Significance
Functional MRI (fMRI) is 25 years old, yet surprisingly its most common statistical methods have not been validated using real data. Here, we used resting-state fMRI data from 499 healthy controls to conduct 3 million task group analyses. Using this null data with different experimental designs, we estimate the incidence of significant results. In theory, we should find 5% false positives (for a significance threshold of 5%), but instead we found that the most common software packages for fMRI analysis (SPM, FSL, AFNI) can result in false-positive rates of up to 70%. These results question the validity of a number of fMRI studies and may have a large impact on the interpretation of weakly significant neuroimaging results.
Keywords: fMRI, statistics, false positives, cluster inference, permutation test
Abstract
The most widely used task functional magnetic resonance imaging (fMRI) analyses use parametric statistical methods that depend on a variety of assumptions. In this work, we use real resting-state data and a total of 3 million random task group analyses to compute empirical familywise error rates for the fMRI software packages SPM, FSL, and AFNI, as well as a nonparametric permutation method. For a nominal familywise error rate of 5%, the parametric statistical methods are shown to be conservative for voxelwise inference and invalid for clusterwise inference. Our results suggest that the principal cause of the invalid cluster inferences is spatial autocorrelation functions that do not follow the assumed Gaussian shape. By comparison, the nonparametric permutation test is found to produce nominal results for voxelwise as well as clusterwise inference. These findings speak to the need of validating the statistical methods being used in the field of neuroimaging.
Since its beginning more than 20 years ago, functional magnetic resonance imaging (fMRI) (1, 2) has become a popular tool for understanding the human brain, with some 40,000 published papers according to PubMed. Despite the popularity of fMRI as a tool for studying brain function, the statistical methods used have rarely been validated using real data. Validations have instead mainly been performed using simulated data (3), but it is obviously very hard to simulate the complex spatiotemporal noise that arises from a living human subject in an MR scanner.
Through the introduction of international data-sharing initiatives in the neuroimaging field (4–10), it has become possible to evaluate the statistical methods using real data. Scarpazza et al. (11), for example, used freely available anatomical images from 396 healthy controls (4) to investigate the validity of parametric statistical methods for voxel-based morphometry (VBM) (12). Silver et al. (13) instead used image and genotype data from 181 subjects in the Alzheimer’s Disease Neuroimaging Initiative (8, 9), to evaluate statistical methods common in imaging genetics. Another example of the use of open data is our previous work (14), where a total of 1,484 resting-state fMRI datasets from the 1,000 Functional Connectomes Project (4) were used as null data for task-based, single-subject fMRI analyses with the SPM software. That work found a high degree of false positives, up to 70% compared with the expected 5%, likely due to a simplistic temporal autocorrelation model in SPM. It was, however, not clear whether these problems would propagate to group studies. Another unanswered question was the statistical validity of other fMRI software packages. We address these limitations in the current work with an evaluation of group inference with the three most common fMRI software packages [SPM (15, 16), FSL (17), and AFNI (18)]. Specifically, we evaluate the packages in their entirety, submitting the null data to the recommended suite of preprocessing steps integrated into each package.
The main idea of this study is the same as in our previous one (14). We analyze resting-state fMRI data with a putative task design, generating results that should control the familywise error (FWE), the chance of one or more false positives, and empirically measure the FWE as the proportion of analyses that give rise to any significant results. Here, we consider both two-sample and one-sample designs. Because two groups of subjects are randomly drawn from a large group of healthy controls, the null hypothesis of no group difference in brain activation should be true. Moreover, because the resting-state fMRI data should contain no consistent shifts in blood oxygen level-dependent (BOLD) activity, for a single group of subjects the null hypothesis of mean zero activation should also be true. We evaluate FWE control for both voxelwise inference, where significance is individually assessed at each voxel, and clusterwise inference (19–21), where significance is assessed on clusters formed with an arbitrary threshold.
In brief, we find that all three packages have conservative voxelwise inference and invalid clusterwise inference, for both one- and two-sample t tests. Alarmingly, the parametric methods can give a very high degree of false positives (up to 70%, compared with the nominal 5%) for clusterwise inference. By comparison, the nonparametric permutation test (22–25) is found to produce nominal results for both voxelwise and clusterwise inference for two-sample t tests, and nearly nominal results for one-sample t tests. We explore why the methods fail to appropriately control the false-positive risk.
Results
A total of 2,880,000 random group analyses were performed to compute the empirical false-positive rates of SPM, FSL, and AFNI; these comprise 1,000 one-sided random analyses repeated for 192 parameter combinations, three thresholding approaches, and five tools in the three software packages. The tested parameter combinations, given in Table 1, are common in the fMRI field according to a recent review (26). The following five analysis tools were tested: SPM OLS, FSL OLS, FSL FLAME1, AFNI OLS (3dttest++), and AFNI 3dMEMA. The ordinary least-squares (OLS) functions only use the parameter estimates of BOLD response magnitude from each subject in the group analysis, whereas FLAME1 in FSL and 3dMEMA in AFNI also consider the variance of the subject-specific parameter estimates. To compare the parametric statistical methods used by SPM, FSL, and AFNI to a nonparametric method, all analyses were also performed using a permutation test (22, 23, 27). All tools were used to generate inferences corrected for the FWE rate over the whole brain.
Table 1.
Parameters tested for the different fMRI software packages, giving a total of 192 (3 × 2 × 2 × 4 × 2 × 2) parameter combinations and three thresholding approaches
| Parameter | Values used |
| fMRI data | Beijing (198 subjects), Cambridge (198 subjects), Oulu (103 subjects) |
| Block activity paradigms | B1 (10-s on off), B2 (30-s on off) |
| Event activity paradigms | E1 (2-s activation, 6-s rest), E2 (1- to 4-s activation, 3- to 6-s rest, randomized) |
| Smoothing | 4-, 6-, 8-, 10-mm FWHM |
| Analysis type | One-sample t test (group activation), two-sample t test (group difference) |
| No. of subjects | 20, 40 |
| Inference level | Voxel, cluster |
| CDT | P = 0.01 (z = 2.3), P = 0.001 (z = 3.1) |
One thousand group analyses were performed for each parameter combination.
Resting-state fMRI data from 499 healthy controls, downloaded from the 1,000 Functional Connectomes Project (4), were used for all analyses. Resting-state data should not contain systematic changes in brain activity, but our previous work (14) showed that the assumed activity paradigm can have a large impact on the degree of false positives. Several different activity paradigms were therefore used, two block based (B1 and B2) and two event related (E1 and E2); see Table 1 for details.
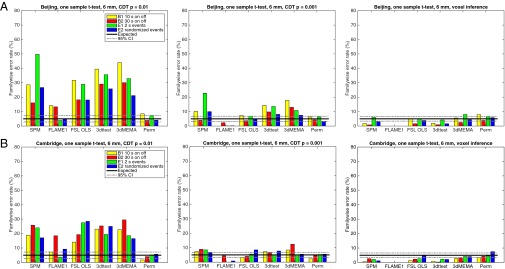
Fig. 1 presents the main findings of our study, summarized by a common analysis setting of a one-sample t test with 20 subjects and 6-mm smoothing [see SI Appendix, Figs. S1–S6 (20 subjects) and SI Appendix, Figs. S7–S12 (40 subjects) for the full results]. In broad summary, parametric software’s FWE rates for clusterwise inference far exceed their nominal 5% level, whereas parametric voxelwise inferences are valid but conservative, often falling below 5%. Permutation false positives are controlled at a nominal 5% for the two-sample t test, and close to nominal for the one-sample t test. The impact of smoothing and cluster-defining threshold (CDT) was appreciable for the parametric methods, with CDT P = 0.001 (SPM default) having much better FWE control than CDT P = 0.01 [FSL default; AFNI does not have a default setting, but P = 0.005 is most prevalent (21)]. Among the parametric software packages, FSL’s FLAME1 clusterwise inference stood out as having much lower FWE, often being valid (under 5%), but this comes at the expense of highly conservative voxelwise inference.
Fig. 1.
Results for one-sample t test, showing estimated FWE rates for (A) Beijing and (B) Cambridge data analyzed with 6 mm of smoothing and four different activity paradigms (B1, B2, E1, and E2), for SPM, FSL, AFNI, and a permutation test. These results are for a group size of 20. The estimated FWE rates are simply the number of analyses with any significant group activation divided by the number of analyses (1,000). From Left to Right: Cluster inference using a cluster-defining threshold (CDT) of P = 0.01 and a FWE-corrected threshold of P = 0.05, cluster inference using a CDT of P = 0.001 and a FWE-corrected threshold of P = 0.05, and voxel inference using a FWE-corrected threshold of P = 0.05. Note that the default CDT is P = 0.001 in SPM and P = 0.01 in FSL (AFNI does not have a default setting).
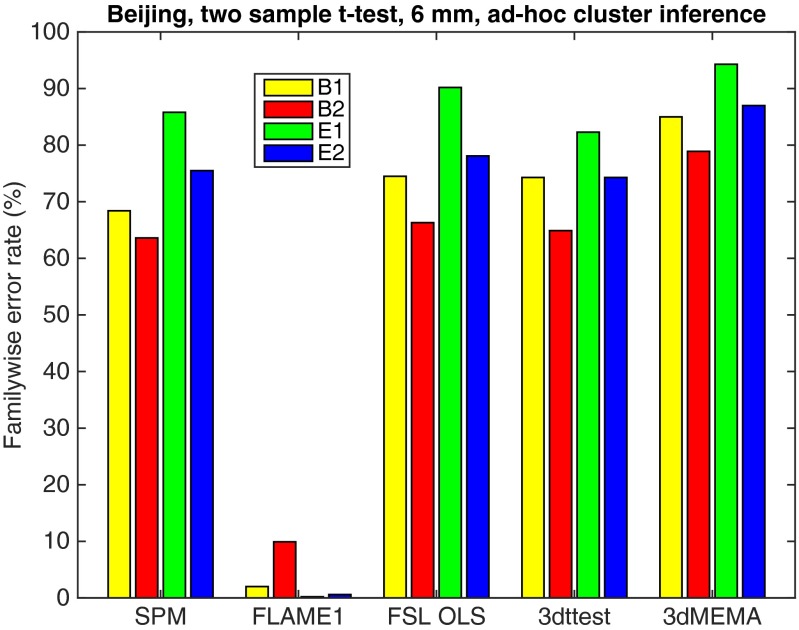
We also examined an ad hoc but commonly used thresholding approach, where a CDT of P = 0.001 (uncorrected for multiple comparisons) is used together with an arbitrary cluster extent threshold of 10 8-mm3 voxels (26, 28). We conducted an additional 1,000 analyses repeated for four assumed activity paradigms and the five different analysis tools (Fig. 2). Although no precise control of false positives is assured, we found this makeshift inference method had FWE ranging 60–90% for all functions except FLAME1 in FSL. Put another way, this “P = 0.001 + 10 voxels” method has a FWE-corrected P value of 0.6–0.9. We now seek to understand the sources of these inaccuracies.
Fig. 2.
Results for two-sample t test and ad hoc clusterwise inference, showing estimated FWE rates for 6 mm of smoothing and four different activity paradigms (B1, B2, E1, and E2), for SPM, FSL, and AFNI. These results were generated using the Beijing data and 20 subjects in each group analysis. Each statistic map was first thresholded using a CDT of P = 0.001 (uncorrected for multiple comparisons), and the surviving clusters were then compared with a cluster extent threshold of 80 mm3 (10 voxels for SPM and FSL which used 2 × 2 × 2 mm3 voxels, three voxels for AFNI, which used 3 × 3 × 3 mm3 voxels). The estimated FWE rates are simply the number of analyses with a significant result divided by the number of analyses (1,000).
Comparison of Empirical and Theoretical Test Statistic Distributions.
As a first step to understand the inaccuracies in the parametric methods, the test statistic values (t or z scores, as generated by each package) were compared with their theoretical null distributions. SI Appendix, Fig. S13, shows the histogram of all brain voxels for 1,000 group analyses. The empirical and theoretical nulls are well matched, except for FSL FLAME1, which has lower variance () than the theoretical null (). This is the proximal cause of the highly conservative results from FSL FLAME1. The mixed-effects variance is composed of intrasubject and intersubject variance (, respectively), and although other software packages do not separately estimate each, FLAME1 estimates each and constrains to be positive. In these null data, the true effect in each subject is zero, and thus the true . Thus, unless FLAME1's is correctly estimated to be 0, it can only be positively biased, and in fact this point was raised by the original authors (29).
In an follow-up analysis on FSL FLAME1 (SI Appendix), we conducted two-sample t tests on task fMRI data, randomly splitting subjects into two groups. In this setting, the two-sample null hypothesis was still true, but . Here, we found cluster false-positive rates comparable to FSL OLS (44.8% for CDT P = 0.01 and 13.8% for CDT P = 0.001), supporting our conjecture of zero between-subject variance as the cause of FLAME1’s conservativeness on completely null resting data.
Spatial Autocorrelation Function of the Noise.
SPM and FSL depend on Gaussian random-field theory (RFT) for FWE-corrected voxelwise and clusterwise inference. However, RFT clusterwise inference depends on two additional assumptions. The first assumption is that the spatial smoothness of the fMRI signal is constant over the brain, and the second assumption is that the spatial autocorrelation function has a specific shape (a squared exponential) (30). To investigate the second assumption, the spatial autocorrelation function was estimated and averaged using 1,000 group difference maps. For each group difference map and each distance (1–20 mm), the spatial autocorrelation was estimated and averaged along x, y, and z. The empirical spatial autocorrelation functions are given in SI Appendix, Fig. S14. A reference squared exponential is also included for each software, based on an intrinsic smoothness of 9.5 mm (FWHM) for SPM, 9 mm for FSL, and 8 mm for AFNI (according to the mean smoothness of 1,000 group analyses, presented in SI Appendix, Fig. S15). The empirical spatial autocorrelation functions are clearly far from a squared exponential, having heavier tails. This may explain why the parametric methods work rather well for a high CDT (resulting in small clusters, more reflective of local autocorrelation) and not as well for a low CDT (resulting in large clusters, reflecting distant autocorrelation). SI Appendix, Fig. S16, shows how the cluster extent thresholds differ between the parametric and the nonparametric methods, for a CDT of P = 0.01. The nonparametric permutation test is valid for any spatial autocorrelation function and finds much more stringent cluster extent thresholds (three to six times higher compared with SPM, FSL, and AFNI).
To better understand the origin of the heavy tails, the spatial autocorrelation was estimated at different preprocessing stages (no preprocessing, after motion correction, after motion correction, and 6-mm smoothing) using the 198 subjects in the Beijing dataset. The resulting spatial autocorrelation functions are given in SI Appendix, Fig. S17. It is clear that the long tails exist in the raw data and become even more pronounced after the spatial smoothing. These long-tail spatial correlations also exist for MR phantoms (31) and can therefore be seen as scanner artifacts.
Spatial Distribution of False-Positive Clusters.
To investigate whether the false clusters appear randomly in the brain, all significant clusters (P 0.05, FWE-corrected) were saved as binary maps and summed together (SI Appendix, Fig. S18). These maps of voxelwise cluster frequency show the areas more and less likely to be marked as significant in a clusterwise analysis. Posterior cingulate was the most likely area to be covered by a cluster, whereas white matter was least likely. As this distribution could reflect variation in the local smoothness in the data, we used group residuals from 1,000 two-sample t tests to estimate voxelwise spatial smoothness (32) (SI Appendix, Fig. S19). The local smoothness maps show evidence of a posterior cingulate “hot spot” and reduced intensity in white matter, just as in the false-positive cluster maps. Notably, having local smoothness varying systematically with tissue type has also been observed for VBM data (13). In short, this suggests that violation of the stationary smoothness assumption may also be contributing to the excess of false positives.
In a follow-up analysis using the nonstationary toolbox for SPM (fmri.wfubmc.edu/cms/software#NS), which provides parametric cluster inference allowing for spatially varying smoothness, we calculated FWE rates for stationary as well as nonstationary smoothness. Use of nonstationary cluster size inference did not produce nominal FWE: relative to the stationary cluster size test, it produced lower but still invalid FWE for a CDT of P = 0.01, and higher FWE for a CDT of P = 0.001 (SI Appendix, Table S2). This inconclusive performance can be attributed to additional assumptions and approximations introduced by the nonstationary cluster size test that can degrade its performance (33, 34). In short, we still cannot rule out heterogeneous smoothness as contributor to the standard cluster size methods’ invalid performance.
Impact on a Non-Null, Task Group Analysis.
All of the analyses to this point have been based on resting-state fMRI data, where the null hypothesis should be true. We now use task data to address the practical question of “How will my FWE-corrected cluster P values change?” if a user were to switch from a parametric to a nonparametric method. We use four task datasets [rhyme judgment, mixed gambles (35), living–nonliving decision with plain or mirror-reversed text, word and object processing (36)] downloaded from OpenfMRI (7). The datasets were analyzed using a parametric (the OLS option in FSL’s FEAT) and a nonparametric method (the randomise function in FSL) using a smoothing of 5-mm FWHM (default option in FSL). The only difference between these two methods is that FSL FEAT-OLS relies on Gaussian RFT to calculate the corrected cluster P values, whereas randomise instead uses the data itself. The resulting cluster P values are given in SI Appendix, Table S3 (CDT of P = 0.01) and SI Appendix, Tables S4 and S5 (CDT of P = 0.001). SI Appendix, Fig. S20, summarizes these results, plotting the ratio of FWE-corrected P values, nonparametric to parametric, against cluster size. All nonparametric P values were larger than parametric (ratio 1). Although this could be taken as evidence of a conservative nonparametric procedure, the extensive simulations showing valid nonparametric and invalid parametric cluster size inference instead suggest inflated (biased) significance in the parametric inferences. For CDT P = 0.01, there were 23 clusters (in 11 contrasts) with FWE parametric P values significant at P = 0.05 that were not significant by permutation. For CDT P = 0.001, there were 11 such clusters (in eight contrasts). If we assume that these mismatches represent false positives, then the empirical FWE for these 18 contrasts considered is 11/18 = 61% for CDT P = 0.01 and 8/18 = 44% for CDT P = 0.001. These findings indicate that the problems exist also for task-based fMRI data, and not only for resting-state data.
Permutation Test for One-Sample t Test.
Although permutation tests have FWE within the expected bounds for all two-sample test results, for one-sample tests they can exhibit conservative or invalid behavior. As shown in SI Appendix, Figs. S3, S4, S9, and S10, the FWE can be as low as 0.8% or as high as 40%. The one-sample permutation FWE varies between site (Beijing, Cambridge, Oulu), but within each site shows a consistent pattern between the two CDTs and even for voxelwise inference. The one-sample permutation test comprises a sign flipping procedure, justified by symmetrically distributed errors (22). Although the voxel-level test statistics appear symmetric and do follow the expected parametric t distribution (SI Appendix, Fig. S13), the statistic values benefit from the central limit theorem and their symmetry does not imply symmetry of the data. We conducted tests of the symmetry assumption on the data for block design B1, a case suffering both spuriously low (Cambridge) and high (Beijing, Oulu) FWE (SI Appendix). We found very strong evidence of asymmetric errors, but with no consistent pattern of asymmetry; that is, some brain regions showed positive skew and others showed negative skew.
Discussion
Using mass empirical analyses with task-free fMRI data, we have found that the parametric statistical methods used for group fMRI analysis with the packages SPM, FSL, and AFNI can produce FWE-corrected cluster P values that are erroneous, being spuriously low and inflating statistical significance. This calls into question the validity of countless published fMRI studies based on parametric clusterwise inference. It is important to stress that we have focused on inferences corrected for multiple comparisons in each group analysis, yet some 40% of a sample of 241 recent fMRI papers did not report correcting for multiple comparisons (26), meaning that many group results in the fMRI literature suffer even worse false-positive rates than found here (37). According to the same overview (26), the most common cluster extent threshold used is 80 mm3 (10 voxels of size 2 × 2 × 2 mm), for which the FWE was estimated to be 60–90% (Fig. 2).
Compared with our previous work (14), the results presented here are more important for three reasons. First, the current study considers group analyses, whereas our previous study looked at single-subject analyses. Second, we here investigate the validity of the three most common fMRI software packages (26), whereas we only considered SPM in our previous study. Third, although we confirmed the expected finding of permutation’s validity for two-sample t tests, we found that some settings we considered gave invalid FWE control for one-sample permutation tests. We identified skewed data as a likely cause of this and identified a simple test for detecting skew in the data. Users should consider testing for skew before applying a one-sample t test, but it remains an important area for developing new methods for one-sample analyses (see, e.g., ref. 38).
Why Is Clusterwise Inference More Problematic than Voxelwise?
Our principal finding is that the parametric statistical methods work well, if conservatively, for voxelwise inference, but not for clusterwise inference. We note that other authors have found RFT clusterwise inference to be invalid in certain settings under stationarity (21, 30) and nonstationarity (13, 33). This present work, however, is the most comprehensive to explore the typical parameters used in task fMRI for a variety of software tools. Our results are also corroborated by similar experiments for structural brain analysis (VBM) (11–13, 39, 40), showing that cluster-based P values are more sensitive to the statistical assumptions. For voxelwise inference, our results are consistent with a previous comparison between parametric and nonparametric methods for fMRI, showing that a nonparametric permutation test can result in more lenient statistical thresholds while offering precise control of false positives (13, 41).
Both SPM and FSL rely on RFT to correct for multiple comparisons. For voxelwise inference, RFT is based on the assumption that the activity map is sufficiently smooth, and that the spatial autocorrelation function (SACF) is twice-differentiable at the origin. For clusterwise inference, RFT additionally assumes a Gaussian shape of the SACF (i.e., a squared exponential covariance function), that the spatial smoothness is constant over the brain, and that the CDT is sufficiently high. The 3dClustSim function in AFNI also assumes a constant spatial smoothness and a Gaussian form of the SACF (because a Gaussian smoothing is applied to each generated noise volume). It makes no assumption on the CDT and should be accurate for any chosen value. As the FWE rates are far above the expected 5% for clusterwise inference, but not for voxelwise inference, one or more of the Gaussian SACF, the stationary SACF, or the sufficiently high CDT assumptions (for SPM and FSL) must be untenable.
Why Does AFNIs Monte Carlo Approach, Unreliant on RFT, Not Perform Better?
As can be observed in SI Appendix, Figs. S2, S4, S8, and S10, AFNI’s FWE rates are excessive even for a CDT of P = 0.001. There are two main factors that explain these results.
First, AFNI estimates the spatial group smoothness differently compared with SPM and FSL. AFNI averages smoothness estimates from the first-level analysis, whereas SPM and FSL estimate the group smoothness using the group residuals from the general linear model (32). The group smoothness used by 3dClustSim may for this reason be too low (compared with SPM and FSL; SI Appendix, Fig. S15).
Second, a 15-year-old bug was found in 3dClustSim while testing the three software packages (the bug was fixed by the AFNI group as of May 2015, during preparation of this manuscript). The bug essentially reduced the size of the image searched for clusters, underestimating the severity of the multiplicity correction and overestimating significance (i.e., 3dClustSim FWE P values were too low).
Together, the lower group smoothness and the bug in 3dClustSim resulted in cluster extent thresholds that are much lower compared with SPM and FSL (SI Appendix, Fig. S16), which resulted in particularly high FWE rates. We find this to be alarming, as 3dClustSim is one of the most popular choices for multiple-comparisons correction (26).
We note that FWE rates are lower with the bug-fixed 3dClustSim function. As an example, the updated function reduces the degree of false positives from 31.0% to 27.1% for a CDT of P = 0.01, and from 11.5% to 8.6% for a CDT of P = 0.001 (these results are for two-sample t tests using the Beijing data, analyzed with the E2 paradigm and 6-mm smoothing).
Suitability of Resting-State fMRI as Null Data for Task fMRI.
One possible criticism of our work is that resting-state fMRI data do not truly compromise null data, as they may be affected by consistent trends or transients, for example, at the start of the session. However, if this were the case, the excess false positives would appear only in certain paradigms and, in particular, least likely in the randomized event-related (E2) design. Rather, the inflated false positives were observed across all experiment types with parametric cluster size inference, limiting the role of any such systematic effects. Additionally, one could argue that the spatial structure of resting fMRI, the very covariance that gives rise to “resting-state networks,” is unrepresentative of task data and inflates the spatial autocorrelation functions and induces nonstationarity. We do not believe this is the case because it has been shown that resting-state networks can be estimated from the residuals of task data (42), suggesting that resting data and task noise share similar properties. We assessed this in our four task datasets, estimating the spatial autocorrelation of the group residuals (SI Appendix, Fig. S21) and found the same type of heavy-tailed behavior as in the resting data. Furthermore, the same type of heavy-tail spatial autocorrelation has been observed for data collected with an MR phantom (31). Finally, another follow-up analysis on task data (see Comparison of Empirical and Theoretical Test Statistic Distributions and SI Appendix, Task-Based fMRI Data, Human Connectome Project, a two-sample t test on a random split of a homogeneous group of subjects) found inflated false-positive rates similar to the null data. Altogether, we find that these findings support the appropriateness of resting data as a suitable null for task fMRI.
The Future of fMRI.
Due to lamentable archiving and data-sharing practices, it is unlikely that problematic analyses can be redone. Considering that it is now possible to evaluate common statistical methods using real fMRI data, the fMRI community should, in our opinion, focus on validation of existing methods. The main drawback of a permutation test is the increase in computational complexity, as the group analysis needs to be repeated 1,000–10,000 times. However, this increased processing time is not a problem in practice, as for typical sample sizes a desktop computer can run a permutation test for neuroimaging data in less than a minute (27, 43). Although we note that metaanalysis can play an important role in teasing apart false-positive findings from consistent results, that does not mitigate the need for accurate inferential tools that give valid results for each and every study.
Finally, we point out the key role that data sharing played in this work and its impact in the future. Although our massive empirical study depended on shared data, it is disappointing that almost none of the published studies have shared their data, neither the original data nor even the 3D statistical maps. As no analysis method is perfect, and new problems and limitations will be certainly found in the future, we commend all authors to at least share their statistical results [e.g., via NeuroVault.org (44)] and ideally the full data [e.g., via OpenfMRI.org (7)]. Such shared data provide enormous opportunities for methodologists, but also the ability to revisit results when methods improve years later.
Materials and Methods
Only publicly shared anonymized fMRI data were used in this study. Data collection at the respective sites was subject to their local ethics review boards, who approved the experiments and the dissemination of the anonymized data. For the 1,000 Functional Connectomes Project, collection of the Cambridge data was approved by the Massachusetts General Hospital partners’ institutional review board (IRB); collection of the Beijing data was approved by the IRB of State Key Laboratory for Cognitive Neuroscience and Learning, Beijing Normal University; and collection of the Oulu data was approved by the ethics committee of the Northern Ostrobothnian Hospital District. Dissemination of the data was approved by the IRBs of New York University Langone Medical Center and New Jersey Medical School (4). The word and object processing experiment (36) was approved by the Berkshire National Health Service Research Ethics Committee. The mixed-gambles experiment (35), the rhyme judgment experiment, and the living–nonliving experiments were approved by the University of California, Los Angeles, IRB. All subjects gave informed written consent after the experimental procedures were explained.
The resting-state fMRI data from the 499 healthy controls were analyzed in SPM, FSL, and AFNI according to standard processing pipelines, and the analyses were repeated for four levels of smoothing (4-, 6-, 8-, and 10-mm FWHM) and four task paradigms (B1, B2, E1, and E2). Random group analyses were then performed using the parametric functions in the three softwares (SPM OLS, FLAME1, FSL OLS, 3dttest, 3dMEMA) as well as the nonparametric permutation test. The degree of false positives was finally estimated as the number of group analyses with any significant result, divided by the number of group analyses (1,000). All of the processing scripts are available at https://github.com/wanderine/ParametricMultisubjectfMRI.
Supplementary Material
Acknowledgments
We thank Robert Cox, Stephen Smith, Mark Woolrich, Karl Friston, and Guillaume Flandin, who gave us valuable feedback on this work. This study would not be possible without the recent data-sharing initiatives in the neuroimaging field. We therefore thank the Neuroimaging Informatics Tools and Resources Clearinghouse and all of the researchers who have contributed with resting-state data to the 1,000 Functional Connectomes Project. Data were also provided by the Human Connectome Project, WU-Minn Consortium (principal investigators: David Van Essen and Kamil Ugurbil; Grant 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research, and by the McDonnell Center for Systems Neuroscience at Washington University. We also thank Russ Poldrack and his colleagues for starting the OpenfMRI Project (supported by National Science Foundation Grant OCI-1131441) and all of the researchers who have shared their task-based data. The Nvidia Corporation, which donated the Tesla K40 graphics card used to run all the permutation tests, is also acknowledged. This research was supported by the Neuroeconomic Research Initiative at Linköping University, by Swedish Research Council Grant 2013-5229 (“Statistical Analysis of fMRI Data”), the Information Technology for European Advancement 3 Project BENEFIT (better effectiveness and efficiency by measuring and modelling of interventional therapy), and the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 7699.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602413113/-/DCSupplemental.
References
- 1.Ogawa S, et al. Intrinsic signal changes accompanying sensory stimulation: Functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci USA. 1992;89(13):5951–5955. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453(7197):869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- 3.Welvaert M, Rosseel Y. A review of fMRI simulation studies. PLoS One. 2014;9(7):e101953. doi: 10.1371/journal.pone.0101953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswal BB, et al. Toward discovery science of human brain function. Proc Natl Acad Sci USA. 2010;107(10):4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Essen DC, et al. WU-Minn HCP Consortium The WU-Minn Human Connectome Project: An overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poldrack RA, Gorgolewski KJ. Making big data open: Data sharing in neuroimaging. Nat Neurosci. 2014;17(11):1510–1517. doi: 10.1038/nn.3818. [DOI] [PubMed] [Google Scholar]
- 7.Poldrack RA, et al. Toward open sharing of task-based fMRI data: The OpenfMRI project. Front Neuroinform. 2013;7(12):12. doi: 10.3389/fninf.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller SG, et al. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15(4):869–877, xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack CR, Jr, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poline JB, et al. Data sharing in neuroimaging research. Front Neuroinform. 2012;6(9):9. doi: 10.3389/fninf.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scarpazza C, Sartori G, De Simone MS, Mechelli A. When the single matters more than the group: Very high false positive rates in single case voxel based morphometry. Neuroimage. 2013;70:175–188. doi: 10.1016/j.neuroimage.2012.12.045. [DOI] [PubMed] [Google Scholar]
- 12.Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage. 2000;11(6 Pt 1):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 13.Silver M, Montana G, Nichols TE. Alzheimer’s Disease Neuroimaging Initiative False positives in neuroimaging genetics using voxel-based morphometry data. Neuroimage. 2011;54(2):992–1000. doi: 10.1016/j.neuroimage.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eklund A, Andersson M, Josephson C, Johannesson M, Knutsson H. Does parametric fMRI analysis with SPM yield valid results? An empirical study of 1484 rest datasets. Neuroimage. 2012;61(3):565–578. doi: 10.1016/j.neuroimage.2012.03.093. [DOI] [PubMed] [Google Scholar]
- 15.Friston K, Ashburner J, Kiebel S, Nichols T, Penny W. Statistical Parametric Mapping: The Analysis of Functional Brain Images. Elsevier/Academic; London: 2007. [Google Scholar]
- 16.Ashburner J. SPM: A history. Neuroimage. 2012;62(2):791–800. doi: 10.1016/j.neuroimage.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Cox RW. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 19.Friston KJ, Worsley KJ, Frackowiak RSJ, Mazziotta JC, Evans AC. Assessing the significance of focal activations using their spatial extent. Hum Brain Mapp. 1994;1(3):210–220. doi: 10.1002/hbm.460010306. [DOI] [PubMed] [Google Scholar]
- 20.Forman SD, et al. Improved assessment of significant activation in functional magnetic resonance imaging (fMRI): Use of a cluster-size threshold. Magn Reson Med. 1995;33(5):636–647. doi: 10.1002/mrm.1910330508. [DOI] [PubMed] [Google Scholar]
- 21.Woo CW, Krishnan A, Wager TD. Cluster-extent based thresholding in fMRI analyses: Pitfalls and recommendations. Neuroimage. 2014;91:412–419. doi: 10.1016/j.neuroimage.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: A primer with examples. Hum Brain Mapp. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE. Permutation inference for the general linear model. Neuroimage. 2014;92:381–397. doi: 10.1016/j.neuroimage.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brammer MJ, et al. Generic brain activation mapping in functional magnetic resonance imaging: A nonparametric approach. Magn Reson Imaging. 1997;15(7):763–770. doi: 10.1016/s0730-725x(97)00135-5. [DOI] [PubMed] [Google Scholar]
- 25.Bullmore ET, et al. Global, voxel, and cluster tests, by theory and permutation, for a difference between two groups of structural MR images of the brain. IEEE Transactions on Medical Imaging. 1999;18(1):32–42. doi: 10.1109/42.750253. [DOI] [PubMed] [Google Scholar]
- 26.Carp J. The secret lives of experiments: Methods reporting in the fMRI literature. Neuroimage. 2012;63(1):289–300. doi: 10.1016/j.neuroimage.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Eklund A, Dufort P, Villani M, Laconte S. BROCCOLI: Software for fast fMRI analysis on many-core CPUs and GPUs. Front Neuroinform. 2014;8:24. doi: 10.3389/fninf.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieberman MD, Cunningham WA. Type I and type II error concerns in fMRI research: Re-balancing the scale. Soc Cogn Affect Neurosci. 2009;4(4):423–428. doi: 10.1093/scan/nsp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woolrich MW, Behrens TE, Beckmann CF, Jenkinson M, Smith SM. Multilevel linear modelling for FMRI group analysis using Bayesian inference. Neuroimage. 2004;21(4):1732–1747. doi: 10.1016/j.neuroimage.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 30.Hayasaka S, Nichols TE. Validating cluster size inference: Random field and permutation methods. Neuroimage. 2003;20(4):2343–2356. doi: 10.1016/j.neuroimage.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Kriegeskorte N, et al. Artifactual time-course correlations in echo-planar fMRI with implications for studies of brain function. Int J Imaging Syst Technol. 2008;18(5-6):345–349. [Google Scholar]
- 32.Kiebel SJ, Poline JB, Friston KJ, Holmes AP, Worsley KJ. Robust smoothness estimation in statistical parametric maps using standardized residuals from the general linear model. Neuroimage. 1999;10(6):756–766. doi: 10.1006/nimg.1999.0508. [DOI] [PubMed] [Google Scholar]
- 33.Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE. Nonstationary cluster-size inference with random field and permutation methods. Neuroimage. 2004;22(2):676–687. doi: 10.1016/j.neuroimage.2004.01.041. [DOI] [PubMed] [Google Scholar]
- 34.Salimi-Khorshidi G, Smith SM, Nichols TE. Adjusting the effect of nonstationarity in cluster-based and TFCE inference. Neuroimage. 2011;54(3):2006–2019. doi: 10.1016/j.neuroimage.2010.09.088. [DOI] [PubMed] [Google Scholar]
- 35.Tom SM, Fox CR, Trepel C, Poldrack RA. The neural basis of loss aversion in decision-making under risk. Science. 2007;315(5811):515–518. doi: 10.1126/science.1134239. [DOI] [PubMed] [Google Scholar]
- 36.Duncan KJ, Pattamadilok C, Knierim I, Devlin JT. Consistency and variability in functional localisers. Neuroimage. 2009;46(4):1018–1026. doi: 10.1016/j.neuroimage.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavlicová M, Cressie NA, Santner TJ. Testing for activation in data from FMRI experiments. J Data Sci. 2006;4(3):275–289. [Google Scholar]
- 39.Scarpazza C, Tognin S, Frisciata S, Sartori G, Mechelli A. False positive rates in voxel-based morphometry studies of the human brain: Should we be worried? Neurosci Biobehav Rev. 2015;52:49–55. doi: 10.1016/j.neubiorev.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Meyer-Lindenberg A, et al. False positives in imaging genetics. Neuroimage. 2008;40(2):655–661. doi: 10.1016/j.neuroimage.2007.11.058. [DOI] [PubMed] [Google Scholar]
- 41.Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: A comparative review. Stat Methods Med Res. 2003;12(5):419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- 42.Fair DA, et al. A method for using blocked and event-related fMRI data to study “resting state” functional connectivity. Neuroimage. 2007;35(1):396–405. doi: 10.1016/j.neuroimage.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eklund A, Dufort P, Forsberg D, LaConte SM. Medical image processing on the GPU—past, present and future. Med Image Anal. 2013;17(8):1073–1094. doi: 10.1016/j.media.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Gorgolewski KJ, et al. NeuroVault.org: A repository for sharing unthresholded statistical maps, parcellations, and atlases of the human brain. Neuroimage. 2016;124(Pt B):1242–1244. doi: 10.1016/j.neuroimage.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.