Significance
During the biogenesis of proteins destined for the secretory pathway, proteins that fail to fold correctly are retained in the endoplasmic reticulum (ER) and targeted for degradation through a quality-control system called “ER-associated protein degradation” (ERAD), but how misfolded proteins are defined has remained unknown. Here we studied the ERAD pathway for misfolded N-glycoproteins, whose ERAD commitment requires the generation of a unique N-glycan structure by the action of the complex of the mannosidase Htm1p and the protein disulfide isomerase Pdi1p (Htm1p–Pdi1p). We found that Htm1p–Pdi1p differentiates the conformations of different N-glycoproteins and preferentially targets proteins trapped in partially structured states. In summary, our study reveals a conformational standard of how ERAD targets the right proteins for degradation.
Keywords: ERAD, ER quality control, mannosidase, N-glycoprotein
Abstract
Our understanding of how the endoplasmic reticulum (ER)-associated protein degradation (ERAD) machinery efficiently targets terminally misfolded proteins while avoiding the misidentification of nascent polypeptides and correctly folded proteins is limited. For luminal N-glycoproteins, demannosylation of their N-glycan to expose a terminal α1,6-linked mannose is necessary for their degradation via ERAD, but whether this modification is specific to misfolded proteins is unknown. Here we report that the complex of the mannosidase Htm1p and the protein disulfide isomerase Pdi1p (Htm1p–Pdi1p) acts as a folding-sensitive mannosidase for catalyzing this first committed step in Saccharomyces cerevisiae. We reconstitute this step in vitro with Htm1p–Pdi1p and model glycoprotein substrates whose structural states we can manipulate. We find that Htm1p–Pdi1p is a glycoprotein-specific mannosidase that preferentially targets nonnative glycoproteins trapped in partially structured states. As such, Htm1p–Pdi1p is suited to act as a licensing factor that monitors folding in the ER lumen and preferentially commits glycoproteins trapped in partially structured states for degradation.
Proteins destined for the secretory pathway enter the endoplasmic reticulum (ER) in unfolded states and generally leave only after they have reached their native conformation. Protein folding in the ER is challenging and often error-prone (1, 2). Thus, the ER must continuously monitor the pool of folding proteins and remove terminally misfolded forms before they induce toxicity. This removal is accomplished through a series of pathways collectively referred to as the “ER-associated protein degradation” (ERAD) machinery (3). Generally, the process of ERAD involves three steps: (i) identification of the misfolded protein, (ii) retrotranslocation and polyubiquitination of the misfolded protein through individual E3 ubiquitin ligase complexes, and (iii) degradation of the misfolded protein by the ubiquitin–proteasome system in the cytosol.
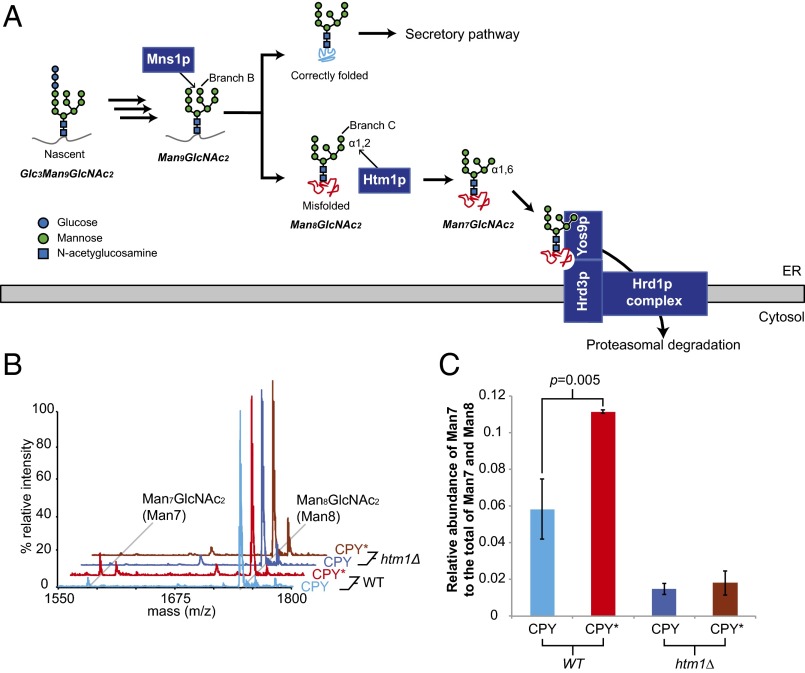
Each ERAD pathway needs to identify substrates accurately to avoid overly promiscuous destruction of functional proteins while preventing the accumulation of toxic forms (1). For the ERAD pathway responsible for the destruction of proteins with misfolded luminal domains (ERAD-L) (4, 5), this process of discrimination depends not only on the folding status of a polypeptide but also on its N-glycosylation state (6–11). Upon entering the ER lumen, nascent proteins acquire en bloc a high-mannose oligosaccharide, Glc3Man9GlcNAc2, appended to a specific subset of asparagine residues (Fig. 1A). The N-glycan then is deconstructed monosaccharide-by-monosaccharide to Man8GlcNAc2 (Man8) in the ER by a series of glycosidases concurrently with the protein-folding process (12, 13). The action of the mannosidase Mns1p, which is responsible for generating Man8, has been proposed to yield a “timer” that universally gives all N-glycoproteins a defined period to fold during which they are immune from ERAD-L (8, 12). Misfolded proteins that are destined for degradation via ERAD-L are further demannosylated by Htm1p (also known as Mnl1p), which removes the α1,2-linked mannose from the C branch of Man8 (14, 15). The resulting Man7GlcNAc2 (Man7) glycan structure with a terminally exposed α1,6-linked mannose serves as a signal for ERAD-L commitment. The Yos9p lectin subsequently binds this terminally exposed α1,6-linked mannose and, in coordination with Hrd3p, queries the misfolded regions on potential substrates (6, 7, 9–11, 16). Only the presence of both the Htm1p-generated N-glycan signal and the misfolded regions on the substrate enables Yos9p–Hrd3p to trigger the downstream retrotranslocation, polyubiquitination, and proteasomal degradation of target proteins (7, 14, 17). The physiological importance of these checkpoints is exemplified by the severe growth defect acquired when the requirement for Hrd3p and Yos9p was bypassed by overexpression of HRD1, which encodes the downstream E3 ubiquitin ligase for this pathway (6).
Fig. 1.
HTM1 mediates N-glycan processing for ERAD-L commitment. (A) Scheme of N-glycan processing and ERAD-L commitment. Upon entering the ER, nascent N-glycoproteins acquire Glc3Man9GlcNAc2 en bloc, which is step-by-step deglycosylated as the nascent polypeptide folds. Mns1p catalyzes the last universal deglycosylation step to generate the Man8GlcNAc2 glycan. Native proteins are targeted for the downstream secretory pathway, and misfolded proteins are targeted by Htm1p, which removes the α1,2-linked mannose at the C branch from the Man8GlcNAc2 glycan. The resulting terminally exposed α1,6-linked mannose and misfolded regions on the polypeptide chain are then read by Yos9p. Along with Hrd3p, Yos9p then permits the downstream retrotranslocation and polyubiquitination through the Hrd1p transmembrane complex for eventual proteasomal degradation. (B) Examples of glycan profiles of CPY and CPY* expressed in wild type (WT) or htm1∆ yeast strains as measured by MALDI-TOF MS after the glycans were released from CPY/CPY* by PNGase F treatment. Each glycan was assigned to the m/z corresponding to its predicted [M+Na]+ value. (C) The relative abundance of Man7 is higher in CPY* than in CPY in wild-type yeast, which is dependent on HTM1. The relative MS peak intensity of Man7, as found in B, is normalized to the total of Man7 and Man8 as a proxy for its relative abundance. Shown are the mean values ± one SD from biological triplicates. The P value was calculated by unpaired t test.
As the first committed step, the generation of the terminally exposed α1,6-linked mannose by Htm1p has the potential to determine which proteins are shunted down the ERAD-L pathway and the stage during their folding process when this shunting occurs. This potential “licensing” role has been supported by in vivo observations that genetic deletion of HTM1, as well as point mutations in its putative active sites, not only retards the degradation of misfolded N-glycoproteins but also decreases substrate-binding by Yos9p (11, 14, 17–19). This retardation of ERAD by the deletion of HTM1 can be circumvented in a genetic background that generates the terminally exposed α1,6-linked mannose (7). The importance of the mannosidase function of Htm1p is further evidenced by the conserved role of HTM1 homologs for the N-glycan–dependent ERAD pathway (20–22). Finally, both in vivo and in vitro studies have provided evidence for the mannosidase activity of Htm1p in generating Man7 (14, 15, 23). However, despite the well-documented role of Htm1p in the N-glycan processing for ERAD-L commitment, it is still unclear whether Htm1p targets misfolded proteins specifically and accurately. This uncertainty mainly results from the challenges in simultaneously monitoring protein conformations and their N-glycosylation states in the highly heterogeneous pool of glycoproteins in the ER. Additionally, only minimal mannosidase activities of Htm1p were observed in previous in vitro studies (15, 23). To advance our understanding of this commitment step, it is critical to develop a reconstituted system for direct investigation of the enzymatic activities of Htm1p against glycoproteins with defined glycosylation and folding states (24).
Here, we reconstituted this ERAD-L commitment step, which consists of a recombinantly expressed Htm1p–Pdi1p complex and glycoprotein substrates with various native and nonnative conformations. Our results reveal that Htm1p–Pdi1p is a glycoprotein-specific mannosidase complex that preferentially demannosylates intrinsically or artificially misfolded proteins but not their native counterparts. Furthermore, among the various nonnative conformations, Htm1p–Pdi1p prefers partially structured proteins over globally unfolded ones. Our findings suggest that Htm1p–Pdi1p monitors the protein-folding states in the ER lumen and preferentially licenses glycoproteins trapped in partially structured states for ERAD-L.
Results
Generation of Man7 in Vivo Is Folding- and HTM1-Dependent.
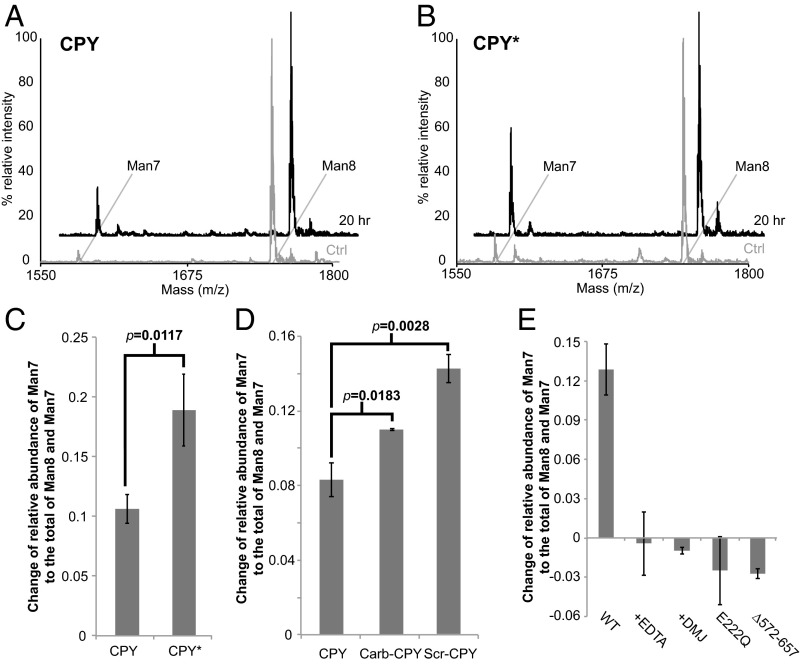
It has been reported that the prototypic ERAD-L substrate CPY* contains higher levels of Man7 than its folding-competent counterpart, procarboxypeptidase Y (CPY) in a wild-type yeast background (11). We first explored whether the higher level of Man7 in CPY* reflects the differences in intrinsic folding competence between CPY* and CPY or instead results passively from the prolonged ER retention of CPY*. Using a previously described overexpression construct (25), we expressed and purified native CPY and CPY* that are retained in the ER by a C-terminal HDEL tag. We used MALDI-TOF MS to determine the glycan profile of CPY and CPY*. This approach provides higher throughput capacity than HPLC-based measurement while still allowing quantitative analysis of neutral glycans (26). As such, it also is amenable to the analysis of mannosidase activities described below. Both CPY and CPY* purified from a wild-type yeast strain (BY4741) carried predominantly Man8 and minor populations of Man7, Man9, and Hex10GlcNAc2 (Hex10), which is likely GlcMan9GlcNAc2 (Fig. S1). The majority of the signal of Man7 disappeared when CPY and CPY* were purified from the htm1∆ strain background (Fig. 1 B and C). When CPY and CPY* were purified from an mns1∆htm1∆ background, the dominant glycan species shifted from Man8 to Man9, further verifying that the majority of Man8 from the wild-type strain is the product of Mns1p, the putative substrate of Htm1p (Fig. S1). Importantly, CPY* carried a significantly higher level of Man7 than CPY (Fig. 1 B and C). Taken together, these observations suggest that, although both are retained in the ER, CPY* and CPY are differentially demannosylated to Man7 in an HTM1-dependent manner. Because CPY and CPY* purified through this HDEL-tagged system are different only at the G255R point mutation on CPY* that disrupts the hydrophobic core on the native structure (27), we conclude that the generation of Man7 in the ER is correlated with the intrinsic folding competence of the underlying protein.
Fig. S1.
Full MS spectra of the glycan profiles on CPY and CPY* purified from different yeast strains. As described in Fig. 1B, glycans of CPY (A) and CPY* (B) expressed in wild type, htm1∆, and mns1∆htm1∆ yeast strains were released by PNGase F for subsequent analysis by MALDI-TOF MS. Each glycan was assigned according to the m/z corresponding to the predicted [M+Na]+ value.
Reconstitution of an Htm1p–Pdi1p Complex.
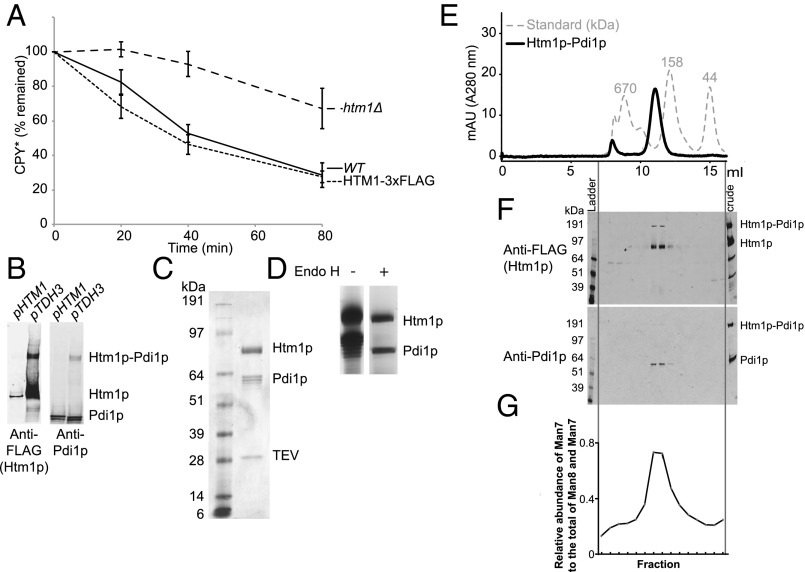
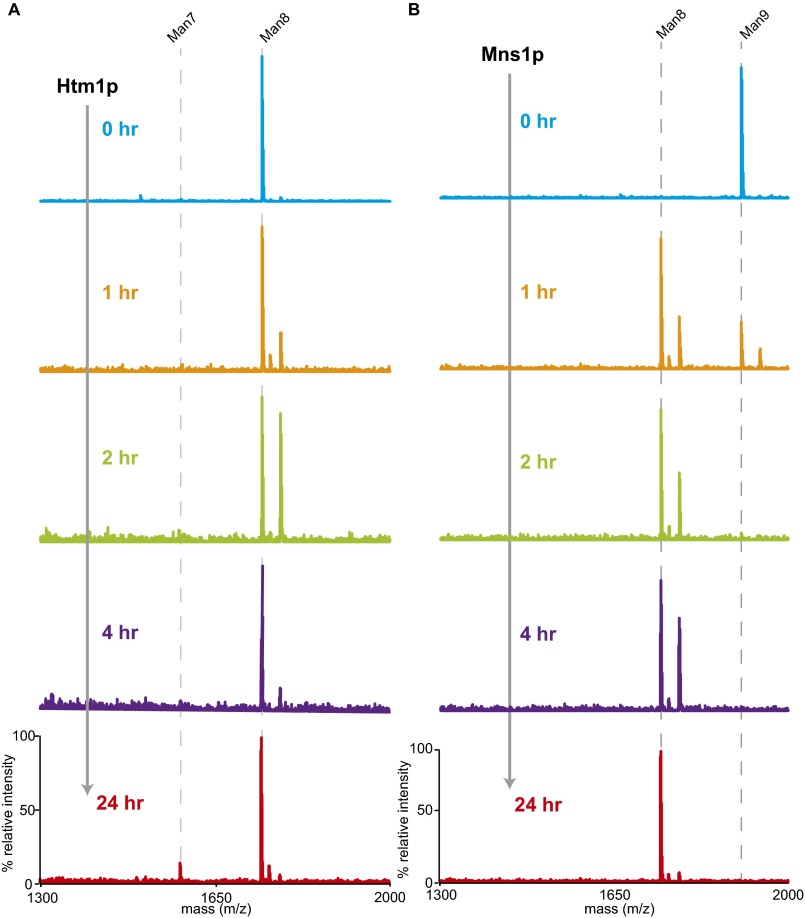
To explore the activity of Htm1p in vitro, we tested the feasibility of producing Htm1p from the native host Saccharomyces cerevisiae. We introduced a 3xFLAG tag, followed by an HDEL tail before the stop codon, to the C terminus of Htm1p for affinity purification. The result of the cycloheximide degradation assay on CPY* suggests that the chromosomally tagged version is similarly functional to wild-type Htm1p (Fig. 2A). Because the endogenous level of Htm1p is low, we enhanced the expression of Htm1p by chromosomally substituting its endogenous promoter with a TDH3 promoter (Fig. 2B). Consistent with findings in a previous study (28), Htm1p readily formed a disulfide complex with endogenous Pdi1p as resolved by nonreducing SDS/PAGE (Fig. 2B). Large-scale immunoprecipitation yielded an Htm1p–Pdi1p complex with nearly 1:1 stoichiometry (Fig. 2C), with a yield of 200 µg of the complex from 3 L of yeast culture. Treatment of the complex with endoglycosidase H confirmed that both Htm1p and Pdi1p were N-glycosylated, indicating that they are correctly targeted to the secretory pathway (Fig. 2D). The complex behaved as a monodisperse species in size-exclusion column chromatography (Fig. 2E), which contained a mixture of both disulfide-linked and noncovalent complexes of Htm1p and Pdi1p (Fig. 2F). We did not detect any monomeric Htm1p or Pdi1p corresponding to their predicted mass (90.2 kDa for Htm1p and 56.0 kDa for Pdi1p). Finally, we observed the mannosidase activity against the most preferred glycoprotein substrate, RBsp (RNase BS protein with the N-terminal S peptide removed), only in the fractions containing the Htm1–Pdi1p complex (Fig. 2G and see below).
Fig. 2.
Recombinant preparation of Htm1–Pdi1p from S. cerevisiae. (A) The percentage of HA-tagged CPY* remaining in wild-type BY4741 (WT), htm1∆, and chromosomally 3xFLAG–HDEL–tagged HTM1 (HTM1-3xFLAG) normalized to time 0 along the time course after the addition of cycloheximide. Shown are the mean values ± one SEM measured by quantitative Western blot from four biological replicates. (B) Expression levels of Htm1p–3xFLAG driven by either the endogenous HTM1 promoter (pHTM1) or the chromosomally inserted TDH3 promoter (pTDH3) as visualized by Western blot against FLAG-tagged Htm1p and Pdi1p after nonreducing SDS/PAGE separation of the same amount of cell pellets. (C) Specific interaction between Htm1p and Pdi1p after immunoprecipitation with anti-FLAG affinity resin followed by TEV protease treatment to release FLAG-tagged Htm1p. Shown is a Coomassie Blue R250-stained gel image after separation by reducing SDS/PAGE. (D) The N-glycosylation states of Htm1p and Pdi1p as checked by endoglycosidase H (Endo H). (E) Superdex 200 10/300GL size-exclusion column chromatography of Htm1p–Pdi1p eluted from anti-FLAG resin by 3xFLAG peptide. (F) Western blot analysis of the distribution of Htm1p–3xFLAG and Pdi1p after nonreducing SDS/PAGE of the fractions separated by the size-exclusion column chromatography. (G) The relative abundance of Man7 in the total of Man7 and Man8 on RBsp after 1-h incubation with the fractions collected from the size-exclusion column chromatography of purified Htm1p–Pdi1p.
Htm1p–Pdi1p Preferentially Demannosylates CPY* and Misfolded CPY Variants.
We first explored the mannosidase activity of reconstituted Htm1p–Pdi1p on CPY and CPY* that we purified from wild-type yeast. After a 20-h reaction, Htm1p–Pdi1p converted a portion of Man8 on CPY and CPY* to Man7 (Fig. 3 A and B). Consistent with our in vivo observation, the increase in Man7 is significantly higher on CPY* than on CPY after incubation with Htm1p–Pdi1p (Fig. 3C). For further validation that Htm1p–Pdi1p preferentially targets nonnative proteins, we reductively denatured CPY to remove its structurally essential disulfide bonds (29). Reduced CPY then was either oxidized under denaturing condition into “scrambled” species with randomly distributed nonnative disulfide bonds (Scr-CPY) or carbamidomethylated to block the reformation of disulfide bonds (Carb-CPY). Htm1p–Pdi1p also demannosylated both Scr-CPY and Carb-CPY more efficiently than the native form, and the yield of Man7 was higher on Scr-CPY than on Carb-CPY (Fig. 3D). Taken together, these results suggest that our reconstituted Htm1p–Pdi1p is an active mannosidase and acts preferentially on intrinsically folding-incompetent CPY* and on artificially misfolded CPY variants rather than on the native CPY.
Fig. 3.
Htm1p–Pdi1p preferentially demannosylates CPY* and nonnative CPY variants. (A and B) Representative glycan profiles of CPY (A) and CPY* (B) before and after 20-h incubation with Htm1p–Pdi1p. CPY or CPY* (2 µM) was incubated with 0.1 µM Htm1p–Pdi1p at 30 °C for 20 h and then was separated from Htm1–Pdi1p by SDS/PAGE for subsequent in-gel glycan release and MALDI-TOF MS analysis. (C) Htm1p–Pdi1p generates more Man7 on CPY* than on CPY after reaction. The mannosidase activity is measured by the change in the relative abundance of Man7 on CPY and CPY* after 20-h incubation with Htm1p–Pdi1p. Shown are the mean values ± one SD from an experiment performed in triplicate. The P value was calculated by unpaired t test. (D) Htm1p–Pdi1p generated more Man7 on Carb-CPY and Scr-CPY than on CPY. Shown are the mean values ± one SD from an experiment performed in triplicate. The P value was calculated by unpaired t test. (E) The mannosidase activity of Htm1p–Pdi1p against CPY* was inhibited by EDTA (+EDTA), 1-deoxymannojirimicin (+DMJ), and mutations including E222Q and ∆572–657. Quantification was carried out as in C. Shown are the mean values ± one SD from an experiment performed in triplicate.
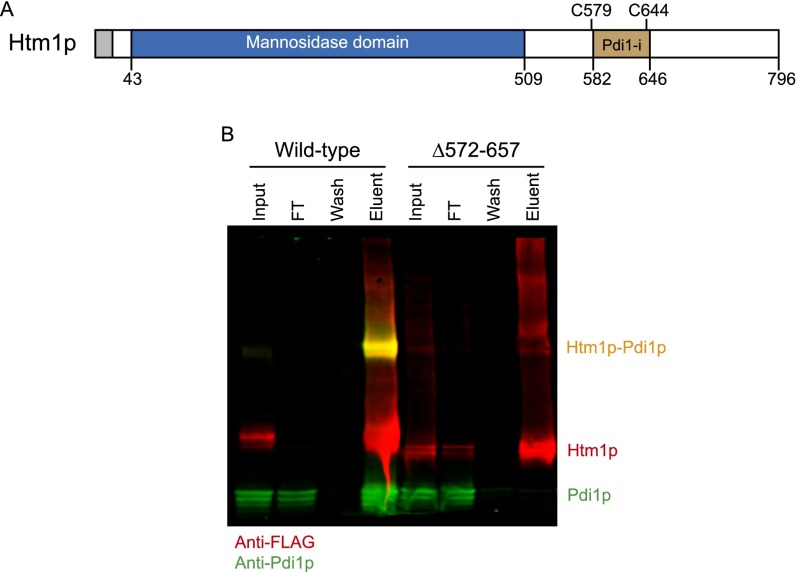
We then wanted to verify whether demannosylation is catalyzed through the predicted mannosidase domain of Htm1p and whether Pdi1p is required for this process. We carried out the reaction in the presence of EDTA, which sequesters the structurally essential Ca2+, and 1-deoxymannojirimycin (DMJ), which inhibits the GH47 mannosidase family to which Htm1p is predicted to belong (30). Both chemicals inhibited the generation of Man7 on CPY* (Fig. 3E). We further verified the predicted mannosidase function of Htm1p with an Htm1 mutant in which one of the putative active site residues, Glu222, is mutated to Gln (E222Q) (14, 31). This mutant indeed was incapable of generating Man7 on CPY* (Fig. 3E). Finally, we made a second Htm1 mutant with a truncation of the amino acyl region from residue 572 to residue 657 (∆572–657), which covers the region necessary for interaction with Pdi1p (Fig. S2A) (14, 23, 28). Immunoprecipitation confirmed that ∆572–657 showed little detectable interaction with Pdi1p (Fig. S2B). Similar to the catalytically dead E222Q, ∆572–657 was incapable of generating Man7 on CPY* (Fig. 3E). Collectively, our results support the notion that the predicted mannosidase domain of Htm1p mediates the demannosylation reaction and that the interaction with Pdi1p is required for this activity.
Fig. S2.
Deletion of the putative Pdi1p-interacting domain of Htm1p abolished the interaction between Htm1p and Pdi1p. (A) Schematic representation of the domain structures of Htm1p. Htm1p consists of the N-terminal conserved GH47 family mannosidase domain and the C-terminal nonconserved region. Amino acyl region 582–646 (Pdi1-i) comprises the previously identified region required for noncovalent interaction. Cys579 (C579) and Cys644 (C644) form disulfide cross-linking with Pdi1p. (B) The ∆572–657 mutation lost specific interaction with Pdi1p. The interaction of Pdi1p with wild-type Htm1p or the ∆572–657 mutation was analyzed by Western blot after immunoprecipitation with anti-FLAG affinity resin followed by 3xFLAG peptide elution to release Htm1p.
The conversion of Man8 to Man7 on CPY* continued during prolonged incubation with Htm1p–Pdi1p (Fig. S3A), but this rate of demannosylation is nonetheless much slower than would be expected from the in vivo half-life of CPY*, which is about 60 min (Fig. 2A). This slow rate could, in principle, be a consequence of CPY having four N-glycans, and thus Htm1p–Pdi1p may act on only a subset of the glycans on CPY*. Indeed, the C-terminal–most N-glycan of CPY* is known to be necessary and sufficient to support its ERAD in vivo (17, 32, 33). However, we found that in vitro Htm1p–Pdi1p demannosylated a CPY* mutant (CPY*1110), in which this ERAD-competent glycosylation site, Asn479, was mutated to Gln, to a level similar to CPY* with all four glycosylation sites (Fig. S3B). A similar level of demannosylation was observed on another CPY* mutant (CPY*0001), in which the other three glycosylation sites (Asn124, Asn198, and Asn279) were all mutated to Gln. The glycosylation site thus is not likely to be the key factor limiting the catalytic activity. Increasing the amount of Htm1p–Pdi1p by fourfold did not increase the yield proportionally, suggesting that the enzyme concentration also is not the major limiting factor (Fig. S3E).
Fig. S3.
Glycosylation sites and concentration of Htm1p–Pdi1p are not the major limiting factors for demannosylation rates. (A) The change in Man7 abundance relative to the sum of Man7 and Man8 on CPY* after 72-h incubation with Htm1p–Pdi1p. Shown are the mean values ± one SD from an experiment performed in triplicate. (B) Htm1p–Pdi1p generated similar levels of Man7 on CPY* and CPY* variants with either the fourth (CPY*1110) or the first three (CPY*0001) glycosylation sites eliminated. Shown are the mean ± one SD from an experiment performed in triplicate. (C) Increasing the concentration of Htm1p–Pdi1p from 0.1 µM to 0.4 µM did not proportionally increase its Man8-to-Man7 mannosidase activity against CPY* after 20-h incubation. Shown are the mean values ± one SD from an experiment performed in triplicate.
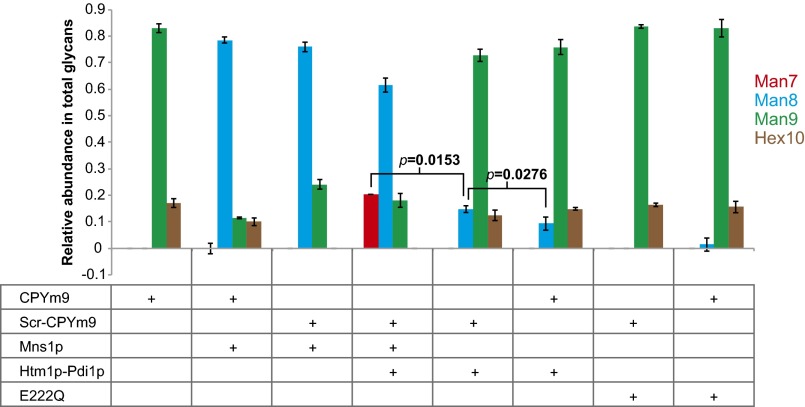
The incomplete reaction may reflect conformational heterogeneity or the aggregation-prone nature of CPY*. Indeed, in our hands, the majority of purified CPY* is in oligomeric forms (Fig. S4), and we observed the formation of visible aggregates from both CPY* and artificially misfolded CPY variants after incubation with Htm1p–Pdi1p for 20 h. A potential consequence of aggregation is that the glycans become inaccessible to the deep active pocket of Htm1p as predicted by the GH47 family mannosidase domain (34). We first explored whether Htm1p–Pdi1p has higher activity against the free Man8 glycan. Htm1p–Pdi1p generated only a limited amount of Man7 after 24 h of reaction (Fig. S5A). In contrast, Mns1p, which similarly contains a GH47 family mannosidase domain, was able to demannosylate Man9 into Man8 completely (Fig. S5B). Then, to assess the accessibility to glycans on CPY, we tested the activity of Mns1p against Man9-carrying CPY (CPYm9) and its scrambled form (Scr-CPYm9) purified from the mns1∆htm1∆ strain background. Mns1p converted most of Man9 on both CPYm9 and Scr-CPYm9 into Man8, suggesting that the glycans are accessible on both native and nonnative forms of CPY (Fig. 4). Mns1p alone did not produce any detectable Man7. Only cotreatment of Mns1p and Htm1p–Pdi1p yielded Man7 at a level similar to that we observed in Fig. 3. We found that Htm1p–Pdi1p alone, without Mns1p, also was capable of demannosylating Man9 on CPYm9 and Scr-CPYm9 into Man8, albeit to a lower extent. Similar to the results in Fig. 3D, the level of Man8 generated by Htm1p–Pdi1p treatment was higher on Scr-CPYm9 than on CPYm9. Collectively, these observations suggest intrinsic differences in substrate specificity between Mns1p and Htm1p–Pdi1p: Although Mns1p is capable of targeting both free glycans and glycoproteins independently of the attached protein conformations, Htm1p–Pdi1p preferentially targets glycans installed on nonnative proteins. In addition, as is consistent with previous in vivo studies (10, 31, 35), our findings suggest that Htm1p–Pdi1p is capable of bypassing the action of Mns1p to demannosylate Man9 directly.
Fig. S4.
Size-exclusion column chromatography analysis of CPY*. Size distribution of CPY* before (CPY* control), immediately after solubilization with 4 M urea, and 20 h after dilution from 4 M urea treatment, as analyzed by Superdex 200 size-exclusion chromatography.
Fig. S5.
Demannosylation of free glycans by Htm1p–Pdi1p and Mns1p. (A) Free Man8 (10 µM) was incubated with 0.1 µM Htm1p–Pdi1p and collected for MS analysis at each indicated time point. Each glycan was assigned to the m/z corresponding to its [M+Na]+ value. Note the occasional presence of a second peak next to the [M+Na]+ peak; this second peak corresponds to the [M+K]+ that does not influence overall quantification. (B) Free Man9 (10 µM) was incubated with 0.1 µM Mns1p and collected for MS analysis at each indicated time point. Each glycan was assigned to the m/z corresponding to its [M+Na]+ value. Note the occasional presence of a second peak next to the [M+Na]+ peak, which corresponds to the [M+K]+ that does not influence overall quantification.
Fig. 4.
Glycans on misfolded CPY are accessible for demannosylation. The relative abundance of each glycan on CPYm9 or Scr-CPYm9 after 24-h incubation with Mns1p and/or Htm1p–Pdi1p or E222Q. Shown are the mean values ± one SD from an experiment performed in triplicate. P values were calculated by unpaired t test.
Htm1p–Pdi1p Preferentially Demannosylates Partially Structured RNase B Variants.
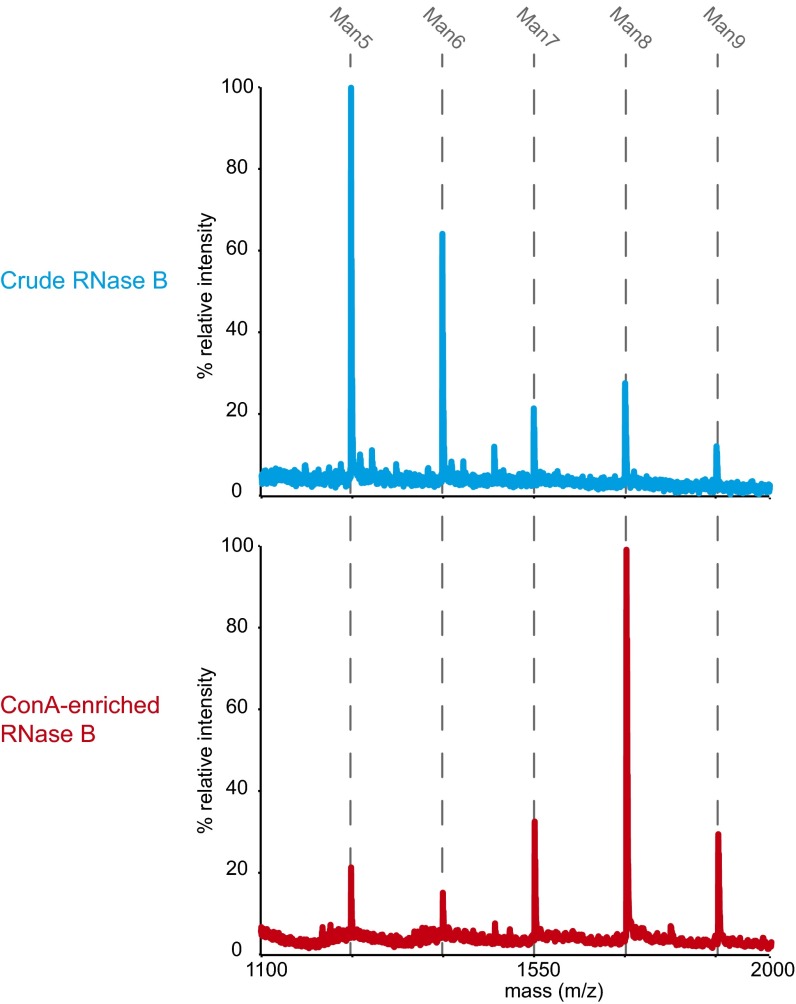
To this point, our findings support two potential models describing how Htm1p–Pdi1p generates the α1,6-linked mannose signal for ERAD-L commitment. In one model, the catalysis by Htm1p–Pdi1p may be intrinsically slow and stochastic, as previously suggested (15, 23), and unfolding facilitates demannosylation by increasing access to the glycan. Alternatively, Htm1p–Pdi1p may preferentially target specific folding states of nonnative proteins, and the aggregation-prone nature of misfolded CPY variants prevents them from populating in these states in vitro. To test these two potential models, we introduced bovine pancreatic ribonuclease B (RNase B) as an alternative substrate. RNase B is identical to the well-characterized classic protein folding substrate RNase A protein except that it contains a single N-glycosylation site (36). The folding of RNase B has been studied extensively, and the proteins can be manipulated to form well-defined, homogeneous states of native, misfolded or unfolded conformations (37–39). Using a concanavalin A (Con A) lectin-based approach (40), we can enrich for Man8-abundant RNase B from commercial sources in biochemical quantities (Fig. S6). To generate a nonnative variant of RNase B, we proteolytically removed the N-terminal S peptide from native RNase B to produce RBsp (Fig. 5A). RBsp is trapped in a compact, disordered, but nonetheless well-behaved state, and it can be readily reconstituted into a native-like RNase BS form by the addition of the S peptide (38).
Fig. S6.
Con A enrichment of Man8-abundant RNase B. Shown are the glycan profiles of RNase B from the commercial source before and after affinity enrichment with Con A lectin column.
Fig. 5.
Htm1p–Pdi1p efficiently demannosylates RBsp. (A) Schematic presentation of the generation of RBsp. Native RNase B was treated with subtilisin in a 100:1 ratio at 4 °C for 24 h to generate RNase BS with a nick between the N-terminal S peptide and the rest of the protein (RBsp). RNase BS then was subjected to precipitation with 10% TCA to remove the soluble S peptide from the precipitated RBsp. (B) Change in the relative abundance of Man7 on RBsp and native RNase B during a time-course incubation with Htm1p–Pdi1p. Shown are the mean values ± one SD from an experiment performed in triplicate. (C) Schematic presentation of the coreaction of RBsp and native RNase B with Htm1p–Pdi1p. RBsp and native RNase B were premixed at a 1:1 ratio, incubated with Htm1p–Pdi1p for 20 min, and subsequently separated by SDS/PAGE for MS analysis of the individual glycan profiles. (D) Change in the relative abundance of Man7 on premixed RBsp and native RNase B after incubation with Htm1p–Pdi1p for 20 min. Shown are the mean values ± one SD from an experiment performed in triplicate.
Remarkably, Htm1p–Pdi1p differentiated between native RNase B and RBsp and efficiently demannosylated only RBsp (Fig. 5B). Demannosylation of Man8 on RBsp into Man7 nearly reached a plateau after 1 h of incubation with Htm1p–Pdi1p (Fig. S7A). We further investigated whether Htm1p–Pdi1p can be trans-activated by RBsp or trans-inhibited by CPY*. We tested these possibilities by incubating Htm1p–Pdi1p with CPY* for 2 h and then adding RBsp. Differences in mass between CPY* (76.7 kDa) and RBsp (13.2 kDa) allow them to be readily separated by SDS/PAGE for subsequent glycan profiling. No obvious differences were observed on demannosylation of RBsp in the presence or absence of CPY*, and vice versa (Fig. S8). We extended this experiment further with a mixture of native RNase B and RBsp, which, given their difference in mass, can be similarly separated by SDS/PAGE by the presence or absence of the S-peptide (Fig. 5C). The presence of RBsp did not induce the demannosylation of native RNase B by Htm1p–Pdi1p (Fig. 5D). Collectively, these data establish that Htm1p–Pdi1p is capable of efficiently demannosylating the specific type of nonnative proteins to which RBsp belongs and that the activity is not trans-regulated in a mixture of different glycoproteins.
Fig. S7.
Mns1p and Htm1p–Pdi1p target different mannoses on RBsp. (A) The abundance Man5 relative to Man9 normalized to the total glycans on RBsp over a16-h time course incubation with Htm1p–Pdi1p. Shown are the mean values ± one SD from an experiment performed in triplicate. (B) The glycan distribution of RBsp before and after incubation with 0.1 µM Mns1p and/or Htm1p–Pdi1p for 16 h. Shown are the mean values ± one SD from an experiment performed in triplicate.
Fig. S8.
Htm1p–Pdi1p is not trans-regulated by CPY* and RBsp. (A) Htm1p–Pdi1p generated similar glycan profiles on RBsp with or without preincubation with CPY* after a 16-h incubation. Shown are the mean values ± one SD from an experiment performed in triplicate. (B) Htm1p–Pdi1p generated similar levels of Man7 on CPY* in the presence or absence of RBsp after a 16-h incubation. Shown are the mean values ± one SD from an experiment performed in triplicate.
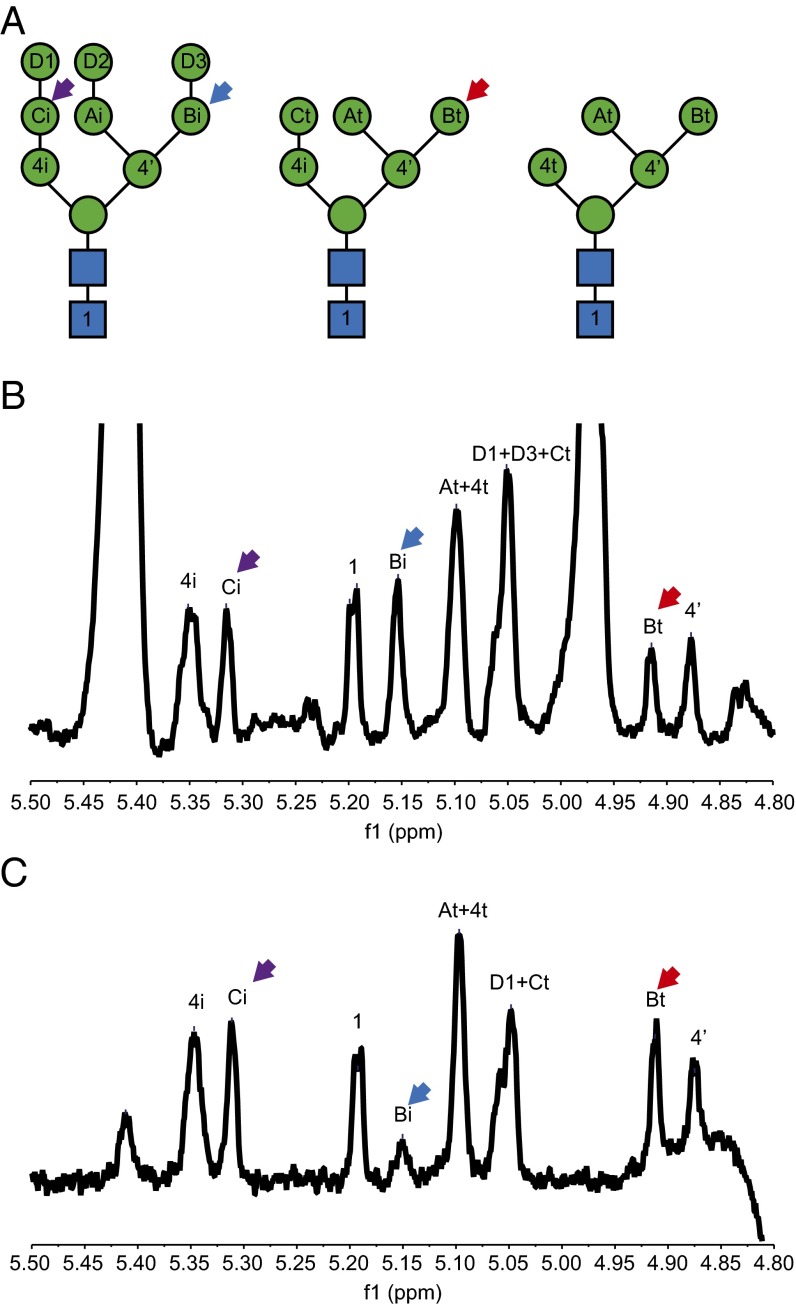
Consistent with the results in Fig. 4, we observed the disappearance of Man9 on RBsp during the reaction with Htm1p–Pdi1p (Fig. S7). The residual Man8 can be attributed to the other two less-abundant Man8 isoforms on RNase B that already have the C branch α1,2-mannose removed (41). Indeed, cotreatment with Mns1p and Htm1p–Pdi1p removed all Man9 and Man8 on RBsp (Fig. S7B), also verifying that Mns1p and Htm1p–Pdi1p target different mannoses on Man9. We further explored whether Htm1p–Pdi1p targets the α1,2-linked mannose on the C branch or the A branch, a question that never has been thoroughly investigated. To obtain a structural view of which mannose is removed by Htm1p–Pdi1p, we performed 1H-NMR analysis on the glycans released from denatured RNase B before and after an overnight reaction with Htm1p–Pdi1p. Indeed, we found that only the α1,2-linked mannose on the C branch was removed after overnight incubation with Htm1p–Pdi1p (Fig. 6), providing structural evidence to support the existing model in which Htm1p–Pdi1p specifically removes the α1,2-mannose on the C branch.
Fig. 6.
1H-NMR analysis of the glycan released from RNase B before and after overnight incubation with Htm1p–Pdi1p. (A) Representative N-glycan structures that cover all potential monosaccharide linkages found on RBsp. The nomenclature and peak assignment are based on previous studies (41). (B and C) The 1H-NMR spectra (D2O, 400 MHz) of the glycans released from RBsp before (B) and after (C) Htm1p–Pdi1p treatment. Note the change in the height of the peaks corresponding to the α1,6-linked mannose on the C branch with (Bi, blue arrow) or without (Bt, red arrow) the terminal α1,2-linked mannose (D3) after incubation with Htm1p–Pdi1p. In contrast, the level of the second α1,2-linked mannose on the A branch (Ci, purple arrow) did not change after Htm1p–Pdi1p treatment. Shown is the region covering the H-1 on the anomeric carbon of each monosaccharide after solvent suppression by the presaturation method.
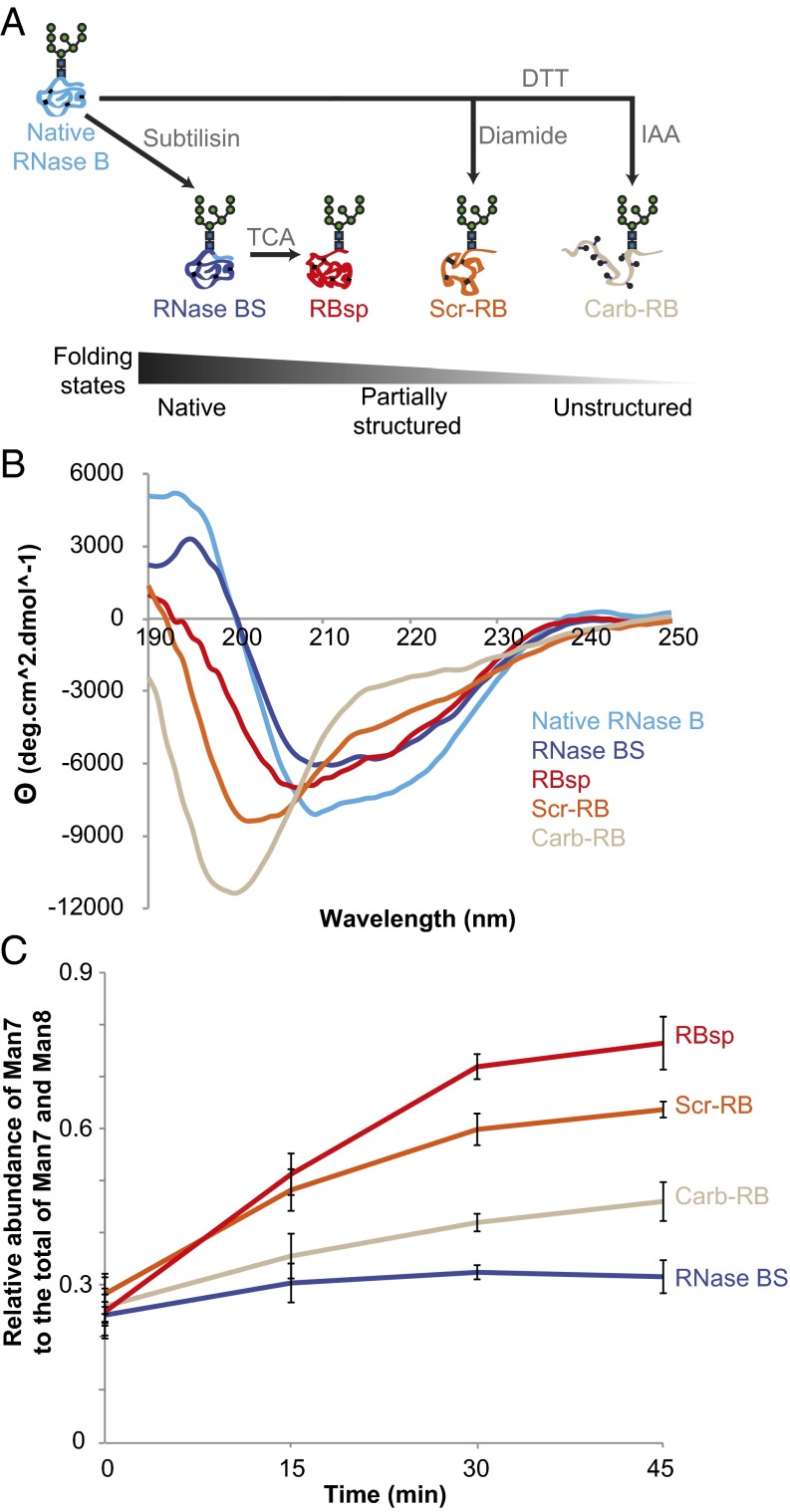
To characterize further how accurately Htm1p–Pdi1p differentiates proteins with different nonnative conformations, we generated three additional variants of RNase B: (i) a subtilisin-nicked noncovalent complex of RBsp and S-peptide (RNase BS); (ii) disulfide-scrambled RNase B (Scr-RB); and (iii) cysteine-carbamidomethylated RNase B (Carb-RB) (Fig. 7A). Consistent with the known properties of these variants (37, 38), our far-UV circular dichroism analysis verified that these RNase B variants cover a range of different conformations (Fig. 7B): RNase BS behaves essentially identically to native RNase B; RBsp is a compact folding intermediate with relatively abundant secondary structure features; Scr-RB consists of random, less compact structures with residual signals from secondary structures; and Carb-RB consists of globally unfolded species with the majority of the signals from random coils. We further confirmed this increasing trend of unfolding from native RNase B to Carb-RB by monitoring the differences in peptide backbone flexibility and glycan accessibility of different RNase B variants through trypsin and peptide N-glycanase F (PNGase F) treatment (Fig. S9 A and B). Of particular note, unlike CPY* and nonnative CPY variants, both Scr-RB and Carb-RB did not oligomerize or aggregate even after overnight incubation with Htm1p–Pdi1p at 30 °C (Fig. S9 C and D).
Fig. 7.
Htm1p–Pdi1p preferentially demannosylates partially structured RNase B variants. (A) Schematic presentation of the generation of RNase B variants. RNase BS and RBsp were generated through subtilisin and TCA treatment. Additionally, RNase B was reductively denatured by DTT in 6 M guanidine hydrochloride and then was either disulfide-scrambled by diamide to produce Scr-RB or cysteine-carbamidomethylated by iodoacetamide (IAA) to produce Carb-RB. (B) Circular dichroism analysis of RNase B variants. (C) The relative abundance of Man7 on RNase B variants during a time-course incubation with Htm1p–Pdi1p. Shown are the mean values ± one SD from an experiment performed in triplicate.
Fig. S9.
Folding states of RNase B variants. (A) Each RNase B variant (10 µM) was treated with 50 ng of trypsin in a total volume of 30 µL at room temperature. At the indicated time points, 10-µL samples were collected and mixed with PMSF to stop proteolysis for subsequent SDS/PAGE analysis. (B) Each RNase B variant (10 µM) was treated with 1 unit of PNGase F in a total volume of 30 µL at 37 °C. At the indicated time points, 10-µL samples were collected and mixed with 1× SDS/PAGE sample buffer to stop deglycosylation for subsequent SDS/PAGE analysis. (C) Size distribution of Scr-RB before (control) and after (Overnight) overnight incubation with Htm1p–Pdi1p at 30 °C, as analyzed by Superdex 200 size-exclusion chromatography. (D) Size distribution of Carb-RB before (control) and after (Overnight) overnight incubation with Htm1p–Pdi1p at 30 °C, as analyzed by Superdex 200 size-exclusion chromatography.
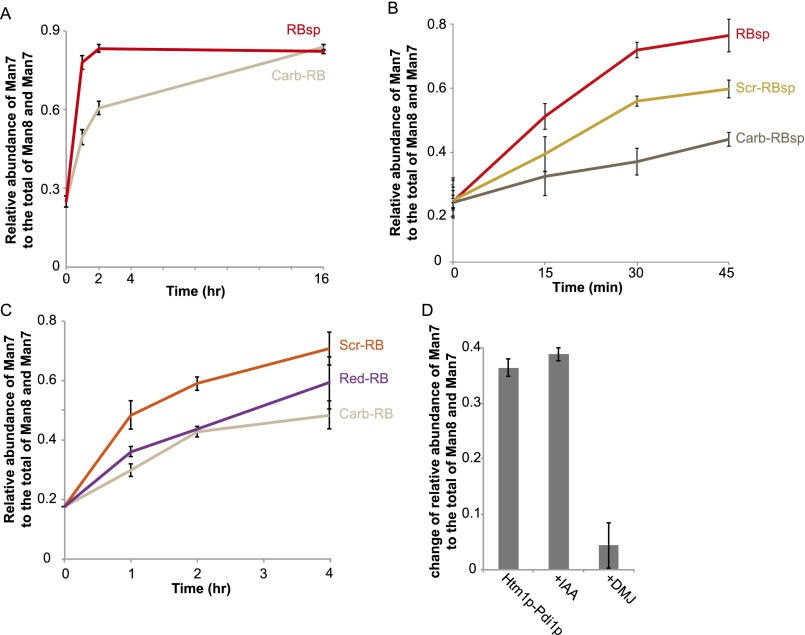
Analysis of the demannosylation of RNase B variants by Htm1p–Pdi1p revealed that Htm1p–Pd1ip was able to distinguish differences in the conformations of these variants (Fig. 7C). Htm1p–Pdi1p showed minimal mannosidase activity on the native-like RNase BS, suggesting that Htm1p–Pdi1p is insensitive to the mild increase in peptide flexibility around the N-glycosylation site of RNase BS (42). For nonnative variants, we observed a clear trend of increasing demannosylation efficiency as one goes from the globally unfolded Carb-RB to the more compact but heterogeneous Scr-RB and finally to the most compact and homogenous form, RBsp. Demannosylation of Carb-RB continued during prolonged incubation (Fig. S10A), suggesting that the lower levels of demannosylation of Carb-RB at earlier time points resulted from slower kinetics. The susceptibility of Scr-RBsp and Carb-RBsp to Htm1p–Pdi1p was similar to that of their full-length counterparts, suggesting that the difference in the kinetics was not caused by the absence of the S peptide per se (Fig. S10B). The slower kinetics against Carb-RB was not caused by alkylation of cysteines, because a fully reduced RNase B (Red-RB) was demannosylated at a rate similar to that of Carb-RB (Fig. S10C). In addition, Htm1p–Pdi1p was not inhibited when its cysteines were first blocked with iodoacetamide before reaction with RBsp, ruling out the possibility of a cysteine-mediated demannosylation mechanism (Fig. S10D). To conclude, our findings demonstrate not only that Htm1p–Pdi1p differentiates nonnative proteins from native ones but also that it prefers nonnative proteins with compact, partially structured conformations over globally unfolded ones. Our findings support our second model, in which Htm1p–Pdi1p is a folding-sensitive mannosidase and preferentially targets nonnative proteins trapped at partially structured states.
Fig. S10.
Additional experiments of the demannosylation of RNase B variants by Htm1p–Pdi1p. (A) The change in the relative abundance of Man7 on RBsp and Carb-RB during the time course of incubation with Htm1p–Pdi1p. Shown are the mean values ± one SD from an experiment performed in triplicate. (B) The changes in the relative abundance of Man7 on Scr-RBsp and Carb-RBsp during the time course of incubation with Htm1p–Pdi1p in comparison with the abundance of Man7 on RBsp. Shown are the mean values ± one SD from an experiment performed in triplicate. (C) The change in the relative abundance of Man7 on Red-RB during the time course of incubation with Htm1p–Pdi1p, in comparison with the abundance of Man7 on Scr–RBsp and Carb–RBsp. Shown are the mean values ± one SD from an experiment performed in triplicate. (D) The change in the relative abundance of Man7 on RBsp after a 30-min incubation with Htm1p–Pdi1p in the absence or presence of 10 mM iodoacetamide (+IAA) or 10 mM 1-deoxymannojirimicin (+DMJ). Shown are the mean values ± one SD from an experiment performed in triplicate.
Discussion
A fundamental question of ERAD is what biochemical properties differentiate an ERAD substrate from a normal protein in the ER (3, 43). The unique requirement of an N-glycan remodeling step for ERAD-L commitment potentially provides a chemical handle for investigating the biochemical basis that determines an ERAD-L substrate. This study provides biochemical evidence supporting a model in which this commitment step by Htm1p–Pdi1p is closely coordinated with the conformations of potential ERAD-L substrates. As such, the resulting terminally exposed α1,6-linked mannose is suitable to be a folding-state mark that flags folding defects in the attached proteins.
Several lines of biochemical evidence support our conclusion that Htm1p–Pdi1p is a folding-sensitive mannosidase. First, we see the mannosidase activity of Htm1p–Pdi1p against misfolded forms of CPY and RNase B variants but not against free Man8, suggesting that, unlike Mns1p, Htm1p–Pdi1p is a glycoprotein-specific mannosidase. Second, we see a marked enhancement of Htm1p–Pdi1p demannosylation activity when native CPY and native RNase B are artificially converted to misfolded forms, suggesting that Htm1p–Pdi1p prefers the nonnative folding states of proteins to their native forms. Third, we see significantly higher rates of Htm1p–Pdi1p activity against partially structured RNase B variants than against their globally unfolded forms, indicating that Htm1p–Pdi1p can distinguish different nonnative states. Our biochemical analysis of the conformational sensitivity of Hmt1p–Pdi1p is supported by our in vivo findings that reveal a higher abundance of steady-state Man7 on ER-retained CPY* than on CPY in wild-type yeast; this finding suggests that the generation of Man7 is not a passive, universal event of prolonged ER retention.
The observed preference for partially folded forms can explain why Htm1p–Pdi1p has higher mannosidase activity against Scr-CPY than against Carb-CPY. On the other hand, the absence of ER chaperones to keep CPY* in partially structured states may explain why Htm1p–Pdi1p is unable to exert its full activity on CPY* in our in vitro system (44). Because proteins populated at partially structured states have the propensity to form aggregates and amyloids (45, 46), this conformational preference of Htm1p–Pdi1p may ensure that proteins on the verge of aggregating are promptly committed for degradation. Additionally, this folding sensitivity of Htm1p–Pdi1p can ensure that nascent, globally unfolded polypeptides are allowed sufficient time for folding. The reconstitution system that we have established here provides a roadmap for future structural investigation of how the recognition of partially structured proteins is achieved.
Interestingly, this conformational preference is reminiscent of that of UDP-glucose glucosyltransferase (UGGT), which reglucosylates partially structured proteins to help their retention in the ER folding cycle (47). The presence of multiple N-glycan–remodeling enzymes with a similar preference for compact intermediates further argues for the broad biological importance of detecting partially structured proteins in the ER. The essential interaction of Htm1p with Pdi1p is also reminiscent of the predicted structure of UGGT, which contains three thioredoxin-linked domains that may contribute to peptide binding (48). Our discovery that Htm1p–Pdi1p can remove the α1,2-linked mannose from branch C on Man9 without the prior action of Mns1p suggests a future direction for applying synthetic Man9-glycoconjugates that were developed for the investigation of UGGT (49) for more detailed analysis of how the N-glycan is used as a potential time indicator by Htm1p–Pdi1p for glycoprotein quality control in the ER.
To conclude, in addition to the previously characterized ERAD-L surveillance step mediated by the Yos9p–Hrd3p complex (6, 7, 16, 50), here we provide evidence for an upstream folding surveillance step that is mediated by Htm1p–Pdi1p. The presence of multiple folding surveillance steps, each with its own unique N-glycan processing or recognition functions, supports a model in which ERAD-L follows a kinetic proofreading mechanism to achieve high fidelity in targeting the right proteins for degradation (6, 7, 51).
Materials and Methods
Yeast Strains.
All yeast transformations were conducted according to standard procedures. For chromosomal overexpression of HTM1 (yJW1819 and yJW1820), the chromosomal HTM1 locus was deleted from BY4741 by a URA3 selection marker. Double-stranded DNA starting with either the endogenous 5′ UTR promoter or a TDH3 promoter for overexpression, followed by the full coding region of wild-type or mutant HTM1, with a C-terminal 3xFLAG-HDEL tag, 3′ UTR of HTM1, and finally a KANMX selection marker was then introduced into the htm1Δ::URA3 strain. A pGAL1-MNS1-3xFLAG::KANMX strain (yJW1832) was generated by the same procedure for the preparation of 3xFLAG-tagged Mns1p that is controlled by a GAL1 promoter.
Plasmids.
The plasmid for GAL1 promoter-driven CPY* expression (pJW1528) was kindly provided by Tom Rapoport, Harvard Medical School, Boston. To generate a native CPY expression plasmid (pJW1529), PCR primers carrying the native G255 codon were used to amplify the G255R region from the CPY* expression plasmid and then were assembled into the same vector backbone by the Gibson method. To generate the CPY*0001 expression plasmid (pJW1530), PCR primers carrying Asn-to-Gln mutations of the first three N-glycosylation sites were used to make mutant fragments of CPY* by PCR amplification, which were assembled into the same 2-μm vector backbone by the Gibson method (52). The same procedure was used to generate the CPY*1110 plasmid (pJW1609).
Antibodies.
Anti-FLAG M2 mouse monoclonal antibody and the antibody-conjugated agarose beads were purchased from Sigma-Aldrich. Anti-yeast Pdi1p rabbit antisera was a gift from Peter Walter, University of California, San Francisco, which was confirmed by recombinantly expressed Pdi1p. IR fluorescence-conjugated secondary antibodies were purchased from LI-COR.
Recombinant Preparation of Htm1p–Pdi1p and Mns1p.
The yJW1820 yeast was grown to the stationary phase in 3 L of yeast extract-peptone-dextrose growth medium at 30 °C. Cells were harvested by centrifuging at 4,000 × g for 5 min, washed with cold deionized water, and stored at −80 °C. All the following purification steps were carried out at 4 °C. Approximately 15 g of cell pellet was mixed with 5 g of dry ice and then was ground by a Proctor Silex E160BY grinder for five 30-s on/off cycles. The ground cell was vacuum-degassed for 5 min to remove residual dry ice and was thawed and resuspended in 25 mL of lysis buffer [20 mM Hepes (pH 7.0), 150 mM NaCl, 2 mM CaCl2, 0.5% (vol/vol) Nonidet P-40, 15% (vol/vol) glycerol, 1× cOmplete EDTA-free protease inhibitor mixture (Roche Diagnostics)]. The slurry was centrifuged twice at 6,000 × g to remove cell debris, nutated for 30 min to solubilize the lipid membrane, and then ultracentrifuged at 60,000 × g for 30 min. The supernatant was collected and transferred into a new tube containing an ∼300-μL bed volume of anti-FLAG M2 affinity agarose beads (Sigma-Aldrich) that had been equilibrated with the lysis buffer. The mixture was nutated for 3 h and then was transferred to an open column. The column was washed with 3 mL of wash buffer 1 [20 mM Hepes (pH 7.0), 150 mM NaCl, 2 mM CaCl2, 1× cOmplete EDTA-free protease inhibitor mixture, 15% (vol/vol) glycerol], 1 mL of wash buffer 2 [20 mM Hepes (pH 7.0), 300 mM NaCl, 2 mM CaCl2, 15% (vol/vol) glycerol], and 1 mL of wash buffer 3 [20 mM Hepes (pH 7.0), 150 mM NaCl, 2 mM CaCl2, 20 mM imidazole, 15% (vol/vol) glycerol]. To release Htm1p–Pdi1p from the affinity beads, the column was closed by an end-cap, and 10 μg of Tobacco etch virus (TEV) protease and 300 μL of wash buffer 3 were added. After overnight incubation without agitation, the cleaved products were eluted with 400 μL of wash buffer 3. All eluent was collected into a bottom-sealed Micro Bio-Spin column (Bio-Rad) containing a 100-μL bed volume of Ni-NTA agarose beads (Qiagen) that had been equilibrated with wash buffer 3. After 30 min of incubation, the spin column was centrifuged at 50 × g for 10 s. The eluent was collected, and the Ni-NTA beads were further eluted two more times with 100 μL wash buffer 3 for full elution. The eluent was buffer-exchanged into HSCG buffer [20 mM Hepes (pH 7.0), 150 mM NaCl, 2 mM CaCl2, 15% (vol/vol) glycerol] on a PD MiniTrap G-25 column (GE Healthcare Life Sciences). Purified Htm1p–Pdi1p was stored in aliquots at −80 °C. Endoglycosidase H (New England Biolabs) treatment was carried out according to the manufacturer’s protocol. For size-exclusion column chromatography, Htm1p–Pdi1p was eluted with 500 µL of 1 mg/mL 3xFLAG peptide (Sigma-Aldrich) in wash buffer 3 instead of with TEV protease. The eluent then was loaded and separated in a Superdex 200 10/300 GL column (GE Healthcare Life Sciences) for subsequent Western blot and mannosidase assays. Protein concentration was determined by Pierce BCA protein assay (Thermo Fisher Scientific).
For Mns1p preparation, yJW1832 was grown in 3-L dextrose-free yeast/peptone with 2% (wt/vol) raffinose at 30 °C to the early stationary phase. Galactose then was added to a final concentration of 2% (wt/vol) to induce expression for 12 h. Then cells were harvested. Cell lysis and affinity purification were carried with the procedure used for Htm1p–Pdi1p.
Recombinant Preparation of CPY and CPY*.
CPY plasmids were introduced into the desired yeast strain, and the transformed cell was grown in 3-L synthetic complete (SC)-Leu with 2% (wt/vol) raffinose to an OD600 of 1.0∼1.2. Galactose was added to a final concentration of 2% (wt/vol) for another 12 h at 30 °C to induce expression. Cells then were harvested and stored at −80 °C. All the purification steps were carried out at 4 °C. For CPY*, 15 g of cells were ground and then resuspended in 25 mL C-lysis buffer [20 mM Hepes (pH 7.0), 500 mM NaCl, 10 mM imidazole, 10 mM Tris(2-carboxyethyl)phosphine (TCEP)⋅HCl, 6 M guanidine hydrochloride, 1% (vol/vol) Nonidet P-40, 1× cOmplete protease inhibitor mixture]. The slurry was nutated for 30 min to solubilize the inclusion body, centrifuged at 6,000 × g for 10 min, and then ultracentrifuged at 60,000 × g for 30 min. The supernatant was collected and loaded on a Ni-NTA column containing a 1-mL bed volume of resins that had been equilibrated with C-lysis buffer. The column was washed with 10 mL C-wash buffer [20 mM Hepes (pH 7.0), 500 mM NaCl, 10 mM imidazole, 10 mM TCEP⋅HCl, 6 M guanidine hydrochloride] and then eluted with C-elution buffer [20 mM Hepes (pH 7.0), 500 mM NaCl, 500 mM imidazole, 6 M guanidine hydrochloride]. The eluent was buffer-exchanged into HSGG buffer [20 mM Hepes (pH 7.0), 150 mM NaCl, 15% (vol/vol) glycerol, 0.1% (wt/vol) octyl-α-glucopyranoside] and stored at −80 °C in aliquots.
The purification of native CPY was performed in the same way as CPY* purification except that guanidine hydrochloride and TCEP⋅HCl were omitted from the C-lysis and C-wash buffer, respectively. To make Scr-CPY, CPY was first reduced with 6 M guanidine hydrochloride and 5 mM DTT at 42 °C for 1 h. N,N,N′,N′-tetramethylazodicarboxamide (diamide) (Santa Cruz Biotechnology) was added to a final concentration of 25 mM. The solution was kept at room temperature for 1 h and then was buffer-exchanged into HSGG buffer. To make Carb-CPY, CPY was first reduced with 6 M guanidine hydrochloride and 5 mM DTT at 42 °C for 1 h. Then iodoacetamide was added to 25 mM. The solution was kept at room temperature in the dark for 1 h and then was buffer-exchanged into HSGG buffer. Ellman’s reagent (Sigma-Aldrich) was used to check the status of cysteines.
Preparation of RNase B.
Crude bovine pancreatic RNase B (Sigma-Aldrich) was first enriched for the Man8GlcNAc2-abundant species. Generally, 50 mg of crude RNase B was dissolved in EQ buffer [20 mM Hepes (pH 7.0), 150 mM NaCl, 5 mM CaCl2, 5 mM MgCl2, 5 mM MnCl2] in a 10 mg/mL concentration and then was mixed with a 5-mL bed volume of EQ buffer-equilibrated Con A Sepharose beads (Sigma-Aldrich) at 4 °C for 2 h. The slurry was subsequently transferred into an open column, washed sequentially with 15 mL EQ buffer, 50 mL high-salt wash buffer [20 mM Hepes (pH 7.0), 500 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM MnCl2], 10 mL EQ buffer, and finally 200 mL low-Glc wash buffer [20 mM Hepes (pH 7.0), 150 mM NaCl, 50 mM Methyl-α-d-glucopyranoside]. The column was first eluted with 100 mL of 200-mM-Glc buffer [20 mM Hepes (pH 7.0), 150 mM NaCl, 200 mM Methyl-α-d-glucopyranoside] into 20 mL per fraction and was further eluted with 100 mL 1 M Glc buffer [20 mM Hepes (pH 7.0), 150 mM NaCl, 1 M Methyl-α-d-glucopyranoside]. The N-glycan profile of RNase B from each step was checked by MALDI-TOF MS. Fractions containing the desired glycan species were pooled together, adjusted to pH 4.0 by acetic acid, and then loaded onto a 5-mL HiTrap-SP FF cation-exchange column (GE Healthcare Life Sciences) that had been equilibrated with acidic buffer [20 mM Hepes (pH 4.0), 150 mM NaCl]. The cation-exchange column was washed with 25 mL acidic buffer and then was eluted with neutral buffer [20 mM Hepes (pH 7.0), 500 mM NaCl]. The eluent was buffer-exchanged into HSG buffer [20 mM Hepes (pH 7.0), 150 mM NaCl, 15% (vol/vol) glycerol] by a PD10 desalting column (GE Healthcare Life Sciences). Purity was checked by size-exclusion column chromatography.
For the preparation of RBsp, 1 mg of RNase B was mixed with 20 μg of freshly dissolved subtilisin (Sigma-Aldrich) in 2 mL HSG buffer. The mixture was kept at 4 °C overnight, and 20 μg more subtilisin was added for another 1-h incubation at 4 °C. The pH of the mixture then was adjusted to 2.0 with 1 M hydrochloric acid for 1 h on ice to destroy subtilisin. Then trichloroacetic acid (TCA) was added to 10% (wt/vol), and the solution was warmed to room temperature to allow RBsp precipitation overnight. The mixture was centrifuged at 15,000 × g for 10 min, and the supernatant was removed. The pellet was dissolved with 9 M deionized urea and then was TCA-precipitated again to remove residual S-peptide completely. The pellet was dissolved with 9 M urea, buffer-exchanged into HSGG buffer, and stored in aliquots at −30 °C. Disulfide-scrambling and carbamidomethylation of RNase B and RBsp were carried out with the procedures used for Scr-CPY and Carb-CPY.
Circular Dichroism Analysis.
RNase B variants were buffer-exchanged into 10 mM sodium phosphate (pH 7.0) to a final concentration of 10 µM. Measurement was conducted on a Jasco J-715 spectrometer in a 1-mm cuvette at 30 °C. The spectrum was recorded over the range of 190–250 nm at a scanning speed of 20 nm/min with a bandwidth of 1.0 nm.
Limited Proteolysis of RNase B Variants.
Trypsin (Sigma-Aldrich) was added to 10 μM RNase B variants to a final concentration of 50 ng/μL at the beginning of the time-course reaction at room temperature. At each time point, an equal amount of sample was withdrawn and mixed with 1/10 volume of phenylmethylsulfonyl fluoride (Sigma-Aldrich) to stop the reaction for reducing SDS/PAGE analysis.
PNGase F Sensitivity of RNase B Variants.
Glycerol-free PNGase F (New England Biolabs) was added to 10 μM RNase B variants to a final concentration of 0.5 units/μL at the beginning of the time-course reaction at room temperature. At each time point, an equal amount of sample was withdrawn and mixed with 4× SDS sample buffer to stop the reaction for reducing SDS/PAGE analysis.
Glycan Profiling by MALDI-TOF MS.
The procedure for N-glycan profiling by MALDI-TOF MS was based on published methods (53, 54) with some modifications. N-glycoproteins were separated by SDS/PAGE and stained by Coomassie Blue R250. Individual gel bands were excised and destained twice with 100 μL of 50% (vol/vol) acetonitrile with 10 mM Na2CO3 at 50 °C for 30 min with vigorous shaking. The gels then were reduced by 50 mM DTT in 20 mM Na2CO3 at 50 °C for 30 min and were alkylated by adding iodoacetamide to a final concentration of 150 mM and incubated in the dark at room temperature for 30 min. The gels were washed twice by 50% (vol/vol) acetonitrile with 10 mM Na2CO3 at room temperature for 30 min, dehydrated by 100% acetonitrile at room temperature for 10 min, and evaporated in a centrifugal evaporator for 10 min. To each dried gel piece, five units of glycerol-free PNGase F (New England Biolabs) in 20 μL of 20 mM Na2CO3 were added, and the mixture was incubated at 37 °C overnight. To extract the released glycans, 100 μL of deionized water was added, and the mixture was sonicated in the water-bath sonicator for 30 min. The solution was collected, and extraction was repeated two more times. To desalt the pooled extract, each sample was loaded into a 20-μL filter tip filled with 10 mg of graphitized carbon (Grace Davidson Discovery Sciences) that had been sequentially washed with 1 mL of acetonitrile and 1 mL of deionized water. After the sample was loaded, the tip-column was washed with 1 mL of deionized water and eluted with 100 μL of 25% (vol/vol) acetonitrile. For free glycans, the reaction solution was loaded directly onto the graphitized carbon tip-column and processed with the same procedure. The eluent was evaporated in the centrifugal evaporator at 60 °C. The dried sample was resuspended with 5 μL of deionized water, and 1 μL of the sample was spotted on the MALDI target plate and vacuum dried. The plate then was washed with pure acetonitrile. Then 1 μL of 5 mg/mL 2.5-dihydroxybenzoic acid (DHB) (Sigma-Aldrich) dissolved in 50% (vol/vol) acetonitrile was spotted onto each sample spot and dried. The mass spectrometric analysis was conducted on a Voyager Elite DE-STR Pro mass spectrometer (Applied Biosystems) or a Shimadzu AXIMA Performance mass spectrometer (Shimadzu) in the positive reflectron mode. For data collected in the Voyager mass spectrometer, the spectrum analysis was carried out with the bundled Data Explorer software to extract the area of the peak matched to each glycan. For spectra collected from the AXIMA Performance mass spectrometer, raw spectra were exported in the mzXML format and were analyzed by mMass software (55) to extract the intensity of the peak matched to each glycan.
Mannosidase Assay.
Unless otherwise specified, glycoprotein substrates in the specified concentration (2 μM for CPY variants, 10 μM for RNase B isoforms) were mixed with 0.1 µM Htm1p–Pdi1p and/or 0.1 µM Mns1p in reaction buffer [20 mM Hepes (pH 7.0), 150 mM NaCl, 2 mM CaCl2, 0.1% (vol/vol) Nonidet P-40]. At each desired time point, EDTA was added to a final concentration of 25 mM to stop the demannosylation reaction. The mixture was separated by reducing SDS/PAGE and then was subjected to N-glycan profiling by MALDI-TOF MS.
NMR Analysis.
For the Man8-abundant RNase B control, 2 mg of RNase B was reduced by 10 mM DTT at 95 °C for 10 min and then was allowed to cool. For the RBsp treated with Htm1p–Pdi1p, 2 mg of RBsp was first incubated overnight with 50 µg of Htm1p–Pdi1p. To both samples, 1,000 units of PNGase F were then added for glycan release at 37 °C overnight. The next day, the reaction mixture was loaded onto a 200-mg graphitized carbon column, washed with 5 mL of water, and then eluted with 1.5 mL of 25% acetonitrile. The eluent was lyophilized and then dissolved by D2O (Sigma-Aldrich) for lyophilization two more times. The dried sample then was dissolved by D2O for NMR measurement. The 1H-NMR was carried out on a 400-MHz Bruker Avance III HD two-channel instrument with a TopSpin v3.5 interface. Spectra were precollected using the default proton method, calibrated to the reference chemical shift of water (4.79 ppm), and then collected with the presaturation method as described by the manufacturer to suppress the signal from water.
Acknowledgments
We thank Tom Rapoport and Alexander Stein (Harvard Medical School) for providing the recombinant CPY plasmids and the detailed purification protocol; Peter Walter (University of California, San Francisco, UCSF) for providing anti-Pdi1p antibody; current and former members of the D.G.F. and J.S.W. laboratories for discussions and laboratory methods; Erin Quan Toyama and Jay Read for the initial tests on Htm1; Shoshana Bar-Nun (Tel Aviv University), Elizabeth Costa, Joshua Dunn, Christina Fitzsimmons, Calvin Jan, Kamena Kostova, Melanie Smith, Lindsey Pack, Dan Santos, and Erin Quan Toyama for critical comments on the manuscript; Yao-ming Huang (Kortemme group, UCSF) for assistance with circular dichroism analysis; and the UCSF Mass Spectrometry Facility, the DeGrado group, and the UCSF NMR laboratory for access to the mass spectrometers and the NMR instrument. This work was supported by a Howard Hughes Medical Institute International Student Research Fellowship (to Y.-C.L.); the Sandler Foundation-UCSF Program in Breakthrough Biomedical Research award (to D.G.F. and J.S.W.); the Howard Hughes Medical Institute (J.S.W.); and NIH Grant U01 GM098254 (to J.S.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608795113/-/DCSupplemental.
References
- 1.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4(3):181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 2.Fewell SW, Travers KJ, Weissman JS, Brodsky JL. The action of molecular chaperones in the early secretory pathway. Annu Rev Genet. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- 3.Smith MH, Ploegh HL, Weissman JS. Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334(6059):1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126(2):361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 5.Vashist S, Ng DTW. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165(1):41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126(2):349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 7.Quan EM, et al. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32(6):870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakob CA, Burda P, Roth J, Aebi M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol. 1998;142(5):1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim W, Spear ED, Ng DTW. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19(6):753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19(6):741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 11.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19(6):765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 13.Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: Recognition and processing in the ER. Trends Biochem Sci. 2010;35(2):74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Clerc S, et al. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184(1):159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gauss R, Kanehara K, Carvalho P, Ng DTW, Aebi M. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol Cell. 2011;42(6):782–793. doi: 10.1016/j.molcel.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 16.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8(8):849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 17.Xie W, Kanehara K, Sayeed A, Ng DT. Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol Biol Cell. 2009;20(14):3317–3329. doi: 10.1091/mbc.E09-03-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakob CA, et al. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001;2(5):423–430. doi: 10.1093/embo-reports/kve089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakatsukasa K, Nishikawa S, Hosokawa N, Nagata K, Endo T. Mnl1p, an alpha-mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J Biol Chem. 2001;276(12):8635–8638. doi: 10.1074/jbc.C100023200. [DOI] [PubMed] [Google Scholar]
- 20.Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299(5611):1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- 21.Oda Y, Hosokawa N, Wada I, Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299(5611):1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- 22.Ninagawa S, et al. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J Cell Biol. 2014;206(3):347–356. doi: 10.1083/jcb.201404075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeiffer A, et al. A complex of Htm1 and the oxidoreductase Pdi1 accelerates degradation of misfolded glycoproteins. J Biol Chem. 2016;291(23):12195–12207. doi: 10.1074/jbc.M115.703256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denic V. No country for old misfolded glycoproteins. Mol Cell. 2011;42(6):715–717. doi: 10.1016/j.molcel.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell. 2014;158(6):1375–1388. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thaysen-Andersen M, Mysling S, Højrup P. Site-specific glycoprofiling of N-linked glycopeptides using MALDI-TOF MS: Strong correlation between signal strength and glycoform quantities. Anal Chem. 2009;81(10):3933–3943. doi: 10.1021/ac900231w. [DOI] [PubMed] [Google Scholar]
- 27.Endrizzi JA, Breddam K, Remington SJ. 2.8-A structure of yeast serine carboxypeptidase. Biochemistry. 1994;33(37):11106–11120. doi: 10.1021/bi00203a007. [DOI] [PubMed] [Google Scholar]
- 28.Sakoh-Nakatogawa M, Nishikawa S, Endo T. Roles of protein-disulfide isomerase-mediated disulfide bond formation of yeast Mnl1p in endoplasmic reticulum-associated degradation. J Biol Chem. 2009;284(18):11815–11825. doi: 10.1074/jbc.M900813200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winther JR, Sørensen P. Propeptide of carboxypeptidase Y provides a chaperone-like function as well as inhibition of the enzymatic activity. Proc Natl Acad Sci USA. 1991;88(20):9330–9334. doi: 10.1073/pnas.88.20.9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallee F, Karaveg K, Herscovics A, Moremen KW, Howell PL. Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases. J Biol Chem. 2000;275(52):41287–41298. doi: 10.1074/jbc.M006927200. [DOI] [PubMed] [Google Scholar]
- 31.Hosomi A, et al. Identification of an Htm1 (EDEM)-dependent, Mns1-independent Endoplasmic Reticulum-associated Degradation (ERAD) pathway in Saccharomyces cerevisiae: Application of a novel assay for glycoprotein ERAD. J Biol Chem. 2010;285(32):24324–24334. doi: 10.1074/jbc.M109.095919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kostova Z, Wolf DH. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J Cell Sci. 2005;118(Pt 7):1485–1492. doi: 10.1242/jcs.01740. [DOI] [PubMed] [Google Scholar]
- 33.Spear ED, Ng DTW. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol. 2005;169(1):73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallée F, et al. Crystal structure of a class I alpha1,2-mannosidase involved in N-glycan processing and endoplasmic reticulum quality control. EMBO J. 2000;19(4):581–588. doi: 10.1093/emboj/19.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chantret I, Kodali VP, Lahmouich C, Harvey DJ, Moore SEH. Endoplasmic reticulum-associated degradation (ERAD) and free oligosaccharide generation in Saccharomyces cerevisiae. J Biol Chem. 2011;286(48):41786–41800. doi: 10.1074/jbc.M111.251371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raines RT. Ribonuclease A. Chem Rev. 1998;98(3):1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 37.Ritter C, Helenius A. Recognition of local glycoprotein misfolding by the ER folding sensor UDP-glucose:glycoprotein glucosyltransferase. Nat Struct Biol. 2000;7(4):278–280. doi: 10.1038/74035. [DOI] [PubMed] [Google Scholar]
- 38.Trombetta ES, Helenius A. Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J Cell Biol. 2000;148(6):1123–1129. doi: 10.1083/jcb.148.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritter C, Quirin K, Kowarik M, Helenius A. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 2005;24(9):1730–1738. doi: 10.1038/sj.emboj.7600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González L, et al. Conformational studies of the Man8 oligosaccharide on native ribonuclease B and on the reduced and denatured protein. Arch Biochem Biophys. 2000;383(1):17–27. doi: 10.1006/abbi.2000.2031. [DOI] [PubMed] [Google Scholar]
- 41.Fu D, Chen L, O’Neill RA. A detailed structural characterization of ribonuclease B oligosaccharides by 1H NMR spectroscopy and mass spectrometry. Carbohydr Res. 1994;261(2):173–186. doi: 10.1016/0008-6215(94)84015-6. [DOI] [PubMed] [Google Scholar]
- 42.Blanchard V, Frank M, Leeflang BR, Boelens R, Kamerling JP. The structural basis of the difference in sensitivity for PNGase F in the de-N-glycosylation of the native bovine pancreatic ribonucleases B and BS. Biochemistry. 2008;47(11):3435–3446. doi: 10.1021/bi7012504. [DOI] [PubMed] [Google Scholar]
- 43.Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: Protein quality control and beyond. J Cell Biol. 2014;204(6):869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153(5):1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eichner T, Radford SE. A diversity of assembly mechanisms of a generic amyloid fold. Mol Cell. 2011;43(1):8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 46.Fink AL. Compact intermediate states in protein folding. Annu Rev Biophys Biomol Struct. 1995;24:495–522. doi: 10.1146/annurev.bb.24.060195.002431. [DOI] [PubMed] [Google Scholar]
- 47.D’Alessio C, Caramelo JJ, Parodi AJ. UDP-GlC:glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin Cell Dev Biol. 2010;21(5):491–499. doi: 10.1016/j.semcdb.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu T, Satoh T, Kato K. Structural insight into substrate recognition by the endoplasmic reticulum folding-sensor enzyme: Crystal structure of third thioredoxin-like domain of UDP-glucose:glycoprotein glucosyltransferase. Sci Rep. 2014;4:7322. doi: 10.1038/srep07322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito Y, Takeda Y, Seko A, Izumi M, Kajihara Y. Functional analysis of endoplasmic reticulum glucosyltransferase (UGGT): Synthetic chemistry’s initiative in glycobiology. Semin Cell Dev Biol. 2015;41:90–98. doi: 10.1016/j.semcdb.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 50.Smith MH, Rodriguez EH, Weissman JS. Misfolded proteins induce aggregation of the lectin Yos9. J Biol Chem. 2014;289(37):25670–25677. doi: 10.1074/jbc.M114.583344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hopfield JJ. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71(10):4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 53.Morelle W, Michalski J-C. Analysis of protein glycosylation by mass spectrometry. Nat Protoc. 2007;2(7):1585–1602. doi: 10.1038/nprot.2007.227. [DOI] [PubMed] [Google Scholar]
- 54.Packer NH, Lawson MA, Jardine DR, Redmond JW. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj J. 1998;15(8):737–747. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]
- 55.Strohalm M, Kavan D, Novák P, Volný M, Havlícek V. mMass 3: A cross-platform software environment for precise analysis of mass spectrometric data. Anal Chem. 2010;82(11):4648–4651. doi: 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]