Significance
RNA viruses encode a unique class of RNA-dependent RNA polymerases (RdRPs) to carry out their fully RNA-based genome replication and transcription. Although the chemical nature of nucleotide addition is essentially shared by all nucleic acid polymerases, the structural and mechanistic details taken by each polymerase class differ to various extents. Here we report seven crystal structures of enterovirus 71 RdRP elongation complex at 2.5–2.8 Å resolution. In these structures the polymerases are poised at various distinct stages to reveal mechanistic details of initial NTP binding, key amino acid side-chain conformational switches during active site closure, and in particular the postcatalysis movement of the RNA duplex on the way to vacate the active site for the next nucleotide addition cycle.
Keywords: RNA-dependent RNA polymerase, nucleotide addition cycle, translocation intermediate, enterovirus 71, crystal structure
Abstract
Viral RNA-dependent RNA polymerases (RdRPs) play essential roles in viral genome replication and transcription. We previously reported several structural states of the poliovirus RdRP nucleotide addition cycle (NAC) that revealed a unique palm domain-based active site closure mechanism and proposed a six-state NAC model including a hypothetical state representing translocation intermediates. Using the RdRP from another human enterovirus, enterovirus 71, here we report seven RdRP elongation complex structures derived from a crystal lattice that allows three NAC events. These structures suggested a key order of events in initial NTP binding and NTP-induced active site closure and revealed a bona fide translocation intermediate featuring asymmetric movement of the template–product duplex. Our work provides essential missing links in understanding NTP recognition and translocation mechanisms in viral RdRPs and emphasizes the uniqueness of the viral RdRPs compared with other processive polymerases.
In recent years, several notable emerging infectious diseases have been caused by RNA viruses, including highly pathogenic avian influenza viruses, Ebola virus, and Middle East respiratory syndrome coronavirus. RNA viruses are quite diverse in virus particle and genome structure and in virus entry and assembly mechanisms. However, they do share fundamental features in their genome replication and transcription, using a virally encoded RNA-dependent RNA polymerase (RdRP) to carry out the biosynthesis of an RNA product directed by an RNA template. Although the genome replication machinery often requires the participation of other factors, typically at the initiation phase of synthesis, the RdRP governs the elongation phase of synthesis that includes thousands of efficient nucleotide addition cycles (NACs). Viral RdRPs vary greatly in size and structural organization, from the ∼50-kDa picornavirus 3Dpol (1, 2), to the ∼100-kDa flavivirus NS5 that contains a naturally fused methyltransferase domain (3), to the ∼250-kDa nonsegmented negative-strand RNA virus L protein harboring at least three enzyme modules (4) and the ∼260-kDa three-subunit PA-PB1-PB2 influenza virus replicase complex (5). On the other hand, all RdRPs share a 50- to 70-kDa polymerase core that forms a unique encircled right-hand structure with palm, fingers, and thumb domains. Among the seven classic RdRP catalytic motifs, A–E are within the most conserved palm domain, and F and G are located in the fingers; they are all arranged similarly around the active site (6–9). The structural conservation of the RdRP polymerase core and the seven motifs form the basis for understanding the common features in viral RdRP catalytic mechanism and for finding intervention strategies targeting these enzymes with possible broad-spectrum potential.
As with other classes of nucleic acid polymerases, the viral RdRP elongation NAC comprises sequential steps of initial NTP binding, active site closure, catalysis, and translocation. In a recent study using CTP and deoxy-CTP analogs in the poliovirus (PV) RdRP elongation complex (EC) crystal-soaking experiments, the polymerase having a guanine base at the +1 template position was successfully trapped at different stages of a single NAC, leading to the proposal of a working NAC model featuring six reference states (10). The model starts with a state 1 (S1) complex with a vacant active site that is in the catalytically open conformation and upon NTP binding proceeds to a state 2 (S2) complex with the active site still in the open conformation. An important conformational change then takes place to position key catalytic residues and two magnesium ions around the priming nucleotide and the substrate NTP to achieve proper geometry of a closed active site for catalysis, yielding state 3 (S3) immediately before and state 4 (S4) immediately after the phosphoryl transfer reaction. As the catalytic geometry starts to disintegrate, the structural changes in the palm domain result in state 5 (S5) with an open conformation active site. State 6 (S6) is then a hypothetical translocation intermediate state that bridges the pretranslocation S5 and the posttranslocation S1 in the next NAC.
The six-state model has provided a framework for understanding the molecular details and the unique features of the viral RdRP elongation NAC. In the active site closure step, the notable backbone conformational changes are limited to motifs A and D in the palm domain. These conformational changes are in drastic contrast to those taken by the well-characterized A-family polymerases that use a large-scale rotational movement of the O-helix–containing fingers domain to achieve the same closure step (11, 12). One implication behind this apparent difference in the active site closure mode is that the mechanisms by which the polymerase selects the correct NTP substrate and induces active site reorganization for catalysis are also quite different. In A-family polymerases, the translocation step is coupled to the postcatalysis reopening of the active site when a conserved tyrosine residue in the O-helix “pushes” the nascent base pair upstream in a motion that is the reverse of that observed during the active site closure (11, 13). However, no intermediate structure between the pre- and posttranslocation states to refine the translocation process further has been captured in A-family polymerases by crystallography. Without an O-helix counterpart, viral RdRPs likely have established unique conserved components to control translocation, and their ECs may experience metastable intermediates that provide valuable details for identifying translocation-related protein components and the details of RNA movement during translocation.
In this study, we obtained a crystal form of the enterovirus 71 (EV71) RdRP EC that allows multiple nucleotide incorporations in NTP-soaking trials. By using natural NTP substrate combinations and controlling the incubation time, we were able to capture EC species that provide previously unidentified mechanistic details for initial NTP binding, active site closure, and, in particular the RNA motion during translocation that shows an asymmetric movement of the two strands in the template–product duplex.
Results
A Unique RdRP EC Lattice That Is Capable of in Situ Elongation for Multiple NACs.
Recently we developed an RNA-mediated crystallization strategy that was highly effective in crystallizing picornaviral RdRP ECs (14). By providing a GC or GU sequence overhanging the upstream end of the template–product duplex to facilitate inter-EC contacts, an EC dimer typically becomes the minimal crystallizing unit, with the two upstream RNA duplexes interacting in the middle and two polymerases facing away from each other. These RNA–RNA interactions play important roles in crystallization but also impose steric constraints for in-crystal soaking experiments designed to go through multiple NACs. By attempting to crystallize EV71 RdRP EC using a combination of RdRP from different viral genotypes and RNA with different lengths of template–product duplex, we have obtained a picornaviral RdRP EC crystal form within which ECs are no longer organized as dimers. Instead, the upstream duplex points toward a spacious solvent channel (Fig. 1A) that may allow multiple incorporation and translocation events to occur in NTP-soaking experiments.
Fig. 1.
A unique EV71 RdRP EC crystal lattice allows multiple nucleotide-incorporation events. (A) 2Fo-Fc electron density map (contoured at 1.5 σ with a radius of 30 Å, in cyan) around the upstream end of one EC (polymerase in green, RNA strands in red and blue) shows a spacious channel to accommodate the growth of RNA duplex. The black dot indicates the center of the map, and symmetry-related neighboring ECs are labeled as Sym1/2/3. (B) RNA sequence flanking the active site of the native EC (C1S1) and those of the other six complexes derived from the native EC in crystal-soaking trials. The template is in cyan, and product is in green. The black box indicates the nucleotides incorporated during soaking. “C” and “S” in complex names stand for “cycle” and “state,” respectively, and the subscript numbers reflect the assigned cycle/state numbers. (C) The isomorphous nature of the EC lattices. C1S1/2 and C3S6 structures (bold-faced in B) were chosen as representatives to indicate the very limited lattice alteration upon three rounds of nucleotide incorporation. The superimposed polymerases (using the traditional least-squares method) are on the right side with the C1S1/2 complex taking a coloring scheme indicated by individual parts of the EC and the C3S6 complex in black; their symmetry-related neighboring ECs are colored in orange (C1S1/2) and purple (C3S6).
The EC in this crystal was obtained after incorporating a (GA)3 hexa-nucleotide sequence into an RdRP–RNA binary complex containing an 8-bp template–primer duplex. The remaining “GGACCU …” template sequence was designed to direct subsequent in-crystal elongation, as may be allowed by this particular lattice (Fig. 1B). When CTP, UTP, and GTP are provided, the EC is expected to incorporate five nucleotides (i.e., CCUGG). However, even with overnight incubations, the EC incorporated three nucleotides (i.e., CCU) at most, indicating a threshold for the crystal lattice to accommodate growth of the template–product duplex. We then explored using this crystal lattice as a platform to capturing important EC states within three consecutive NACs. By combining an incubation time scanning strategy (15, 16) and the use of different NTP combinations in the native EC-soaking trials, we obtained seven representative EC structures distributed in the first and the third NACs (Fig. 1B and Tables 1 and 2). The resulting crystal lattices were nearly isomorphous based on unit cell dimensions and intercomplex packing modes (Tables 1 and 2 and Fig. 1C). This observation is drastically different from the output of the soaking experiments using a PV RdRP EC crystal that also allows multiple nucleotide incorporation (14); in that lattice (named “PV_r5”) every translocation event resulted in obvious translational movement between RdRPs within the aforementioned EC dimer as the growing RNA duplexes collided with each other.
Table 1.
X-ray diffraction data collection and structure refinement statistics (set 1)
| NAC state* (PDB ID code) | ||||
| C1S1 (5F8G) | C1S1/2 (5F8H) | C1S2/3 (5F8I) | C1S4 (5F8J) | |
| Data collection† | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a, b, c, Å | 63.1, 77.6, 153.6 | 63.3, 77.1, 149.9 | 63.7, 77.3, 149.7 | 63.4, 76.6, 149.3 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution, Ň | 60.0–2.78 (2.88–2.78) | 60.0–2.45 (2.54–2.45) | 60.0–2.50 (2.59–2.50) | 60.0–2.66 (2.76–2.66) |
| Rmerge | 0.117 (0.50) | 0.062 (0.51) | 0.060 (0.48) | 0.073 (0.50) |
| I/σI | 13.6 (3.2) | 21.4 (3.3) | 20.1 (2.9) | 17.7 (2.5) |
| Completeness, % | 97.6 (98.4) | 99.7 (99.9) | 99.6 (100.0) | 98.1 (96.4) |
| Redundancy | 5.4 (5.5) | 4.7 (4.8) | 4.3 (4.3) | 4.9 (4.9) |
| Refinement | ||||
| Resolution, Å | 2.78 | 2.45 | 2.50 | 2.66 |
| No. reflections | 19,258 | 27,772 | 26,095 | 20,430 |
| Rwork/Rfree§, % | 19.3/24.2 | 19.8/24.4 | 19.9/24.4 | 18.3/22.8 |
| No. atoms | ||||
| Protein/RNA | 3,681/471 | 3,681/471 | 3,681/471 | 3,685/491 |
| Ligand/ion/water | –/ 1/115 | 5/2/141 | 40/3/144 | 25/3/111 |
| B-factors | ||||
| Protein/RNA | 42.6/53.2 | 52.3/60.8 | 57.5/72.2 | 50.4/62.8 |
| Ligand/ion/water | –/18.6/38.9 | 36.3/44.4/49.8 | 53.0/53.2/55.5 | 43.6/39.1/48.1 |
| RMSD | ||||
| Bond lengths, Å | 0.008 | 0.008 | 0.008 | 0.008 |
| Bond angles, ° | 1.13 | 1.13 | 1.12 | 1.17 |
| Ramachandran statistics¶ | 91.2/8.3/0.2/0.2 | 93.1/6.6/0.0/0.2 | 92.9/6.9/0.0/0.2 | 92.6/6.9/0.2/0.2 |
Coding scheme: C, cycle; S, state; subscript numbers reflect the cycle and state numbers: x/(x+1) indicates an NAC species between state x and the next reference state. Soaking strategy: C1S1/2: CTP for 4 min; C1S2/3 – CTP for 4 min, and then transfer to UTP for 10 min; C1S4: CTP for 5 min 10 s.
One crystal was used for data collection for each structure.
Values in parentheses are for highest-resolution shell.
5% of data are taken for the Rfree set, and the same Rfree set is applied for all structures.
Values are in percentage and are for most favored, additionally allowed, generously allowed, and disallowed regions in Ramachandran plots, respectively.
Table 2.
X-ray diffraction data collection and structure refinement statistics (set 2)
| NAC state* (PDB ID code) | |||
| C3S1 (5F8L) | C3S4/5 (5F8M) | C3S6 (5F8N) | |
| Data collection† | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c, Å | 62.3, 76.7, 151.2 | 63.6, 76.7, 150.1 | 63.7, 77.6, 151.4 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution, Ň | 60.0–2.81 (2.91–2.81) | 60.0–2.83 (2.93–2.83) | 60.0–2.47 (2.56–2.47) |
| Rmerge | 0.102 (0.50) | 0.065 (0.50) | 0.047 (0.51) |
| I/σI | 18.6 (4.0) | 18.0 (2.6) | 23.1 (2.6) |
| Completeness, % | 99.9 (100.0) | 95.8 (96.1) | 95.1 (94.8) |
| Redundancy | 6.4 (6.6) | 3.7 (3.8) | 4.1 (3.9) |
| Refinement | |||
| Resolution, Å | 2.81 | 2.83 | 2.47 |
| No. reflections | 18,195 | 17,637 | 25,817 |
| Rwork/Rfree§, % | 20.0/23.8 | 19.3/23.2 | 19.4/23.5 |
| No. atoms | |||
| Protein/RNA | 3,677/468 | 3,681/465 | 3,677/360 |
| Ligand/ion/water | –/1/96 | 20/3/78 | 20/1/138 |
| B-factors | |||
| Protein/RNA | 50.4/73.5 | 61.6/79.4 | 60.2/70.4 |
| Ligand/ion/water | –/46.9/45.2 | 56.3/49.2/49.8 | 54.5/42.2/58.5 |
| RMSD | |||
| Bond lengths, Å | 0.009 | 0.009 | 0.008 |
| Bond angles, ° | 1.16 | 1.20 | 1.13 |
| Ramachandran statistics¶ | 91.4/8.3/0.0/0.2 | 91.7/7.8/0.2/0.2 | 91.7/7.8/0.2/0.2 |
Coding scheme is the same as in Table 1. Soaking strategy: C3S1: CTP for 16 h; C3S4/5: CTP for 5 min, and then transfer to CTP/UTP/GTP for 20 min; C3S6: CTP/UTP for 16 h.
One crystal was used for data collection for each structure.
Values in parentheses are for highest-resolution shell.
5% of data are taken for the Rfree set, and the same Rfree set is applied for all structures.
Values are in percentage and are for most favored, additionally allowed, generously allowed, and disallowed regions in Ramachandran plots, respectively.
A Plausible Order of Events During Initial NTP Binding and Active Site Closure.
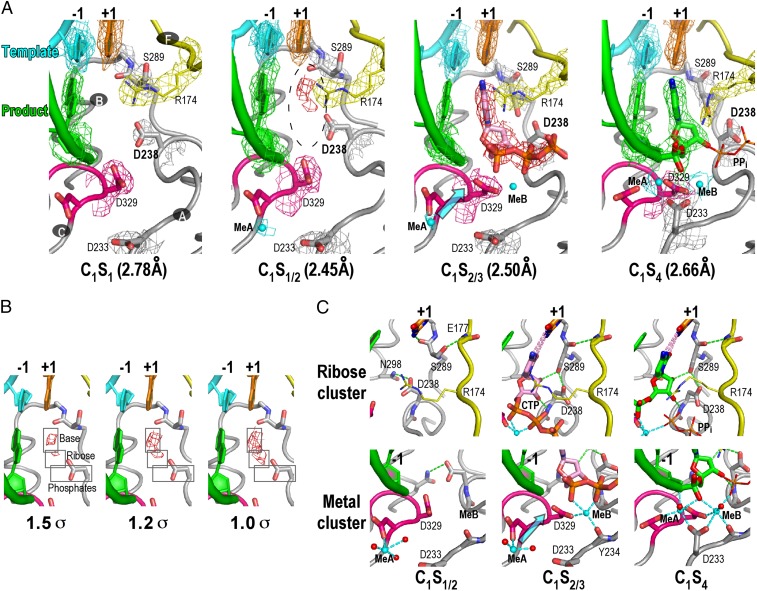
The isomorphous feature of the EV71 RdRP EC lattice and the fact that the EC can enter the third NAC mean the three consecutive NACs occurred in an environment free of constraints brought by lattice variation, providing a valid platform for time-resolved NTP-soaking experiments. We tested CTP soaking with various incubation times and obtained three representative states in the first NAC (denoted “C1” for “cycle 1”; Table 1). In the prior PV RdRP EC work, a 2′,3′-dideoxy CTP (ddCTP)-derived structure showed clear density for the entire ddCTP molecule, whereas the RdRP conformation remained essentially unchanged around the active site. This structure was assigned as the reference state 2 (S2) to represent a fully open conformation active site with a bound NTP. However, such a state was not observed in all of the CTP-soaked EV71 RdRP EC C1 structures. If the active site conformation remained fully open, only medium-level density for the CTP base moiety and weak-level density of the ribose was evident (C1S1/2 structure, CTP not modeled; Fig. 2 A and B). This observation indicates that the template–NTP substrate base-pairing appears to be sampled before the rearrangement of NTP ribose, triphosphate, and the surrounding active site motifs toward the in-line catalytic geometry. Defined density next to residue D330 (the second aspartate in the RdRP hallmark motif C sequence XGDD) with an interacting distance of 2.1 Å to a side-chain oxygen indicated a bound magnesium ion (“metal A” or MeA according to consensus nomenclature) that is required for subsequent catalysis.
Fig. 2.
CTP-derived sequential EC structures demonstrate a plausible order of precatalysis NAC events. (A) Four-cycle 1 (C1) NAC structures arranged in a sequential order with composite simulated-annealing (SA) omit electron density maps contoured at 1.2 σ. C1S1 represents the starting state 1 with a vacant active site; C1S1/2 shows evidence of initial NTP binding via base-paring; C1S2/3 shows the initial conformational change around D238 that is triggered by ribose hydroxyls during active site closure; C1S4 represents the bona fide catalytically closed state 4 with 11 of 12 Mg2+ ion contacts intact. Coloring scheme: template in cyan (+1 nucleotide in orange), product in green, palm in gray (YGDD sequence in magenta), ring finger in yellow, and metal ions in cyan. (B) Electron density around the NTP site in the C1S1/2 structure indicates that base-paring interactions and placement of the NTP ribose likely form before the correct alignment of triphosphate moiety. (C) The change in the interaction network around NTP ribose (ribose cluster) and catalytic metal sites (metal cluster) demonstrates the order of events upon NTP binding and active site closure. Red spheres indicate water molecules coordinating with Mg2+.
If clear triphosphate density was visible, then key conformational changes around the CTP ribose 2′ and 3′ hydroxyl groups had already occurred, but the active site had not yet fully closed, leading to an intermediate state we call “S2/3” (C1S2/3 structure; Fig. 2A). The conserved motif A residue D238 experienced a hallmark side-chain rotamer change to accommodate the ribose hydroxyls and established a hydrogen-bonding network with the ribose and motif B residue S289 (Fig. 2C). As in the C1S1/2 structure, metal A still resided several Ångstroms away from its catalytic position, whereas metal B was observed within coordination-distance range of the CTP phosphates and the motif A residue D329. D329 (the first aspartate in sequence XGDD) is one of two universally conserved aspartic acid residues in polymerases following the two-metal-ion catalytic mechanism (17). However, this structure is not in the fully closed catalytically competent state because the other universal aspartic acid residue located at position 233 in motif A is not in place. As characterized in the PV RdRP EC study, D233 features the most notable backbone movement during active site closure to achieve coordination with both metal ions. The capture of EV71 RdRP in a partially closed state, S2/3, provides further evidence that the likely order of events during active site closure is as originally suggested by the PV study (10). In such a proposal, the precise placement of ribose 2′ and 3′ hydroxyls triggers the reorganization around the ribose, including residue D238, which in turn induces the movement of the D233 region within the same motif for metal ion coordination and catalysis.
A fully closed postcatalysis state in the EV71 structure was obtained also (C1S4 structure; Fig. 2A) in which the details of the active site were essentially identical to the S4 structures seen in the PV study. Relative to their location in the C1S2/3 structure, the D233 side-chain carboxylate group moved about 3.8 Å and rotated ∼120°, and metal A moved 5.5 Å to be coordinated simultaneously by D233, D329, and D330 in the catalytically competent conformation (Fig. 2C).
Postcatalysis Complexes in Two NACs Provide the NTP Selection Details by +1 Template Purine Bases.
Many DNA-dependent polymerase ECs contain a preinsertion site (the “E-site” where “E” stands for “entry”) in which the +1 template nucleotide is poised to allow initial NTP binding (13, 18–20). This site allows the nascent base pair to form without establishing the stacking interactions with the −1 base pair. To achieve active site closure, an NTP repositioning step needs to take place to move the NTP into the insertion site (the “A-site” where “A” stands for “addition”), and this repositioning usually is accompanied by relatively large conformational changes in the vicinity of the active site (11–13, 18). These features in general permit the NTP selection process to occur at two distinct sites that have different sets of interactions, possibly improving nucleotide selection fidelity. In contrast, in viral RdRP ECs the +1 template nucleotide is prestacked on the −1 base pair, and as a result the initial NTP binding is to a site nearly identical to the catalytic insertion site. Active site closure involves only limited backbone shifts in motifs A and D with several hallmark side-chain rotamer changes as mentioned above. The NTP selection by the RdRP EC therefore is structurally less complicated, because the initial NTP binding and subsequent catalysis occurs in very similar protein environments. In addition to the C1S1 native structure and C1S4 structure that show the details of CTP selection with a guanine base at the template +1 position (denoted “+1G:C”), we obtained C3S1 and C3S4/5 structures showing UTP selection with a +1 adenine base (+1A:U) (Fig. 3). When comparing the NTP-free S1 structures to the NMP-incorporated S4 and S4/5 structures by a maximum likelihood superimposition of the polymerase molecules (21), the placement of the +1 template base was subjected only to very subtle movement toward the major groove side (Fig. 3A). The polymerase active site conformation is essentially identical in the two product structures, with key residues D238, D233, S289, G290, and R174 following the same conformational switches for both NTPs. These residues, together with motif F residue K159 sitting on the major groove side and residue I176 that stacks onto the template base from the downstream side, define a compact substrate pocket for the final precatalysis fidelity checkpoint. The 2′ and 3′ hydroxyls of the nascent NMP ribose were precisely placed toward D238, with the positional deviation of the both hydroxyl oxygen atoms in C1S4 and C3S4/5 complexes being within 0.2 Å. This observation suggests that the establishment of the interaction network around the NTP ribose hydroxyls is a key determinant for active site closure and likely precedes and triggers the structural rearrangements around the catalytic metal ions.
Fig. 3.
Watson–Crick base-pair geometry as a major determinant of NTP selection. (A) Interaction details for NTP selection directed by +1 guanine (C1S1 and C1S4) and adenine (C3S1 and C3S4/5). All polymerase molecules were superimposed, and the structures are shown as individual panels with the S1 complexes on the left and S4/S4/5 complexes on the right. The coloring scheme is the same for each complex and is the same as in Fig. 2 with the product RNA in green, template RNA in cyan, and the +1 template nucleotide in orange. The ribose C4′ atoms of the +1 nucleotides are shown as spheres to aid the structural comparison with the alignment grids. Key interactions around the ribose cluster upon active site closure, and base-pairing hydrogen-bonding interactions at the +1 position are indicated by green and pink dashed lines, respectively. K159- and R174-related interactions are shown in purple. (B) A comparison of the metal cluster of the two product complexes. Mg2+–oxygen distances greater than 2.5 Å are shown in green text to highlight the disintegration of the ion-coordination interactions in the postcatalysis S4/5 complex. Red spheres indicate water molecules coordinating with Mg2+.
The two postcatalysis structures reported here are nearly identical in polymerase active site conformation. However, only the C1S4 structure contains all 12 coordination partners for the two metal ions, with the 11 of these found within Mg2+ coordination distances (Fig. 3B). Therefore, we consider C1S4 a bona fide postcatalysis S4 structure. In contrast, the coordination geometry has begun to disintegrate in the C3S4/5 structure, and it therefore should be considered an intermediate between S4 and S5, even if the protein active site conformation remains closed. Note that two water molecules participate in Mg2+ coordination in the C1S4 structure. The water molecule coordinating metal A is about 3.5 Å away from the 3′ hydroxyl oxygen. In the norovirus RdRP-RNA-CTP crystal structure, a structurally equivalent water molecule coordinates the metal A (a Mn2+ in that case) and was suggested to serve as the general base in the proposed two-proton transfer mechanism (22, 23). The water molecule coordinating metal B in the C1S4 structure is about 3.0–3.1 Å away from two phosphate oxygen atoms of the pyrophosphate, and a structurally equivalent water molecule has not been observed in RdRP structures. Along with the previously proposed motif D lysine (K359 in PV RdRP) (24) and motif F arginine (R174 in PV RdRP) (10) residues, this water molecule also may be a candidate to protonate the β-phosphate of the substrate NTP as a general acid.
A Previously Unidentified Translocation Intermediate Suggests an Asymmetric Movement of Template–Product RNA Duplex.
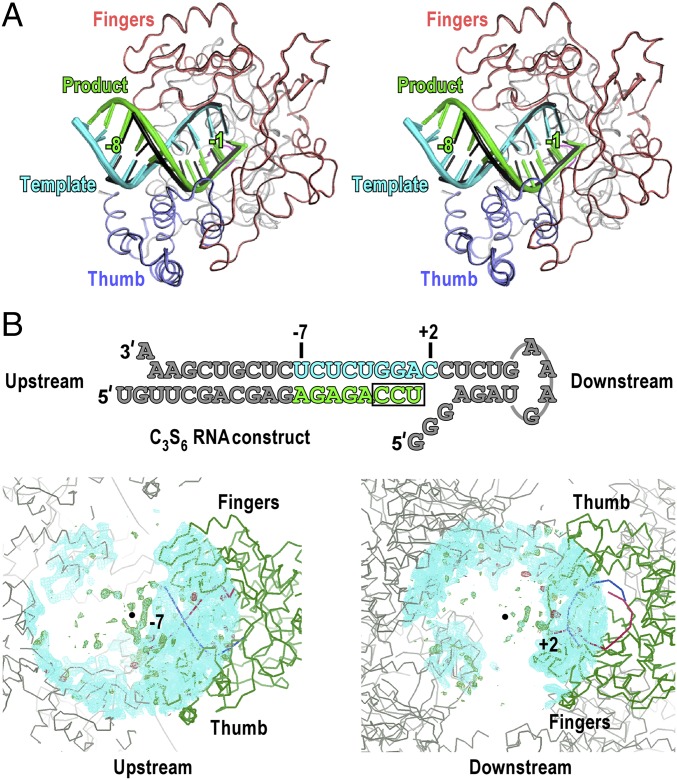
Translocation has the net effect of changing the footprint of the polymerase on nucleic acid by one nucleotide register and represents an important postcatalysis event that completes the NAC. Local, if not major, conformational changes must take place to achieve translocation, and higher-energy intermediates may exist between the pre- and posttranslocation states. However, such translocation intermediates often are difficult to capture by experimental biology because of their short-lived nature. Compared with in-solution approaches, in-crystal polymerase synthesis may increase the possibility of capturing such intermediates by providing extra constraints through the packing environment of each polymerase. Indeed, the time scale for a single NAC in the current lattice is on the order of minutes (Tables 1 and 2), at least 100-fold slower than that measured in the solution studies in PV RdRP (25, 26). Ideally, the EV71 RdRP EC lattice reported here could allow the capture of such intermediates in all three NACs. To date, we have not successfully obtained any translocation intermediate within the first or the second NAC, even after numerous rounds of attempts. However, with overnight incubation in the presence of CTP and UTP, the EC could be paused at a clear translocation-intermediate state in which all regions of the template–product RNA duplex except for the backbone region from position −3 to position +1 of the template strand had undergone movement in the upstream direction (Fig. 4A, C3S6). To the best of our knowledge, this is the first reported crystallography-derived polymerase EC intermediate that demonstrates the global motion of the template–product duplex during translocation, and it is conceptually different from the crystal structure of yeast RNA polymerase II (Pol II) EC in complex with α-amanitin, in which only the intermediate conformation of the downstream DNA is observed (27). Note that the intermediate RNA conformation in our C3S6 structure mostly likely represents a naturally occurring state rather than an artifact induced by crystal-packing constraints. In the previous work describing the method used in crystallizing picornaviral polymerase EC, we demonstrated that the polymerase with consistent global conformation maintains the native RNA conformation from the active site to about position −7, despite various extents of RNA conformational distortion occurring further upstream (14). The polymerase global conformation of the C3S6 complex is highly consistent with the other six complexes presented in this study (Fig. 4A). The RNA regions upstream of position −7 and downstream of position +2 are largely disordered in the C3S6 structure (Fig. 4B). Although a portion of RNA backbone electron density of the disordered upstream RNA is somewhat traceable, defined intercomplex contacts are lacking at both ends of the RNA construct. Taken together, the consistent polymerase conformation, the absence of defined crystal contacts at both ends of the RNA constructs, and the ability of polymerase to absorb the conformational distortion induced at the upstream end of its RNA collectively support the validity of the C3S6 structure.
Fig. 4.
The C3S6 complex is a translocation intermediate, has a global polymerase conformation consistent with other complexes, and is free of defined intercomplex interactions at either end of its RNA constructs. (A) Stereo-pair images of all seven complexes with their polymerases superimposed. The C3S6 complex is shown in black, and the other six complexes are colored as the C1S1/2 complex in Fig. 1C. The +2 template nucleotides in all complexes are omitted for clarity. The −1 and −8 position labels correspond to the regular translocational positions defined by the six nontranslocation-intermediate complexes. (B, Upper) The complete sequence of the RNA constructs in the C3S6 complex is shown with gray font indicating unresolved nucleotides in the structure. (Lower Left) A portion of the unresolved upstream RNA backbone in the C3S6 structure is somewhat traceable. (Lower Right) The downstream RNA is essentially disordered. Electron densities for defined crystal contacts are lacking at both ends of the RNA construct. Cyan mesh indicates the 2Fo-Fc electron density map contoured at 1 σ; green/red mesh indicates the Fo-Fc electron density map contoured at 3 σ. Green ribbons indicate polymerase; red/blue ribbons indicate template/product RNA. Black dots indicate the center of the map. The map radius is 30 Å.
In comparison with the C3S4/5 pretranslocation structure within the same NAC, in the C3S6 structure the entire product strand moved in the upstream direction (Fig. 5). With all base-paring hydrogen bonds maintained between the two strands, the product strand phosphates moved about a half-register on average, with some variation (Fig. 5 B and C). The product riboses and bases also moved in the same direction but with larger variation. The upstream-most nucleotide resolved in the structure is the −7 position, whose base almost reached the −8 position of the pretranslocation structure, but the downstream-most +1 base has moved by only about one fifth of a register (Fig. 5 B and C). The template strand lagged behind, with its backbone phosphates at positions −2 to +1 remaining locked at their pretranslocational positions, whereas the −7 to −4 phosphates moved upstream by less than a half-register on average (Fig. 5). Globally assessed, the template–product RNA duplex had undergone an asymmetric movement with the product strand preceding the template strand and the upstream region leading the downstream region.
Fig. 5.
A previously unidentified translocation intermediate supports the asymmetric movement of the template–product duplex during translocation. (A) Stereo-pair images of the translocation intermediate complex C3S6 with a composite SA omit electron density map (contoured at 1.2 σ) of RNA, pyrophosphate (PPi), and residue D238 overlaid. The coloring scheme is as in Fig. 3A. The C3S4/5 pretranslocation (dark gray) and C3S1 posttranslocation (brown) complexes are shown with polymerases superimposed on the intermediate complex for comparisons. (B) RNA-only comparison of the translocation intermediate (Left: template in cyan; Right: product in green) and the pretranslocation complex (dark gray) in the same NAC. (C) Schematic illustration of the different protein–RNA interactions in the pretranslocation C3S4/5, intermediate C3S6, and posttranslocation C3S1 complexes. RNA movement of the C3S6 structure was estimated using the base, ribose, and phosphate components of the pre- and posttranslocation complexes as references. The zigzagged red symbol indicates the irregular backbone conformation of the template −2 position. Phosphate, ribose, and base are shown as circles, pentagons, and blocks, respectively. Solid arrows indicate hydrogen bonding, electrostatic, or hydrophobic interactions. Gray fonts indicate weaker interactions (judged by distance) compared with interactions involving the same residue in other structures. Underlining indicates a change of interaction partner(s) for a polymerase residue when switching from the posttranslocation complex to the intermediate complex. Strikethroughs indicate nonexistent interactions.
With the aim of understanding this apparent strand asymmetry in viral RdRP translocation, we compared the details of the protein–RNA interactions from the active site to position −7 in all three C3 structures (Fig. 5C). Except for the interactions with the newly incorporated UMP in the C3S4/5 structure, the interactions are largely the same in the pre- and posttranslocation states. Interactions between the product strand and the polymerase are fairly evenly distributed from −7 to −1 without being concentrated in any one region. In contrast, polymerase interactions with the template strand are clustered in two distinct regions around the −5 to −3 and −2 to +1 nucleotides, with the latter having the most extensive contacts. Motifs B, F, and G converge at the −2 to +1 region and create a sharp turn in the template strand (Fig. 5A) that also is observed in DNA-dependent RNA polymerases (28, 29). As a result, the +2 template base is fully unstacked from the upstream nucleotides and is tucked into a small surface pocket created by the index and ring fingers (Fig. 5A) (14). Motif G residues T114 and S115 pack against the template strand +1/+2 backbone linkage and likely serve as a control point for template strand movement during translocation (9). At the upstream end of this interaction cluster there is an unusual and conserved backbone conformation for the ribose–phosphate linkage of the template strand −2 nucleotide (14). Together, these two interactions appear to lock the −2 to +1 sequence in place, specifically preventing this region of the template from moving while the remainder of the product–template RNA duplex undergoes translocation in an asymmetric fashion. To achieve the final posttranslocation state, i.e., S1 of the next NAC, the special backbone conformations at the S6 template +1/+2 linkage and the −2 position need to transition to the corresponding regions one base step downstream, and the energy to achieve the requisite conformational changes is likely an important contributor to the ultimate energy barrier for translocation.
We next compared the pretranslocation C3S4/5 structure with the translocation-intermediate C3S6 structure. Although the interactions between the template strand and the polymerase remained largely unchanged, more than half of the residues participating in product strand contacts changed their interaction modes by interacting with a different site, losing original interactions or establishing additional interactions (Fig. 5C). This finding is consistent with the observation that the interactions between the polymerase and the product RNA are relatively nonintensive and evenly distributed. We therefore propose that the product RNA strand may be capable of sliding back and forth between its pre- and posttranslocation states. In contrast, the movement of the template RNA is controlled more stringently by the polymerase, in particular around the −2 to +2 region. The backbone conformational changes required to translocate the +1/+2 junction and the −2 ribose–phosphate linkage are likely accompanied or assisted by local polymerase conformational changes that also require energy, making the final step in translocation rate-limiting and largely irreversible (Fig. 5C).
Discussion
An Improved View of Viral RdRP Elongation NAC.
Using the advantages provided by a particular EV71 RdRP EC lattice, the current study makes important advances toward the understanding of the viral RdRP elongation NAC (Fig. 6). The current NAC working model starts with an S1 complex in the absence of NTP substrate with an open active site; after diffusing into the NTP entry channel formed between the motif F in the ring finger and motifs A and D in the palm, the NTP establishes its initial interaction through base-paring with the +1 template base and stacking with the −1 priming base (S1/2), leading to the rearrangement around the NTP ribose and motif A residue D238 and finally bringing the NTP triphosphate, two divalent metal ions, and another motif A residue, D233, into place to achieve the catalytically closed conformation (S3) as observed in a norovirus polymerase–RNA–CTP complex structure (23). Immediately after the phosphoryl transfer reaction (S4), the catalytic geometry around the metal ions starts to disintegrate, and the active site reopens (S5); translocation begins with the less-restrained product RNA and the upstream region of the template–product duplex (S6) and then finally needs to overcome the conformational transition around the −2 to +2 region of the template strand to become the posttranslocation S1 complex of the next NAC (Fig. 6A).
Fig. 6.
Working models for viral RdRP elongation NAC and translocation. (A) The NAC model shown in a circle format. Previously reported native PV RdRP EC and three of its derivative structures obtained by CTP, 3′-deoxy-CTP (3dCTP), or ddCTP soaking serve as the reference states 1, 2, 4, and 5 (gray fonts; PDB ID codes are listed). All seven structures reported in the current study obtained by natural NTP soaking were assigned at corresponding positions in the cycle. A norovirus (NV) polymerase–RNA–CTP complex exhibiting a precatalysis closed-conformation active site represents reference state 3. (B) A schematic free energy diagram for translocation. The pretranslocation state could establish fast equilibrium with the S6 intermediate state, and the subsequent transition to the posttranslocation state 1 of the next NAC is rate-limiting. Empty triangles indicate the interactions needed to maintain the irregular backbone conformation of the template −2 position and the +1/+2 bend of the template. These interactions include those between the motif G T114–S115 backbone and the +1/+2 junction of the template strand backbone and those between pinky finger K127 and R188 side chain and the template strand backbone phosphates stabilizing the irregular conformation of the ribose–phosphate linkage at the −2 position. These interactions must be broken (indicated by the unlocked symbol) during the final step toward the posttranslocation state. P, product; T, template; T1/T2, transition states.
On NTP Selection and Fidelity Control by Viral RdRP.
Viral RdRPs typically have misincorporation rates in the range of 10−4–10−5 (30–33) and therefore ought to be considered as medium-fidelity polymerases compared with high- and low-fidelity representatives (26, 34, 35). Therefore, the NTP selection by viral RdRP, in particular for its EC, is expected to be reasonably stringent. Because a preinsertion site is missing in viral RdRP ECs, the NTP selection occurs in a limited space in an induced-fit manner with only local conformational changes. It has been proposed that the precise recognition of the equivalent geometry of the Watson–Crick base pairs may be the most important factor in NTP selection by polymerases (36). Although we have not obtained structural data for all four RNA base pairs at the RdRP +1 position, the +1G:C and +1A:U product complexes (C1S4 and C3S4/5) strongly support this proposal, because the spatial placement of the +1 nucleotides, the edges of the +1 base pair, and the shape of the active site complementing the +1 nucleotides are highly analogous (Fig. 3A). As suggested by the C1 structures in the current study, the three sequential events of initial base-pairing, ribose hydroxyl-induced conformational change around D238 (hereafter the “ribose–Asp switch”), and the alignment of NTP triphosphate and D233 around the two metal ions (hereafter the “metal–Asp switch”) likely provide the fidelity check points. The precise placement of the nascent NMP ribose hydroxyl groups in the two product complexes emphasizes the importance of the ribose–Asp switch in substrate selection. We propose that the RdRP specific ribose–Asp switch is a positive contributor to the overall fidelity for viral RdRPs and to some extent may compensate for the loss of one fidelity checkpoint resulting from the lack of a preinsertion site in these polymerases.
On the Translocation Mechanism by Polymerases.
Two major theories have been proposed to describe the central mechanism of polymerase translocation, namely the Brownian ratchet model and the power stroke model (11, 37–39). The Brownian ratchet model features a fast equilibrium between the pre- and posttranslocation states, and it allows possible intermediates in the absence of the next incoming NTP, but the presence of a bound NTP strongly stabilizes the polymerase in the posttranslocation state. The power stroke model emphasizes the correlation between pyrophosphate release and the required conformational changes to convert the pretranslocation state to the posttranslocation state in a single and largely irreversible step. Although we are not trying to reconcile these two models here, our structures suggest that the RdRP translocation process uses essential features of both. As suggested by the C3S6 structure, the postcatalysis product strand is subject to Brownian motion (fast equilibrium) between the pre- and posttranslocation states because of the evenly distributed interactions with the polymerase (Figs. 5 and 6B). In contrast, the movement of the template strand −2 to +2 region toward the upstream direction requires overcoming the sharp backbone turn at +1/+2 and the special conformation of the −2 phosphate linkage. This movement is likely the rate-limiting step during translocation and therefore is largely irreversible. Therefore, the newly established protein interactions at the −2 to +2 region in the posttranslocation state could be regarded as the ratchet to prevent back-translocation (Fig. 6B). In the C3S6 structure, pyrophosphate is present with a refined occupancy of 0.55, comparable to the 0.66–0.72 occupancies seen in the pretranslocation structures (Figs. 3A and 5A), but pyrophosphate is totally absent both in the S1 structures in this study and in previously determined picornavirus polymerase EC S1 structures (10, 14). Thus, the release of pyrophosphate may coincide with the final upstream movement of the template −2 to +2 region, although whether and how the pyrophosphate release and the conformational changes controlling the templates strand movement are mechanically related is not obvious.
It is plausible that the asymmetric movement of the two RNA strands observed in the current study also may be a general feature of other classes of polymerases. Although the special backbone conformation of the template −2 ribose–phosphate linkage is observed only in viral RdRP–RNA complexes (2, 10, 14, 23), the sharp turn at the +1/+2 junction of the template strand is shared by several classes of polymerases (28, 29, 40, 41). The polymerases need to create the active site at the downstream end of the +1 site, and therefore the helical trace of the template strand backbone must deform around the +1/+2 junction, making the postcatalysis translocation around the junction less fluid. Very interestingly, the eukaryotic RNA Pol II initiation complex (IC) has been captured with a template–product duplex adopting a similar asymmetric conformation when the transcript length is 4–5 nt (42). Note that these Pol II IC complexes are not translocation intermediates, because the asymmetric conformation was achieved through slippage-mode movement of the product strand in the downstream direction. As a result, the −1 and −2 (“i-1” and “i-2” in Pol II nomenclature) nucleotides in the product strand seem to form mismatches with the −1 and +1 nucleotides in the template strand. These obvious conformational dynamics of the template–product duplex may be explained in part by the intrinsic instability of a DNA-dependent RNA polymerase IC with a short transcript. In fact, a similar conformation has not been observed in Pol II IC structures with longer transcripts or in EC structures that have established intensive transcript–template and transcript–polymerase interactions (29, 42–44). Therefore, the mechanism for achieving a similar asymmetric template–product conformation in these Pol II ICs appears to be different from that suggested by the translocation intermediate in the current study. More high-resolution translocation-intermediate structures are needed to refine further the understanding of the polymerase translocation in general. As mentioned, capturing translocation intermediates by crystallography is somewhat serendipitous, but perhaps molecular dynamics approaches could serve as a valid tool for testing whether other classes of polymerases also exhibit asymmetric RNA-strand movements during translocation.
Materials and Methods
Cloning and Protein Expression.
The EV71 3Dpol gene within the DNA clone of SK-EV006-LPS1 (GenBank accession no. AB550335.1, genotype B) was cloned into a pET26b-Ub vector (45). The resulting plasmid was transformed into Escherichia coli strain BL21(DE3) pCG1 (kindly supplied by Craig Cameron, Pennsylvania State University, State College, PA) for expression of 3Dpol with a C-terminal hexa-histidine tag as described previously (3, 45). 3Dpol was produced as a ubiquitin-fused protein, and the ubiquitin was cleaved in vivo by a coexpressed ubiquitin-specific carboxyl terminal protease Ubp1 to produce the 3Dpol with homogenous native N-terminal glycine residue. Cells were grown at 25 °C overnight in LB medium with 50 µg/mL kanamycin (KAN50) and 17 µg/mL chloramphenicol (CHL17) until the OD600 was 1.0. The overnight culture was used to inoculate 1 L of LB medium with KAN50 and CHL17 to reach an initial OD600 around 0.025. The cells were grown at 37 °C to an OD600 of 0.6 and then were cooled to room temperature. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.5 mM, and the cells were grown for an additional 11–12 h before harvesting.
Purification of EV71 3Dpol.
Cell lysis, subsequent purification, and storage procedures were as described previously (3), except that a HiTrap Q HP column (GE Healthcare) was used in the second chromatographic purification step. This column was equilibrated with a buffer containing 50 mM NaCl, 25 mM Tris (pH 8.5), 0.1 mM EDTA, 20% (vol/vol) glycerol, and 0.02% (wt/vol) NaN3, and the protein was eluted by a linear gradient to 600 mM NaCl. The final buffer condition for protein storage was 300 mM NaCl, 5 mM Tris (pH 7.5), 0.02% (wt/vol) NaN3, and 5 mM Tris(2-carboxyethyl)phosphine. The molar extinction coefficient for 3Dpol protein was calculated based on protein sequence using the ExPASy ProtParam program (www.expasy.org/tools/protparam.html). The yield is typically in the range of 8–15 mg of pure protein per liter of bacterial culture.
RNA Preparation.
The template strand RNA to assemble the r5 construct (14) solely used in the current study was obtained by in vitro T7 RNA polymerase transcription using a parental plasmid pRAV23 (kindly supplied by Jeffrey Kieft, University of Colorado Denver, Denver, and Robert Batey, University of Colorado Boulder, Boulder, CO) and approaches modified from protocols described previously (10, 46, 47). The 10mer RNA primer (P10) was purchased from Integrated DNA Technologies. The procedures for the self-annealing of the template strand, the subsequent annealing with P10, and the r5 construct storage were as previously described (14).
EC Assembly, Purification, and Storage.
EC assembly, purification, and storage was carried out using the protocols in the PV work (10), except that KCl was provided at a concentration of 70 mM, the NaCl concentration was reduced to 40 mM for the assembly reaction, and the Hepes pH was increased to 7.0 for both the assembly reaction and complex storage.
EC Crystallization and NTP Soaking of the EC Crystals.
The EC crystals were grown by sitting-drop vapor diffusion at 16 °C using 7.8 mg/mL EC sample. Crystals grew in 1–2 wk with a precipitant solution containing 0.17 M ammonium sulfate, 0.085 M Mes (pH 6.5), 25.5% (wt/vol) PEG 5000 monomethyl ether, and 15% (vol/vol) glycerol. NTP-soaking experiments were done under the precipitant solution using 5 mM NTP and 10 mM MgCl2. For each NAC complex obtained, the NTP combination and incubation time are listed in Tables 1 and 2.
Crystallographic Data Processing and Structure Determination.
All final diffraction data for all crystals were collected at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U1 at 100 Kelvin [wavelengths: 0.9791 Å for Protein Data Bank (PDB) entries 5F8G and 5F8L; 0.9789 Å for PDB entries 5F8H, 5F8I, 5F8M, and 5F8N; and 0.9792 Å for PDB entry 5F8J]. Data (100–150°) were typically collected in 0.5° oscillation steps. Reflections were integrated, merged, and scaled using HKL2000 or D*Trek v9.9 (48, 49). The initial structure solution was obtained using the molecular replacement program PHASER (50) using coordinates derived from the PV EC structure (PDB ID code 3OL6) as the search model (10). Manual model building and structure refinement were done using Coot and Phenix, respectively (51, 52). The 3,500-K composite simulated-annealing omit 2Fo-Fc electron density maps were generated using CNS (53). In this process, the entire asymmetric unit was first divided into small boxes, each including a fraction not exceeding 5% of the model. For each box, the model in the box was omitted for calculating the corresponding omit maps. The composite map then was generated by stitching all the individual omit maps together in order to make the entire map, not only a specific region, less model-biased. Unless otherwise indicated, all polymerase superimpositions were done using the maximum likelihood-based structure superimpositioning program THESEUS (21).
Acknowledgments
We thank Dr. Olve Peersen for helpful discussions and valuable input on the manuscript content; Dr. Zhiyong Lou for providing the cloning material for the EV71 polymerase gene; Dr. Craig Martin and Dr. Zhongzhou Chen for critical reading of the manuscript; Dr. Hanzhong Wang, Dr. Bo Zhang, and Dr. Huimin Yan for help in initiating this EV71 RdRP project; Wei Shi for contributions in optimizing the EC assembly reaction condition; Liu Deng for laboratory assistance; the Shanghai Synchrotron Radiation Facility (beamline BL17U1, Shanghai, China) and the Beijing Synchrotron Radiation Facility (beamline 3W1A, Beijing, China) synchrotrons for access to beamlines; and the Core Facility and Technical Support of the Wuhan Institute of Virology for access to instruments. This work was supported by National Key Basic Research Program of China Grant 2013CB911100, National Natural Science Foundation of China Grant 31370198, and the Hundred Talents Program of the Chinese Academy of Sciences.
Footnotes
References
- 1.Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23(17):3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrer-Orta C, et al. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J Biol Chem. 2004;279(45):47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 3.Lu G, Gong P. Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013;9(8):e1003549. doi: 10.1371/journal.ppat.1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang B, et al. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell. 2015;162(2):314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pflug A, Guilligay D, Reich S, Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516(7531):355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 6.Gorbalenya AE, et al. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol. 2002;324(1):47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruenn JA. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31(7):1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.te Velthuis AJ. Common and unique features of viral RNA-dependent polymerases. Cell Mol Life Sci. 2014;71(22):4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Liu W, Gong P. A structural overview of RNA-dependent RNA Polymerases from the Flaviviridae family. Int J Mol Sci. 2015;16(6):12943–12957. doi: 10.3390/ijms160612943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong P, Peersen OB. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 2010;107(52):22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116(3):393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998;17(24):7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Temiakov D, et al. Structural basis for substrate selection by t7 RNA polymerase. Cell. 2004;116(3):381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 14.Gong P, Kortus MG, Nix JC, Davis RE, Peersen OB. Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PLoS One. 2013;8(5):e60272. doi: 10.1371/journal.pone.0060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura T, Zhao Y, Yamagata Y, Hua YJ, Yang W. Watching DNA polymerase η make a phosphodiester bond. Nature. 2012;487(7406):196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu RS, Murakami KS. Watching the bacteriophage N4 RNA polymerase transcription by time-dependent soak-trigger-freeze X-ray crystallography. J Biol Chem. 2013;288(5):3305–3311. doi: 10.1074/jbc.M112.387712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: Nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119(4):481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116(6):803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 20.Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448(7150):163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 21.Theobald DL, Wuttke DS. THESEUS: Maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22(17):2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castro C, et al. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci USA. 2007;104(11):4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamyatkin DF, et al. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem. 2008;283(12):7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- 24.Castro C, et al. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16(2):212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong P, Campagnola G, Peersen OB. A quantitative stopped-flow fluorescence assay for measuring polymerase elongation rates. Anal Biochem. 2009;391(1):45–55. doi: 10.1016/j.ab.2009.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43(18):5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15(8):811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 28.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298(5597):1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 29.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292(5523):1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 30.Campagnola G, McDonald S, Beaucourt S, Vignuzzi M, Peersen OB. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J Virol. 2015;89(1):275–286. doi: 10.1128/JVI.01574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward CD, Stokes MA, Flanegan JB. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J Virol. 1988;62(2):558–562. doi: 10.1128/jvi.62.2.558-562.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graci JD, et al. Mutational robustness of an RNA virus influences sensitivity to lethal mutagenesis. J Virol. 2012;86(5):2869–2873. doi: 10.1128/JVI.05712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84(19):9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): An archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res. 2001;29(22):4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umar A, Kunkel TA. DNA-replication fidelity, mismatch repair and genome instability in cancer cells. FEBS. 1996;238(2):297–307. doi: 10.1111/j.1432-1033.1996.0297z.x. [DOI] [PubMed] [Google Scholar]
- 36.Sloane DL, Goodman MF, Echols H. The fidelity of base selection by the polymerase subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 1988;16(14A):6465–6475. doi: 10.1093/nar/16.14.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guajardo R, Sousa R. A model for the mechanism of polymerase translocation. J Mol Biol. 1997;265(1):8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 38.Gelles J, Landick R. RNA polymerase as a molecular motor. Cell. 1998;93(1):13–16. doi: 10.1016/s0092-8674(00)81140-x. [DOI] [PubMed] [Google Scholar]
- 39.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272(24):15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 40.Tahirov TH, et al. Structure of a T7 RNA polymerase elongation complex at 2.9 Å resolution. Nature. 2002;420(6911):43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 41.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260(5106):352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD. Initiation complex structure and promoter proofreading. Science. 2011;333(6042):633–637. doi: 10.1126/science.1206629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16(6):955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 44.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127(5):941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gohara DW, et al. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3Dpol) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr Purif. 1999;17(1):128–138. doi: 10.1006/prep.1999.1100. [DOI] [PubMed] [Google Scholar]
- 46.Wu J, Lu G, Zhang B, Gong P. Perturbation in the conserved methyltransferase-polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J Virol. 2015;89(1):249–261. doi: 10.1128/JVI.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batey RT, Kieft JS. Improved native affinity purification of RNA. RNA. 2007;13(8):1384–1389. doi: 10.1261/rna.528007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 10):1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 50.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]