Abstract
Background aims
To develop a treatment option for Philadelphia chromosome—positive acute lymphoblastic leukemia (Ph+ALL) resistant to tyrosine kinase inhibitors (TKIs), we evaluated the anti-leukemic activity of T cells non-virally engineered to express a CD19-specific chimeric antigen receptor (CAR).
Methods
A CD19.CAR gene was delivered into mononuclear cells from 10 mL of blood of healthy donors through the use of piggyBac-transposons and the 4-D Nucleofector System. Nucleofected cells were stimulated with CD3/CD28 antibodies, magnetically selected for the CD19.CAR, and cultured in interleukin-15–containing serum-free medium with autologous feeder cells for 21 days. To evaluate their cytotoxic potency, we co-cultured CAR T cells with seven Ph+ALL cell lines including three TKI-resistant (T315I–mutated) lines at an effector-to-target ratio of 1:5 or lower without cytokines.
Results
We obtained ~ 1.3 × 108 CART cells (CD4+, 25.4%; CD8+, 71.3%), co-expressing CD45RA and CCR7 up to ~80%. After 7-day co-culture, CAR T cells eradicated all tumor cells at the 1:5 and 1:10 ratios and substantially reduced tumor cell numbers at the 1:50 ratio. Kinetic analysis revealed up to 37-fold proliferation of CART cells during a 20-day culture period in the presence of tumor cells. On exposure to tumor cells, CAR T cells transiently and reproducibly upregulated the expression of transgene as well as tumor necrosis factor–related apoptosis-inducing ligand and interleukin-2.
Conclusions
We generated a clinically relevant number of CAR T cells from 10 mL of blood through the use of piggyBac-transposons, a 4D-Nulcleofector, and serum/xeno/tumor cell/virus-free culture system. CAR T cells exhibited marked cytotoxicity against Ph+ALL regardless of T315I mutation. PiggyBac-mediated CD19-specific T-cell therapy may provide an effective, inexpensive and safe option for drug-resistant Ph+ALL.
Keywords: piggyBac-transposon, CAR, Ph+ALL, tyrosine kinase inhibitor, T315I
Introduction
Philadelphia chromosome–positive (Ph+) acute lymphoblastic leukemia (ALL) characterized by the existence of a BCR-ABL fusion gene is observed in 20–30% of adult patients and in 2–3% of pediatric patients with ALL (1). Although the introduction of BCR-ABL tyrosine kinase inhibitors (TKIs) into conventional chemotherapy has improved the outcomes (1), TKI resistance, mainly because of the emergence of mutations in the BCR-ABL kinase domain, remains a problem in a substantial proportion of patients with Ph+ALL (2). In particular, T315I–mutated leukemic clones are resistant to all approved TKIs (imatinib, nilotinib and dasatinib). Consequently, the outcome of patients with T315I mutations is very poor (3). An alternative, effective and safe option is needed for patients with TKI-resistant Ph+ALL.
Adoptive immunotherapy with the use of T cells expressing a chimeric antigen receptor (CAR) targeting CD19 (CD19.CAR) is a novel approach for the treatment of B-cell malignancies. Several clinical trials have proved that adoptive transfer of T cells retrovirally or lentivirally engineered to express the CD19.CAR is effective for the treatment of refractory/relapsed chronic lymphocytic leukemia and follicular lymphoma. Although when effective, this treatment eliminates normal B cells and can cause severe cytokine release syndrome, both adverse events are manageable (4–8). More recently, two groups demonstrated successful treatment of relapsed B-cell precursor ALL (not including Ph+ALL) through the use of CD19.CAR-modified T cells (9,10). Brentjens et al. (9) reported five adult patients in whom CD19.CAR-modified T cells induced complete remission with clearance of minimal residual disease. In a report of Grupp et al. (10), two pediatric patients achieved complete hematologic remission after transfer of CD19.CAR-modified T cells. One patient continued in complete molecular remission for at least 11 months after treatment, whereas the other relapsed at 2 months.
To save the labor, reduce the cost and improve the safety of CAR-modified T-cell therapy, we developed a non-viral gene-transfer method into T cells through the use of the piggyBac-transposon system (11–17). In the present study, we transferred a CD19.CAR gene into T cells with the use of the piggyBac-transposon system in combination with a 4D-Nucleofector system and expanded CD19.CAR-modified T cells under serum/animal-derived component-free, tumor cell/virus-infected cell-free culture conditions to facilitate the regulatory approval of CD19.CAR-modified T-cell therapy. The transgenic T cells exerted anti-leukemic activity against TKI-resistant Ph+ALL cells as well as TKI-sensitive Ph+ALL cells.
Methods
Plasmids
The piggyBac-transposase plasmid (pCMV-piggyBac) and the transposon plasmid for CD19.CAR (pIRII-CAR.CD19–28-ξ), have been described previously (11,15,18). Briefly, pIRII-CAR.CD19–28-ξ encodes a single chain variable fragment from an anti-CD19 antibody, likened to the hinge–CH2CH3 domain of human immunoglobulin (Ig)G1, the endodomains of a CD28 molecule and a T-cell receptor ξ chain. Both vectors are transcriptionally regulated by the cytomegalovirus (CMV) immediate early gene enhancer/ promoter sequence.
Blood donors and cell lines
Peripheral blood mononuclear cells (PBMCs) were obtained from three healthy adult volunteers with informed consent, approved by the Institutional Review Board of Shinshu University School of Medicine.
We used seven different Ph+ALL cell lines in this study: four cell lines, SU-Ph2 (19), TCC-Y (20), KOPN57bi (21) and KOPN30bi (21), were sensitive to TKIs, and three cell lines, SU/SR (19), TCC-Y/sr (20) and SK-9 (22), were resistant to TKIs as the result of a BCR-ABL T315I mutation. SU/SR and TCC-Y/sr were derived from SU-Ph2 and TCC-Y, respectively, by culture with imatinib (19,20). All seven Ph+ALL cell lines were maintained in Roswell Park Memorial Institute (RPMI) 1640 Medium, GlutaMAX (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Hyclone Laboratories, Logan, UT, USA). A myeloid leukemia cell line, K562, was also maintained in 10% serum-containing RPMI 1640.
Gene transfer into T cells and expansion of transgenic T cells
Unstimulated PBMCs obtained from 10 mL of peripheral blood of healthy donors were nucleofected with a pIRII-CAR.CD19–28-ξ plasmid (5 µg) and a pCMV-piggyBac plasmid (5 µg) through the use of the 4D-Nucleofector Device (Program EO-115) and P3 Primary Cell 4D-Nucleofector X Kit (Lonza, Basel, Switzerland). Nucleofected cells were maintained in serum-free and animal-derived component-free T-cell culture medium (TexMACS Medium; Miltenyi Biotec, Auburn, CA, USA) supplemented with recombinant human interleukin (IL)-15 (5 ng/ mL, Miltenyi Biotec) at 37°C in a humidified 5% CO2 incubator. The following day, cells were transferred and cultured in 24-well culture plates coated with CD3 monoclonal antibody (mAb) and CD28 mAb (Miltenyi Biotec) for 4 days. Six days after stimulation, cells were labeled with biotin-conjugated goat anti-human IgG (H+L) (Jackson ImmunoResearch, West Grove, PA, USA), which bound to the hinge-CH2CH3 domain of human IgG1 of the CD19.CAR, then selected for the CD19.CAR with Anti-Biotin MicroBeads (Miltenyi Biotec) and MACS Column (Miltenyi Biotec). The negatively selected cells, consisting of almost all CD19.CAR-negative activated T cells, were irradiated and plated as feeder cells. The positively selected cells were restimulated on CD3/CD28 mAb-coated wells with autologus feeder cells in TexMACS medium containing 5 ng/mL of IL-15 for 4 days, then transferred to a G-Rex 10 device (Wilson Wolf Manufacturing Inc, New Brighton, MN, USA) with 30 mL of IL-15–containing TexMACS for a further 10 days. IL-15–containing TexMACS was half-changed every 4 or 5 days during the culture period. The number of viable cells was determined by means of trypan blue exclusion test with the use of a hemocytometer at the indicated points. Twenty-one days after the start of culture, the final product was cryopreserved at −80°C for further studies (CAR T cells). As controls, non-transfected PBMCs were concurrently stimulated on CD3/CD28 mAb-coated plates and cultured in IL-15–containing TexMACS for 21 days (mock T cells).
Flow cytometric analysis
With the use of the BD FACSCalibur with BD Cell-Quest Pro software [Becton, Dickinson and Company (BD), Franklin Lakes, NJ, USA], we analyzed the surface markers of the expanded CAR T cells by use of allophycocyanin (APC)-conjugated CD3 mAb, phycoerythrin (PE)-conjugated CD4 mAb, APC-conjugated CD8 mAb, APC-conjugated CD45RO mAb, APC-conjugated CD45RA mAb, PE-conjugated CD56 mAb and PE-conjugated CD62L mAb, PE-conjugated CCR7 mAb (all mAbs were purchased from Miltenyi Biotec). The expression of CAR on T cells was examined by staining with APC-conjugated CD3 mAb and fluorescein isothiocyanate (FITC)-conjugated goat anti-human IgG (H+L) (Jackson ImmunoResearch). The relative fluorescence intensity (RFI) was determined by calculation of the ratio of mean fluorescence intensity for specific staining to that for control staining. The expression of tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) receptors on Ph+ALL cells were assessed by means of staining with APC-conjugated CD19 mAb (Miltenyi Biotec) and PE-conjugated mAb against DR4, or DR5 (purchased from Biolegend, San Diego, CA, USA). The expression of TRAIL on T cells was examined by means of staining with PE-conjugated CD253 (TRAIL) mAb (Biolegend). APC-, FITC- and PE-conjugated mouse isotype-matched IgG (Miltenyi Biotec or Biolegend) were used as controls in each analysis.
Cytotoxicity assay
To determine if CAR T cells were able to lyse target cells, we performed a 4-hour cytotoxicity assay with the use of the LDH Cytotoxicity Detection Kit (Takara Bio Inc, Otsu, Japan) according to the manufacturer’s protocol. As targets, we used CD19-positive Ph+ALL (SU/SR) cells and CD19-negative leukemia (K562) cells at effector to target cell (E:T) ratios from 4:1 to 1:4. As controls, we used mock T cells.
Analysis of karyotype and T-cell clonality of CAR T cells
To detect aberrant T-cell growth in the expanded CAR T cells, we analyzed the karyotype by G-banding and T-cell receptor (TCR) gene rearrangement in TCR-β and TCR-γ chains by multiplex polymerase chain reaction (PCR) as reported previously (14,23).
Quantification of integrated plasmids in CAR T cells
To investigate the integrated gene copy number of pIRII-CAR.CD19–28-ξ and pCMV-piggyBac plasmids in each T-cell product, we performed a real-time quantitative PCR (qPCR) assay targeting sequences specific for the promoter of CD 19.CAR and the piggy-Bac, respectively, in both plasmids, with the use of two sets of primers and a fluorogenic TaqMan probe containing fluorescein amidite (FAM) at its 5′-end and a quencher (minor groove binder, MGB) at its 3′-end: for the promoter of CD19.CAR, forward primer, 5′-ACGTGACTTTTAAGATTTAACTCATACGA-3’; reverse primer, 5′-CAATGAATAATATGGCTAAT GGCCA-3′; probe, FAM-CTTGTTATAGATAA GATCTTC-MGB; for the piggyBac, forward primer, 5′-AGGAACACAGACCAACGGAGTAC-3′; reverse primer, 5′-TGCACAGGCTTTGATAACTCCTT -3′; probe, FAM-ACTCGGTGAATACTACG-MGB.
We extracted DNA from 1.0 × 106 live cells, which were thawed and determined by trypan blue exclusion test, from each of the CAR T-cell products and mock T-cell products of three donors. qPCR reactions were run on an ABI 7900HT (Applied Biosystems, Foster City, CA, USA). Each assay was designed against a six-point standard curve constructed through the use of pIRII-CAR.CD19–28-ξ and pCMV-piggyBac plasmids, respectively, at concentrations ranging from 106 to 10 copies. The threshold cycle value for each sample was plotted on the standard curve, and the copy number of CD19.CAR and the piggyBac DNA was calculated and expressed as the copy number per cell. We confirmed the negativity in our assay by qPCR reactions by use of the opposite pair of the plasmids and the primer/probe sets and the use of DNA from mock T-cell lines. The detection limit of the qPCR assay was 1.0 × 10−4 of copies per cell.
Co-culture experiments
Anti-leukemic effects of CAR T cells on Ph+ALL cells were evaluated in co-culture experiments. Briefly, either CAR T cells or mock T cells were added to 5 × 105 Ph+ALL cells at an E:T ratio of 1:5 in wells of a 48-well culture plate containing RPMI 1640 plus 10% fetal bovine serum without cytokines and cultured for 20 days. At the indicated points, the number of viable cells was determined by trypan blue exclusion test, and the percentages of CAR T cells and Ph+ALL cells were determined with the use of APC-conjugated CD3 mAb and PE-conjugated CD19 mAb by flow cytometry. In concurrent experiments, we added Ph+ALL cells again to the co-cultures at the E:T ratio of 1:5 on day 10 and then observed them until day 20. As control, mock T cells, CAR T cells or Ph+ALL cells were cultured alone.
Additionally, we serially collected the culture supernatants at the indicated points and measured the concentrations of interferon (IFN)-γ and IL-2 by enzyme-linked immunosorbent assay (ELISA; Mitsubishi Chemical Medicine, Tokyo, Japan).
To test anti-leukemic effects of CAR T cells at lower E:T ratios, we also performed a 7-day co-culture experiment at ratios of 1:10, 1:50 and 1:100.
Quantification of BCR/ABL transcripts
To examine the clearance of Ph+ALL cells at the molecular level, we used real-time qPCR for the minor BCR/ABL messenger RNA (mRNA) according to the procedure reported by Gabert et al. (24). The primers and a fluorogenic TaqMan-probe containing FAM at its 5′-end and a quencher (TAMRA) at its 3′-end were as follows: forward primer, 5′-CTGGCCCAACGATGGCGA-3′; reverse primer, 5′-CACTCAGACCCTGAGGCTCAA-3′; probe 5′-FAM-CCCTTCAGCGGCCAGTAGCATCTGA-TAMRA-3′. The quantity of the BCR-ABL mRNA was normalized through the use of the level of β-glucuronidase mRNA.
Statistical analysis
The results from three donors are expressed as mean ± standard deviation. To determine the significance of difference between two independent groups, we used the unpaired t-test, and statistical significance was defined as P < 0.05.
Results
Generation of T cells modified to express CD19.CAR with piggyBac-transposons in serum/xeno-free and tumor cell/virus-infected cell-free culture
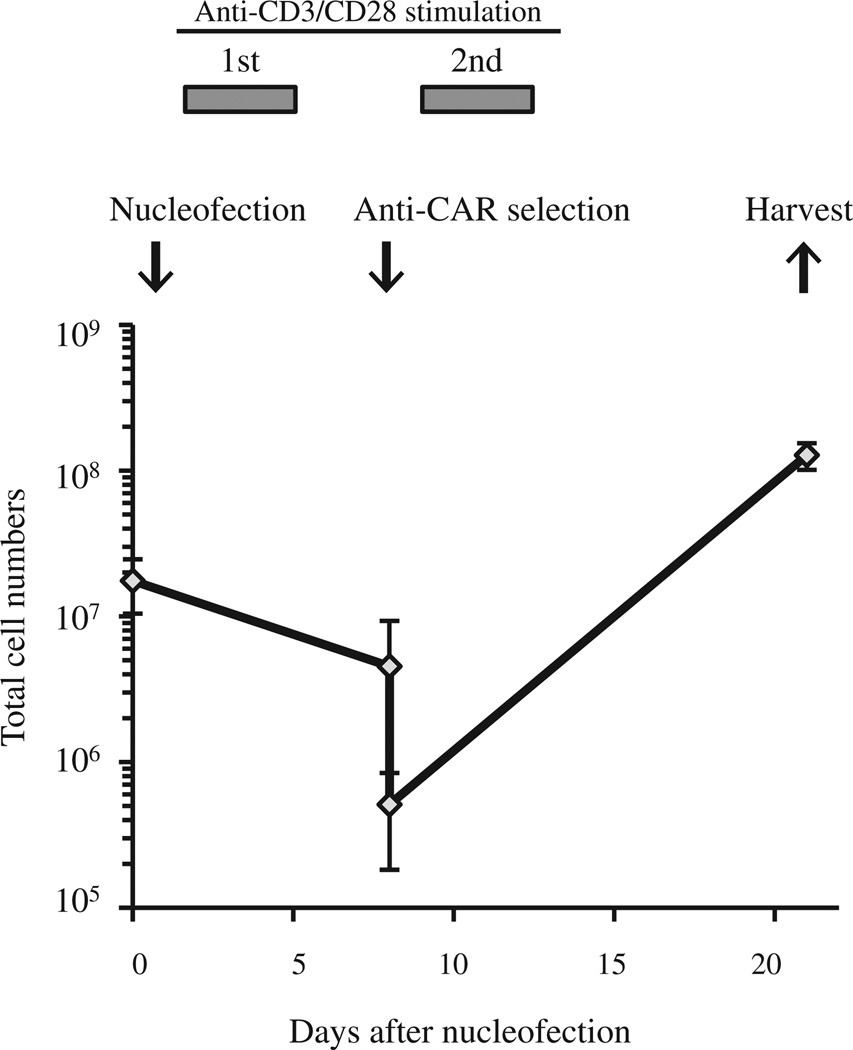
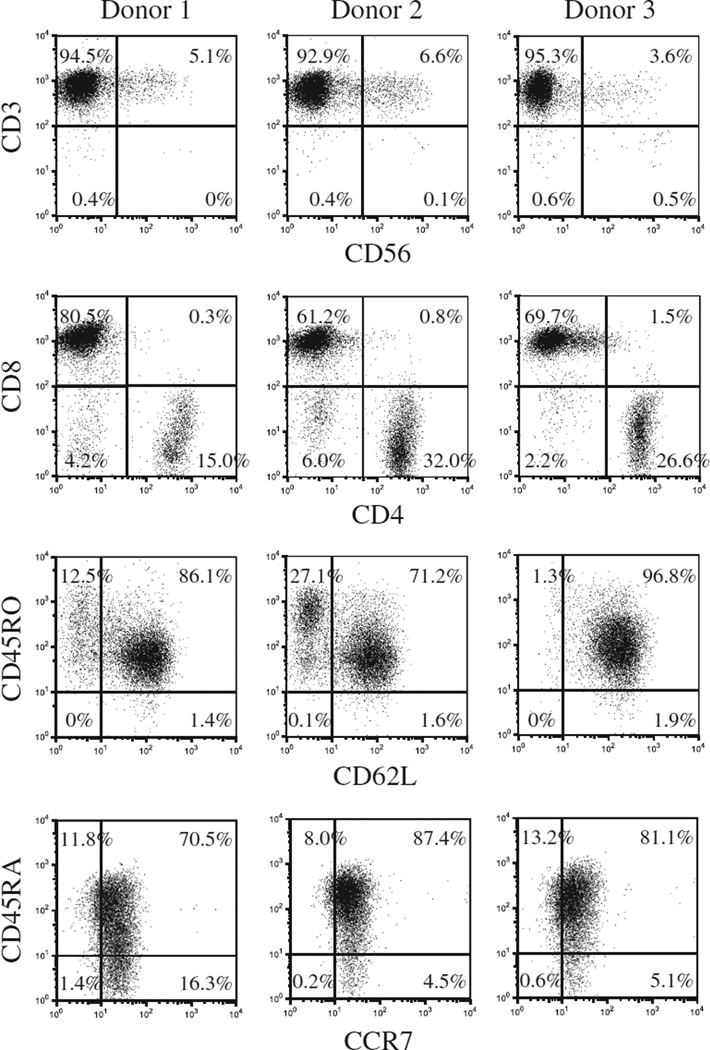
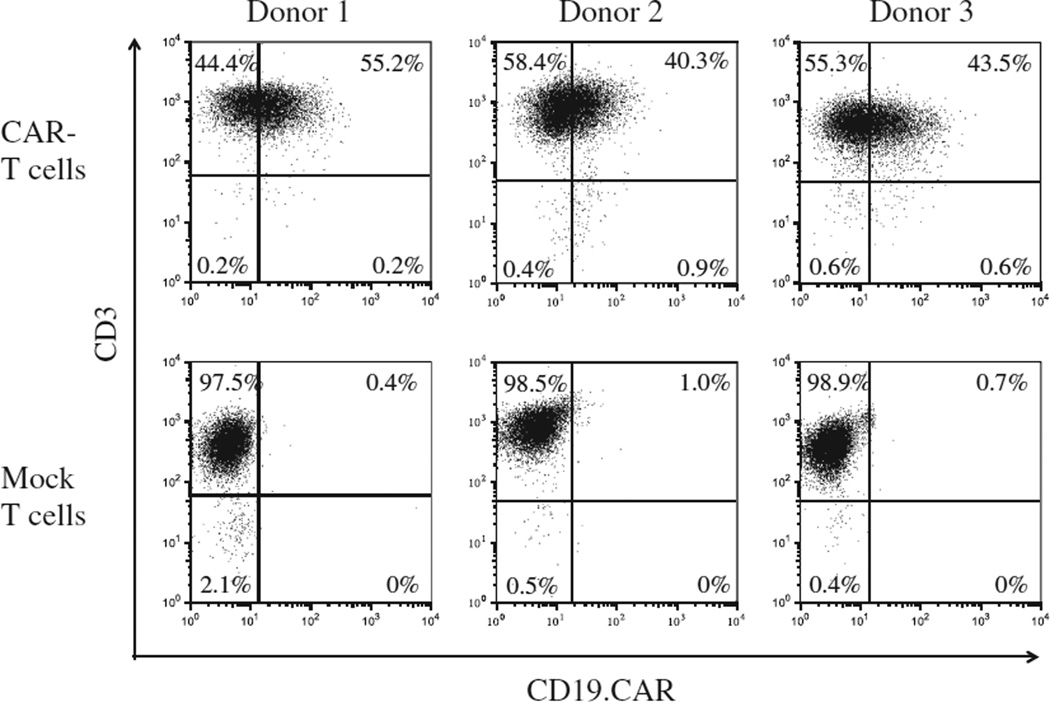
We previously reported non-viral delivery of CAR genes into unstimulated T cells with the use of the piggyBac-transposon system and the first-generation Nucleofector system consisting of a Nucleofector II Device and Human T Cell Nucleofector Kit (Lonza) and efficient expansion of CAR-modified T cells with artificial K562 cells or autologous Epstein-Barr virus (EBV)-infected lymphoblastic B cells (LCLs) in fetal bovine serum–containing culture condition (14,15). In the present study, we modified the gene-transfer and T-cell culture methods to adapt to current Good Manufacturing Practice (cGMP) and improve safety. We first introduced a CD19.CAR gene into PBMCs with piggyBac-transposons with the use of the cGMP-compliant 4D-Nulcleofector and then expanded the transgenic T cells in a culture system that was free of animal serum, tumor-derived feeder cells, and virus-infected cells. As shown in Figure 1, an average of 1.75 ± 0.71 × 107 PBMCs from 10 mL of blood of healthy donors was nucleofected with a CD19.CAR-containing piggyBac-transposon and the piggyBac-transposase on a separate plasmid, then stimulated with CD3/CD28 mAbs. Six days after nucleofection, 4.54 ± 0.45 × 106 cells were obtained and then selected for expression of the CD19.CAR. An average of 5.12 ±3.19 × 105 CD19.CAR-positive cells were restimulated with CD3/CD28 mAbs together with irradiated CD19.CAR-negative autologous cells as feeders. Twenty-one days after the start of culture, we obtained a total of 1.28 ± 0.27 × 108 cells, of which 99.3% ± 0.4% were positive for CD3, 25.4% ± 9.1% were positive for CD4, and 71.3% ± 9.4% were positive for CD8, respectively (Figure 2). There were less than 1% CD3− CD56+ NK cells. Most of the T cells were positive for CD45RA, CD45RO, CD62L and CCR7 (90.7% ± 7.3%, 98.3% ± 0.3%, 86.7% ± 12.5% and 88.3% ± 4.1%, respectively; Figure 2). In particular, 79.7% ± 8.5% of T cells co-expressed CD45RA and CCR7. An average of 46.7% ± 7.7% of CD3+ cells expressed the CD19.CAR on their cell surface (Figure 3). Notably, there was no significant difference in CAR expression on T cells once selected through the use of MACS microbeads and cultured for 3 or 4 weeks after piggyBac-based gene transfer with the use of two Nucleofector devices [47.9 % ± 15.5% by Nucleofector II system (14) versus 46.7% ± 7.7% by 4D-Nulclofector system (in this study), P = 0.91].
Figure 1.
Generation of piggyBac-mediated CD19-specific T cells in serum/xeno/tumor cell/virus-free culture. Mononuclear cells from 10 mL of blood of healthy donors were nucleofected with pIRII-CAR.CD19-28-ξ and pCMV-piggyBac with the use of the 4D-Nucleofector Device and immediately transferred into IL-15–containing TexMACS medium. On day 1, nucleofected cells were stimulated with the use of CD3/CD28 mAbs for 4 days. Six days after the stimulation, cells were selected with the use of an anti-CAR antibody and restimulated with CD3/CD28 mAbs for 4 days, then cultured for another 10 days in IL-15–containing TexMACS with autologous feeder cells. Twenty-one days after the gene transfer, cells were harvested, analyzed and cryopreserved. Data are presented as mean ± standard deviation of experiments from three donors.
Figure 2.
Immunophenotype of CD19.CAR-modified cells. Surface antigens on generated cells were determined by flow cytometry with the use of CD3, CD4, CD8, CD56, CD45RO, CD62L, CD45RA and CCR7 mAbs. Results from three donors are shown.
Figure 3.
CD19.CAR expressed on T cells. Expression of CD19.CAR on T cells genetically modified by piggyBac and expanded (CAR T cells) was determined by flow cytometry with the use of goat anti-human IgG (H+L) antibody and anti-CD3 mAb. Non–gene-transferred T cells (mock T cells) were also analyzed as controls. Results from three donors are shown.
The final cell product (CAR T cells) showed superior lysis of CD19-positive Ph+ALL cells, at least at the 2:1 ratio, but not CD19-negative K562 cells in two donors tested (supplementary Figure 1).
T cells genetically modified by piggyBac-transposons and then cultured for 21 days had a normal karyotype according to G-banding analysis (data not shown) and revealed oligoclonality or polyclonality according to multiplex PCR analysis targeting T-cell receptor genes (supplementary Figure 2). We detected an average of 5.5 ± 1.2 copies of CD19.CAR gene and 2.1 ± 0.4 copies of piggyBac gene per cell in the CAR T-cell lines, although we might have excessively detected the copy numbers because we calculated them per live cell, which were determined by dye-exclusion test, but possibly contaminated with plasmid-containing dead cells.
CAR T cells eliminate Ph+ALL cells in co-cultures
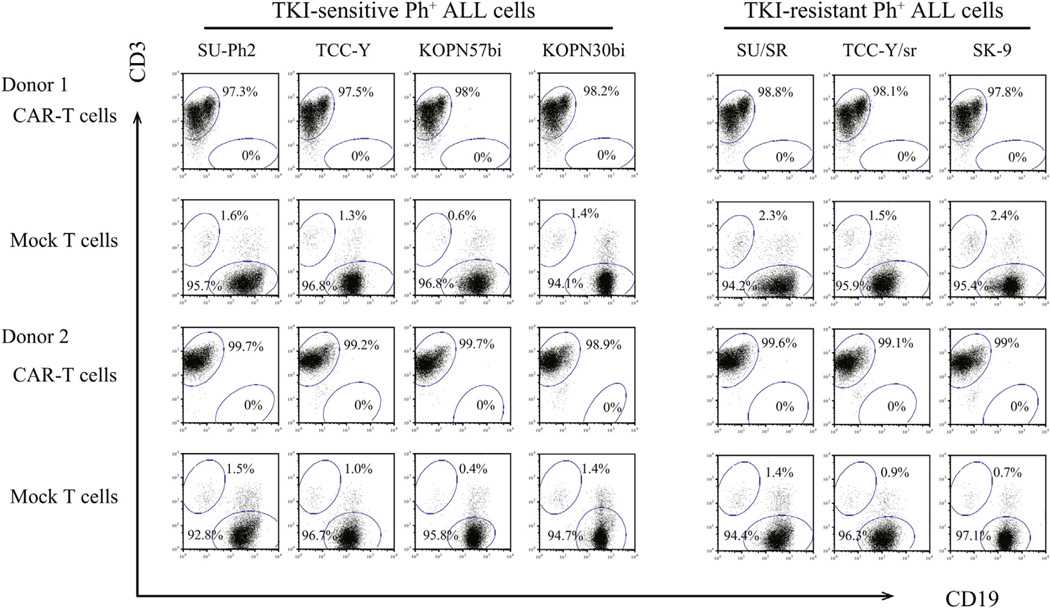
We examined the anti-proliferative effects of CAR T cells on TKI-sensitive and TKI-resistant Ph+ALL cell lines: SU-Ph2, TCC-Y, KOPN57bi and KOPN30bi were sensitive to TKIs, and SU/SR, TCC-Y/sr and SK-9 were resistant to TKIs because of T315I mutations (19–22). Either CAR T cells or mock T cells were added to each Ph+ALL cell line at an E:T ratio of 1:5, then cultured in the absence of cytokines for 7 days. As shown in Figure 4, CAR T cells almost completely eliminated the three TKI-resistant Ph+ALL cells and the four TKI-sensitive Ph+ALL cells (CD19+cells in the co-culture, <0.1%), whereas the Ph+ALL cells survived in co-cultures with mock T cells (CD19+ cells, 94.9–97.2%). At the E:T ratio of 1:5, CAR T cells failed to influence the proliferation of K562 cells, which did not express CD19 on their cell surface (data not shown).
Figure 4.
Anti-leukemic activity of CAR T cells against Ph+ALL cells Either of CAR T cells or mock T cells was co-cultured with seven different Ph+ALL cell lines at an E:T ratio of 1:5. Seven days later, co-cultured cells were stained with CD3-APC mAb and CD19-PE mAb and analyzed by flow cytometry. Representative results from two donors are shown.
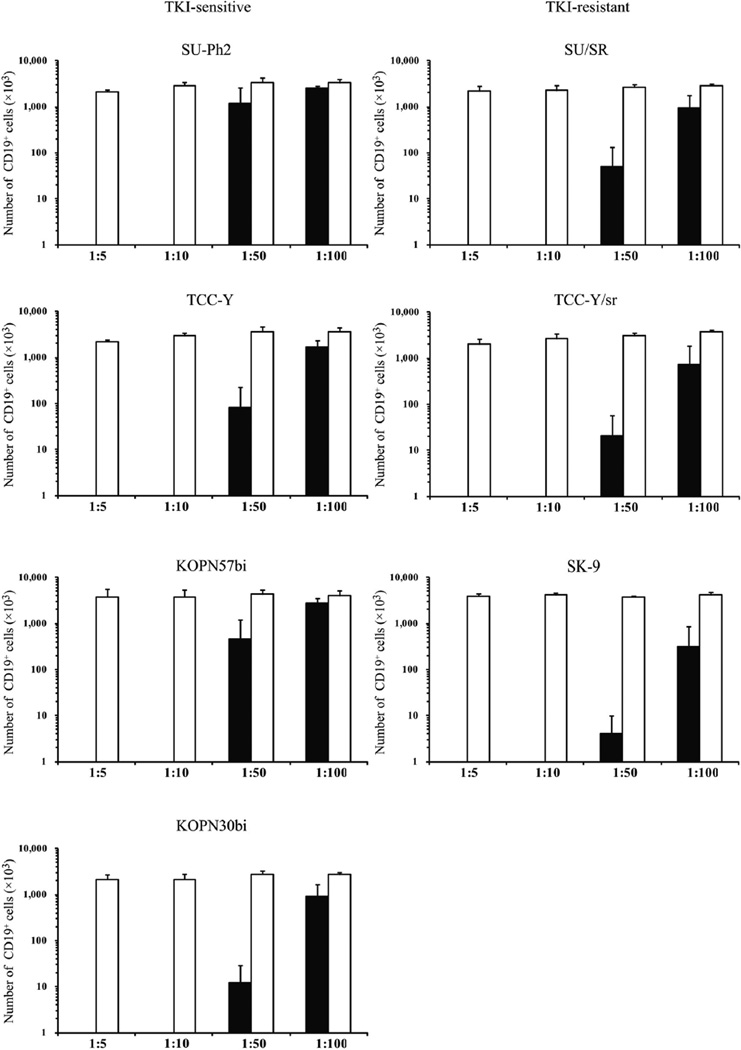
To elucidate whether smaller numbers of CAR T cells can suppress Ph+ALL cells, we performed 7-day co-culture experiments at lower E:T ratios (1:10, 1:50 and 1:100). As shown in Figure 5, flow cytometric analysis revealed that all seven Ph+ALL cell lines with CAR T cells decreased below one-thousandth of those with mock T cells in the co-cultures at 1:10 regardless of TKI sensitivity. Moreover, we observed substantial reduction in all but the SU-Ph2 cell line in the co-cultures at 1:50 and a decreasing trend in four cell lines (KOPN30bi and three TKI-resistant cell lines) in the co-cultures at 1:100.
Figure 5.
Co-cultures of CD19-specific CAR T cells with Ph+ALL cells at low E:T ratios. CAR T cells (closed bar) or mock T cells (open bar) were co-cultured with each Ph+ALL cell line at E:T ratios of 1:5, 1:10, 1:50 and 1:100. After the 7-day co-culture, viable cells were counted and analyzed by means of flow cytometry with the use of CD3-APC/CD19-PEmAbs. Number of Ph+ALL (CD19+) cells (mean ± standard deviation from three donors) of CD19-positive/CD3-negative cells in each co-culture is shown.
We validated elimination of Ph+ALL cells by use of CAR T cells at a molecular level with the use of real-time qPCR targeting BCR-ABL mRNA in the 1:5 and 1:10 co-cultures with four tested Ph+ALL cell lines (SU-Ph2, SU/SR, KOPN57bi and SK-9) (supplementary Figure 3).
CAR T cells proliferate in response to Ph+ALL cells
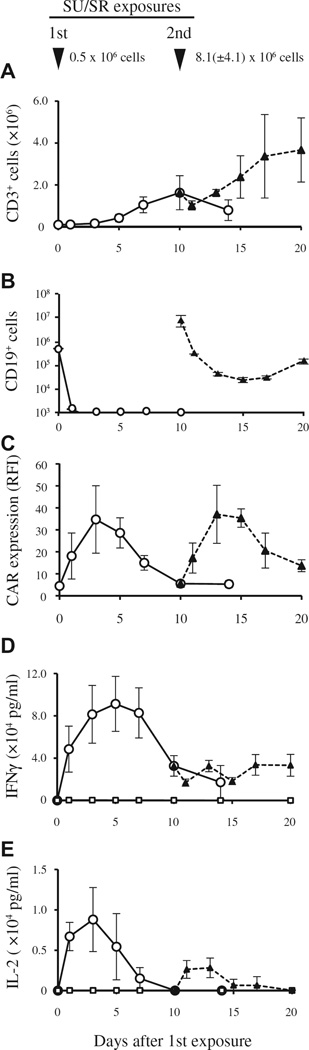
Next, we examined the kinetics of CAR T-cell proliferation after antigen stimulation once or twice with live TKI-resistant SU/SR cells at an E:T ratio of 1:5. Ten days after the co-culture of 1 × 105 CAR T cells with 5 × 105 SU/SR cells, the number of CAR T cells expanded approximately 16-fold with sustained growth inhibition of SU/SR cells, despite the lack of exogenous cytokines in the medium (CAR T cells, 1.63 ± 0.81 × 106; SU/SR cells, <1.0 × 104; Figure 6A,B). Interestingly, the RFI of CD19.CAR expression on CAR T cells increased approximately 4-fold after 20–24 hours of co-culture with SU/SR cells (RFI, 4.45 ± 1.09 on day 0 versus 18.1 ± 10.5 on day 1) and reached approximately 8-fold on day 3 (RFI, 34.7 ± 15.3), returning to basal levels on day 10 (RFI, 5.52 ± 0.25), as shown in Figure 6C.
Figure 6.
Kinetics of CD19-specific CAR T cells after encounter with a TKI-resistant Ph+ALL cell line (SU/SR). CAR T cells (1 × 105) were exposed to SU/SR cells (5×105) once on day 0 or twice on days 0 and 10 at an E:T ratio of 1:5 in the absence of cytokines. CAR T cells or Ph+ALL cells were planted alone as controls. Data are presented as mean ± standard deviation of experiments from three donors. (A, B) Change of cell numbers of CAR T cells (A) and SU/SR cells (B) were determined by use of a combination with trypan blue exclusion test and flow cytometric analysis with the use of CD3-APC and CD19-PE mAbs. CD3+ cells represent CART cells and CD19+ cells represent SU/SR cells. Open circles (○) indicate the cell numbers after the first SU/SR exposure; closed triangles (▲) indicate cell numbers after the second SU/SR exposure. (C) Change of CAR expression on CAR T cells. The intensity of CAR expression is shown as RFI, determined by calculating the ratio of mean fluorescence intensity for specific staining to that for control staining. Open circles (○) indicate RFI after the first SU/SR exposure; closed triangles (▲) indicate RFI after the second SU/SR exposure. (D, E) Cytokine release by CAR T cells. The concentrations of IFN-γ and IL-2 in the supernatant of the co-cultures were determined by means of ELISA, as shown in (D) and (E), respectively. Open circles (○) indicate the cytokine concentrations after the first SU/SR exposure; closed triangles (▲) after the second SU/SR exposure; open squares (□) after no SU/SR exposure (CAR T cells alone).
In concurrent co-cultures, proliferating CAR T cells were re-exposed to SU/SR cells (8.1 ± 4.1 × 106) at an E:T ratio of 1:5 on day 10 and showed a further increase in CD19.CAR expression (up to 8-fold compared with baseline) (RFI, from 4.52 ± 1.11 to 37.1 ± 13.2 on day 13; Figure 6C), marked cytotoxicity on day 11 (SU/SR cells, from 8.1 ± 4.1 × 106 to 2.88 ± 0.16 × 105; Figure 6B) and further proliferation in the absence of added cytokines (CAR T cells, from 1.63 ± 0.81 × 106 to 3.67 ±0.15 × 106 on day 20; Figure 6A).
INF-γ and IL-2 production by CAR T cells
To evaluate cytokine release from CAR T cells in response to Ph+ALL cells, we measured the levels of IFN-γ and IL-2 in the supernatants of co-cultures by means of ELISA. As shown in Figure 6D, abundant levels of IFN-γ were detected in the 1:5 ratio co-culture of CAR T cells with SU/SR cells from day 1 through day 20 (peak level, 91,300 ± 21,247 pg/mL on day 5), compared with the culture of CAR T cells alone (peak level, 319 ± 280 pg/mL on day 1). The addition of SU/SR cells at the 1:5 ratio on day 10 had little effect on IFN-γ production by CAR T cells on day 14 [32,500 ± 4391 pg/mL compared with T cells that did not receive a second stimulation (29,867 ± 8062 pg/mL), P = 0.706]. SU/SR cells produced no IFN-γ (data not shown). By contrast, as shown in Figure 6E, the IL-2 level in the 1:5 co-culture of CAR T cells with SU/SR cells was elevated to the maximum level on day 3 (8807 ± 3237 pg/mL), and the addition of SU/SR cells on day 10 induced a second IL-2 production (2830 ± 992 pg/mL on day 14), albeit to a lesser extent than the first IL-2 production. CAR T cells (Figure 6E) or SU/SR cells alone produced no IL-2 (data not shown).
Expression of TRAIL and TRAIL receptors
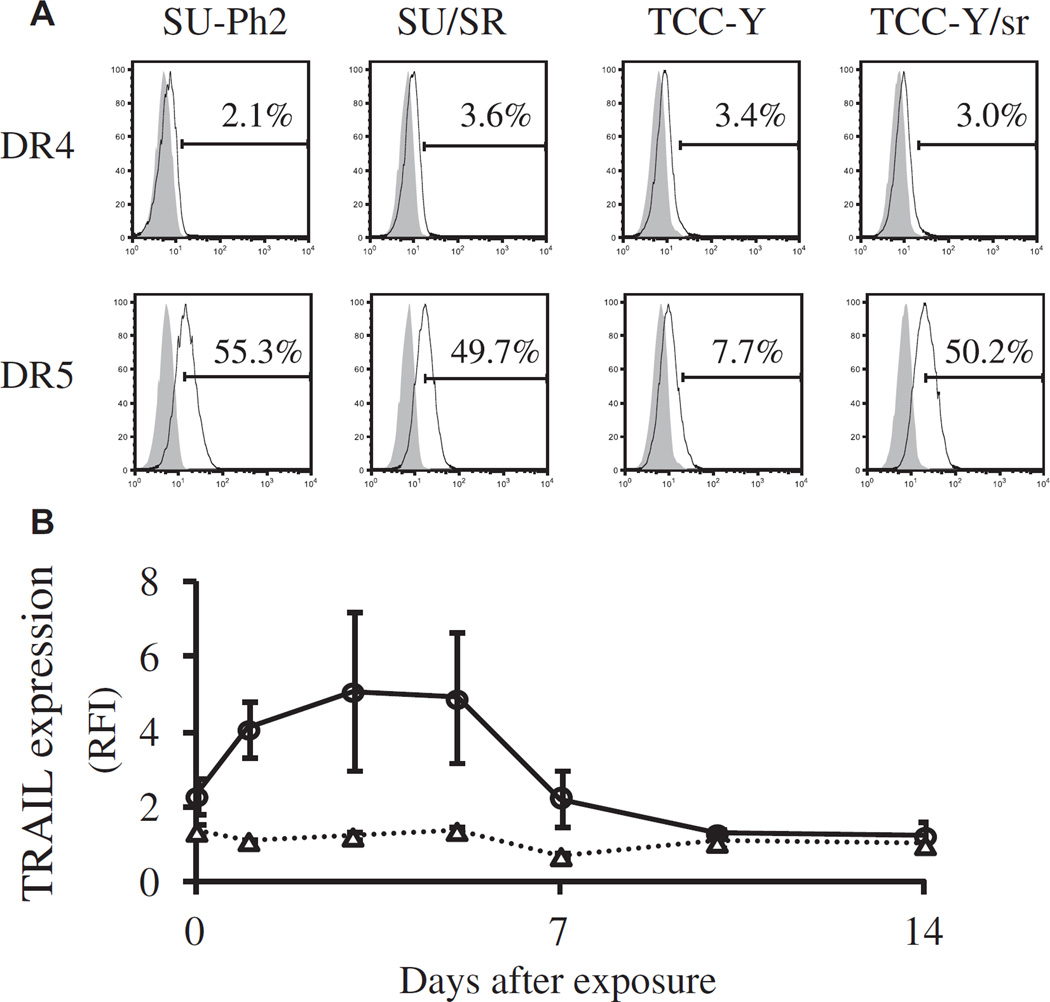
TRAIL is a pro-apoptotic member of the tumor necrosis factor (TNF) superfamily that also includes TNF-α and Fas ligand. TRAIL induces apoptosis in a variety of tumor cells by interacting with the cell-surface receptors DR4 and DR5. We previously reported that primary Ph+ALL cells and cell lines are sensitive to soluble TRAIL because of their broad expression of the TRAIL receptors DR4 and/or DR5, which is partly regulated by the BCR-ABL signaling pathway (21,25). In this study, we investigated the expression of TRAIL in CAR T cells after stimulation with Ph+ALL cells. We first examined whether the T315I BCR-ABL mutation influenced DR4 and/or DR5 expression on Ph+ALL cells through the use of two pairs of parent and daughter cell lines in which the daughters had acquired T315I mutations after culture in imatinib (19,20). As shown in Figure 7A, the acquisition of T315I mutation in SU/SR and TCC-Y/ sr did not affect their expression of DR4, whereas expression of DR5 was greater in TCC-Y/sr than in the parental (TCC-Y) cell line but unaffected in SU/ SR cells. Next, we serially analyzed TRAIL expression on the cell surface of CAR T cells in the 1:5 co-culture with SU/SR cells. Whereas CAR T cells modestly expressed TRAIL before the co-culture (RFI, 2.31 ± 0.49), levels increased from day 1 of co-culture, reached the maximum intensity on day 3 (RFI, 5.10 ± 2.10) and returned to the basal level by day 7 (RFI, 2.22 ± 0.75), as shown in Figure 7B. On the other hand, we did not observe any change in TRAIL expression on the cell surface of mock T cells. The transient increase of TRAIL expression in CAR T cells was found mainly in CD8-positive T cells (the positivity for CD4 and CD8 of TRAIL-expressing T cells was 7.1% ± 3.0% and 93.0% ± 2.5% on day 3, respectively).
Figure 7.
Expression of TRAIL and TRAIL receptors. (A) Expression of death receptors DR4 and DR5 on four Ph+ALL cell lines were determined by flow cytometry. We used two pairs of parent and daughter cell lines in which the daughters had acquired T315I mutations after culture in imatinib (SU-Ph2 and SU/SR; TCC-Y and TCC-Y/sr). (B) Change of TRAIL expression on CAR T cells compared with mock T cells after antigen stimulation with Ph+ALL. CAR T cells were co-cultured with SU/SR cells at an E:T ratio of 1:5. Expression of TRAIL on generated mock T cells and CAR T cells was determined by means of flow cytometry, with the use of anti–CD253 (TRAIL)-PE mAbs and anti–CD3-APC mAbs. The change of TRAIL expression is shown as RFI, determined by calculating the ratio of mean fluorescence intensity for specific staining to that for control staining. Open triangles (△) and open circles (○) indicate the RFI of TRAIL expression in mock T cells and CAR T cells, respectively. Data are presented as mean ± standard deviation of experiments from three donors.
Discussion
We previously reported a DNA plasmid-based gene-transfer method into T cells with the use of the piggyBac-transposon system in combination with the first-generation Nucleofector system (Nucleofector II Device and Human T Cell Nucleofector Kit), which is simple, inexpensive, and safe to handle (11–17). However, gene delivery by the first-generation Nucleofector system does not fulfill cGMP requirements. In the present study, we modified our gene-transfer method to facilitate regulatory approval. We transferred a CD19.CAR gene into PBMCs with the use of piggyBac-transposons and the second-generation Nucleofector system (4D-Nucleofector Device and P3 Primary Cell 4D-Nucleofector X Kit), for which a cGMP certificate is available. Consequently, we elucidated that the 4D-Nucleofector system could mediate efficient gene delivery comparable to the Nucleofector II system, in combination with piggyBac-transposons.
We previously reported a culture system for the expansion of piggyBac-transfected CAR-expressing T cells with the use of autologous EBV-LCLs or K562 cells genetically engineered to express the costimulatory molecules CD80, CD83, CD86 and 4-1BBL as feeder cells and fetal bovine serum–containing culture medium (14,15). However, the inclusion of animal-derived components, tumor cell lines, and live virus–infected cells all introduce a potential risk for infection or unexpected immune reaction when used in the manufacture of clinical-grade cell therapy products. In the present study, we modified the T-cell culture method to improve safety by use of serum/xeno-free TexMACS medium and irradiated autologous activated T cells as feeder cells. We also used IL-15 as T-cell growth factor because IL-15 can mediate the most efficient piggyBac gene transfer and piggyBac-transgenic T-cell growth among IL-2, IL-4, IL-7 and IL-15, as reported previously (11). With the use of these gene-transfer and T-cell culture methods, we obtained a total of 1.28 ± 0.26 × 108 CAR T cells (CD3+ cells, 99.3% ± 0.4%) from 10 mL of blood (Figures 1 and 2). In the clinical study described by Brentjens et al. (9), 1.5 to 3.0 × 106 CD19.CAR-modified T cells per kilogram were administered in adults with ALL. Given these values, we would be able to obtain sufficient numbers of piggyBac-mediated CAR T cells from 10 to 20 mL of blood in adults and from 10 mL of blood or less in children.
In a previous report (14) and in this study, transgene expression in piggyBac-mediated CAR-modified T cells demonstrated a modest intensity (Figure 3) (14). Nevertheless, the piggyBac-mediated T cells exerted a marked cytotoxicity in the co-cultures with Ph+ALL cells at E:T ratios of 1:5 and 1:10 (Figures 4 and 5). Further analysis of transgene expression revealed that CD19.CAR expression was transiently but reproducibly and repeatedly upregulated on encounter with CD19 on Ph+ALL cells (Figure 6C). This suggests that the relatively modest CD19.CAR expression on T cells genetically modified with piggyBac transposons would not attenuate anti-tumor activity mediated by the CD19.CAR.
Our piggyBac-modified CAR T cells exhibited anti-leukemic activity: very low E:T ratios of below 1:5. To test the hypothesis that a small number of CAR T cells could expand after the encounter with Ph+ALL cells, we examined the kinetics of CAR T cells in the co-cultures with Ph+ALL cells in the absence of cytokines. CAR T cells were able to proliferate up to 37-fold within 3 weeks in response to repeated exposure to Ph+ALL cells while maintaining their cytotoxic function (Figure 6A,B). More recently, Yang et al. (26) reported that the frequency of a CD8+CD45RA+CCR7+ subset of the infused CD19.CAR T-cell products correlates with overall in vivo expansion in patients with lymphoma and showed that the combination of IL-7 and IL-15 plays a crucial role in increasing the number of CD8+CD45RA+CCR7+ CAR T cells, also co-expressing CD62L and CD45RO, enhancing their persistence and anti-tumor activity in preclinical models (26). Remarkably, our CD8-dominant (~71%) CAR T cells co-expressed CD45RA and CCR7 up to ~ 80%, although cultured with IL-15 alone (Figure 2). The major subset (CD8+CD45RA+CCR7+) within our CAR T-cell products appears to contribute to anti-leukemic activity at very low E:T ratios in this study. Additionally, we observed that CAR T cells transiently and reproducibly produced IL-2 after the encounter with Ph+ALL cells (Figure 6E), suggesting that the proliferative response of CAR T cells to Ph+ALL cells rely partly on autocrine IL-2 secretion.
We previously showed that Ph+ALL cell lines and primary leukemic cells of patients with Ph+ALL are insensitive to Fas ligand but sensitive to soluble TRAIL, in a manner dependent on expression of death-inducing receptors DR4 and/or DR5 on the leukemic cells, which is positively regulated at least in part by the BCR-ABL signaling pathway (21,25). Soluble TRAIL effectively induced cell death in Ph+ALL cell lines refractory to the first-generation TKI imatinib, although T315I–mutated cell lines were not tested (21). To investigate the relation of the TRAIL/TRAIL receptor axis in our therapeutic model for Ph+ALL, we evaluated DR4 and DR5 expression on T315I–mutated Ph+ALL cells (19,20) and TRAIL expression on CAR T cells. The T315I mutation did not affect the expression of death receptors on Ph+ALL cells (Figure 7A). TRAIL expression in CART cells but not mock T cells transiently increased from day 1 to day 5 after encounter with Ph+ALL cells (Figure 7B), as observed for the expression of the CD19.CAR transgene. The perforin/granzyme pathway is thought to play a crucial role in the cytolytic mechanism of CD19-specific CAR T cells (7), whereas the significance of the TRAIL pathway remains unclear. Unfortunately, we were unable to elucidate the role of CD19 antigen-specific transient increase of TRAIL expression in CAR T cells. These findings are, however, interesting in view of the high sensitivity of Ph+ALL cells to CAR T cells.
Acquired and/or intrinsic drug resistance is a great concern in patients treated with TKI. Acquired TKI resistance is dependent on mutations in the BCR-ABL kinase domains. In particular, patients who acquired the T315I mutant will no longer respond to the first-/second-generation TKI. Other mechanisms of resistance independent of BCR-ABL include pharmacokinetic factors that reduce the availability of TKIs and activation of alternative signaling pathways (27). More recently, the third-generation TKI ponatinib was developed to overcome BCR-ABL-mutations including T315I. In a phase 2 trial of posatinib monotherapy, however, among 32 patients with Ph+ALL who had TKI resistance (in whom 22 had T315I mutation) or TKI-associated unacceptable side effects, the major hematologic response rate was 41%, and the estimated rate of a sustained response of at least 12 months was only 8% (28). These results suggest that an alternative or combined therapy is required even in patients who receive the third-generation or newer TKI treatment. Although allogeneic stem cell transplantation is considered the best treatment option for patients with TKI-resistant Ph+ALL (1,29,30), the outcome depends on the residual tumor burden at the time of transplantation. In this study, we demonstrate that CAR T cells exert profound CD19-specific cytotoxicity against all seven Ph+ALL cell lines and that mutations in BCR-ABL kinase domain do not impart resistance to T-cell killing (Figures 4 and 5). Hence, CD19-sepecific T-cell therapy may be a promising pretransplant option in patients with TKI-refractory Ph+ALL.
Finally, we successfully generated a clinically relevant number of CAR T cells, which can exert marked cytotoxicity against drug-resistant Ph+ALL, from 10 mL of blood of healthy donors with the use of a piggyBac-transposon, a 4D-Nulcleofector and a serum/xeno/tumor cell/virus-free culture system. However, we have some problems to be solved in this study: (i) we need to generate CAR T cells from leukemic patients treated with cytotoxic agents and/ or TKIs who are probably poor responders, (ii) we have no comparative data on the anti-leukemic potency of CAR T cells between retro/lentiviral and piggyBac gene modifications, (iii) we used an anti-CH2CH3 antibody, which is not compliant to cGMP, for selection of CAR-expressing T cells and culture plates inappropriate to cGMP, and (iv) we observed relatively high copy number integration of CD19.CAR-transposon plasmids (~5 copies per cell) with residual piggyBac-transposase plasmids (~2 copies per cell), which may be a potential risk of unexpected T-cell transformation, re-transposition of a transgene, or rejection of infused T cells in patients if expressing piggyBac protein. Regarding the safety concerns, co-delivery of a suicide gene or use of piggyBac-transposase mRNA may be helpful (11,13,31). All things considered, clinical-grade DNA plasmids are inexpensive and easy to manufacture relative to viral vectors; therefore, the piggyBac-transposon system in combination with our T-cell culture system would increase the cost-benefit and safety of CAR-modified T-cell therapy, thereby facilitating regulatory approval. PiggyBac-mediated CD19-specific T-cell therapy appears to be a promising option for drug-resistant Ph+ALL.
Supplementary Material
Acknowledgments
This work was supported by JSPS KAKENHI grant Nos. 23591533, 24791050 and 26461574, Takeda Science Foundation and Japan Leukemia Research Fund.
Footnotes
Disclosure of interests: The authors have no commercial, proprietary, or financial interest in the products or companies described in this article.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jcyt.2014.05.022
References
- 1.Fielding AK. How I treat Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2010;116:3409–3417. doi: 10.1182/blood-2010-01-242750. [DOI] [PubMed] [Google Scholar]
- 2.Soverini S, Colarossi S, Gnani A, Rosti G, Castagnetti F, Poerio A, et al. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of Philadelphia-positive patients: by the GIMEMA Working Party on Chronic Myeloid Leukemia. Clin Cancer Res. 2006;12:7374–7379. doi: 10.1158/1078-0432.CCR-06-1516. [DOI] [PubMed] [Google Scholar]
- 3.Nicolini FE, Mauro MJ, Martinelli G, Kim DW, Soverini S, Müller MC, et al. Epidemiologic study on survival of chronic myeloid leukemia and Ph(+) acute lymphoblastic leukemia patients with BCR-ABL T315I mutation. Blood. 2009;114:5271–5278. doi: 10.1182/blood-2009-04-219410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116:4099–4102. doi: 10.1182/blood-2010-04-281931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brentjens RJ, Rivière I, Park JH, Davila ML, Wang X, Stefanski J, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011;118:4817–4828. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kochenderfer JN, Dudley ME, Feldman SA, Wilson WH, Spaner DE, Maric I, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119:2709–2720. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5:177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakazawa Y, Huye LE, Dotti G, Foster AE, Vera JF, Manuri PR, et al. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J Immunother. 2009;32:826–836. doi: 10.1097/CJI.0b013e3181ad762b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galvan DL, Nakazawa Y, Kaja A, Kettlun C, Cooper LJ, Rooney CM, et al. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J Immunother. 2009;32:837–844. doi: 10.1097/CJI.0b013e3181b2914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manuri PV, Wilson MH, Maiti SN, Mi T, Singh H, Olivares S, et al. piggyBac transposon/transposase system to generate CD19-specific T cells for the treatment of B-lineage malignancies. Hum Gene Ther. 2010;21:427–437. doi: 10.1089/hum.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakazawa Y, Huye LE, Salsman VS, Leen AM, Ahmed N, Rollins L, et al. PiggyBac-mediated cancer immunotherapy using EBV-specific cytotoxic T-cells expressing HER2-specific chimeric antigen receptor. Mol Ther. 2011;19:2133–2143. doi: 10.1038/mt.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huye LE, Nakazawa Y, Patel MP, Yvon E, Sun J, Savoldo B, et al. Combining mTor inhibitors with rapamycin-resistant T cells: a two-pronged approach to tumor elimination. Mol Ther. 2011;19:2239–2248. doi: 10.1038/mt.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha S, Nakazawa Y, Huye LE, Doherty JE, Galvan DL, Rooney CM, et al. PiggyBac transposon system modification of primary human T cells. J Vis Exp. 2012;69:e4235. doi: 10.3791/4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakazawa Y, Saha S, Galvan DL, Huye L, Rollins L, Rooney CM, et al. Evaluation of long-term transgene expression in piggyBac-modified human T lymphocytes. J Immunother. 2013;6:3–10. doi: 10.1097/CJI.0b013e3182791234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 19.Hirase C, Maeda Y, Takai S, Kanamaru A. Hypersensitivity of Ph-positive lymphoid cell lines to rapamycin: possible clinical application of mTOR inhibitor. Leuk Res. 2009;33:450–459. doi: 10.1016/j.leukres.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Shiotsu Y, Kiyoi H, Ishikawa Y, Tanizaki R, Shimizu M, Umehara H, et al. KW-2449, a novel multikinase inhibitor, suppresses the growth of leukemia cells with FLT3 mutations or T315I–mutated BCR/ABL translocation. Blood. 2009;114:1607–1617. doi: 10.1182/blood-2009-01-199307. [DOI] [PubMed] [Google Scholar]
- 21.Uno K, Inukai T, Kayagaki N, Goi K, Sato H, Nemoto A, et al. TNF-related apoptosis-inducing ligand (TRAIL) frequently induces apoptosis in Philadelphia chromosome-positive leukemia cells. Blood. 2003;101:3658–3667. doi: 10.1182/blood-2002-06-1770. [DOI] [PubMed] [Google Scholar]
- 22.Okabe S, Tauchi T, Ohyashiki K. Establishment of a new Philadelphia chromosome-positive acute lymphoblastic leukemia cell line (SK-9) with T315I mutation. Exp Hematol. 2010;38:765–772. doi: 10.1016/j.exphem.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda K, Nakazawa Y, Yanagisawa R, Honda T, Ishii E, Koike K. Detection of T-cell receptor gene rearrangement in children with Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis using the BIOMED-2 multiplex poly-merase chain reaction combined with GeneScan analysis. Clin Chim Acta. 2011;412:1554–1558. doi: 10.1016/j.cca.2011.04.036. [DOI] [PubMed] [Google Scholar]
- 24.Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N, et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia: a Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda I, Inukai T, Zhang X, Kikuchi J, Furukawa Y, Nemoto A, et al. BCR-ABL regulates death receptor expression for TNF-related apoptosis-inducing ligand (TRAIL) in Philadelphia chromosome-positive leukemia. Oncogene. 2013;32:1670–1681. doi: 10.1038/onc.2012.186. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Zhang M, Ramos C, Durett A, Liu E, Dakhova O, et al. Closely-related T-memory stem cells correlate with in-vivo expansion of CAR.CD19-T cells in patients and are preserved by IL-7 and IL-15. Blood. 2014;123:3750–3759. doi: 10.1182/blood-2014-01-552174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravandi F, O’Brien S, Thomas D, Faderl S, Jones D, Garris R, et al. First report of phase 2 study of dasatinib with hyper-CVAD for the frontline treatment of patients with Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia. Blood. 2010;116:2070–2077. doi: 10.1182/blood-2009-12-261586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortes JE, Kim DW, Pinilla-Ibarz J, le Coutre P, Paquette R, Chuah C, et al. A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N Engl J Med. 2013;369:1783–1796. doi: 10.1056/NEJMoa1306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fielding AK, Goldstone AH. Allogeneic haematopoietic stem cell transplant in Philadelphia-positive acute lymphoblastic leukaemia. Bone Marrow Transplant. 2008;41:447–453. doi: 10.1038/sj.bmt.1705904. [DOI] [PubMed] [Google Scholar]
- 30.Nicolini FE, Basak GW, Soverini S, Martinelli G, Mauro MJ, Müller MC, et al. Allogeneic stem cell transplantation for patients harboring T315I BCR-ABL mutated leukemias. Blood. 2011;118:5697–5700. doi: 10.1182/blood-2011-07-367326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bire S, Ley D, Casteret S, Mermod N, Bigot Y, Rouleux-Bonnin F. Optimization of the piggyBac transposon using mRNA and insulators: toward a more reliable gene delivery system. PLoS One. 2013;8:e82559. doi: 10.1371/journal.pone.0082559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.