Abstract
As the cost of next-generation sequencing has decreased, library preparation costs have become a more significant proportion of the total cost, especially for high-throughput applications such as single-cell RNA profiling. Here, we have applied novel technologies to scale down reaction volumes for library preparation. Our system consisted of in vitro differentiated human embryonic stem cells representing two stages of pancreatic differentiation, for which we prepared multiple biological and technical replicates. We used the Fluidigm (San Francisco, CA) C1 single-cell Autoprep System for single-cell complementary DNA (cDNA) generation and an enzyme-based tagmentation system (Nextera XT; Illumina, San Diego, CA) with a nanoliter liquid handler (mosquito HTS; TTP Labtech, Royston, UK) for library preparation, reducing the reaction volume down to 2 µL and using as little as 20 pg of input cDNA. The resulting sequencing data were bioinformatically analyzed and correlated among the different library reaction volumes. Our results showed that decreasing the reaction volume did not interfere with the quality or the reproducibility of the sequencing data, and the transcriptional data from the scaled-down libraries allowed us to distinguish between single cells. Thus, we have developed a process to enable efficient and cost-effective high-throughput single-cell transcriptome sequencing.
Keywords: miniaturization, single cell, library, scale-down
Introduction
Novel technologies for single-cell next-generation sequencing (NGS) offer new opportunities for understanding variations among the genomes, epigenomes, and transcriptomes of seemingly identical cells.1–6 Although early studies on gene expression on the single-cell level focused on small sets of selected transcripts, single-cell RNA sequencing offers unbiased exploration of the variability and heterogeneity among the transcriptomes of different cells. Individual mammalian cells are estimated to contain an average of 1 million messenger RNA (mRNA) molecules, adding up to approximately 10 pg of total RNA.7 The relative proportions of different transcripts are highly variable depending on the cell type and environment.8 To study the transcriptional differences between cell types, the influence of the cellular environment on transcriptional profiles, and the potential subpopulations present within cell types, there is a need to develop methods that can be feasibly used to analyze large numbers of single-cell transcriptomes.
With the rapidly decreasing cost of sequencing, library preparation costs have become an increasingly significant fraction of the total cost of NGS, especially in applications that require analysis of large numbers of libraries, such as single-cell RNA sequencing. The ability to decrease reaction volumes without compromising library quality would result in lower reagent costs. Historically, bottlenecks to this miniaturization process include the inability of standard liquid handlers to accurately dispense volumes under 2 µL and the relatively large volumes needed for the physical nucleic acid shearing steps used in many RNA-seq protocols. We have overcome these barriers by using the mosquito HTS liquid handler (TTP Labtech, Royston, UK) (SF1), which accurately dispenses volumes between 25 nL and 1.2 µL using true-positive displacement technology in conjunction with an enzyme-based fragmentation method (Nextera XT; Illumina, San Diego, CA), which can be performed in extremely low-reaction volumes.
Here we present a high-throughput workflow in which single-cell RNA-seq libraries are prepared using the C1 single-cell Auto Prep System (Fluidigm) combined with the mosquito liquid handler to simultaneously decrease the reaction volume and increase the number of reactions. This system allows us to significantly decrease library preparation costs and increase throughput. To establish the quality of the libraries, we applied this system to the analysis of human embryonic stem cells differentiated in vitro to two early stages of pancreatic differentiation. We analyzed the resulting single-cell RNA-seq data to determine the reproducibility of this system and its ability to distinguish not only between cells at different stages of differentiation but also between individual cells within each stage.
Materials and Methods
Cell Culture and Differentiation
All cell cultures were maintained in vitronectin-coated plates at 37 °C and 5% CO2. WA09 human embryonic stem cells (hESCs) were maintained in undifferentiated conditions in E8 media (GIBCO, Carlsbad, CA). To obtain definitive endoderm cells (stage 1), we initially induced primitive streak formation by treatment of cells for 24 h with 100 ng/mL ActivinA (Stemgent, Cambridge, MA), 2 µM CHIR99021 GSK-3 inhibitor (TOCRIS, Bristol, UK), 10 ng/mL BMP4 (R&D Systems, Tustin, CA), 10 µM LY294002 (Calbiochem, San Diego, CA), and 20 ng/mL FGF2 (Millipore, San Diego, CA). After primitive streak commitment, cells were treated for 2 days with 100 ng/mL ActivinA, 10 µM LY294002, 20 ng/mL FGF2, and 100 nM dorsomorphin (Axon Medchem, Reston, VA). For stage 1 differentiation, cells were grown in CDM-PVA media, which contains 250 mL DMEM-F12 (GIBCO, Carlsbad, CA) and 250 mL IMDM (GIBCO) mixed 1:1, 1% Glutamax, 1% concentrated lipids (GIBCO), 450 mM Monohioglycerol (Sigma, St. Louis, MO), 1% insulin-transferrin-selenium ITS supplement (GIBCO), and 1 mg/mL polyvinylalcohol (Sigma). To obtain posterior foregut (stage 2), cells were cultured for 3 days in advanced Dulbecco’s modified Eagle’s medium (Advanced-DMEM) supplemented with 10 µM SB431542 (Selleckchem, Houston, TX), 2 µM retinoic acid (Sigma), 100 nM dorsomorphin (Axon Medchem), and 50 ng/mL FGF10 (Peprotech, Rocky Hill, NJ).
Cells were collected and dissociated to a single-cell suspension using Cell Dissociation Reagent (GIBCO) and resuspended in culture media. An average cell concentration of 250,000 cells/mL was loaded into Fluidigm C1 Single-Cell Auto Prep Arrays for mRNA-Seq, using medium and small chips for the stage 1 and stage 2 cells, respectively.
Single-Cell Library Generation
The C1 Single-Cell Auto Prep System (Fluidigm) was used to perform SMARTer (Clontech, Mountain View, CA) cDNA generation and amplification. Prior to loading the single-cell suspension onto the C1 chips, we stained the cells with the LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells (Life Technologies, Carlsbad, CA). After loading, we visualized each microchamber in the C1 chip to identify those chambers that contained a single live (Calcein+/Ethidium homodimer−) cell. We selected two cells from stage 1 (cell A and cell B) and two cells from stage 2 (cell C and cell D), with an average cDNA concentration of 0.4 ng/µL in an approximate output volume of 15 µL (ST1). DNA concentration was quantified using the Qubit dsDNA High Sensitivity Kit (Life Technologies), according to the manufacturer’s instructions. The resulting cDNAs were diluted to a final concentration of 0.1 ng/µL and then converted to Illumina sequencing libraries using the Nextera XT (Illumina) kit using protocols specifically designed for the mosquito HTS (TTP Labtech) (ST2). We generated libraries in three different final reaction volumes (2 µL, 4 µL, and 8 µL) using Axygen (Palo Alto, CA) 384 plates. Dual indexing was performed using the Nextera XT Index Kit v2 Set A (96 Indexes) to enable multiplexing of libraries.
PCR
PCR reactions were performed using a CFX384 Real Time System C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA) in 384 PCR hard-well microplates (Axygen). PCR cycling conditions were as follows: 72 °C for 3 min; 95 °C for 30 s; 12 cycles of 95 °C for 10 s, 55 °C for 30 s, and 72 °C for 60 s; 72 °C for 5 min; and a hold step at 10 °C.
Cleanup
Samples were pooled using the mosquito HTS, taking 500 nL of each single-library prep from the destination plate. A total of 48 independent single-cell libraries were pooled and subjected to bead cleanup using AMPure XP beads (Beckman Coulter, Brea, CA). Specifically, 24 µL total of pooled PCR-amplified libraries was mixed with 21.6 µL AMPure XP beads (a 0.9x DNA/bead ratio), according to the Nextera XT DNA Library Prep Kit (Illumina) protocol. After two washes with 80% ethanol, the beads were air-dried and then resuspended in 20 µL TE buffer. For the unpooled single-cell libraries shown in SF2, 2 µL of each single-cell library was diluted with 8 µL TE buffer and then mixed with 9 µL AMPure XP beads. After two washes with 80% ethanol, dried beads were carefully resuspended in a final volume of 6 µL TE buffer and analyzed with BioAnalyzer in High-Sensitivity DNA Chips (Agilent Technologies, Santa Clara, CA) (see SF2 for quality controls in pooled and single-cell libraries).
Sequencing
The 48 pooled libraries were sequenced on one lane of an Illumina HiSeq 2500 at an average total read depth of 5.6 million reads per sample (ST3). Average read length was 100 bps, paired-end mode, high-throughput run, and dual indexing with v3 chemistry.
Data Preprocessing
The raw reads were trimmed using cutadapt (1.8.1)9 and mapped onto the human genome (version Hg19) using STAR (version 2.3.0).10 Normalization and differential expression was performed using DESeq.11 To avoid artifacts that could result from differences in the depth of sequencing, 1.5 million uniquely mapped reads were randomly selected from each library for further analysis. The data were then filtered by removing the transcripts that had zero counts in all samples.
Data Analysis
Pearson correlation, hierarchical clustering, principal component analysis (PCA), Venn diagrams, coefficient of variation analysis, and CLICK12 were used to analyze the processed data. Calculations, dendrograms, and PCA plots were performed in R (3.2.0).13
Results
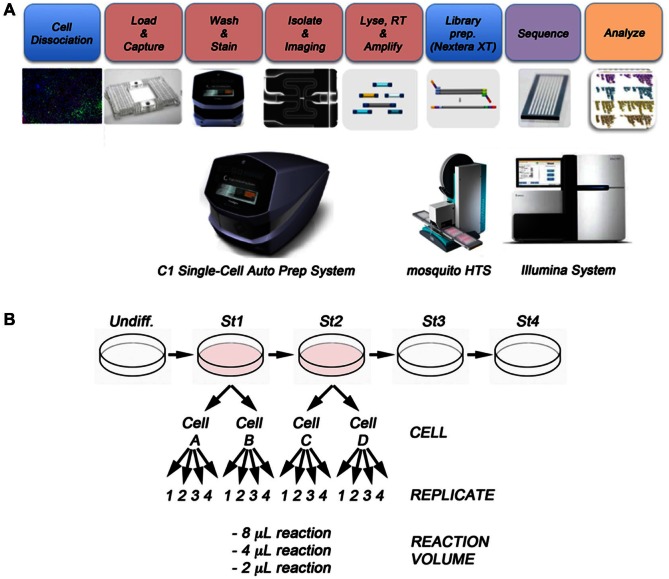
In this work, we combined three state-of-the-art technologies to develop a novel workflow for automated single-cell RNA-seq library generation at extremely low reaction volumes. In this workflow, we used the Fluidigm C1 Single-Cell Auto Prep System to generate cDNA from single cells, followed by library preparation using the Nextera XT DNA Library Prep Kit (Illumina) on the mosquito HTS liquid handler (TTP) ( Fig. 1A ). Given the efficient conversion of mRNA to cDNA afforded by the Fluidigm C1 System, the efficiency of the Nextera XT kit, and the low-volume liquid handling capabilities of the mosquito HTS, we were able to perform multiple technical replicates at three different reaction volumes from each of four individual cells ( Fig. 1B ).
Figure 1.
Experimental scheme. (A) After differentiation, the cell cultures were dissociated to single-cell suspensions and loaded onto a Fluidigm C1 Single-Cell Auto Prep Array for mRNA-Seq. On the arrays, the cells were lysed, and reverse transcription of the mRNA and PCR amplification of the cDNA were performed using the C1 Single-Cell Auto Prep System (Fluidigm). Libraries were then prepared using the Nextera XT DNA Library Prep Kit and mosquito HTS liquid handler (TTP). For the final PCR reactions, we used a Bio-Rad 384 Thermal Cycler. Libraries were pooled and sequenced on an Illumina HiSeq 2500. (B) WA09 human embryonic stem cells were differentiated in vitro to the pancreatic lineage. Cells from stages 1 and 2 were collected and analyzed using the procedures outlined in A. Two independent cells from stage 1 (cell A and cell B) and two cells from stage 2 (cell C and cell D) were selected and yielded with similar cDNA concentrations (mean [SD] concentration = 0.38 [0.04] ng/µL). For library preparation, we tested 2-µL, 4-µL, and 8-µL final volume reactions, with four replicates per reaction volume.
In our experimental design, we incorporated two levels of biological replication (two individual cells from each of two stages of in vitro pancreatic differentiation), as well as extensive technical replication ( Fig. 1B ). For each cell, we used the cDNA generated by the Fluidigm C1 System to prepare libraries in quadruplicate at three different final reaction volumes (8 µL, 4 µL, and 2 µL), using the Nextera XT kit on the mosquito HTS liquid handler. We sequenced each sample to an average depth of 5.6 million paired-end reads per sample (ST3), as it has been reported that single-cell expression estimates stabilized at relatively low read depths.2
Technical Reproducibility in Library Construction
For each library, we randomly selected 1.5 million uniquely mapped reads to avoid artifacts due to variations in sequencing depth. After DEseq normalization of the data set, we calculated the Pearson correlation coefficients between each set of replicates for each cell at each reaction volume ( Table 1 , ST4). In this analysis, we found that nearly all of the mean correlation coefficients were >0.936 ( Table 1 ) for each cell at each reaction volume, both with and without down-sampling, indicating that the reproducibility was extremely high. We also noted that the correlation coefficients between different reaction volumes for a given cell were nearly as high ( Table 1 ). We noticed two exceptions to these extremely high correlations. First, the 2-µL reactions for cell D had a mean correlation coefficient of 0.943 for all reads and 0.940 for the down-sampled reads ( Table 1 ). We believe these low correlation coefficients can be attributed to a low total read count of replicate 4 for the 2-µL reactions for cell D (ST3) and a lower correlation coefficient of 0.900 to 0.903 compared with the other cell D libraries (ST4). In routine RNA sequencing analysis pipelines, such a library would be discarded during initial quality control assessment of the RNA-seq data. However, in our study, we retained this sample to demonstrate the performance of suboptimal libraries. Second, we found that the correlation coefficients for the cell C libraries were between 0.936 and 0.943 after down-sampling ( Table 1 ). In this case, there were no outliers (ST3). However, we noted that the cDNA concentration for cell C was lower than that for the other cells (ST1), suggesting the possibility that the higher variability among the cell C libraries may have resulted from a less efficient cDNA generation step.
Table 1.
Average Pearson Correlation Values for DESeq Normalized Data from Technical Replicates within and between Reaction Volumes, with and without Down-Sampling by Randomly Selecting 1.5 Million Uniquely Mapped Reads (UMRs) from Each Library.
| 2 µL |
4 µL |
8 µL |
2 vs 4 µL |
2 vs 8 µL |
4 vs 8 µL |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Cell A | 0.973 | 0.016 | 0.981 | 0.012 | 0.982 | 0.011 | 0.969 | 0.002 | 0.969 | 0.002 | 0.975 | 0.001 | |
| All data | Cell B | 0.980 | 0.012 | 0.985 | 0.009 | 0.985 | 0.009 | 0.976 | 0.001 | 0.976 | 0.001 | 0.980 | 0.001 |
| Cell C | 0.939 | 0.036 | 0.947 | 0.032 | 0.951 | 0.030 | 0.922 | 0.002 | 0.924 | 0.003 | 0.931 | 0.004 | |
| Cell D | 0.943 | 0.042 | 0.971 | 0.018 | 0.973 | 0.016 | 0.941 | 0.025 | 0.942 | 0.026 | 0.962 | 0.002 | |
| Cell A | 0.959 | 0.025 | 0.951 | 0.029 | 0.956 | 0.026 | 0.939 | 0.004 | 0.943 | 0.003 | 0.939 | 0.004 | |
| 1.5 UMR | Cell B | 0.958 | 0.025 | 0.953 | 0.028 | 0.957 | 0.026 | 0.941 | 0.003 | 0.943 | 0.003 | 0.940 | 0.003 |
| Cell C | 0.943 | 0.034 | 0.943 | 0.034 | 0.936 | 0.038 | 0.923 | 0.004 | 0.918 | 0.006 | 0.918 | 0.006 | |
| Cell D | 0.940 | 0.040 | 0.944 | 0.034 | 0.944 | 0.034 | 0.923 | 0.013 | 0.922 | 0.014 | 0.926 | 0.004 | |
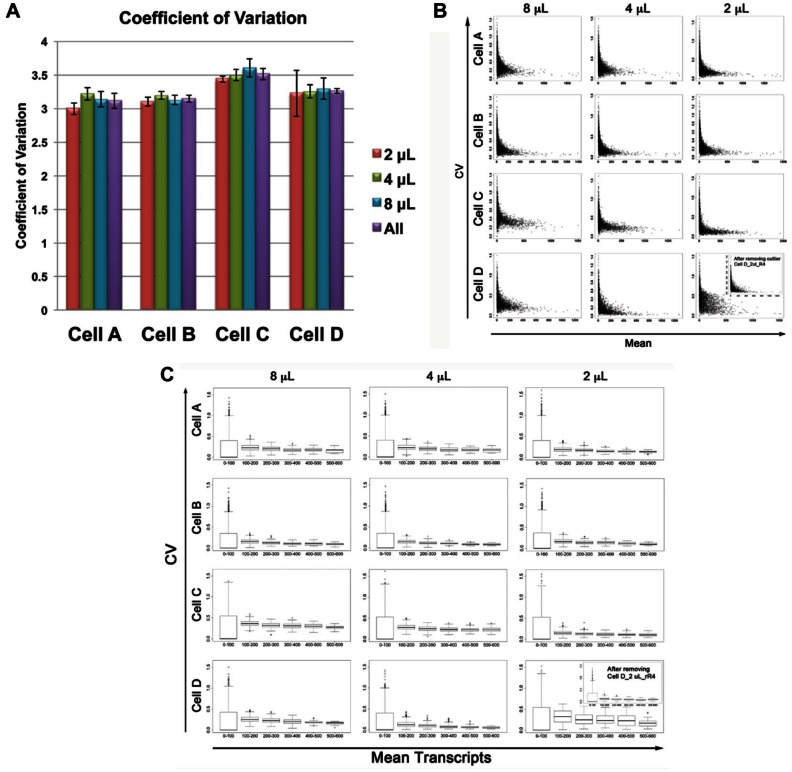
To further determine whether there was an effect of reaction volume on the reproducibility of the library preparation process, we calculated the coefficient of variation (CV) for each library, using the number of counts per cell. The CVs were very similar, with the only statistically significant difference in CV being found between the 2-µL and 4-µL reaction volumes for cell A, and in this comparison, the lower reaction volume was the one with the lower CV ( Fig. 2A , ST5). We note that the standard error of the mean for the 2-µL reactions for cell D is higher than for the other categories. We believe this is again is due to the low sequencing depth of the cell D 2-µL replicate 4 library.
Figure 2.
Coefficients of variation (CVs) for each reaction volume for each cell. (A) Table of mean CVs calculated from DESeq normalized data. For each cell, t tests were performed to compare the CVs between each pair of reaction volumes. Significant differences (p < 0.05) are indicated by an asterisk. (B) Scatterplots of CV versus mean transcript counts. (C) Box-and-whisker plots of CV versus mean transcript counts, dividing the data into six windows: 0 to 100 counts, 101 to 200 counts, 201 to 300 counts, 301 to 400 counts, 401 to 500 counts, and >500 counts.
To examine whether the CV for low-expressed transcripts compared with highly expressed transcripts is different for the different reaction volumes, we plotted the CV against the mean transcript expression level ( Fig. 2B , C ). These results indicate that the reproducibility among technical replicates that pass basic quality control metrics is very high and not influenced by the reaction volume.
Clustering to Evaluate Technical Reproducibility in the Context of Biological Variance
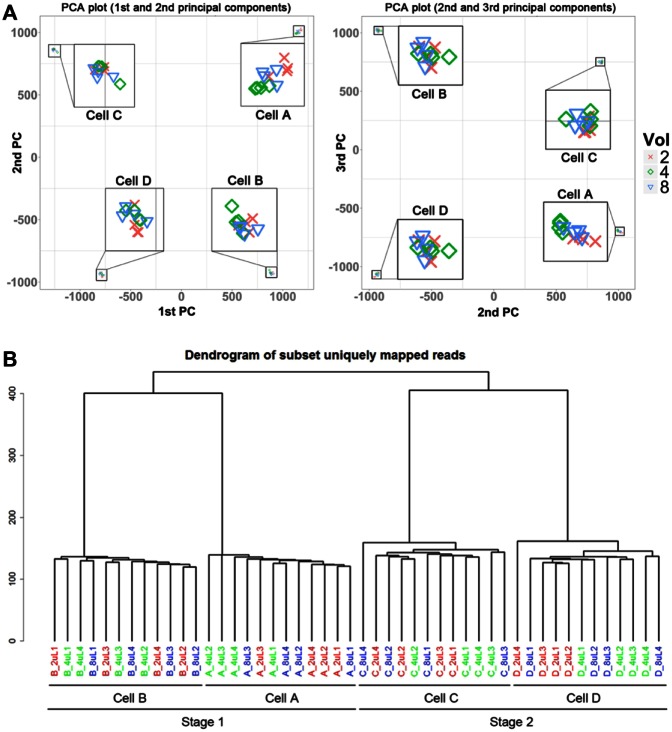
We explored the relationships among the libraries using two unsupervised clustering methods: 2D PCA ( Fig. 3A ) and hierarchical clustering ( Fig. 3B ). Using both methods, we could easily distinguish between the libraries from each of the four cells, and as expected, the stage 1 cells (A and B) separated from the stage 2 cells (C and D) along the first principal component ( Fig. 3A ) and at the first branchpoint of the dendrogram ( Fig. 3B ; plate localization of each independent library is shown in SF3). Importantly, the libraries did not cluster according to reaction volume, even within a single cell.
Figure 3.
Clustering analysis. (A) Principal component analysis (PCA) for libraries. The data for the first and second principal components (PCs) are shown on the left, and the second and third PCs are shown on the right. (B) Hierarchical clustering of all libraries. Euclidian distance with complete linkage was used to construct the dendogram. Data were normalized using DESeq. Red denotes 2-µL reactions, green denotes 4-µL reactions, and blue denotes 8-µL reactions.
Complexity and Sensitivity of Libraries from Different Reaction Volumes
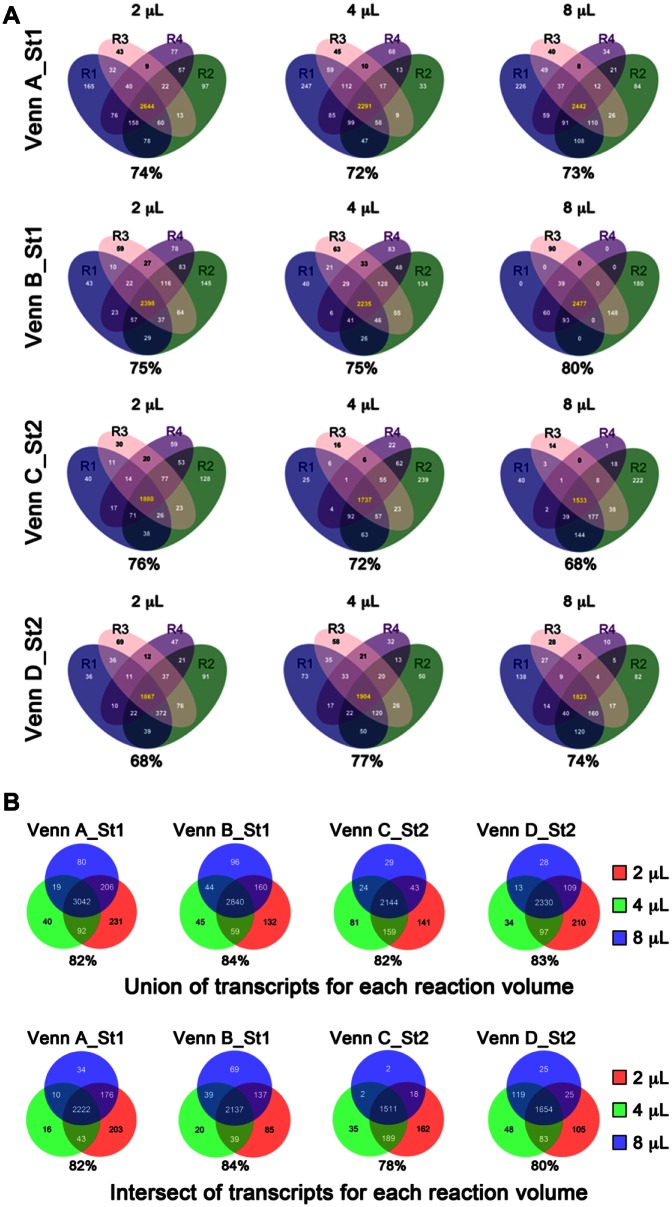
A potential concern with decreasing the reaction volume for library sample preparation is that we could introduce sampling error, which could result in decreased detection of low-expressed transcripts, thus decreasing the complexity and sensitivity of the libraries. If there was higher sampling error in the lower reaction volumes, we would expect that the intersection in detected transcripts for the four replicates of the 2-µL libraries would be lower than for the 4-µL or 8-µL libraries. We therefore determined the overlap in detected (read count >10) transcripts among the four replicates for each reaction volume for each cell ( Fig. 4A , ST6, set 4–6 comparisons) and found that there was a significant difference in the percentage of overlapping transcripts only between the 2-µL and 8-µL reaction volumes for cell C. In inspecting the percent overlaps for this comparison, we found that the percent overlap for the 2-µL reactions was actually better (higher) than that for the 8-µL reactions. We also examined the percentage of overlapping transcripts compared across reaction volumes for each cell. We did this in three ways: taking the union of overlapping transcripts among the four replicates for each reaction volume for each cell and then inspecting the overlaps between the 2-µL, 4-µL, and 8-µL libraries ( Fig. 4B , top); taking the intersect of overlapping transcripts among the four replicates for each reaction volume for each cell and then inspecting the overlaps between the 2-µL, 4-µL, and 8-µL libraries ( Fig. 4B , bottom); and determining the percent overlap for all pairs of libraries within each cell across reaction volumes (ST6, set 1–3 comparisons). In some cases, the overlaps between the 2-µL libraries and the higher reaction volume libraries were slightly lower than the other comparisons, but overall, the overlaps were again very similar.
Figure 4.
Venn diagrams. (A) Venn diagrams displaying the overlap in detected transcripts (>10 counts) for the four replicate libraries for each reaction volume in each cell. The percentage of transcripts in the common region of intersection (i.e., R1 ∩ R2 ∩ R3 ∩ R4) compared to all transcripts (i.e., R1 U R2 U R3 U R4) is shown below each Venn diagram. (B) Venn diagrams displaying the overlap in detected transcripts among the different reaction volumes, using the union (top) or intersect (bottom) of detectable genes in the four replicates. The percentage of transcripts in the common region of intersection (i.e., 2 µL ∩ 4 µL ∩ 8 µL) compared with all transcripts (i.e., 2 µL U 4 µL U 8 µL) is shown below each Venn diagram.
We also compared the distribution of transcript counts among libraries by tallying the number of transcripts in the following bins: 1 to 9 counts (mean [SD]: 1661.7 [87.0]), followed by 10 to 99 counts (mean [SD]: 1623.3 [367.5]) and 100 to 999 counts (mean [SD]: 835.3 [170.6]). Low detectable transcripts were observed in >1000 counts (mean [SD]: 8.7 [6.8]) (ST7). The numbers of detected transcripts present in each bin were quite similar for each cell (SF4). The only clear difference in transcript count distribution was seen in the 10- to 99-count bin, for which the stage 1 cells contained a markedly higher number of transcripts than the stage 2 cells (SF4). Thus, we saw cell type–associated, but not reaction volume–associated, differences in transcript count distribution.
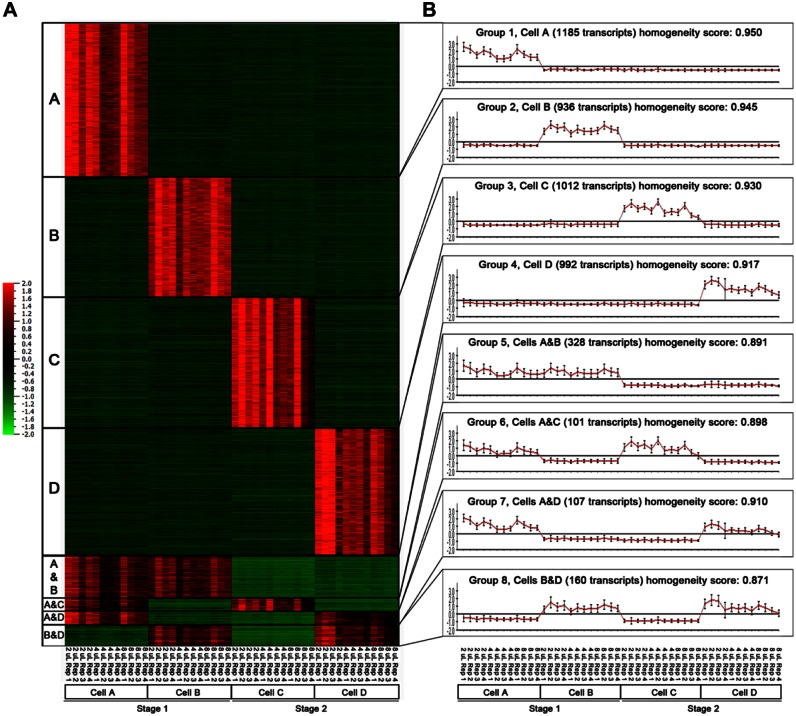
Differential Expression Analysis
To explore our ability to identify differences in the gene expression profiles of single cells, we used DESeq10 to identify transcripts that were differentially expressed between each pair of cells and then applied CLICK analysis11 to identify groups of coexpressed transcripts ( Fig. 5 ). As shown in the heatmap ( Fig. 5A ) and average expression graphs ( Fig. 5B ), groups 1 to 4 consist of transcripts that are specifically expressed in each of the four single cells, while groups 5 to 8 contain transcripts that are expressed in different combinations of two of four cells. We note that there are no systematic differences in gene expression patterns according to reaction volume (ST3).
Figure 5.
Global single-cell gene expression analysis by RNA-seq. (A) Heatmap displaying differentially expressed genes (4821 genes, adjusted p < 0.00001, absolute log2 fold-change >4, maximum count >20) between cells A, B, C, and D, clustered using CLICK. (B) Graphs displaying the mean expression values for the transcripts in each CLICK cluster in each library. All replicate libraries for each cell were included in these analyses.
Discussion
For many research and clinical applications, it is desirable to perform NGS on large numbers of samples. In particular, taking full advantage of single-cell transcriptome analysis would require analysis of hundreds, if not thousands, of cells per experiment to detect and characterize rare subpopulations of cells. With the decreasing cost of sequencing, library preparation is becoming an increasingly significant factor in the total cost of the final experiment. In general, the protocols provided by manufacturers of commercially available library preparation kits produce more material than is needed for the subsequent sequencing procedure, resulting in significant reagent waste. This is largely due to the fact that manual pipetting and commonly used liquid handling systems do not reliably dispense low volumes. Therefore, sophisticated liquid handling systems specifically designed to accurately dispense sub-microliter volumes of reagents would both increase throughput and decrease costs, thus enabling experiments involving larger numbers of samples than are currently feasible. Here, we report the sequential application of two such systems to single-cell transcriptome analysis. First, we used the Fluidigm C1 Single-Cell Auto Prep System, which uses specifically designed microfluidic chips to capture single cells in individual microchambers and carry out the reverse transcription and second-strand cDNA synthesis reactions in volumes of 4.5 to 135 nL using the Clontech SMARTer Universal Low Input RNA Kit. We then used the mosquito HTS liquid handler (TTP Labtech) to complete the NGS library preparation process using the Nextera XT Library Preparation Kit (Illumina). The mosquito HTS uses positive displacement to accurately dispense 25-nL to 1.2-µL volumes, enabling us to scale down the Nextera XT reaction volumes to 2 µL, using as little as 20 pg of input cDNA (compared with a reaction volume of 10 µL and cDNA input amount of 125–375 pg recommended by Fluidigm). This translates to a reduction in input cDNA of 5-fold and a cost savings of over 4-fold compared with the protocol recommended by the manufacturer and results in a library preparation cost of less than $1.50 per single cell. There are several publications in which tagmentation technology has been shown to produce high-quality genomic or cDNA libraries from very low-input material.13–18 Some of those technology descriptions start with as little as 20 pg of prokaryotic or mouse genomic DNA.15 Novel technologies for mammalian single-cell RNA-seq using the Fluidigm C1 Single-Cell Auto Prep System combined with tagmentation library preparation use as little as 250 pg of input amplified DNA and reaction volumes of 12.5 µL.2 In all these descriptions, the reaction volume exceeds by far the largest volume described in this article and implies manually generated libraries. Lamble et al.19 reported generation of sequencing libraries from 12.5 ng genomic DNA using the Nextera XT Kit in a reaction volume of 6.25 µL. The resulting libraries had an average insert size of 250 to 300 bp, similar to our libraries (SF2), with excellent accuracy and genomic coverage. However, since this previous study included only genomic DNA sequencing of bacterial species, it did not address many issues pertinent to mammalian transcriptome sequencing, including library complexity and reproducibility. Descriptions by Shapland et al.20 demonstrate Nextera libraries in 500 nL for synthesized genes using an Echo acoustic dispensing system, but our work is the first study to report the combination of extremely low input (20 pg cDNA) with low reaction volume (2 µL) for mammalian single-cell transcriptome sequencing using multidispensing liquid handler systems.
This study was designed to determine not only whether scaling down reaction volumes would affect the reproducibility of transcriptome sequencing results but also whether there would be effects on library complexity or the ability to distinguish between different cell types and different individual cells ( Fig. 5 ). We used the Fluidigm C1 system to generate cDNA from four single cells representing two stages of in vitro pancreatic differentiation of human embryonic stem cells, with two cells from the definitive endoderm stage (stage 1) and two cells from the posterior foregut stage (stage 2). The amount of cDNA generated from each single cell was sufficient to perform quadruplicate experiments at three reaction volumes: 2 µL, 4 µL, and 8 µL. Correlation analysis showed extremely high reproducibility among libraries generated from the same cDNA sample, both among libraries produced in a reaction volume of 2 µL and between libraries produced in reaction volumes of 2 µL, 4 µL, and 8 µL. We noted that there was slightly higher variability among replicate libraries at all reaction volumes for the stage 2 cells and postulate that this might be due to a more diverse population of RNAs present at this later stage of differentiation, which would make the data more sensitive to library complexity. We therefore evaluated library saturation by looking at the percentage of duplicated reads, as well as library complexity by inspecting the representation of RNAs at different levels of expression and the overlap in the number of measurable transcripts among replicate libraries. The results of these analyses showed no reaction volume–associated differences in transcript count distribution but did reveal a higher number of transcripts in the 10- to 99-count in the stage 1 cells compared with the stage 2 cells (SF4).
The inclusion of two cells from each of two stages of differentiation in this study allowed us to assess the technical variability of the transcriptome sequencing process performed in different reaction volumes in the context of the biological variability between single cells both within and between cell populations. From unsupervised clustering and principal component analyses, it is clear that the technical variability was far lower than the variability between the single cells, even single cells from the same stage of differentiation, resulting in clear separation among all four cells, and the ability to clearly identify groups of coexpressed genes that displayed cell-specific expression.
Although not the focus of this article, we have applied this miniaturized process to generation of single-cell libraries on a high-throughput level, generating 384 single-cell libraries in one experiment, including automation of all the upstream and downstream processes. Upstream processes include transfer of the cDNA from the C1 Fluidigm chip to a 384-well plate and quantification and normalization of each cDNA sample, while downstream processes include bead purification, quantification, and normalization of each library. We note that a second bead purification of the pooled libraries further reduces the amount of adaptor dimers (SF5).
Taken together, our results indicate that the application of a nanoliter-scale liquid handling system enables automated library preparation for single-cell transcriptome sequencing at markedly lower reaction volume without compromising reproducibility, quality, or complexity of the resulting libraries. This technical advance will significantly decrease both the cost and labor required for these studies, making analysis of hundreds to thousands of single cells feasible. The ability to carry out large-scale studies will allow for detailed studies aimed at detecting transcriptional differences between cell populations collected at multiple time points or exposed to different experimental conditions, as well as identifying rare subpopulations of cells.
Acknowledgments
The authors acknowledge Sanford Consortium for Regenerative Medicine, particularly the Genomics core for providing access to the C1 Fluidigm Autoprep system. We are grateful to Sonal Naik for providing technical and experimental support for the C1 single-cell experiments. We thank the staff at the Institute for Genomic Medicine Facility at UCSD for sequencing the single-cell RNA-seq libraries. Special thanks to Thomas Touboul for providing oversight and expert guidance in the pancreatic differentiation experiments. Computations resources were provided through an allocation from XSEDE.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH/NCATS UH2 TR000906, NIH/NHLBI 1U01 HL126494, and the UCSD Department of Reproductive Medicine. Computational resources were part of an allocation to the Laurent laboratory from the Extreme Science and Engineering Discovery Environment (XSEDE). This work was performed under an approved UCSD ESCRO/IRB protocol. Funding for open-access charge was provided by the UCSD Department of Reproductive Medicine. HCA was supported by the Women’s Reproductive Health Research grant K12 HD001259, and by a grant from the Howard and Georgeanna Jones Foundation for Reproductive Medicine.
References
- 1. Sandberg R. Entering the Era of Single-Cell Transcriptomics in Biology and Medicine. Nat. Methods 2014, 11, 22–24. [DOI] [PubMed] [Google Scholar]
- 2. Shalek A. K., Satija R., Shuga J., et al. Single-Cell RNA-Seq Reveals Dynamic Paracrine Control of Cellular Variation. Nature 2014, 510, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farlik M., Sheffield N. C., Nuzzo A., et al. Single-Cell DNA Methylome Sequencing and Bioinformatic Inference of Epigenomic Cell-State Dynamics. Cell Rep. 2015, 10, 1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Urich M. A., Nery J. R., Lister R., et al. MethylC-Seq Library Preparation for Base-Resolution Whole-Genome Bisulfite Sequencing. Nat. Protoc. 2015, 10, 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Macosko E. Z., Basu A., Satija R., et al. Highly Parallel Genome-Wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Macaulay I. C., Haerty W., Kumar P., et al. G&T-Seq: Parallel Sequencing of Single-Cell Genomes and Transcriptomes. Nat. Methods 2015, 12, 519–522. [DOI] [PubMed] [Google Scholar]
- 7. Islam S., Zeisel A., Joost S., et al. Quantitative Single-Cell RNA-Seq with Unique Molecular Identifiers. Nat. Methods 2014, 11, 163–166. [DOI] [PubMed] [Google Scholar]
- 8. Sanchez A., Golding I. Genetic Determinants and Cellular Constraints in Noisy Gene Expression. Science 2013, 342, 1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martin M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet.journal 2011, 17(1):10–12. https://cutadapt.readthedocs.org/en/stable/. [Google Scholar]
- 10. Dobin A., Davis C. A., Schlesinger F., et al. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. http://doi.org/10.1093/bioinformatics/bts635 and https://github.com/alexdobin/STAR/releases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anders S., Huber W. Differential Expression Analysis for Sequence Count Data. Genome Biol. 2010, 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sharan R., Maron-Katz A., Shamir R. CLICK and EXPANDER: A System for Clustering and Visualizing Gene Expression Data. Bioinformatics 2003, 19, 1787–1799. http://bioinformatics.oxfordjournals.org/content/19/14/1787 and http://acgt.cs.tau.ac.il/expander/. [DOI] [PubMed] [Google Scholar]
- 13. R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, 2013, Vienna, Austria: http://www.R-project.org/. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 14. Caruccio N. Preparation of Next-Generation Sequencing Libraries Using Nextera Technology: Simultaneous DNA Fragmentation and Adaptor Tagging by In Vitro Transposition. Methods Mol. Biol. 2011, 733, 241–255. [DOI] [PubMed] [Google Scholar]
- 15. Parkinson N. J., Maslau S., Ferneyhough B., et al. Preparation of High-Quality Next-Generation Sequencing Libraries from Picogram Quantities of Target DNA. Genome Res. 2012, 22, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marine R., Polson S. W., Ravel J., et al. Evaluation of a Transposase Protocol for Rapid Generation of Shotgun High-Throughput Sequencing Libraries from Nanogram Quantities of DNA. Appl. Environ. Microbiol. 2011, 77, 8071–8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adey A., Shendure J. Ultra-Low-Input, Tagmentation-Based Whole-Genome Bisulfite Sequencing. Genome Res. 2012, 22, 1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gertz J., Varley K. E., Davis N. S., et al. Transposase Mediated Construction of RNA-Seq Libraries. Genome Res. 2012, 22, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamble S., Batty E., Attar M., et al. Improved Workflows for High Throughput Library Preparation Using the Transposome-Based Nextera System. BMC Biotechnol. 2013, 13, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shapland E. B., Holmes V., Reeves C. D., et al. Low-Cost, High-Throughput Sequencing of DNA Assemblies Using a Highly Multiplexed Nextera Process. ACS Synth. Biol. 2015, 4, 860–866. [DOI] [PubMed] [Google Scholar]