Significance
RNA helicases can remodel substrate RNA–protein complexes in diverse ways. However, these activities have typically been studied using noncognate helicase-substrate systems, because the in vivo targets of many helicases are unknown. We investigated how the Brr2 protein disrupts its cognate U4/U6 di-small nuclear RNA–protein complex (di-snRNP), a reaction required for spliceosome catalytic activation. Our results suggest that Brr2 acts like a switch, initiating displacement of U6 snRNA, thereby inducing it to adopt an alternative conformation incompatible with base pairing to U4 snRNA. This mechanism explains how one U4/U6-bound protein, pre-mRNA processing factor 3 (Prp3), is displaced, whereas Prp31 and small nuclear ribonucleoprotein-associated protein 1 (Snu13) are released in complex with U4 snRNA. This unprecedented mode of RNP disruption minimizes requirements for reassembly of the U4/U6 di-snRNP after splicing.
Keywords: pre-mRNA splicing, RNA helicase, RNP remodeling, small nuclear ribonucleoprotein particle, spliceosome catalytic activation
Abstract
The Brr2 RNA helicase disrupts the U4/U6 di-small nuclear RNA–protein complex (di-snRNP) during spliceosome activation via ATP-driven translocation on the U4 snRNA strand. However, it is unclear how bound proteins influence U4/U6 unwinding, which regions of the U4/U6 duplex the helicase actively unwinds, and whether U4/U6 components are released as individual molecules or as subcomplexes. Here, we set up a recombinant Brr2-mediated U4/U6 di-snRNP disruption system, showing that sequential addition of the U4/U6 proteins small nuclear ribonucleoprotein-associated protein 1 (Snu13), pre-mRNA processing factor 31 (Prp31), and Prp3 to U4/U6 di-snRNA leads to a stepwise decrease of Brr2-mediated U4/U6 unwinding, but that unwinding is largely restored by a Brr2 cofactor, the C-terminal Jab1/MPN domain of the Prp8 protein. Brr2-mediated U4/U6 unwinding was strongly inhibited by mutations in U4/U6 di-snRNAs that diminish the ability of U6 snRNA to adopt an alternative conformation but leave the number and kind of U4/U6 base pairs unchanged. Irrespective of the presence of the cofactor, the helicase segregated a Prp3-Prp31-Snu13-U4/U6 RNP into an intact Prp31-Snu13-U4 snRNA particle, free Prp3, and free U6 snRNA. Together, these observations suggest that Brr2 translocates only a limited distance on the U4 snRNA strand and does not actively release RNA-bound proteins. Unwinding is then completed by the partially displaced U6 snRNA adopting an alternative conformation, which leads to dismantling of the Prp3-binding site on U4/U6 di-snRNA but leaves the Prp31- and Snu13-binding sites on U4 snRNA unaffected. In this fashion, Brr2 can activate the spliceosome by stripping U6 snRNA of all precatalytic binding partners, while minimizing logistic requirements for U4/U6 di-snRNP reassembly after splicing.
RNA helicases form a large family of nucleotide triphosphate-dependent enzymes, many of which can unwind RNA duplexes in vitro (1, 2). However, in vivo, these enzymes typically act on RNA–protein complexes (RNPs) and can also exhibit other activities, including RNA annealing (3, 4), RNA clamping (5), displacement of RNA-bound proteins (6), or displacement of RNA-bound RNPs (7). Often, the precise functions of RNA helicases are unclear because their in vivo substrates are mostly unknown. Consequently, the RNP remodeling activities of RNA helicases have typically been studied using model RNPs that these helicases would not encounter naturally (6, 7).
Eight superfamily (SF) 2 RNA helicases are required for pre-mRNA splicing by the spliceosome (8). During every splicing reaction, these enzymes drive and control multiple compositional and conformational RNP remodeling events that accompany the initial assembly of an inactive spliceosome, its catalytic activation, its splicing catalysis, and its disassembly (8, 9). The most profound helicase-mediated rearrangements occur during spliceosome activation. In the precatalytic spliceosome, the U4 and U6 small nuclear (sn) RNAs are paired via two regions (stem I and stem II) (10) and are bound by several U4/U6 proteins (di-snRNP). During activation, this U4/U6 di-snRNP is disrupted by the Brr2 helicase (11–14), which liberates U6 snRNA and leads to release of U4 snRNA and U4/U6-bound proteins (15). U6 snRNA can then engage in alternative interactions with the pre-mRNA and U2 snRNA and form an internal stem loop (ISL) (16) that is essential for splicing catalysis (17–19).
Brr2 is a stable subunit of the U5 snRNP and an unusual member of the Ski2-like subfamily of SF2 helicases, with a tandem array of similarly structured active (N-terminal) and inactive (C-terminal) helicase cassettes (14). During U4/U6 di-snRNP disruption, Brr2 engages a single-stranded region of U4 snRNA preceding stem I (Fig. 1A) and translocates in a 3′-to-5′ direction on the U4 strand, excluding U6 snRNA and thereby unwinding the RNAs (20–22). Brr2 is extensively regulated both intra- and intermolecularly (14, 20, 23–28). The two helicase cassettes are preceded by an approximately 450-residue N-terminal region (NTR), which autoinhibits the enzyme via conformational clamping and substrate competition (28). Furthermore, a C-terminal Jab1 domain of the Prp8 protein can bind to the helicase (25, 26) and inhibit Brr2 in trans by inserting a C-terminal tail into its RNA-binding tunnel (25). Upon removal of the tail, the globular portion of the Jab1 domain is converted into a strong Brr2 activator (25, 27).
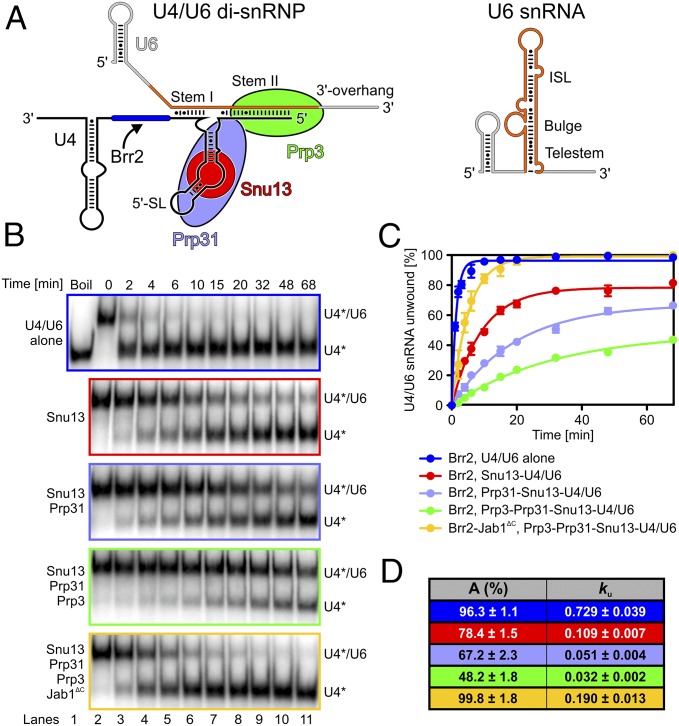
Fig. 1.
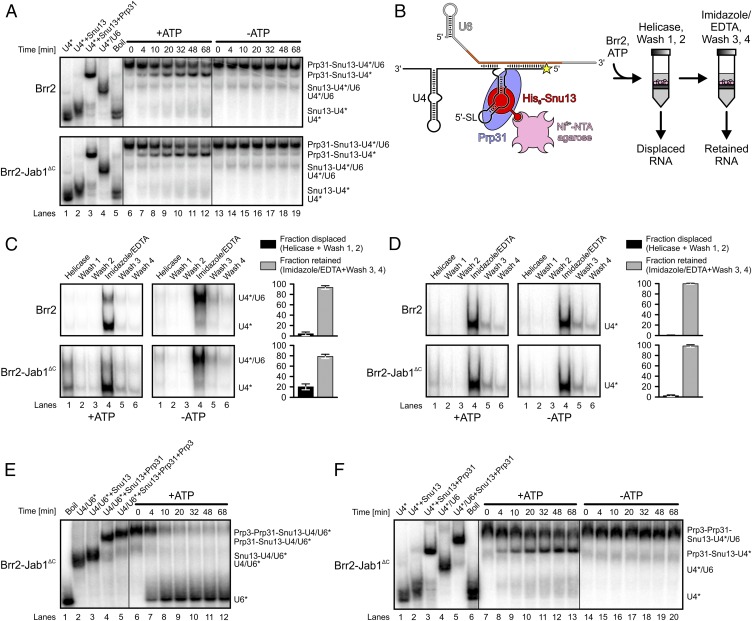
U4/U6-bound proteins modulate Brr2- and Brr2-Jab1∆C–mediated U4/U6 unwinding. (A) Scheme of the investigated U4/U6 di-snRNP (Left) and a possible U6 snRNA product (Right). Blue rectangle, Brr2 entry site; orange, U6 snRNA regions involved in both states. (B) Gel analysis of Brr2- or Brr2-Jab1∆C–mediated unwinding in the presence of the indicated proteins. Time points are indicated above the gels, and bands are identified on the right. *Radiolabel. (C) Quantification of the data in B. (D) Data were fit to a single exponential equation [fraction unwound = A{1 − exp(−kut)}; A, amplitude of the reaction; ku, apparent first-order rate constant for unwinding; t, time]. Data represent mean ± SEM of at least three independent experiments.
Among the U4/U6 proteins, small nuclear ribonucleoprotein-associated protein 1 (Snu13), pre-mRNA processing factor 31 (Prp31), and Prp3 bind in the vicinity of a three-way junction (3WJ) of the U4/U6 duplex, formed by stem I, stem II, and an intervening U4 5′ stem loop (5′SL) (22, 29–31) (Fig. 1A). They bind U4/U6 di-snRNA in a hierarchical manner, with Snu13 required for Prp31 binding (29, 32); both proteins together enhance binding of Prp3 (30, 32). Snu13 and Prp31 bind the U4 5′SL and maintain additional contacts to stems I and II, whereas Prp3 binds stem II and a U6 3′-overhang (22, 29–33). However, it is presently not known how U4/U6-bound proteins influence Brr2-mediated U4/U6 unwinding, which regions of the U4/U6 di-snRNA Brr2 actively unwind, and how U4/U6 components segregate upon Brr2-mediated U4/U6 di-snRNP disruption. Here, we have addressed these questions using in vitro RNA unwinding and RNP disruption assays with recombinantly reconstituted helicase machineries and RNP substrates. Our results suggest that initial active U4/U6 duplex unwinding by Brr2 triggers snapping of U6 snRNA into an alternative conformation, which leads to release of isolated Prp3 and a Prp31–Snu13–U4 snRNA complex. This unusual two-step mechanism allows Brr2 to displace intact U4 snRNP from U6 snRNA during spliceosome activation, although it translocates on the U4 strand.
Results
U4/U6-Bound Proteins Attenuate Brr2-Mediated U4/U6 Unwinding.
A recent cryo-electron microscopic (cryo-EM) structure of the yeast U4/U6•U5 tri-snRNP showed Brr2 bound to the Prp8 Jab1 domain (but not to other regions of Prp8) in a state ready to unwind the U4/U6 duplex (22), with the C-terminal Jab1 tail and the autoinhibitory NTR displaced from the Brr2 RNA-binding tunnel and the helicase cassettes, respectively. We recapitulated this active state in our experiments by using a truncated version of yeast Brr2, which lacks the first 270 autoinhibitory residues (“Brr2” in the following), and a truncated yeast Prp8 Jab1 domain, which lacks the last 16 inhibitory residues (Jab1ΔC). As substrates, we used full-length wild-type (wt) or mutant yeast U4/U6 di-snRNA alone or bound to various combinations of yeast Snu13 (full-length), Prp312–388 (“Prp31” in the following), and Prp3296–469 (“Prp3” in the following). The Prp31 and Prp3 constructs contained the major U4/U6 di-snRNA–binding portions of the proteins.
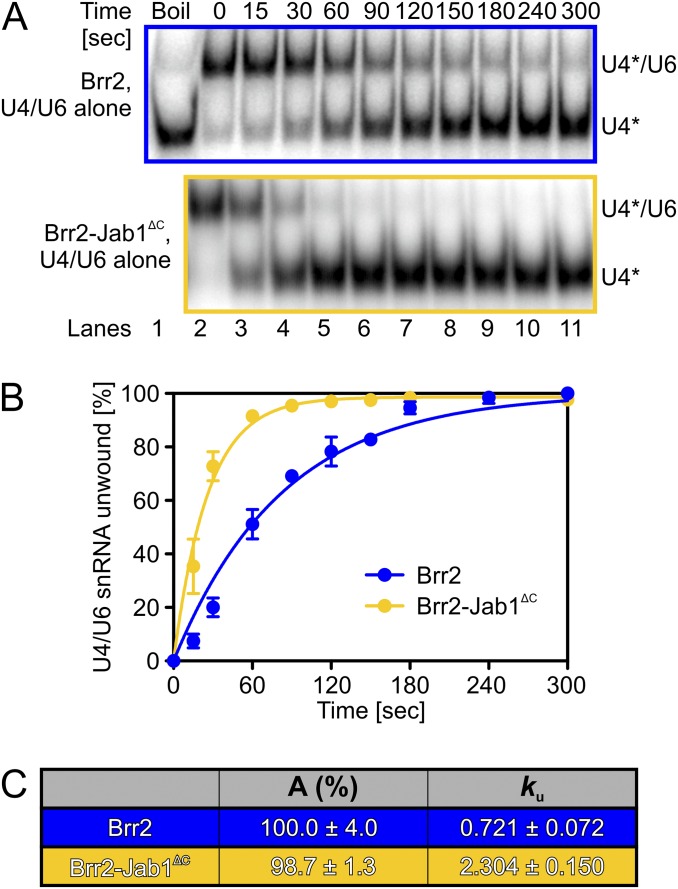
To investigate whether U4/U6-bound proteins modulate Brr2-mediated U4/U6 unwinding, we conducted systematic gel-based RNA unwinding and RNP disruption analyses in vitro. We used U4/U6 di-snRNA with U4 radiolabeled, preincubated alone or with Snu13, Prp31, and/or Prp3. After additional preincubation with Brr2, the reactions were started by the addition of ATP and Mg2+ (“ATP” in the following). Compared with the RNAs alone, addition of Snu13 reduced the rate of Brr2-mediated U4/U6 unwinding 6.7-fold, Prp31 led to an additional 2.1-fold decrease, and Prp3 gave a further 1.6-fold reduction (Fig. 1 B–D). However, a preassembled Brr2–Jab1ΔC complex exhibited a 5.9-fold faster unwinding rate for the Prp3-Prp31-Snu13-U4/U6 RNP compared with Brr2 alone (Fig. 1 B–D). Thus, although proteins that are bound at regions of U4/U6, toward which the Brr2 helicase is translocating during U4/U6 unwinding, substantially attenuate the helicase reaction, Jab1ΔC restores efficient Brr2-mediated U4/U6 unwinding to a large extent in the context of these proteins. As previously shown for full-length Brr2 (25), Jab1ΔC also enhanced the rate of unwinding for the truncated version of Brr2 in the absence of U4/U6-bound proteins (Fig. S1).
Fig. S1.
Jab1∆C enhances Brr2-mediated U4/U6 unwinding in the absence of U4/U6-bound proteins. (A) Gel analysis of Brr2- or Brr2-Jab1∆C–mediated unwinding of U4/U6 di-snRNA. Time points are indicated above the gels, and bands are identified on the right. *Radiolabel. (B) Quantification of the data presented in A. (C) Data were fit to a single exponential equation [fraction unwound = A{1 − exp(−ku t)}]. Data represent mean ± SEM of at least three independent experiments.
Mutations in Stem II and the U6 3′-Overhang Inhibit Brr2-Mediated U4/U6 Unwinding.
Three scenarios could be envisaged for Brr2-mediated U4/U6 di-snRNP disruption.
First, Brr2 might translocate along the entire U4 strand, actively unwinding all U4/U6 base-paired regions and the intervening U4 5′SL. Here, Snu13, Prp31, and Prp3 could hinder progression of Brr2 on the U4 strand, explaining their inhibitory effects on Brr2-mediated U4/U6 unwinding.
Second, Brr2 could unwind stem I, disengage from the substrate, and reengage at and unwind stem II, which would leave the U4 5′SL intact.
Third, Brr2 might only actively unwind stem I (or part of it), thereby inducing U6 snRNA to adopt an ISL-containing structure (34) (Fig. 1A) or a 3WJ structure suggested as an alternative for isolated U6 (35), both of which are mutually exclusive with U4/U6 pairing.
In the latter two cases, attenuation of Brr2-mediated U4/U6 unwinding by Snu13, Prp31, and Prp3 may reflect the overall stabilizing effect of the proteins on the U4/U6 duplex.
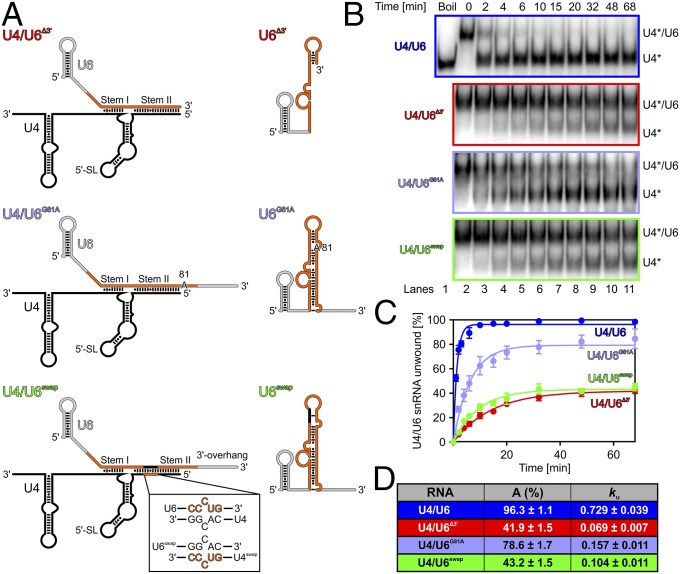
To distinguish between these scenarios, we tested Brr2-mediated unwinding of WT U4/U6 di-snRNA or U4/U6 variants that are impaired in U6 ISL/3WJ formation without influencing the number and kind of U4/U6 base pairs (Fig. 2A). The substrate variants are not expected to influence U4/U6 unwinding significantly by a mechanism that involves active unwinding of stem II (first and second scenarios) but could impair a mechanism that relies on snapping of U6 into an alternative conformation (third scenario). Brr2-mediated unwinding of U4/U6∆3′, lacking the U6 3′-overhang involved in ISL/3WJ formation (Fig. 2A), was 10.6-fold slower compared with unwinding of WT U4/U6 di-snRNA (Fig. 2 B–D). Notably, this manipulation only affects a region that is single-stranded in U4/U6 and does not lead to changes in the identity, number, or sequence of base pairs in stem I or stem II. This result is most likely not due to the removal of an alternative entry site for the helicase, because Brr2 does not unwind the RNAs by engaging the U6 single-stranded 3′-overhang, even when acting on isolated U4/U6 di-snRNA in vitro (20). Consistently, Brr2-mediated unwinding of a U4/U6G81A variant, in which only one nucleotide was changed in the U6 3′-overhang that is expected to disrupt a base pair in the alternative U6 structures (Fig. 2A), was 4.6-fold slower compared with unwinding of WT U4/U6 di-snRNA (Fig. 2 B–D). We also tested a U4/U6 variant (U4/U6swap) in which the U6 3′-overhang was unaltered and short regions were swapped between the RNAs to reduce the number of Watson–Crick base pairs in the ISL/3WJ forms of U6 snRNA, although only changing the orientation of some base pairs in U4/U6 (Fig. 2A). Unwinding of U4/U6swap di-snRNA was 7.0-fold slower than unwinding of WT U4/U6 (Fig. 2 B–D). Decreased unwinding was not caused by spontaneous reannealing of any of the variant U4 and U6 RNAs (Fig. S2). These results support the idea that alternative folding of the U6 snRNA favors unwinding by Brr2 (third scenario).
Fig. 2.
Brr2-mediated U4/U6 unwinding involves U6 conformational switching. (A) Schemes of U4/U6 mutant di-snRNAs and an expected mutant U6 snRNA product. (Inset) Regions U414–10 and U671–67 exchanged in U4/U6swap. (B) Gel analysis of Brr2-mediated WT and mutant U4/U6 di-snRNA unwinding. Time points are indicated above the gels, and bands are identified on the right. *Radiolabel. (C) Quantification of the data in B. (D) Data were fit to a single exponential equation [fraction unwound = A{1 − exp(−kut)}]. Data represent mean ± SEM of at least three independent experiments.
Fig. S2.
Reannealing controls of WT and mutant U4 and U6 snRNAs. Gels monitoring the incubation of isolated WT or mutant U4 and U6 snRNAs (lanes 2–11) show that none of the snRNA combinations spontaneously anneal. C, control showing the migration of the respective wt or mutant duplexes (lane 1). Time points are indicated above the gels, and bands are identified on the right. *Radiolabel.
U6 Structures Beyond the ISL Are Required for Conformational Switching.
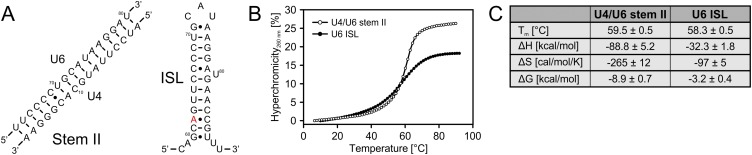
During spliceosome activation, U6 snRNA adopts a conformation containing the ISL but lacking the telestem that can form in isolated U6 (Fig. 1A). To test if the U6 ISL is sufficient for conformational switching after stem I has been unwound by Brr2, we compared thermodynamic stabilities of synthetic oligonucleotides comprising U4/U6 stem II (U41–17, U664–80) and the U6 ISL (U658–89). UV melting analyses revealed similar melting temperatures (Tms) for the two constructs (Tmstem II = 59.5 °C, TmU6 ISL = 58.3 °C) (Fig. S3). However, van’t Hoff analysis of the thermal melting profiles showed a higher enthalpy of transition (ΔH) for stem II than for U6 ISL (ΔHstem II = −88.8 kcal/mol, ΔHU6 ISL = −32.3 kcal/mol), which translates into higher stability (ΔG) of stem II than U6 ISL at 298 K (ΔGstem II = −8.9 kcal/mol, ΔGU6 ISL = −3.2 kcal/mol). Full-length U6 and U4/U6 constructs exhibited complex melting behavior, which prevented determination of thermodynamic parameters. Although the experimentally determined stabilities are closer than previously predicted (ΔGstem II = −28.1 kcal/mol, ΔGU6 ISL = −6.7 kcal/mol) (36), they still suggest that additional structures forming in isolated U6 are required for spontaneous stem II disruption.
Fig. S3.
Thermodynamic stabilities. (A) Sequences of U4/U6 stem II and U6 ISL. The exchange of residue A62 for G (red in the ISL structure) leads to a cold-sensitive phenotype and reduced U4/U6 levels due to hyperstabilization of the ISL. (B) UV melting curves. Hyperchromicity was calculated from absorbance at 260 nm and plotted vs. temperature for U4/U6 stem II and U6 ISL at 1 μM in unwinding buffer. Representative heating curves are shown. Heating and cooling curves showed reversible folding and unfolding. (C) Thermodynamic data were determined by van’t Hoff analysis from two datasets of four melting curves each. ΔH and ΔS were determined at Tm, and ΔG was extrapolated to 298 K.
Brr2 and Brr2-Jab1ΔC Detach an Intact Prp31–Snu13–U4 snRNA Complex from U6 snRNA.
Switching of U6 snRNA into its ISL/3WJ conformation would sequester U6 portions involved in formation of stem I, stem II, and parts of the U6 3′-overhang, although the U4 5′SL would remain undisturbed (Fig. 1A). Binding of Prp3 to U4/U6 requires an intact stem II and the U6 3′-overhang (30), whereas Snu13 and Prp31 bind stably to the U4 5′SL alone (29). We thus reasoned that U4/U6 di-snRNP disruption by Brr2-mediated unwinding of stem I followed by U6 conformational switching would lead to displacement of Prp3 from the RNAs, whereas Snu13 and Prp31 may remain bound to U4 snRNA; conversely, active unwinding of the U4 5′SL would lead to the additional displacement of Snu13 and Prp31.
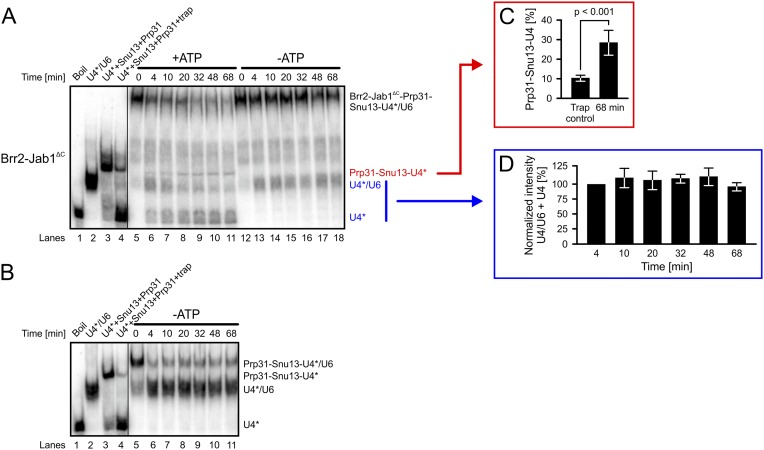
We first conducted combined unwinding and electrophoretic mobility shift assays (6) using a Prp31–Snu13–U4/U6 di-snRNA complex as a substrate (Fig. 3A). Brr2 liberated only minute amounts of free U4 snRNA from a preformed Prp31–Snu13–U4/U6 complex, irrespective of the presence of Jab1ΔC (Fig. 3A, lanes 6–12). Instead, a predominant band corresponding to U4 snRNA in complex with both proteins accumulated in a time-dependent manner. Neither Brr2 nor Brr2-Jab1∆C led to U4/U6 unwinding or displacement of any subunits in the absence of ATP (Fig. 3A, lanes 13–19). We confirmed these results in additional assays with large amounts of a U4 5′SL oligo as a trap, which efficiently prevented rebinding of Prp31 (SI Results and Fig. S4).
Fig. 3.
Subunit segregation upon Brr2-mediated U4/U6 di-snRNP disruption. (A) Gel analysis of Brr2-mediated (Upper) or Brr2-Jab1ΔC–mediated (Lower) disruption of Prp31-Snu13-U4/U6 RNP. Time points are indicated above the gels, and bands are identified on the right. *Radiolabel. Lanes 1–5, migration of RNAs and RNPs; lanes 6–19, Brr2-mediated (Upper) or Brr2-Jab1ΔC–mediated (Lower) Prp31-Snu13-U4/U6 RNP disruption in the presence (lanes 6–12) or absence (lanes 13–19) of ATP. (B) Column-based disruption assay. (C) Brr2-mediated (Upper) or Brr2-Jab1ΔC–mediated (Lower) disruption of Prp31-Snu13-U4/U6 RNP in a column-based disruption assay. Bands are identified on the right. *Radiolabel. Lane 1 (Helicase), elution after incubation with Brr2 or Brr2-Jab1ΔC; lanes 2, 3, 5, and 6 (Wash 1/2/3/4), additional wash steps; lane 4 (Imidazole/EDTA), elution by imidazole/EDTA. Bar graphs illustrate quantification of U4 snRNA fractions liberated during incubation with Brr2 or Brr2-Jab1ΔC (Fraction displaced, black bars; lanes 1–3) or during imidazole/EDTA elution (Fraction retained, gray bars; lanes 4–6). For quantification, U4 snRNA displaced in the experiments without ATP was subtracted from the U4 snRNA displaced in the experiments with ATP. Data represent mean ± SEM of at least three independent experiments. (D) Identical experiments as in C but using a Prp31-Snu13-U4 snRNP. (E and F) Gel analysis of Brr2-Jab1ΔC–mediated disruption of Prp3-Prp31-Snu13-U4/U6 di-snRNP. (Due to increased amounts of heparin required to resolve the band shifts, which reduced RNA engagement by isolated Brr2, disruption of the Prp3-Prp31-Snu13-U4/U6 RNP by Brr2 alone was too slow to be reliably monitored.) Time points are indicated above the gels, and bands are identified on the right. *Radiolabel. (E) Substrate RNP with radiolabeled U6 snRNA. Lanes 1–5, migration of RNAs and RNPs; lanes 6–12, time course in the presence of ATP. (F) Substrate complex with radiolabeled U4 snRNA. Lanes 1–6, migration of RNAs and RNPs; lanes 7–13 and 14–20, time courses in the presence or absence of ATP, respectively.
Fig. S4.
Gel analysis of Brr2-mediated U4/U6 di-snRNP disruption. (A) Experiment is as in Fig. 3A but with smaller amounts of Snu13 and a large excess of U4 5′SL trap. Lanes 1–3, migration of RNAs and RNPs; lane 4, trapping control; lanes 5–18, time courses of Brr2-Jab1ΔC–mediated Prp31-Snu13-U4/U6 di-snRNP disruption in the presence (lanes 5–11) or absence (lanes 12–18) of ATP. Under these conditions, Prp31-Snu13-U4/U6 migrated as a complex with Brr2-Jab1ΔC on the gels. (B) Control experiment showing that the high concentration of U4 5′SL trap disrupts part of the Prp31-Snu13-U4/U6 RNP. Lanes 1–3, migration of RNAs and RNPs; lane 4, trapping control; lanes 5–11, time course of Prp31-Snu13-U4/U6 di-snRNP disruption upon addition of U4 5′SL trap. Time points are indicated above the gels, and bands are identified on the right. *Radiolabel. (C) Quantification of U4 and Prp31-Snu13-U4 band intensities of lanes 4 and 11 of A, showing that more relative Prp31-Snu13-U4 RNP is formed during Brr2-Jab1ΔC–mediated unwinding than in the trap control experiment. Significance was assessed using an unpaired Student’s t test. (D) Combined U4/U6 and U4 in lanes 6–11 of A normalized to band intensities of U4/U6 and U4 in lane 6. Data represent mean ± SEM of four independent experiments.
As a further test, we conducted column-based disruption assays in which we immobilized a Prp31–Snu13–U4/U6 di-snRNA complex via His6-tagged Snu13 and added Brr2 or Brr2-Jab1ΔC and ATP to initiate on-beads unwinding (Fig. 3B). We then monitored the liberated fraction of U4 snRNA in the eluate and the retained fraction of U4 snRNA by subsequent elution with imidazole/EDTA. If the reaction was accompanied by disruption of the Prp31–Snu13–U4/U6 di-snRNA interaction, free U4 snRNA was expected in the first eluate. Only minute amounts of U4 snRNA were eluted when ATP was omitted during incubation with Brr2 or Brr2-Jab1ΔC (Fig. 3C, Right), showing that mere binding of the helicase does not induce protein displacement. A total of 4 ± 4% of U4 snRNA was liberated during on-column incubation with Brr2 and ATP (Fig. 3C, Upper Left, lanes 1–3), whereas 96 ± 4% of U4 snRNA was retained on the beads (Fig. 3C, Upper Left, lanes 4–6). At the same time, the imidazole/EDTA-eluted fraction revealed that 66 ± 7% of the U4/U6 di-snRNA duplex had been unwound during the incubation (Fig. 3C, Upper Left, lanes 4–6). Upon incubation with the Brr2–Jab1∆C complex, 20 ± 6% of U4 snRNA was initially released (Fig. 3C, Lower Left, lanes 1–3), whereas 80 ± 6% of U4 snRNA was retained on the column (Fig. 3C, Lower Left, lanes 4–6) and 70 ± 1% of U4/U6 di-snRNA was unwound under these conditions (Fig. 3C, Lower Left, lanes 4–6). Essentially no U4 snRNA was displaced in an ATP-dependent manner by Brr2 or the Brr2–Jab1∆C complex (0 ± 1% and 2 ± 2%, respectively) when we immobilized a Prp31–Snu13–U4 snRNA complex on the beads (Fig. 3D, lanes 1–3). Similar results were again obtained using a FRET-based U4/U6 disruption assay (SI Results and Fig. S5). Together, these results suggest that Brr2 or Brr2-Jab1∆C can unwind U4/U6 di-snRNA when bound to Snu13 and Prp31 without displacement of the proteins from U4 snRNA.
Fig. S5.
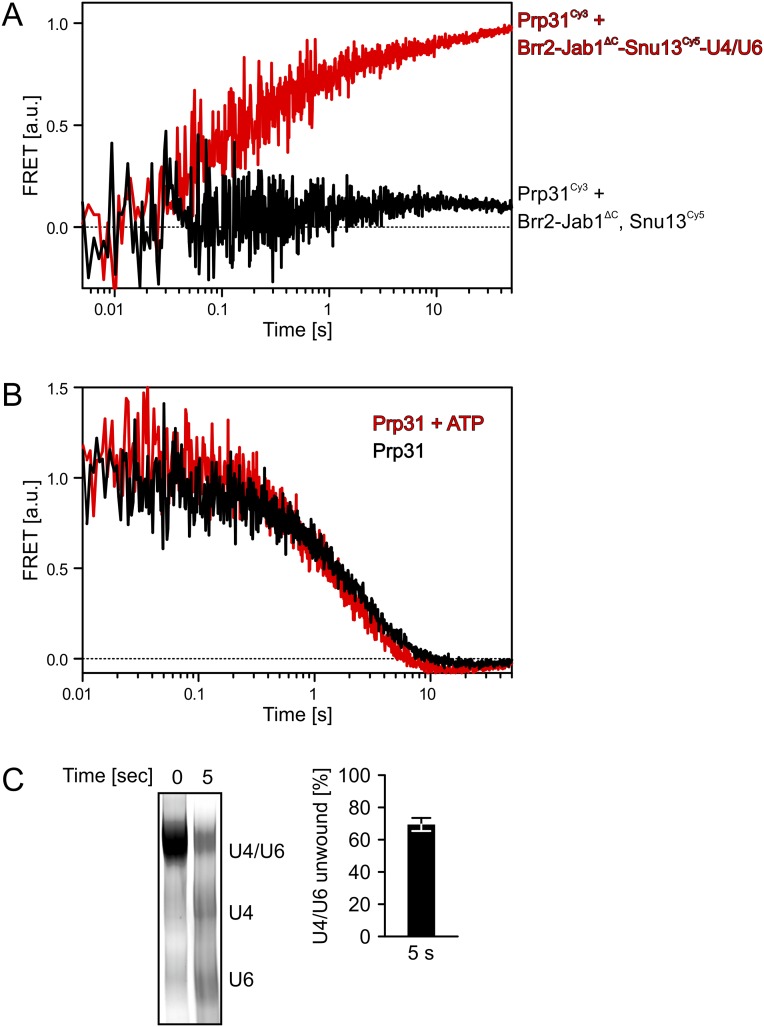
FRET-based assay monitoring Brr2-mediated U4/U6 di-snRNP disruption using a stopped-flow apparatus. (A) Binding of Prp31Cy3 to Brr2-Jab1ΔC-Snu13Cy5-U4/U6 (red curve) gave rise to FRET signal, whereas no signal was observed upon mixing Prp31Cy3, Snu13Cy5, and Brr2-Jab1ΔC in the absence of U4/U6 di-snRNA (black curve). a.u., arbitrary units. (B) Addition of a fivefold excess of unlabeled Prp31 to a Brr2-Jab1ΔC-Prp31Cy3-Snu13Cy5-U4/U6 RNP led to a time-dependent decrease of the FRET signal, representing spontaneous dissociation of Prp31Cy3 and its replacement by unlabeled Prp31 (black curve). Prp31Cy3 dissociation rates were similar in the presence of ATP (FRET decrease rate in the absence of ATP = 0.40 ± 0.007 per second; FRET decrease rate in the presence of ATP = 0.55 ± 0.009 per second). For each experiment, four traces were averaged and fit to a single exponential equation to obtain the FRET reduction rates and their respective SEs. (C) Brr2-Jab1ΔC–dependent disruption of a Prp31Cy3-Snu13Cy5-U4/U6 RNP within 5 s. (Left) Gel monitoring disruption under conditions identical to the conditions used for the FRET assays. Due to the large volumes and high RNA concentrations, disruption was monitored by methylene blue staining of the gel. (Right) Quantification of the fraction of Prp31Cy3-Snu13Cy5-U4/U6 RNP disrupted after 5 s. Due to possible differential staining of single- and double-stranded RNAs, quantification was based on the reduction of the U4/U6 band. Data represent mean ± SEM of four independent experiments.
Brr2-Jab1ΔC-Mediated U4/U6 Di-snRNP Disruption Involves Displacement of Prp3.
To test the effects of Brr2-Jab1∆C on Prp3 during U4/U6 di-snRNP disruption, we conducted combined unwinding/displacement experiments using a preformed Prp3-Prp31-Snu13-U4/U6 RNP as a substrate. Monitoring disruption of a complex with radiolabeled U6 snRNA revealed that Brr2-Jab1∆C led to liberation of free U6 snRNA (Fig. 3E, lanes 6–12). Monitoring a complex with radiolabeled U4 snRNA showed that, concomitantly, Brr2-Jab1ΔC elicited the release of a Prp31-Snu13-U4 snRNP (Fig. 3F, lanes 7–13). Again, disruption of the Prp3-Prp31-Snu13-U4/U6 particle was ATP-dependent (Fig. 3F, lanes 14–20). These results indicate that during Brr2-Jab1ΔC–mediated U4/U6 di-snRNP disruption, Prp3 is detached from U6 snRNA and does not remain associated with the Prp31-Snu13-U4 snRNA portion.
SI Results
Additional Gel-Based Disruption Assays.
Initial gel-based disruption assays included an excess of nonradioactive U4 snRNA trap over Snu13 to sequester possibly dissociated proteins, which prevented Snu13 binding to the maximally expected radioactive U4 snRNA product, but not the formation of a Prp31–Snu13–U4 snRNA complex. Larger amounts of U4 snRNA trap sequestered the helicase and interfered with unwinding. We therefore conducted the above experiments including larger amounts of unlabeled U4 5′SL (U4 residues 20–53), which lacks the Brr2-binding U4 3′–single-stranded region. U4 5′SL prevented rebinding of 90 ± 1% of Snu13 and Prp31 to radiolabeled U4 snRNA (Fig. S4 A and B, compare lanes 3 and 4). Thus, if Prp31–Snu13–U4 snRNA complex formation was due to rebinding of the proteins after their displacement, we would expect a maximum of 10 ± 1% Prp31-Snu13-U4 snRNA product relative to free U4. Instead, we observed 28 ± 3% relative Prp31–Snu13–U4 snRNA complex after 68 min during Brr2-Jab1∆C–mediated U4/U6 di-snRNP disruption (Fig. S4A, lane 11; quantification in Fig. S4C). Formation of the Prp31-Snu13-U4 RNP was again ATP-dependent (Fig. S4A, lanes 12–18). The large amounts of trap led to some initial Brr2-independent liberation of free U4/U6 di-snRNA (Fig. S4 A, lanes 5–11 and 12–18 and Fig. S4B, lanes 5–11), which is efficiently unwound by Brr2-Jab1∆C (Fig. S1). The amount of U4/U6 di-snRNA due to this side reaction fully accounts for the U4 snRNA product formed during unwinding by Brr2-Jab1∆C (Fig. S4D).
Monitoring U4/U6 Disruption via FRET.
Because Snu13 and Prp31 do not stably interact in the absence of U4 or U4/U6 (29), we monitored whether Brr2-mediated U4/U6 disruption leads to spatial separation of the proteins, which would indicate their displacement from the RNAs. We generated variants of Snu13 and Prp31 bearing single modifiable cysteine residues that allowed labeling with Cy3 and Cy5 dyes. Labeling did not alter the RNA-binding properties of the proteins. We then tested binding, dissociation, and possible Brr2-mediated displacement using a stopped-flow apparatus. Due to the omission of glycerol, heparin, and BSA in this setup, reactions were considerably faster than in the gel-based assays. As expected from Snu13 and Prp31 binding close to each other on U4/U6, addition of Prp31Cy3 to Snu13Cy5–U4/U6 or Brr2–Jab1ΔC–Snu13Cy5–U4/U6 complexes gave rise to a FRET signal (Fig. S5A). In the absence of ATP, addition of a fivefold excess of unlabeled Prp31 to a Brr2-Jab1ΔC-Prp31Cy3-Snu13Cy5-U4/U6 RNP led to a time-dependent reduction of the FRET signal, representing spontaneous dissociation of Prp31Cy3 and its replacement by unlabeled Prp31 (Fig. S5B). Notably, Prp31Cy3 dissociation rates were very similar in the presence of ATP (Fig. S5B), although Brr2-Jab1ΔC unwound 69 ± 5% of U4/U6 di-snRNA within 5 s under ATP conditions (Fig. S5C). These results again suggest that Brr2-Jab1ΔC–mediated U4/U6 disruption does not involve active displacement of either protein from the RNAs.
Discussion
An Unprecedented Mode of RNP Disruption by an RNA Helicase.
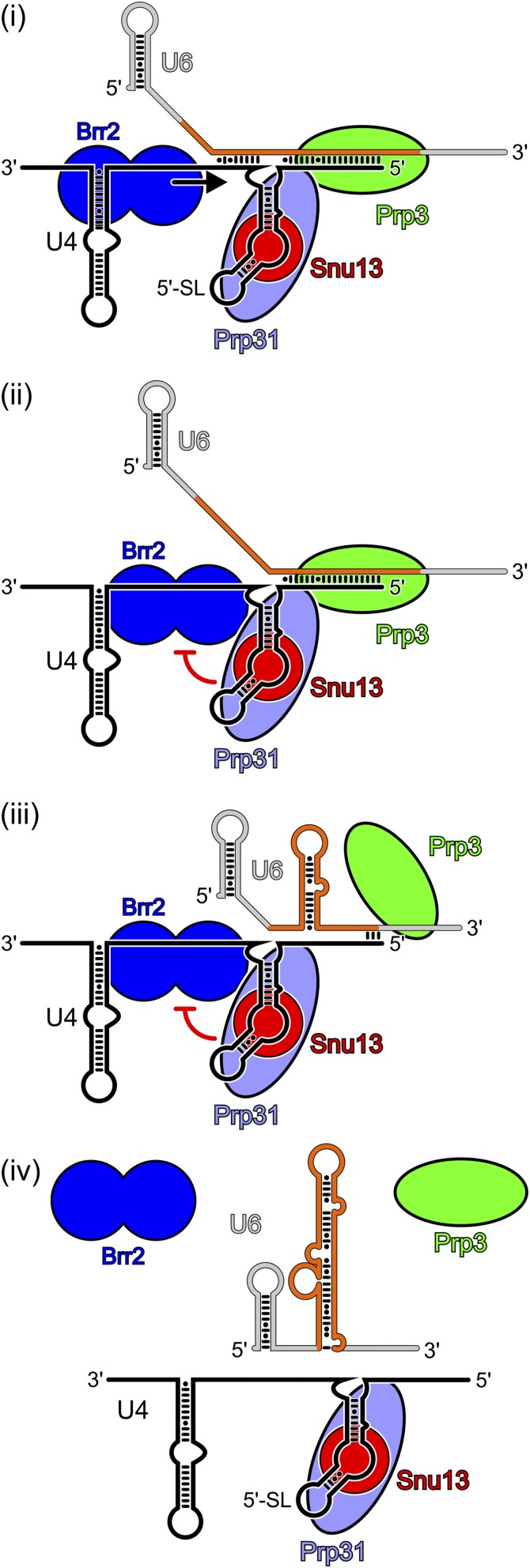
Using a recombinant system, we have investigated U4/U6 di-snRNP disruption by the Brr2 RNA helicase. This reaction constitutes the key event of spliceosome catalytic activation and provides a model system to study RNP disruption mechanisms by an RNA helicase using a cognate enzyme/substrate pair. Our results suggest that Brr2-mediated U4/U6 disruption takes advantage of the conformational bistability of U6 snRNA (Fig. S6). We propose that Brr2 actively unwinds only part of U4/U6, starting at stem I, which initiates zippering of U6 snRNA into an alternative conformation. Consequently, U6 snRNA detaches from U4 and the Prp3 protein is released, although Snu13 and Prp31 remain associated with U4 snRNA (see also SI Discussion).
Fig. S6.
Model of Brr2-mediated U4/U6 di-snRNP disruption. (i) Brr2 binds to a single-stranded region of U4 snRNA and unwinds U4/U6 stem I (black arrow). (ii) Kink in U4 snRNA and/or the tightly bound Snu13 and Prp31 proteins prevents Brr2 from translocating into the U4 5′SL (red symbol). (iii) Unwound U6 snRNA regions induce formation of an alternative U6 structure, dismantling the Prp3-binding site. (iv) Brr2 disengages from U4 snRNA, yielding a Prp31–Snu13–U4 snRNA complex, free Prp3, and free U6 snRNA.
Our experiments probed key aspects of this model. First, results from rational RNA mutagenesis demonstrated that Brr2-mediated U4/U6 unwinding is strongly influenced by RNA variants, in which U4/U6 base-paired regions were unaltered or in which only the sequence and orientation of base pairs, but not their number or nature, were changed. Although these findings argue against an active, Brr2-mediated unwinding of the entire U4/U6 stem II, they are easily reconciled by the destabilizing effects that the mutations have on alternative U6 conformations. Second, our model of Brr2-mediated U4/U6 disruption implies that the U4 5′SL is not unwound in the process. Consistently, our results suggest that proteins Snu13 and Prp31, which bind at the U4 5′SL, remain associated with U4 during U4/U6 unwinding. Finally, our model predicts that Prp3 is detached as an isolated protein during Brr2-mediated U4/U6 disruption, as its binding site on U4/U6 is dismantled (30), which is again borne out by our experiments. Our results also suggest that although it increases the activity of Brr2, the Jab1∆C cofactor does not alter the mode of Brr2-mediated U4/U6 di-snRNP disruption.
The relative stabilities of U4/U6 stem II and the U6 ISL indicate that the latter is not sufficient to explain spontaneous unwinding of stem II after Brr2-mediated disruption of stem I. Thus, a more complex U6 structure (e.g., involving the U6 telestem) could aid in conformational switching in our in vitro system (Fig. 1A). Indeed, telestem mutations that are expected to destabilize the ISL and induce an alternative U6 structure give rise to enhanced spontaneous U4/U6 annealing (36), and destabilization of the telestem in the human system promotes U4/U6 formation (37). Telestem formation is prevented in yeast U4/U6 (38), presumably by the coaxially stacked U4/U6 stems I and II intervening between its branches (22, 31, 39). Thus, in our in vitro system, U6 conformational switching could be initiated by telestem formation upon Brr2-mediated unwinding of U4/U6 stem I. Such early formation of the telestem during U4/U6 unwinding would provide an additional explanation for the importance of the U6 3′-overhang in the process.
Although U6∆3′ and U6swap mutations are expected to destabilize U6 secondary structure elements to different extents, they led to a comparable reduction of Brr2-mediated U4/U6 unwinding, and U6G81A had a fairly large effect considering that it is expected to disrupt only a single base pair in isolated U6. These findings could again be explained by a more complex 3D structure of isolated U6 required for conformational switching and involving the mutated residues. In addition, differences in the stabilities of U4/U6 and isolated U6 structures may be small so as to allow both U4/U6 unwinding during splicing and reannealing after splicing. Even a minor disturbance of isolated U6 would then be expected to exhibit a comparatively large effect in our in vitro system. This notion is in agreement with cold-sensitive mutations and some of their cis-acting suppressor mutations that introduce or remove single Watson–Crick base pairs in U6 (40) or U4/U6 (41) and that significantly alter cellular U4/U6 levels.
Previously, it has been shown that some SF2 RNA helicases can detach intact RNPs from a hybridized RNA strand. Both the nontranslocating DEAD-box protein, DED1, from yeast, and the vaccinia virus NHP-II protein, a 3′-to-5′ processive DExH helicase (42), were shown to displace U1 snRNP from a cRNA strand with single-stranded overhangs, which was bound to U1 snRNP by a combination of RNA–RNA and RNA–protein interactions (7). However, both of these proteins mediate U1 snRNP detachment through ATP-driven activities on the single-stranded regions contained in the cRNA (7). Brr2, on the other hand, disrupts the U4/U6 di-snRNP by engaging and translocating on the U4 snRNA strand (20–22), yet our results indicate that U4 snRNA is part of the RNP (Prp31-Snu13-U4 snRNA) that Brr2 liberates, a hitherto unobserved activity for an RNA helicase.
U4/U6 Disruption and Reannealing During Splicing.
Cross-linking immunoprecipitation analyses in vivo uncovered cross-links of Brr2 to regions of stem I and the upper part of the U4 5′SL, with the latter likely reflecting contacts involving the leading edge of the helicase while its active site is still positioned on stem I (21). However, these analyses failed to detect any Brr2 contacts to the lower parts of the U4 5′SL, stem II, or the U6 3′-overhang. Thus, a similar unusual U4/U6 disruption mechanism as we see in vitro likely also applies during Brr2-mediated spliceosome activation in vivo.
Partially different and additional factors will influence Brr2-mediated U4/U6 disruption in vivo. Binding of the Sm-like (LSm) proteins to the very 3′-end of U6 in the U4/U6 di-snRNP (43) (i.e., close to the U6 sequence forming the 3′-branch of the telestem in isolated U6) might modulate telestem formation (38). Furthermore, in the activated spliceosome, U6 snRNA forms the ISL but not the telestem, because neighboring elements base-pair with U2 snRNA, forming helices Ia and Ib preceding the ISL and helix II following the ISL (44). Unlike oligos complementary to either branch of the telestem, which favor U4/U6 formation by disrupting the telestem (37), U2 interacts with both telestem branches concomitantly, thus likely promoting the U6 ISL conformation. Furthermore, the cryo-EM structure of a poststep I spliceosome directly revealed that helix Ia/Ib and neighboring regions can engage in higher order interactions with the U6 ISL, and that this RNA interaction network rests on, and is thus likely stabilized by, a positively charged cavity of the Prp8 protein (19). Thus, U6/U2 interactions in the context of Prp8 might promote U4/U6 unwinding by supporting U6 conformational switching in vivo. This notion is supported by a U4 allele, which leads to cold-sensitive growth due to hyperstabilization of U4/U6, whose effects are suppressed by changes to several Prp8 residues (45, 46). The only two Prp8 suppressor variants in which residues are affected that directly contact U6 or U2 in the available spliceosome structure are Prp8K611R and Prp8D1094A/N/V (19), which may act via increased affinity to U6/U2 (stronger or more direct contacts by Prp8K611R, removal of a negative charge in Prp8D1094A/N/V).
Following each round of splicing, U4/U6 di-snRNP has to be regenerated, which, in yeast, requires the Prp24 protein (47). Prp24 may have a second function in disrupting U6/U2 interactions after splicing (48, 49). During U4/U6 annealing, three RNA recognition motifs of Prp24 interlock with the U6 ISL, asymmetrical bulge, and telestem (48), giving rise to an electropositive groove, which could mediate U4/U6 stem I annealing (36, 48). Subsequent formation of U4/U6 stem II might then lead to displacement of Prp24 and complete U4/U6 assembly (49). Cold-sensitive mutations in U6 or U4, which hyperstabilize the U6 ISL or weaken U4/U6 stem II, respectively, cause reduced levels of U4/U6 di-snRNA, and numerous suppressor mutations have been identified (40, 41). Several cis- and trans-acting suppressors map to Prp24-U6 contacts and function by destabilizing Prp24–U6 interactions and promoting U6/U2 interactions (48, 49). These findings suggest that apart from normally being required to deliver U6 to the spliceosome, release of U6 from Prp24 is another important function of the U4/U6 interaction (49).
Together with our results, the above observations reveal interesting similarities between the RNP remodeling factors Brr2 and Prp24. Both seem to initiate only the remodeling of their respective substrates. The remodeling events are then driven to completion by conformational switches in the participating RNAs (i.e., snapping of U6 into the ISL structure in the case of Brr2 and formation of U4/U6 stem II in the case of Prp24). Unlike Prp24, Brr2 requires ATP hydrolysis to initiate its reaction, indicating that the U4/U6 state is more stable than the isolated U6 state. Both Prp24- and Brr2-mediated reactions are also modulated by additional factors. Brr2-mediated U4/U6 disruption is influenced by U4/U6-bound proteins, and U6 conformational switching during U4/U6 unwinding may be aided by emerging U6/U2 interactions on Prp8; Prp24-mediated U4/U6 annealing is enhanced by the LSm proteins (36, 50–53) and possibly Prp3 (30).
The major RNP remodeling events during splicing offer many possibilities for regulation [e.g., during alternative splicing (54)], but they also burden the cell with the requirement to reassemble spliceosomal subunits after each round of splicing. Although de novo assembly of snRNPs in vivo is supported by the SMN and PRMT5 complexes (55, 56) and the NUFIP/R2TP system (57), it is not clear to what extent these machineries also mediate snRNP reassembly after splicing. The mode of U4/U6 di-snRNP disruption by Brr2 suggested here efficiently supports the build-up of the spliceosome’s active site. At the same time, it is economical because it leaves the U4 snRNP intact, possibly circumventing U4 snRNP reassembly via the above machineries and thus minimizing the logistic demands for U4/U6 di-snRNP reassembly after each splicing reaction.
SI Discussion
After release of U6 snRNA by Brr2-mediated unwinding of stem I and conformational switching of U6, Brr2 might still remain associated with U4 and, upon further ATP-driven translocation on U4, would encounter bound Prp31 and Snu13. Several mechanisms can be envisaged by which Prp31 and Snu13 could counteract their own displacement from U4 snRNA. Single-molecule analyses have shown that the coaxial stack of U4/U6 stems I and II with the U4 5′SL in a perpendicular orientation as seen in cryo-EM structures of U4/U6•U5 tri-snRNPs (22, 31, 58) is also adopted by isolated U4/U6 di-snRNA and is stabilized by binding of Snu13, Prp31, and a Prp3/Prp4 complex (39). U4/U6 di-snRNA thus exhibits a kink between stem I and the U4 5′SL, which could prevent or delay Brr2 from translocating into the U4 5′SL and favor its dissociation before it can displace Prp31 and Snu13. However, as shown here, Brr2 also does not seem to dislodge Prp31 and Snu13 from U4 snRNA alone, where this conformational restraint most likely does not exist. Because Prp31 and Snu13 bind U4 snRNA cooperatively (29, 30, 39), the two proteins together might present a stable roadblock that prevents translocation of Brr2 into the U4 5′SL. At the point when the helicase encounters these physical obstacles, it apparently has unwound a sufficient fraction of U4/U6 to allow U6 snRNA to switch into an alternative conformation. In either scenario, Prp31 and Snu13 acts as a gatekeeper that prevents Brr2 from translocating into the U4 5′SL.
Materials and Methods
Recombinant Snu13, Prp31, Prp3, and Jab1ΔC were produced in Escherichia coli, and Brr2 was produced via a recombinant baculovirus in insect cell culture. Proteins were purified by affinity, ion exchange, and size exclusion chromatography. RNAs were produced by in vitro transcription using T7 RNA polymerase or chemically synthesized. RNA binding, unwinding, and RNP disruption were analyzed with 5′-[32P]–labeled RNAs by nondenaturing PAGE or by stopped-flow/fluorescence assays. Unwinding by Brr2 was initiated by addition of ATP and terminated at various time points with stop buffer containing EDTA and SDS. The stop buffer used in EMSA and protein displacement assays did not contain SDS to maintain protein–RNA interactions. Detailed materials and methods can be found in SI Materials and Methods.
SI Materials and Methods
Protein Production and Purification.
A synthetic gene encoding yeast Brr2271-end was cloned in-frame with an N-terminal His10-tag and a tobacco etch virus (TEV) protease cleavage site into a modified pFL vector. Tn7 transposition was used to transfer the expression construct into an EMBacY baculovirus genome, maintained as a bacterial artificial chromosome (bacmid) in E. coli. Sf9 cells (Invitrogen) were transfected with the purified recombinant bacmid, and the first virus generation (V0) was harvested and further amplified in Sf9 cells (Invitrogen). A high-titer virus generation (V1) was used for large-scale expression in High Five Cells (Invitrogen). The cell pellet of an 800-mL culture was resuspended in 50 mM Hepes (pH 8.0), 20% (vol/vol) glycerol, 600 mM NaCl, 10 mM imidazole, 1.5 mM MgCl2, 0.05% Nonidet P-40, and 2 mM β-mercaptoethanol supplemented with EDTA-free protease inhibitor (Roche). Cells were lysed by sonication for 30 min in a Sonopuls Ultrasonic Homogenizer HD 3100 (Bandelin; amplitude of 50%, pulse on for 0.5 s, pulse off for 2.0 s). Brr2 was affinity-captured on a 5-mL HisTrap FF column (GE Healthcare) equilibrated in 50 mM Hepes (pH 8.0), 20% (vol/vol) glycerol, 600 mM NaCl, 10 mM imidazole, and 2 mM β-mercaptoethanol, and eluted with a linear gradient from 10 to 250 mM imidazole. Brr2 used in RNA displacement assays was treated with TEV protease during overnight dialysis at 4 °C against 50 mM Hepes (pH 8.0), 10% (vol/vol) glycerol, 600 mM NaCl, 2 mM β-mercaptoethanol, and 15 mM imidazole to remove the N-terminal His10-tag, and subsequently separated from the His-tagged TEV protease and the cleaved His10-tag by recycling over a 5-mL HisTrap FF column. Brr2 used in all other experiments was not treated with TEV protease. The sample was then diluted to a final concentration of 70–80 mM NaCl and loaded on a HiPrep Heparin FF 16/10 column (GE Healthcare) equilibrated with 50 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 5% (vol/vol) glycerol, and 2 mM DTT. The protein was eluted with a linear gradient from 0.05 to 1.5 M NaCl and further purified by gel filtration on a 16/60 or 26/60 Superdex 200 column (GE Healthcare) run with 40 mM Tris⋅HCl (pH 8.0), 20% (vol/vol) glycerol, 200 mM NaCl, and 2 mM DTT.
DNA constructs encoding for yeast Snu13 and Prp3296–469 were cloned into a pETM-11 vector (EMBL) for production of the targets as N-terminal His6-tagged fusion proteins. DNA encoding yeast Prp312–388 was cloned into a pGEX-6P vector (GE Healthcare) for production of the target as an N-terminal, PreScission-cleavable GST fusion protein. The Snu13 and Prp31 plasmids were transformed into E. coli BL21 (DE3)-T1R cells. The Prp3 plasmid was transformed into E. coli BL21 (DE3)-RIL cells. Proteins were produced at 18 °C for 2 d using autoinduction media (59).
For purification of Prp3296–469, a cell pellet of a 400-mL culture was resuspended and incubated for 1 h at 4 °C in 20 mM Tris⋅HCl (pH 7.0), 5% (vol/vol) glycerol, 300 mM NaCl, 30 mM imidazole, 10 mM MgCl2, 5 mM β-mercaptoethanol, 2 μg/mL DNaseI, and 10 μg/mL lysozyme supplemented with EDTA-free protease inhibitor pills. Cells were lysed by sonication for 15 min in a Sonopuls Ultrasonic Homogenizer HD 3100 (amplitude of 60%, pulse on for 2.0 s, pulse off for 1.0 s). Prp3 was affinity-captured on a 5-mL HisTrap FF column equilibrated in 20 mM Tris⋅HCl (pH 7.0), 5% (vol/vol) glycerol, 300 mM NaCl, 30 mM imidazole, 10 mM MgCl2, and 5 mM β-mercaptoethanol, and eluted with a linear gradient from 30 to 500 mM imidazole. A wash step with 1 M LiCl was included during the affinity chromatography to remove nucleic acids. Prp3 was further purified by gel filtration on a 16/60 Superdex 200 column equilibrated in 20 mM Tris⋅HCl (pH 7.0), 150 mM NaCl, and 2 mM DTT.
For purification of Snu13, a cell pellet of a 1-L culture was resuspended in 50 mM Hepes (pH 7.0), 500 mM NaCl, 15 mM imidazole, 0.05% Tween-20, 2 mM DTT, 5 μg/mL DNaseI, 10 μg/mL RNaseA, and 0.2 mg/mL lysozyme supplemented with EDTA-free protease inhibitor, and cells were lysed by sonication for 22 min in a Sonopuls Ultrasonic Homogenizer HD 3100 (amplitude of 80%, pulse on for 1.0 s, pulse off for 2.5 s). Snu13 was affinity-purified on a 5-mL HisTrap FF column equilibrated in 50 mM Hepes (pH 7.0), 500 mM NaCl, 15 mM imidazole, and 2 mM DTT. The protein was eluted with a linear gradient from 15 to 300 mM imidazole. The N-terminal His6-tag was cleaved with TEV protease during overnight dialysis at 4 °C against 50 mM Hepes (pH 7.0), 500 mM NaCl, 10 mM imidazole, and 2 mM DTT, and then run again over a 5-mL HisTrap FF column. Snu13 used in on-column RNA displacement assays was not treated with TEV protease to keep the His6-tag. The sample was then diluted to a final concentration of 120 mM NaCl for ion exchange chromatography on a HiPrep Heparin FF 16/10 column equilibrated in 50 mM Hepes (pH 7.0), 100 mM NaCl, and 2 mM DTT. The protein was eluted with a linear gradient from 0.1 to 1 M NaCl and further purified by gel filtration on a 16/60 or 26/60 Superdex 75 (GE Healthcare) column equilibrated in 10 mM Hepes (pH 7.0), 200 mM NaCl, and 2 mM DTT.
For purification of Prp31, a cell pellet of a 1-L culture was resuspended in 50 mM Hepes (pH 7.5), 600 mM NaCl, 5% (vol/vol) glycerol, 0.02% Triton X-100, 2 mM DTT, 5 μg/mL DNaseI, 10 μg/mL RNaseA, and 0.2 mg/mL lysozyme supplemented with EDTA-free protease inhibitor, and cells were lysed by sonication for 22 min in a Sonopuls Ultrasonic Homogenizer HD 3100 (amplitude of 80%, pulse on for 1.0 s, pulse off for 2.5 s). GST-Prp31 was affinity-captured using glutathione-Sepharose beads (GE Healthcare) equilibrated in 50 mM Hepes (pH 7.5), 600 mM NaCl, 5% (vol/vol) glycerol, and 2 mM DTT, and eluted in 50 mM Hepes (pH 7.75), 600 mM NaCl, 5% (vol/vol) glycerol, 2 mM DTT, and 20 mM glutathione. The N-terminal GST-tag was cleaved with PreScission protease during overnight dialysis at 4 °C against buffer containing 50 mM Hepes (pH 7.5), 200 mM NaCl, 5% (vol/vol) glycerol, and 2 mM DTT. Glutathione-Sepharose beads were used to separate the cleaved protein from the GST-tag and the GST-tagged PreScission protease. The sample was loaded on a HiPrep Heparin FF 16/10 column equilibrated in 50 mM Hepes (pH 7.5), 200 mM NaCl, 5% (vol/vol) glycerol, and 2 mM DTT; eluted with a linear gradient from 0.2 to 1 M NaCl in buffer containing 50 mM Hepes (pH 7.5), 5% (vol/vol) glycerol, and 2 mM DTT; and further purified by gel filtration on a 16/60 or 26/60 Superdex 75 column equilibrated in 10 mM Hepes (pH 7.5), 200 mM NaCl, 5% (vol/vol) glycerol, and 2 mM DTT.
RNA Production and Purification.
U4 and U6 snRNAs and mutants thereof were in vitro-transcribed from PCR-amplified, linearized DNA fragments derived from recombinant pUC19 vectors containing the cloned U4, U6, or mutant genes for 4 h at 37 °C in 200 mM Tris⋅HCl (pH 7.5), 75 mM MgCl2, 10 mM spermidine trihydrochloride, 50 mM NaCl, 100 mM NTPs (Jena Bioscience), 0.02 U/μL RNasin (moloX), 0.005 U/μL pyrophosphatase (Thermo Scientific), and 1.6 U/μL T7 RNA polymerase. In vitro-transcribed RNAs were treated with 0.02 U/μL DNaseI (Thermo Scientific) for 15 min at 37 °C. RNAs were then diluted in 35 mM Bis-Tris-Propane (pH 6.9), 50 mM NaCl, and 0.2 mM EDTA; loaded on a MonoQ 10/100 GL column (GE Healthcare) equilibrated in 35 mM Bis-Tris-Propane (pH 6.9), 200 mM NaCl, and 0.2 mM EDTA; and eluted with a linear gradient from 0.2 to 1.2 M NaCl. The sample was further purified by phenol-chloroform extraction using Roti-Aqua-P/C/I RNA extraction mix (Roth). Extracted RNAs were mixed with 10% (vol/vol) 3 M sodium acetate (pH 5.3) and precipitated with 1 vol of isopropanol for 20 min at −20 °C. The RNA pellets were washed with 70% (vol/vol) ethanol and dissolved in water.
UV Melting Analyses.
Oligonucleotides for UV melting analyses were chemically synthesized. Absorbance versus temperature profiles were recorded at 250, 260, 270, and 280 nm on a Cary 100 spectrophotometer (Agilent) equipped with a multiple cell holder and a Peltier temperature control element. Samples were prepared at 1 μM RNA in unwinding buffer [40 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 0.5 mM MgCl2] and measured in quartz cuvettes with a path length of 1 cm. Data were collected for two complete heating and cooling cycles (5–90–5 °C) at a rate of 0.7 °C for 1 min. Melting transitions were reversible at all four different wavelengths. Thermodynamic parameters were derived from two-state van’t Hoff analysis of melting curves as described elsewhere (60).
RNA Labeling and Assembly of U4/U6 Di-snRNA and Di-snRNP.
U4 and U6 snRNAs and mutants thereof were dephosphorylated with 0.27 U/μL Fast Alkaline Phosphatase (Thermo Scientific) in the manufacturer’s buffer for 1 h at 37 °C and purified by phenol-chloroform extraction and isopropanol precipitation. Sixty picomoles of U4 RNAs were 5′–end-labeled with [γ-32P]-ATP (PerkinElmer) using T4 polynucleotide kinase in the manufacturer’s buffer (New England Biolabs) for 1 h at 37 °C. The labeling reaction was then loaded on a G25 spin column (GE HealthCare), and the labeled U4 RNA was recovered. Duplex annealing was carried out in 40 mM Tris (pH 7.5) and 50 mM NaCl in the presence of a fivefold molar excess of U6 RNA. The annealing mixture was incubated for 5 min at 80 °C and cooled to 70 °C, when the reaction was supplied with 10 mM MgCl2. The reaction was further cooled to 25 °C over 1 h. The RNA duplex was separated from free U4 and U6 RNAs via 6% native PAGE run for 1 h at 200 V. The RNA duplex band was cut from the gel and eluted overnight in buffer containing 50 mM Tris (pH 7.5), 400 mM NaCl, 100 mM EDTA, and 0.5% SDS. Duplex RNA was then phenol-chloroform–extracted and isopropanol-precipitated. The duplex RNA pellets were washed with 70% (vol/vol) ethanol and subsequently dissolved in 40 mM Tris (pH 7.5) and 50 mM NaCl. The same steps were performed when U6 or mutant RNAs were 5′–end-labeled with [γ-32P]-ATP.
U4/U6 di-snRNP assembly was initiated by incubation of U4/U6 di-snRNA with Snu13. Prp31, Prp3, and Brr2 alone or preassembled with Jab1ΔC were added sequentially. Each incubation step was performed at 30 °C for 3 min.
Unwinding Assays.
Helicase assays were performed at 30 °C for 0–68 min with 20 nM Brr2 alone or preassembled with 50 nM Jab1ΔC, 1.75 nM U4/U6 di-snRNA or mutants thereof, and optionally with 50 nM Snu13, 50 nM Prp31, and 50 nM Prp3 in unwinding buffer [40 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 8% (vol/vol) glycerol, 0.5 mM MgCl2, 15 ng/μL acetylated BSA, 1 U/μL RNasin, 1.5 mM DTT]. The reactions were initiated by addition of 1.7 mM ATP/MgCl2. Aliquots were taken at the indicated time points, and reactions were stopped by addition of 1 vol of stop buffer [40 mM Tris⋅HCl (pH 7.4), 50 mM NaCl, 25 mM EDTA, 1% SDS, 10% (vol/vol) glycerol, 0.05% xylene cyanol, 0.05% bromophenol blue]. Samples were run on a 6% nondenaturing PAGE gel at 4 °C at 200 V for 1 h. RNA bands were visualized by autoradiography using a phosphor-imager (Molecular Dynamics) and quantified with Image Quant 5.2 software (GE Healthcare). Data were fit to a single exponential equation [fraction unwound = A{1 − exp(−ku t)}; A, amplitude of the reaction; ku, apparent first-order rate constant of unwinding; t, time] using GraphPad Prism.
Gel-Based Protein Displacement Assays.
Protein displacement assays were performed at 30 °C for 0–68 min with 400–800 nM Brr2 alone or preassembled with 1 μM Jab1ΔC, with 1.75 nM U4/U6 di-snRNA, 200 nM Snu13, 1 μM Prp31, and optionally 4 μM Prp3 in 40 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 8% (vol/vol) glycerol, 0.5 mM MgCl2, 10–67 ng/μL heparin, 15–67.5 ng/μL acetylated BSA, 0–360 ng/μL yeast tRNA, 1 U/μL RNasin, and 1.5 mM DTT. To reduce unspecific binding and to resolve the band shifts well, the concentrations of heparin, acetylated BSA, and yeast tRNA were increased when more proteins were present in the assays. A total of 225 nM U4 snRNA or 1 μM U4 5′SL (U420–53) trap was used in experiments containing radiolabeled U4 snRNA. Experiments containing U4 5′SL trap were performed with 30 nM Snu13 and 1.5 μM Brr2-Jab1ΔC. Experiments performed with radiolabeled U6 snRNA were done without U6 snRNA trap, because no rebinding of any of the proteins to U6 snRNA could be observed. The reactions were initiated by adding 1.7 mM ATP/MgCl2 alone or premixed with trap RNA or with trap RNA alone (control). Aliquots were taken at the indicated time points, and reactions were stopped with 1 vol of stop buffer without SDS. Samples were loaded on a 4% nondenaturing PAGE gel containing 5% (vol/vol) glycerol and run at 170 V for 3–4 h. RNA bands were visualized as described above.
Column-Based Disruption Assays.
All steps were performed at 30 °C. To monitor RNA displacement, 1.75 nM U4/U6 di-snRNA (U4 radiolabeled) was incubated with 200 nM His6-tagged Snu13 and 1 μM Prp31 for 3 min in a total volume of 20 μL in unwinding buffer. Subsequently, the complex was immobilized for 6 min in 25 μL of Ni2+-NTA agarose beads (Qiagen) preequilibrated in unwinding buffer and loaded on Pierce Micro-Spin Columns (Thermo Scientific). The on-column unwinding reaction was performed for 5 min with 20 μL of 1.7 mM ATP/MgCl2 and 2.4 μM Brr2 alone, or optionally preassembled with 2.5 μM Jab1ΔC in unwinding buffer. Elution of nondisplaced U4 snRNA and U4/U6 di-snRNA was performed for 5 min in 20 μL of 300 mM imidazole and 25 mM EDTA in unwinding buffer. Immobilization, unwinding, and elution steps were interspersed with two wash steps with 20 μL of unwinding buffer supplemented with 1 μM Prp31 to counteract spontaneous complex dissociation. After each incubation step, the solutions were recovered by centrifugation at 2,000 × g for 10 s in a 5415D tabletop centrifuge (Eppendorf). Fifteen microliters of the recovered solutions was mixed with 10 μL of stop buffer, and the samples were run on a 6% nondenaturing PAGE at 4 °C at 200 V for 1 h. RNA bands were visualized and quantified described as above.
Protein Labeling with Fluorescent Dyes.
We exchanged a serine at position 93 of Snu13 for a cysteine because none of the other cysteine residues in WT Snu13 was accessible for labeling. In addition, we produced a Prp3135–388 variant in which all cysteine residues had been exchanged with serines (C230S, C257S, C307S), except for an accessible cysteine at position 36. Snu13 and Prp3135–388 variants were produced and purified like the WT proteins, except that the final buffer did not contain DTT. One hundred fifty micromolar proteins were preincubated with 500 μM Tris(2-carboxyethyl)phosphine hydrochloride (Sigma–Aldrich) for 10 min at 37 °C in labeling buffer [50 mM Hepes (pH 6.8), 200 mM NaCl]. Labeling was performed with 50 μM Cy3 or Cy5 maleimide monoreactive dye (GE Healthcare) in labeling buffer for 3.5 h. The reaction was stopped with 6 mM 2-mercaptoethanol (Merck). Labeled proteins were separated from the fluorescent dyes via gel filtration on a 10/300 Superdex 75 column (GE Healthcare) equilibrated in labeling buffer.
FRET-Based Disruption Assays.
FRET-based stopped-flow measurements were performed on a SX-20 stopped-flow spectrometer (Applied Photophysics) at 30 °C in buffer containing 40 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, and 0.5 mM MgCl2. Cy3 fluorescence was excited at 530 nm, and its emission was measured after passing a 550-nm cutoff filter (Applied Photophysics). Cy5 was excited via FRET from Cy3 if labeled Snu13 and Prp3135–388 were bound to U4/U6 di-snRNA. Cy5 fluorescence was monitored after passing a 645-nm cutoff filter (Applied Photophysics).
U4/U6 di-snRNP was assembled from 250 nM U4/U6 di-snRNA, 500 nM labeled Snu13, 250 nM labeled Prp3135–388, 50 nM Brr2, and 50 nM Jab1ΔC. Brr2-Jab1ΔC–mediated disruption reactions were initiated by rapid mixing of 60 μL of the assembled complex with 60 μL of 1.7 mM ATP/MgCl2 and 1.25 μM nonlabeled Prp31. The fivefold excess of nonlabeled Prp31 over labeled Prp31 was used to avoid rebinding of the labeled Prp31 variant. Fluorescence change time courses were averaged from at least four individual traces. Time-dependent FRET decrease rates were obtained by fitting the data to a single exponential equation [FRET = A{1 − exp(−ku t)}; A, amplitude of the reaction; kF, apparent first-order rate constant of FRET decrease; t, time) using GraphPad Prism.
Acknowledgments
This work was funded by Deutsche Forschungsgemeinschaft Grant SFB 740 (to M.C.W.) and Bundesministerium für Bildung und Forschung Grant 05K10KEC (to M.C.W.). M.T. was supported by a fellowship from the Boehringer Ingelheim Fonds. K.F.S. was supported by a Dahlem International Network PostDoc grant from the Freie Universität Berlin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524616113/-/DCSupplemental.
References
- 1.Tanner NK, Linder P. DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol Cell. 2001;8(2):251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E. RNA helicases at work: Binding and rearranging. Trends Biochem Sci. 2011;36(1):19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44(41):13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 4.Halls C, et al. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365(3):835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballut L, et al. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12(10):861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 6.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291(5501):121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 7.Bowers HA, et al. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12(5):903–912. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92(3):315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 9.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334(6179):213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- 11.Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA. 1998;95(8):4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8(15):847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 13.Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5(7):959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos KF, et al. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci USA. 2012;109(43):17418–17423. doi: 10.1073/pnas.1208098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agafonov DE, et al. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol Cell Biol. 2011;31(13):2667–2682. doi: 10.1128/MCB.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat Struct Biol. 2002;9(6):431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- 17.Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol. 2004;11(12):1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- 18.Fica SM, et al. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503(7475):229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science. 2015;349(6253):1191–1198. doi: 10.1126/science.aac8159. [DOI] [PubMed] [Google Scholar]
- 20.Mozaffari-Jovin S, et al. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev. 2012;26(21):2422–2434. doi: 10.1101/gad.200949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn D, Kudla G, Tollervey D, Beggs JD. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 2012;26(21):2408–2421. doi: 10.1101/gad.199307.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen TH, et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature. 2016;530(7590):298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell. 2006;23(3):389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeder C, Kutach AK, Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol. 2009;16(1):42–48. doi: 10.1038/nsmb.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mozaffari-Jovin S, et al. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science. 2013;341(6141):80–84. doi: 10.1126/science.1237515. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen TH, et al. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure. 2013;21(6):910–919. doi: 10.1016/j.str.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mozaffari-Jovin S, et al. Novel regulatory principles of the spliceosomal Brr2 RNA helicase and links to retinal disease in humans. RNA Biol. 2014;11(4):298–312. doi: 10.4161/rna.28353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Absmeier E, et al. The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev. 2015;29(24):2576–2587. doi: 10.1101/gad.271528.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, et al. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316(5821):115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, et al. A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing. eLife. 2015;4:e07320. doi: 10.7554/eLife.07320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan R, et al. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351(6272):466–475. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 32.Nottrott S, Urlaub H, Lührmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: A biochemical role for U4/U6 proteins. EMBO J. 2002;21(20):5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultz A, Nottrott S, Hartmuth K, Lührmann R. RNA structural requirements for the association of the spliceosomal hPrp31 protein with the U4 and U4atac small nuclear ribonucleoproteins. J Biol Chem. 2006;281(38):28278–28286. doi: 10.1074/jbc.M603350200. [DOI] [PubMed] [Google Scholar]
- 34.Karaduman R, Fabrizio P, Hartmuth K, Urlaub H, Lührmann R. RNA structure and RNA-protein interactions in purified yeast U6 snRNPs. J Mol Biol. 2006;356(5):1248–1262. doi: 10.1016/j.jmb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Dunn EA, Rader SD. Secondary structure of U6 small nuclear RNA: Implications for spliceosome assembly. Biochem Soc Trans. 2010;38(4):1099–1104. doi: 10.1042/BST0381099. [DOI] [PubMed] [Google Scholar]
- 36.Didychuk AL, Montemayor EJ, Brow DA, Butcher SE. Structural requirements for protein-catalyzed annealing of U4 and U6 RNAs during di-snRNP assembly. Nucleic Acids Res. 2016;44(3):1398–1410. doi: 10.1093/nar/gkv1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brow DA, Vidaver RM. An element in human U6 RNA destabilizes the U4/U6 spliceosomal RNA complex. RNA. 1995;1(2):122–131. [PMC free article] [PubMed] [Google Scholar]
- 38.Mougin A, Gottschalk A, Fabrizio P, Lührmann R, Branlant C. Direct probing of RNA structure and RNA-protein interactions in purified HeLa cell’s and yeast spliceosomal U4/U6.U5 tri-snRNP particles. J Mol Biol. 2002;317(5):631–649. doi: 10.1006/jmbi.2002.5451. [DOI] [PubMed] [Google Scholar]
- 39.Hardin JW, Warnasooriya C, Kondo Y, Nagai K, Rueda D. Assembly and dynamics of the U4/U6 di-snRNP by single-molecule FRET. Nucleic Acids Res. 2015;43(22):10963–10974. doi: 10.1093/nar/gkv1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fortner DM, Troy RG, Brow DA. A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes Dev. 1994;8(2):221–233. doi: 10.1101/gad.8.2.221. [DOI] [PubMed] [Google Scholar]
- 41.Shannon KW, Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991;5(5):773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- 42.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403(6768):447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 43.Zhou L, et al. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature. 2014;506(7486):116–120. doi: 10.1038/nature12803. [DOI] [PubMed] [Google Scholar]
- 44.Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71(5):803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 45.Kuhn AN, Li Z, Brow DA. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol Cell. 1999;3(1):65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
- 46.Kuhn AN, Brow DA. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics. 2000;155(4):1667–1682. doi: 10.1093/genetics/155.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghetti A, Company M, Abelson J. Specificity of Prp24 binding to RNA: A role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1(2):132–145. [PMC free article] [PubMed] [Google Scholar]
- 48.Montemayor EJ, et al. Core structure of the U6 small nuclear ribonucleoprotein at 1.7-Å resolution. Nat Struct Mol Biol. 2014;21(6):544–551. doi: 10.1038/nsmb.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burke JE, Butcher SE, Brow DA. Spliceosome assembly in the absence of stable U4/U6 RNA pairing. RNA. 2015;21(5):923–934. doi: 10.1261/rna.048421.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Achsel T, et al. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18(20):5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryan DE, Stevens SW, Abelson J. The 5′ and 3′ domains of yeast U6 snRNA: Lsm proteins facilitate binding of Prp24 protein to the U6 telestem region. RNA. 2002;8(8):1011–1033. doi: 10.1017/s1355838202026092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rader SD, Guthrie C. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA. 2002;8(11):1378–1392. doi: 10.1017/s1355838202020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verdone L, Galardi S, Page D, Beggs JD. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr Biol. 2004;14(16):1487–1491. doi: 10.1016/j.cub.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 54.Wahl MC, Lührmann R. SnapShot: Spliceosome dynamics II. Cell. 2015;162(2):456–e1. doi: 10.1016/j.cell.2015.06.061. [DOI] [PubMed] [Google Scholar]
- 55.Kolb SJ, Battle DJ, Dreyfuss G. Molecular functions of the SMN complex. J Child Neurol. 2007;22(8):990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 56.Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582(14):1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 57.Bizarro J, et al. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res. 2015;43(18):8973–8989. doi: 10.1093/nar/gkv809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agafonov DE, et al. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016;351(6280):1416–1420. doi: 10.1126/science.aad2085. [DOI] [PubMed] [Google Scholar]
- 59.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 60.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26(9):1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]