT datasets to be analyzed, and this is a moving target. As CT scanning systems improve, they will likely generate ever‐larger datasets. As a rule of thumb, however, the amount of RAM available should be at least twice the size of the CT dataset being analyzed. Furthermore, even in cases where the analysis of very large CT datasets is impossible with available computing resources, we recommend acquiring the highest‐resolution data that one can afford. Data can always be downsized for analysis, and the larger dataset may be more useful for future research.
What if the dataset is too large to be opened by the image processing software or becomes too burdensome for the system's processing capabilities?
Although we advocate using the best data possible, we realize that this is not always feasible for many researchers. Large datasets can pose a number of problems, including simply loading them into the image‐processing software. Recommended steps for dealing with overwhelming data are outlined in Fig. 4 and discussed here. As a first step, 3D visualization applications usually allow the user to crop excess canvas (typically air) surrounding the specimen before the data are fully imported, which will reduce file size without loss of resolution or quality of the data. If this is not an option, image‐processing applications such as imageJ can be used as well. Cropping the rostrum (while preserving a full archival dataset) is also an option at this point in data processing.
Figure 4.
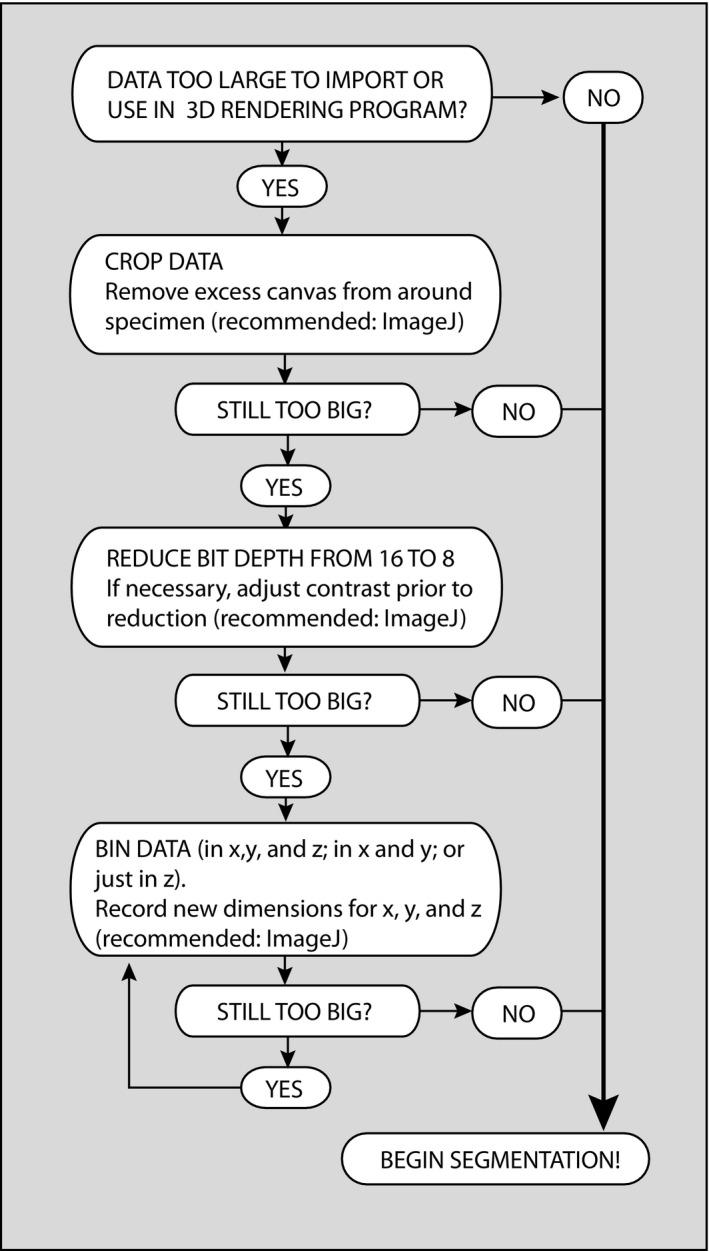
Flow chart illustrating preferred procedures for reducing CT data size.
Although we discourage their use, if the data are still too large to open or easily manipulate, then options that result in data loss may need to be taken. TIFF images can be downsized from 16‐ to 8‐bit. This reduces the number of gray values in the data, but does not change their spatial resolution. If one is planning on adjusting the grayscale contrast prior to segmentation (e.g. for a scan of a fossil exhibiting poor contrast between bone and matrix), this should be done prior to downsizing to 8‐bit format to minimize information loss. Conversion to JPEGs (8‐bit) is not recommended because this is a ‘lossy’ compression that reduces the quality of the data. Besides having a smaller initial number of grayscale values, JPEG images must be decompressed and recompressed each time they are opened and resaved, resulting in further reduction in the number of gray values.
In more extreme cases, one can reduce the data size by lowering the resolution (i.e. by increasing the voxel size/reducing voxel number). This is best accomplished in applications such as imageJ that allow the data to be binned in X, Y, and Z dimensions. Binning is a process in which multiple voxels are combined into a single voxel and is preferable to simpler methods of reduction (such as discarding every other slice) because it accounts for the lost data, and accordingly will mitigate artifacts engendered by missing data. Note that if one chooses to bin data on X and Y, but not Z, then the aspect of the voxels will be changed (the altered parameters will need to be used when loading the resized data into image‐processing software). In all cases of data modification one should keep an archival set of the unaltered slices as a reference.
Once the data are loaded, what is the process for constructing an endocast?
The process of constructing a digital endocast (i.e. segmenting the space of interest and rendering it in 3D) will vary based on the software and especially the nature of the dataset. The general goal, however, should always be to construct the endocast in the most objective way possible. Data segmentation does not precede the scientific process, but is very much a part of that process. A segmentation is a hypothesis and thus needs to be repeatable.
Segmentation at the most basic level involves assigning pixels in a region of interest (ROI) with a digital label. These label fields are essentially 1‐bit image overlays that gain volume when adjacent tomograms are tagged with the same label, resulting in tagged voxel models that resemble sculptures made of bricks until rendered by the software. The most objective criteria on which to base segmentation are usually the grayscale values. Grayscale values in CT data reflect the density and elemental composition of a material and, ideally, the various tissues and fluids represented in a dataset will be defined by a unique range of grayscale values (Fig. 2). If the contrast of grayscales between various components of the data (e.g. between the grays of bone and air for scans of an osteological specimen) is sufficient and consistent throughout the dataset, then one can define a threshold value that will separate those components.
In the best‐case scenario, the true threshold between two components can be specified by the half‐maximum height protocol that calculates the threshold value as the mean of the maximum and minimum grayscale values in the pixels spanning a boundary (see Coleman & Colbert, 2007). Most programs that enable segmentation offer threshold tools, or ‘magic wand’ tools for which a threshold can be specified. Because of its objectivity and relative speed, particularly with osteological specimens, threshold selection should be the initial step in the construction of every endocast (Figs 2 and 5). There are, however, several cases where a calculated threshold value will fail. These most commonly include the situation where the grayscale values between bone and the material filling the endocranial space (i.e. air, tissue or matrix) are broadly overlapping and thus no unique threshold value can accurately describe the boundary. This overlap can result from natural causes (as is the case in many fossils), differential staining or from artifacts in the CT data. Another common problem with defining a threshold relates to a partial volume effect, where the thickness of a material (either bone or endocranial fill) is not sufficient to capture the average gray value for that material seen elsewhere (Fig. 5), necessitating the use of local thresholds, or definition of the boundary using other tools.
Figure 5.
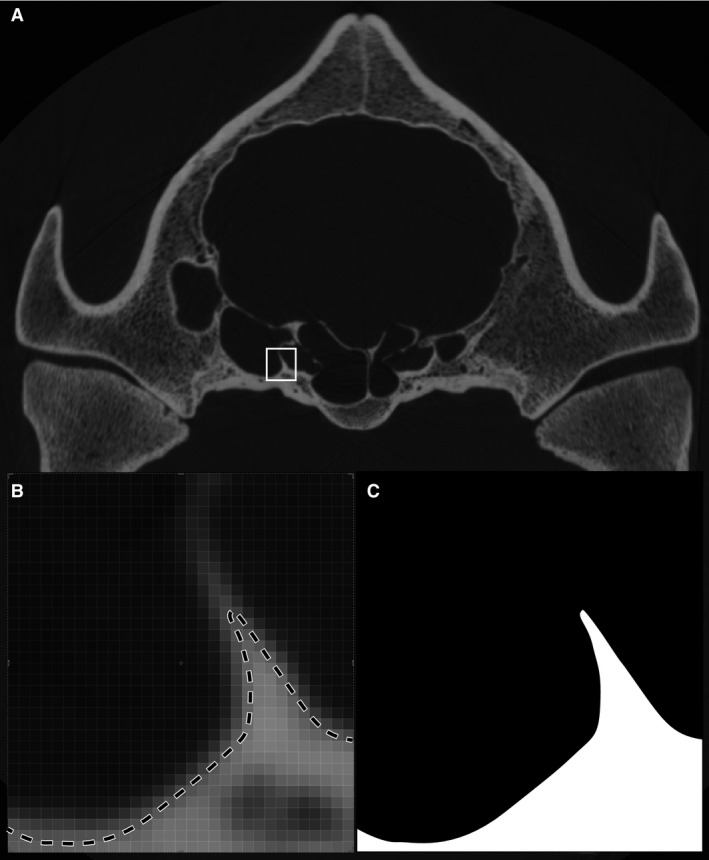
In some cases there is no single threshold value that will correctly select the surface of interest because of partial volume effects. (A) In this example the specimen (Tapirus pinchaque: MVZ 124091) has regions of thin and thick bone. (B). Use of the half‐maximum thresholding protocol will correctly define the surface location in regions of thick bone but not in regions where the bone is too thin to reach the true CT value for bone. (C) Accordingly, the threshold will drop out surfaces defined by thin bone.
If the product of the threshold segmentation needs to be refined (which is almost always the case), or if the threshold segmentation largely failed, segmentation can proceed using the more subjective freehand tools, such as the lasso or paintbrush. Of these tools, the lasso is generally preferred for its higher degree of accuracy in‐and‐around complex structures. However, different programs offer a variety of segmentation tools and, at some level, the choice of tool comes down to personal preference. Some high‐end rendering software packages enable interpolation between selections on non‐contiguous slices. Interpolation can save considerable time and effort as it requires a selection to be defined only every few slices. Although this method has the potential to miss fine‐scale features, it generally results in a cleaner 3D surface than when the selection is hand‐drawn for each slice. Interpolation is most effective when the feature of interest is relatively uncomplicated (e.g. the internal surface of the calvarium). Segmentation in three orthogonal planes also typically results in a cleaner surface by providing a useful way to observe the full complexity of anatomical structures.
With respect to foramina and fenestrae, how should the anatomical limits of a cranial endocast be defined?
The cranial cavity of most skeletally mature vertebrates, including birds, is largely enclosed by bone, but in all vertebrates the cranial cavity communicates with the exterior through several large openings. Posteriorly the foramen magnum opens into the spinal canal, and anteriorly the cranial cavity is confluent with the interorbital space and nasal cavity. Opening laterally, a series of perforations convey the cranial nerves, primary cerebral vessels, and secondary neurovascular bundles. Defining limits to the endocast during segmentation can be difficult to achieve with precision and objectivity, but failure to do so can significantly affect the shape and volume of the endocast. As a means to increase objectivity and ease comparisons between datasets, we advise stopping the endocast segmentation at the first slice in which the margin of a foramen or fenestra is encountered in two of the orthogonal planes. This approach creates two intersecting lines when viewed in the third plane. These two lines form two of three sides of a right triangle that can be objectively completed in the third plane (Fig. 6). Although the product of this approach is sometimes jagged and not necessarily aesthetically pleasing, it does provide a relatively accurate and standardized approximation of the 2D opening even when that opening lies at a marked angle to the sectional planes.
Figure 6.

Suggested approach for drawing the anatomical cutoff point for openings into the endocranial space such as the foramen magnum. Sagittal slice through skull of Eurypyga helias, in which the endocast has been segmented in the coronal and horizontal planes (blue). The red dotted line indicates where the third line should be drawn to complete this cutoff.
The rostral end of the endocast can be especially challenging. Unlike the condition in crown mammals where the boundary between the cranial and nasal cavities is clearly indicated by the ossified cribriform plate, this transition is often broadly confluent in most other vertebrate lineages (including the majority of mammalian and avian stem taxa). In crown birds, this space typically is enclosed to varying degrees by ossifications of the orbital (laterosphenoid and orbitosphenoid) and ethmoid cartilages (mesethmoid). Thus, the anterior opening can be segmented using the approach outlined above. In many non‐avian dinosaurs, however, the olfactory bulbs leave a depression on the internal surface of the frontals but their ventral and ventrolateral margins are not defined by bone. Instead of trying to guess the ventral dimensions of this structure, we recommend defining a plane to close the ventral surface based on the lower limits of the ossified margins. This will underestimate the volume of the olfactory bulbs, and thus the endocast as a whole, but it is a conservative and objective approach.
It is often desirable to segment and study the morphology of the neurovascular canals communicating with the cranial cavity. For example, the assessment of the relative importance of certain sensory modalities requires information about cranial nerve bundle thickness (Hall et al. 2009; Walsh & Milner, 2011b). In such cases, the digital cast of the cranial cavity should be terminated at the opening into the cranial cavity using the above‐described approach. Individual neurovascular canals can then be segmented distal to this point as separate partitions. Keep in mind that it is easy to combine partitions but often labor‐intensive to partition a segment into components (although some 3D visualization packages include 3D segmentation tools). We therefore encourage the use of multiple partitions in the initial segmentation.
Can a cranial endocast be partitioned to reflect different regions of the brain?
In taxa with a high BEC index where a close correlation exists between the shape of the braincase‐dural interface and the underlying brain, it may be possible to meaningfully partition that space into some combination of the regions comprising the brain (cerebrum, cerebellum, olfactory bulbs, optic lobes, medulla, etc.). The great challenge in this endeavor lies in the relationships of these regions. In life, cell layers contact one another in complex patterns deep within the brain, but endocasts can provide only surface morphology. How these neurological regions relate to the surrounding skeleton is an active area of research (Walsh & Milner, 2011a; Balanoff et al. 2013; Kawabe et al. 2015; Morhardt et al. 2012) and represents an important step forward in integrating endocast, neuroanatomical, and neurophysiological data. It is probably too early to articulate a detailed ‘best practices’ for subdividing the endocast into functional regions because approaches targeting a restricted area of the tree are likely to differ from those aimed at the important task of broad phylogenetic comparisons. We encourage researchers to explore different means of partitioning the endocast, and to do so in the spirit of promoting objectivity and comparability in endocast research, using homologous osteological landmarks whose impressions are readily visible on the endocranial surface.
How should a completed endocast be visualized?
Because an endocast is a reflection of surface morphology with no internal structure, it is best visualized as an isosurface. In the context of endocasts, an isosurface is a 3D surface corresponding to a 3D contour of a particular grayscale value (e.g. the value separating the endocast segment from material outside that segment) within the selected region of interest.
Isosurfaces can be saved as polygon files in various formats (STL and PLY are commonly used) that can be read by most 3D visualization programs and also distributed as 3D PDFs. These files also can be used for rapid prototyping (i.e. ‘3D printing’). The size of these files is largely dependent upon the number of polygons defining the surface and can be reduced by decimation (i.e. reducing the number of polygons). When decimating a polygon rendering to reduce file size, it is important to pay attention to how image quality and surface detail are affected. Overly decimated renderings can begin to look like cubist paintings, which can obscure potentially informative anatomical features.
In addition to presenting the isosurface as a file that can be manipulated, one can present visualizations of the endocast either as still images or 3D PDFs (for conventional publication) or animations. Such animations typically depict the endocast rotating around its anatomical axes, and can be generated in most 3D visualization programs.
What are the benefits and costs of smoothing an isosurface?
In addition to tools for smoothing voxel segmentations, 3D visualization programs generally offer a number of options for smoothing polygon surfaces. This process removes or at least reduces the stair‐stepping artifacts that characterize many raw endocasts, especially those constructed from fossil specimens in which segmentation has been done manually or where the interslice spacing exceeds the resolution in the x–y plane. Smoothing increases the aesthetic quality of the endocast and, by reducing the stair stepping, may actually produce a shape and volume that is more biologically realistic. However, it should be recognized that too much smoothing may produce misleading volumetric values and/or obscure potentially important morphological details. Very thin structures, such as nerve and vessel canals or semicircular canals of the inner ear, can be distorted or lost entirely due to smoothing. Generally speaking, smoothing is likely to have a greater impact on surface area measurements than on volumetric analyses, but it is clear that sensitivity analyses are needed to test the implications of smoothing, especially with regard to the overall size of the endocast (smaller specimens and smaller structures likely being more sensitive to smoothing effects). Until there is an empirical understanding of the effects of smoothing, we recommend that researchers find a balance between the benefits and potential problems of smoothing, and explicitly report their smoothing parameters.
How should measurements be taken from the endocast?
Most 3D visualization software includes tools for taking both volumetric and linear measurements. Volumes and surface areas are generally obtained from the isosurface polygon renderings (see above regarding the potential effects of smoothing). To help standardize the volumes of cranial endocasts, these should be calculated without the casts of the neurovascular canals as these have the potential to add a large amount to the total volume. Some software packages can take linear measurements from the stack of 2D slices or linear and curved surface measurements from the 3D volume rendering. Both methods are effective, but taking measurements using the 2D slices can be more difficult if the specimen was scanned in a non‐orthogonal plane. At any rate, taking measurements from the 3D rendering is more intuitive to most workers and since because there are no internal structures to measure within an endocast, the 3D option is generally preferred.
What other data should be acquired while in possession of the specimen?
Before returning a scanned specimen to its archival collection, it is often desirable to collect associated data. While the possibilities for additional data collection are nearly endless, body size is a variable commonly compared with endocast volume, often to estimate an encephalization quotient (e.g. Jerison, 1973; Domínguez Alonso et al. 2004; Balanoff et al. 2013). Body mass at death may be recorded directly but this information is comparatively rare for most museum specimens. More often, body mass is estimated from one or more skeletal dimensions. Body mass in crown‐group birds (both flying and flightless) scales closely with the minimum circumference of the femoral shaft (R 2 = 0.9527; Field et al. 2013) but other estimators may perform better within more exclusive clades (Field et al. 2013; Smith, in press). The relationship between any skeletal metric and body mass includes inherent statistical uncertainty that should be reported as a statistical prediction interval (Elzanowski et al. 2012; Field et al. 2013). This practice will make subsequent calculations, such as encephalization quotients, more conservative and less prone to over‐ or underestimation due to statistical chance (Campione & Evans, 2012; Field et al. 2013).
When dealing with fossils it is important to recognize that all skeletal dimensions have some relationship with body mass, so even fragmentary fossil material may yield useful measurements for body mass estimation (e.g. Longrich et al. 2011); however, it may not be statistically defensible to apply equations derived from skeletons of the crown group to stem‐group lineages. Fortunately for researchers of stem‐group birds, a general statistical conversion has been derived to facilitate the estimation of body mass in these non‐avian bipeds (Campione et al. 2014).
What details of the scan should be reported in the methods section of a publication?
Publishing observations and analysis of digital endocasts requires that certain parameters of the original scan be reported to enable replication of the work and allow interpretation of the results. These data should minimally include the make and model of the scanner, date of the scan, the lab in which it was scanned, the power of the X‐ray beam (kV, mA), the size of the CT slices in pixels (e.g. 1024 × 1024), the number of slices, the voxel dimensions (mm or μm), the catalogue number of the specimen, and its sex (if known). Other useful pieces of information that should be included if known include exposure time, frame averaging, number of views, filter type, and any corrections applied during reconstruction. Although not standard practice at this time, we advocate including a sample of the primary data in the form of one or more CT slices through the endocranium to provide the reader insight into the quality of the segmented data.
How, where, and what data should be publicly archived?
Publication of endocasts and other forms of CT data raises important but complex questions regarding public access to the data, and how and where those data should be archived. We do not propose a resolution of this issue here, but we would like to provide information on the currently available possibilities and make some general comments about how we might best move forward.
Determining which data should be archived is often dependent on the requirements of the journal in which the endocast is being published. However, because the use of CT data to construct endocasts is a relatively new technique, few journals have any stringent guidelines in place at this time. This situation may change as these studies become more prevalent. Although rapidly changing, authors currently have a large say in what data are shared with the public, ranging from the full CT dataset to a table of measured endocranial volumes. Ideally, the type of study should dictate what data are shared. Morphological studies (e.g. pure description or phylogenetic analyses) should try to make available the CT slices (at least in a reduced, compressed format) and the polygon model of the endocast. If the journal does not offer an outlet for such data, a digital library such as DigiMorph.org or Datadryad.org may be used. For comparative analyses of endocranial volumes, a table of volumes likely would be sufficient. In some cases, full datasets and 3D models cannot be included because the lending institution asserts copyright and will not permit these data to be shared. It is therefore critical that permission is granted from the lending institution prior to scanning their specimen(s) or sharing proprietary data. This situation is one that will only be resolved through discussion and creation of clear and unambiguous guidelines, but as copyright laws are not universal we suspect that such guidelines will be slow to appear.
Ideally, the journal supports 3D PDFs as part of the primary publication; however, few journals currently offer this as an option. The most widely available recourse is to share digital data within the supplementary information. 3D models and movies of image stacks that have been reduced in size and compressed are typically small enough to be included as supplementary data. This outlet may not always be a viable option, however, if you are planning to upload a full, unreduced CT dataset because many journals have strict restrictions on supplemental file sizes. In recent years, a number of online servers have become available that are able to host large amounts of digital data associated with publications (e.g. datadryad.org). Some funding bodies (e.g. the UK Natural Environment Research Council) also operate open access archives for datasets generated by their grant funding, and such archiving is often a condition of the grant award.
It is clear that the interests of science are generally served by making data available so that results can be tested, evaluated, and replicated. This is both logistically and legally complicated in the case of CT data and their derivative products. It is hoped that the scientific community can come together to establish not only standards for data dissemination, but also the necessary funding to support this dissemination.
Conclusions
The construction and study of endocasts is almost certain to continue its rapid ascent within the broader neuroanatomical community. In addition to achieving a more meaningful understanding of the spatial relationships between the endocast and the 3D morphology of underlying neural structures, a major challenge researchers collectively face is to make our endocast data as replicable and as broadly comparable as possible. This is especially critical as endocasts become integrated into synthetic analyses aimed at establishing evolutionary patterns across the chordate tree.
There are several points in the process of endocast construction and analysis where human error and biologically uninformative variation might be introduced. These range from the relatively simple, such as how and where to define the limits of the endocast, to more complex computation and rendering issues whose implications for evolutionary studies need to be studied using formal sensitivity analyses. Such issues include relative error rates when taking measurements on isosurface models vs. volumetric renderings, the influence of data reduction and data resolution on the endocast, as well as the repeatability of endocast reconstruction. It is our intent that this paper stimulates discussion and empirical testing of methods, so that scientific consistency can be achieved throughout this research community. It is hoped that the suggestions and best practices discussed here will help address some of the common questions regarding endocast reconstruction, encourage researchers to construct endocasts that can be easily repurposed for comparative analyses, and stimulate conversation regarding future development of best practices for endocast construction, analysis and data dissemination.
Author contributions
AMB and GSB took leadership in organizing and writing the manuscript drafts based on the collective activities of the NESCent working group. AMB, MWC, DTK contributed figures. AMB, DTK, NAS led the NESCent working group out of which this manuscript grew. AMB, GSB, MWC, JAC, DJF, PMG, DTK, RR, NAS, CT, SW, LMW contributed to the writing of the manuscript and participated as active members of the NESCent working group session.
Acknowledgements
This paper is a product of the Catalysis meeting ‘A Deeper Look into the Avian Brain: Using Modern Imaging to Unlock Ancient Endocasts’, funded by The National Evolutionary Synthesis Center (NESCent, NSF EF‐0905606) and organized by AMB, DTK and NAS. Candace Brown, Stephanie Ribson, and Danielle Wiggins provided logistical support at NESCent. Nate Kley allowed use of the iodine‐stained specimen in Fig. 2, and UTCT provided CT data used in Fig. 5. Lyn Merrill helped with image processing. Robin M. Jones provided graphical assistance. Mark Norell (AMNH, Division of Paleontology) and Joel Cracraft, Paul Sweet, Tom Trombone and Peter Capainola (AMNH, Division of Ornithology) allowed access to specimens. Richie Abel, Dmitri Grinev and Andrew Ramsay advised on some radiographic issues. Eric Ekdale and Ted Macrini provided helpful suggestions during the review process. Research for this paper was funded by NSF DEB‐1457181 grant to AMB, GSB, and PMG, NSF EAGER 1450850 to PMG, and NSF IBN‐0343744, IOB‐0517257, and IOS‐1050154 to LMW and RCR.
References
- Abràmoff MD, Magalhães PJ, Ram SJ (2004) Image processing with ImageJ. Biophot Int 11, 36–43. [Google Scholar]
- Atala A, Yoo JJ (2015) Essentials of 3D Biofabrication and Translation. New York: Academic Press. [Google Scholar]
- Balanoff AM, Rowe T (2007) Osteological description of an embryonic skeleton of the extinct elephant bird, Aepyornis (Palaeognathae: Ratitae). J Vertebr Paleontol 27, 1–53. [Google Scholar]
- Balanoff AM, Xu X, Kobayashi Y, et al. (2009) Cranial osteology of the theropod dinosaur Incisivosaurus gauthieri (Theropoda: Oviraptorosauria). Am Mus Novit 3651, 1–35. [Google Scholar]
- Balanoff AM, Bever GS, Ikejiri T (2010) The braincase of Apatosaurus (Dinosauria: Sauropoda) based on computed tomography of a new specimen with comments on variation and evolution in sauropod neuroanatomy. Am Mus Novit 3677, 1–32. [Google Scholar]
- Balanoff AM, Bever GS, Rowe TB, et al. (2013) Evolutionary origins of the avian brain. Nature 501, 93–96. [DOI] [PubMed] [Google Scholar]
- Balanoff AM, Bever GS, Norell MA (2014) Reconsidering the avian nature of the oviraptorosaur brain (Dinosauria: Theropoda). PLoS ONE 9, e113559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever GS, Bell CJ, Maisano JA (2005) The ossified braincase and cephalic osteoderms of Shinisaurus crocodilurus (Squamata, Shinisauridae). Palaeontol Electron 8, 1–36. [Google Scholar]
- Bever GS, Brusatte SL, Balanoff AM, et al. (2011) Variation, variability, and the origin of the avian endocranium: insights from the anatomy of Alioramus altai (Theropoda: Tyrannosauroidea). PLoS ONE 6, e23393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever GS, Brusatte SL, Carr TD, et al. (2013) The braincase anatomy of the Late Cretaceous dinosaur Alioramus (Theropoda: Tyrannosauroidea). Bull Am Mus Nat Hist 376, 1–72. [Google Scholar]
- Bourke JM, Porter WR, Ridgely RC, et al. (2014) Breathing life into dinosaurs: tackling challenges of soft‐tissue restoration and nasal airflow in extinct species. Anat Rec 297, 2148–2186. [DOI] [PubMed] [Google Scholar]
- Boyd AA, Motani R (2008) Three‐dimensional re‐evaluation of the deformation removal technique based on ‘jigsaw puzzling’. Palaeontol Electron, 11, 7A. [Google Scholar]
- Campione NE, Evans DC (2012) A universal scaling relationship between body mass and proximal limb bone dimensions in quadrupedal terrestrial tetrapods. BMC Biol 10, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campione NE, Evans DC, Brown CM, et al. (2014) Body mass estimation in non‐avian bipeds using a theoretical conversion to quadruped stylopodial proportions. Methods Ecol Evol 5, 913–923. [Google Scholar]
- Carlson WD, Rowe T, Ketcham RA, et al. (2003) Applications of high‐resolution X‐ray computed tomography in petrology, meteoritics and palaeontology. Geol Soc Spec Publ 215, 7–22. [Google Scholar]
- Carlson KJ, Stout D, Jashashvili T, et al. (2011) The endocast of MH1, Australopithecus sediba . Science 333, 1402–1407. [DOI] [PubMed] [Google Scholar]
- Carril J, Tambussi CP, Degrange FJ, et al. (2015) Comparative brain morphology of neotropical parrots inferred from virtual 3D endocasts. J Anat. doi: 10.1111/joa.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapla ME, Nowacek DP, Rommel SA, et al. (2007) CT scans and 3D reconstructions of Florida manatee (Trichechus manatus latirostris) heads and ear bones. Hear Res 228, 123–135. [DOI] [PubMed] [Google Scholar]
- Coates MI (1999) Endocranial preservation of a Carboniferous actinopterygian from Lancashire, UK, and the interrelationships of primitive actinopterygians. Philos Trans R Soc Lond B 354, 435–462. [Google Scholar]
- Colbert MW, Racicot R, Rowe T (2005) Anatomy of the cranial endocast of the bottlenose dolphin, Tursiops truncatus, based on HRXCT. J Mamm Evol 12, 195–207. [Google Scholar]
- Coleman MN, Colbert MW (2007) CT thresholding protocols for taking measurements on three‐dimensional models. Am J Phys Anthropol 133, 723–725. [DOI] [PubMed] [Google Scholar]
- Conroy GC, Vannier MW (1985) Endocranial volume determination of matrix‐filled skull using high‐resolution computed tomography. In: Hominid Evolution: Past, Present and Future (ed. Tobias P.), pp. 419–426. New York: Alan R. Liss. [Google Scholar]
- Corfield JR, Wild JM, Cowan BR, et al. (2008) MRI of postmortem specimens of endangered species for comparative brain anatomy. Nat Protoc 3, 597–605. [DOI] [PubMed] [Google Scholar]
- Corfield JR, Wild JM, Parsons S, et al. (2012) Morphometric analysis of telencephalic structure in a variety of neognath and paleognath bird species reveals regional differences associated with specific behavioral traits. Brain Behav Evol 80, 181–195. [DOI] [PubMed] [Google Scholar]
- Cunningham JA, Rahman IA, Lautenschlager S, et al. (2014) A virtual world of paleontology. TREE 29, 347–357. [DOI] [PubMed] [Google Scholar]
- Degrange FJ, Tambussi CP, Taglioretti ML, et al. (2015) A new Mesembriornithinae (Aves, Phorusrhacidae) provides new insights into the phylogeny and sensory capabilities of terror birds. J Vertebr Paleontol 35, e912656. [Google Scholar]
- Domínguez Alonso P, Milner AC, Ketcham RA, et al. (2004) The avian nature of the brain and inner ear of Archaeopteryx . Nature 430, 666–669. [DOI] [PubMed] [Google Scholar]
- Dupret V, Sanchez S, Goujet D, et al. (2014) A primitive placoderm sheds light on the origin of the jawed vertebrate face. Nature 507, 500–503. [DOI] [PubMed] [Google Scholar]
- Edinger T (1926) The brain of Archaeopteryx . J Nat Hist 18, 151–156. [Google Scholar]
- Edinger T (1941) The brain of Pterodactylus . Am J Sci 239, 665–682. [Google Scholar]
- Edinger T (1951) The brains of the Odontognathae. Evolution 5, 6–24. [Google Scholar]
- Ekdale EG (2010) Ontogenetic variation in the bony labyrinth of Monodelphis domestica (Mammalia: Marsupialia) following ossification of the inner ear cavities. Anat Rec 293, 1896–1912. [DOI] [PubMed] [Google Scholar]
- Ekdale EG (2011) Morphological variation in the ear region of Pleistocene Elephantimorpha (Mammalia, Proboscidea) from central Texas. J Morphol 272, 452–464. [DOI] [PubMed] [Google Scholar]
- Ekdale EG (2013) Comparative anatomy of the bony labyrinth (inner ear) of placental mammals. PLoS ONE 8, e66624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdale EG, Racicot RA (2015) Anatomical evidence for low frequency sensitivity in an archaeocete whale: comparison of the inner ear of Zygorhiza kochii with that of crown Mysticeti. J Anat 226, 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdale EG, Rowe T (2011) Morphology and variation within the bony labyrinth of zhelestids (Mammalia, Eutheria) and other therian mammals. J Vertebr Paleontol 31, 658–675. [Google Scholar]
- Elzanowski A, Bieńkowska‐Wasiluk M, Chodyń R, et al. (2012) Anatomy of the coracoid and diversity of the Procellariiformes (Aves) in the Oligocene of Europe. Palaeontology 55, 1199–1221. [Google Scholar]
- Emery NJ, Clayton NS (2005) Evolution of the avian brain and intelligence. Curr Biol 15, R946–R950. [DOI] [PubMed] [Google Scholar]
- Evans DC (2005) New evidence on brain‐endocranial cavity relationships in ornithischian dinosaurs. Acta Palaeontol Pol 50, 617–622. [Google Scholar]
- Evans DC (2006) Nasal cavity homologies and cranial crest function in lambeosaurine dinosaurs. Paleobiology 32, 109–125. [Google Scholar]
- Evans DC, Ridgely R, Witmer LM (2009) Endocranial anatomy of lambeosaurine hadrosaurids (Dinosauria: Ornithischia): a sensorineural perspective on cranial crest function. Anat Rec 292, 1315–1337. [DOI] [PubMed] [Google Scholar]
- Farke AA (2010) Evolution, homology, and function of the supracranial sinuses in ceratopsian dinosaurs. J Vertebr Paleontol 30, 1486–1500. [Google Scholar]
- Farris JS (1983) The logical basis of phylogenetic analysis In: Advances in Cladistics (eds Platnick NI, Funk VA.), pp. 7–36. New York: Columbia University Press. [Google Scholar]
- Fedorov A, Beichel R, Kalpathy‐Cramer J, et al. (2012) 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30, 1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field DJ, Lynner C, Brown C, et al. (2013) Skeletal correlates for body mass estimation in modern and fossil flying birds. PLoS ONE 8, e82000–e82013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa J (2004) Evolution of the brain in Theropoda (Dinosauria). Unpub. PhD diss. University of Texas at Austin, pp. 1–381. [Google Scholar]
- Franzosa J, Rowe T (2005) Cranial endocast of the Cretaceous theropod dinosaur Acrocanthosaurus atokensis . J Vertebr Paleontol 25, 859–864. [Google Scholar]
- Gauthier J (1986) Saurischian monophyly and the origin of birds. Mem Calif Acad Sci 8, 1–55. [Google Scholar]
- Gauthier J, Kluge AG, Rowe T (1988) Amniote phylogeny and the importance of fossils. Cladistics 4, 105–209. [DOI] [PubMed] [Google Scholar]
- George ID, Holliday CM (2013) Trigeminal nerve morphology in Alligator mississippiensis and its significance for crocodyliform facial sensation and evolution. Anat Rec 296, 670–680. [DOI] [PubMed] [Google Scholar]
- Georgi JA, Sipla JS (2008) Comparative and functional anatomy of balance in aquatic reptiles and birds In: Sensory Evolution on the Threshold: Adaptations in Secondarily Aquatic Vertebrates (eds. Thewissen JGM, Nummela S.), pp. 233–256. Berkeley: University of California Press. [Google Scholar]
- Georgi JA, Sipla JS, Forster CA (2013) Turning semicircular canal function on its head: dinosaurs and a novel vestibular analysis. PLoS ONE 8, e58517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gignac PM, Kley NJ (2014) Iodine‐enhanced micro‐CT imaging: methodological refinements for the study of the soft‐tissue anatomy of post‐embryonic vertebrates. J Exp Zool (Mol Dev Evol) 322B, 166–176. [DOI] [PubMed] [Google Scholar]
- Giles S, Friedman M (2014) Virtual reconstruction of endocast anatomy in early ray‐finned fishes (Osteichthyes, Actinopterygii). J Paleontol 88, 636–651. [Google Scholar]
- Hall MI, Iwaniuk AN, Gutíerrez‐Ibáñez C (2009) Optic foramen morphology and activity pattern in birds. Anat Rec 292, 1827–1845. [DOI] [PubMed] [Google Scholar]
- Hopson JA (1979) Paleoneurology In: Biology of the Reptilia, vol. 9 (eds Gans C, Northcutt RG, Ulinski P.), pp. 39–146. London: Academic Press. [Google Scholar]
- Hurlburt GR (1996) Relative brain size in recent and fossil amniotes: determination and interpretation. Unpub Ph.D. Diss., University of Toronto. [Google Scholar]
- Hurlburt GR, Ridgely RC, Witmer LM (2013) Relative size of brain and cerebrum in tyrannosaurid dinosaurs: an analysis using brain‐endocast quantitative relationships in extant alligators In: Tyrannosaurid Paleobiology (eds Parrish JM, Molnar RE, Currie PJ, Koppelhus EB.), pp. 1–21. Bloomington: Indiana University Press. [Google Scholar]
- Iwaniuk AN, Hurd PL (2005) The evolution of cerebrotypes in birds. Brain Behav Evol 65, 215–230. [DOI] [PubMed] [Google Scholar]
- Iwaniuk AN, Wylie DRW (2006) The evolution of stereopsis and the Wulst in caprimulgiform birds: a comparative analysis. J Comp Physiol A 192, 1313–1326. [DOI] [PubMed] [Google Scholar]
- Iwaniuk AN, Heesy CP, Hall MI, et al. (2007) Relative Wulst volume is correlated with orbit orientation and binocular visual field in birds. J Comp Physiol A 194, 267–282. [DOI] [PubMed] [Google Scholar]
- Janensch W (1935) Die Schädel der Sauropoden Brachiosaurus, Barosaurus und Dicraeosaurus aus den Tendaguruschichten Deutsch‐Ostafrikas (Anfang). Palaeontographica Suppl 2, 145–248. [Google Scholar]
- Jarvis ED, Yu J, Rivas MV, et al. (2013) Global view of the functional molecular organization of the avian cerebrum: mirror images and functional columns. J Comp Neurol 521, 3614–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerison HJ (1968) Brain evolution and Archaeopteryx . Nature 219, 1381–1382. [DOI] [PubMed] [Google Scholar]
- Jerison HJ (1969) Brain evolution and dinosaur brains. Am Nat 103, 575–588. [Google Scholar]
- Jerison HJ (1973) Evolution of the Brain and Intelligence. New York: Academic Press. [Google Scholar]
- Jerison HJ (1977) The theory of encephalization. Ann N Y Acad Sci 299, 146–160. [DOI] [PubMed] [Google Scholar]
- Kawabe S, Shimokawa T, Miki H, et al. (2013) Variation in avian brain shape: relationship with size and orbital shape. J Anat 223, 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe S, Masuda S, Tsunekawa N, et al. (2015) Ontogenetic shape change in the chicken brain: implications for paleontology. PLoS ONE 10, e0129939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketcham RA, Carlson WD (2001) Acquisition, optimization and interpretation of X‐ray computed tomographic imagery: applications to the geosciences. Comput Geosci 27, 381–400. [Google Scholar]
- Kirk EC, Daghighi P, Macrini TE, et al. (2014) Cranial anatomy of the Duchesnean primate Rooneyia viejaensis: new insights from high resolution computed tomography. J Hum Evol 74, 82–95. [DOI] [PubMed] [Google Scholar]
- Knoll F, Witmer LM, Ortega F, et al. (2012) The braincase of the basal sauropod dinosaur Spinophorosaurus and 3D reconstructions of the cranial endocast and inner ear. PLoS ONE 7, e30060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll F, Ridgely RC, Ortega F, et al. (2013) Neurocranial osteology and neuroanatomy of a Late Cretaceous titanosaurian sauropod from Spain (Ampelosaurus sp.). PLoS ONE 8, e54991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotrschal K, Van Staaden MJ, Huber R (1998) Fish brains: evolution and environmental relationships. Rev Fish Biol Fisheries 8, 373–408. [Google Scholar]
- Ksepka DT, Balanoff AM, Walsh S, et al. (2012) Evolution of the brain and sensory organs in Sphenisciformes: new data from the stem penguin Paraptenodytes antarcticus . Zool J Linn Soc 166, 202–219. [Google Scholar]
- Kundrát M (2007) Avian‐like attributes of a virtual brain model of the oviraptorid theropod Conchoraptor gracilis . Naturwiss 94, 499–504. [DOI] [PubMed] [Google Scholar]
- Kundrát M, Janáček J (2007) Cranial pneumatization and auditory perceptions of the oviraptorid dinosaur Conchoraptor gracilis (Theropoda, Maniraptora) from the Late Cretaceous of Mongolia. Naturwiss 94, 769–778. [DOI] [PubMed] [Google Scholar]
- Larsson HCE, Sereno PC, Wilson JA (2000) Forebrain enlargement among nonavian theropod dinosaurs. J Vertebr Paleontol 20, 615–618. [Google Scholar]
- Lautenschlager S, Rayfield EJ, Altangerel P, et al. (2012) The endocranial anatomy of Therizinosauria and its implications for sensory and cognitive function. PLoS ONE 7, e52289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenschlager S, Witmer LM, Altangerel P, et al. (2014) Cranial anatomy of Erlikosaurus andrewsi (Dinosauria, Therizinosauria): new insights based on digital reconstruction. J Vertebr Paleontol 34, 1263–1291. [Google Scholar]
- Longrich NR, Tokaryk T, Field DJ (2011) Mass extinction of birds at the Cretaceous‐Paleogene (K–Pg) boundary. Proc Natl Acad Sci U S A 108, 15253–15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZX, Ruf I, Schultz JA, et al. (2010) Fossil evidence on evolution of inner ear cochlea in Jurassic mammals. Proc Biol Sci 278, 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrini TE, Rougier GW, Rowe T (2007a) Description of a cranial endocast from the fossil mammal Vincelestes neuquenianus (Theriiformes) and its relevance to the evolution of endocranial characters in therians. Anat Rec 290, 875–892. [DOI] [PubMed] [Google Scholar]
- Macrini TE, Rowe T, VandeBerg JL (2007b) Cranial endocasts from a growth series of Monodelphis domestica (Didelphidae, Marsupialia): a study of individual and ontogenetic variation. J Morphol 268, 844–865. [DOI] [PubMed] [Google Scholar]
- Macrini TE, Flynn JJ, Croft DA, et al. (2010) Inner ear of a notoungulate placental mammal: anatomical description and examination of potentially phylogenetically informative characters. J Anat 216, 600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisey JG (2004) Morphology of the braincase in the broadnose sevengill shark Notorynchus (Elasmobranchii, Hexanchiformes), based on CT scanning. Am Mus Novit 3429, 1–52. [Google Scholar]
- Marino L, Uhen MD, Pyenson ND, et al. (2003) Reconstructing cetacean brain evolution using computed tomography. Anat Rec B New Anat 272, 107–117. [DOI] [PubMed] [Google Scholar]
- Marsh OC (1880) Odontornithes: A Monograph on the Extinct Toothed Birds of North America: with Thirty‐four Plates and Forty Woodcuts. 18.
- Martin CM, Roach VA, Nguyen N, et al. (2013) Comparison of 3D reconstructive technologies used for morphometric research and the translation of knowledge using a decision matrix. Anat Sci Educ 6, 393–403. [DOI] [PubMed] [Google Scholar]
- Milner AC, Walsh SA (2009) Avian brain evolution: new data from Palaeogene birds (Lower Eocene) from England. Zool J Linn Soc 155, 198–219. [Google Scholar]
- Miyashita T, Arbour VM, Witmer LM, et al. (2011) The internal cranial morphology of an armoured dinosaur Euoplocephalus corroborated by X‐ray computed tomographic reconstruction. J Anat 219, 661–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morhardt AC, Ridgley RC, Witmer LM (2012) From endocast to brain: assessing brain size and structure in extinct archosaurs using gross anatomical brain region approximation (GABRA). J Vert Paleontol 32(suppl), 145. [Google Scholar]
- Norell MA, Makovicky PJ, Bever GS, et al. (2009) A review of the Mongolian Cretaceous dinosaur Saurornithoides (Troodontidae: Theropoda). Am Mus Novit 3654, 1–63. [Google Scholar]
- Northcutt RG (1977) Elasmobranch central nervous system organization and its possible evolutionary significance. Am Zool 17, 411–429. [Google Scholar]
- Northcutt RG (2002) Understanding vertebrate brain evolution. Integr Comp Biol 42, 743–756. [DOI] [PubMed] [Google Scholar]
- Olori JC (2010) Digital endocasts of the cranial cavity and osseous labyrinth of the burrowing snake Uropeltis woodmasoni (Alethinophidia: Uropeltidae). Copeia 2010, 14–26. [Google Scholar]
- Osborn HF (1912) Crania of Tyrannosaurus and Allosaurus . Mem Am Mus Nat Hist 1, 1–30. [Google Scholar]
- Picasso MBJ, Tambussi C, Dozo MT (2009) Neurocranial and brain anatomy of a Late Miocene eagle (Aves, Accipitridae) from Patagonia. J Vertebr Paleontol 29, 831–836. [Google Scholar]
- Picasso MBJ, Tambussi CP, Degrange FJ (2010) Virtual reconstructions of the endocranial cavity of Rhea americana (Aves, Palaeognathae): postnatal anatomical changes. Brain Behav Evol 76, 176–184. [DOI] [PubMed] [Google Scholar]
- Pradel A, Langer M, Maisey JG, et al. (2009) Skull and brain of a 300‐million‐year‐old chimaeroid fish revealed by synchrotron holotomography. PNAS 106, 5224–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel A, Tafforeau P, Maisey JG, et al. (2011) A new Paleozoic Symmoriiformes (Chondrichthyes) from the Late Carboniferous of Kansas (USA) and cladistic analysis of early chondrichthyans. PLoS ONE 6, e24938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel A, Didier D, Casane D, et al. (2013) Holocephalan embryo provides new information on the evolution of the glossopharyngeal nerve, metotic fissure and parachordal plate in gnathostomes. PLoS ONE 8, e66988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz K, Gauthier J (1992) Phylogenetic taxonomy. Annu Rev Ecol Syst 23, 449–480. [Google Scholar]
- Racicot RA, Berta A (2013) Comparative morphology of porpoise (Cetacea: Phocoenidae) pterygoid sinuses. J Morphol 274, 49–62. [DOI] [PubMed] [Google Scholar]
- Racicot RA, Colbert MW (2013) Morphology and variation in porpoise (Cetacea: Phocoenidae) cranial endocasts. Anat Rec 296, 979–992. [DOI] [PubMed] [Google Scholar]
- Racicot RA, Rowe T (2014) Endocranial anatomy of a new fossil porpoise (Odontoceti: Phocoenidae) from the Pliocene San Diego of California. J Paleontol 88, 652–663. [Google Scholar]
- Rahman IA, Smith SY (2014) Virtual paleontology: computer‐aided analysis of fossil form and function. J Paleontol 88, 633–635. [Google Scholar]
- Reiner A, Yamamoto K, Karten HJ (2005) Organization and evolution of the avian forebrain. Anat Rec 287A, 1080–1102. [DOI] [PubMed] [Google Scholar]
- Rosset A, Spadola L, Ratib O (2004) OsiriX: an open‐source software for navigating in multidimensional DICOM images. J Digit Imaging 17, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T, Carlson W, Bottorff W (1995) Thrinaxodon: Digital Atlas of the Skull. CD‐ROM, 2nd edn Austin: University of Texas Press. [Google Scholar]
- Rowe T, Kappelman J, Carlson WD (1997) High‐resolution computed tomography: a breakthrough technology for earth scientists. Geotimes 42, 23–27. [Google Scholar]
- Rowe TB, Eiting TP, Macrini TE, et al. (2005) Organization of the olfactory and respiratory skeleton in the nose of the gray short‐tailed opossum Monodelphis domestica . J Mamm Evol 12, 303–336. [Google Scholar]
- Rowe TB, Macrini TE, Luo Z‐X (2011) Fossil evidence on origin of the mammalian brain. Science 332, 955–957. [DOI] [PubMed] [Google Scholar]
- Sales MAF, Schultz CL (2014) Paleoneurology Teyumabaita sulcognathus (Diapsida: Archosauromorpha) and the sense of smell in rhynchosaurs. Palaeontol Electron 17, 1–10. [Google Scholar]
- Sampson SD, Witmer LM (2007) Craniofacial anatomy of Majungasaurus Crenatissimus (Theropoda: Abelisauridae) from the Late Cretaceous of Madagascar. J Vertebr Paleontol 27, 32–104. [Google Scholar]
- Sereno PC, Wilson JA, Witmer LM, et al. (2007) Structural extremes in a cretaceous dinosaur. PLoS ONE 2, e1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silcox MT, Benham AE, Bloch JI (2010) Endocasts of Microsyops (Microsyopidae, Primates) and the evolution of the brain in primitive primates. J Hum Evol 58, 505–521. [DOI] [PubMed] [Google Scholar]
- Smith NA (in press) Evolution of body mass in the Pan‐Alcidae (Aves, Charadriiformes): the effects of combining neontological and paleontological data. Paleobiology. [Google Scholar]
- Smith NA, Clarke JA (2012) Endocranial anatomy of the Charadriiformes: sensory system variation and the evolution of wing‐propelled diving. PLoS ONE 7, e49584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snitting D (2008) A redescription of the anatomy of the Late Devonian Spodichthys Buetleri Jarvik, 1985 (Sarcopterygii, Tetrapodomorpha) from East Greenland. J Vertebr Paleontol 28, 637–655. [Google Scholar]
- Spoor F, Zonneveld F (1998) Comparative review of the human bony labyrinth. Am J Phys Anthropol 107, 211–251. [DOI] [PubMed] [Google Scholar]
- Sues H‐D, Averianov AO, Ridgely RC, et al. (2015) Titanosauria (Dinosauria, Sauropoda) from the upper cretaceous bissekty formation of Uzbekistan. J Vertebr Paleontol 35, e889145. [Google Scholar]
- Sutton MD (2008) Tomographic techniques for the study of exceptionally preserved fossils. Proc Biol Sci 275, 1587–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau P, Boistel R, Boller E, et al. (2006) Applications of X‐ray synchrotron microtomography for non‐destructive 3D studies of paleontological specimens. Appl Phys A 83, 195–202. [Google Scholar]
- Tahara R, Larsson HCE (2011) Cranial pneumatic anatomy of Ornithomimus edmontonicus (Ornithomimidae: Theropoda). J Vertebr Paleontol 31, 127–143. [Google Scholar]
- Tallman M, Amenta N, Delson E, et al. (2014) Evaluation of a new method of fossil retrodeformation by algorithmic symmetrization: cranial of papionins (Primates, Cercopithecidae) as a test case. PLoS ONE 9, e100833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Linden A, Verhoye M, Van Audekerke J (1998) Non‐invasive in vivo anatomical studies of the oscine brain by high resolution MRI microscopy. J Neurosci 81, 45–52. [DOI] [PubMed] [Google Scholar]
- Van Dongen P (1998) Brain size in vertebrates In: The Central Nervous System of Vertebrates (eds Nieuwenhuys R, Donkelaar HJ, Nicholson C.), pp. 2099–2134. Berlin: Springer. [Google Scholar]
- Walsh S, Milner A (2011a) Halcyornis toliapicus (Aves: Lower Eocene, England) indicates advanced neuromorphology in Mesozoic Neornithes. J Syst Palaeontol 9, 173–181. [Google Scholar]
- Walsh S, Milner A (2011b) Evolution of the avian brain and senses In: Living Dinosaurs: The Evolutionary History of Modern Birds (eds. Dyke G, Kaiser G.), pp. 282–306. Chichester: John Wiley & Sons Ltd. [Google Scholar]
- Walsh SA, Barrett PM, Milner AC, et al. (2009) Inner ear anatomy is a proxy for deducing auditory capability and behaviour in reptiles and birds. Proc Biol Sci 276, 1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh SA, Iwaniuk AN, Knoll MA, et al. (2013) Avian cerebellar floccular fossa size is not a proxy for flying ability in birds. PLoS ONE 8, e67176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh SA, Luo Z‐X, Barrett PM (2014) Modern imaging techniques as a window to prehistoric auditory worlds In: Insights from Comparative Hearing Research (eds Köppl C, Manley G.), pp. 227–261. Springer Handbook of Auditory Research 49. Berlin: Springer Verlag. [Google Scholar]
- Witmer LM (1995) The extant phylogenetic bracket and the importance of reconstructing soft tissues in fossils In: Functional Morphology in Vertebrate Paleontology (ed. Thomason J.), pp. 19–33. New York: Cambridge University Press. [Google Scholar]
- Witmer LM, Ridgely RC (2008a) Structure of the brain cavity and inner ear of the centrosaurine ceratopsid Pachyrhinosaurus based on CT scanning and 3D visualization In: A New Horned Dinosaur from an Upper Cretaceous Bone Bed in Alberta (ed. Currie PJ.), pp. 117–144. Ottawa: National Research Council Research Press. [Google Scholar]
- Witmer LM, Ridgely RC (2008b) The paranasal air sinuses of predatory and armored dinosaurs (Archosauria: Theropoda and Ankylosauria) and their contribution to cephalic architecture. Anat Rec 291, 1362–1388. [DOI] [PubMed] [Google Scholar]
- Witmer LM, Ridgely RC (2009) New insights into the brain, braincase, and ear region of tyrannosaurs (Dinosauria, Theropoda), with implications for sensory organization and behavior. Anat Rec 292, 1266–1296. [DOI] [PubMed] [Google Scholar]
- Witmer LM, Ridgely RC (2010) The Cleveland tyrannosaur skull (Nanotyrannus or Tyrannosaurus): new findings based on CT scanning, with special reference to the braincase. Kirtlandia 57, 61–81. [Google Scholar]
- Witmer LM, Chatterjee S, Franzosa J, et al. (2003) Neuroanatomy of flying reptiles and implications for flight, posture and behaviour. Nature 425, 950–953. [DOI] [PubMed] [Google Scholar]
- Witmer LM, Ridgely RC, Dufeau DL, et al. (2008) Using CT to peer into the past: 3D visualization of the brain and ear regions of birds, crocodiles, and nonavian dinosaurs In: Anatomical Imaging: Towards a New Morphology (eds Endo H, Frey R.), pp. 67–88. Tokyo: Springer‐Verlag. [Google Scholar]
- Zelenitsky DK, Therrien F, Ridgely RC, et al. (2011) Proc Biol Sci 278, 3625–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollikofer CPE, Ponce de León MS (2005) Virtual Reconstruction: A Primer in Computer‐Assisted Paleontology and Biomedicine. New York: Wiley & Sons. [Google Scholar]
- Zollikofer CPE, de Léon MSP, Lieberman DE, et al. (2005) Virtual cranial reconstruction of Sahelanthropus tchadensis . Nature 434, 755–759. [DOI] [PubMed] [Google Scholar]


