Significance
Oxidative DNA damage can be cytotoxic or mutagenic to cells. NEIL1 (Nei-like 1) is a DNA repair glycosylase guarding the mammalian genome against various oxidized DNA bases; yet how NEIL1 recognizes and catalyzes the removal of its substrates remains poorly understood. Here we integrate crystal structures of a NEIL1/double-stranded DNA complex, computational simulations, and biochemical analyses and show that NEIL1 promotes tautomerization of thymine glycol—a cognate substrate—for efficient substrate recognition and excision. Such tautomerism-dependent substrate recognition and catalysis is, to the best of our knowledge, reported for the first time in an enzyme-catalyzed reaction.
Keywords: base-excision repair, substrate recognition, enzyme catalysis, glycosylase, QM/MM
Abstract
NEIL1 (Nei-like 1) is a DNA repair glycosylase guarding the mammalian genome against oxidized DNA bases. As the first enzymes in the base-excision repair pathway, glycosylases must recognize the cognate substrates and catalyze their excision. Here we present crystal structures of human NEIL1 bound to a range of duplex DNA. Together with computational and biochemical analyses, our results suggest that NEIL1 promotes tautomerization of thymine glycol (Tg)—a preferred substrate—for optimal binding in its active site. Moreover, this tautomerization event also facilitates NEIL1-catalyzed Tg excision. To our knowledge, the present example represents the first documented case of enzyme-promoted tautomerization for efficient substrate recognition and catalysis in an enzyme-catalyzed reaction.
DNA oxidation damage can be induced by both endogenous and environmental reactive oxygen species. Such oxidized DNA bases are primarily recognized and removed by the base-excision repair (BER) pathway, which is initiated by a lesion-specific DNA glycosylase (1–4). Based on sequence homology and structural motifs, glycosylases that cleave oxidation damage are grouped into two families: the helix–hairpin–helix (HhH) family and the Fpg/Nei family (5, 6); the latter is named after the prototypical bacterial members formamidopyrimidine DNA glycosylase (Fpg) and endonuclease eight (Nei).
NEIL1 (Nei-like 1) is one such Fpg/Nei family glycosylase that guards the mammalian genome against oxidation damage (7–10). NEIL1 is bifunctional in that it catalyzes both the hydrolysis of the N-glycosylic bond linking a base to a deoxyribose (glycosylase activity) and the subsequent cleavage of the DNA 3′ to the newly created apurinic/apyrimidinic site (lyase activity) (7–10). The N terminus contains the glycosylase domain of NEIL1, and the C terminus is intrinsically disordered (8, 11). Whereas the C terminus is dispensable for both glycosylase and lyase activities in vitro, it interacts with many proteins in vivo and is required for efficient DNA repair activity inside the cells (12, 13). NEIL1 is also unique among the three human NEIL proteins in that it is increased in an S-phase–specific manner and carries out prereplicative repair of oxidized bases in the human genome (8, 12). Moreover, increasing literature has further emphasized the importance of NEIL1’s cellular repair activity, as NEIL1 deficiency has led to multiple abnormalities and is associated with severe human diseases, including cancer (14–19). Additionally, emerging evidence has also implicated a role of NEIL1 in active DNA demethylation (20–22).
NEIL1 is capable of removing a wide array of oxidized pyrimidines and purines; representative substrates of extensive investigations include thymine glycol (Tg), 5-hydroxyuracil (5-OHU), 5-hydroxycytosine (5-OHC), dihydrothymine (DHT), and dihydrouracil (DHU), as well as the formamidopyridines (FapyA and FapyG), spiroiminodihydantoin (Sp), and guanidinohydantoin (Gh) (23–27). Among these lesions, Tg is the most common pyrimidine base modification produced under oxidative stress and ionizing radiation (28)—arising from oxidation of thymine and 5-methylcytosine—and is also a preferred substrate of NEIL1 (3). When left unrepaired, Tg severely blocks replicative polymerases in continued DNA synthesis and elicits cytotoxic lethal consequences (29). Interestingly, Tg can be differentially processed by two naturally existing NEIL1 forms: one with a Lys at position 242, which is encoded in the genome, and the other with an Arg242 residue, due to RNA editing of NEIL1 pre-mRNA; the former removes Tg from duplex DNA ∼30 times faster than the latter (30). Subsequent binding analysis using synthetic Tg analogs shows that binding affinities are similar for the two NEIL1 forms, hinting that the editing effect is beyond substrate affinity (31). Although the crystal structure of NEIL1 protein is determined (32), a complex structure of NEIL1 bound to duplex DNA has not been reported. Recent studies have determined several complex structures of MvNei1 (mimivirus Nei1), a viral ortholog of human NEIL1 (33, 34). However, MvNei1 and NEIL1 have apparent differences in their 3D structures and enzymatic properties (5). Hence, structures of human NEIL1 with bound substrates are desired to understand the mechanism of substrate recognition and excision by NEIL1.
Here we report the structures of human NEIL1 bound to dsDNA containing either a tetrahydrofuran (THF), a stable abasic site analog, or a Tg. Our structures show a flexible loop that adopts distinct conformations in the free protein, THF-, and Tg-bound structures. Combining crystallographic observations with quantum mechanics/molecular mechanics (QM/MM) simulations and biochemical investigations, we reveal that NEIL1 promotes tautomeric shifts of Tg in its active site, so as to allow optimal substrate binding. Moreover, the enzyme-promoted tautomerization also facilitates the NEIL1-mediated Tg excision reaction. Such tautomerism-dependent substrate recognition and catalysis is reported, to the best of our knowledge, for the first time in base-excision DNA repair.
Results
Crystal Structures of NEIL1/dsDNA Complexes.
We first identified a truncated NEIL1 lacking 95 amino acids at the C-terminal domain (CTD) for use in complex structure determination. This NEIL1CΔ95 truncation retained similar in vitro repair activities compared with the WT protein; its free protein structure (solved to 1.48 Å) further supported that additional truncations at the CTD have minimal impact on the structure of NEIL1’s glycosylase domain (SI Appendix, Fig. S1) [with rmsd ∼0.23 Å to the previously determined NEIL1CΔ56 structure; Protein Data Bank (PDB) 1TDH]. Hence, NEIL1CΔ95 was used for structural characterizations of NEIL1/dsDNA complexes.
We solved the structures of NEIL1 bound to dsDNA containing either a THF (a stable abasic site analog) or a Tg (a preferred substrate of NEIL1) (Fig. 1). For the THF-bound complexes, we used both the WT NEIL1 and a catalytically deficient NEIL1 variant (P2G); an overlay of the two structures showed that this mutation introduced minimal perturbation to the recognition of THF (SI Appendix, Fig. S2 A and B). Thus, this NEIL1 P2G mutation was further used for the Tg-bound structures.
Fig. 1.
Crystal structures of NEIL1 bound to dsDNA containing THF and Tg, respectively. (A) Overall view of NEIL1 bound to a THF-containing duplex, with the flipped THF shown in purple. (B) Accommodation of THF in the active site of NEIL1. Density map (2Fobs − Fcal) of THF is shown. Hydrogen bonds are shown in green dashed lines. The red dashed line shows the distance between the α-amino group of Pro2 and C1 atom of THF. (C) Overall view of NEIL1 bound to a Tg-containing duplex, with the flipped Tg shown in purple. (D) Overlay of the apo and THF-bound NEIL1 structures, highlighting the conformational change of Arg242. In the Tg-bound structure, Arg242 in the flexible lesion recognition loop flips over, and the polar side chain of Arg242 points to the Tg base. Tyr244 resides approximately in the same location in both apo (gray) and Tg-bound (green) structures. (E) Overlay of the lesion recognition in the apo (gray), Tg (green)-, and THF (cyan)-bound structures. The same angle as in D is shown here for comparison.
NEIL1 Uses a Flexible Lesion Recognition Loop to Allow Substrate Binding.
In all complex structures, DNA is bound in the DNA binding cleft of NEIL1 and is bent by ∼42°, similar to complex structures of EcoNei (Escherichia coli Nei) (35). Both Tg and THF are flipped out from the duplex, with Met81, Arg118, and Phe120 filling the void of DNA. A loop region (residues 240–252, between helices αG and αH) displays major conformational changes among the free protein, THF- and Tg-bound NEIL1 structures (Fig. 1 D and E and SI Appendix, Fig. S2C). In the absence of DNA, the loop adopts an “open” conformation. Upon binding to the THF-containing DNA, the entire loop moved ∼10 Å toward the flipped THF moiety, capping the THF and forming a “closed” conformation. In this conformation, the loop is kinked by ∼90° (Fig. 1E); such conformational change is at least partially driven by Tyr244, which caps THF by stacking upon the deoxyribose ring via its aromatic side chain (Fig. 1B). In the Tg structure, although the loop is partially disordered, a different closed conformation was observed. Instead of displacement of the entire loop seen in the THF structure, the difference in the Tg structure was mainly the flip-over of Arg242 and Gly243 with respect to the loop in the open conformation (Fig. 1D). As a result, the polar side chain of Arg242 now points right toward the extrahelical Tg base (see section below), whereas in the free protein structure, Arg242 points to an opposite direction and is fully solvent exposed. As a reference, Tyr244 resides in approximately the same location both in the open conformation and this second closed one (Fig. 1D). We also superimposed our NEIL1 structures onto the bacterial Fpg complex structures (36–38) and found that this flexible loop region corresponds to the so-called “lesion recognition loop” of Fpg proteins, typically composed of the α-F-β9/10 loop (SI Appendix, Fig. S2D). It was thought that NEIL1 lacked such a lesion recognition loop (5). Although the loop region between helices αG and αH is much shorter in NEIL1, our structures revealed that NEIL1 uses such a flexible loop to allow recognition of both THF and Tg.
Computational Optimizations of the Tg Crystal Structure Suggest That NEIL1 Uses a Tautomerization-Dependent Substrate Recognition Mechanism.
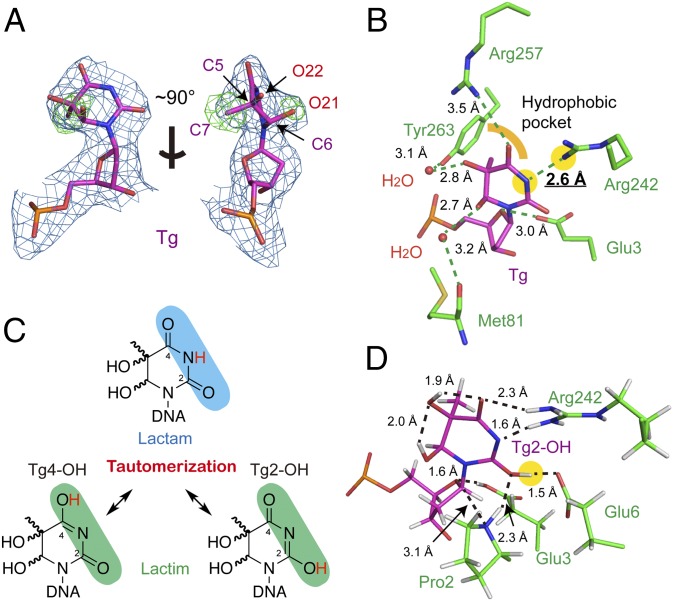
The excellent electron density of the flipped Tg allowed us to unambiguously assign its (5R, 6S) stereogeometry in the refined structure (Fig. 2A and SI Appendix, Fig. S3A). The 5-methyl group is tucked into a hydrophobic pocket formed by Tyr177, Phe256, Leu260, and Tyr263, whereas the two hydroxyl groups, on the opposite side of the Tg base, point toward a more open pocket with multiple water molecules in the structure (SI Appendix, Fig. S3B). Through these well-ordered water molecules, several indirect hydrogen bonds between the two hydroxyl groups and the main chain atoms of Met81 and Tyr263 were formed (Fig. 2B). At the Watson–Crick interface, Tg is nicely accommodated in the compact active site of NEIL1. For instance, Arg242, residing in the lesion recognition loop and exposed to solvent in the free protein structure, now flips over and points toward N3 of Tg; additionally, Arg257 also interacts with O4 of Tg (Fig. 2B).
Fig. 2.
Tautomerization-dependent recognition of Tg. (A) Electron density map of Tg. The blue 2Fobs − Fcal map is contoured at 1.2σ and the green Fobs − Fcal omit map—by removing the 5-methyl (C7) and 6-hydroxyl group (O21) of Tg—is contoured at 3.0σ [omit maps removing 5-hydroxyl group (O22) are shown in SI Appendix, Fig. S3A]. (B) The active-site pocket of Tg-bound NEIL1. Hydrogen bonds are shown in green dashed lines. The hydrophobic pocket surrounding the 5-methyl group of Tg is indicated by a yellow curve. The N3 of Tg and Nη of Arg242 is highlighted with yellow background. (C) Tg tautomers in the lactam (Upper) and lactim (Lower) forms. (D) Optimized structure of the Tg-bound NEIL1 active site. Due to the Tg2-OH tautomer, a new hydrogen bond was observed between 2-OH of Tg and Glu6. Key distances are marked in black (in angstroms), and the 2-OH group of Tg is highlighted with yellow background.
A closer inspection at the Tg recognition pocket revealed seemingly unfavorable interactions between the Tg base and the nearby Arg242: the guanidine group (Nη) of Arg242 is ∼2.6 Å away from the N3 position (as an –NH group in the canonical keto form, or “lactam”) of the Tg base, both of which being proton donors and hence are expected to be repulsive to each other. Then how would NEIL1 favorably bind Tg in its active site? Deprotonation of either Arg242 or Tg base would generate a normal donor/acceptor pair; however, the very high pKa values of both Arg242 (>12 in water) and Tg (much higher than thymine) (SI Appendix, Detailed Materials and Methods, section XI) make deprotonation energetically demanding. Alternatively, Tg base could undergo keto–enol conversion to become a “lactim” tautomer (Fig. 2C), which would then make the N3 position a hydrogen bond acceptor and hence allow favorable interactions with Arg242. Two lactim tautomers could be formed: one has the proton migrated to O2 (termed the “2-OH” lactim) and the other to O4 (the “4-OH” form) (Fig. 2C). To investigate whether or not tautomerization would allow favorable Tg binding, we performed all-atom QM(density functional theory)/MM simulations for the Tg-bound crystal structure (SI Appendix, Table S1). Both lactim tautomers were found to be more stable than the lactam form, with the 2-OH lactim structure being energetically the most favorable (Table 1). More specifically, one single tautomerization event stabilized the overall structure by ∼9.6 kcal/mol. In this optimized 2-OH lactim structure, both N3 and O4 atoms, being hydrogen bond acceptors, favorably interact with the guanidine group of Arg242, whereas the 2-OH group now forms an additional hydrogen bond to the carboxylate of Glu6 (Fig. 2D). Based on our calculations, all these interactions were found to be essential to maintain the conformation of the Tg-bound active site. As a matter of fact, loss of such interactions as in the lactam form destabilized the active site and led to an average structure that lacked all of the key features of the crystal structure (SI Appendix, Fig. S4). Therefore, our computational studies propose a mechanism in which NEIL1 requires Tg to undergo keto–enol conversion in its active site to achieve optimal substrate binding.
Table 1.
QM/MM calculated tautomerization energy of Tg when bound by NEIL1
| Tautomer | NEIL1 242R | NEIL1 242K |
| Tg (lactam) | 0.0* | 0.0 |
| Tg4-OH | −6.2 | −7.8 |
| Tg2-OH | −9.6 | −10.8 |
Unit: kcal/mol.
The corresponding minimized energy of lactam Tg–NEIL1 was set as the reference for calculating tautomerization energy of Tg4-OH and Tg2-OH in each system (R242 or K242). ΔEtau = E(X) − E(Tg), X = Tg4-OH or Tg2-OH.
Computational Simulations Suggest That Tautomerization Promotes NEIL1-Catalyzed Tg Excision.
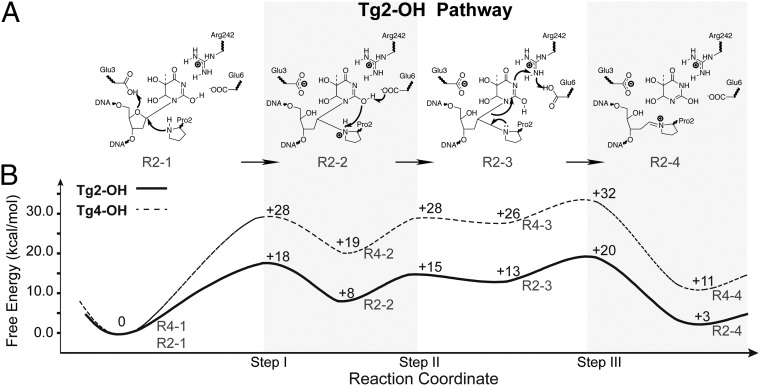
To investigate potential influence of the proposed tautomerization events on NEIL1-catalyzed Tg excision, we performed QM/MM calculations to simulate the base-excision reaction. We first established that a ribose-protonated reaction pathway—instead of a nucleobase-protonated pathway—represents a reasonable mechanism (Fig. 3A and SI Appendix, Figs. S5 and S6), consistent with observations from a recent study (19). The reaction pathway starting from the most stable initial 2-OH lactim structure was also found to be more energetically favorable than those starting from the 4-OH tautomer or lactam form, with the highest free-energy barrier being ∼18 kcal/mol for the excision reaction (Fig. 3B). The 2-OH lactim tautomer contributes to multiple aspects of the catalytic process by: (i) preactivating Tg. The 2-OH lactim nucleoside is ∼20.5 kcal/mol higher in energy than the lactam form in the absence of protein (SI Appendix, Table S4). Therefore, before the excision reaction, tautomeric shift of Tg into the 2-OH lactim form preactivates the substrate for the subsequent enzymatic reaction. (ii) Enabling proton transfer during Tg excision. In intermediate structure R2-2, the 2-OH group of Tg serves to deprotonate the positively charged Pro2, transferring a proton to the carboxylate of Glu6 (Fig. 3, step II). Additionally, in intermediate structure R2-3, the N3 atom of Tg abstracts one proton from Arg242, which in turn accepts one proton from Glu6 (Fig. 3, step III). (iii) Facilitating release of free Tg base. The cleaved base (intermediate structure R2-4), which now has an –NH group at position 3, is repulsive to the side chain of Arg242 and thus could be “expelled” from the active site, completing the glycosylase function of NEIL1. Therefore, in the proposed tautomerization-dependent mechanism, this tautomerization event plays important roles in the entire base-excision reaction. We concluded that tautomerism of Tg not only allowed optimal substrate binding, but also promoted base excision of Tg.
Fig. 3.
Proposed mechanisms and computational verification. (A) Ribose-protonated pathway initiated with the NEIL1–R242–Tg2-OH structure. Along this pathway, we list the reactant (R2-1), two intermediate states (R2-2 and R2-3), and the product (R2-4). (B) The quantitative characterization of the above proposed mechanisms using QM/MM umbrella sampling (see SI Appendix, Detailed Materials and Methods, Section IX for more details): NEIL1–R242–Tg2-OH ribose-protonated pathway (full line), and NEIL1–R242–Tg4-OH ribose-protonated pathway (SI Appendix, Scheme S1) (dashed line). Numbers along the curves correspond to relative free energies of the transition states and intermediate states (including reactant and product).
A Naturally Existing NEIL1 Variant Also Supports the Tautomerization-Based Mechanism.
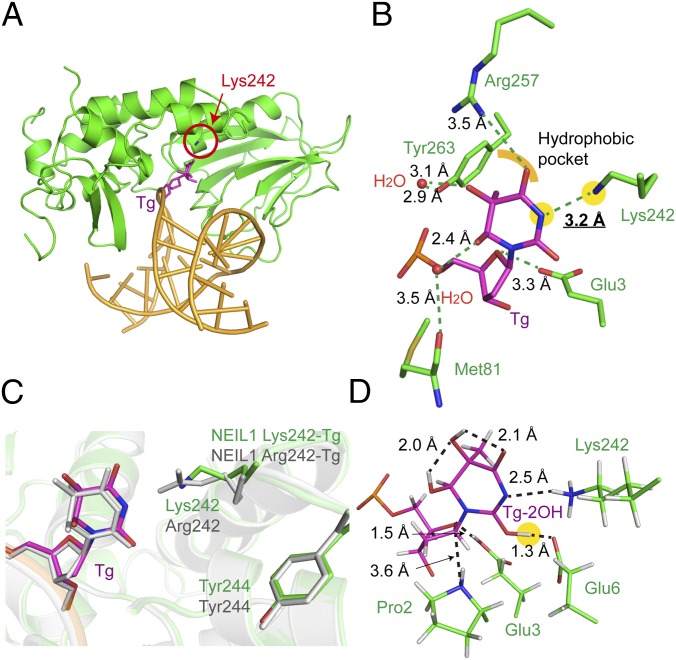
To further demonstrate such tautomerization-dependent readout and excision of Tg, we extended our study to a naturally existing NEIL variant, NEIL1 Lys242. Lys242 is the form of NEIL1 encoded in the genome; RNA editing of NEIL1 pre-mRNA results in the edited Arg242 form, which was the NEIL1 protein studied in the sections above. The unedited Lys242 form was reported to be active toward Tg as well; in fact, the Tg excision activity of this form is ∼30-fold faster (30). We also confirmed that the Lys242 form cleaves Tg much faster than the Arg242 form (Table 2). If such a tautomerization-dependent Tg recognition and excision mechanism is also occurring to the unedited form, then the side chain of Lys would be expected to point toward the Watson–Crick interface of Tg and be in close contact with the N3 position, as observed in the NEIL1 Arg242 Tg structure. To test this hypothesis, we crystallized the complex structure of NEIL1 Lys242 bound to Tg-containing DNA (Fig. 4A). The NEIL1 Lys242 Tg structure is nearly identical to the NEIL1 Arg242 Tg structure (SI Appendix, Fig. S7A); Tg is flipped into the same binding pocket and recognized by similar interactions as those found in the Arg242 Tg structure (Fig. 4 A–C). Remarkably, the side chain of Lys242 extends right toward the Watson–Crick interface of Tg, and its ε-amino group is ∼3.2 Å away from the N3 of Tg. To further support a tautomerization-dependent mechanism for NEIL1 Lys242, we again performed QM/MM calculations for Lys242-mediated Tg binding and excision reaction (Fig. 4D and SI Appendix, Fig. S6A). Our results showed that NEIL1 Lys242 again preferentially recognizes the 2-OH lactim tautomer structure (∼10.8 kcal/mol more stable than the lactam structure) (Table 1). Additionally, this tautomerization event was also found to promote the excision reaction for the unedited protein.
Table 2.
Rate constants (kobs) of Tg removal by NEIL1 with varying residues at position 242
| NEIL1 residues | kobs (min−1) |
| NEIL1 242R | 0.0224 ± 0.004 |
| NEIL1 242K | 1.901 ± 0.11 |
| NEIL1 242A* | 0.0015 ± 0.0005 |
| NEIL1 242E* | 0.0011 ± 0.0003 |
| NEIL1 242Q* | 0.00096 ± 0.0003 |
Rate constants in min−1 measured under single-turnover conditions at 16 °C.
Slow reaction rates were determined based on initial rate rather than complete fitting of the curve.
Fig. 4.
Structure of unedited NEIL1 (Lys242) bound to Tg. (A) Overall view of unedited NEIL1 bound to dsDNA containing Tg. Lys242 is highlighted in a red circle. (B) The active-site pocket of unedited NEIL1 bound to Tg. Hydrogen bonds are shown in green dashed lines. The N3 of Tg and Nζ of Lys242 is highlighted with yellow background. (C) Superposition of unedited (green) and edited (gray) NEIL1–Tg structures. The positions of the flipped base are almost identical, and both Arg242 and Lys242 point toward Tg. (D) Optimized structure of the Tg-bound NEIL1 (Lys242) active site. A hydrogen bond between 2-OH of Tg and Glu6 was also observed in this structure. Key distances are marked in black (in angstroms), and the 2-OH group is highlighted with yellow background.
Finally, we performed single turnover kinetics experiments to investigate the roles of key amino acid residues that are important for the tautomerization-based recognition and excision of Tg (Table 2). Because both NEIL1 Lys242 and Arg242 forms were capable of promoting Tg tautomerization, we sought to introduce mutations that have either neutral or acidic side chains so as to disrupt such tautomerization events. For all of the five mutants we tested, Tg excision reactions were significantly impeded, with the slowest mutant Gln242 being ∼15-fold and >1,000-fold slower than Arg242 and Lys242, respectively. Interestingly, the lyase activity of NEIL1 was found to be very similar for Arg242 and Lys242 (SI Appendix, Fig. S8 A and B); even for the Ala242 NEIL1 mutant, its lyase activity appeared to be unaffected, hinting that such tautomerization is important for base-excision activity of Tg but not for the lyase function of NEIL1. This notion is further supported by the crystal structure of NEIL1 Lys242 bound to THF-containing DNA, where Lys242 points away from THF (but overlaps well with that of Arg242 in the THF-containing structure) (SI Appendix, Fig. S7B). Additionally, our simulation of the excision reaction also confirmed Glu6 as an important residue for catalysis. Consistent with our proposed catalytic mechanism, E6A NEIL1 was found to be severely hindered in Tg excision (SI Appendix, Fig. S8 C and D). Thus, this biochemical evidence further supported the essential roles of the amino acid residues in the Tg excision function of NEIL1.
Discussion
NEIL1 is an Fpg/Nei family DNA glycosylase that functions importantly in protecting mammalian genomes from oxidation damage. In this study, we reported several complex structures of NEIL1 bound to THF and Tg. These structures identified a 13-amino acid residue loop that plays important roles in the recognition of both THF and Tg. For oxoG-recognizing Fpg proteins, the similar lesion recognition loop is much longer, although the lesion recognition loops of Bacillus stearothermophilus Fpg (BstFpg) and Lactococcus lactis Fpg were either visible or disordered in different structures (36, 38, 39); for Arabidopsis thaliana Fpg which does not excise oxoG, its corresponding loop was found to be in one uniform conformation in the apo and THF-bound structures (40). For the Tg-recognizing MvNei1, only one conformation was observed for the corresponding loop in the apo, THF- and Tg-bound structures of MvNei1, which mostly resembles the THF-bound NEIL1 structure (33, 34). Nevertheless, our structures revealed that the lesion recognition loop of NEIL1 can adopt multiple conformations, and this flexibility is important for optimal substrate binding.
Our study showed that recognition of Tg by NEIL1 involves multiple interactions with the Tg base. Besides the cis (5R, 6S) isomer used in this study, Tg can also exist in the other (5S, 6R) isomer; both diastereoisomers are efficiently removed by NEIL1 (41, 42). In our structures, the two hydroxyl groups were recognized by water-mediated hydrogen bonds instead of direct interactions with the protein; such a feature might provide flexibility to the accommodation of both stereoisomers in the binding pocket. More importantly, our computational studies suggested that interactions on the Watson–Crick interface of Tg contribute primarily to its binding by NEIL1; these interactions are achiral and are expected to be retained for the (5S, 6R) isomer.
A unique feature of Tg recognition by NEIL1 is that NEIL1 promoted a tautomeric shift of Tg in its active site so as to achieve favorable binding. Such a tautomerization-dependent mechanism was first hinted at by the NEIL1 Arg242 Tg structure, then proposed by our computational simulations, and eventually corroborated by the NEIL1 Lys242 Tg structure and additional biochemical evidence. Although such protein-mediated tautomerization stabilized the NEIL1/DNA complex by ∼9.6 kcal/mol, tautomerization of the free Tg nucleotide is nonspontaneous and endoergic (SI Appendix, Table S4). The energy barrier of tautomerization can be affected by the conjugation system of a base: for Tg, oxidation at its C5–C6 double bond reduces its aromatic character and effectively lowers the energy barrier. Additionally, disruption of the aromaticity redistributes the electron density of the heteroatoms (O2, N3, and O4 of Tg) on the Watson–Crick interface, rendering O2 more nucleophilic and hence Tg more capable of tautomerization (SI Appendix, Fig. S9 C and D). In comparison, tautomerization is more energetically demanding for a regular T. Thus, it is tempting to speculate that such differences in the chemical nature of nucleobases could be potentially used by NEIL1 to discriminate a cognate substrate Tg from a nonsubstrate T: even if a regular thymine base were accidentally flipped into the active site of NEIL1, the difficulty of its undergoing tautomerization would result in unfavorable interactions with NEIL1 and hence fast rejection from the active site. Alternatively, it is also possible that an accidentally flipped thymine could be differentially recognized by NEIL1. The use of different binding pockets to discriminate a flipped base has precedence in the case of hOGG1, which uses two distinct pockets for recognizing oxoG and G (43).
The tautomerization-dependent substrate readout mechanism of NEIL1 is also reminiscent of oxoG recognition by the Fpg proteins (38, 39). Unlike a regular guanosine, oxoG has a protonated N7 due to the presence of a carbonyl group at position 8. The keto form of oxoG is more stable than its enol form and is important for optimal oxoG recognition when bound by glycosylases. For Tg, although the keto form is also more stable in solution, tautomeric shifts of Tg into the lactim form render the entire NEIL1/dsDNA complex more stable. Interestingly, an overlay of the Tg structures with the oxoG-bound BstFpg structure (PDB ID code 1R2Y) reveals that N3 of Tg overlaps well with N7 of oxoG (SI Appendix, Fig. S10). Thus, it appears that these glycosylases—recognizing different oxidation damage—can both read out the protonation states of flipped DNA bases so as to achieve optimal substrate binding.
Our results also showed that tautomerization effectively promoted the NEIL1-catalyzed Tg excision reaction. Both the two NEIL1 variants (Arg242 and Lys242) promoted Tg excision, with the unedited form being ∼30- to 50-fold more efficient in removing Tg. Interestingly, in vitro binding experiments using noncleavable Tg analogs show that the two NEIL1 variants bind to Tg-containing DNA with very similar affinity (31); hence, it is tempting to speculate that specific step(s) during catalysis could be responsible for this observed difference in Tg excision. In fact, our simulations of the glycosylation reaction revealed that both Arg and Lys chemically participated in the catalysis. Given that Lys has a lower pKa value compared with Arg, it is anticipated that Lys would be a better proton donor for the Tg base during the glycosidic bond cleavage step (refer to intermediate structure R2-3 in Fig. 3). It should be noted that the accuracy of our current calculations does not permit free energy comparisons as precisely as to a few kcal/mol.
Besides Tg, NEIL also catalyzes the excision of a variety of modified DNA bases, including oxidized purines and pyrimidines. Whether or not NEIL1 also promotes tautomeric shifts of these substrates for efficient recognition and catalysis remains an open question at the moment. An ∼30- to 50-fold rate difference in Tg excision is observed between NEIL1 Arg242 and Lys242; yet unlike Tg, reaction rates are comparable for the two NEIL1 variants for both Gh and Sp (30). Therefore, it appears from this biochemical evidence that amino acid residues at position 242 may have different effects on the apparent reaction rates for different NEIL1 substrates. Additionally, the chemical structures of these NEIL1 substrates differ greatly, and such differences also influence their ability to shift into different tautomeric states. Thus, future structural and computational studies of these substrates bound to NEIL1 will be necessary to reveal whether or not NEIL1 uses different mechanisms for its recognition and excision.
Existing literature has documented tautomerization events of DNA bases that lead to important biological outcomes. One such example is the rare tautomer hypothesis of spontaneous mutagenesis, which states that mutations can arise through the formation of high-energy tautomeric forms of DNA bases at low frequency. Indeed, C:A and G:T mismatched pairs, in which one of the bases shifts to a tautomeric state, can adopt the canonical Watson–Crick geometry both in crystals (in the insertion site of a high-fidelity DNA polymerase) and in solution (44, 45). In another example, tautomerization (and base shifting) explains the mutagenicity of FapyG and FapyA, which cause transversion mutations during replication (46). However, in these cases, tautomerized DNA bases (whether regular or modified) are confined to the context of DNA base pairs instead of being recognized by proteins. Thus, to the best of our knowledge, tautomeric shifts of Tg by NEIL1 represent the first example of enzyme-promoted tautomerization, which allow efficient recognition and catalysis of substrates in an enzyme-catalyzed reaction.
Materials and Methods
The human NEIL1s in the pET30a(+) vector were transformed and expressed in E. coli BL21 (DE3) cells overnight. Purification was taken on nickel-nitrilotriacetic acid affinity chromatography and size exclusion chromatography. Protein–DNA complex (10 mg/mL) was crystallized with reservoir solution at 4 °C. The models of NEIL1–DNA complexes were solved by molecular replacement using 1DTH as a model (PhaserMR) and refined using Refmac5. Refinement statistics are shown in SI Appendix, Table S1. Detailed materials and methods (including the sections of molecular simulation) are in SI Appendix.
Supplementary Material
Acknowledgments
The authors thank Dr. Meng, Dr. Y. Q. Song, H. Zeng, and X. T. Shu for technical assistance; and the Shanghai Synchrotron Radiation Facility (BL17U), and the National Center for Protein Science Shanghai (BL19U) for assistance with X-ray data collection. This work was supported by the National Basic Research Foundation of China Grant 2014CB964900 (to C.Y.) and the National Natural Science Foundation of China Grants 31270838 and 21522201 (to C.Y.), and Grant 21573006 (to Y.Q.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.B. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5ITQ, 5ITT, 5ITR, 5ITU, 5ITY, and 5ITX).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604591113/-/DCSupplemental.
References
- 1.Lindahl T. My journey to DNA repair. Genomics Proteomics Bioinformatics. 2013;11(1):2–7. doi: 10.1016/j.gpb.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18(1):27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalhus B, Laerdahl JK, Backe PH, Bjørås M. DNA base repair--recognition and initiation of catalysis. FEMS Microbiol Rev. 2009;33(6):1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- 5.Wallace SS. DNA glycosylases search for and remove oxidized DNA bases. Environ Mol Mutagen. 2013;54(9):691–704. doi: 10.1002/em.21820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004;14(1):43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1(7):517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 8.Hazra TK, et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99(6):3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morland I, et al. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: An alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30(22):4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takao M, et al. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002;277(44):42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 11.Hegde ML, et al. The disordered C-terminal domain of human DNA glycosylase NEIL1 contributes to its stability via intramolecular interactions. J Mol Biol. 2013;425(13):2359–2371. doi: 10.1016/j.jmb.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegde ML, et al. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc Natl Acad Sci USA. 2013;110(33):E3090–E3099. doi: 10.1073/pnas.1304231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegde PM, et al. The C-terminal domain (CTD) of human DNA glycosylase NEIL1 is required for forming BERosome repair complex with dna replication proteins at the replicating genome: DOMINANT NEGATIVE FUNCTION OF THE CTD. J Biol Chem. 2015;290(34):20919–20933. doi: 10.1074/jbc.M115.642918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinmura K, et al. Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis. 2004;25(12):2311–2317. doi: 10.1093/carcin/bgh267. [DOI] [PubMed] [Google Scholar]
- 15.Vartanian V, et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci USA. 2006;103(6):1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan MK, et al. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst) 2009;8(7):786–794. doi: 10.1016/j.dnarep.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori H, et al. Deficiency of the oxidative damage-specific DNA glycosylase NEIL1 leads to reduced germinal center B cell expansion. DNA Repair (Amst) 2009;8(11):1328–1332. doi: 10.1016/j.dnarep.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canugovi C, et al. Endonuclease VIII-like 1 (NEIL1) promotes short-term spatial memory retention and protects from ischemic stroke-induced brain dysfunction and death in mice. Proc Natl Acad Sci USA. 2012;109(37):14948–14953. doi: 10.1073/pnas.1204156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadeghian K, et al. Ribose-protonated DNA base excision repair: A combined theoretical and experimental study. Angew Chem Int Ed Engl. 2014;53(38):10044–10048. doi: 10.1002/anie.201403334. [DOI] [PubMed] [Google Scholar]
- 20.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Müller U, Bauer C, Siegl M, Rottach A, Leonhardt H. TET-mediated oxidation of methylcytosine causes TDG or NEIL glycosylase dependent gene reactivation. Nucleic Acids Res. 2014;42(13):8592–8604. doi: 10.1093/nar/gku552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schomacher L, et al. Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat Struct Mol Biol. 2016;23(2):116–124. doi: 10.1038/nsmb.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43(50):15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 24.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47(27):7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazra TK, et al. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002;277(34):30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 26.Vik ES, et al. Biochemical mapping of human NEIL1 DNA glycosylase and AP lyase activities. DNA Repair (Amst) 2012;11(9):766–773. doi: 10.1016/j.dnarep.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49(8):1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33(1):1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 29.Clark JM, Beardsley GP. Functional effects of cis-thymine glycol lesions on DNA synthesis in vitro. Biochemistry. 1987;26(17):5398–5403. doi: 10.1021/bi00391a027. [DOI] [PubMed] [Google Scholar]
- 30.Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc Natl Acad Sci USA. 2010;107(48):20715–20719. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onizuka K, Yeo J, David SS, Beal PA. NEIL1 binding to DNA containing 2′-fluorothymidine glycol stereoisomers and the effect of editing. ChemBioChem. 2012;13(9):1338–1348. doi: 10.1002/cbic.201200139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doublié S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci USA. 2004;101(28):10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imamura K, Wallace SS, Doublié S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J Biol Chem. 2009;284(38):26174–26183. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imamura K, Averill A, Wallace SS, Doublié S. Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA. J Biol Chem. 2012;287(6):4288–4298. doi: 10.1074/jbc.M111.315309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zharkov DO, et al. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21(4):789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struct Biol. 2002;9(7):544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 37.Serre L, Pereira de Jésus K, Boiteux S, Zelwer C, Castaing B. Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 2002;21(12):2854–2865. doi: 10.1093/emboj/cdf304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003;278(51):51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 39.Pereira de Jésus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: Comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005;33(18):5936–5944. doi: 10.1093/nar/gki879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duclos S, et al. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair (Amst) 2012;11(9):714–725. doi: 10.1016/j.dnarep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McTigue MM, Rieger RA, Rosenquist TA, Iden CR, De Los Santos CR. Stereoselective excision of thymine glycol lesions by mammalian cell extracts. DNA Repair (Amst) 2004;3(3):313–322. doi: 10.1016/j.dnarep.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Miller H, et al. Stereoselective excision of thymine glycol from oxidatively damaged DNA. Nucleic Acids Res. 2004;32(1):338–345. doi: 10.1093/nar/gkh190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434(7033):612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci USA. 2011;108(43):17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW, Al-Hashimi HM. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature. 2015;519(7543):315–320. doi: 10.1038/nature14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gehrke TH, et al. Unexpected non-Hoogsteen-based mutagenicity mechanism of FaPy-DNA lesions. Nat Chem Biol. 2013;9(7):455–461. doi: 10.1038/nchembio.1254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.