Abstract
Plant genomes produce a variety of small RNAs that function in distinct, yet overlapping, genetic and epigenetic silencing pathways. However, the abundance and diversity of small RNA classes varies in different plant species, suggesting co-evolution between environmental adaptations and gene silencing mechanisms. Small RNA biogenesis in plants is well understood, but we are just beginning to uncover their intricate regulation and activity. Here, we discuss the biogenesis of plant small RNAs, such as microRNAs, secondary small-interfering RNAs and heterochromatic small-interfering RNAs, and their diverse cellular and developmental functions, including reproductive transitions, genomic imprinting and paramutation. We also discuss the diversification of small RNA-directed silencing pathways through the expansion of RNA-dependent RNA polymerases, Dicer and Argonaute proteins.
Introduction
Small RNAs are involved in plant development, reproduction and genome reprogramming, and the large variety of small RNA pathways in plants is likely to contribute to their phenotypic plasticity. It is generally accepted that these pathways evolved as a cellular defense mechanism against RNA viruses and transposable elements, and later adapted to regulate the expression of endogenous genes. This is consistent with the fact that most small RNA classes have a recognized role in defense responses as well as in epigenetic regulation, but their relative importance and overlap varies between plant species1. Most plant small RNAs are produced as 21 to 24-nucleotide RNA molecules as a result of the activity of DICER-LIKE (DCL) proteins2,3, which relies on the formation of double-stranded RNA (dsRNA) intermediates from either hairpin precursors, derived from overlapping sense and antisense transcripts or from the synthesis of dsRNA from ssRNA by RNA-DEPENDENT RNA POLYMERASEs (RDRs). Processed small RNA duplexes are loaded onto ARGONAUTE (AGO) proteins to target coding or non-coding RNAs (ncRNAs) by sequence complementarity. Depending on the nature of the target transcript and AGO protein involved, this process might lead to target cleavage and degradation, translational repression or recruitment of additional co-factors.
In this review we discuss recent findings and the current understanding of the origin and biogenesis of small RNAs in plants, and the molecular pathways contributing to their diversification and function. The duplication of genes encoding DCL and RDR proteins resulted in the diversification of small RNAs4,5, whereas the diversification of AGO proteins resulted in the development of distinct gene silencing processes based on differential AGO affinities to small RNA duplexes6 (Box 1). Endogenous small RNAs in plants can be divided into several major classes: microRNAs (miRNAs), hairpin-derived small-interfering RNAs (hp-siRNAs), natural antisense siRNAs (natsiRNAs), secondary siRNAs and heterochromatic siRNAs (hetsiRNAs). All small RNAs in plants are modified at the 3′ end by 2′-O-methylation, including miRNA which lack this modification in animals. 2′-O-methylation is essential to confer stability and protection from 3′ uridylation and degradation. In plants, miRNAs are involved in post-transcriptional gene silencing (PTGS) by transcript cleavage or translational repression, and might trigger secondary siRNA production from Pol II-derived cleaved transcripts. While many small RNAs are involved in PTGS, the majority of siRNAs in plants are associated with RNA-directed DNA methylation (RdDM) and transcriptional gene silencing (TGS). Once established, TGS is maintained by 24-nucleotide (nt) hetsiRNAs, which regulate important epigenetic mechanisms such as imprinting and paramutation. Many small RNA biogenesis pathways have been genetically characterized in Arabidopsis thaliana, as these mutations are viable. In plants with larger genomes, hetsiRNAs play essential roles during reproductive transitions such as meiosis, gametogenesis and embryogenesis, likely associated with the more repetitive nature of the genome.
Box 1. Sorting small RNA onto Argonaute proteins.
The ARGONAUTE (AGO) protein family in plants has diversified extensively giving rise to plant-specific AGO proteins155. The identity of the 5′ terminal nucleotide of plant small RNAs has a strong effect on the loading of small RNAs to specific AGO proteins, which determines their activity. This bias might be intrinsic to all AGOs, as in organisms with only one AGO (such as Schizosaccharomyces pombe) the protein has a strong bias towards small RNAs with 5′ uracil (see the figure, part a). In Arabidopsis thaliana, AGO1 and AGO10 also bind small RNAs with a 5′ uracil, whereas AGO2, AGO4, AGO6, AGO7 and AGO9 prefer adenines and AGO5 has a bias for cytosines111,156,157 (see the figure, part a). This 5′ nucleotide specificity is determined by a structural loop lining the sRNA-binding pocket in the MID domain of AGO proteins158. An additional layer of complexity is particularly obvious for microRNA (miRNA) loading, which is affected also by the imperfect complementarity and bulges that characterize miRNA duplex structures. For example, AGO10 is a critical regulator of shoot apical meristem (SAM) specification because of its preferential interaction with miR166, which is mediated by one internal base mismatch flanked by two paired bases within the mature duplex159–161 (see the figure, part b). This prevents the loading of miR166 onto AGO1 and silencing of the class III HD-ZIP transcription factors in the SAM159,160. Another example is miR390, which is preferentially loaded onto AGO7 (see the figure, part b), leading to the production of trans-acting small interfering RNAs (tasiRNAs) from TAS3 transcripts67,162. AGO7 association with miR390 involves preference for a 5′-A, but also a mismatch at position 11 of the miRNA duplex163. Importantly during the AGO7–miR390 interaction, AGO7-mediated cleavage of the complementary strand is required to establish a functional silencing complex163 (see the figure, part b).
The miR393b duplex provides another interesting example of miRNA sorting in plants, as its guide strand is loaded onto AGO1, whereas the other strand, miR393b*, is loaded onto AGO2 and plays an essential role in mediating antibacterial defense164. Mechanistic insight into this type of small RNA sorting was recently reported, showing that the 15th nucleotide of a miR165 duplex directs miRNA loading onto both AGO1 and AGO2 through their PIWI domain (see the figure, part b). In contrast, AGO2 preferential binding to miR396* requires base pairing at position 11 and 15 of the duplex, whereas AGO1 tolerates mismatches at these central positions165. The importance of this mechanism was nicely illustrated using the miR165–miR165* duplex, where removing the 15th nucleotide mismatch in artificial miR165 stem-loops led to loading onto AGO2 instead of AGO1, down-regulation of miR165 target genes and partial suppression of the adaxialized phenotype, which is characteristic of ago1 mutant alleles165.

The biogenesis of small RNAs in plants
Processing of dsRNA into small RNAs requires the activity of Dicer enzymes. The minimal functioning Dicer is found in budding yeasts (though not in Saccharomyces cerevisiae), and is composed of a ribonuclease III domain and a dsRNA-binding domain (dsRBD), but lacks the DExD box helicase and PIWI/ARGONAUTE/ZWILLE (PAZ) domains found in higher eukaryotes4,7. The PAZ domain binds the 2-nucleotide 3′ overhang of dsRNAs, and is connected to the catalytic domain through an α-helical structure that acts as a ruler to determine small RNA size. The four DCL proteins in A. thaliana have been well characterized2,3, indicating that the duplication of DCL genes has occurred early in plant evolution.
The synthesis of dsRNAs by RDRs contributed significantly to the expansion, diversification and evolution of small RNA functions. RDR genes are found in RNA viruses, plants, fungi, protists, and some lower animals, but are absent in flies, mice, and humans. In contrast to DCLs, the RDR family seems to be more complex and many RDR genes remain uncharacterized5. Generally there are three main pathways responsible for the biogenesis of the vast majority of small RNAs in plants: one for the biogenesis of miRNAs, one for the biogenesis of 21 and 22 nucleotide secondary siRNAs and another for the biogenesis of 24 nucleotide hetsiRNAs (Fig. 1).
Figure 1. Main pathways for biogenesis of endogenous small RNAs in plants.
a. Genes encoding microRNAs (miRNAs; left) are transcribed by RNA Polymerase II (Pol II) and fold into hairpin-like structures called primary (pri)-miRNAs, which are processed by DICER-LIKE 1 (DCL1) into a shorter stem-loop structure called precursor (pre)-miRNAs. Pre-miRNAs are processed again by DCL1 into the mature miRNA duplex. During miRNA processing, DCL1 is assisted by several proteins (reviewed in 8). MiRNAs are involved in post-transcriptional gene silencing (PTGS) by mediating mRNA cleavage or translational repression. Longer Pol II-derived hairpins, termed hairpin-derived small-interfering RNAs (hp-siRNAs; middle), might originate from inverted repeats, and are originally processed by all DCLs. These hairpins might evolve into miRNAs, and are often designated as proto-MIRs. Natural-antisense small-interfering RNAs (natsiRNA; right) are produced from dsRNAs originating from overlapping transcription (cis-natsiRNA) or highly complementary transcripts originated from different loci (trans-natsiRNA)175–177. The biogenesis and function of natsiRNAs is still largely unclear. b. The precursors of secondary siRNAs are transcribed by Pol II, and may originate from non-coding loci, protein-coding genes and transposable elements. These transcripts are converted into double-stranded RNA (dsRNA) by RNA-DEPENDENT RNA POLYMERASE 6 (RDR6), and processed by DCL2 and DCL4 to produce siRNAs of 22- or 21-nucleotide (nt) in length, respectively. Secondary siRNAs are mostly involved in PTGS, but can also initiate RNA-directed DNA methylation (RdDM) at specific loci. They are subdivided into trans-acting siRNAs (tasiRNA)34,66,162,178, phased siRNA (phasiRNA)65 or epigenetically-activated siRNA (easiRNAs) 77,179. c. Heterochromatic siRNAs are derived from transposable elements and repeats located at pericentromeric chromatin. Their biogenesis requires Pol IV transcription and the synthesis of dsRNA by RDR2, which is subsequently processed into 24-nucleotide long siRNAs by DCL3. These small RNAs are involved in maintaining RdDM-mediated transcriptional gene silencing (TGS) (reviewed in 31).
The origin and biogenesis of microRNAs
Plant miRNAs, typically 20 to 22 nucleotides in length, are endogenous genes transcribed by RNA Polymerase II (Pol II) into long primary microRNAs (pri-miRNAs), which are single-stranded and polyadenylated RNA molecules that fold into hairpin-like structures (Fig. 1a). The pri-miRNAs are cleaved by DCL1 into a smaller stem-loop structure called precursor microRNAs (pre-miRNAs), which are subsequently processed again by DCL1 to produce the mature miRNA duplexes consisting of the active miRNA strand and its complementary strand miRNA. MiRNA biogenesis pathways are particularly well described in A. thaliana where several DCL1 partners have been characterized as required for pri-miRNA processing (reviewed in 8).
MicroRNAs mediate PTGS through mRNA cleavage or translation repression, and play essential roles in plant development. Null dcl1 alleles in A. thaliana are embryonic lethal9, whereas different hypomorphic alleles give rise to variable defects in integument, ovule, and floral development, and also have maternal effects10. DCL1 knockdown in rice also results in defects in plant growth and in shoot, root, and leaf development, and ultimately leads to developmental arrest11. fuzzy tassel (fzt) mutants in maize have a broad range of vegetative and reproductive defects, and are hypomorphic dcl1 alleles, as in these plants the abundance of some miRNAs is more dramatically reduced than others12. The vast majority of MIRNA genes are species- or family-specific, suggesting rapid evolution and a high turnover rate13,14.
A subtype of miRNAs in A. thaliana and rice are the relatively rare, longer miRNAs, which are 23 to 25-nucleotide in length, are processed by DCL3 and function in transcriptional gene silencing (TGS)15–17. Furthermore, in A. thaliana, DCL4 was also found to process a class of newly evolved miRNAs14,18,19. These observations have led to the idea that pre-miRNA recognition by different DCL proteins might reflect the evolution of MIRNA genes: Some of these newer, DCL4-procesed pre-miRNAs are long and exhibit complementarity with their targets that extends beyond that of the mature miRNA, suggesting that MIRNA genes arose from inverted duplications of their target genes14,20,21 and that DCL2, DCL3 and DCL4 processed these longer hairpins (or proto-MIRNA) into small RNAs of different sizes (hp-siRNA) (Fig. 1a). During evolution, the near-perfect sequence complementarity in proto-MIRNA hairpins has decreased and consequently refined into smaller transcripts that are now processed by DCL1 as a single miRNA duplex22. Another, similar origin for miRNAs has been proposed from miniature inverted-repeat transposable elements (MITEs) that, when transcribed, create hairpin RNAs resembling proto-MIRNAs23,24.
The origin and biogenesis of small-interfering RNAs
Long dsRNAs that are the precursors of siRNAs can arise from the hybridization of sense and antisense transcripts, from the fold-back of an inverted-repeat sequence, from the hybridization of unrelated RNA molecules with sequence complementarity or, most commonly, following synthesis by RDRs (reviewed in 1). Long dsRNA molecules can be synthesized by RDRs with or without initial priming25,26, resulting in the amplification of a primary, small RNA-mediated silencing-triggering signal. The three major clades of eukaryotic RDRs are RDRα, RDRβ, and RDRγ; the RDRα clade is present in the fungal, plant and animal kingdoms, whereas RDRβ has been found in animals and fungi and RDRγ only in plants and fungi5,27. In A. thaliana, the RDRα clade is composed of RDR1, RDR2, and RDR6, while the RDRγ clade is composed of RDR3, RDR4, and RDR5. The RDRγ clade remains functionally uncharacterized in plants, but the presence and expression of at least one of its members in several other plant genomes and many fungi suggests functional significance5,27. Interestingly, efficient antiviral defense and viral siRNA biogenesis were detected in rdr1, rdr2 and rdr6 triple mutants in A. thaliana28, indicating that RDR3, RDR4 and RDR5 might represent alternative pathways for antiviral defense. In addition, in the fission yeast Schizosaccharomyces pombe, RDRγ is involved in transcriptional gene silencing29.
Endogenous siRNAs in plants are primarily processed by DCL2, DCL3 and DCL4, and have been categorized into secondary siRNAs (Fig. 1b) and hetsiRNAs (Fig. 1c). Secondary siRNAs include different subclasses such as trans-acting small interfering RNAs (tasiRNAs), phased small interfering RNAs (phasiRNAs), epigenetically-activated small interfering RNAs (easiRNAs) and natsiRNAs. The most abundant small RNAs are the 24-nucleotide hetsiRNAs, which mediate transcriptional silencing of transposons and pericentromeric repeats through RdDM (reviewed in 30,31). The biogenesis of hetsiRNAs requires transcription by Pol IV followed by dsRNA synthesis by RDR2 and processing by DCL3 (Fig. 1c). By contrast, 21 and 22-nucleotide secondary siRNAs such as tasi-, phasi- and easiRNA are produced by DCL4 and DCL2, respectively, following Pol II transcription and dsRNA synthesis by RDR6 (Fig. 1b). DCL2 is often regarded as a backup for DCL4, as the former is recruited to dsRNA following the deletion of latter32. RDR1 activity is mostly associated with the amplification of exogenous, virus-derived small RNAs, being part of the main plant antiviral RNAi system together with DCL2 and DCL428,33. Additional processing of siRNA requires SUPPRESSOR OF GENE SILENCING 3 (SGS3), which functions together with RDR634, and dsRNA binding protein 4 (DRB4), which interacts with DCL4 in the production of endogenous and exogenous 21-nucleotide siRNAs35.
Small RNA modifications
Small RNA modifications can regulate their abundance and function, thus contributing to regulation of gene silencing. In plants, these modifications have been observed primarily at the 3′ end and are essential to confer stability and prevent small RNA degradation. Further mechanistic insight into these pathways in A. thaliana has suggested that protective RNA modifications are bypassed in certain tissues, cell types or growth conditions, thereby promoting small RNA diversity.
2′-O-methylation and uridylation
After processing, eukaryotic small RNA duplexes such as siRNAs, miRNAs and piwi-interacting RNA (piRNAs) can be modified by 2′-O-methylation, 3′ uridylation or adenylation, and adenosine deamination36. 2′-O-methylation of the terminal 3′ nucleotide is important for miRNA and siRNA stability, because unmethylated small RNAs are signaled for degradation by 3′uridylation37,38. Plant small RNAs are 2′-O-methylated at the 3′ terminal by HUA HENHANCER 1 (HEN1)37–39 to prevent uridylation by the nucleotidyl transferase HEN1 SUPPRESSOR 1 (HESO1) 40,41; uridylation is a signal for degradation via SMALL RNA DEGRADING NUCLEASE 1 (SDN1)42 (Fig. 2). Loss of 2′-O-methylation activity in A. thaliana results in severe developmental defects, likely because essential miRNAs are depleted39,43. Small RNA 3′ uridylation was also observed in the algae Chlamydomonas reinhardtii and requires the nucleotidyl transferase MUT6844, whereas in humans and Caenorhabditis elegans, many enzymes have been shown to uridylate miRNAs in a sequence-specific manner45. In human cells, poly-uridylation of pre-miRNAs is performed by terminal uridylyltransferase 4 (TUT4). The recruitment of TUT4 to pre-miRNAs by the RNA binding protein Lin-28 destabilizes the pre-miRNAs and reduces the levels of mature miRNAs46,47. In contrast to Lin-28-dependent poly-uridylation, Lin-28-independent mono-uridylation by TUTs is required for the processing of certain pre-miRNAs in human cells48. In C. elegans, the uridylation of some siRNAs restricts them to binding only the AGO protein CSR-1 thus reducing their abundance, which is required for proper chromosome segregation49 and the recognition of self from non-self mRNA in the germline50,51.
Figure 2. 2′-O-methylation, uridylation and degradation of miRNAs in A. thaliana.
MicroRNA (miRNA) duplexes are 2′-O-methylated at both 3′ ends by HUA ENHANCER 1 (HEN1), which protects them from uridylation and degradation (left). HEN SUPRESSOR 1 (HESO1) and UTP:RNA URYDILTRANSFERASE 1 (URT1) are nucleotidyl transferases that uridylate unprotected 3′ ends of small RNAs, triggering their degradation by the 3′-5′ exonucleases SMALL RNA DEGRADING NUCLEASE (SDN; middle). ARGONAUTE 1 (AGO1) recruits HESO1 during mRNA target recognition and cleavage in order to polyuridylate and degrade the 3′ of cleaved target transcripts52. Thus, the 3′ methylation of miRNAs loaded onto AGO1 serves to protect them from HESO1 activity. Recent studies have shown that URT1 also interacts with AGO1 to establish mono-uridylation of particular miRNAs53,54 (left), and this process may produce 22-nucleotide miRNA variants that are able to form functional RNA-induced silencing complexes and trigger post-transcriptional gene silencing (PTGS)54 (see Fig. 3). HESO1 and URT1 have been shown to act both independently and synergistically, perhaps reflecting their different affinities for 3′ terminal nucleotides in vitro. HESO1 has preference for tailing 3′-uracil, whereas URT1 prefers 3′-adenine54. Although these features explain how these enzymes act synergistically at non-3′-uracil miRNA targets (URT1 forms substrates for HESO1), it does not fully account for their substrate preferences found in vivo53,54.
Although it is clear that these modifications play an essential role in regulating small RNA biogenesis and function, it remains poorly understood how these factors are recruited to miRNA processing complexes in plants. In A. thaliana, polyuridylation and degradation of the guide strand of miRNA duplexes requires loading onto AGO1, which has been shown to interact directly with HESO138. This suggests that the primary role of 2′-O-methylation is to protect miRNAs from the AGO1-associated HESO1 activity that also uridylates 3′ ends of cleaved target transcripts and leads to their degradation52 (Fig. 2). Recent studies have shown that HESO1 acts on most miRNAs, whereas the monouridylation of certain miRNAs requires another nucleotidyl transferase, UTP: RNA URIDYLYLTRANSFERASE (URT1)53,54. HESO1 seems to be more processive than URT1, perhaps because of their different substrate preference, which depends on the 3′ terminal nucleotide of the small RNAs: URT1 prefers 3′ adenine whereas HESO1 has strong preference for 3′ uridine, thus explaining why it polyuridylates its substrates. These enzymes also act synergistically to uridylate some miRNAs, as HESO1 acts on some monouridylated small RNAs derived from URT1 activity54 (Fig. 2). Uridylation of miRNAs by URT1 and HESO1 is generally associated with reduced efficiency of target gene cleavage. An exception to this is the monouridylation of miR170 and miR171a by URT1; the resulting 22-nucleotide miRNA variants lead to the production of secondary siRNAs from their target transcripts and to efficient gene silencing38,54 (see below). This indicates that tailed miRNAs loaded onto AGO complexes could be non-canonically functional.
Other types of miRNA tailings were found in mutants lacking both HESO1 and URT1 activities, including non-uridine nucleotides53, suggesting that other small RNA modification pathways might exist in plants. There are eight nucleotidyl transferases that remain uncharacterized in A. thaliana, and these could have a role in miRNA modification or in processes utilizing secondary or heterochromatic siRNAs.
Novel small RNA modifications in plants
Recent efforts to discover novel small RNA modifications have identified base modifications by comparing mismatches between genomic sequences and sequencing reads of small RNAs, as some modifications result in preferential nucleotide misincorporation by reverse transcriptases during cDNA synthesis55. Such analyses revealed frequent A-to-G, G-to-A and U-to-C substitutions in A. thaliana and rice small RNA data56,57. U-to-C substitutions were also detected in small RNAs from rice anthers58, and are likely the result of reverse transcriptase misincorporation at modified U, as editing enzymes responsible for U-to-C conversion have not been identified in plants. Such U-to-C mismatches in small RNA reads could be caused by pseudouridine, which is abundant in structured RNAs and is required for the stabilization and function of tRNAs and rRNAs59. Pseudouridylation in eukaryotic small RNAs has not been directly observed, but was recently reported in mRNAs in yeast and humans60,61. Putative functions for pseudouridine might be the stabilization and transport of small RNA duplexes, as pseudouridylation is essential for tRNA biogenesis and nuclear export in yeast62,63.
Secondary siRNA biogenesis and control
PTGS in plants can be amplified when miRNA-mediated cleavage or aberrant processing of particular transcripts leads to the formation of dsRNA by RDR proteins, which is subsequently processed by DCLs into secondary siRNAs. This powerful silencing machinery is conserved within the plant kingdom, but notably, it has widely differing targets in different plant species such as A. thaliana, rice, maize and soybean, including mRNAs, ncRNAs and repeat-derived RNAs. Depending on the precursor mRNA, secondary siRNAs have been classified into different subclasses, such as tasiRNAs and phasiRNAs. While tasiRNA have been demonstrated to act in trans, phasiRNAs are secondary siRNAs of unknown function but identified as phased, and could therefore include coding and non-coding transcripts64.
The biogenesis of tasiRNAs and phasiRNAs
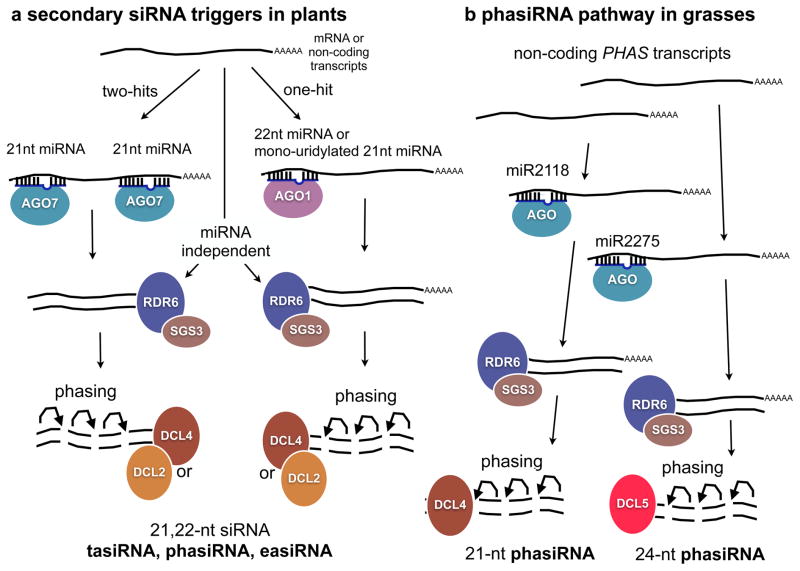
The production of secondary, RDR-dependent small RNAs such as tasi- and phasiRNAs, requires transcript targeting by miRNAs65 (Fig. 3). Targeting by two 21-nucleotide miRNAs at independent target sites along the transcript (“two-hits”), or targeting by a single 22-nucleotide miRNA (“one-hit”) can trigger the production of secondary RNA from transcripts34,66,67 (Fig. 3a). In addition, the structure of particular miRNA duplexes may also influence siRNA biogenesis regardless of miRNA length68. Cleaved transcripts serve as templates for dsRNA synthesis by RDR6 and the production of 21- and 22-nucleotide siRNAs by DCL4 and DCL2 respectively, which can function in trans to target other transcripts. Secondary siRNAs are often “phased” so that their first nucleotide occurs every 21 or 22 nucleotides from the miRNA cleavage site65,66,69. This is because DCL2 and DCL4 digest the dsRNA processively (Fig. 3), and might even interact directly with RDR6. However, phasing can be difficult to detect when there are multiple miRNA cleavage sites, or multiple related template RNAs.
Figure 3. Triggers of secondary siRNA biogenesis.
a. Plant microRNAs (miRNAs) target transcripts for cleavage or translational repression, and also trigger the production of secondary small-interfering RNAs (siRNAs) from mRNAs, non-coding RNAs and transposable elements. The most accepted mechanism for the biogenesis of trans-acting siRNA (tasiRNA), phased siRNA (phasiRNA) and epigenetically activated siRNA (easiRNA) relies on two distinct pathways. One consists a two-hit system utilizing two 21-nucleotide (nt) miRNAs per transcript, and requires the activity of an RNA-inducing silencing complex comprising ARGONAUTE 7 (AGO7). The second pathway consists of a one-hit system that usually involves a 22-nt miRNA loaded on AGO1, or 22-nt miRNA variants that are produced from monouridylation of 21-nt miRNAs (see Fig. 2). Both pathways are routed towards RNA-DEPENDENT RNA POLYMERASE 6 (RDR6)-mediated dsRNA synthesis aided by SUPPRESSOR OF GENE SILENCING 3 (SGS3), and processing of 21 and 22-nucleotide siRNAs by DICER-LIKE 4 (DCL4) and DCL2, respectively. RNA Polymerase II (Pol II)-derived transcripts might also produce miRNA-independent secondary siRNA via interactions with other RNA processing machineries such as the spliceosome85, or during RNA decay100,101, but these pathways are not fully understood. b. An additional phasiRNA biogenesis pathway was found in monocot plants such as maize and rice, which involves the transcription of non-coding PHAS transcripts form intergenic loci. Two miRNAs (miR2118 and miR2275) were found involved in cleavage of PHAS transcripts by an unknown AGO protein. These cleavage products are converted into dsRNA by RDR6 and SGS3, and processed into 21- and 24-nucleotide phasiRNAs by DCL4 and DCL5, respectively (reviewed in 65).
Secondary siRNAs are relatively rare in somatic cells of wild-type A. thaliana, even though many mRNA targets generate secondary siRNAs in this species69. In contrast, other plant genomes such as rice and maize contain thousands of tasi- and phasiRNA-generating loci that encode large families of ncRNAs65. TasiRNA biogenesis and functions have been well studied in A. thaliana, which has only four families of TAS genes. TAS1a/b/c and TAS2 loci are targeted by miR173, while miR390 targets the TAS3a/b/c, and miR828 triggers TAS4-derived tasiRNAs18,34,66. TAS3 is the most well conserved TAS locus, as it is also present in moss, rice, maize, and gymnosperms65. TAS3-derived siRNAs are designated tasi-ARFs, and their production requires a two-hit system with miR390 that is exclusively loaded to a specialized Argonaute protein (AGO7), to regulate auxin-related developmental responses (see Box 1 and Box 2).
Box 2. Intercellular movement and transgenerational inheritance of small RNAs.
Different small RNA classes have been associated with non-cell autonomous signaling involving short (cell-to-cell) and long (between organs) distance movement of small RNAs166,167. Intercellular movement has broad implications for cell-to-cell communication and transgenerational inheritance of epigenetic signals78,150. Spatiotemporal coordination of cell fate decisions and tissue patterning in multicellular organisms also depend on intercellular communication by small RNAs. For example, adaxial-abaxial (upper-lower) patterning of lateral organs in plants requires two mobile small RNAs with opposing functions: miR165 and miR166 (produced in the lower side of the leaf) restrict the accumulation of the transcription factors HD-ZIP III to the upper ventral domain while tasi-ARFs (produced in the upper side) are able to diffuse and create a gradient to restrict their targets AUXIN RESPONSE FACTOR 3 and 4 (ARF3 and ARF4) to the lower ventral domain (reviewed in 168). Non-cell-autonomous small RNAs are also required for radial patterning of root tissues, which is regulated in part by the transcription factors SHORT ROOT (SHR) and SCARECROW (SCR) that act together to induce the expression of miR165 and miR166169. Upon induction in the endodermal cell layer, miR165 and miR166 move into the central vascular cylinder and target the HD-ZIP III transcripts, creating a radial gradient that is important for the differentiation of the central vascular tissues169.
Considering the primary evolutionary role of small RNAs as a genome defense mechanism against viruses and transposons, several studies have attempted to define the molecular requirements for biogenesis and systemic transmission of silencing signals in several organisms. In plants, siRNAs from reporter transgenes as well as endogenous sequences are able to spread systemically and induce gene silencing in recipient cells, but several questions remain regarding the biogenesis of small RNAs in response to environmental cues and the transgenerational inheritance of newly acquired epigenetic states (reviewed in 170). In the Arabidopsis thaliana male and female gametophytes, small RNA movement has been proposed to follow epigenome reprogramming, which results in transcriptional re-activation of transposable elements in germline companion cells78,150. These small RNA-directed mechanisms could provide surveillance of and protection against transposon activity during meiosis and epigenomic reprogramming in the germline78,150, or in the seed after fertilization171. Additional sources of transposon-derived siRNAs during reprogramming in the gametophytes were also associated with genomic imprinting through targeted DNA demethylation and RdDM172–174. From these studies, it is clear that the high degree of reprogramming observed during plant gametogenesis provides numerous possibilities for establishing novel and beneficial epigenetic states by mobile small RNAs.
In maize anthers, phasiRNAs are produced from ncRNA precursors designated as PHAS ncRNA (Fig. 3b). There are thousands of PHAS loci in grass genomes, which are transcribed by Pol II, capped and polyadenylated, resembling protein-coding and trans-acting siRNA precursors (TAS) in this respect. In both rice and maize, internal cleavage directed by miR2118 triggers the production of 21-nucleotide phasiRNAs, whereas miR2275-directed cleavage triggers the production of 24-nucleotide phasiRNAs70–72. It is hypothesized that a complex containing homologs of RDR6 and SGS3 recognizes the 3′ end of cleaved PHAS transcripts and synthesizes dsRNA from the polyA tail to the cleavage site73. The dsRNAs are subsequently processed by DCL4 and DCL5 to generate 21- and 24-nucleotide phasiRNAs, respectively74. Both DCL4 and DCL5 in grasses have phased activity, generating populations of regularly spaced siRNAs from each PHAS precursor65. Although the expression of non-coding PHAS loci is anther-specific in grasses, the miR2118-482 superfamily is conserved in dicots and triggers phasiRNA production from coding NB-LRR genes in legumes and solanaceous species64,75,76, and in some gymnosperms as well64. NB-LRR disease resistance genes encode innate immunity receptors, and these phasiRNAs in legumes and Solanaceae appear to be beneficial for plant-microbial interactions and plant immunity. It is likely that this elaborate defense mechanism was lost in grasses, to be replaced by a variety of non-coding PHAS loci producing different types of anther-specific phasiRNAs65.
easiRNAs are produced from active retrotransposons
miRNAs are also able to trigger secondary siRNA biogenesis from transcriptionally re-activated transposable elements using a similar genetic pathway to tasiRNAs77. Re-activation of some transposons and easiRNA biogenesis occurs in wild-type A. thaliana pollen during epigenetic reprogramming78, in cell cultures79 or under stress conditions80. In DNA methylation mutants such as DECREASE IN DNA METHYLATION 1 (DDM1) and DNA METHYLTRANSFERASE 1 (MET1)78,81, as many as 2500 transposons are activated, and subsequently targeted by more than 50 miRNAs77. These miRNAs have well known functions in plant development, and are highly conserved, although targeting does not always result in easiRNA production77. It is possible therefore that miRNAs evolved originally to target transposons, and only subsequently adopted other functions, such as gene regulation and triggering the processing of tasi- and phasiRNA from non-coding precursors. This hypothesis is consistent with a transposon origin of miRNA precursors82 (see above), as transposable elements-derived proto-miRNA could of course target related transposons.
RNA interference and splicing
Secondary siRNA biogenesis pathways may also interact with additional cellular pathways that silence transposable elements and transgenes, such as RNA splicing. Recent work in yeast and flies have shown that stalled spliceosomes at weak splice sites and suboptimal introns could function as a signal to trigger RNAi, thus representing a way to discriminate between transposons or precursors of small RNAs and protein-coding transcripts83,84. In fact, previous studies in A. thaliana have shown that intron splicing is a potent suppressor of RDR6-dependent PTGS85, and notably this requires SERRATE (SE) and the nuclear cap-binding complex ABA HYPERSENSITIVE 1 (ABH1), which are components of the miRNA biogenesis pathway86–88. However, SE function during intron splicing seems to be miRNA-independent, while it is unclear whether ABH1 and miRNAs are directly involved in splicing or else in the expression of specific splicing factors. It is possible that splicing suppresses PTGS by removing potential miRNA target sites located within introns, but this hypothesis awaits further investigation. Interestingly, splicing factors have also been associated with TGS, such as the SR45 protein that is required to establish RdDM and 24-nucleotide siRNA processing at FLOWERING WAGENINGEN (FWA) transgenes89. However, Pol IV-dependent transcripts involved in RdDM activity appear to be unspliced90, supporting the idea that SR45 participates in RdDM indirectly, perhaps by regulating the splicing of RdDM components, as has been proposed in S. pombe91–94.
RNA interference and RNA decay
RNA decay and PTGS are functionally linked, as RDR6-mediated PTGS of transgenes and endogenous genes is enhanced in mutants of non-sense mediated decay (NMD), decapping (which triggers RNA decay) and exosome factors in A. thaliana95–101. The interplay between RNAi and RNA decay was also observed in the fission yeast and fruit flies, where heterochromatic silencing of transgenes and transposons is promoted in the absence of the exosome102,103.
In A. thaliana, loss of the NMD factor 5′ to 3′ exoribonuclease 4 (XRN4, also known as EIN5) enhances PTGS of thousands of endogenous genes upon viral infection104. This is mediated by the production of secondary siRNAs, which were designated virus-activated siRNAs (vasiRNAs) and, interestingly, their biogenesis relies on RDR1, DCL4 and AGO2, which is identical to the genetic pathway responsible for the production and activity of viral siRNAs28,33,105. Given that vasiRNAs are only observed upon infection with viruses lacking RNAi suppressors, vasiRNAs could represent an additional layer of antiviral defense104, but this interesting idea needs further experimental evidence.
The decapping complex comprising DCP1, DCP2 and VARICOSE (VCS), which co-localize in processing bodies (P-bodies), is also associated with PTGS101. The link between P-bodies and PTGS remains unclear, as RDR6-mediated PTGS occurs in distinct cytoplasmic siRNA-bodies106. However, a possible interaction between P-bodies and siRNA-bodies was recently proposed101, providing a direct subcellular connection between RNA decay and secondary siRNA biogenesis.
The switch to transcriptional silencing
Barbara McClintock coined the term “Changes of Phase” to describe how transposons switched between active and inactive forms107, and recently there has been a lot of interest in finding distinctive features of transposon transcripts that trigger an epigenetically heritable state of TGS through RdDM. As discussed above, many retrotransposons in A. thaliana give rise to abundant 21 to 22-nucleotide easiRNAs when transcriptionally active, and recent evidence indicates that this could represent the entry point for TGS108 through establishment of RDR6- and AGO6-mediated DNA methylation109 (Fig. 4a). A PTGS-to-TGS transition would occur when Pol II transcription is replaced by the plant-specific RNA polymerases Pol IV and V, switching from 21 or 22-nucleotide to 24-nucleotide siRNA production and epigenetic silencing by RdDM108,110 (Fig. 4b). Instead of using RDR6 and DCL2 or DCL4, Pol IV transcripts are processed by RDR2 and DCL3 into 24-nucleotide siRNAs, which are loaded onto AGO4, AGO6 or AGO9 to reinforce DNA methylation31,111,112 (Fig. 4b). Of course, transcriptional silencing is only achieved when RdDM is able to spread into the promoter of the retrotransposon, which is found in the upstream long terminal repeat (LTR). This switch occurs immediately in rdr6 and dcl4 mutants indicating that 21nt siRNA might actually inhibit the production of 24nt siRNA77,110. An example is provided by the retrotransposon Evadé (EVD)110 that generates increasing levels of RDR6-dependent dsRNA during multiple transposition events, which could eventually saturate the 21 and 22-nucleotide siRNA biogenesis enzymes DCL2 and DCL4, instead leading to DCL3 generating 24-nucleotide siRNAs from transcribed regions of the retrotransposon (Fig. 4a). In contrast to EVD, other retrotransposons are able to switch from producing 21-nucleotide to 24-nucleotide siRNAs following transient somatic re-activation by heat-stress, and independently of transposition80. The mechanisms responsible for these switches in siRNA biogenesis will require further investigation.
Figure 4. The transition from silencing by PTGS to silencing by TGS in transgenes, epialleles and active transposons.
a. Post-transcriptional gene silencing (PTGS) by miRNAs is likely the major pathway triggering biogenesis of secondary 21 and 22-nucleotide (nt) siRNAs, in a process involving RNA-DEPENDENT RNA POLYMERASE 6 (RDR6), SUPPRESSOR OF GENE SILENCING 3 (SGS3), DICER-LIKE 4 (DCL4) and DCL2 (see Fig. 3). 21- and 22-nt siRNAs are required for the establishment of RNA-directed DNA methylation (RdDM) at particular transposable elements and epialleles, which at least at some loci requires the activity ARGONAUTE 6 (AGO6)109. This pathway is able to target nascent Pol II transcripts and recruit the DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) to establish DNA methylation in all sequence contexts (1), but this interplay is not fully understood. An alternative pathway was proposed for transgenes and active retrotransposons, perhaps depending on their variable copy number and transcription levels. The accumulation of long dsRNA molecules might saturate both the DCL2 and DCL4 processing pathways, resulting in functional compensation by DCL3, which instead produces 24-nt siRNAs for the establishment of RdDM via AGO4110. b. CHG (H denotes A, C or T) methylation previously established by DRM2, is recognized by the histone methyltransferase KRYPTONITE (KYP), which reinforces the repressed chromatin state of methylated DNA by establishing the dimethylation of histone 3 Lys 9 (H3K9me2) 122 (2). A complete PTGS-to-TGS switch occurs when SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1) binds H3K9me2 and recruits RNA Polymerase IV (Pol IV) to initiate the biogenesis of 24-nt siRNAs through RDR2 and DCL3123 (3). RdDM consolidation is achieved by the recruitment of Pol V to unmethylated DNA by SU(VAR)3–9 HOMOLOG 2 (SUVH2) and SUVH9124 (4). This is followed by the recruitment of AGO4, mediated by sequence complementarity between the 24-nt siRNAs and the Pol V-nascent transcripts, and by the conserved GW/WG motif (also known as Ago hook) present in the carboxy-terminal region of the Pol V subunit NRPE1. Then AGO4 is able to recruit DRM2 to establish additional DNA methylation de novo (reviewed in 112 and 31).
Nuclear functions of small RNAs
RdDM is an important RNAi-mediated epigenetic pathway in plants. It is involved in transcriptional silencing of transposons and repetitive sequences31,112, and relies on a specialized transcriptional machinery that requires the plant-specific RNA polymerases Pol IV and Pol V113. Pol IV transcripts are rapidly processed into dsRNAs by RDR2, which are subsequently processed into 24-nucleotide siRNAs by DCL3 and exported to the cytoplasm. Once in the cytoplasm, they are incorporated into AGO4- (and other AGO-) containing complexes, and imported back to the nucleus to target nascent transcripts transcribed by Pol V at the same loci (Fig. 4b). While AGO4 is the most abundant AGO protein involved in RdDM, the related proteins AGO6 and AGO9 are also loaded with 24-nucleotide siRNAs and appear to be only partially redundant with AGO4, having specific functions (see below) 109,111,114. In a plausible model, AGO4 is thought to recruit (in part) the DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) to establish de novo DNA methylation at cytosines in all sequence contexts (CG, CHG and CHH, where H is A, C or T) (reviewed in 112 and 31) (Fig. 4b).
Paramutation
Once established, DNA methylation can be passed on to other alleles in repulsion by a mechanism known as paramutation. First discovered in maize in the 1950s and 60s, paramutation has been more recently found in many other organisms115,116. During paramutation in maize, trans-homolog interactions between otherwise identical alleles can lead to heritable epigenetic changes mediated by small RNA116,117. Similarly, the PAI2 gene in A. thaliana is epigenetically silenced in trans by siRNAs derived from an inverted duplication of PAI genes at another genomic locus, although silencing is not maintained in its absence118,119. In maize, mutations in one of several components of the RdDM pathway are defective in paramutation, including RDR2, Pol IV and V and chromatin remodelers related to the A. thaliana DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1) and CLASSY 1 (CLSY1), which are also required for TGS120. Recent work also clarified that small RNA production is essential but not sufficient for paramutation, as DNA methylation also contributes to the strength of paramutation121. In A. thaliana, CHG methylation recruits the histone methyltransferase KRYPTONITE (KYP), which is responsible for the dimethylation of histone 3 Lys 9 (H3K9me2)122. This mark recruits the CHG chromomethylase CMT3, and both are required to initiate and maintain PAI2 silencing118,119. Although a role in maize has yet to emerge, H3K9me2 in A. thaliana is recognized by the SAWADEE HOMEODOMAIN HOMOLOGUE 1 (SHH1), which recruits Pol IV and initiates siRNA biogenesis for the maintenance of gene silencing123 (Fig. 4b). Thus higher levels of DNA methylation and H3K9me2 might result in stronger siRNA-mediated paramutation via Pol IV recruitment to paramutagenic alleles. As for paramutable alleles, it is still unclear how DNA methylation is established de novo in the presence of siRNAs, as in A. thaliana, Pol V recruitment also seems to require pre-methylated DNA124. One possibility is that the establishment of retrotransposon silencing in A. thaliana requires another RdDM pathway that involves 21 to 22-nucleotide siRNAs — the precursors of which are transcribed by Pol II from transposon LTRs — that are targeted to the chromatin by AGO6109,110 (Fig. 4a). A similar model was proposed to explain stepwise TGS of the hypomethylated FWA epiallele by virus-induced gene silencing (VIGS), which also involves 21 and 22-nucleotide wsiRNAs, Pol V and DRM2125. This progressive silencing of transposable elements and epialleles by RdDM is reminiscent of paramutation.
Heterosis, polyploidy and hybrid incompatibility
Hybridization between different species (interbreeding) and the resulting interspecific allopolyploids are particularly common in plants126. Crops such as wheat, cotton, and canola are interspecific allopolyploids, whereas maize and sorghum are maintained as intraspecific hybrids. In both cases, maintaining heterozygosity is a key factor contributing to enhanced growth phenotypes, also known as hybrid vigor or heterosis127. The role of small RNAs during hybridization has been extensively studied in many plant species (reviewed in 128 and 129), including stable cultivated Arabidopsis spp. allopolyploids generated by crossing tetraploid A. thaliana with A. arenosa, which resembles the natural allotetraploid A. suecica. Small RNAs, particularly miRNAs and tasiRNAs, were recognized as important genetic regulators of Arabidopsis allopolyploids130. Also in introgression lines derived from cultivated and wild relatives of tomato, miR395 was associated with beneficial transgressive phenotypes associated with salt tolerance131. Paramutation is likely involved in this process as well, but previous studies in maize, rice, tomato and A. thaliana have reported contradictory evidence in this respect. In maize, loss of hetsiRNAs in mop1 mutants (ortholog to RDR2 in A. thaliana) does not influence hybrid vigor in reciprocal intraspecific crosses132. In fact, down-regulation of 24-nucleotide siRNAs in F1 hybrids seems to be a common observation at loci where both parents show differential accumulation of small RNAs129. These loci include genes and promoter regions that are often occupied by transposable elements, thus the loss of 24-nucleotide siRNAs could result in lower levels of DNA methylation and up-regulation of genes responsible for heterosis phenotypes. Intraspecific hybridization between ecotypes of A. thaliana had strikingly contrasting results, as increased levels of DNA methylation were observed in reciprocal crosses while 24-nucleotide siRNAs levels were unchanged133. In tomato, small RNAs were more abundant in introgression lines than in each parent and are associated with suppression of target genes and DNA hypermethylation131, reminiscent of paramutation. Although these reports suggest a role for small RNAs during hybridization of genetically and epigenetically distinct genomes, their importance for growth and vigor remains largely unknown.
Epigenetic variation, polyploidy and small RNA diversity can ultimately lead to strong hybridization barriers in wide crosses, where transposon activity and genomic imprinting seem to respond in a parent- and dosage-dependent manner134. A simple illustration of small RNA-derived hybrid incompatibility in A. thaliana, is the truncated duplication of the essential gene FOLT1, which segregated within natural strains135. This truncated copy is able to produce siRNAs and trigger heritable silencing of active full-length copies elsewhere in the genome in trans, resulting in allelic incompatibilities in hybrid lines135. Similar observations in maize136,137 suggest that segregation of epialleles and duplicate alleles may also be relevant in hybrid crops.
DNA damage repair
RNAi-mediated DNA damage repair occurs in plants138, which goes in line with previous studies in the fungus Neurospora crassa139 and S. pombe140, in which rDNA-derived small RNAs and centromeric small RNAs, respectively, are induced upon DNA damage. Furthermore, both Dicer and AGO mutants in S. pombe are synthetic lethal with the key homologous repair protein Rad51 and have DNA damage response phenotypes141. Both AGO2 and AGO9 have a detectable effect on DNA repair efficiency in A. thaliana138,142, where 21- and 24-nucleotide double-strand-break-induced siRNAs (diRNAs) are induced in the vicinity of double strand breaks (DSBs) and are produced by DCL2, DCL3 and DCL4138. The additional requirement of RDRs and Pol IV for diRNA biogenesis also suggests that de novo transcription and dsRNA amplification mechanisms are involved in DSB repair, although small amounts of diRNAs provide sufficient repair capacity in the absence of RDRs138. Notably, a role for small RNAs and AGO in DSB repair and specifically in homologues recombination was also found in human cells, Drosophila melanogaster, and S. pombe141,143–145, suggesting an important and conserved role for small RNAs in DSB repair, possibly recruiting other protein complexes to DSB sites138,146.
Small RNA in meiosis and gametogenesis
Some AGO proteins are preferentially expressed in reproductive tissues and enriched in germline cells147–150, with specialized functions in chromosome segregation and cell fate specification. Small RNA sorting into different AGO proteins also contributes to functional diversity (Box 1), as unlike miRNAs, secondary siRNA duplexes have perfect complementarity between guide and passenger strands, so that sorting into different AGOs must rely exclusively on their 5′ terminal nucleotides. In rice and maize, phasiRNAs derived from thousands of non-coding precursors accumulate in meiotic and pre-meitoic cells. In A. thaliana, loss of heterochromatin in the vegetative nucleus is accompanied by accumulation of retrotransposon and other transposon easiRNAs in sperm cells78. In each case, intercellular transport is implicated in germline accumulation of small RNAs78 (Box 2). The potential functions of these small RNAs in meiosis and gametogenesis remain enigmatic but are now being explored.
A recent study showed that 21-nucleotide phasiRNAs in maize anthers are expressed in meiocytes, and decline during gametophytic development, whereas 24-nucleotide phasiRNAs accumulate throughout meiosis and remain abundant in mature pollen72 (Fig. 5a). A subset of 21-nucleotide phasiRNAs is loaded onto the MEIOSIS ARRESTED AT LEPTOTENE 1 (MEL1) in rice151, while its closest ortholog in maize is AGO5c, which seems to be expressed in a coordinated fashion with 21-nucleotide phasiRNAs in anthers72. By contrast, a binding partner of 24-nucleotide phasiRNAs has not yet been characterized, but transcriptional profiling suggests that AGO18b is the most promising candidate in maize, as it seems to be a recently evolved AGO, only found in monocot species72. Similarly in A. thaliana sperm cells, AGO1 (which binds most miRNAs and some secondary siRNAs) is largely replaced by its close homolog AGO5149. Null ago5 mutants are fertile in A. thaliana152, but mutations in MEL1 lead to early meiotic arrest and male sterility153. MEL1 localizes to the cytoplasm of pre-meiotic cells153, and like AGO5 in A. thaliana, shows selective binding of 5′ cytosine of small RNAs151. mel1 mutants have abnormal tapetum and aberrant pollen mother cells (PMC) that arrest in early meiosis153, suggesting that the subset of 21-nucleotide phasiRNAs bound to MEL1151 are crucial for male fertility. The function of phasiRNAs in monocot plant species remains a mystery as they have no obvious targets in the genomes72, but their peculiar accumulation dynamics observed in maize anthers is reminiscent of mammalian pachytene piRNAs, and might illustrate a possible convergent evolution of small RNAs in male gametogenesis154.
Figure 5. Small RNA functions in meiosis and cell fate specification.
a. In grass anthers, two distinct small RNA classes are produced from non-coding PHAS transcripts: 21-nucleotide (nt) phasiRNAs are produced upon cleavage of PHAS transcripts by miR2118, whereas miR2275 triggers 24-nucleotide phasiRNA biogenesis from a different subset of PHAS loci (reviewed in 65). The spatiotemporal dynamics of phasiRNA biogenesis was recently described throughout anther development in maize72, showing a distinct and mostly non-overlapping accumulation patterns for both phasiRNA classes, which nicely coincides with the expression of their respective miRNA triggers. 21-nucleotide phasiRNAs are essentially pre-meiotic, whereas 24-nucleotide phasiRNAs peak during meiosis and decrease during pollen development. The function of these male-specific small RNAs remains unknown, but their different size and accumulation patterns suggest distinct biological activities. A subset of 21-nt phasiRNAs in rice is loaded onto the MEIOSIS ARRESTED AT LEPTONENE1 (MEL1) protein151, which is the ortholog of AGO5 in Arabidopsis thaliana. The mel1 mutants arrest during early meiotic stages, and produce dysfunctional pollen mother cells (PMCs) that appear frequently in developing anthers. b. ARGONAUTE (AGO) functions in meiosis, cell specification and chromosome segregation. (Left) In the female gametophyte, AGO104 in maize and AGO9 in A. thaliana were associated with non-cell-autonomous regulation of meiosis and germline specification, but the molecular pathways responsible for that are still unclear148,150. Despite both being expressed in companion cells, AGO104 and AGO9 are involved in epigenetic silencing of transposable elements in the megaspore mother cells (MMC), perhaps through RdDM activity and mobile small RNA148,150. (Right) Importantly, ago104 mutants also produce viable unreduced diploid gametes, indicating that AGO104 has a role meiotic chromosome segregation and establishing a direct link between small RNA regulation and apomixis148. Top image in panel a adapted from 72. Right Image in panel b reproduced from 148 (arrowheads indicate micronuclei in abnormal tetrads).
Although phasiRNA and easiRNA biogenesis have only been reported in anthers and pollen, AGO5 and MEL1 are also expressed in ovules where other small RNA pathways have critical roles in reproduction. In maize, AGO104 (ortholog of AGO9 in A. thaliana) accumulates specifically in ovule somatic cells surrounding female meiocytes, and is involved in non-CG DNA methylation in heterochromatin148. In ago104 mutants chromosome segregation is blocked during meiosis I and diploid female gametes arise at high frequencies148 (Fig. 5b), whereas the formation of triads and microspores with multiple nuclei was also observed during male meiosis148. In A. thaliana, AGO9 binds 24-nucleotide siRNAs111 and silences transposable elements in the egg cell150. AGO104 and AGO9 are active in somatic cells and regulate cell fate specification in a non-cell autonomous manner. AGO104 represses somatic cell fate in germ cells148; conversely, AGO9 prevents sub-epidermal cells from adopting a megaspore-like identity150 (Fig. 5b). These findings demonstrate a crucial role for small RNAs and epigenetic regulation during sexual reproduction in higher plants, and highlight an important link to apomictic development. Thus understanding these mechanisms might provide an excellent opportunity to use apomixis as a fast and efficient way to fix hybrid genotypes in crop species.
Conclusions and future perspectives
The diversification and specialization of gene silencing networks in plants is likely reflecting an important role for small RNAs in adaptation to a sessile life style. However, it remains unclear the contribution of most small RNA classes upon biotic and abiotic stress, as well as the transgenerational inheritance and stability of acquired small RNA-based responses. Most of our current understating of small RNA activity in plants comes from their prominent functions in plant development, starting from an essential role during the first embryonic divisions up until the regulation of meiosis and gametogenesis. Despite the extensive functional diversity in different plant species, the several pathways for small RNA biogenesis and function are evolutionary related, relying on tissue-specific expression patterns and sophisticated mechanisms to sort small RNA duplexes onto specific AGO proteins. We have been able to depict the complex molecular mechanisms involved in small RNA biogenesis and function in plants, but a complete understanding of the specificities and interplay between the different gene silencing machineries operating in plant cells will remain difficult until we are able to profile small RNAs in isolated cell-types and single cells. These future challenges are well underway, and will provide important new insight into small RNA-based gene regulation in a variety of cellular, developmental and transgenerational contexts.
Online summary.
Functional diversification and expansion of silencing pathways in plants relies on duplication of Dicer and Argonaute proteins.
The main small RNA classes in plants are microRNAs (miRNAs), 21 to 22-nucleotide secondary small-interfering RNAs (siRNAs) and 24-nucleotide heterochromatic siRNAs (hetsiRNAs).
All small RNAs in plants are modified at their 3′ end by 2′-O-methylation, including miRNAs, which lack this modification in animals. This modification confers stability and protection from degradation.
Plant miRNAs are mainly involved in post-transcriptional gene silencing (PTGS) by transcript cleavage or translational repression, and also trigger secondary siRNA production from Pol II transcripts.
Secondary siRNAs are produced as 21 and 22-nucleotide small RNAs involved in cleavage or translational repression or target transcripts in cis and trans. They are also able to initiate transcriptional gene silencing (TGS) by establishing DNA methylation at particular loci.
The majority of siRNAs in plants are 24-nucleotide heterochromatic siRNAs and are involved in silencing repeats and transposable elements by RNA-directed DNA methylation (RdDM).
Small RNAs in plants are involved in reproductive transitions such as meiosis and gametogenesis, and regulate important epigenetic mechanisms such as genomic imprinting and paramutation.
Acknowledgments
We thank Jean-Sébastien Parent for critically reading the manuscript, and all the members of the Martienssen lab for insightful discussions. Research in the Martienssen laboratory is supported by the National Institute of Health (NIH) grant R01 GM067014 and Gordon and Betty Moore Foundation (HHMI-GBMF).
Glossary
- Wide crosses
Crosses of related species or genera that naturally do not sexually reproduce with each other.
- Intraspecific hybrids
Genetically divergent plants from the same species
- Introgression lines
Population containing genetic material derived from similar species or wild relatives. They are generally produced through successive backcrossing and selection of single introgressed genomic segments from one of the parental lines.
- Dicer-like (DCL)
Plant orthologs of other eukaryotic ribonucleose III Dicer enzymes; required for small RNA processing.
- Argonaute (AGO)
The main effector proteins of gene silencing, which bind small RNA duplexes and promote small RNA-mediated target recognition and cleavage.
- RNA-induced silencing complex
A protein complex that includes Argonaute proteins and small RNAs. The small RNAs hybridize to complementary target RNAs, which then undergo cleavage or translational repression, or recruit other factors, such as chromatin modifiers.
- Paramutation
An inter-chromosomal sensing mechanism that initiates heritable epigenetic changes in trans. Small RNAs are often involved in this process by mediating RNA-directed DNA methylation (RdDM).
- Transgressive phenotypes
Phenotypes in a hybrid progeny that are either superior or inferior to both parents. Transgressive phenotypes might facilitate hybrid specialization and are particularly important in crops when hybrid yields are higher than those of each parent.
- piwi-interacting RNAs (piRNAs)
Large class of small RNAs produced in animal cells, which form functional silencing complexes by loading onto piwi proteins. piRNA complexes are mainly involved in the post-transcriptional gene silencing of retrotransposons in the germline.
- Exosome
Multi-protein complex involved in 3′ to 5′ degradation of RNA molecules such as mRNAs or rRNAs.
- Processing bodies (P-bodies)
Cytoplasmic foci that have essential roles in most mRNA decay mechanisms, including decapping and non-sense mediated decay, as well as in storing processed mRNAs to postpone their translation.
- siRNA-bodies
Cytoplasmic foci in plant cells where RDR-DEPENDENT RNA POLYMERASE 6 (RDR6) and SUPPRESSOR OF GENE SILENCING 3 (RDR3) synthesize double-stranded RNA from single-stranded RNA.
- Epiallele
A genetic locus where transcriptional activity is regulated by epigenetic silencing marks such as DNA methylation and histone modifications.
- Interspecific allopolyploids
Polyploid organisms with two or more sets of genetically distinct chromosomes, which results from the cross between different species.
- Apomixis
Natural ability of certain plants species to reproduce asexually through seed, producing offspring genetically identical to the parental plant.
Biographies
Filipe Borges is a postdoctoral fellow in Rob Martienssen’s laboratory at the Cold Spring Harbor Laboratory, New York, USA. He is studying reprogramming and transgenerational silencing of transposable elements in pollen by single-cell genomics and epigenomics, with a focus on the evolution of epigenetic regulation in eukaryotic systems. His training and graduate research at the Gulbenkian Institute in Lisbon, Portugal, focused on understanding the molecular mechanisms regulating germline specification in Arabidopsis thaliana, and developing new tools for purification of plant cells by fluorescence-activated cell sorting.
Robert A. Martienssen leads the plant biology group at Cold Spring Harbor Laboratory, where he focuses on epigenetic mechanisms that shape and regulate the genome and their impact on development and inheritance. His work on transposons, or ‘jumping genes’, in plants and in fission yeast revealed a link between heterochromatin and RNA interference. His laboratory currently focuses on mechanistic aspects of germline reprogramming and epigenetic inheritance, including DNA methylation, histone replacement and modification, and RNA interference.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- 2.Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nature Genetics. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 3.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially Redundant Functions of Arabidopsis DICER-like Enzymes and a Role for DCL4 in Producing trans-Acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee K, Campos H, Kolaczkowski B. Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol. 2013;30:627–641. doi: 10.1093/molbev/mss263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willmann MR, Endres MW, Cook RT, Gregory BD. The Functions of RNA-Dependent RNA Polymerases in Arabidopsis. Arabidopsis Book. 2011;9:e0146. doi: 10.1199/tab.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nature Rev Genet. 2010;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg DE, Nakanishi K, Patel DJ, Bartel DP. The inside-out mechanism of Dicers from budding yeasts. Cell. 2011;146:262–276. doi: 10.1016/j.cell.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- 9.Nodine MD, Bartel DP. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010;24:2678–2692. doi: 10.1101/gad.1986710. Demonstrates that a miRNA (miR156) plays an essential role in the early development of A. thaliana embryos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- 11.Liu B, et al. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005;139:296–305. doi: 10.1104/pp.105.063420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson BE, et al. The dicer-like1 Homolog fuzzy tassel Is Required for the Regulation of Meristem Determinacy in the Inflorescence and Vegetative Growth in Maize. Plant Cell. 2014;26:4702–4717. doi: 10.1105/tpc.114.132670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. Plant Cell. 2011;23:431–442. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahlgren N, et al. High-Throughput Sequencing of Arabidopsis microRNAs: Evidence for Frequent Birth and Death of MIRNA Genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chellappan P, et al. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. 2010;38:6883–6894. doi: 10.1093/nar/gkq590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vazquez F, Blevins T, Ailhas J, Boller T, Meins F. Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 2008;36:6429–6438. doi: 10.1093/nar/gkn670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu L, et al. DNA Methylation Mediated by a MicroRNA Pathway. Mol Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben Amor B, et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19:57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen E, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nature Genetics. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 21.Fahlgren N, et al. MicroRNA Gene Evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell. 2010;22:1074–1089. doi: 10.1105/tpc.110.073999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Axtell MJ, Westholm JO, Lai EC. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piriyapongsa J, Jordan IK. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA. 2008;14:814–821. doi: 10.1261/rna.916708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Jiang WK, Gao LZ. Evolution of microRNA genes in Oryza sativa and Arabidopsis thaliana: an update of the inverted duplication model. PLoS ONE. 2011;6:e28073. doi: 10.1371/journal.pone.0028073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moissiard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zong J, Yao X, Yin J, Zhang D, Ma H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups. Gene. 2009;447:29–39. doi: 10.1016/j.gene.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Ruiz H, et al. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell. 2010;22:481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nature Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nature Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 31.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nature Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 32.Parent JS, Bouteiller N, Elmayan T, Vaucheret H. Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 2015;81:223–232. doi: 10.1111/tpj.12720. [DOI] [PubMed] [Google Scholar]
- 33.Wang XB, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:484–489. doi: 10.1073/pnas.0904086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiraguri A, et al. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 36.Kim YK, Heo I, Kim VN. Modifications of Small RNAs and Their Associated Proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhai J, et al. Plant MicroRNAs Display Differential 3′ Truncation and Tailing Modifications That Are ARGONAUTE1 Dependent and Conserved Across Species. Plant Cell. 2013;25:2417–2428. doi: 10.1105/tpc.113.114603. This study addresses the prevalence, conservation and biological significance of truncated and uridylated miRNA variants in plants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y, et al. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren G, Chen X, Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr Biol. 2012;22:695–700. doi: 10.1016/j.cub.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ibrahim F, et al. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci USA. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyman SK, et al. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Heo I, et al. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 49.van Wolfswinkel JC, et al. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 50.Shirayama M, et al. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev Cell. 2013;27:664–671. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 52.Ren G, et al. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc Natl Acad Sci USA. 2014;111:6365–6370. doi: 10.1073/pnas.1405083111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, et al. Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3′ Tailing of Small RNAs in Arabidopsis. PLoS Genet. 2015;11:e1005091. doi: 10.1371/journal.pgen.1005091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tu B, et al. Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in Arabidopsis. PLoS Genet. 2015;11:e1005119. doi: 10.1371/journal.pgen.1005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryvkin P, et al. HAMR: high-throughput annotation of modified ribonucleotides. RNA. 2013;19:1684–1692. doi: 10.1261/rna.036806.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ebhardt HA, et al. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iida K, Jin H, Zhu JK. Bioinformatics analysis suggests base modifications of tRNAs and miRNAs in Arabidopsis thaliana. BMC Genomics. 2009;10:155. doi: 10.1186/1471-2164-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan J, Zhang H, Zheng Y, Ding Y. Comparative expression profiling of miRNAs between the cytoplasmic male sterile line MeixiangA and its maintainer line MeixiangB during rice anther development. Planta. 2015;241:109–123. doi: 10.1007/s00425-014-2167-2. [DOI] [PubMed] [Google Scholar]
- 59.Kierzek E, et al. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42:3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwartz S, et al. Transcriptome-wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simos G, et al. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 63.Hellmuth K, et al. Cloning and characterization of the Schizosaccharomyces pombe tRNA:pseudouridine synthase Pus1p. Nucleic Acids Res. 2000;28:4604–4610. doi: 10.1093/nar/28.23.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 67.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A Two-Hit Trigger for siRNA Biogenesis in Plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 68.Manavella PA, Koenig D, Weigel D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci USA. 2012;109:2461–2466. doi: 10.1073/pnas.1200169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ronemus M, Vaughn MW, Martienssen RA. MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell. 2006;18:1559–1574. doi: 10.1105/tpc.106.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson C, et al. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009;19:1429–1440. doi: 10.1101/gr.089854.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arikit S, Zhai J, Meyers BC. Biogenesis and function of rice small RNAs from non-coding RNA precursors. Curr Opin Plant Biol. 2013;16:170–179. doi: 10.1016/j.pbi.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 72.Zhai J, et al. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci USA. 2015;112:3146–3151. doi: 10.1073/pnas.1418918112. Comprehensive spatiotemporal characterization of the biogenesis pathways and dynamics of two functionally uncharacterized, male-specific phasiRNA clusters in maize. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song X, et al. Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J. 2012;71:378–389. doi: 10.1111/j.1365-313X.2012.05001.x. [DOI] [PubMed] [Google Scholar]
- 74.Song X, et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012;69:462–474. doi: 10.1111/j.1365-313X.2011.04805.x. [DOI] [PubMed] [Google Scholar]
- 75.Shivaprasad PV, et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arikit S, et al. An Atlas of Soybean Small RNAs Identifies Phased siRNAs from Hundreds of Coding Genes. Plant Cell. 2014;26:4584–4601. doi: 10.1105/tpc.114.131847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Creasey KM, et al. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature. 2014;508:411–415. doi: 10.1038/nature13069. Shows that secondary siRNAs are produced from epigenetically activated transposable elements and that miRNAs are involved in this process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. Shows that transposons are expressed and active in the vegetative nuclei of pollen, producing secondary siRNAs that accumulate in the neighboring gametes, potentially reinforcing transgenerational epigenetic silencing of transposons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanurdzić M, et al. Epigenomic Consequences of Immortalized Plant Cell Suspension Culture. PloS Biol. 2008;6:e302. doi: 10.1371/journal.pbio.0060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ito H, et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472:115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 81.May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005;1:e79. doi: 10.1371/journal.pgen.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roberts JT, Cardin SE, Borchert GM. Burgeoning evidence indicates that microRNAs were initially formed from transposable element sequences. Mob Genet Elements. 2014;4:e29255. doi: 10.4161/mge.29255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dumesic PA, et al. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell. 2013;152:957–968. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Z, et al. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157:1353–1363. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Christie M, Croft LJ, Carroll BJ. Intron splicing suppresses RNA silencing in Arabidopsis. Plant J. 2011;68:159–167. doi: 10.1111/j.1365-313X.2011.04676.x. [DOI] [PubMed] [Google Scholar]
- 86.Laubinger S, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Christie M, Carroll BJ. SERRATE is required for intron suppression of RNA silencing in Arabidopsis. Plant Signal Behav. 2011;6:2035–2037. doi: 10.4161/psb.6.12.18238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raczynska KD, et al. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2014;42:1224–1244. doi: 10.1093/nar/gkt894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ausin I, Greenberg MVC, Li CF, Jacobsen SE. The splicing factor SR45 affects the RNA-directed DNA methylation pathway in Arabidopsis. Epigenetics. 2012;7:29–33. doi: 10.4161/epi.7.1.18782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li S, et al. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome es. 2015;25:235–245. doi: 10.1101/gr.182238.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bayne EH, et al. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322:602–606. doi: 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bernard P, Drogat J, Dheur S, Genier S, Javerzat JP. Splicing factor Spf30 assists exosome-mediated gene silencing in fission yeast. Mol Cell Biol. 2010;30:1145–1157. doi: 10.1128/MCB.01317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chinen M, Morita M, Fukumura K, Tani T. Involvement of the spliceosomal U4 small nuclear RNA in heterochromatic gene silencing at fission yeast centromeres. J Biol Chem. 2010;285:5630–5638. doi: 10.1074/jbc.M109.074393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kallgren SP, et al. The proper splicing of RNAi factors is critical for pericentric heterochromatin assembly in fission yeast. PLoS Genet. 2014;10:e1004334. doi: 10.1371/journal.pgen.1004334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gy I, et al. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19:3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 97.Moreno AB, et al. Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants. Nucleic Acids Res. 2013;41:4699–4708. doi: 10.1093/nar/gkt152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thran M, Link K, Sonnewald U. The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis. Plant J. 2012;72:368–377. doi: 10.1111/j.1365-313X.2012.05066.x. [DOI] [PubMed] [Google Scholar]
- 99.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 100.Zhang X, et al. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science. 2015;348:120–123. doi: 10.1126/science.aaa2618. [DOI] [PubMed] [Google Scholar]
- 101.Martínez de Alba AE, et al. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015;43:2902–2913. doi: 10.1093/nar/gkv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bühler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 103.Yamanaka S, et al. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493:557–560. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cao M, et al. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:14613–14618. doi: 10.1073/pnas.1407131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang XB, et al. The 21-Nucleotide, but Not 22-Nucleotide, Viral Secondary Small Interfering RNAs Direct Potent Antiviral Defense by Two Cooperative Argonautes in Arabidopsis thaliana. Plant Cell. 2011;23:1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumakura N, et al. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Letters. 2009;583:1261–1266. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 107.McClintock B. The suppressor-mutator system of control of gene action in maize. Carnegie Institution of Washington Year Book. 1958;57:415–429. [Google Scholar]
- 108.Nuthikattu S, et al. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 2013;162:116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McCue AD, et al. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 2015;34:20–35. doi: 10.15252/embj.201489499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mari-Ordonez A, et al. Reconstructing de novo silencing of an active plant retrotransposon. Nature Genetics. 2013;45:1029–1039. doi: 10.1038/ng.2703. A study of proliferation dynamics and molecular pathways involved in restoring transcriptional silencing of a functionally active retrotransposon in A. thaliana. [DOI] [PubMed] [Google Scholar]
- 111.Havecker ER, et al. The Arabidopsis RNA-Directed DNA Methylation Argonautes Functionally Diverge Based on Their Expression and Interaction with Target Loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. Shows that the functional diversification of AGO4, AGO6 and AGO9 and their preference for certain RdDM-mediating siRNAs relies on cell- and tissue-specific gene expression patterns. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nature Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 114.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nature Rev Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- 116.Hollick JB. Paramutation: a trans-homolog interaction affecting heritable gene regulation. Curr Opin Plant Biol. 2012;15:536–543. doi: 10.1016/j.pbi.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 117.Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nature Rev Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- 118.Luff B, Pawlowski L, Bender J. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol Cell. 1999;3:505–511. doi: 10.1016/s1097-2765(00)80478-5. [DOI] [PubMed] [Google Scholar]
- 119.Melquist S, Luff B, Bender J. Arabidopsis PAI gene arrangements, cytosine methylation and expression. Genetics. 1999;153:401–413. doi: 10.1093/genetics/153.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Erhard KF, Hollick JB. Paramutation: a process for acquiring trans-generational regulatory states. Curr Opin Plant Biol. 2011;14:210–216. doi: 10.1016/j.pbi.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 121.Belele CL, et al. Specific tandem repeats are sufficient for paramutation-induced trans-generational silencing. PLoS Genet. 2013;9:e1003773. doi: 10.1371/journal.pgen.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Du J, et al. Dual Binding of Chromomethylase Domains to H3K9me2-Containing Nucleosomes Directs DNA Methylation in Plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. This study establishes the first direct link between H3K9me2 and Pol IV recruitment to methylated DNA via SHH1, as an essential step to initiate and/or maintain RdDM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Johnson LM, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507:124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bond DM, Baulcombe DC. Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2015;112:917–922. doi: 10.1073/pnas.1413053112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Washburn JD, Birchler JA. Polyploids as a ‘model system’ for the study of heterosis. Plant Reprod. 2014;27:1–5. doi: 10.1007/s00497-013-0237-4. [DOI] [PubMed] [Google Scholar]
- 127.Chen ZJ. Genomic and epigenetic insights into the molecular bases of heterosis. Nature Rev Genet. 2013;14:471–482. doi: 10.1038/nrg3503. [DOI] [PubMed] [Google Scholar]
- 128.Ng DWK, Lu J, Chen ZJ. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr Opin Plant Biol. 2012;15:154–161. doi: 10.1016/j.pbi.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 129.Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES. The role of epigenetics in hybrid vigour. Trends Genet. 2013;29:684–690. doi: 10.1016/j.tig.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 130.Ha M, et al. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA. 2009;106:17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC. Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 2011;31:257–266. doi: 10.1038/emboj.2011.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Barber WT, et al. Repeat associated small RNAs vary among parents and following hybridization in maize. Proc Natl Acad Sci USA. 2012;109:10444–10449. doi: 10.1073/pnas.1202073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shen H, et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell. 2012;24:875–892. doi: 10.1105/tpc.111.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Josefsson C, Dilkes B, Comai L. Parent-Dependent Loss of Gene Silencing during Interspecies Hybridization. Curr Biol. 2006;16:1322–1328. doi: 10.1016/j.cub.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 135.Durand S, Bouché N, Perez Strand E, Loudet O, Camilleri C. Rapid Establishment of Genetic Incompatibility through Natural Epigenetic Variation. Curr Biol. 2012;22:326–331. doi: 10.1016/j.cub.2011.12.054. [DOI] [PubMed] [Google Scholar]
- 136.Settles AM, Baron A, Barkan A, Martienssen RA. Duplication and suppression of chloroplast protein translocation genes in maize. Genetics. 2001;157:349–360. doi: 10.1093/genetics/157.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Slotkin RK, Freeling M, Lisch D. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nature Genetics. 2005;37:641–644. doi: 10.1038/ng1576. First example of a transposon-derived inverted repeat that is able to establish heritable epigenetic silencing of active transposons in the maize genome. [DOI] [PubMed] [Google Scholar]
- 138.Wei W, et al. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. Demonstrates the involvement of small RNAs and AGO2 in DSB repair in A. thaliana. [DOI] [PubMed] [Google Scholar]
- 139.Lee HC, et al. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA Interference Guides Histone Modification during the S Phase of Chromosomal Replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zaratiegui M, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Oliver C, Santos JL, Pradillo M. On the role of some ARGONAUTE proteins in meiosis and DNA repair in Arabidopsis thaliana. Front Plant Sci. 2014;5:177. doi: 10.3389/fpls.2014.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Michalik KM, Böttcher R, Förstemann K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012;40:9596–9603. doi: 10.1093/nar/gks711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Francia S, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chiolo I, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gao M, et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014;24:532–541. doi: 10.1038/cr.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kapoor M, et al. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics. 2008;9:451. doi: 10.1186/1471-2164-9-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Singh M, et al. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell. 2011;23:443–458. doi: 10.1105/tpc.110.079020. This paper describes a genetic screen in maize that uncovered ago104 mutants that mimic apomixis by forming viable unreduced gametes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Borges F, Pereira PA, Slotkin RK, Martienssen RA, Becker JD. MicroRNA activity in the Arabidopsis male germline. J Exp Bot. 2011;62:1611–1620. doi: 10.1093/jxb/erq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Olmedo-Monfil V, et al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. Describes a non-cell-autonomous function of small RNAs and AGO9 during differentiation and specification of female gametes in A. thaliana. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Komiya R, et al. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014;78:385–397. doi: 10.1111/tpj.12483. [DOI] [PubMed] [Google Scholar]
- 152.Tucker MR, et al. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development. 2012;139:1399–1404. doi: 10.1242/dev.075390. [DOI] [PubMed] [Google Scholar]
- 153.Nonomura KI, et al. A Germ Cell Specific Gene of the ARGONAUTE Family Is Essential for the Progression of Premeiotic Mitosis and Meiosis during Sporogenesis in Rice. Plant Cell. 2007;19:2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fu Q, Wang PJ. Mammalian piRNAs: Biogenesis, function, and mysteries. Spermatogenesis. 2014;4:e27889. doi: 10.4161/spmg.27889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Poulsen C, Vaucheret H, Brodersen P. Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures. Plant Cell. 2013;25:22–37. doi: 10.1105/tpc.112.105643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mi S, et al. Sorting of Small RNAs into Arabidopsis Argonaute Complexes Is Directed by the 5′ Terminal Nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. First study showing that biased small RNA sorting into Argonaute proteins in Arabidopsis is regulated by the 5′ terminal nucleotides of small RNA duplexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 158.Frank F, Hauver J, Sonenberg N, Nagar B. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J. 2012;31:3588–3595. doi: 10.1038/emboj.2012.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhu H, et al. Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to Regulate Shoot Apical Meristem Development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. Demonstrated that the preference of AGO10 for miR165 and miR166 relies on the structure of the mature small RNA duplex, and is essential to promote shoot apical meristem development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu Q, et al. The ARGONAUTE10gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 2009;58:27–40. doi: 10.1111/j.1365-313X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- 161.Ji L, et al. ARGONAUTE10 and ARGONAUTE1 Regulate the Termination of Floral Stem Cells through Two MicroRNAs in Arabidopsis. PLoS Genet. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 163.Endo Y, Iwakawa H-O, Tomari Y. Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 2013;14:652–658. doi: 10.1038/embor.2013.73. Reports the extensive characterization of structural determinants in the conserved miR390 mature duplex, and their importance for preferential loading onto AGO7 and the formation of a functional RISC complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang X, et al. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell. 2011;42:356–366. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang X, et al. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nature Comms. 2014;5:5468. doi: 10.1038/ncomms6468. This paper provides further mechanistic insight and functional significance for sorting both guide and passenger strands of miRNA duplexes into AGO1 and AGO2, respectively. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chitwood DH, et al. Pattern formation via small RNA mobility. Genes Dev. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Molnar A, et al. Small Silencing RNAs in Plants Are Mobile and Direct Epigenetic Modification in Recipient Cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 168.Skopelitis DS, Husbands AY, Timmermans MCP. Plant small RNAs as morphogens. Curr Opin Cell Biol. 2012;24:217–224. doi: 10.1016/j.ceb.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 169.Carlsbecker A, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sarkies P, Miska EA. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nature Rev Mol Cell Biol. 2014;15:525–535. doi: 10.1038/nrm3840. [DOI] [PubMed] [Google Scholar]
- 171.Mosher RA, et al. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 172.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vu TM, et al. RNA-directed DNA methylation regulates parental genomic imprinting at several loci in Arabidopsis. Development. 2013;140:2953–2960. doi: 10.1242/dev.092981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 2010;24:1010–1021. doi: 10.1101/gad.1882810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Felippes FF, Weigel D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Rep. 2009;10:264–270. doi: 10.1038/embor.2008.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.McCue AD, Nuthikattu S, Reeder SH, Slotkin RK. Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA. PLoS Genet. 2012;8:e1002474. doi: 10.1371/journal.pgen.1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]