Abstract
The viscosity of genomic DNA can interfere with digital PCR systems that partition samples into oil droplets or microfluidic wells. Restriction digestion may reduce the viscosity, but the process is labor-intensive, and the buffer can alter the conditions for PCR. DNA fragmentation using the QIAshredder (a biopolymer spin column) is faster, may result in more predictable and uniformly-sized fragments, and avoids the need for restriction buffers that can inhibit downstream PCR. In 10 separate head-to-head experiments comparing aliquots of DNA processed using the QIAshredder to those digested with RsaI or BsaJI prior to droplet digital PCR, we found that the copy numbers measured from the QIAshredded DNA tended to be greater than those measured from the digested DNA (average of 1.35-fold compared with BsaJI; P < 0.0001), even for inputs as high as 1.8 µg or dilution down to the single copy level.
Keywords: digital PCR, Droplet Digital PCR, DNA, QIAshredder, restriction enzyme, BsaJI, RsaI
With increasing recognition of the advantages of digital PCR (1–3) and the recent increase in commercially-available platforms, digital PCR has become the method of choice for a number of molecular biology applications. One requirement of digital PCR is that the sample must be equally distributed across a large number of partitions, which is usually accomplished by dispersion into oil droplets (Quantalife/Bio-Rad, RainDance Technologies) or microfluidic wells (Fluidigm, Life Technologies, Formulatrix). However, with higher inputs of genomic DNA (>66 ng/20 µl for Bio-Rad), the viscosity of the DNA changes the average volume of oil droplets, which interferes with the accuracy of concentration measurements (4), and viscosity could also impair equal partitioning into microfluidic wells. Therefore, some companies recommend (Bio-Rad, RainDance) or encourage (Life Technologies) restriction digestion of DNA prior to partitioning. However, restriction digestion is labor-intensive, and the buffer can alter the conditions for subsequent PCR.
The QIAshredder (QIAGEN, Valencia, CA)(5,6) is a microcentrifuge spin column containing a biopolymer that filters out insoluble particles, decreases viscosity, and is reported to shear DNA (QIAGEN technical support). Assuming equal likelihood of breakage along the DNA, a fragment size of >23,000 bp, and a qPCR amplicon of <230 bp, the breakage should occur within the amplicon <1% of the time. Use of the QIAshredder saves time (<5 min, versus >1 h for digestion), may result in more uniformly-sized fragments, and avoids the need for restriction buffers that may inhibit downstream PCR. We hypothesized that QIAshredded DNA would perform at least as well as digested DNA when used for droplet digital PCR.
To test this hypothesis, droplet digital PCR was performed using the QX-100 system (Bio-Rad, Hercules, CA) on aliquots of unprocessed, QIAshredded, or restriction-digested genomic DNA. To increase generalizability, we tested different cell sources [peripheral blood mononuclear cells (PBMCs) or the HIV-infected 8E5 cell line] (7), different methods of DNA extraction (TRI Reagent, Molecular Research Center, Inc., Cincinnati, OH or QIAamp DNA Blood Mini Kits, QIAGEN), different restriction enzymes (BsaJI, RsaI), and different PCR targets [telomere reverse transcriptase (TERT), HIV]. One aliquot of DNA was processed using the QIAshredder, and concentrations of the QIAshredded and unshredded DNA were measured by UV spectrophotometry (ND-1000 Spectrophotometer, NanoDrop Technologies, Inc., Wilmington, DE). Aliquots of unshredded and QIAshredded DNA were diluted in water (to 60 ng/µl or 200 ng/µl), while a separate aliquot of unshredded DNA was diluted to an identical concentration in a restriction digest containing water, 10× buffer, and enzyme. Digestion was performed with RsaI or BsaJI (New England Biolabs, Ipswich, MA) according to previously published protocols (4,8), and the enzyme was heat-inactivated.
Aliquots of unprocessed, QIAshredded, and digested DNA were visualized by 0.8% agarose gel electrophoreses. Prior to PCR, each DNA preparation was serially diluted in order to test a range of DNA inputs and dilute potential inhibitors. DNA at each dilution was combined with master mix [containing ddPCR Supermix for Probes (Bio-Rad), primer/probe, +/−water] in a final volume of 70 µl. Three 20 µl aliquots were used to make droplets, and PCR was performed for TERT (PBMCs, 8E5 cells) or HIV (8E5 cells) (9). Each 20 µl reaction contained 10 µl of 2× ddPCR Supermix and 1 µl of 20× human TERT TaqMan copy number reference assay (Life Technologies, Grand Island, NY) or 20× HIV oligos (final concentrations: 250 nM probe, 900 nM each primer) (9). PCR conditions included 95°C for 10 min, then 45 cycles of 94°C for 30 s and 59°C for 1 min, then 98°C for 10 min. Droplets were analyzed using the QX-100, and the measured copies were then compared with the number of copies expected from the DNA input. Measured concentrations (copies/20 µl) and droplet counts (positive, total) from the three DNA preparations were analyzed in pairwise fashion using the Wilcoxon signed rank test.
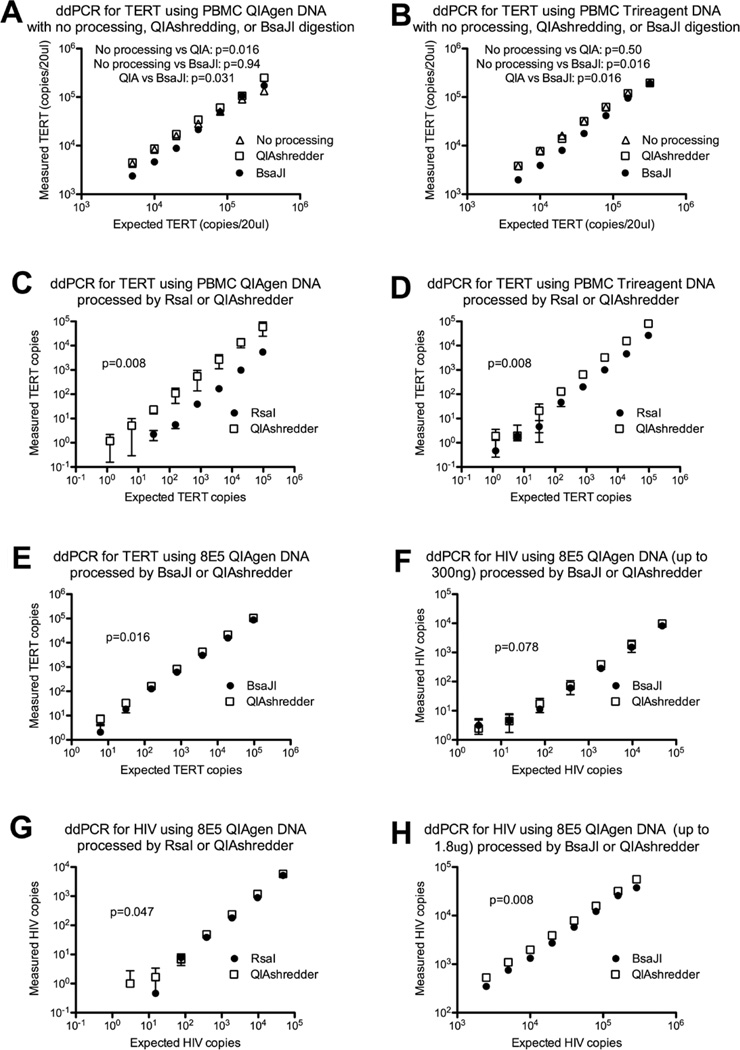
The average DNA concentration after QIAshredding was 99.64% of the unshredded DNA concentration. By agarose gel electrophoresis, the unprocessed and QIAshredded DNA showed a single band at >23 kb, which was replaced in the digested samples by a smear and smaller band(s). For both the TRI Reagent and QIAGEN extractions, the BsaJI-digested DNA tended to perform worse than the unprocessed DNA (lower copies/20 µl and numbers of positive droplets) at lower DNA inputs, but this difference decreased, resolved, or reversed at higher DNA inputs (Figure 1A–B). For QIAGEN but not TRI Reagent extracts, the QIAshredded DNA performed better than unprocessed DNA [higher copies/20 µl (P = 0.016), trended towards more positive droplets (P = 0.078)], and the effect was stronger at higher DNA inputs (Figure 1A and B). Compared with the digested DNA, the QIAshredded DNA produced higher measured copies/20 µl (average QIA/BsaJI = 1.35, P < 0.0001; QIA/RsaI = 5.98, P < 0.0001) (Figure 1), reflecting higher numbers of positive droplets (QIA/BsaJI = 1.25, P = 0.0001; QIA/RsaI = 6.31, P = 0.031) and somewhat lower numbers of total droplets (QIA/BsaJI = 0.98, P = 0.045; QIA/RsaI = 0.86, P = 0002).
Figure 1. Copy numbers as measured by droplet digital PCR in unprocessed, QIAshredded, or restriction-digested genomic DNA.
Genomic DNA was isolated using TRI Reagent (B,D) or QIAGEN kits (A,C,E–H) from peripheral blood mononuclear cells (PBMCs) (A–D) or 8E5 cells containing one HIV provirus (E-H). One aliquot of each DNA sample was passed through a QIAshredder and diluted to a final concentration of 60 ng/µl or 200 ng/µl, while other aliquots of unshredded DNA were diluted to an identical concentration in water (no processing) or added to a restriction digest reaction containing RsaI (C,D, G) or BsaJI (A–B, E–F,H). Each DNA sample was serially diluted, and DNA inputs ranging from 0.00384 ng to 1800 ng per 20 µl reaction were combined with master mix containing 2× PCR buffer, 20× primer/probe, +/− water in a final volume of 70 µl. Three 20 µl replicates were used to make droplets in which PCR was performed for the housekeeping gene telomerase reverse transcriptase (TERT) (A–E) or for HIV (F–H). Droplets were analyzed using the QX-100 system, and the measured concentrations were used to calculate the copies per 20 µl reaction. The measured copy numbers (y-axis) for the unprocessed DNA (open triangles), QIAshredded DNA (open squares), and digested DNA (black circles) were plotted against the expected copy numbers (x-axis) as predicted from the cell equivalents corresponding to the DNA input. All results are plotted on a log scale. Error bars represent the standard deviation of three replicate measurements. For each experiment, the copy numbers from the shredded and digested DNA were compared using the Wilcoxon signed rank test (P values). The results for QIAshredded versus BsaJI-digested PBMC DNA in Figure 1A–B are representative of two additional experiments using PBMCs from another donor.
In 10 experiments, the QIAshredded DNA yielded more positive droplets than DNA digested with RsaI or BsaJI. The mechanism is unclear and may be multifactorial. If the QIAshredder does fragment DNA, the difference from the unprocessed DNA is not easily discernible by conventional gel electrophoresis. Given that the average size of the QIAshredded DNA was larger than the digested DNA and that the QIAshredded DNA produced fewer total droplets, the higher copy numbers are likely not attributable to a lower viscosity. At the same time, the QIAshredded DNA yielded more positive droplets than the unprocessed QIAGEN DNA, and the difference was greater at higher DNA inputs, suggesting that the QIAshredder columns may still do something to improve partitioning or remove inhibitors, though the effect may be extraction and concentration dependent. The difference between the QIAshredded and digested DNA may also reflect inhibitors in the digested DNA, since the digested DNA tended to perform worse than the unprocessed DNA at lower DNA inputs. This difference diminished or reversed at higher DNA inputs, suggesting that digestion may also have a favorable effect by reducing viscosity and/or improving partitioning of concentrated DNA.
While no processing may be necessary for lower inputs of DNA, the QIAshredder should be considered as an alternative to digestion with either BsaJI or RsaI for higher concentrations of QIAGEN DNA. Use of the QIAshredder saves time and labor, which could reduce overall costs. Future experiments should test other methods of DNA fragmentation, such as acoustic shearing, and should examine whether QIAshredded DNA offers advantages for other digital PCR platforms (such as those based on partitioning in microfluidic wells) or even real-time or conventional PCR.
Method summary
We compared the copy numbers as measured by droplet digital PCR in aliquots of unprocessed, QIAshredded, or restriction-digested genomic DNA. In 10 experiments using DNA from different cell sources (PBMC, 8E5 cells), different methods of DNA extraction (solubility, silica columns), different restriction enzymes (RsaI, BsaJI), different DNA inputs (0.00384–1800 ng), and different PCR targets [telomerase reverse transcriptase (TERT), HIV], we found that use of the QIAshredder saved time (<5min, versus >1h) and yielded copy numbers that were similar to or greater than those obtained with restriction digestion.
Supplementary Material
Acknowledgments
The authors thank the blood donors and the Blood Systems Research Institute for PBMCs. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: 8E5/LAV from Dr. Thomas Folks. This work was supported by the U.S. Department of Veterans Affairs [1 IK2 CX000520-01 (to S.Y.), I01 BX000192 (to J.W.)], the National Institute of Allergy and Infectious Diseases at the National Institutes of Health [R56AI091573 (J.W., S.Y.), U19AI096109 (to J.W.)], and the Swiss National Science Foundation [PBZHP3_147260 (to P.K.)]. This paper is subject to the NIH Public Access Policy.
Footnotes
Supplementary material for this article is available at www.BioTechniques.com/article/114159.
Author contributions
S.Y. and J.W. conceived and developed the experiments. S.Y., Ph.K., and Pe.K. executed the experiments. S.Y. and J.W. analyzed data. S.Y. wrote the first draft of the manuscript; Ph.K., Pe.K., P.L., and J.W. edited the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Simmonds P, Balfe P, Ludlam CA, Bishop JO, Brown AJ. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J. Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- 3.Vogelstein B, Kinzler KW. Digital PCR. Proc. Natl. Acad. Sci. USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinclair K, McKechnie VM. DNA extraction from stamps and envelope flaps using QIAamp and QIAshredder. J. Forensic Sci. 2000;45:229–230. [PubMed] [Google Scholar]
- 6.Castella V, Dimo-Simonin N, Brandt-Casadevall C, Mangin P. Forensic evaluation of the QIAshredder/QIAamp DNA extraction procedure. Forensic Sci. Int. 2006;156:70–73. doi: 10.1016/j.forsciint.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 7.Folks TM, Powell D, Lightfoote M, Koenig S, Fauci AS, Benn S, Rabson A, Daugherty D, et al. Biological and biochemical characterization of a cloned Leu-3- cell surviving infection with the acquired immune deficiency syndrome retrovirus. J. Exp. Med. 1986;164:280–290. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE. 2013;8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yukl SA, Shergill A, Ho T, Killian M, Girling V, Epling L, Li P, Wong LK, et al. The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV+ patients on ART: implications for viral persistence. J Infect Dis. 2013;208:1212–1220. doi: 10.1093/infdis/jit308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.