Significance
Despite widespread reports on deciphering the sequences of all kinds of genomes, most of these reconstructed genomes rely on a comparison of short DNA sequencing reads to a reference sequence, rather than being independently reconstructed. This method limits the insights on genomic differences to local, mostly small-scale variation, because large rearrangements are likely overlooked by current methods. We have de novo assembled the genome of a common strain of Arabidopsis thaliana Landsberg erecta and revealed hundreds of rearranged regions. Some of these differences suppress meiotic recombination, impacting the haplotypes of a worldwide population of A. thaliana. In addition to sequence changes, this work, which, to our knowledge is the first comparison of an independent, chromosome-level assembled A. thaliana genome, revealed hundreds of unknown, accession-specific genes.
Keywords: de novo assembly, Arabidopsis, PacBio sequencing, inversions, gene absence/presence polymorphisms
Abstract
Resequencing or reference-based assemblies reveal large parts of the small-scale sequence variation. However, they typically fail to separate such local variation into colinear and rearranged variation, because they usually do not recover the complement of large-scale rearrangements, including transpositions and inversions. Besides the availability of hundreds of genomes of diverse Arabidopsis thaliana accessions, there is so far only one full-length assembled genome: the reference sequence. We have assembled 117 Mb of the A. thaliana Landsberg erecta (Ler) genome into five chromosome-equivalent sequences using a combination of short Illumina reads, long PacBio reads, and linkage information. Whole-genome comparison against the reference sequence revealed 564 transpositions and 47 inversions comprising ∼3.6 Mb, in addition to 4.1 Mb of nonreference sequence, mostly originating from duplications. Although rearranged regions are not different in local divergence from colinear regions, they are drastically depleted for meiotic recombination in heterozygotes. Using a 1.2-Mb inversion as an example, we show that such rearrangement-mediated reduction of meiotic recombination can lead to genetically isolated haplotypes in the worldwide population of A. thaliana. Moreover, we found 105 single-copy genes, which were only present in the reference sequence or the Ler assembly, and 334 single-copy orthologs, which showed an additional copy in only one of the genomes. To our knowledge, this work gives first insights into the degree and type of variation, which will be revealed once complete assemblies will replace resequencing or other reference-dependent methods.
Landsberg erecta (Ler) is presumably the second-most-used strain of Arabidopsis thaliana after the reference accession Columbia (Col-0). It is broadly known as Ler-0, which is an abbreviation for its accession code La-1 and a mutation in the ERECTA gene. In 1957, George Rédei, at the University of Missouri–Columbia, irradiated La-1 samples, which were provided by Friedrich Laibach and were collected in Landsberg an der Warthe (now called Gorzów Wielkopolski), Poland, where Ler-0–related genotypes are still present (1). Some seeds of the original batch were irradiated with X-rays, resulting, among others, in the isolation of the erecta (er) mutant (2, 3). Will Feenstra received this mutant from George Rédei in 1959 because he was interested in its erect growth habit and introduced it as the standard strain in the Department of Genetics at Wageningen University. There, he started a mutant induction program that was later continued by Jaap van der Veen and Maarten Koornneef. Mutants from this program as well as parental lines of recombinant inbred lines were mainly distributed as Ler-0 lines to other laboratories, reflecting the increasing interest in A. thaliana. Some descendants of Ler-0 were later renamed to Ler-1 and -2 to identify genotypes used in different laboratories, but most likely all derived from the original mutant isolated by Rédei, and we will collectively refer to them as “Ler.”
First comparative analyses of the Ler genome included cytogenetic studies using pachytene cells and in situ hybridization (4, 5), suggesting a large inversion on the short arm of chromosome 4, as well as differences between 5S rDNA clusters compared with the genome of Col-0 (4). The first large-scale analysis of the Ler genome sequence was published together with the Col-0 reference sequence in 2000 (6). Within 92 Mb of random shotgun dideoxy sequencing reads, 25,274 SNPs and 14,570 indels were identified. Although this was a severe underestimation (7–11), the authors already observed that many of the large indels contained entire active genes, half of which were found at different loci in the genome of Ler, whereas others were entirely absent (6).
The advent of next-generation sequencing greatly expanded the knowledge of natural genetic diversity in A. thaliana (reviewed in refs. 12 and 13). However, genome-wide studies on gene absence/presence polymorphisms were not repeated, because short-read analyses focused on small-scale changes only. To resolve large variation, reference-guided assemblies (8, 9, 14) and structural variation-identifying tools (15) were introduced. However, such methods mostly reveal local differences, which do not include the complement of large-scale rearrangements including inversions or transpositions.
So far, two de novo assemblies of Ler have been published. The first was based on Illumina short-read data and resulted in an assembly with an N50 of 198 kb, showing similar performance as a reference-guided assembly (8). The second was based on a set of previously released Pacific Bioscience’s single-molecule, real-time sequencing (PacBio) data (16) and assembled the genome into 38 contigs with an N50 of 11.2 Mb (17). This drastic improvement outlines the potential of long-read sequencing technologies such as PacBio sequencing (18) to overcome the limitations of short-read methods because long reads can span many of the repetitive regions, which are presumably the most common reason for assembly breaks in short-read assemblies. Alternatively, low-fold long-read sequencing could be combined with cheaper short-read sequencing, either by using the short reads for error-correcting the long reads (19) or for integrating long-read information into short-read assemblies or vice versa (20).
Despite the unprecedented contiguity of this long-read assembly, both earlier studies focused on methodological aspects and did not perform any whole-genome comparisons of the Ler genome or gene annotations, and, as a consequence, comprehensive reports on large-scale rearrangements and nonreference genes are still sparse. We have generated an advanced de novo assembly of Ler consisting of 117 Mb arranged into five sequences representing the five chromosomes, which is based on a combination of short-read assembly, long-read-based gap closure, and scaffolding based on genetic maps. This chromosome-scale assembly and its comparison with the reference assembly revealed features that are typically not analyzed within next-generation sequencing assemblies, including the location of a polymorphic rDNA cluster and centromeric repeats, as well as the exact makeup of all large rearrangements including a 1.2-Mb inversion on the short arm of chromosome 4. This inversion suppresses meiotic recombination in Ler and Col-0 hybrids, and we show that this suppression introduced genetically isolated inversion haplotypes into the worldwide population of A. thaliana. De novo gene annotation revealed hundreds of copy-number polymorphisms as well as novel genes that are entirely absent in one or the other genome. Finally, we report on variation in different Ler genomes, suggesting that some Ler lines feature unexpected footprints of an additional mutagenesis event.
Results
Karyotype-Resolving Assembly of A. thaliana Ler.
We deeply sequenced the genome of Ler using Illumina libraries of different insert sizes (SI Appendix, Table S1). Combined with recently generated data (8), we obtained an initial assembly with an N50 of 7.5 Mb using ALLPATHS-LG (21) and SSPACE (22). For further improvement, we generated long-read data from 10 PacBio SMRTcells. However, during the course of this project, Pacific Biosciences released sequencing reads of Ler with higher quality (16), which we used for gap closure (20) and scaffolding (23) to generate an assembly with an N50 of 12.8 Mb consisting of 65 scaffolds >50 kb. Following the POPSEQ approach (24), we anchored 31 of these scaffolds to two public genetic maps (25, 26), which allowed us to generate five chromosome-representing sequences using stretches of Ns as indication of assembly breaks (Table 1 and SI Appendix, SI Materials and Methods and Fig. S1). Because some of the short scaffolds were too small and did not overlap with enough markers, we introduced an additional seven scaffolds with a combined length of 1.4 Mb into the reconstructed chromosomes based on homology information from Col-0. The final assembly consisted of five pseudomolecules and 25 unplaced scaffolds (including scaffolds representing the organelle genomes) with a combined length of 118.9 Mb, including <2 Mb of ambiguous bases.
Table 1.
Assembly statistics and comparison with the reference sequence
| Feature | Col-0 | Ler |
| Chromosome scaffolds | 5 | 5 |
| Unplaced scaffolds | 0 | 25 |
| Assembly length | 119,146,348 | 118,890,721 |
| Ambiguous bases | 185,738 | 1,777,652 |
| Gene no. | 27,416 | 27,170 |
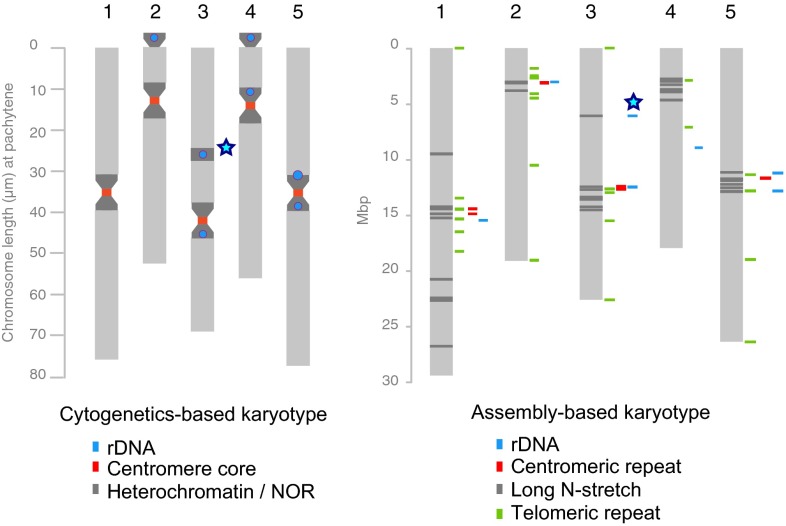
Within the assembly, we found sequence similarity to telomeric repeats at five of eight chromosome ends without heterochromatic nuclear organizing region (i.e., shorter arms of chromosomes 2 and 4) and centromeric repeat sequences and rDNA clusters within most of the pericentromeric regions (Fig. 1). Telomeric repeats were also found as interstitial repeats near or in the pericentromeric regions, which is similar to their location in the Col-0 genome (27). Interestingly, we found a few rDNA copies in the upper arm of chromosome 3, revealing the location of a (not fully assembled) rDNA cluster, which is in agreement with earlier findings of a Ler-specific rDNA locus in this region (4). To test the assembly quality further, we analyzed nucleotide-binding leucine-rich repeat (NB-LRR) gene loci, which include regions that are known to be structurally diverse between accessions and are challenging to assemble because of high levels of local repeats (28). For 154 of the 159 genes, we could identify the orthologous regions in the Ler assembly. Only 10 (6%) of these regions included ambiguous sequences, which would indicate failures in sequence assembly. In contrast, 49 (31%) of them revealed differences >100 bp, corroborating their strong divergence.
Fig. 1.
Chromosome-level sequence assembly reveals the karyotype and the arrangement of structural features of the A. thaliana Ler genome. Ideogram of the Ler genome shows idealized chromosomes at pachytene and chromosomal marks, which were revealed with cytogenetics including heterochromatic clusters (dark gray), clusters of centromeric repeats (red), and rDNA (blue) (4, 5; modified from ref. 56). On the right is an illustration of the assembly, including five chromosomal sequences. The colored bars next to the chromosomes indicate the location of sequence similarity to telomeric repeat sequence (green), centromeric repeat (red), and rDNA (blue) as well as major gaps in the sequence (dark gray). The blue star marks a Ler-specific rDNA cluster, which was earlier identified by cytogenetics (4) and was also found in the assembly.
Finally, we used Ler wild-type RNA-sequencing (RNA-seq) reads from two public datasets (9, 29) and homology to Col-0 protein sequences to annotate 27,170 protein-coding genes in the assembly of Ler (compared with 27,416 protein-coding genes in the reference annotation) (SI Appendix, SI Materials and Methods).
The Complement, Divergence, and Impact of Nonallelic Rearrangements.
In contrast to resequencing, full-length assemblies facilitate direct identification of large-scale genomic rearrangements, including transpositions and inversions. These types of higher-order differences constitute a second layer of genetic variation, because local differences (e.g., SNPs) can be found in colinear as well as in rearranged regions (Fig. 2A). However, because resequencing studies typically cannot distinguish between rearranged and nonrearranged regions, this distinction was so far not possible (30).
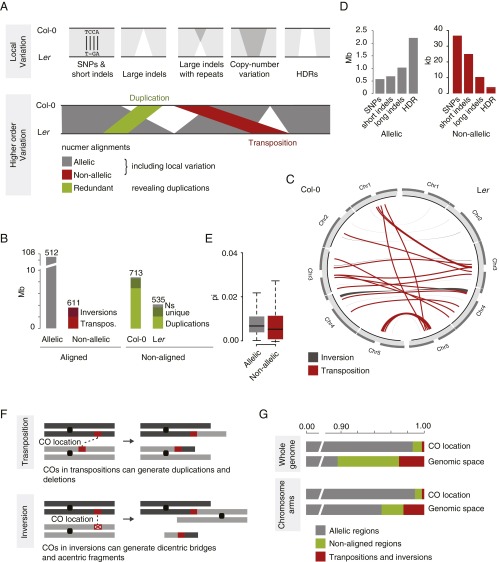
Fig. 2.
Higher-order sequence variation. (A) Schematic of local (Upper) and higher-order (Lower) sequence variation as revealed by a whole-genome alignment. Local sequence divergence does not only include small-scale variation like SNPs and small indels, but also structural variation like large indels and HDRs. Higher-order variation includes transpositions and inversions, which do not reside in the orthologous regions in the other genome. Both colinear (allelic) and rearranged (nonallelic) regions can harbor local variation. (B) Amount of aligned and nonaligned regions in a nonredundant whole-genome alignment of Col-0 and Ler. Aligned regions can be separated into colinear (gray) and rearranged regions [inversions and transpositions (transpos.); red]. Nonaligned regions, typically residing in the breaks between allelic and nonallelic regions, are shown for Col-0 and Ler separately, including the amount of putatively duplicated regions. (C) Location of transpositions and inversions. (D) Genomic space involved in different types of local sequence variation, separately shown for allelic and nonallelic regions. (E) Sequence divergence in allelic and nonallelic alignments. (F) Schematic examples for the consequences of meiotic recombination (CO) events in transposed (Upper) and inverted (Lower) regions. Chromosome arm exchange in nonallelic regions can lead to extreme chromosomal rearrangements. (G) Distribution of the location of 362 CO events in respect to their occurrence in allelic (gray), nonaligned (green), and nonallelic (red) regions in contrast to the genomic fractions of these regions; shown are complete genome (Upper) and only chromosome arms (Lower).
A whole-genome alignment of the Ler and Col-0 genome assemblies (31) revealed 512 colinear (allelic) and 611 rearranged (nonallelic) regions comprising ∼107.6 and 3.6 Mb (Fig. 2B and SI Appendix, SI Materials and Methods). Among the nonallelic regions, we identified 47 inversions and 383 and 181 inter- and intrachromosomal transpositions (Dataset S1). Most of the transpositions resided in pericentromeric regions, whereas inversions were also found in chromosome arms (Fig. 2C). Nearly 40% of the transposed sequences overlapped with transposable elements (TEs); however, only a minor fraction of the inversion sequences were related to TEs. The inversion breakpoints overlapped with 7 and 10 genes in the Ler and Col-0 assemblies, which did not have syntenic orthologs in the other assembly, suggesting that these genes have been deleted by the inversion events.
The by far largest sequence difference between the two assemblies was an inversion of 1.2 Mb on chromosome 4, which we confirmed by PCR (SI Appendix, Figs. S2 and S3). The latter inversion was already described by Fransz et al. (5) and was found to be associated with a polymorphic, heterochromatic knob on the short arm of the Col-0 chromosome, presumably due to inverting parts derived from the pericentromere (5, 32). This inversion could also be observed between Col-0 and the closely related plant Arabidopsis lyrata, further corroborating that the Col-0 allele is the derived form (33). Moreover, 7.9 Mb of the Col-0 and 4.1 Mb of the Ler assembly were not aligned to any homologous region of the other genome at all. This sequence space was separated into 713 and 535 regions, respectively (Fig. 2B). Even though not aligned in a strict one-to-one whole-genome alignment, large portions of these unaligned regions showed extensive similarity to regions in the other genome, suggesting that most of these regions originated from duplication events (Fig. 2 A and B) (SI Appendix, SI Materials and Methods).
To compare sequence divergence in allelic and nonallelic regions, we searched for local differences, including SNPs and small and large indels, as well as highly divergent regions (HDRs) (Fig. 2A, SI Appendix, SI Materials and Methods, and Dataset S2). Approximately 4.5 Mb (4.2%) of the 107.6 Mb of allelic sequence were polymorphic, with the majority of this variation organized in long indels and HDRs (Fig. 2D). Approximately 39% of the long indels had flanking (short or tandem) repeat sequences, indicating that a large proportion of the indel mutations were introduced by homology-dependent events, whereas only 16% of them highly overlapped with TEs. Even though rearranged (nonallelic) sequence harbored less large local differences, presumably due to their short length [average lengths: transpositions, 4 kb; inversions, 9 kb (without the 1.2 Mb inversion); allelic regions, 209 kb], the average pairwise difference in the alignments of nonallelic regions was still similar compared to the differences in allelic regions (Fig. 2 D and E).
To avoid recombination between nonallelic regions, meiotic recombination requires pairing of homologous sequence between allelic regions (11), implying that rearranged regions should be suppressed for meiotic recombination in heterozygotes (Fig. 2F). To test this hypothesis, we overlapped the precise location of 362 crossover (CO) events from Col-0/Ler hybrids previously collected from different studies (34) with allelic and nonallelic regions (SI Appendix, SI Materials and Methods). Only five of the CO events did not reside in colinear, allelic regions (expected: 35 CO events), of which four COs were located in nonaligned regions, and only one CO was mapped to a transposition (Fig. 2G). This highly significant underrepresentation of COs in nonallelic regions provides evidence that rearranged regions are in fact suppressed for meiotic recombination (P = 1.96e-06, χ2 test).
The Effects of Inversions on Natural Haplotypes.
Inverted regions were most drastically suppressed for meiotic recombination and in consequence genetic exchange between the two alleles of an inversion is expected to be minimized (35). Despite this strong impact, no population-wide analysis of inversions in Arabidopsis and only few inversions have been reported so far (5, 8, 36).
Two inversions between Col-0 and Ler were >100 kb, including a 170-kb inversion on chromosome 3 and the 1.2-Mb inversion on chromosome 4. Overlapping the regions of the inversion with meiotic recombination frequency data (26) showed locally reduced recombination rates co-occurring with both of these two large inversions (Fig. 3A and SI Appendix, Fig. S4). The effect of the chromosome 4 inversion extended its recombination suppression into the heterochromatic pericentromere and was in agreement with early reports of reduced recombination specific to hybrids of these accessions (26, 37).
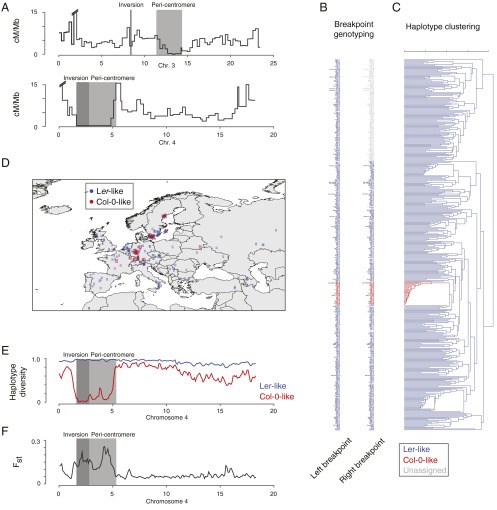
Fig. 3.
Impact of large-scale inversions on meiotic recombination and haplotype diversity in a worldwide collection of A. thaliana accessions. (A) Male meiotic recombination frequencies across chromosomes 3 and 4 contrasted with the location of the two large-scale inversions (dark gray boxes) and the pericentromeric regions (light gray boxes) [recombination data generated by Giraut et al. (26)]. Recombination frequency was measured between markers with an average distance of 316 kb. Both inversions co-occur with locally reduced recombination frequencies. The interval harboring the inversion on chromosome 3, however, showed residual recombination activity, which does not imply recombination in the inverted region, but might arise from recombination in 111 kb of noninverted sequence in this interval. (B) The names of 409 accessions colored by the inferred chromosome 4 inversion allele (blue, Ler allele; red, Col-0 allele) as assessed on the left and right breakpoints of the inversion. The accessions were ordered after their occurrence in the haplotype clustering shown in C. (C) Haplotype clustering based on 9,198 SNPs located within the chromosome 4 inversion, revealing two distinct clusters, which perfectly matched the two chromosome 4 inversion alleles. (D) Distribution of the accession origins in central Europe, colored by their respective chromosome 4 inversion alleles. (E) Haplotype diversity within the accessions carrying a Col-0–like (red) or Ler-like allele (blue) of the chromosome 4 inversion. (F) Population differentiation (Fst) between these two groups of accessions. Inversion and pericentromere shown with dark and light gray boxes.
Selection acting on one of the alleles of an inverted region can have dramatic effect on its allele frequency and haplotype divergence across the entire inverted regions. To estimate the impact of these large inversions on the population of A. thaliana, we genotyped a worldwide selection of 409 accessions using public whole-genome sequencing data (15, 30, 38) (SI Appendix, SI Materials and Methods). For this process, we simultaneously aligned the short reads against the Col-0 and Ler reference sequences and calculated the ratio of alignments to either of the inversion breakpoints to assign the respective allele (SI Appendix, Fig. S5). Surprisingly, only sequencing reads of Ler matched the Ler inversion breakpoints of the 170-kb inversion on chromosome 3, whereas the reads of all other accessions matched the Col-0 breakpoints, suggesting that this inversion was either specific to the La-1 accession or was introduced during the mutagenesis leading to the Ler genotype. In contrast, genotyping for the 1.2-Mb inversion revealed 26 accessions as carriers of the Col-0 allele and 383 accessions as carriers of the Ler allele (Fig. 3B). Most accessions could be characterized at the distal inversion breakpoint; however, some accessions showed an additional rearrangement on the proximal breakpoint complicating short read alignments. Despite this complication, none of the accessions showed contradicting genotypes at the two breakpoints.
The relatedness of the accessions was estimated by using 20,408 SNPs from within the inversion and revealed a perfectly separated subclade, which matched the assignment of the Col-0–like inversion allele during the breakpoint genotyping (Fig. 3C and SI Appendix, SI Materials and Methods). This finding suggested that suppressed recombination and genetic exchange separated the population in two distinct groups, in particular because this separation was not mirrored by geographic isolation (Fig. 3D). However, this separation was not absolute: Across the 20,408 sites, we found 13 (0.06%) sites with shared variation between both groups (whereas 20,306 sites were polymorphic only within the Ler-like inversion accessions, and 89 sites were polymorphic only within the Col-0-like accessions). Although this very low level of shared variation still could indicate the existence of rare (double) recombination events between two inversion alleles, such patterns might also be explained by gene conversions (11) or accumulation of false-positive SNP prediction within the set of shared SNPs.
The minor Col-0 inversion allele mostly co-occurred together with the Ler-like allele across its entire distribution range in Central and Northern Europe, and even in recently invaded North America (39). Although the Ler-like genotypes showed genetic diversity within the inversion region, which was not different from the rest of the genome, the diversity among the Col-0–like genotypes was greatly reduced within the inversion (Fig. 3E and SI Appendix, Fig. S6) and extended across the low recombining pericentromere. The drastic reduction in diversity most likely reflects a bottleneck event as a result of the inversion generating the derived Col-0 allele, which was maintained by the suppression of recombination in inversion heterozygotes.
Taken together, these findings suggest that the inversion was introduced by a single event, and, in consequence, population differentiation between the two groups was not uniform across the chromosome. Wright’s fixation index (FST) was elevated in the inversion, and, again, the effect was extended into the pericentromeric region (Fig. 3F and SI Appendix, Fig. S7).
Finally, we assessed patterns of genic selection in the inversion by calculating Ka/Ks to explain the relatively high allele frequency of the inversion allele. One of the genes in the inversions, RLP46, a LRR-receptor domain-containing gene with function in defense response, was among the four genes with the highest Ka/Ks values across the entire genome. However, the numbers of nonsynonymous and synonymous changes were not high enough to prove a significant difference from neutral evolution, preventing a final conclusion as to whether selection of a gene in the inversion helped to retain the inverted allele or whether the inversion simply drifted into the population.
Genic Copy Number Variation Between Two A. thaliana Lines.
To assess the amount of genes that are specific to Ler or Col-0, we first calculated groups of orthologous genes between the two genomes (SI Appendix, SI Materials and Methods). This process revealed initial sets of 212 and 240 single-copy genes that appeared specific to either of the genomes. These gene sets were then further filtered to exclude the possibility that their accession-specific occurrence was due to failures in the assemblies, annotations, or assignment of orthologs (SI Appendix, SI Materials and Methods). This conservative approach led to 40 unique genes specific for Ler and 63 genes specific for Col-0. Using the genome of the close relative A. lyrata (36, 40) as outgroup, we found that the majority of these polymorphic genes (77%) evolved via deletions within the genome in which they are absent, rather than by a spontaneous appearance in the genome in which they are present (Fig. 4A). In fact, the number of genes is presumably underestimated because the A. lyrata assembly might not be complete and might lack some genes, leading to a false categorization of genes.
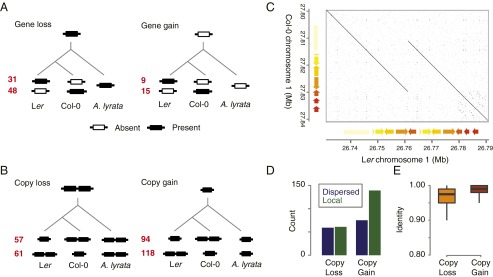
Fig. 4.
Gene absence/presence polymorphisms between Col-0 and Ler. (A) Amount of single-copy, polymorphic genes in Col-0 and Ler. The genes were separated by the presence (Left) or absence (Right) of an ortholog in the related species A. lyrata. (B) Amount of single-copy genes with one additional copy in Col-0 and Ler. Cases were separated by the presence of one or two orthologs in the genome of A. lyrata as in A. (C) Dot plot (57) of an example of a local duplication event coping multiple genes in the genome of Ler. Identically colored arrows indicate similarity between underlying gene loci. (D) Amount of local or dispersed gene copies separately shown for copy loss or gain events as defined in B. (E) Sequence identity between gene copies separately shown for copy loss or gain events as defined in B.
Single-copy gene loss is an extreme type of modification of the gene content of a genome, because no additional copy can replace the function of the absent gene. In contrast, small gene families with multiple copies are expected to be at least partially functionally redundant and, therefore, more variable for copy number. In fact, 330 unique one-to-one orthologous gene pairs showed one additional copy in either the Ler or Col-0 genome (again, after strict filtering for artifacts) (SI Appendix, SI Materials and Methods). In 151 of the cases, the additional gene was found in the genome of Col-0, whereas in 179 of the cases, the additional copy was found in Ler. In contrast to accession-specific genes, copy-number variation did not primarily evolve through deletion events; instead, copy loss events were underrepresented (36%) compared with acquiring new copies by novel duplication (64%) (again, gain and loss events were distinguished by using A. lyrata as an outgroup) (Fig. 4B). These additional genes resided in 134 duplicated regions in Col and 148 duplicated regions in Ler, implying that some of the duplicated genes were introduced by the same duplication event (Fig. 4C). Interestingly, although the loss of additional copies was independent of the location, the gain of additional copies was clearly enriched for copies residing close to the original gene (Fig. 4D), suggesting that novel genes primarily evolved by local duplication events. This result was further supported by differences in the similarity between new copies and the original genes. Gained copies were much more similar to each other compared with copies, where the other genome lost one copy, corroborating that gene gains predominantly result from recent events (Fig. 4E).
Performing a Gene Ontology enrichment analysis, we found that polymorphic single-copy genes are enriched for signaling and signal transduction-related pathways, whereas genes with copy-number changes were enriched for defense-related categories, protein polymerization, and translation.
Polymorphisms Between Diverse Ler Lines.
Earlier comparisons of different Ler lines have revealed a polymorphic premature stop codon in the HUA2 gene (41). This allele, hua2-5, was specific only to some, but not all, Ler lines; could not be identified in any of 29 other A. thaliana accessions (41); and still has not been identified in any of the hundreds of other accessions released by the 1001 Genomes Project (www.1001genomes.org). This result suggested that the hua2-5 mutation occurred after the original La-1 mutagenesis and, because it was not found in some flowering time mutants isolated in the 1960s but was present in mutants isolated in the early 1970s at the Genetics Department in Wageningen, this mutation most likely occurred in that laboratory (41).
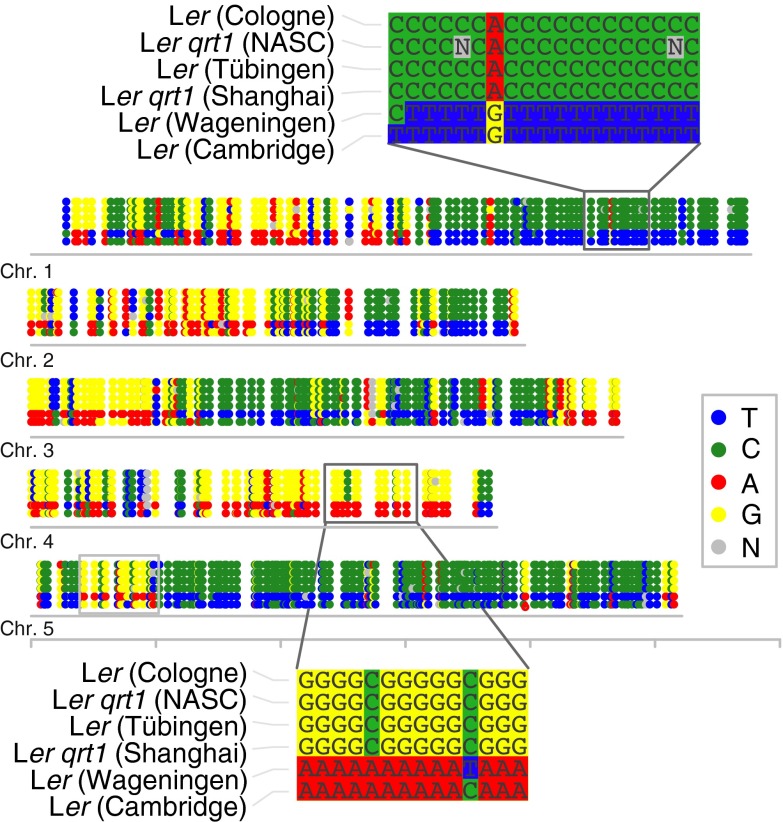
We have reanalyzed six different datasets, all of which have been labeled as whole-genome sequencing data of Ler (8–11) (SI Appendix, Table S2). We aligned the reads of all sets against the Ler reference sequence, and, after stringent filtering, we identified only a marginal amount of structural variations between these lines, including five deletions and two duplication events using read copy and read pair analysis. However, we also identified an unexpected amount of 723 single-nucleotide variations, including the hua2-5 mutation, which was present in two of the six lines. Surprisingly, most of the other polymorphic sites were not specific to a single genome as expected for inbred strains distributed to different laboratories, but were associated to the hua2-5 mutation. Nearly all of these mutations were C→T or G→A mutations and were found in large clusters throughout the entire genome (Fig. 5). Because such mutations and their clustering are characteristic for ethyl methanesulfonate (EMS) mutations, it is likely that the hua2-5 mutation (and all other associated mutations) resulted from an EMS mutagenesis.
Fig. 5.
SNPs between six Ler genomes from different laboratories. Location and type of SNPs distinguishing six genomes published as the genome of Ler. Genome-wide visualization revealed large blocks of C→T and G→A mutations specific to two Ler lines.
Discussion
We have used a combination of short Illumina reads, long PacBio reads, and genetic maps to generate a chromosome-level assembly of A. thaliana Ler. Despite the unprecedented contiguity that we achieved by combining different data types, integration was not straightforward. In contrast, de novo assemblies based only on long reads are easier to perform and start to become a powerful alternative (17, 42), although they do not assemble entire chromosomes yet. Independent of the assembly type, the amount of rearranged sequence that was revealed by comparing two assemblies underlines the general importance of chromosome-level assemblies, in particular because rearrangements have been recognized as confounding factors in genome-wide screens (11, 43) and are essential to fully understand the segregation and evolution of natural haplotypes.
Recent meiotic recombination estimates suggested that high levels of sequence divergence themselves are not inhibitory for meiotic recombination (26, 44, 45), which is in agreement with a positive correlation of ancestral recombination frequency and regions with high sequence divergence (46). However, this finding refers to local sequence divergence without considering rearrangements like transpositions or inversions. In fact, by analyzing CO events, which were observed in Col-0/Ler hybrids, in respect to their occurrence in colinear and rearranged regions, we could show that nonallelic regions have significantly reduced levels of meiotic recombination, despite no obvious difference in local sequence divergence.
Such cross-specific suppression of recombination was described for individual regions before including the 1.2-Mb inversion on the upper arm chromosome 4 (5, 26, 37), which was found between Col-0 and Ler, as well as Ws and Ler. Similar suppression patterns were found in a 2.2-Mb region on the upper arm of chromosome 3 within the offspring of Bay-0 and Sha (47) [and were later also observed in crosses between Col-0 and Sha (45, 48, 49)], a 2- to 3-Mb region on the upper arm of chromosome 5 specific to RRS7 and a 0.2-Mb region on the lower arm of chromosome 1 specific to Bay-0 (49). Recently, local suppression of recombination on the lower arm of chromosome 4 between a cross of Col-0 and Ws-2 led to the identification of a 1.8-Mb inversion, which perfectly matched the region without any CO events (36).
Using a 1.2-Mb inversion on the upper arm of chromosome 4 as an example, we studied the effects of such inversion-mediated suppression of meiotic recombination on the global population of A. thaliana. Suppression of genetic exchange between these two inversion alleles introduced a new genetically isolated haplotype into the global population. Although accessions carrying the Col-0–like allele showed a clear concentration in Germany and Sweden, it was surprising that these genotypes were rather widely distributed. Other examples of widely distributed polymorphisms include deletions in FRIGIDA (50), which could be associated with different selective pressures. We could not identify similar signals for any of the alleles in the derived form of the inversion, which, nevertheless, does not exclude a possible selective advantage of this inversion allele, but might point to a more complex scenario of selection.
In addition to rearrangements, an even larger portion of the genome was not assigned to any orthologous region in the whole-genome alignment. Many of these regions resulted from duplication events, which had a drastic impact on the gene content of the genomes. Gene absence/presence polymorphisms have been connected to phenotypic variation (1) and genetic incompatibilities (51–54) before, but the genome-wide extent of hundreds of polymorphic genes in both genomes was surprisingly high, even though we only looked at low-copy differences. The functional classes, which were enriched among the duplicated genes, included defense-related genes, which can have a selective advantage when duplicated because they can evolve race-specific resistances exemplified by the RPP1 locus where Ler has a 70-kb additional sequence including several strain-specific genes (1, 55).
Even in an organism with so many resequenced genomes as A. thaliana, a single chromosome-level genome assembly enabled us to analyze a second layer of genetic variation, which so far could not be considered in most short-read-based genome-wide studies. These regions showed a drastic impact on natural haplotypes and introduced great variability into the gene space, giving a first glimpse of the degree of natural variation, which can be revealed once more chromosome-level de novo genome assemblies become available.
Materials and Methods
To assemble the genome of the A. thaliana Ler genome, we used ALLPATH-LG (21) and SSPACE-ShortRead (22) for an Illumina short-read contig assembly and scaffolding. Integration of public PacBio sequencing data (16) was performed with PBJelly (20) to close gaps and SSPACE-LongRead (23) to connect scaffolds. Higher-order scaffolding was based on two public genetic maps with 676 and 386 markers with a location on the scaffolds (25, 26). Additional integration of seven scaffolds (combined length of 1.4 Mb) based on synteny assumptions led to the final assembly of five chromosome-representing scaffolds. Finally, PacBio data were used to correct 3.5-Mb ambiguous (N) bases. For gene annotation, we used AUGUSTUS, including alignment from public RNA-seq data.
Supplementary Material
Acknowledgments
We thank Erik Wijnker for help with interpretation of meiotic recombination suppression in non-allelic regions; Petra Tänzler for help with DNA extractions; Avraham Levy and Shay Shilo for sharing their recombination data; and the 1001 Genomes Project for releasing their data, allowing us to search for natural variants of the hua2-5 allele. This work was supported by Spanish Ministry of Economy and Competitiveness Centro de Excelencia Severo Ochoa 2013-2017 Grant SEV-2012-0208. L.Z. was supported by the International PhD scholarship program of La Caixa at CRG.
Footnotes
The authors declare no conflict of interest.
Data deposition: Raw sequencing data have been deposited in the NCBI Sequence Read Archive [accession nos. SRX1567556 (180-bp fragment library), SRX1567557 (8-kb jumping library), SRX1567558 (20-kb jumping library), and SRX1567559 (raw reads with unknown insert size)]. The Whole Genome Shotgun project has been deposited at DNA Data Bank of Japan/European Nucleotide Archive/GenBank (accession no. LUHQ00000000; the version described in this paper is version LUHQ01000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607532113/-/DCSupplemental.
References
- 1.Alcázar R, et al. Analysis of a plant complex resistance gene locus underlying immune-related hybrid incompatibility and its occurrence in nature. PLoS Genet. 2014;10(12):e1004848. doi: 10.1371/journal.pgen.1004848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rédei GP. Single loci heterosis. Z Vererbungsl. 1962;93(1):164–170. [Google Scholar]
- 3.Rédei GP. A heuristic glance at the past of Arabidopsis genetics. In: Koncz C, Chua NH, Schell J, editors. Methods in Arabidopsis Research. World Scientific; Singapore: 1992. pp. 1–15. [Google Scholar]
- 4.Fransz P, et al. Cytogenetics for the model system Arabidopsis thaliana. Plant J. 1998;13(6):867–876. doi: 10.1046/j.1365-313x.1998.00086.x. [DOI] [PubMed] [Google Scholar]
- 5.Fransz PF, et al. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell. 2000;100(3):367–376. doi: 10.1016/s0092-8674(00)80672-8. [DOI] [PubMed] [Google Scholar]
- 6.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 7.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317(5836):338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 8.Schneeberger K, et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci USA. 2011;108(25):10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477(7365):419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu P, et al. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 2012;22(3):508–518. doi: 10.1101/gr.127522.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wijnker E, et al. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife. 2013;2(2):e01426. doi: 10.7554/eLife.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneeberger K, Weigel D. Fast-forward genetics enabled by new sequencing technologies. Trends Plant Sci. 2011;16(5):282–288. doi: 10.1016/j.tplants.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Hollister JD. Genomic variation in Arabidopsis: Tools and insights from next-generation sequencing. Chromosome Res. 2014;22(2):103–115. doi: 10.1007/s10577-014-9420-1. [DOI] [PubMed] [Google Scholar]
- 14.Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18(12):2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43(10):956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 16.Kim KE, et al. Long-read, whole-genome shotgun sequence data for five model organisms. Sci Data. 2014;1:140045. doi: 10.1038/sdata.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berlin K, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 2015;33(6):623–630. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 18.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 19.Koren S, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30(7):693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.English AC, et al. Mind the gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012;7(11):e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 2011;108(4):1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 23.Boetzer M, Pirovano W. SSPACE-LongRead: Scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15(1):211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mascher M, et al. Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ) Plant J. 2013;76(4):718–727. doi: 10.1111/tpj.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singer T, et al. A high-resolution map of Arabidopsis recombinant inbred lines by whole-genome exon array hybridization. PLoS Genet. 2006;2(9):e144. doi: 10.1371/journal.pgen.0020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giraut L, et al. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7(11):e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uchida W, Matsunaga S, Sugiyama R, Kawano S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet Syst. 2002;77(1):63–67. doi: 10.1266/ggs.77.63. [DOI] [PubMed] [Google Scholar]
- 28.Guo Y-L, et al. Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol. 2011;157(2):757–769. doi: 10.1104/pp.111.181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen H, et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell. 2012;24(3):875–892. doi: 10.1105/tpc.111.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long Q, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45(8):884–890. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium The complete sequence of a heterochromatic island from a higher eukaryote. Cell. 2000;100(3):377–386. [PubMed] [Google Scholar]
- 33.Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43(5):476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shilo S, Melamed-Bessudo C, Dorone Y, Barkai N, Levy AA. DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis. Plant Cell. 2015;27(9):2427–2436. doi: 10.1105/tpc.15.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkpatrick M. How and why chromosome inversions evolve. PLoS Biol. 2010;8(9):e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowan BA, Patel V, Weigel D, Schneeberger K. Rapid and inexpensive whole-genome genotyping-by-sequencing for crossover localization and fine-scale genetic mapping. Genes Genomes Genet. 2015;5(3):385–398. doi: 10.1534/g3.114.016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drouaud J, et al. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 2006;16(1):106–114. doi: 10.1101/gr.4319006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz RJ, et al. Patterns of population epigenomic diversity. Nature. 2013;495(7440):193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platt A, et al. The scale of population structure in Arabidopsis thaliana. PLoS Genet. 2010;6(2):e1000843. doi: 10.1371/journal.pgen.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rawat V, et al. Improving the annotation of Arabidopsis lyrata using RNA-Seq data. PLoS One. 2015;10(9):e0137391. doi: 10.1371/journal.pone.0137391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doyle MR, et al. HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J. 2005;41(3):376–385. doi: 10.1111/j.1365-313X.2004.02300.x. [DOI] [PubMed] [Google Scholar]
- 42.VanBuren R, et al. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature. 2015;527(7579):508–511. doi: 10.1038/nature15714. [DOI] [PubMed] [Google Scholar]
- 43.Qi J, Chen Y, Copenhaver GP, Ma H. Detection of genomic variations and DNA polymorphisms and impact on analysis of meiotic recombination and genetic mapping. Proc Natl Acad Sci USA. 2014;111(27):10007–10012. doi: 10.1073/pnas.1321897111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barth S, Melchinger AE, Devezi-Savula B, Lübberstedt T. Influence of genetic background and heterozygosity on meiotic recombination in Arabidopsis thaliana. Genome. 2001;44(6):971–978. doi: 10.1139/g01-094. [DOI] [PubMed] [Google Scholar]
- 45.Ziolkowski PA, et al. Juxtaposition of heterozygosity and homozygosity during meiosis causes reciprocal crossover remodeling via interference. eLife. 2015;4:e03708. doi: 10.7554/eLife.03708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39(9):1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 47.Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 x Shahdara recombinant inbred line population: S powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet. 2002;104(6-7):1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- 48.Simon M, et al. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics. 2008;178(4):2253–2264. doi: 10.1534/genetics.107.083899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salomé PA, et al. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity (Edinb) 2012;108(4):447–455. doi: 10.1038/hdy.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toomajian C, et al. A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 2006;4(5):e137. doi: 10.1371/journal.pbio.0040137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bikard D, et al. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323(5914):623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- 52.Vlad D, Rappaport F, Simon M, Loudet O. Gene transposition causing natural variation for growth in Arabidopsis thaliana. PLoS Genet. 2010;6(5):e1000945. doi: 10.1371/journal.pgen.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith LM, Bomblies K, Weigel D. Complex evolutionary events at a tandem cluster of Arabidopsis thaliana genes resulting in a single-locus genetic incompatibility. PLoS Genet. 2011;7(7):e1002164. doi: 10.1371/journal.pgen.1002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chae E, et al. Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell. 2014;159(6):1341–1351. doi: 10.1016/j.cell.2014.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alcázar R, García AV, Parker JE, Reymond M. Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci USA. 2009;106(1):334–339. doi: 10.1073/pnas.0811734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koornneef M, Fransz P, de Jong H. Cytogenetic tools for Arabidopsis thaliana. Chromosome Res. 2003;11(3):183–194. doi: 10.1023/a:1022827624082. [DOI] [PubMed] [Google Scholar]
- 57.Krumsiek J, Arnold R, Rattei T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23(8):1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.