Significance
The importance of the Mps1 (monopolar spindle 1) protein kinase stems from its regulatory role in many important processes, among which are centrosome duplication and the spindle assembly checkpoint. Both of these processes help prevent mitotic errors that may lead to tumorigenesis, making it of great importance to elucidate how Mps1 is targeted to regulate two such spatially distinct processes. This study presents data that distinguish the separation of those two functions in the cell cycle and potentially help in the understanding of the different mechanisms by which Mps1 might influence tumorigenesis.
Keywords: Mps1/TTK, centrosome, centriole, kinetochore, TPR
Abstract
Faithful segregation of chromosomes to two daughter cells is regulated by the formation of a bipolar mitotic spindle and the spindle assembly checkpoint, ensuring proper spindle function. Here we show that the proper localization of the kinase Mps1 (monopolar spindle 1) is critical to both these processes. Separate elements in the Mps1 N-terminal extension (NTE) and tetratricopeptide repeat (TPR) domains govern localization to either the kinetochore or the centrosome. The third TPR (TPR3) and the TPR-capping helix (C-helix) are each sufficient to target Mps1 to the centrosome. TPR3 binds to voltage-dependent anion channel 3, but although this is sufficient for centrosome targeting of Mps1, it is not necessary because of the presence of the C-helix. A version of Mps1 lacking both elements cannot localize to or function at the centrosome, but maintains kinetochore localization and spindle assembly checkpoint function, indicating that TPR3 and the C-helix define a bipartite localization determinant that is both necessary and sufficient to target Mps1 to the centrosome but dispensable for kinetochore targeting. In contrast, elements required for kinetochore targeting (the NTE and first two TPRs) are dispensable for centrosomal localization and function. These data are consistent with a separation of Mps1 function based on localization determinants within the N terminus.
The correct segregation of chromosomes during cellular division is crucial for ensuring genomic stability, and when unregulated, it can lead to a gain or loss of genetic material known as aneuploidy (1), a hallmark of many cancers (2). To regulate chromosome segregation, an elaborate checkpoint has evolved to prevent the onset of anaphase when chromosomes are not properly attached to the mitotic spindle (3). This spindle assembly checkpoint (SAC) has several modular steps (4), and its core function is to prevent mitotic errors by ensuring anaphase does not proceed until each pair of sister chromatids is properly attached to and aligned by microtubules from each mitotic spindle pole. This is achieved by inhibiting the anaphase-promoting complex/cyclosome (APC/C)-mediated degradation of cyclin B and securin (5, 6). A collection of proteins including BubR1 (mitotic checkpoint serine/threonine kinase B) and Mad2 (mitotic arrest deficient 2-like protein 1) prevents Cdc20 (cell-division cycle 20 homolog) from activating the APC/C (7). Mps1 (monopolar spindle 1), another core SAC protein, is a dual-specificity kinase whose activity is necessary for the recruitment of BubR1 and Mad2 to the kinetochore and the conversion of Mad2 from an inactive conformation to an active state that inhibits Cdc20 (8–10).
Mps1 has also been implicated in centrosome function. It was first identified in Saccharomyces cerevisiae, where it is essential for spindle pole body duplication (11), and has been found to localize to centrosomes and regulate centrosome duplication in mice and humans (12–15). Although human Mps1 phosphorylates the centriole component Centrin 2 (Cetn2) and recruits it to centrosomes (16, 17), one difficulty in assigning a centrosomal function to Mps1 has been the possibility that centrosomal phenotypes caused by manipulation of Mps1 function might be attributed to a failure of the SAC in the previous mitosis. However, failure to regulate the centrosomal pool of Mps1 can lead to centrosome amplification (18–21), and extra centrosomes in mitosis can lead to chromosomal segregation errors invisible to the SAC (22). Therefore, it is critical to elucidate the mechanisms regulating Mps1 recruitment both to kinetochores and to the centrosome to have a complete understanding of how Mps1 can influence processes that are likely to be important in tumorigenesis.
Recently, a centrosome localization domain (CLD) was found in human Mps1 (amino acids 53–175), based on homology among vertebrate Mps1 proteins (23). This domain interacts with a novel centrosomal component voltage-dependent anion channel 3 (VDAC3), which is required for Mps1 localization to the centrosome (23). This CLD partially overlaps with a region in the N terminus of Mps1 that folds into a tetratricopeptide repeat (TPR) motif and is required for kinetochore localization and function of Mps1 (24–26). Mutations resulting in improper folding of the first two TPRs lead to a loss of Mps1 at the kinetochore (25), and an N-terminal extension (NTE) preceding the TPRs is responsible for Mps1 dimerization (25, 26); however, the contribution of the third TPR to Mps1 localization and the relationship between the TPRs and the CLD remain to be determined.
We sought to examine the extent of correlation between our previously defined CLD and the newly defined TPR motifs in Mps1. Here we provide evidence that although the NTE and the first two TPRs are responsible for targeting the kinase to the kinetochore during mitosis, the third TPR and its C-terminal capping helix control the centrosomal pool of Mps1. Removing TPR3 and the C-helix impairs centrosome targeting and function without interfering with kinetochore localization or SAC function, whereas removing the NTE and TPR1/2 impairs kinetochore targeting and SAC function without preventing centrosome localization and function. Removal of kinetochore targeting elements also greatly enhances centrosome targeting, suggesting they negatively regulate centrosome targeting of Mps1. This separation of Mps1 function at centrosomes and in the SAC indicates that Mps1-associated centrosomal phenotypes are not the result of SAC errors in the previous mitosis, but represent a bona fide centrosomal function.
Results
Mps1 Amino Acids 136–248 Are Required for VDAC3 Interaction and Centrosomal Localization.
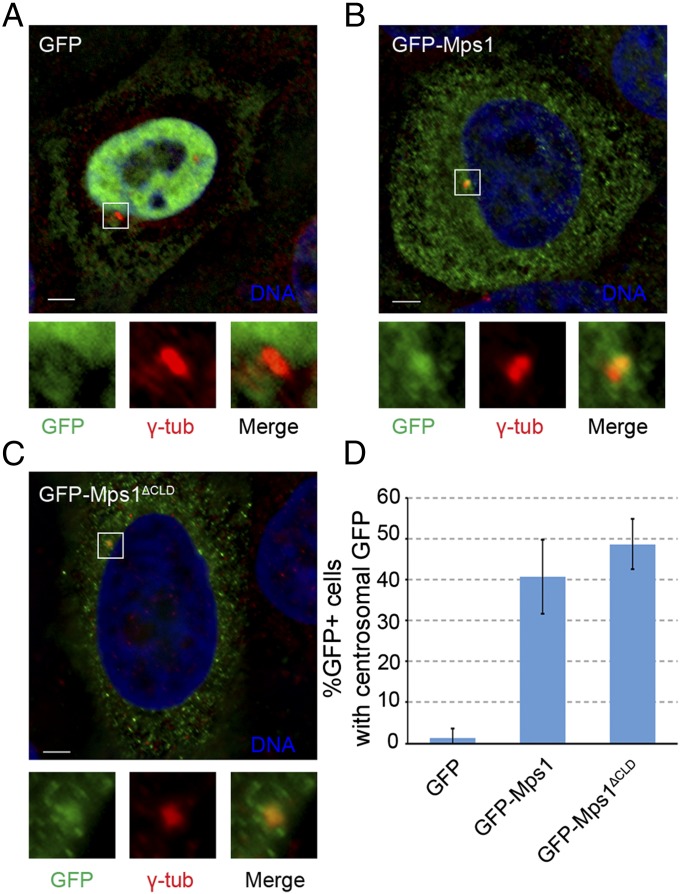
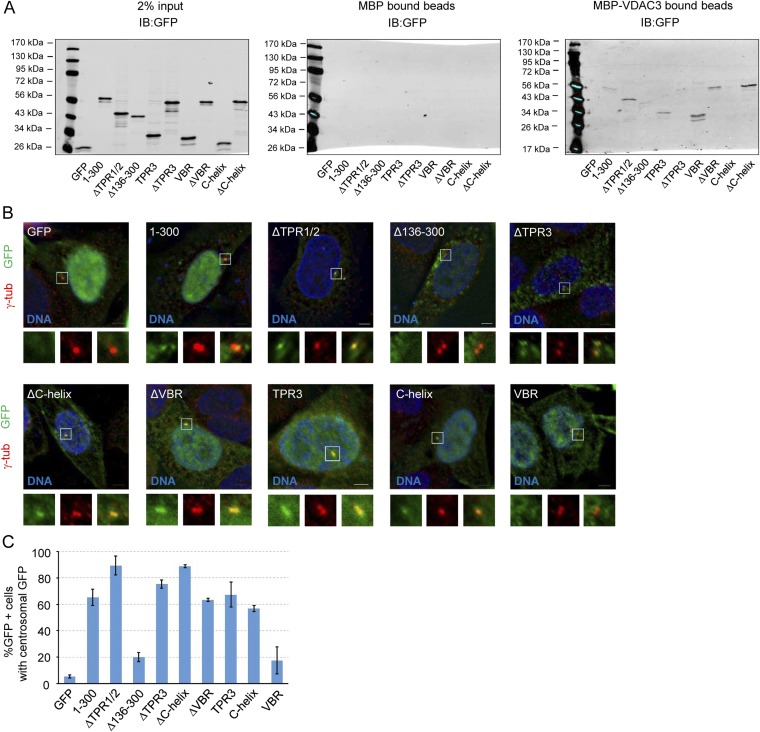
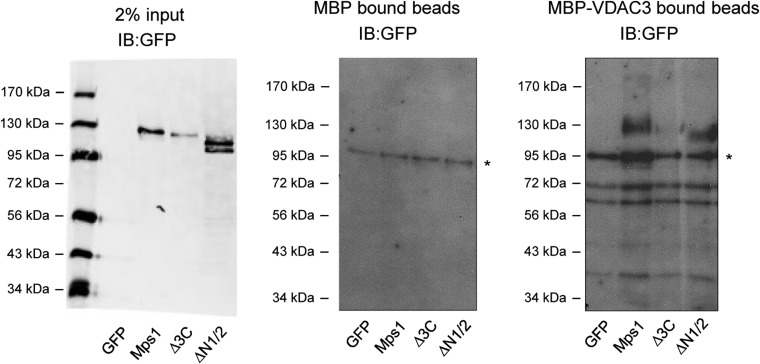
VDAC3 is required for centrosomal targeting of Mps1, and we previously characterized the Mps1 CLD as a VDAC3-binding element (23). However, we subsequently found that in S-phase arrested HeLa cells there was no apparent difference in GFP fluorescence at centrosomes between wild-type GFP-Mps1 and GFP-Mps1∆CLD (GFP accumulated at centrosomes in 40.7 ± 9.0% and 48.7 ± 6.1% of GFP+ cells, respectively) (Fig. 1). Thus, although the CLD is sufficient to target GFP to centrosomes, it is not necessary for the centrosomal targeting of full-length Mps1. This suggested the presence of additional centrosome targeting information in Mps1 and prompted us to expand our analysis of the Mps1 amino terminus. Using the crystal structures of the Mps1 TPRs (24–26) as a guideline, various truncations were generated in the first 300 amino acids of Mps1, which has previously been shown to be sufficient for centrosomal and kinetochore localization (15). The ability of these truncation constructs to bind VDAC3 and localize to centrosomes was then assessed (summary of results in Fig. 2A). To investigate binding to VDAC3, lysates from HEK293 cells expressing GFP-tagged constructs were incubated with amylose resin bound to bacterially expressed maltose binding protein (MBP) or MBP-VDAC3. Loss of the first two TPRs (ΔTPR1/2) did not prevent VDAC3 binding, whereas loss of TPR3 through amino acid 300 (∆136–300) greatly reduced binding to MBP-VDAC3 (Fig. S1A). Interestingly, ∆TPR1/2 consistently showed enhanced VDAC3 binding compared with Mps11–300 (Fig. S1A), although the magnitude of this effect was variable. Both TPR3 and amino acids 218–248 (an unstructured region we have termed the VDAC3 binding region, or VBR) could independently bind to VDAC3, whereas the TPR capping helix (C-helix) could not (Fig. S1A). However, removal of TPR3 from 1 to 300 (∆TPR3) greatly reduced VDAC3 binding, whereas removal of the VBR (∆VBR) had little effect (Fig. S1A), suggesting TPR3 is the functional VDAC3 binding element in the Mps1 N terminus.
Fig. 1.
The CLD is not necessary for Mps1 centrosomal localization. (A–C) Representative images of S-phase arrested HeLa cells transfected with GFP (A), GFP-Mps1 (B), or GFP-Mps1ΔCLD (C), showing DNA (blue), GFP (green), and centrosomes (γ-tub, red). In this and subsequent figures, lower panels depict approximate fourfold digital magnification of boxed regions. (Scale bars, 5 µm.) (D) Percentage of GFP+ cells in which GFP colocalizes with γ-tubulin; mean ± SD (SD) of n = 3 independent experiments of n = 50 cells for each experiment.
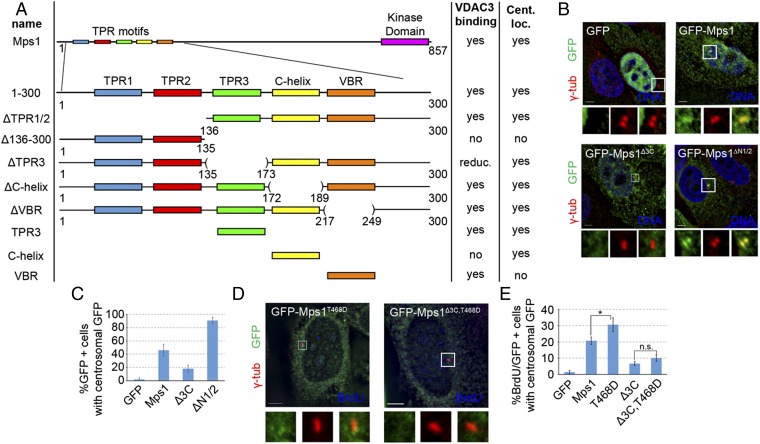
Fig. 2.
The third TPR and capping helix are necessary for Mps1 targeting to the centrosome. (A) Schematic summary of fragments, their interaction with VDAC3, and localization to centrosomes (see also Fig. S1). Numbers indicate amino acid positions. C-helix, capping helix; VBR, VDAC3-binding region; reduc., reduced binding; Cent. Loc., centrosomal localization. (B) Representative images of S-phase arrested HeLa cells transfected with the indicated internal truncation constructs, showing DNA (blue), GFP (green), and centrosomes (γ-tub, red). (C) Percentage of GFP+ cells where GFP colocalizes with γ-tubulin; mean ± SD of n = 3 independent experiments of n = 50 cells for each experiment. (D) Representative images of asynchronously growing HeLa cells transfected with indicated construct and stained for BrdU to identify S-phase cells, showing BrdU (blue), GFP (green), and centrosomes (γ-tub, red). (E) Percentage of cells positive for BrdU and GFP in which GFP signal colocalizes with γ-tubulin; mean ± SD of n = 3 independent experiments of n = 50 cells for each experiment. (Scale bars, 5 µm.)
Fig. S1.
TPR3 and downstream elements are required for VDAC3 binding and centrosomal localization. (A) In vitro pulldown assay between indicated constructs expressed in HEK293 cells and either MBP (Center) or MBP-VDAC3 (Right). MBP or MBP-VDAC3 were expressed in Escherichia coli and bound to amylose resin, and the ability of each construct to bind to the amylose resin was assessed by immunoblotting with an antibody against GFP. (Left) Two percent of the amount of lysate added to the amylose resin. Size of molecular markers for each blot is indicated in kilodaltons. (B) Representative images of asynchronous HeLa cells transfected with the indicated constructs, showing DNA (blue), GFP (green), and centrosomes (γ-tub, red). In this and subsequent figures, lower panels depict approximate fourfold digital magnification of boxed regions. (Scale bar, 5 µm.) (C) Quantification of the percentage of GFP+ cells where GFP signal colocalizes with γ-tubulin at centrosomes, presented as the mean ± SD (SD) of n = 3 independent experiments of n = 50 cells for each experiment; error bars indicate SD (SD).
To investigate the centrosome localization of truncated fragments, their colocalization with the centrosome marker γ-tubulin was assessed, using rigorous and objective criteria, as previously described (18) (see, for example, Fig. S3). Removing the first two TPRs did not hinder centrosomal targeting, and as with VDAC3 binding, ΔTPR1/2 showed enhanced centrosomal targeting, with 89.3 ± 7.0% of GFP+ cells showing centrosomal GFP signal compared with 65.3 ± 6.1% for GFP-Mps11–300 (Fig. S1 B and C). Similar to VDAC3 binding, multiple elements downstream of TPR2 could confer centrosomal localization. Both TPR3 and the C-helix are each able to independently target GFP to centrosomes, whereas the VBR could not (Fig. S1 B and C).
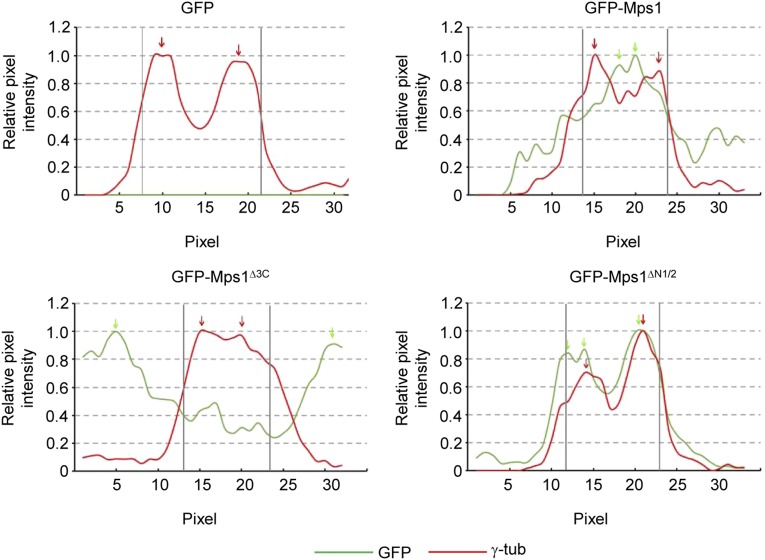
Fig. S3.
The third TPR and capping helix are necessary for Mps1 targeting to the centrosome. Line scan intensity profiles of indicated images from Fig. 2B. As previously described (18), the pixel intensity of both GFP and γ-tubulin is measured along a 5-µm line drawn through the centrosomal γ-tubulin. The reported intensities are normalized to the maximum intensity along the line. GFP is scored as centrosomally localized in a cell if all GFP peaks within 80% of the maximal GFP signal along the line lie within two pixels of the γ-tubulin peaks along the line. Green and red arrows indicate peaks of intensity above 80% of the maximum intensity measured for GFP and γ-tubulin, respectively. Vertical gray bars indicate two-pixel intervals for the γ-tubulin peaks.
The Third TPR and C-Helix Are the Mps1 Centrosomal Determinant.
To refine the centrosomal targeting motif of Mps1, we generated a variety of internal truncations in the full-length Mps1 protein. As observed in the Mps11–300 fragment, deletion of TPR3 and the C-helix prevented the binding of Mps1 to VDAC3, even though the VBR is still present in the GFP-Mps1∆3C construct (Fig. S2), confirming TPR3 as the functional VDAC3 binding element within Mps1. Deletion of TPR3 and the C-helix also prevented efficient localization to the centrosome, as GFP-Mps1Δ3C accumulated at centrosomes in only 18.0 ± 5.6% of S-phase arrested cells (Fig. 2 B and C and Fig. S3). Although it is possible that loss of TPR3 and the C-helix renders the VBR inaccessible to VDAC3, the VBR does not confer centrosomal localization (Fig. S1 B and C), its removal has little effect on VDAC3 binding (Fig. S1A), and its presence is not sufficient for binding of Mps1 to VDAC3 (Fig. S2). Therefore, we have not considered the VBR further. As seen with the N-terminal fragments, deletion of the first two TPRs and the preceding NTE increased the number of GFP+ cells with centrosomal GFP by approximately twofold compared with wild-type Mps1 (90.7 ± 5.0% vs. 46.0 ± 8.7%, respectively) (Fig. 2 B and C).
Fig. S2.
Noncentrosomal Mps1Δ3C cannot interact with VDAC3. In vitro pulldown assay between indicated construct expressed in HEK293 cells and either MBP (Center) or MBP-VDAC3 (Right). Binding to MBP or MBP-VDAC3 was assessed as in Fig. S1. (Left) Two percent of the amount of lysate added to the amylose resin. Size of molecular markers for each blot is indicated in kilodaltons. An asterisk indicates a nonspecific band.
Because Mps1 centrosomal levels are tightly regulated by the proteasome-dependent degradation of Mps1 at centrosomes (18, 20, 27), we tested whether the reduced centrosomal accumulation of GFP-Mps1∆3C could be the result of enhanced degradation. However, proteasome inhibition increases the centrosomal levels of the Mps1 targeting factor VDAC3 (23), which would complicate the analysis. Therefore, we introduced into Mps1∆3C the T468D mutation that prevents degradation of centrosomal Mps1 by mimicking Cdk2 phosphorylation of the centrosomal Mps1 degradation signal (18). Because the Cdk2-regulation of Mps1 degradation occurs during S-phase, we focused our analysis on S-phase cells from asynchronously growing HeLa cells after bromodeoxyuridine (BrdU) labeling. Compared with cells expressing wild-type GFP-Mps1, there was a significant increase in the percentage of BrdU+ cells with GFP signal at centrosomes in cells expressing GFP-Mps1T468D. However, there was no significant difference between cells expressing GFP-Mps1∆3C,T468D and those expressing GFP-Mps1∆3C (Fig. 2 D and E). Because preventing the degradation of GFP-Mps1Δ3C does not increase its accumulation at centrosomes, this supports the suggestion that TPR3 and the C-helix act as a targeting motif for the recruitment of Mps1 to the centrosome, at least in part through the interaction with the centrosomal protein VDAC3.
Mps1 Localization to the Centrosome Is Critical for Its Centrosomal Functions.
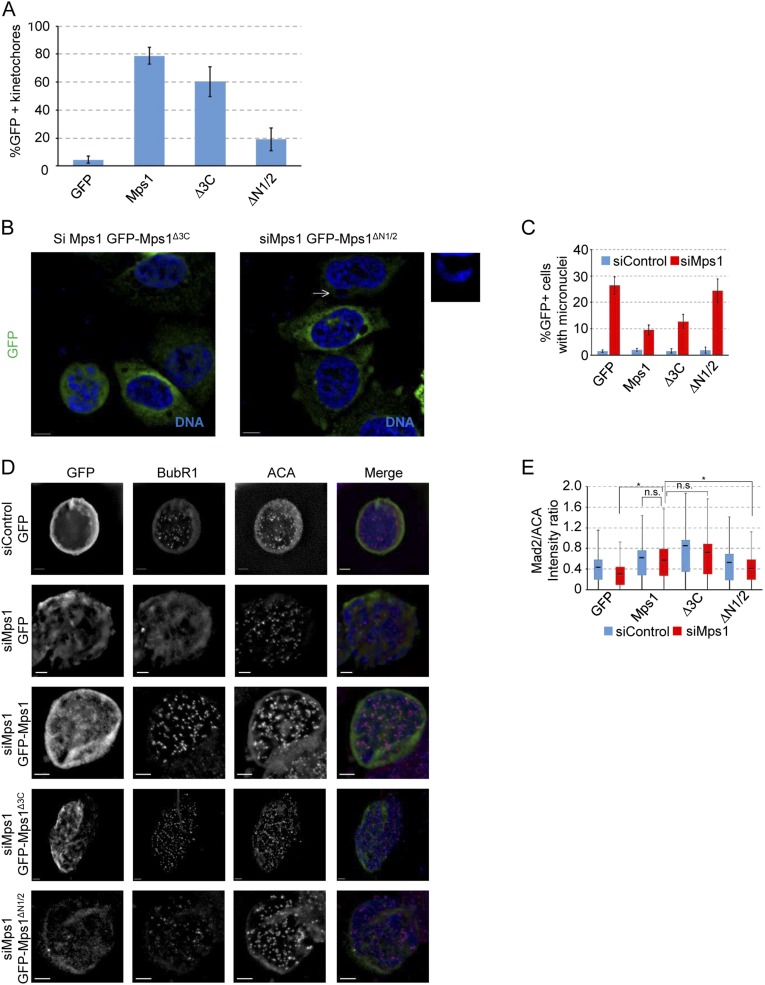
To determine whether the various internal truncation constructs can rescue the centrosomal functions of Mps1, we performed siRNA-mediated depletion of endogenous Mps1, followed by expression of siRNA-resistant Mps1 (sirMps1) constructs. Western blot analysis confirmed efficient knockdown of endogenous Mps1 (Fig. S4A). Surprisingly, we observed an increased amount of the transfected constructs in the presence of siRNA targeting the endogenous Mps1 compared with that in cells transfected with control siRNA (Fig. S4A), which we assume reflects a competitive advantage for Mps1-depleted cells that express an Mps1 construct. Centrosomal localization was not affected by knockdown of endogenous Mps1 (Fig. S4B), indicating that interaction with endogenous Mps1 was not required for the ability of these constructs to bind to centrosomes. Roughly 20% of Mps1-depleted S-phase cells expressing GFP alone have unduplicated centrioles. Although a roughly fivefold increase, this effect is lower than seen in nontransformed cells (23), perhaps because rapidly cycling HeLa cells are more likely to enter S-phase before Mps1 is depleted below the low threshold required for centrosome duplication (13). Regardless, this phenotype is fully rescued by wild-type GFP-sirMps1, but not by GFP-sirMps1∆3C (Fig. 3 A and B). As expected, GFP-sirMps1∆N1/2 that hyperaccumulates at centrosomes could rescue the centriole duplication phenotype (Fig. 3 A and B).
Fig. S4.
Expression and localization of siRNA-resistant GFP-Mps1 constructs. (A) HeLa cells were transfected with either siControl (siRNA against Lamin A/C) or siMps1 for 24 h before being transfected with the indicated siRNA-resistant construct for 48 h. For the final 24 h, cells were arrested in S-phase by HU treatment. Cells were lysed, separated via SDS/PAGE, and blotted with indicated antibodies. Total transfected protein amount was assayed by GFP immunoblot; knockdown of endogenous Mps1 was confirmed by Mps1 (N1) immunoblot, and alpha tubulin (α-tub) served as a loading control. (B) Percentage of S-phase–labeled (by BrdU-incorporation) GFP+ cells with GFP signal colocalizing with γ-tubulin at centrosomes, presented as the mean ± SD of n = 3 independent experiments of n = 100 cells for each experiment.
Fig. 3.
Mps1 localization to the centrosome is critical for its centrosomal functions. (A) Representative images of asynchronously growing HeLa cells transfected with indicated constructs and stained for BrdU to identify S-phase cells, showing BrdU (blue), GFP (green), centrioles (Cetn2, red), and centrosomes (γ-tub, magenta). (B) Percentage of cells positive for BrdU and GFP with two or fewer centrioles, as judged by Cetn2 foci; mean ± SD of n = 3 independent experiments of n = 100 cells for each experiment. (C) Immunoblot analysis of HEK293 cells transfected with either siControl (siRNA against Lamin A/C) or siMps1 for 24 h before being transfected with indicated siRNA-resistant constructs for 48 h. For the final 24 h, cells were arrested in S-phase with HU. Cells were lysed, separated via SDS/PAGE, and blotted with anti-GFP to assess transfected protein level, and anti-pT686 to assess Mps1 activity, with alpha tubulin (α-tub) serving as a loading control. An asterisk signifies a nonspecific band. (Scale bars, 5 µm.)
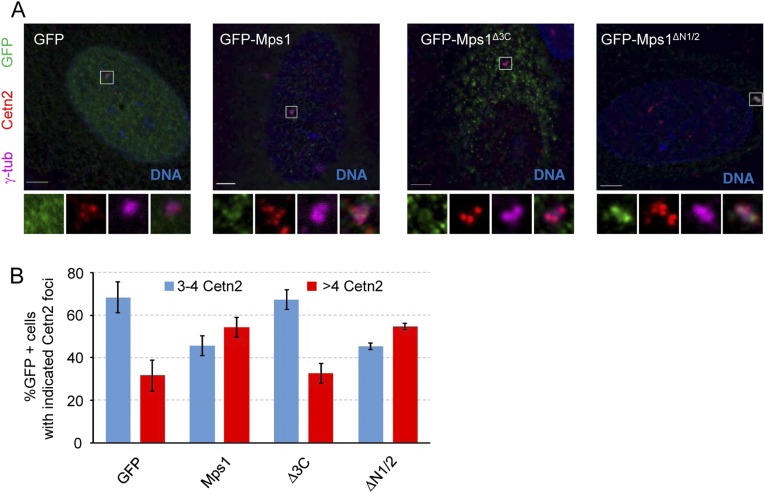
To further assess the centrosomal function of the internal truncation mutants, we tested whether they were competent to modulate the centriole reduplication phenotype that normally occurs in U2OS cells during a prolonged S-phase arrest. Overexpression of wild-type Mps1 has previously been shown to accelerate this reduplication (13, 14, 18, 20, 28), and after a short S-phase arrest of 24 h, GFP-Mps1 increases the percentage of S-phase-arrested U2OS cells with more than four centrioles (as indicated by Cetn2 foci) compared with GFP alone (Fig. S5). In contrast, there was no statistically significant difference in the percentage of cells with more than four centrioles between cells expressing the noncentrosomal Mps1 (GFP-Mps1∆3C) and cells overexpressing GFP alone (32.7 ± 4.7% and 31.7 ± 7.2%, respectively) (Fig. S5). Despite its enhanced centrosomal accumulation, GFP-Mps1∆N1/2 did not accelerate centriole reduplication any more than did wild-type Mps1 (55.0 ± 2.0% vs. 54.3 ± 4.7%, respectively) (Fig. S5).
Fig. S5.
Noncentrosomal Mps1 does not accelerate centriole reduplication. (A) Representative images of U2OS cells transfected with the indicated constructs after a 24-h S-phase arrest. At 24 h after transfection, S-phase arrest was achieved by adding HU for 24 h; 48 h after transfection was considered to be t = 0 of S-phase arrest and was followed by addition of fresh HU for a further 24 h. DNA (blue), GFP (green), centrioles (Cetn2, red), centrosomes (γ-tub, magenta). (Scale bar, 5 µm.) (B) Percentage of GFP+ cells with indicated number of Cetn2 foci, presented as the mean ± SD of n = 3 independent experiments of n = 100 cells for each experiment.
Because deletion of the third TPR and C-helix might result in a nonfunctioning Mps1 variant, we analyzed cells expressing these constructs via Western blot, using the pT686 antibody specific for T686-phosphorylated Mps1, an auto-phosphorylation event that marks fully active Mps1 (25, 29). When normalized to the expression level of each construct, there was no difference between the pT686 staining of either GFP-Mps1∆3C or GFP-Mps1∆N1/2 and wild-type GFP-Mps1 (Fig. 3C). In addition, neither GFP-Mps1∆3C nor GFP-Mps1∆N1/2 prevented centriole assembly, a phenotype caused by overexpression of catalytically inactive Mps1 (13, 14, 28), further supporting the conclusion that the internal truncation mutants are functional.
Noncentrosomal Mps1 Can Still Localize and Function at Kinetochores in Mitosis.
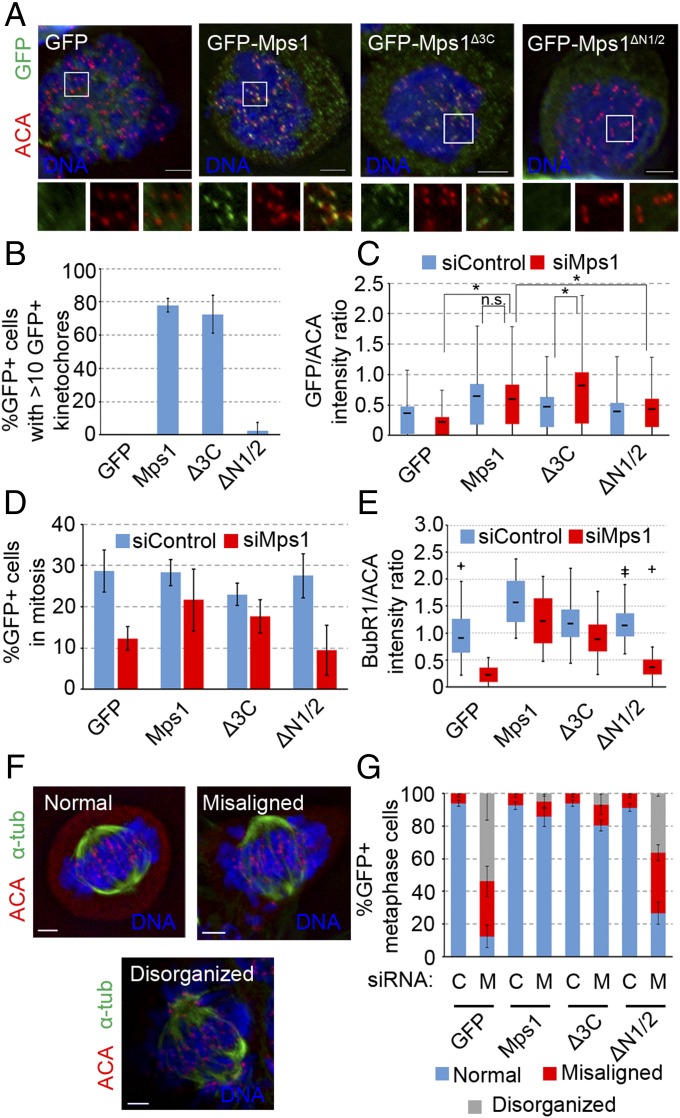
To examine the ability of GFP-Mps1∆3C and GFP-Mps1∆N1/2 to function in the spindle assembly checkpoint, we arrested HeLa cells expressing these constructs in mitosis, using the microtubule poison nocodazole. The noncentrosomal GFP-Mps1∆3C localized to kinetochores as effectively as GFP-Mps1, as judged by the percentage of GFP+ nocodazole-treated cells in which a majority of kinetochores were GFP+ (72.5 ± 11.5% compared with 78.0 ± 4.0%, respectively) (Fig. 4 A and B). In contrast, GFP-Mps1∆N1/2 only satisfied these criteria in 2.5 ± 5.0% of cells (Fig. 4 A and B). This conforms with previous data showing that deletions of the NTE or mutations in the first two TPRs prevent kinetochore targeting (25, 26). There was a slight, yet significant, decrease in the percentage of GFP+ kinetochores in individual cells expressing GFP-Mps1∆3C (60.3 ± 10.5%) compared with that in cells expressing wild-type Mps1 (78.8 ± 5.9%) (Fig. S6A). However, this may be a result of competition with endogenous Mps1, as GFP-Mps1∆3C intensity increases at kinetochores when the endogenous Mps1 is depleted by siRNA (Fig. 4C). As expected, kinetochore levels of GFP-Mps1∆N1/2 were significantly lower than levels of either GFP-Mps1 or GFP-Mps1∆3C (Fig. 4C).
Fig. 4.
Mps1 localization to the centrosome is dispensable for its kinetochore functions. (A) Representative images of HeLa cells expressing the indicated constructs treated with nocodazole for 16 h showing DNA (Hoechst, blue), GFP (green), autoimmune anticentromere antibody (ACA; red). (B) Percentage of cells from A in which a majority of kinetochores are GFP+, as judged by colocalization of GFP and ACA on at least 10 of 20 randomly chosen kinetochores; mean ± SD of n = 3 independent experiments of n = 100 cells for each experiment. (C) Box and whisker plot of GFP intensity normalized to that of ACA (GFP/ACA) at kinetochores. Box indicates 50% of intensity measurements from the 25th to the 75th percentile of n = 300 kinetochores (20 kinetochores from each of five nocodazole-treated cells from n = 3 independent experiments), and whiskers extend up and down to the statistical maximum and minimum. Line in box indicates the mean of the data set. *P ≤ 0.05 from Student’s t test (two-tailed), n.s., not significant (P > 0.05). (D) Percentage of GFP+ cells with condensed mitotic chromatin after 16-h nocodazole treatment; mean ± SD of n = 3 independent experiments of n = 200 cells for each experiment. (E) Box and whisker plot of BubR1 intensity normalized to that of ACA (BubR1/ACA) at kinetochores as in C, with n = 30 kinetochores from n = 3 independent experiments. +, statistical outliers (greater than 1.5× the interquartile range). (F) Representative images of metaphase phenotypes: DNA (blue), α-tubulin (green), centromeres/kinetochores (ACA, red). (G) Percentage of GFP+ cells with indicated metaphase phenotype after release from double thymidine block into MG132; mean ± SD of n = 3 independent experiments of n ≥ 35 cells for each experiment. (Scale bars, 5 µm.)
Fig. S6.
Noncentrosomal Mps1 rescues mitotic phenotypes associated with loss of Mps1. (A) Percentage of GFP+ kinetochores, presented as the mean ± SD of n = 3 independent experiments of n = 100 kinetochores, or 20 kinetochores from five cells in each experiment. (B) Representative images of HeLa cells transfected with the indicated constructs and treated with nocodazole for 16 h, showing DNA (blue) and GFP (green). Arrow indicates a micronucleus magnified in smaller panel. (Scale bar, 10 µm.) (C) Percentage GFP+ interphase cells with micronuclei after 16 h nocodazole treatment. Presented is the mean ± SD of n = 3 independent experiments of n = 100 cells for each experiment. (D) Representative images of HeLa cells transfected with the indicated constructs and treated with nocodazole overnight. From left to right by column shown in grayscale: GFP, BubR1, ACA, and a merged image showing DNA (blue), GFP (green), BubR1 (red), and ACA (magenta). (Scale bar, 5 µm.) (E) Box and whisker plot of Mad2 intensity normalized to that of ACA (Mad2/ACA) at kinetochores. Box and whisker plots represent the same statistical data points as in Fig. 4C. *P ≤ 0.05 from Student’s t test (two-tailed), n.s., not significant (P > 0.05).
We next treated cells with nocodazole for 16 h to assess the ability of the mutant proteins to support the function of the SAC when endogenous Mps1 is depleted. We first noticed an accumulation of micronuclei in nearly 50% of interphase Mps1-depleted cells. Both GFP-sirMps1 and noncentrosomal GFP-sirMps1∆3C efficiently rescued the micronuclei phenotype, whereas the kinetochore-deficient GFP-sirMps1∆N1/2 did not (Fig. S6 B and C). The appearance of micronuclei can arise from a failure of the SAC to properly delay anaphase in the presence of unaligned chromosomes (13). Indeed, there was more than a twofold decrease in the number of mitotic cells on Mps1 depletion, indicating that Mps1-depleted cells cannot activate the SAC to arrest cells in mitosis in response to nocodazole (Fig. 4D). This could not be rescued by GFP alone or the kinetochore-deficient GFP-sirMps1∆N1/2 (Fig. 4D), but checkpoint arrest could be restored by either GFP-sirMps1 or GFP-sirMps1∆3C (for which the minor decrease in mitotic index in Mps1-depleted cells was not statistically significant) (Fig. 4D). GFP-sirMps1∆3C also behaved similarly to full-length Mps1 in its ability to support recruitment of BubR1 (Fig. 4E) and Mad2 (Fig. S6E) to kinetochores in response to nocodazole. The nonkinetochore GFP-sirMps1∆N1/2 negligibly rescued BubR1 recruitment (Fig. 4E) and partially supported Mad2 recruitment (Fig. S6E), but because it failed to support mitotic arrest, we conclude it could not support SAC function to the threshold required for mitotic arrest. BubR1 recruitment is critical for chromosome alignment in metaphase (30), and indeed, Mps1 depletion led to major chromosome alignment defects; these defects were rescued by GFP-sirMps1 and GFP-siRMps1∆3C, but not GFP-alone or GFP-sirMps1∆N1/2 (Fig. 4 F and G).
Discussion
The kinase Mps1 has many functions in the cell cycle, including its well-defined role in recruiting kinetochore components such as BubR1 and Mad2 to unattached kinetochores in the SAC (9) and its role at the centrosome to regulate centriole duplication (13). According to previous studies, localization of Mps1 to kinetochores and centrosomes seemed to be controlled by overlapping regions in the N terminus, the TPR domains, and the CLD (23–26). However, data shown here suggest the two localization determinants are in fact composed of separate elements in the N-terminal TPR domain.
Interestingly, although the first two Mps1 TPRs share a striking similarity in structure to the first two TPRs in the mitotic kinases Bub1 (budding uninhibited by benzimidazoles 1 homolog) and BubR1, Mps1 TPR3 is quite divergent from that in Bub1 and BubR1 (24). The capping helix of Mps1 folds back on the TPR folds in Mps1, whereas those in Bub1 and BubR1 extend away from the TPR domains (24). These factors may explain why the third TPR and capping helix act as a centrosomal determinant in Mps1, but not in Bub1 and BubR1, and why removal of TPR1 and TPR2 enhances centrosome targeting. The third TPR and the largely unstructured VBR were each able to interact with VDAC3, and perhaps the orientation of the capping helix (26) serves to align TPR3 and the VBR to generate a VDAC3 binding surface. Unstructured TPR flanking regions have similarly been reported to cooperate with the TPR in the binding of hsp90 to cyclophilin 40 (31). However, the VBR is not necessary or sufficient for centrosomal targeting, is not required for binding of the N-terminal fragment to VDAC3, and is not sufficient to promote binding of VDAC3 to Mps1∆3C. Thus, although it remains possible that the VBR contributes to VDAC3 binding, our data suggest that TPR3 and the C-helix constitute a bipartite localization determinant required for centrosome localization and function. We have also noted that the C-helix is able to localize to the centrosome without interacting with VDAC3, raising the possibility that the C-helix may help to target Mps1 to the centrosome through a protein or proteins other than VDAC3.
The NTE and first two TPRs, although being necessary for kinetochore localization and function, were not only dispensable for centrosome targeting, but their removal actually enhanced centrosomal targeting. The mechanism whereby these elements might inhibit centrosomal targeting remains unknown, but given that their removal also enhanced the VDAC3 binding of the N-terminal fragment, perhaps they serve to modulate the interaction of Mps1 with centrosomal VDAC3. Given that the NTE has been implicated in the dimerization that is necessary for kinetochore targeting (26), another intriguing possibility could be a preference for monomeric Mps1 at centrosomes.
This study illustrates that a separation of Mps1 function is achieved through localization changes throughout the cell cycle. With Mps1 playing regulatory roles in both centriole duplication and the SAC, the kinase provides an interesting link between centrosome homeostasis and mitotic checkpoint activity to achieve mitotic fidelity. A similar link has been seen with separase and cohesin, which localize to and regulate the separation of both centrioles and chromosomes (32–34), which points to cells using many of the same proteins in different mechanisms to achieve the same ultimate goal of faithful cell division. This study also provides evidence that previously described centrosomal functions of Mps1 are not indirect effects of a SAC failure in a previous mitosis, but definitive centrosomal functions based on a different localization in a different cell cycle stage.
Materials and Methods
Plasmids.
Previously described plasmids used in this study include pHF36 (pECE-GFP-Mps1), pHF140 (pECE-GFP-Mps1T468D), pHF273 (pKM596-VDAC3), and pHF286 (pECE-GFP) (12, 13, 18, 23). The vector pMAL-c2X (New England Biolabs) was used to express MBP in bacteria. Plasmids created for this study are as follows: mammalian expression constructs, pHF330 (GFP-Mps1∆CLD), pHF331 (GFP-Mps11–300), pHF332 (GFP-Mps1136–300), pHF333 (GFP-Mps11–135), pHF334 (GFP-Mps11–300∆TPR3), pHF335 (GFP-Mps11–300∆VBR), pHF336 (GFP-Mps11–300∆C-helix), pHF337 (GFP-Mps1136–171), pHF338 (GFP-Mps1218–248), pHF339 (GFP-Mps1172–188), pHF340 (GFP-sirMps1), pHF341 (GFP-sirMps1∆3C), pHF342 (GFP-sirMps1∆N1/2), pHF343 (GFP-Mps1T468D), pHF344 (GFP-Mps1Δ3C,T468D), and pHF345 (GFP-sirMps1 KD). The Gene-Tailor site directed mutagenesis kit (Invitrogen) was used to create an internal truncation of the CLD (amino acids 52–177) in pHF36 to create pHF330. To create GFP-tagged constructs pHF331, pHF332, pHF333, pHF337, pHF338, and pHF339, PCR products were amplified and inserted into pHF286 (pECE-GFP containing the β globin intron downstream of the SV40 promoter), using In-Fusion HD cloning kit (Clontech). pHF331 was then used as a template for creating pHF334, pHF335, and pHF336 by PCR amplification, using outward-facing BsmBI-flanked primers adjacent to respective deleted domains, followed by digestion with BsmBI and ligation of custom complimentary overhangs. pHF340 and pHF343 were created by inserting a 492-bp DNA fragment encoding the β globin intron downstream of the SV40 promoter in pHF149 (GFP-siRMps1) and pHF140 (GFP-Mps1T468D, respectively (18). pHF341 and pHF342 were created by using pHF340, and pHF344 by using pHF343, respectively, as PCR templates for outward-facing BsmBI-flanked primers adjacent to respective deleted domains, followed by digestion with BsmBI and ligation of custom complimentary overhangs. The siRNA-resistant pHF345 (GFP-siRMps1KD) was created by PCR amplification from plasmid pHF56 (GFP-Mps1KD) (13) with primers to introduce silent mutations into the siRNA binding site of Mps1 (base pairs 1,360–1,384). All constructs were validated by sequence analysis, and PCR primer sequences are available by request.
Cell Culture.
HEK293, HeLa S3, and U2OS cells were cultured in DMEM supplemented with 10% (vol/vol) FBS (Atlanta Biologicals), 100 U/mL penicillin G, and 100 µg/mL streptomycin (HyClone) in the presence of 5% (vol/vol) CO2. S-phase arrest was achieved using 4 mM hydroxyurea (HU; Sigma) for 24 h. S-phase cells in asynchronous cultures were labeled by incorporation of BrdU during a 4-h treatment with 40 µM BrdU (Sigma). Centriole reduplication assays were performed as previously described (13), using cells treated with 4 mM HU for 48 h to achieve a prolonged S-phase arrest (24 h arrest after an initial 24-h treatment). For mitotic checkpoint-induced arrest, asynchronous cells were treated with 250 ng/mL nocodazole (Sigma) for 16 h. For metaphase arrest, cells were synchronized by double thymidine (2 µM) block, as described previously (30). Rescue constructs were transfected concurrent with the first block, and cells were released into fresh DMEM for 9 h, at which point 20 µM MG132 was added for 1 h.
DNA and siRNA Transfections.
Mammalian expression constructs were transfected using JetPrime (PolyPlus) in HeLa and HEK293 and FuGENE6 (Roche) in U2OS cells. Lipofectamine RNAiMax (Invitrogen) was used to deliver siRNAs directed against Mps1 (siMps1: nucleotides 1,360–1,384; Invitrogen) and Lamin A/C (siControl; Dharmacon) at a final concentration of 30 nM.
Cytology.
The following primary antibodies were used for indirect immunofluorescence: rabbit polyclonal anti–γ-tubulin, 1:200 (Sigma); goat polyclonal anti–γ-tubulin, 1:50 (Santa Cruz Biotechnology); affinity purified rabbit anti-Cetn2, 1:4,000 (17); rat monoclonal anti-BrdU, 1:250 (Abcam); mouse monoclonal anti-GFP, 1:250 (Invitrogen); rabbit polyclonal anti-GFP, 1:500 (Abcam); human anti-centromere serum (HCT-0100), 1:500 (ImmunoVision); mouse monoclonal anti-BubR1, 1:100 (Active Motif); mouse anti-α-tubulin DM1A, 1:1,000 (Sigma); and rabbit polyclonal anti-Mad2 serum, 1:200 (Covance). Secondary antibodies for indirect immunofluorescence were donkey anti-rabbit, donkey anti-mouse, donkey anti-human, donkey anti-goat, or donkey anti-rat conjugated with Alexa 350 (1:200), Alexa 488 (1:1,000), Alexa 594 (1:1,000), Alexa 750 (1:200; Invitrogen), or IRDye 800 (1:200; Rockland), and DNA was stained with Hoechst 33342 (1:1,000; Sigma). Indirect immunofluorescence was performed as previously described (23). Briefly, cells were fixed with either 4% (wt/vol) formaldehyde (Ted Pella) in PBS supplemented with 0.5 mM MgCl2 and 0.2% Triton X-100 for 10 min at room temperature or methanol for 10 min at −20 °C. All primary antibody staining was performed overnight at 4 °C in blocking buffer containing 5% (vol/vol) FBS, 0.2 M glycine, 0.5 mM MgCl2, and 0.1% Triton X-100 in PBS. All secondary antibodies and Hoechst were incubated in blocking buffer for 1 h at room temperature. For visualizing BrdU, methanol-fixed cells were stained with other primary and secondary antibodies before being fixed with methanol again, treated with 2 N HCl for 30 min at room temperature, neutralized with Tris at pH 8.0, and stained with the anti-BrdU antibody for 1 h at 37 °C. All images were acquired at ambient temperature, using an Olympus IX-81 microscope with a 60× or 100× Plan Apo oil immersion objective (1.4 numerical aperture) and a QCAM Retiga ExiFast 1394 camera, and were analyzed using Slidebook software (Intelligent Imaging Innovations). Kinetochore intensities were calculated as previously described (35), with one exception: subtracted background intensities were measured with an identically sized circle inside the cells at a centromere-negative location.
Immunoblotting and Pulldown Assays.
Cells were lysed in lysis buffer (50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, and 1% Nonidet P-40) supplemented with 1× Halt Protease inhibitor mixture (Invitrogen) and 1 mM AEBSF-HCl (Invitrogen). Protein concentrations were determined via Bradford assay and samples to SDS/PAGE before being transferred to nitrocellulose. The following primary antibodies were used for immunoblotting: rabbit anti-GFP, 1:2,000 (Sigma); rabbit anti-p686 Mps1, 1:1,000 (kind gift from P. Eyers, University of Liverpool, Liverpool, UK); mouse anti-Mps1 N1, 1:1,000 (Invitrogen); and mouse anti–α-tubulin DM1A, 1:10,000 (Sigma). Secondary antibodies were Alexa680-conjugated donkey anti-rabbit (Invitrogen), IRDye800-conjugated donkey anti-mouse (Rockland), and HRP-conjugated goat anti-rabbit (GE Healthcare), all at 1:10,000 dilutions. Immunoblots were imaged and analyzed using the Odyssey imaging system (Li-COR) or the Super Signal West Femto Chemiluminescent substrate (Thermo Scientific). Pull-down assays were performed as previously described (23), with the exception that bait-bound beads were incubated with cell lysates for 2 h at 4 °C.
Acknowledgments
This work was supported in part by NIH R01 GM077311 (to H.A.F.) and Pelotonia Graduate and Undergraduate Fellowships from the Ohio State University Comprehensive Cancer Center (to J.R.M and K.J.B, respectively).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607421113/-/DCSupplemental.
References
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 3.Vleugel M, Hoogendoorn E, Snel B, Kops GJPL. Evolution and function of the mitotic checkpoint. Dev Cell. 2012;23(2):239–250. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 5.Hwang LH, et al. Budding yeast Cdc20: A target of the spindle checkpoint. Science. 1998;279(5353):1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 6.Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: An effector of the Mad2-dependent spindle checkpoint. Science. 1998;279(5353):1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 7.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154(5):925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190(1):25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maciejowski J, et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190(1):89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tipton AR, et al. Monopolar spindle 1 (MPS1) kinase promotes production of closed MAD2 (C-MAD2) conformer and assembly of the mitotic checkpoint complex. J Biol Chem. 2013;288(49):35149–35158. doi: 10.1074/jbc.M113.522375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: Novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114(4):745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106(1):95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 13.Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci USA. 2003;100(25):14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanai M, et al. Physical and functional interaction between mortalin and Mps1 kinase. Genes Cells. 2007;12(6):797–810. doi: 10.1111/j.1365-2443.2007.01091.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu S-T, et al. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14(4):1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dantas TJ, et al. Calcium-binding capacity of centrin2 is required for linear POC5 assembly but not for nucleotide excision repair. PLoS One. 2013;8(7):e68487. doi: 10.1371/journal.pone.0068487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C-H, Kasbek C, Majumder S, Yusof AM, Fisk HA. Mps1 phosphorylation sites regulate the function of centrin 2 in centriole assembly. Mol Biol Cell. 2010;21(24):4361–4372. doi: 10.1091/mbc.E10-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasbek C, et al. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18(11):4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasbek C, Yang C-H, Fisk HA. Mps1 as a link between centrosomes and genomic instability. Environ Mol Mutagen. 2009;50(8):654–665. doi: 10.1002/em.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasbek C, Yang C-H, Fisk HA. Antizyme restrains centrosome amplification by regulating the accumulation of Mps1 at centrosomes. Mol Biol Cell. 2010;21(22):3878–3889. doi: 10.1091/mbc.E10-04-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, et al. Phosphorylation of Mps1 by BRAFV600E prevents Mps1 degradation and contributes to chromosome instability in melanoma. Oncogene. 2013;32(6):713–723. doi: 10.1038/onc.2012.94. [DOI] [PubMed] [Google Scholar]
- 22.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majumder S, Slabodnick M, Pike A, Marquardt J, Fisk HA. VDAC3 regulates centriole assembly by targeting Mps1 to centrosomes. Cell Cycle. 2012;11(19):3666–3678. doi: 10.4161/cc.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S, et al. Characterization of spindle checkpoint kinase Mps1 reveals domain with functional and structural similarities to tetratricopeptide repeat motifs of Bub1 and BubR1 checkpoint kinases. J Biol Chem. 2012;287(8):5988–6001. doi: 10.1074/jbc.M111.307355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thebault P, et al. Structural and functional insights into the role of the N-terminal Mps1 TPR domain in the SAC (spindle assembly checkpoint) Biochem J. 2012;448(3):321–328. doi: 10.1042/BJ20121448. [DOI] [PubMed] [Google Scholar]
- 26.Nijenhuis W, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J Cell Biol. 2013;201(2):217–231. doi: 10.1083/jcb.201210033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivas V, Kitagawa M, Wong J, Liao PJ, Lee SH. The tumor suppressor Cdkn3 is required for maintaining the proper number of centrosomes by regulating the centrosomal stability of Mps1. Cell Rep. 2015;13(8):1569–1577. doi: 10.1016/j.celrep.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 28.Mattison CP, et al. Mps1 activation loop autophosphorylation enhances kinase activity. J Biol Chem. 2007;282(42):30553–30561. doi: 10.1074/jbc.M707063200. [DOI] [PubMed] [Google Scholar]
- 29.Tyler RK, et al. Phosphoregulation of human Mps1 kinase. Biochem J. 2009;417(1):173–181. doi: 10.1042/BJ20081310. [DOI] [PubMed] [Google Scholar]
- 30.Dou Z, et al. Dynamic localization of Mps1 kinase to kinetochores is essential for accurate spindle microtubule attachment. Proc Natl Acad Sci USA. 2015;112(33):E4546–E4555. doi: 10.1073/pnas.1508791112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratajczak T, Carrello A. Cyclophilin 40 (CyP-40), mapping of its hsp90 binding domain and evidence that FKBP52 competes with CyP-40 for hsp90 binding. J Biol Chem. 1996;271(6):2961–2965. doi: 10.1074/jbc.271.6.2961. [DOI] [PubMed] [Google Scholar]
- 32.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400(6739):37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 33.Beauchene NA, et al. Rad21 is required for centrosome integrity in human cells independently of its role in chromosome cohesion. Cell Cycle. 2010;9(9):1774–1780. doi: 10.4161/cc.9.9.11524. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura A, Arai H, Fujita N. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J Cell Biol. 2009;187(5):607–614. doi: 10.1083/jcb.200906019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elowe S, Hümmer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21(17):2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]