Abstract
Macrophages are important for tissue development, homeostasis as well as immune response upon injury or infection. For a long time they were only seen as one uniform group of phagocytes with a common origin and similar functions. However, this view has been challenged in the last decade and revealed a complex diversity of tissue resident macrophages. Here, we want to present the current view on macrophage development and tissue specification and we will discuss differences as well as common patterns between heterogeneous macrophage subpopulations.
Keywords: macrophage, erythro-myeloid progenitor, tissue specification, pulse labeling models, mononuclear phagocytes
Among the cells of the immune system, macrophages are professional phagocytes defined as mobile cells with the capacity to engulf and digest pathogens, particles and dead cells (1). Phagocytes were first described one century ago by Ellie Metchnikoff in the star fish larvae (2), and he described these cells as “the police” of the organism by fighting against invading pathogens and restricting injuries (3), but also as the “janitor” of the organism, since phagocytes ensure the “clean” removal of used epidermal pigmented cells in the star fish, thereby leading to renewal of the epithelium. Thus, phagocytes have dual functions, as immune sentinels but also as regulators of tissue homeostasis. In vertebrates, many professional phagocytes belong to innate immunity system, including neutrophils and mononuclear cells like dendritic cells, circulating monocytes and tissue macrophages (4). These mononuclear myeloid cells were hierarchically organized in developmental pathway named “the mononuclear phagocyte system” (MPS) in 1969, based on their similar morphology, putative common origin, overlapping immune functions and repopulation kinetics (5). According to the MPS, the most immature cells are the bone marrow promonocytes, which divide and give rise to circulating immature cells, or monocytes. Under favorable conditions and in anatomical locations where phagocytosis is needed, monocytes leave the blood stream and mature into tissue phagocytes or macrophages (5). The paradigm that circulating monocytes are the sole precursors of tissue macrophages (6–9) was challenged by results from bone marrow chimera, parabiotic mice and adoptive transfer experiments (7,10,11). Subsequent kinetic studies of the spleen macrophages led Van Furth to revise the MPS framework and he proposed a dual origin, with half of myeloid cells renewing from blood and the other half being produced locally within the spleen (12). This seminal work raised the question whether tissue resident macrophages could originate from another source than circulating monocytes and self-maintain in tissues.
Here, we will summarize the recent findings in the field of mammalian tissue resident macrophage biology and give an overview about our current understanding on the functions and the development of tissue resident macrophages, as well as their heterogeneity across various organs. The molecular mechanisms underlying macrophage self-renewal and tissue macrophage functions in response to infection and tissue injury will not be discussed, as reviewed elsewhere (13,14).
Functions of tissue resident macrophage in homeostasis
In mammals, tissue resident macrophages are found all over the body in all organs and serous membranes, which surround organs and the body cavities, like the well-studied macrophages of the peritoneal cavity (15,16). Long elongated processes or dendrites extend from their cell bodies and build up a 3D network-like structure throughout each organ (17–19), thereby allowing constant surveillance or scavenging of their tissue (20–22). In general, activation of macrophages leads to an immediate retraction of the elongated processes and a change in cell morphology (19,23,24). This can be accompanied by the migration of macrophages to the injury site or source of inflammation in their host tissue (25,26). Upon activation, macrophages actively phagocyte intruding pathogens or necrotic and apoptotic cells after tissue injury, release bio-active molecules like cytokines and chemokines and they can also serve as antigen presenting cells to activate the adaptive immune response (27).
Macrophages play homeostatic functions such as scavenging of macromolecules, debris, necrotic or apoptotic cells, and invading pathogens, clearance of senescent cells as well as the production of bioactive molecules, both concurring to tissue development. During mouse development, macrophages are involved in clearance of apoptotic and senescent cells during organogenesis, in the brain, limb and lung (28–30) and they scavenge and digest the nuclei released by maturing erythroblast (pyeroncyte) (31). Fetal macrophages are important during branching morphogenesis (such as in the mammary gland, pancreas, lung testis and kidney) (32–37) and critical regulators of blood and lymphatic vessel morphogenesis and maturation during fetal and postnatal development (32,38–40).
While macrophages have similar properties across different tissues in regards to their phagocytic abilities and trophic functions (11), they also have highly specific functions depending on their anatomical location. Tissue of residence accounts for the first source of heterogeneity in phenotype and function(s) among different tissue macrophages populations during steady-state For example bone osteoclasts, brain microglia, liver Kupffer cells or lung alveolar macrophages share common functions, however, they are also highly adapted to their organ-specific purpose: osteoclasts in the bone are important for the continuous resorption and restructuring of the bone mass (41,42), whereas microglia in the CNS are highly adapted to support the neuronal network and neuronal circuit development under steady-state conditions (28,43,44). In the liver, Kupffer cells are important for the uptake of blood particles and dying red blood cells from the circulation and iron recycling while lung alveolar macrophages are critical for the uptake of surfactant and removal of particles form the alveoli (45–47) (Table 2). This functional tissue specialization is reflected at the gene expression and epigenetic (enhancer landscapes) of different tissue resident macrophages (48) (49).
Table 2.
Diversity of EMP-derived tissue macrophages in adulthood
| Brain | Epidermis | Liver | Lung | Spleen | Peritoneum | Heart | |
|---|---|---|---|---|---|---|---|
| Surface markers | F4/80+ CD45low CD11b+ CX3CR1+ Siglec-H+ P2YR12+ |
F4/80bright CD45+ CD11b+ Epcam+ MHCII+ |
F4/80bright CD45+ CD11b+ Tim4+ MHCII+ |
F4/80bright CD45+ CD11blow Siglec-F+ CD64+ CD11c+ CX3CR1gfp |
F4/80bright CD45+ CD11blow CD68+ CD64+ MHCII+ CX3CR1+ |
F4/80bright CD45+ CD11b+ Tim4+ MerTK+ |
F4/80bright CD45+ CD11b+ CD14+ MerTK+ CX3CR1+ MHCII+ |
| GF requirements | CSF1, IL-34, TGFβ | CSF1, IL-34, TGFβ | CSF1 | GM-CSF, CSF1 | CSF1 | CSF1 | CSF1 |
| Transcription factor for tissue adaption | Mef2c Smad Irf8 |
Id2 Runx3 |
Lxr-α Rxr-α |
Jun PPARγ C/EBPβ Bach2 |
Spi-C C/EBPβ |
Gata6 RAR-α/β C/EBPβ |
Not known |
| Physiological function within tissues | removal of apoptotic cells | ||||||
| Synaptic pruning; Immune quiescence | Tolerance induction | RBC recycling; Hemoglobin clearance and heme degradation | Surfactant removal; Clearing of inhaled particles | RBC recycling; T reg differentiation | Maintenance of B1 cell pool in the gut | Angiogenesis Fibrosis Immune quiescence Maintaining the cardiac stem cell niche | |
| Function under challenge within tissues | phagocytosis of dead cells and pathogens | ||||||
| Cytokine release to recruit further immune cells and blood-brain barrier break down; | Antigen presentation in draining lymph nodes | Cytokine release to initiate of acute phase proteins | Immune suppression in asthma | Type I interferon production against parasites | Clearing Nematode infection; NO production | Angiogenesis; Fibrosis | |
A second source of heterogeneity is found among different macrophages populations within the same organ or tissue. Beside the differences due to their anatomical location, distinct macrophage subsets within the same organ can coexist next to each other, and display different phenotypes and functions, as seen in the lung or spleen. Within the spleen, macrophages are highly diverse in phenotype and specialized function. They are found in the three regions of the spleen: the red and white pulp, as well as the marginal zone at the interface between both. Red pulp macrophages are efficient in phagocytizing aged erythrocytes that arrive with the circulating blood and they play a major role in iron metabolism (50). Marginal zone macrophages scavenge antigens and play an important role in retaining B cells in the marginal zone (51), while marginal metallophilic macrophages are known to scavenge viral antigens and release type I interferon (52). Finally, white pulp tingible body macrophages are efficient phagocytes for apoptotic B cells that are left over from germinal center reactions in the spleen (14,53).
Origin of tissue resident macrophages
Tissue macrophages are present in the developing embryo prior to the detection of circulating monocytes (54) and before the emergence of the first hematopoietic stem cells (HSCs) in the aorta-gonado-mesonephro (AGM) region (55). Since this early observation, three hematopoietic waves that can produce macrophages have been described: a first wave of “primitive” progenitors and second wave of “definitive” progenitors are generated in the extra-embryonic yolk sac, and a third “definitive” wave of macrophages from fetal and adult HSCs (56,57). While Palis and colleagues characterized two waves of YS progenitors with macrophage potential in vitro (58), Bertrand and colleagues identified two progenitor types that coexist and can give rise to macrophages in the mouse YS in vivo: monopotent macrophage-restricted progenitors and multipotent myeloid and erythroid progenitors (59). However, the contribution of the different hematopoietic waves to tissue macrophages in adulthood remained unclear.
The yolk sac (YS) origin of macrophages was first proposed by Alliot et al (60). In this pioneering study it was shown that microglial progenitors are found in the brain rudiment of the developing embryo around 8.0 dpc (days post coitum), shortly after the detection of first hematopoietic progenitors with myeloid potential around 7.5 dpc in the YS. These YS progenitors are capable of giving rise to microglial cells on astrocytic feeder monolayers, whereas hematopoietic progenitors isolated from the embryo proper around 9.0 dpc, failed to differentiate into microglia (60). Nevertheless, there was no direct formal proof that YS progenitors give rise to microglial cells or to other tissue macrophages in the developing embryo and adult.
To understand hematopoietic waves in vivo, a non-invasive pulse labelling system based on the Cre/loxP system was utilized, which used a tamoxifen-inducible MER Cre-MER recombinase gene (Cre recombinase fused to two mouse estrogen receptor sequences) under control of the Runx1 promoter (61). This study labeled for the first time hematopoietic progenitors in the early embryo and YS, and followed these cells during development until adulthood. Tamoxifen injection at 7.25-7.5 dpc in the Runx1MER-Cre-MER strain showed that microglia derive from YS progenitors (62). It was proposed that YS macrophages invade the embryo proper and colonize the central nervous system. However, labeling was also detected in bona fide fetal and adult HSCs, albeit to a lesser extent, hampering interpretation of the data. Another caveat of the study is that the knock-in Runx1-MER-Cre-MER animals are heterozygous for Runx1, and Runx1 gene-dosage is critical for definitive hematopoiesis. Runx1 haploinsufficiency results in a dramatic change in the temporal and spatial distribution of HSCs, leading to their early appearance in the AGM region and also their ectopic emergence in the YS (63).
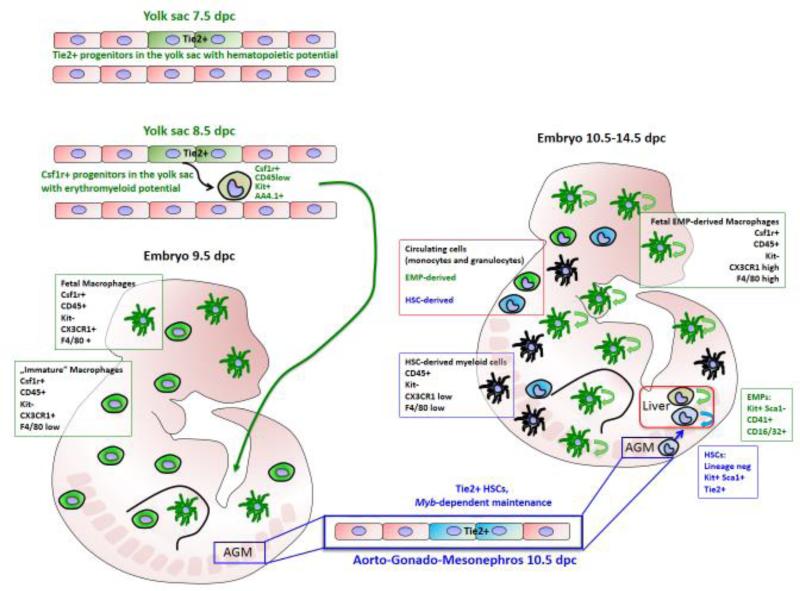
The respective contribution of HSCs and YS progenitors to tissue macrophages remained unclear, especially outside the central nervous system. During embryonic development, two distinct subsets of myeloid cells can be characterized in CX3CR1GFP/+ reporter mice (64), based on kinetics of emergence, phenotype and genome-wide gene expression profile (Figure 1). Both populations are bona fide myeloid populations as revealed by analysis of embryos lacking Pu.1 (65), a master regulator of the macrophage lineage (66). CD45+ CX3CR1high CD11b+ F4/80bright macrophages were detected in all embryonic tissues starting from 9.5 dpc while a second myeloid cell population, described as a CX3CR1+ CD45+ CD11bhigh F4/80low, appeared in most tissues (except the brain) around 12.5 dpc (65), when the definitive HSC-derived hematopoiesis in the fetal liver starts and the first HSCs begin to differentiate into hematopoietic lineages. These two myeloid populations can be genetically distinguished by their dependency on the transcription factor Myb. Myb is required for HSC maintenance and self-renewal (67,68), and Myb-deficient embryos have a complete loss of hematopoietic progenitors in the embryo proper but not in the YS (67,69). F4/80low myeloid cells in the different embryonic tissues are generated in a Myb-dependent manner from HSCs. In contrast, fetal F4/80high macrophages developed independently of Myb and in the absence of fetal HSCs (Table 1) (65). The presence of “phenotypic” fetal monocytes in the fetal liver suggests the existence of definitive progenitors with myeloid potential in Myb-deficient embryos (70). Pulse-labeling with tamoxifen in Csf1rMER-iCre-MER animals at 8.5 dpc showed that Csf1r-expressing cells present in the YS generate fetal F4/80bright macrophages and adult tissue resident macrophages (65). These YS-derived F4/80bright tissue macrophages are found in most adult organs and are maintained within their tissue of residency without further input from bone marrow HSC and progenitors (Figure 1) as evidenced in non-irradiation chimeras and BM progenitor fate mapping animals (Flt3-Cre) (65). Subsequent studies using parabiotic animals, genotoxic depletion of macrophages or CX3CR1-based fate mapping systems confirmed the self-maintenance of tissue macrophages with no contribution of monocytic or CCR2+ bone marrow progenitors (71,72) and extended them to a subset of cardiac macrophages, at least in young animals (73,74). Fate mapping models (S100A4-Cre) with high labeling efficiency in hematopoietic stem and progenitor cells in the bone marrow and circulating monocytes, but a low labeling in tissue resident macrophages from lung, spleen and brain further exclude a contribution of circulating monocytes and HSC-derived hematopoiesis as a source for tissue resident macrophages (71).
Figure 1. Embryonic development of resident tissue macrophages from erythromyeloid-progenitors in the yolk sac.
The first progenitors of tissue resident macrophages arise from Tie2+ hemogenic/endothelial progenitors in the blood islands of the yolk sac (YS) as early as 7.5 days post coitum (dpc). These Tie2+ progenitors give rise to Myb-independent erythromyeloid progenitors (EMP), which are first detectable around 8.5 dpc in the YS and characterized by the expression of Csf1r. The EMP migrates to the embryo proper and gives rise to resident tissue macrophages, which are seeding embryonic tissues from 9.5 dpc onwards. These EMP-derived tissue macrophages actively proliferate within their developing tissues. Beside the EMP-derived tissue macrophages, there is another wave of myeloid cells developing after Myb-dependent hematopoietic stem cells (HSCs) emerge in the embryo starting at 10.5 dpc in the AGM (aorta-gonado-mesonephro) region, and their myeloid progeny can be detected between 10.5 and 14.5 dpc. In contrast to the proliferating self-renewing tissue macrophages, these cells are renewed by differentiation from proliferating HSCs in the fetal liver.
Table 1.
Comparison of YS-EMP-derived tissue macrophages and HSC-derived myeloid cells/macrophages
| Myb-independent tissue macrophages | HSC-derived myeloid cells/macrophages | |
|---|---|---|
| Progenitor | erythromyeloid progenitor (EMP) | hematopoietic stem cells (HSC) |
| Progenitor surface phenotype | CD45low Kit+ AA4.1+ CD16/32+ CD41+ Sca1neg | LSK-SLAM (Linneg Sca1+ Kit+CD150+CD48−) |
| Hematopoietic niche | Yolk sac/fetal liver | AGM region and large embryonic arteries/ fetal liver; Bone marrow |
| Macrophage Surface markers | F4/80high CD11blow | F4/80low CD11bhigh |
| Common markers (gene expression pattern) | Csf1r, CX3CR1 | CCR2, Flt3 |
| Maintenance in the adult | Local self-renewal, proliferation, depending on Csf1r-signaling (Il-34 or Csf1) | Replacement by circulating HSC-derived progenitors |
| Transcription factors for differentiation | Pu.1, Runx1 | Pu.1, Runx1, c-Myb, Gata-2 (?) |
| Transcription factor for tissue adaption | Different factors depending on tissue (see table 2) | Not described yet |
| Physiological function within tissues | Tissue homeostasis, removal of apoptotic cells and secretion of growth factors, stem cell survival | Not described yet |
| Function under challenge within tissues | Tissue inflammation, phagocytosis of dead cells and pathogens, antigen presentation Tissue repair, phagocytosis |
Tissue inflammation, secretion of pro-inflammatory cytokines, antigen presentation |
These approaches do not allow yet to conclude on the YS progenitor type (primitive or definitive) that gives rise to fetal HSC-independent macrophages and it remains to be elucidated whether all YS-derived tissue macrophages originate from the same precursor/progenitor. The YS Csf1r-expressing cells labeled with a tamoxifen pulse at 8.5 dpc in Csf1rMER-iCre-MER animals are Csf1r+ AA4.1+ ckit+ CD45low progenitors. Functionally defined as erythro-myeloid progenitors (EMPs), they first appear in the YS at the 16-18 somite pairs stage and are a common progenitor for resident tissue macrophages (70) (Figure 1). EMPs and HSCs have distinct differentiation and repopulation potentials and can be identified by distinct cell surface phenotypes (70,75). EMPs are identified as kithighCD41+CD16/32+ cell population around 9.5 dpc (Table 1) and they develop within the YS vasculature (blood islands and remodeled vascular plexus) in a Runx-dependent manner (76) and not in the embryo proper, the placenta or vitelline and umbilical vessels. YS-EMPs are generated from 8.5 until 11.5 dpc and they seed the fetal liver where they expand and differentiate into macrophages, erythrocytes, monocytes, granulocytes, and mast cells (70,75). YSEMP-derived Kit+ progenitor cells and short-lived YS-EMP-derived monocytes and granulocytes are found until at least 16.5 dpc in the fetal liver, in accordance with reports that EMP-derived hematopoiesis is not only necessary but also sufficient to support survival of embryos lacking HSCs until the time of birth (77). EMPs and HSCs differentiate from distinct populations of hemogenic endothelial cells (77), and the developmental lineages of HSC-derived myeloid cells and YS-EMP-derived myeloid cells are disconnected from each other (70) (Table 1). YS-EMPs and tissue resident macrophages were only labeled in Tie2Mer-iCre-Mer mice when tamoxifen was applied before 9.5 dpc, and did not equilibrate with HSC labeling efficiency (Figure 1).
Immature macrophages present in the YS at 8.5 dpc could also be labeled inCsf1rMER-iCre-MER embryos, and as such, this pulse-labeling strategy does not allow to rule out a contribution of “primitive” macrophage progenitors to tissue resident macrophages. Brain microglia is labeled more efficiently than other tissue resident macrophages when tamoxifen is applied at E7.5 in Runx1MER-iCre-MER, Tie2MER-iCre-MER and KitMER-iCre-MER (73,80,92). These findings, together with the comparison of the labeling efficiency over time of microglia and macrophages in other tissues after a single tamoxifen pulse (62,65,70), has led some authors to hypothesize that they could originate from different YS progenitors. Therefore, more investigations are required to demonstrate that the differences observed between microglia and other tissue resident macrophages correspond to different YS progenitor populations. Collectively, these findings suggest that (i) “primitive” progenitors can contribute to and only microglia pools or that (ii) colonization of the brain is performed only in a very limited time window from YS-EMPs, and these two hypotheses are not mutually exclusive. In favor of the latter, pulse-labeling at E8.5 only allows to label YS-EMPs in a narrow time window following the pulse, while YS-EMPs continue to emerge from the YS until at least 11.5 dpc (70,75,78) and expand in the fetal liver after colonization (70). In the central nervous system, it was shown that microglia enter the neuroectoderm between 9.0 dpc and 9.5 dpc (60,62,79), while the colonization of other tissues by macrophages is not yet fully defined.
From EMP to tissue resident macrophages
Further investigations tracking the differentiation and migration of YS-EMPs in vivo might be necessary to completely identify the differentiation pathway of the progenitors. It is currently not known what are the differentiation steps followed by YS-EMPs during their differentiation into mature tissue macrophages. Myeloid progenitors/precursors derived from YS-EMPs were described in ex vivo culturing systems (79). These intermediate progenitors seemed to have different myeloid differentiation levels characterized by gradual upregulation of mature macrophage/myeloid markers like CX3CR1 and down regulation of immature macrophage markers like CD31 (59,79).
In regards to the migration of YS-EMPs into the embryo, most available data supports the importance of blood circulation. This hypothesis is supported by the analysis of blood-circulation-deficient animals like the cardiac Na+-Ca2+ exchanger (Ncx1) knockout mice. Ncx1-deficient animals die in utero at 9.5 dpc due to heart failure and lack of blood circulation (80,81) and they show an accumulation of macrophages in the YS, but a decrease of microglial cells in the developing neuroectoderm (62). Similarly, YS-EMPs are not detected in the fetal liver in Ncx1-deficient embryos, but increase massively in the YS (75). Thus, an established blood circulation is needed for the migration of macrophage progenitors to the embryo proper. However, once in the embryo proper, the progenitors or their daughter cells do not necessarily need any established vascularization to colonize tissues, as microglia are found in the brain before the neuroectoderm is vascularized and further support angiogenesis or new vessel formation in the brain parenchyma (32).
Further work is also necessary to establish whether tissue seeding is performed by EMPs, EMP-derived myeloid precursors or by their mature progeny, the macrophages. In favor of the latter, before or prior to their entrance into the CNS, myeloid cells express mature microglial/macrophage markers like F4/80 and CX3CR1 (60,62,79), thereby suggesting that colonization in the brain is performed by macrophages. It will also be interesting to identify the cues and factors triggering the entrance of tissue macrophages into tissues. In the case of brain colonization, Csf1 and CXCL12 are secreted from neural progenitors in the developing brain parenchyma and play a role in the recruitment of macrophages to the developing cortex in mice (32,82), while loss of vascular endothelial growth factor (Vegf) and classical chemokine receptor signaling does not impact brain colonization (32,79). It was recently proposed that senescent cells in the limb could be responsible for macrophage recruitment at 12.5 dpc (30), however, CX3CR1gfp F4/80bright macrophages are found in the limb as early as 10.5 dpc (65).
Albeit the identification of the YS-EMP as a major source for F4/80bright tissue macrophages, there are still many open questions concerning the development of tissue macrophages. It is not clear yet how the YS-EMP migrate from the YS to the embryo proper and how this cell differentiates into different “varieties” of tissue macrophages, each of them highly adapted to their specific function. Furthermore, the signals recruiting resident macrophages to the tissue remain unclear.
Extrinsic cues involved in maintenance of EMP-derived macrophages
It was proposed that YS-derived macrophages would be replaced late during gestation by HSC-derived fetal monocytes in the lung, skin and gastro-intestinal tract. While available data does not yet fully support this model for the lung alveolar macrophages and epidermal Langerhans cells from young adults (83,84), it has now been clearly characterized for the lamina propria macrophages in the small intestine and colon (85,86). Within the intestine, YS-derived macrophage numbers decrease progressively until weaning, after which all lamina propria macrophages are continuously replenished by infiltrating CCR2+ Ly6Chigh monocytes (85).
A hallmark of embryonic tissue macrophages after entrance into the tissue is that they disseminate and proliferate, resulting in a massive increase in their cell number in the developing tissue, as shown for Kupffer cells and microglia (60,87). This “burst of proliferation” is also well described for Langerhans cells after entrance in the epidermis at 18 dpc (18). The proliferation rate of Langerhans cells is stable at 5% throughout adulthood, further indicating that Langerhans cells maintain themselves in the tissue by local proliferation rather than recruitment of new progenitors, under physiological conditions of the skin (18,88). The decline of proliferation is seen for all adult tissue macrophages, and their proliferation never stops completely during adulthood. Kupffer cells labeled by injection of latex beads still show the same labeling efficiency with beads 3 months after injection and have a low mitotic index under physiological conditions (89). Microglial cells have also been shown to be a stable cell population over life time with no contribution from circulating progenitors (10,90,91) and can repopulate by local proliferation after genetic depletion (92). In the lung and heart, EMP-derived macrophages appear to be replaced during aging (70,73,74). In specific settings where tissue macrophages are depleted, such as lethal irradiation or Listeria infection, monocytes can replace tissue macrophages (92,93). However, the functional consequences of macrophage replacement for tissue homeostasis require further investigation. All these results point towards a new paradigm for tissue macrophage homeostasis, with long-lived tissue resident macrophages with a low proliferation rate and nearly no contribution of circulating progenitors. A long life span demands the maintenance of resident macrophages in the tissues and in the aging organism. Therefore, the host tissues have to provide “cues and factors” which support and help to maintain the “resident” tissue macrophage population.
One of the key factors important for the maintenance and also differentiation of myeloid progenitors is Colony stimulating factor 1 (Csf1). Discovered in the 1970s in the Jackson Laboratories, Csf1op/op mice harbor a spontaneous point mutation in the locus of the Csf1 gene (94). Csf1op/op mice have severe osteopetrosis and a compromised hematopoietic compartment, especially the monocytic/macrophage lineage, where differentiated cells are severely decreased (95). The loss of differentiated myeloid cells cannot be rescued by bone marrow transplantation in Csf1op/op mice, indicating a defect in the microenvironment of the hematopoietic compartment rather than a cell-intrinsic defect of myeloid cells (96). Csf1 binds to a 165kDa surface glycoprotein which is encoded by the c-fms protooncogene and is expressed on nearly all murine macrophages, the Csf1 receptor (Csf1r) (97). Detailed characterization of macrophage populations in Csf1op/op demonstrated that not all tissue resident macrophages are affected by the loss of Csf1 (98). Epidermal Langerhans cells and brain microglia remained mostly unaffected by the loss of Csf1, whereas osteoclasts are massively reduced in these animals (99,100) (Table 2). Csf1r-deficient (Csf1r−/−) animals phenocopy the osteopetrotic pathology, as well as the reduced fertility and life span found in Csf1op/op mice, but have a severe reduction of microglia and Langerhans cells (62,101). Csf1r is widely expressed in the hematopoietic system, in embryonic and adult tissue macrophages, as well as bone marrow derived myeloid cells (102). Csf1 was considered to be the only ligand of the receptor, but the discrepancy between the phenotype of Csf1op/op and Csf1r −/− suggested the existence of another ligand. In a systematic functional screen, interleukin-34 (Il-34) improved monocyte viability, whereas other cell types remained unaffected (103). Il-34 binds to Csf1r and triggers downstream signaling upon binding in a similar manner as known for Csf1. Il-34-deficient mice have reduced numbers of microglia and Langerhans cells while other tissue macrophage populations are unaffected (104,105) (Table 2). Il-34 is released by “stromal” cells, such as neurons in the CNS and keratinocytes in the epidermis. Both ligands, Csf1 and IL-34 share the same receptor and it is not yet clear how and if they can trigger different activation cascades upon Csf1r binding. A recent study revealed that Il-34 and Csf1 are structurally similar but do not share a common sequence (106). Both can support the survival and maintenance of myeloid cell lines in vitro, but they trigger different activation responses in regards to cytokine release. IL-34 induces a stringer activation of the signaling cascade downstream of Csf1r, but this activation lasts much shorter compared to Csf1 binding (107). Further investigations are needed to elucidate if Csf1 and IL-34 play completely independent functions on tissue resident macrophages or if their effects are complementary. Yet, it remains unclear why tissue resident macrophages are dependent on one of these two Csf1r ligands depending on their tissue.
Depending on the tissue of residence, a complexity of secreted factors from the surrounding cells maintains the macrophage pool. Transforming growth factor beta (TGFβ) was long known to differentiate hematopoietic stem cells and bone marrow myeloid progenitors into cells with a Langerhans cell-like phenotype in vitro (108). Mice deficient for TGFβ have an epidermis completely devoid of Langerhans cells even before the autoimmune phenotype is established in these animals (109). Deletion of the TGF beta receptor (Tgfbr) in epidermal Langerhans cells led to a reduced number of Langerhans cells during the first week of life, albeit the initial seeding of the epidermis at birth was normal. It was proposed that Langerhans cells leave the epidermis and adopt a migratory phenotype without TGFβ signal. These studies indicate that TGFβ is not important for the colonization of the epidermis with tissue resident macrophages, but rather important to maintain the macrophages in a resting state within the epidermis (110) (Table 2).
Among environmental factors shaping tissue macrophages at steady-state, products of intestinal commensals could contribute to tissue macrophage maintenance and function in the mouse brain. Housing of mice under complete sterile conditions (also termed ‘germ-free’) leads to a pronounced immature phenotype of microglia, including upregulation of several surface proteins such as F4/80 and increased numbers in different brain regions, in line with altered expression of genes regulating the cell cycle (111). Notably, such macrophage alterations were found to be extremely plastic in adult mice, as eradication of intestinal bacteria by antibiotic treatment drives microglia to acquire an immature status. Vice versa, recolonization of mice harboring a reduced flora with complex microbiota leads to microglia ‘maturation’, indicating a great plasticity of the gut-microglia connection (112).
Transcription factors in tissue macrophage development and specialization
Various transcription factors have been identified in myeloid cell development and as discussed above, most efforts were focused during the last 20 years in describing the detailed development of myeloid cells in the bone marrow (113–117). While the detailed differentiation steps of tissue macrophages from their YS progenitor are not yet fully understood, transcription factors important for tissue specific macrophage differentiation and diversification are starting to emerge (48).
The most studied transcription factor in macrophage differentiation is Pu.1 (Spi1, Sfpi1). Pu.1 belongs to the ets transcription factor family, which is characterized by the ETS DNA binding domain (118). Expression of Pu.1 is restricted to hematopoietic cells (mostly B cells and myeloid cells). Animals deficient for the transcription factor Pu.1 show multiple defects in hematopoiesis and die either prenatally or a few days after birth (66,119). Further analysis of the hematopoietic defects by McKercher et al revealed a loss of B cells and mature myeloid cells, but normal erythrocytes and megakaryocytes, which revealed a key role of Pu.1 in myeloid differentiation and the development of myeloid progenitors. Pu.1-deficient HSCs have a homing and maintenance defect in the fetal liver and Pu.1-deficient embryos can be rescued by wild-type HSC transplantation in utero. Pu.1-deficient mice completely lack tissue resident macrophages from bone marrow and YS origin (65) but an earlier report found Csf1r-expressing cells but no F4/80+ cells in the absence of Pu.1 (120). However, the activity of Pu.1 is not an ”on-or-off” signal. High concentrations of Pu.1 trigger development of macrophages, whereas low levels favor B cell development (121). Gradients of Pu.1 are also involved in the lineage decision of myeloid progenitors, where low levels of Pu.1 lead to the development of granulocytic progenitors and high levels of Pu.1 trigger macrophage differentiation (122). Pu.1 also plays a role beyond myeloid differentiation, in mature macrophage functions. In vitro knockdown of Pu.1 in bone marrow derived macrophages leads to a reduction in proliferation, which is associated with downregulation of Csf1r on the cell surface (123). Pu.1 is not only able to regulate macrophage-specific genes like Csf1r. Chromatin immunoprecipitation (ChIP) for Pu.1 reveals its association with binding sites all over the genome in adult macrophages (124). Pu.1 binding to the enhancer motifs or promoter regions in macrophages is most often associated with monomethylation of lysine 4 in histone 3, which leads to an opening of the chromatin structure at these positions and allows further recruitment of a secondary set of transcription factors, for example liver X receptors (LXRs) or Nfκb. Therefore, Pu.1 is also important in mature macrophages to modulate the chromatin landscape and orchestrate other transcription factors to their binding sides, which might be induced by external stimuli and are important for the functional properties of macrophages (125).
Whereas Pu.1 activity is essential in all tissue macrophages for their development and for their function, other transcription factors are found to play a role in specific subsets of tissue macrophages. Another member of the Ets transcription factor family is Spi-C (Spic). Spi-C was initially found in a yeast-two hybrid-screen with a cDNA library of LPS-stimulated B cells and high expression was described in mature B cells, whereas macrophages only expressed lower levels of Spi-C (126). However, Spi-C is highly expressed in one specific subset of tissue resident macrophages, the red pulp macrophages (RPMs) of the spleen. This specialized subset of macrophages is important for the phagocytosis of old erythrocytes from the blood stream and to maintain iron homeostasis. Spic-deficient mice showed a selective loss of RPMs in the spleen, whereas B cells and other myeloid cells like monocytes and dendritic cells were not affected. Furthermore, these mice showed a disturbance in iron homeostasis with an accumulation of iron specifically in the red pulp of the spleen, indicating the importance of RPMs for maintaining iron homeostasis in this organ. Additional studies confirmed the tissue-specific expression and significance of Spi-C for homeostasis (48,127). Spi-C is a transcription factor crucial for the development of one single subset of tissue macrophages and further indicates that myeloid cells adapt specified transcriptional programs after they reach their organ of residency. This indicates that tissue macrophage subsets acquire a specified transcriptional program after reaching the target tissue to develop specific properties required in these organs.
Peritoneal macrophages are one of the best-studied tissue macrophages over the last decades. Two functionally distinct macrophage subsets are found in the peritoneal cavity: large peritoneal macrophages (LPMs) and small peritoneal macrophages (SPMs), with LPMs largely outnumbering SPMs under physiological conditions (16). Both subsets express the macrophage markers CD11b and F4/80 and phagocytize bacteria injected in vivo. However, LPMs are more efficient than SPMs in apoptotic cell clearance (128) and both subsets show a differential expression of other surface markers like MHC class II and respond differently to various stimuli in vitro (16). Several studies suggest that LPMs are derived from an embryonic source and are not maintained by monocytic turn over (72,129). Beside functional differences, C/EBPβ (Cebpb) was found to be highly expressed in LPMs and at a lower level in SPMs. Cebpb-deficient mice show a massive decrease in LPM generation, but show also in lung alveolar macrophages (128). Transplanted Cebpb-sufficient SPMs into Cebpb-deficient mice can differentiate into LPMs in the peritoneal cavity, but this differentiation was not observed after transplantation of SPMs into wildtype animals, which suggests another differentiation pathway for LPMs under steady state conditions. While C/EBPβ is important in general for monocyte and macrophage activation upon different stimuli. C/EBPβ is also a transcription factor that controls the differentiation and function of these two specific subsets of tissue macrophages.
Several recent studies further characterized the macrophage compartment of the peritoneal cavity. Two independent studies identified GATA-6 as a major transcription factor inducing peritoneal macrophage specific gene expression in LPMs (130,131). GATA-6 is a zinc finger transcription factor involved in different lineages from mesoderm during embryonic development like precardiac mesoderm or the primitive gut (132). GATA-6 is highly and specifically expressed in LPMs, whereas SPMs and other tissue macrophages do not express it (130,131). Gata6-deficiency in myeloid cells leads to reduced LPMs numbers, altered gene expression and loss of proliferation capacities in these cells. Gata-6-deficient LPMs failed to regulate peritoneal B1 cells (130), which are the main producers of IgM antibodies and continuously migrate to the lamina propria of the gut to give rise to IgA producing B cells (133–135). These two studies concluded that Gata-6 links peritoneal-specific identity (gene expression profile), function (regulation of peritoneal B1 cells) and renewal (proliferation) of LPMs.
Tissue-specific adaption of macrophages seems to occur in nearly each tissue, at the phenotypic, genetic and epigenetic levels (Table 2). To understand how the tissue environment and the different genetic programs of macrophages could lead to unique macrophage identities and specific functions in each tissue, two studies analyzed different subsets of tissue macrophage populations in different sets of high-throughput sequencing data, like RNA sequencing (RNA-seq, transcriptome) or chromatin immunoprecipitation sequencing (ChIP-seq, chromatin structure and modifications). Tissue resident macrophages revealed distinct gene expression patterns depending on their tissue of residence and they have different enhancer landscapes, especially active enhancers (48,49). As expected from their different developmental and cellular origin, resident tissue macrophages have a distinct chromatin landscape signature from bone marrow-derived myeloid cells like monocytes and neutrophils (48). Among tissue macrophages, there was a striking variability in active enhancers depending on the tissue of residency. Bone-marrow-derived macrophages, which can repopulate various tissues after tissue macrophage depletion by irradiation and bone marrow transplantation, can adopt a tissue-specific chromatin landscape similar to host macrophages. When transplanted into the alveolar cavity, peritoneal macrophages “adopt” a lung-like phenotype and gene expression profile. Based on these, tissue environment was proposed to be responsible for the differences in gene expression and cell identity between macrophages in different tissues. Nevertheless, tissue microenvironment alone cannot account for the diversity of macrophage identities within each tissue. Within the peritoneal cavity, two subsets of macrophages co-exist, the large peritoneal macrophages and small peritoneal macrophages. These two macrophage populations are thought to share the same origin and are located in the same anatomical structure, however their chromatin structure and the enhancers they use are quite different (49).
Collectively, both the developmental pathway and the tissue microenvironment contribute to create a unique chromatin structure in each adult macrophage subset, which is highly adaptable depending on the physiological status and the organ. It will be most interesting to investigate whether adult tissue macrophages bear in the chromatin landscape marks from their common origin and whether they carry a “basic” genetic program that persists until adulthood.
Summary and Outlook: What are the missing links in the macrophage puzzle?
In the last years, many studies using new genetic mouse models and high-throughput gene expression analyses have provided insights into how macrophages develop and that their heterogeneity can be partially driven by their tissue environment, respectively. Albeit many questions are still open, it is now demonstrated that most tissue resident macrophages are generated from yolk sac progenitors but do not develop or renew from blood monocytes and hematopoietic stem cells. Many challenges lie in the identification of the missing steps in their developmental pathway(s) and its transcriptional control, as further differentiation steps as well as migratory pathways of macrophage precursors in the developing embryo remain undefined. Our knowledge on macrophage specialization to their host tissues has dramatically increased, especially in terms of their intrinsic and extrinsic (epi)genetic programs playing a role in this process in different tissues. However, our understanding of how all these different layers are integrated to give rise to the newly defined heterogeneity of resident macrophage is still limited, in particular when addressing macrophage functions. Because macrophages were most often considered to have activation properties and functions specific for the stimulus but independent of their location, macrophage functions in homeostasis and disease were transposed from one tissue to another. It is thus an exiting scientific time to reinvestigate macrophage functions more carefully, in regards to their developmental origin and their cell-intrinsic tissue specialization, to identify the environmental factors supporting or driving tissue macrophage specialization and maintenance, and to characterize their contribution to tissue homeostasis and function, but also to disease pathogenesis and tissue repair, distinguishing them from infiltrating monocyte-derived macrophages in inflamed tissues.
Highlights.
-Macrophage subsets are heterogeneous in their developmental hematopoietic origin
-Erythromyeloid progenitors give rise to tissue macrophages persisting until adulthood
-Tissue macrophages have homeostatic functions depending on their anatomic location
-Tissue adaption of macrophages occurs on phenotypic, epigenetic and genetic levels
Acknowledgement
K.K. was funded by a research fellowship by the DFG (German Research Foundation). M.P. was supported by the BMBF-funded competence network of multiple sclerosis (KKNMS), the Sobek Foundation, the DFG (SFB 992, FOR1336, PR 577/8-1), the Fritz-Thyssen Foundation and the Gemeinnützige Hertie Foundation (GHST). E.G.P is supported by the Institut Pasteur and Agence Nationale de la Recherche (Laboratoire d'Excellence Revive, Investissement d'Avenir; ANR-10-LABX-73).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gordon S. Elie Metchnikoff: Father of natural immunity. Eur J Immunol. 2008 Dec 1;38(12):3257–64. doi: 10.1002/eji.200838855. [DOI] [PubMed] [Google Scholar]
- 2.METSCHNIKOFF E. Memoirs: Researches on the Intracellular Digestion of Invertebrates. Q J Microsc Sci. 1884;2(93):89–111. [Google Scholar]
- 3.Tauber AI. Metchnikoff and the phagocytosis theory. Nat Rev Mol Cell Biol. 2003 Nov;4(11):897–901. doi: 10.1038/nrm1244. [DOI] [PubMed] [Google Scholar]
- 4.Silva MT, Correia-Neves M. Neutrophils and macrophages: the main partners of phagocyte cell systems. Microb Immunol. 2012;3:174. doi: 10.3389/fimmu.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull World Health Organ. 1972;46(6):845–52. [PMC free article] [PubMed] [Google Scholar]
- 6.Godleski JJ, Brain JD. The origin of alveolar macrophages in mouse radiation chimeras. J Exp Med. 1972 Sep 1;136(3):630–43. doi: 10.1084/jem.136.3.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy DW, Abkowitz JL. Kinetics of Central Nervous System Microglial and Macrophage Engraftment: Analysis Using a Transgenic Bone Marrow Transplantation Model. Blood. 1997 Aug 1;90(3):986–93. [PubMed] [Google Scholar]
- 8.Virolainen M. Hematopoietic origin of macrophages as studied by chromosome markers in mice. J Exp Med. 1968 May 1;127(5):943–52. doi: 10.1084/jem.127.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volkman A, Gowans JL. The Origin of Macrophages from Bone Marrow in the Rat. Br J Exp Pathol. 1965 Feb;46(1):62–70. [PMC free article] [PubMed] [Google Scholar]
- 10.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FMV. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007 Dec;10(12):1538–43. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 11.Parwaresch MR, Wacker HH. Origin and kinetics of resident tissue macrophages. Parabiosis studies with radiolabelled leucocytes. Cell Tissue Kinet. 1984 Jan;17(1):25–39. doi: 10.1111/j.1365-2184.1984.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 12.Furth R van Dulk MMD. Dual origin of mouse spleen macrophages. J Exp Med. 1984 Nov 1;160(5):1273–83. doi: 10.1084/jem.160.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentek R, Molawi K, Sieweke MH. Tissue macrophage identity and self-renewal. Immunol Rev. 2014 Nov;262(1):56–73. doi: 10.1111/imr.12224. [DOI] [PubMed] [Google Scholar]
- 14.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol. 2013 Oct;14(10):986–95. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohn ZA. Determinants of Infection in the Peritoneal Cavity. Yale J Biol Med. 1962 Aug;35(1):12–28. [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosn EEB, Cassado AA, Govoni GR, Fukuhara T, Yang Y, Monack DM, et al. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc Natl Acad Sci. 2010 Feb 9;107(6):2568–73. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chopin M, Nutt SL. Establishing and maintaining the Langerhans cell network. Semin Cell Dev Biol. 2015 May;41:23–9. doi: 10.1016/j.semcdb.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Chorro L, Sarde A, Li M, Woollard KJ, Chambon P, Malissen B, et al. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J Exp Med. 2009 Dec 21;206(13):3089–100. doi: 10.1084/jem.20091586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005 May 27;308(5726):1314–8. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 20.Del Rio-Hortega P. El “tercer elemento” de los centros nerviosus. I. La microglia en estado normal. II. Intervencion de la microglia en los procesos patologicos (Celulas en bastoncito y cuerpos granuloadiposos). III. Naturaleza probable de la microglia. Bol Soc Espan Biol. 1919;9:68–120. [Google Scholar]
- 21.Kupffer C. Ueber Sternzellen der Leber. Arch Für Mikrosk Anat. 1876 Dec 1;12(1):353–8. [Google Scholar]
- 22.Langerhans P. Ueber die Nerven der menschlichen Haut. Arch Für Pathol Anat Physiol Für Klin Med. 1868 Sep 1;44(2-3):325–37. [Google Scholar]
- 23.Ling EA, Wong WC. The origin and nature of ramified and amoeboid microglia: a historical review and current concepts. Glia. 1993 Jan;7(1):9–18. doi: 10.1002/glia.440070105. [DOI] [PubMed] [Google Scholar]
- 24.McWhorter FY, Wang T, Nguyen P, Chung T, Liu WF. Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci. 2013 Oct 22;110(43):17253–8. doi: 10.1073/pnas.1308887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005 Jun;8(6):752–8. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 26.Nishibu A, Ward BR, Jester JV, Ploegh HL, Boes M, Takashima A. Behavioral Responses of Epidermal Langerhans Cells In Situ to Local Pathological Stimuli. J Invest Dermatol. 2006 Jan 26;126(4):787–96. doi: 10.1038/sj.jid.5700107. [DOI] [PubMed] [Google Scholar]
- 27.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011 Nov 1;11(11):723–37. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011 Sep 9;333(6048):1456–8. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 29.Li MO, Sarkisian MR, Mehal WZ, Rakic P, Flavell RA. Phosphatidylserine receptor is required for clearance of apoptotic cells. Science. 2003 Nov 28;302(5650):1560–3. doi: 10.1126/science.1087621. [DOI] [PubMed] [Google Scholar]
- 30.Muñoz-Espín D, Cañamero M, Maraver A, Gómez-López G, Contreras J, Murillo-Cuesta S, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013 Nov 21;155(5):1104–18. doi: 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida H, Kawane K, Koike M, Mori Y, Uchiyama Y, Nagata S. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature. 2005 Sep 29;437(7059):754–8. doi: 10.1038/nature03964. [DOI] [PubMed] [Google Scholar]
- 32.Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, et al. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 2010 Aug 5;116(5):829–40. doi: 10.1182/blood-2009-12-257832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blackwell TS, Hipps AN, Yamamoto Y, Han W, Barham WJ, Ostrowski MC, et al. NF-κB signaling in fetal lung macrophages disrupts airway morphogenesis. J Immunol Baltim Md 1950. 2011 Sep 1;187(5):2740–7. doi: 10.4049/jimmunol.1101495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higashi K, Naito M, Takeya M, Ando M, Araki S, Takahashi K. Ontogenetic development, differentiation, and phenotypic expression of macrophages in fetal rat lungs. J Leukoc Biol. 1992 May;51(5):444–54. doi: 10.1002/jlb.51.5.444. [DOI] [PubMed] [Google Scholar]
- 35.Ingman WV, Wyckoff J, Gouon-Evans V, Condeelis J, Pollard JW. Macrophages promote collagen fibrillogenesis around terminal end buds of the developing mammary gland. Dev Dyn Off Publ Am Assoc Anat. 2006 Dec;235(12):3222–9. doi: 10.1002/dvdy.20972. [DOI] [PubMed] [Google Scholar]
- 36.DeFalco T, Bhattacharya I, Williams AV, Sams DM, Capel B. Yolk-sac-derived macrophages regulate fetal testis vascularization and morphogenesis. Proc Natl Acad Sci U S A. 2014 Jun 10;111(23):E2384–93. doi: 10.1073/pnas.1400057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banaei-Bouchareb L, Gouon-Evans V, Samara-Boustani D, Castellotti MC, Czernichow P, Pollard JW, et al. Insulin cell mass is altered in Csf1op/Csf1op macrophage-deficient mice. J Leukoc Biol. 2004 Aug;76(2):359–67. doi: 10.1189/jlb.1103591. [DOI] [PubMed] [Google Scholar]
- 38.Kubota Y, Takubo K, Shimizu T, Ohno H, Kishi K, Shibuya M, et al. M-CSF inhibition selectively targets pathological angiogenesis and lymphangiogenesis. J Exp Med. 2009 May 11;206(5):1089–102. doi: 10.1084/jem.20081605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon EJ, Rao S, Pollard JW, Nutt SL, Lang RA, Harvey NL. Macrophages define dermal lymphatic vessel calibre during development by regulating lymphatic endothelial cell proliferation. Dev Camb Engl. 2010 Nov;137(22):3899–910. doi: 10.1242/dev.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rymo SF, Gerhardt H, Wolfhagen Sand F, Lang R, Uv A, Betsholtz C. A two-way communication between microglial cells and angiogenic sprouts regulates angiogenesis in aortic ring cultures. PloS One. 2011;6(1):e15846. doi: 10.1371/journal.pone.0015846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blair HC, Teitelbaum SL, Ghiselli R, Gluck S. Osteoclastic bone resorption by a polarized vacuolar proton pump. Science. 1989 Aug 25;245(4920):855–7. doi: 10.1126/science.2528207. [DOI] [PubMed] [Google Scholar]
- 42.Teitelbaum SL. Bone Resorption by Osteoclasts. Science. 2000 Sep 1;289(5484):1504–8. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 43.Wake H, Moorhouse AJ, Miyamoto A, Nabekura J. Microglia: actively surveying and shaping neuronal circuit structure and function. Trends Neurosci. 2012 Dec 20; doi: 10.1016/j.tins.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Zhan Y, Paolicelli RC, Sforazzini F, Weinhard L, Bolasco G, Pagani F, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci. 2014 Mar;17(3):400–6. doi: 10.1038/nn.3641. [DOI] [PubMed] [Google Scholar]
- 45.Terpstra V, Berkel TJC van. Scavenger receptors on liver Kupffer cells mediate the in vivo uptake of oxidatively damaged red blood cells in mice. Blood. 2000 Mar 15;95(6):2157–63. [PubMed] [Google Scholar]
- 46.Forbes A, Pickell M, Foroughian M, Yao L-J, Lewis J, Veldhuizen R. Alveolar macrophage depletion is associated with increased surfactant pool sizes in adult rats. J Appl Physiol Bethesda Md 1985. 2007 Aug;103(2):637–45. doi: 10.1152/japplphysiol.00995.2006. [DOI] [PubMed] [Google Scholar]
- 47.Dong Q, Wright JR. Degradation of surfactant protein D by alveolar macrophages. Am J Physiol. 1998 Jan;274(1 Pt 1):L97–105. doi: 10.1152/ajplung.1998.274.1.L97. [DOI] [PubMed] [Google Scholar]
- 48.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014 Dec 4;159(6):1312–26. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014 Dec 4;159(6):1327–40. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ganz T. Macrophages and systemic iron homeostasis. J Innate Immun. 2012;4(5-6):446–53. doi: 10.1159/000336423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karlsson MCI, Guinamard R, Bolland S, Sankala M, Steinman RM, Ravetch JV. Macrophages control the retention and trafficking of B lymphocytes in the splenic marginal zone. J Exp Med. 2003 Jul 21;198(2):333–40. doi: 10.1084/jem.20030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eloranta ML, Alm GV. Splenic marginal metallophilic macrophages and marginal zone macrophages are the major interferon-alpha/beta producers in mice upon intravenous challenge with herpes simplex virus. Scand J Immunol. 1999 Apr;49(4):391–4. doi: 10.1046/j.1365-3083.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- 53.Bronte V, Pittet MJ. The spleen in local and systemic regulation of immunity. Immunity. 2013 Nov 14;39(5):806–18. doi: 10.1016/j.immuni.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naito M, Takahashi K, Nishikawa S. Development, differentiation, and maturation of macrophages in the fetal mouse liver. J Leukoc Biol. 1990 Jul 1;48(1):27–37. doi: 10.1002/jlb.48.1.27. [DOI] [PubMed] [Google Scholar]
- 55.Cline MJ, Moore M a. S. Embryonic Origin of the Mouse Macrophage. Blood. 1972 Jun 1;39(6):842–9. [PubMed] [Google Scholar]
- 56.McGrath KE, Palis J. Hematopoiesis in the yolk sac: more than meets the eye. Exp Hematol. 2005 Sep;33(9):1021–8. doi: 10.1016/j.exphem.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 57.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008 Feb 22;132(4):631–44. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Dev Camb Engl. 1999 Nov;126(22):5073–84. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 59.Bertrand JY, Jalil A, Klaine M, Jung S, Cumano A, Godin I. Three pathways to mature macrophages in the early mouse yolk sac. Blood. 2005 Nov 1;106(9):3004–11. doi: 10.1182/blood-2005-02-0461. [DOI] [PubMed] [Google Scholar]
- 60.Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res. 1999 Nov 18;117(2):145–52. doi: 10.1016/s0165-3806(99)00113-3. [DOI] [PubMed] [Google Scholar]
- 61.Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007 Apr 26;446(7139):1056–61. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- 62.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate Mapping Analysis Reveals That Adult Microglia Derive from Primitive Macrophages. Science. 2010 Nov 5;330(6005):841–5. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai Z, de Bruijn M, Ma X, Dortland B, Luteijn T, Downing RJ, et al. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity. 2000 Oct;13(4):423–31. doi: 10.1016/s1074-7613(00)00042-x. [DOI] [PubMed] [Google Scholar]
- 64.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000 Jun;20(11):4106–14. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schulz C, Perdiguero EG, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. A Lineage of Myeloid Cells Independent of Myb and Hematopoietic Stem Cells. Science. 2012 Apr 6;336(6077):86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 66.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996 Oct 15;15(20):5647–58. [PMC free article] [PubMed] [Google Scholar]
- 67.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, et al. A functional cmyb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991 May 17;65(4):677–89. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 68.Soza-Ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci U S A. 2010 Oct 5;107(40):17304–8. doi: 10.1073/pnas.1004640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. [2012 Oct 29];Initiation of adult myelopoiesis can occur in the absence of c-Myb whereas subsequent development is strictly dependent on the transcription factor. doi: 10.1038/sj.onc.1203660. Publ Online 14 July 2000 Doi101038sjonc1203660 [Internet]. 2000 Jul 14;19(30). Available from: http://www.nature.com/onc/journal/v19/n30/full/1203660a.html. [DOI] [PubMed]
- 70.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015 Feb 26;518(7540):547–51. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, et al. Tissue-Resident Macrophages Self-Maintain Locally throughout Adult Life with Minimal Contribution from Circulating Monocytes. Immunity. 2013 Apr 18;38(4):792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity. 2013 Jan 24;38(1):79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014 Jan 16;40(1):91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Molawi K, Wolf Y, Kandalla PK, Favret J, Hagemeyer N, Frenzel K, et al. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med. 2014 Oct 20;211(11):2151–8. doi: 10.1084/jem.20140639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, et al. Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep. 2015 Jun 17; doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frame JM, Fegan KH, Conway SJ, McGrath KE, Palis J. Definitive Hematopoiesis in the Yolk Sac Emerges from Wnt-Responsive Hemogenic Endothelium Independently of Circulation and Arterial Identity. Stem Cells Dayt Ohio. 2015 Sep 29; doi: 10.1002/stem.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen S-K, Tvrdik P, Peden E, Cho S, Wu S, Spangrude G, et al. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell. 2010 May 28;141(5):775–85. doi: 10.1016/j.cell.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sheng J, Ruedl C, Karjalainen K. Most Tissue-Resident Macrophages Except Microglia Are Derived from Fetal Hematopoietic Stem Cells. Immunity. 2015 Aug 18;43(2):382–93. doi: 10.1016/j.immuni.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 79.Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013 Mar;16(3):273–80. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 80.Cho CH, Kim SS, Jeong MJ, Lee CO, Shin HS. The Na+ -Ca2+ exchanger is essential for embryonic heart development in mice. Mol Cells. 2000 Dec 31;10(6):712–22. doi: 10.1007/s10059-000-0712-2. [DOI] [PubMed] [Google Scholar]
- 81.Cho C-H, Lee S-Y, Shin H-S, Philipson KD, Lee CO. Partial rescue of the Na+-Ca2+ exchanger (NCX1) knock-out mouse by transgenic expression of NCX1. Exp Mol Med. 2003 Apr 30;35(2):125–35. doi: 10.1038/emm.2003.18. [DOI] [PubMed] [Google Scholar]
- 82.Arnò B, Grassivaro F, Rossi C, Bergamaschi A, Castiglioni V, Furlan R, et al. Neural progenitor cells orchestrate microglia migration and positioning into the developing cortex. Nat Commun. 2014;5:5611. doi: 10.1038/ncomms6611. [DOI] [PubMed] [Google Scholar]
- 83.Hoeffel G, Wang Y, Greter M, See P, Teo P, Malleret B, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med. 2012 Jun 4;209(6):1167–81. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013 Sep 23;210(10):1977–92. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol. 2014 Oct;15(10):929–37. doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, Margalit R, et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med. 2007 Jan 22;204(1):171–80. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Naito M, Hasegawa G, Takahashi K. Development, differentiation, and maturation of Kupffer cells. Microsc Res Tech. 1997 Nov 15;39(4):350–64. doi: 10.1002/(SICI)1097-0029(19971115)39:4<350::AID-JEMT5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 88.Ghigo C, Mondor I, Jorquera A, Nowak J, Wienert S, Zahner SP, et al. Multicolor fate mapping of Langerhans cell homeostasis. J Exp Med. 2013 Aug 26;210(9):1657–64. doi: 10.1084/jem.20130403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bouwens L, Baekeland M, De Zanger R, Wisse E. Quantitation, tissue distribution and proliferation kinetics of Kupffer cells in normal rat liver. Hepatol Baltim Md. 1986 Aug;6(4):718–22. doi: 10.1002/hep.1840060430. [DOI] [PubMed] [Google Scholar]
- 90.Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch U-K, Mack M, et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10(12):1544–53. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 91.Goldmann T, Wieghofer P, Müller PF, Wolf Y, Varol D, Yona S, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci. 2013 Nov;16(11):1618–26. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- 92.Bruttger J, Karram K, Wörtge S, Regen T, Marini F, Hoppmann N, et al. Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. [2015 Jul 21];Immunity [Internet] doi: 10.1016/j.immuni.2015.06.012. Available from: http://www.sciencedirect.com/science/article/pii/S1074761315002563. [DOI] [PubMed]
- 93.Blériot C, Dupuis T, Jouvion G, Eberl G, Disson O, Lecuit M. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity. 2015 Jan 20;42(1):145–58. doi: 10.1016/j.immuni.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 94.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, et al. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990 May 31;345(6274):442–4. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 95.Marks SC, Lane PW. Osteopetrosis, a new recessive skeletal mutation on chromosome 12 of the mouse. J Hered. 1976 Feb;67(1):11–8. doi: 10.1093/oxfordjournals.jhered.a108657. [DOI] [PubMed] [Google Scholar]
- 96.Marks SC, Seifert MF, McGuire JL. Congenitally osteopetrotic (oplop) mice are not cured by transplants of spleen or bone marrow cells from normal littermates. Metab Bone Dis Relat Res. 1984;5(4):183–6. doi: 10.1016/0221-8747(84)90027-4. [DOI] [PubMed] [Google Scholar]
- 97.Sherr CJ, Rettenmier CW, Sacca R, Roussel MF, Look AT, Stanley ER. The c-fms proto-oncogene product is related to the receptor for the mononuclear phagocyte growth factor, CSF 1. Cell. 1985 Jul;41(3):665–76. doi: 10.1016/s0092-8674(85)80047-7. [DOI] [PubMed] [Google Scholar]
- 98.Cecchini MG, Dominguez MG, Mocci S, Wetterwald A, Felix R, Fleisch H, et al. Role of colony stimulating factor-1 in the establishment and regulation of tissue macrophages during postnatal development of the mouse. Dev Camb Engl. 1994 Jun;120(6):1357–72. doi: 10.1242/dev.120.6.1357. [DOI] [PubMed] [Google Scholar]
- 99.Felix R, Cecchini MG, Hofstetter W, Elford PR, Stutzer A, Fleisch H. Impairment of macrophage colony-stimulating factor production and lack of resident bone marrow macrophages in the osteopetrotic op/op mouse. J Bone Miner Res Off J Am Soc Bone Miner Res. 1990 Jul;5(7):781–9. doi: 10.1002/jbmr.5650050716. [DOI] [PubMed] [Google Scholar]
- 100.Naito M, Hayashi S, Yoshida H, Nishikawa S, Shultz LD, Takahashi K. Abnormal differentiation of tissue macrophage populations in “osteopetrosis” (op) mice defective in the production of macrophage colony-stimulating factor. Am J Pathol. 1991 Sep;139(3):657–67. [PMC free article] [PubMed] [Google Scholar]
- 101.Dai X-M, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002 Jan 1;99(1):111–20. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 102.Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wainwright BJ, et al. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003 Feb 1;101(3):1155–63. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- 103.Lin H, Lee E, Hestir K, Leo C, Huang M, Bosch E, et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008 May 9;320(5877):807–11. doi: 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- 104.Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity. 2012 Dec 14;37(6):1050–60. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13(8):753–60. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma X, Lin WY, Chen Y, Stawicki S, Mukhyala K, Wu Y, et al. Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R. Structure. 2012 Apr 4;20(4):676–87. doi: 10.1016/j.str.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 107.Chihara T, Suzu S, Hassan R, Chutiwitoonchai N, Hiyoshi M, Motoyoshi K, et al. IL-34 and MCSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ. 2010 Dec;17(12):1917–27. doi: 10.1038/cdd.2010.60. [DOI] [PubMed] [Google Scholar]
- 108.Strobl H, Riedl E, Scheinecker C, Bello-Fernandez C, Pickl WF, Rappersberger K, et al. TGF-beta 1 promotes in vitro development of dendritic cells from CD34+ hemopoietic progenitors. J Immunol. 1996 Aug 15;157(4):1499–507. [PubMed] [Google Scholar]
- 109.Borkowski TA, Letterio JJ, Farr AG, Udey MC. A Role for Endogenous Transforming Growth Factor β1 in Langerhans Cell Biology: The Skin of Transforming Growth Factor β1 Null Mice Is Devoid of Epidermal Langerhans Cells. J Exp Med. 1996 Dec 1;184(6):2417–22. doi: 10.1084/jem.184.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kel JM, Girard-Madoux MJH, Reizis B, Clausen BE. TGF-β Is Required To Maintain the Pool of Immature Langerhans Cells in the Epidermis. J Immunol. 2010 Sep 15;185(6):3248–55. doi: 10.4049/jimmunol.1000981. [DOI] [PubMed] [Google Scholar]
- 111.Khosravi A, Yáñez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014 Mar 12;15(3):374–81. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015 Jul;18(7):965–77. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006 Jan 6;311(5757):83–7. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 114.Auffray C, Fogg DK, Narni-Mancinelli E, Senechal B, Trouillet C, Saederup N, et al. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J Exp Med. 2009 Mar 16;206(3):595–606. doi: 10.1084/jem.20081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009 Apr 17;324(5925):392–7. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hettinger J, Richards DM, Hansson J, Barra MM, Joschko A-C, Krijgsveld J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013 Aug;14(8):821–30. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- 117.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of Monocytes, Macrophages, and Dendritic Cells. Science. 2010 Feb 5;327(5966):656–61. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990 Apr 6;61(1):113–24. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 119.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994 Sep 9;265(5178):1573–7. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 120.Lichanska AM, Browne CM, Henkel GW, Murphy KM, Ostrowski MC, McKercher SR, et al. Differentiation of the Mononuclear Phagocyte System During Mouse Embryogenesis: The Role of Transcription Factor PU.1. Blood. 1999 Jul 1;94(1):127–38. [PubMed] [Google Scholar]
- 121.DeKoter RP, Singh H. Regulation of B Lymphocyte and Macrophage Development by Graded Expression of PU.1. Science. 2000 May 26;288(5470):1439–41. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 122.Jin H, Li L, Xu J, Zhen F, Zhu L, Liu PP, et al. Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1 expression. Blood. 2012 May 31;119(22):5239–49. doi: 10.1182/blood-2011-12-398362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Celada A, Borràs FE, Soler C, Lloberas J, Klemsz M, Beveren C van, et al. The transcription factor PU.1 is involved in macrophage proliferation. J Exp Med. 1996 Jul 1;184(1):61–9. doi: 10.1084/jem.184.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010 May 28;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010 Mar 26;32(3):317–28. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 126.Bemark M, Mårtensson A, Liberg D, Leanderson T. Spi-C, a novel Ets protein that is temporally regulated during B lymphocyte development. J Biol Chem. 1999 Apr 9;274(15):10259–67. doi: 10.1074/jbc.274.15.10259. [DOI] [PubMed] [Google Scholar]
- 127.Haldar M, Kohyama M, So AY-L, Kc W, Wu X, Briseño CG, et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 2014 Mar 13;156(6):1223–34. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cain DW, O'Koren EG, Kan MJ, Womble M, Sempowski GD, Hopper K, et al. Identification of a tissue-specific, C/EBPβ-dependent pathway of differentiation for murine peritoneal macrophages. J Immunol Baltim Md 1950. 2013 Nov 1;191(9):4665–75. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Melnicoff MJ, Horan PK, Breslin EW, Morahan PS. Maintenance of peritoneal macrophages in the steady state. J Leukoc Biol. 1988 Nov;44(5):367–75. doi: 10.1002/jlb.44.5.367. [DOI] [PubMed] [Google Scholar]
- 130.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014 May 8;157(4):832–44. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rosas M, Davies LC, Giles PJ, Liao C-T, Kharfan B, Stone TC, et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014 May 9;344(6184):645–8. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996 Jul 10;177(1):309–22. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 133.Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol. 2011 Jan;11(1):34–46. doi: 10.1038/nri2901. [DOI] [PubMed] [Google Scholar]
- 134.Kroese FG, Butcher EC, Stall AM, Lalor PA, Adams S, Herzenberg LA. Many of the IgA producing plasma cells in murine gut are derived from self-replenishing precursors in the peritoneal cavity. Int Immunol. 1989;1(1):75–84. doi: 10.1093/intimm/1.1.75. [DOI] [PubMed] [Google Scholar]
- 135.Kroese FG, Ammerlaan WA, Kantor AB. Evidence that intestinal IgA plasma cells in mu, kappa transgenic mice are derived from B-1 (Ly-1 B) cells. Int Immunol. 1993 Oct;5(10):1317–27. doi: 10.1093/intimm/5.10.1317. [DOI] [PubMed] [Google Scholar]