Significance
Bacterial symbionts profoundly influence the biology of their animal hosts, yet complex interactions between animals and their resident bacteria often make it challenging to characterize the molecules and mechanisms. Simple model systems can reveal fundamental processes underlying interactions between eukaryotes and their associated microbial communities and provide insight into how bacteria regulate animal biology. In this study we isolate and characterize bacterial molecules that regulate multicellular development in the closest living relatives of animals, the choanoflagellate. We find that multiple bacterially derived lipids converge to activate, enhance, and inhibit choanoflagellate multicellular development.
Keywords: choanoflagellates, bacteria, host–microbe, sulfonolipid, multicellularity
Abstract
In choanoflagellates, the closest living relatives of animals, multicellular rosette development is regulated by environmental bacteria. The simplicity of this evolutionarily relevant interaction provides an opportunity to identify the molecules and regulatory logic underpinning bacterial regulation of development. We find that the rosette-inducing bacterium Algoriphagus machipongonensis produces three structurally divergent classes of bioactive lipids that, together, activate, enhance, and inhibit rosette development in the choanoflagellate Salpingoeca rosetta. One class of molecules, the lysophosphatidylethanolamines (LPEs), elicits no response on its own but synergizes with activating sulfonolipid rosette-inducing factors (RIFs) to recapitulate the full bioactivity of live Algoriphagus. LPEs, although ubiquitous in bacteria and eukaryotes, have not previously been implicated in the regulation of a host–microbe interaction. This study reveals that multiple bacterially produced lipids converge to activate, enhance, and inhibit multicellular development in a choanoflagellate.
The foundational event in animal origins—the transition to multicellularity (1–3)—occurred in oceans filled with diverse bacteria (4–7). There is a growing appreciation that specific bacteria direct diverse animal developmental processes, including light organ development in the Hawaiian bobtail squid and immune system development and maturation in organisms as diverse as cnidaria and mammals (8–20). However, the multicellularity of animals and the complex communities of bacteria with which they often interact hinder the complete characterization of many host–microbe dialogues.
Choanoflagellates, a group of microbial eukaryotes that are the closest living relatives of animals (21–24), promise to help illuminate the mechanisms by which bacteria influence animal development. As did cells in the first animals, choanoflagellates use a distinctive collar of actin-filled microvilli surrounding a flow-generating apical flagellum to capture bacteria as prey (25–27). Indeed, choanoflagellate-like cells likely formed the basis for the evolution of animal epithelial cells that today provide a selective barrier for mediating interactions with bacteria (27–29).
In many choanoflagellates, including Salpingoeca rosetta, a developmental program can be initiated such that single cells develop into multicellular rosettes. Importantly, rosette development does not occur through cell aggregation. Instead, as in the development of an animal from a zygote, rosettes develop from a single founding cell that undergoes serial rounds of oriented cell division, with the sister cells remaining stably adherent (Fig. 1). The orientation of the nascently divided cells around a central focus, the production of extracellular matrix, and the activity of a C-type lectin called Rosetteless, ultimately result in the formation of spherical, multicellular rosettes (30–32). Rosettes resemble morula-stage embryos, and the transition to multicellularity in S. rosetta evokes ancestral events that spawned the first animals (26, 27, 33).
Fig. 1.
Stages of rosette development in S. rosetta. During rosette development, a single founding cell undergoes serial rounds of cell division, resulting in a structurally integrated rosette. Importantly, rosette development does not involve cell aggregation. Shown are a single cell (A), a pair of cells (B), a 4-cell rosette (C), an 8-cell rosette (D) and a 16-cell rosette (E).
The initiation of rosette development was recently found to be induced by a coisolated environmental bacterium, Algoriphagus machipongonensis (phylum Bacteroidetes) (34, 35). The ecological relevance of the interaction between A. machipongonensis (hereafter, Algoriphagus) and S. rosetta is evidenced by the coexistence of these organisms in nature (35) and the predator–prey relationship between choanoflagellates and bacteria (25, 36). Indeed, rosettes likely have a fitness advantage over single cells in some environments, as multicellular choanoflagellates are predicted to produce increased flux of water past each cell (37), and prey capture studies reveal that rosettes collect more bacterial prey/cell/unit time than do single cells (38). However, in other environments, rosette development would likely reduce fitness as rosettes have reduced motility relative to single cells. Therefore, we hypothesize that choanoflagellates use bacterially produced molecules to identify environments in which rosette development might provide a fitness advantage.
The simplicity of the interaction between S. rosetta and Algoriphagus, in which both members can be cultured together or independently, offers a biochemically tractable model for investigating the molecular bases of bacteria–eukaryote interactions. Using rosette development as a bioassay, the first rosette-inducing molecule, Rosette Inducing Factor-1 (RIF-1), was isolated from Algoriphagus. The observation that RIF-1 fails to fully recapitulate the bioactivity of the live bacterium (Fig. 2 A and C) raised the possibility that additional molecules might be required (35). To gain a more complete understanding of the molecules and regulatory logic by which bacteria regulate rosette development, we set out to identify the minimal suite of molecules produced by Algoriphagus that are necessary and sufficient to regulate rosette development in S. rosetta.
Fig. 2.
Maximal rosette development requires lipid cofactor interactions. (A) When treated with media that lack necessary bacterial signals (Media Control), S. rosetta does not produce rosettes. In contrast, when treated with live Algoriphagus, Algoriphagus-conditioned media, OMVs from Algoriphagus, or bulk lipids extracted from Algoriphagus, rosettes develop at maximal (∼90% cells in rosettes) or near-maximal levels. (B) A heat map depicts the rosette-inducing activity of Algoriphagus lipid fractions used to treat SrEpac, either in isolation or in combination, at a final lipid concentration of 2 μg/mL Sulfonolipid-enriched fraction 11 was the only fraction sufficient to induce rosette development when tested alone (30% of cells in rosettes). Tests of each of the lipid fractions in combination revealed previously unidentified inhibitory and enhancing activity. Fractions 4 and 5 decreased rosette development (to 12% and 8%, respectively) in fraction 11-treated cells, whereas fraction 7 increased rosette development to 65%. (C) The RIF mix (solid square) and purified RIF-2 (solid circle) induced rosette development at micromolar concentrations. (Inset) RIF-1 (open circle) is active at femtomolar to nanomolar concentrations, but induces 10-fold lower levels of rosette development than RIF-2. The long gray box in the main graph indicates the range of concentrations at which RIF-1 is active and the range of its rosette-inducing activity. Rosette development was quantified 24 h after induction. Minor ticks on x axis are log-spaced.
Results
A Newly Identified Sulfonolipid Activates the Rosette Development Pathway.
To identify the minimal set of Algoriphagus molecules required for full rosette induction, we used a bioassay based on a coculture of S. rosetta with the non-rosette–inducing prey bacterium Echinocola pacifica (SI Appendix, SI Materials and Methods). This culture, called “SrEpac” (for S. rosetta + E. pacifica) (39), reproducibly yields high percentages of cells in rosettes (>80%) in response to live Algoriphagus, outer membrane vesicles (OMVs) isolated from Algoriphagus-conditioned medium (SI Appendix, SI Text), and bulk lipids extracted from Algoriphagus (Fig. 2A and SI Appendix, Fig. S1). In addition, incubation of SrEpac with the only previously known Rosette Inducing Factor, the sulfonolipid RIF-1, results in low but reproducible levels of rosette development (∼1.5% of cells in rosettes; Fig. 2C), consistent with previous results using S. rosetta grown with different bacteria (35).
Because bulk lipids extracted from Algoriphagus elicit the same rosette development response as live bacteria (Fig. 2A), we began by fractionating a bulk lipid extraction by reversed-phase high-performance liquid chromatography (HPLC) and testing the resulting 15 lipid fractions in SrEpac (SI Appendix, SI Materials and Methods). Only fraction 11 was sufficient to induce rosette development, whereas all other lipid fractions lacked rosette-inducing activity at all concentrations tested (Fig. 2B). To further separate and isolate the active molecules in fraction 11, we performed a subsequent round of reversed-phase HPLC and tested the resulting subfractions for activity in SrEpac. The rosette-inducing activity tracked with one subfraction (hereafter, “RIF mix”) that induced rosette development in 23.5% of cells (Fig. 2C and SI Appendix, Fig. S2). Structural analysis by NMR, high-resolution mass spectrometry (HRMS), and tandem mass spectrometry (MSMS) revealed that the RIF mix contained RIF-1 and two structurally related but previously uncharacterized sulfonolipids with approximate molecular weights of 605 Da and 593 Da (SI Appendix, Figs. S2–S17). Sulfonolipids are a largely uncharacterized class of molecules that are structurally similar to sphingolipids, a diverse group of molecules based on sphingoid bases that play structural roles in cell membranes and important nonstructural roles in signal transduction (40). Although sulfonolipids have been reported to contribute to the gliding motility of Bacteroidetes bacteria (41, 42), almost nothing is known regarding their potential roles as signaling molecules.
Additional activity-guided fractionation by HPLC allowed us to isolate pure samples of RIF-1 (35, 43) and of the 605-Da sulfonolipid. Purified RIF-1 induced maximal (∼1.5%) rosette development at femtomolar to nanomolar concentrations (Fig. 2C, Inset). In contrast, the purified 605-Da sulfonolipid (hereafter “RIF-2”) elicited sevenfold higher levels of rosette development (10.5% of cells in rosettes; Fig. 2C) than RIF-1, although at micromolar concentrations. The planar structure of RIF-2 (Fig. 3A) was determined by 1D and 2D NMR (SI Appendix, Table S1 and Figs. S3–S17) and was found to closely resemble RIF-1 with the exception of slight structural variations of the capnoid base, which contains a double bond at C-4 and a hydroxyl group at C-6.
Fig. 3.
Structural similarities and differences among RIFs, an inactive sulfonolipid, and the inhibitory capnine IOR-1. (A) The 3D structure of RIF-1 (43), compared with the proposed molecular structure of RIF-2, and (B) the structure of an inactive Algoriphagus sulfonolipid, Sulfobacin F (43). Shared features of Algoriphagus sulfonolipids include a fatty acid chain (shown in gray) and a capnoid base (shown in black). Distinguishing features between RIF-1 and RIF-2 (highlighted in red) include a double bond at position 4 and a hydroxyl group at position 6. The tight structure–activity relationships of RIF-1 and RIF-2 suggest a restricted set of interactions between these molecules and a binding target. No features are shared by RIF-1 and RIF-2 to the exclusion of Sulfobacin F. (C) The IOR-1 capnine antagonizes the rosette-inducing activity of RIFs. Like the capnoid base of RIF sulfonolipids, IOR-1 is composed of a sulfonic acid head group and a branched chain containing two -OH groups. Furthermore, the carbon chain length and branching pattern of IOR-1 matches the capnoid base in RIF-1 and -2. The similarities between IOR-1 and the RIFs raise the possibility that IOR-1 competitively inhibits RIF binding to a target receptor.
The remaining 593-Da sulfonolipid in the RIF mix is produced by Algoriphagus at low levels (approximately one-fifth the amount of RIF-2) and elutes closely to RIF-2 during fractionation. Although HRMS and high-resolution tandem mass spectrometry (HRMSMS) data suggest that this molecule is a sulfonolipid similar to RIF-1, low levels of production and coelution with RIF-2 prevented us from fully isolating and characterizing the activity of the 593-Da sulfonolipid (SI Appendix, Figs. S2 and S6). However, because the combination of RIF-2 and the 593-Da sulfonolipid induced rosettes at levels indistinguishable from those of RIF-2 alone (SI Appendix, Fig. S18), we infer that the rosette-inducing activity of the RIF mix is largely the product of RIF-2. Nonetheless, we note that the maximal level of rosette development induced by the RIF mix (Fig. 2C) is greater than the sum of purified RIF-1 + RIF-2 for reasons that we do not yet understand.
The discovery of RIF-2 revealed that RIF-1 is not the sole Algoriphagus-derived determinant of rosette development in S. rosetta. However, even the RIF mix, which contains both RIF-1 and RIF-2, failed to recapitulate the full level of rosette induction elicited by either intact Algoriphagus or bulk lipids extracted from Algoriphagus. Therefore, we hypothesized that additional molecular cues are required to fully potentiate the rosette-inducing activities of RIF-1 and -2.
Lipid Cofactors Inhibit and Enhance RIF Activity.
To identify potential cofactors of the RIFs, we mixed each of the 15 lipid fractions isolated from Algoriphagus in pairwise combinations and tested the mixtures at several concentrations in SrEpac (Fig. 2B and SI Appendix, SI Materials and Methods). We observed two types of cofactor activity: enhancing activity in fraction 7 and, unexpectedly, inhibitory activity in fractions 4 and 5. Importantly, the activities of these cofactor-containing fractions were evident only when tested in combination with fraction 11, which contained both RIF-1 and RIF-2.
The inhibitory activity observed in fractions 4 and 5 is the first example of a compound(s)—either isolated from Algoriphagus or commercially available—that specifically reduces levels of rosette development at concentrations that do not otherwise inhibit growth (SI Appendix, Table S2). Therefore, we used bioactivity-guided fractionation in the presence of RIF-2 to determine the molecular basis for inhibition. HRMS and NMR experiments, together with total synthesis (44), allowed us to propose the absolute structure for the 351-Da molecule (hereafter referred to as Inhibitor of Rosettes-1 (IOR-1); Fig. 3C). Comprehensive methods detailing IOR-1 isolation and structure determination, along with dose–response curves of IOR-1 in the presence of the RIF mix and RIF-2, are described in ref. 44.
Nanomolar concentrations of IOR-1 completely inhibit the ability of RIF-2 to induce rosette development and reduce rosette development in the presence of the RIF mix (SI Appendix, Fig. S19). IOR-1 is a capnine lipid that resembles the capnoid backbone of RIFs (Figs. 3 A and C). Thus, we hypothesize that IOR-1 antagonizes rosette development by competitively binding a RIF-2 target receptor. Because the RIF mix induces low levels of rosette formation in the presence of IOR-1, we infer that the combined effects of the RIFs are sufficient to partially overcome the presence of IOR-1.
In contrast to the inhibitory activity associated with IOR-1, lipid fraction 7 from Algoriphagus greatly enhanced rosette development when used in combination with the RIF-containing fraction 11 (Fig. 2B). Notably, fraction 7 did not contain any sulfonolipids, the only class of molecules previously known to regulate rosette development. After separating the components of fraction 7 by HPLC, we treated SrEpac with each subfraction in combination with the RIF mix and quantified the level of rosette development. The subfractions that enhanced rosette development in the presence of the RIF mix contained one or both of two lysophosphatidylethanolamines (LPEs) with molecular weights of 451 Da and 465 Da (hereafter referred to as LPE 451 and LPE 465, respectively; Fig. 4A).
Fig. 4.
LPEs synergize with RIFs to enhance rosette development. (A) The structures of LPE 451 and LPE 465 as determined by NMR and tandem mass spectrometry. LPE 451 and LPE 465 differ by only one methyl group (highlighted in red). (B) The addition of 2 μM LPE mix increases the maximal percentage of cells in rosettes in RIF-2–treated SrEpac from 10.5% (solid circle) to 52% (open circle) and the maximal inducing activity of the RIF mix from 23.5% (solid square) to 82% (open square) of cells in rosettes. Minor ticks on x axis are log-spaced.
As this class of molecules is well known, literature precedence allowed us to confirm the core LPE structure by NMR and tandem mass spectrometry (Fig. 4A and SI Appendix, Figs. S20–S27). We performed an olefin metathesis on the most active LPE fractions (45) to determine that the major species present (in both LPE 451 and LPE 465) contain a double bond between carbons 9 and 10, which is common for fatty acid chains of this length (SI Appendix, Fig. S28). Due to the difficulties associated with purifying these types of molecules, we were unable to completely exclude other LPE isoforms (which can differ in double-bond location or position on the glycerol backbone); however, multiple iterations of bioassay-guided fractionation consistently yielded a fraction from the purification process (hereafter, the “LPE mix”) in which 98% of the fraction was composed of LPEs 465 and 451 with the remaining 2% of the subfraction containing trace amounts of other structurally related LPE analogs. Importantly, no commercially available LPEs tested in combination with the RIF mix either activated or enhanced rosette development (SI Appendix, Table S2). Therefore, we infer that LPE 451, LPE 465, or both are responsible for the synergistic RIF-enhancing activity of the LPE mix. Furthermore, as with the RIFs (43) and IOR-1 (44), it appears that the enhancing activity of the LPEs results from a highly specific structure–activity relationship.
LPEs belong to a large and diverse class of deacylated phospholipids, called lysophospholipids, that include structural components of cellular membranes as well as biologically active lipid mediators (46, 47). Although LPEs are found in most bacterial and eukaryotic cell membranes and present in somewhat elevated concentrations in many marine and estuarine bacteria (48), little is known about how and in what contexts LPEs might act as signaling molecules (47, 49).
To characterize how LPEs regulate rosette development, we started by investigating the concentrations at which the LPE mix displayed maximal enhancing activity. In contrast with the 10.5% of cells in rosettes induced by 2 μM RIF-2 alone, treatment of SrEpac with 2 μM RIF-2 and micromolar concentrations of the LPE mix increased rosette development fivefold to 53% (Fig. 4B and SI Appendix, Fig. S19). Furthermore, maximal levels of rosette development elicited by the RIF mix + the LPE mix matched those induced by the Algoriphagus lipid extract (Fig. 2A and Fig. 4B).
Finally, we observed that LPEs also influence RIF potency. In bioassays in which the concentration of the LPE mix was held stable at 2 μM and the RIF mix or RIF-2 was titrated, the sensitivity of S. rosetta to the RIFs increased such that 25-fold less RIF mix and 3-fold less RIF-2 was required to achieve half-maximal induction (Fig. 4B). These results reveal that the rosette-inducing activity of Algoriphagus can be largely recapitulated with specific representatives from just two different classes of lipids: sulfonolipids and LPEs.
LPEs Promote a Previously Unidentified Maturation Step in Rosette Development.
Rosettes induced by live Algoriphagus bacteria or OMVs, lipid-rich vesicles isolated from Algoriphagus-conditioned media that fully recapitulate the inducing activity of live bacteria, are remarkably resistant to shear and can range in size from 4 cells, the minimum number of cells required to confirm the organized polarity of a rosette (SI Appendix, SI Materials and Methods), to as many as 50 cells. Because the rosette-inducing activity of OMVs is stable, highly reproducible, and equivalent to that of live bacteria, we used it as a positive control for the study of rosette cell number. Within just 22 h after treatment, OMV-induced rosettes were resistant to shear introduced by pipetting, and the median cell number per rosette was 8 cells, although some grew to as large as 16 cells/rosette (Fig. 5A). In contrast, treatment with purified RIF-2 resulted in rosettes that were sensitive to mechanical disruption; after exposure to shear, the median cell number per rosette was significantly smaller (four cells/rosette) than that induced by OMVs released by Algoriphagus (eight cells/rosette; Fig. 5A). Furthermore, the size frequency distribution for RIF-2–induced rosettes was restricted to small rosettes, ranging from the minimum size of 4 cells up to 8 cells, compared with Algoriphagus- and OMV-induced cultures in which larger rosettes of 10–16 cells were frequently observed. Because the combinatorial activity of RIF-2 + LPE mix resulted in elevated percentages of cells in rosettes, we hypothesized that LPEs might promote rosette stability and therefore protect larger rosettes when exposed to shear. Indeed, the median cell number (seven cells/rosette) and size frequency distribution of SrEpac induced by RIF-2 + LPE mix was statistically indistinguishable from OMV-induced cultures (Fig. 5A).
Fig. 5.
LPEs promote proper rosette development and maturation. (A) Frequency distribution of rosette size in SrEpac incubated with OMVs, RIF-2, and RIF-2 + LPE mix after exposure to shear stress by pipetting. Rosettes induced by RIF-2 alone contained fewer cells on average and reached a smaller maximal size than rosettes induced with OMVs isolated from Algoriphagus-conditioned media. The addition of the LPE mix to RIF-2 increased the median rosette size and frequency distribution to levels that recapitulated induction by OMVs. Rosette size was assessed 22 h after induction (n = 139 for each condition). Violin box plots show the median cell number (white circle), 75% quartile (thick line), and range excluding outliers (thin line). Surrounding the box plot is a kernel density trace, plotted symmetrically to show rosette size frequency distribution. P values (unpaired t tests) were calculated using GraphPad Prism v6 for Mac, GraphPad Software. (B–E) Rosette morphology, cell packing, and localization of Rosetteless protein (a marker of rosette development) in rosettes induced by (B) OMVs, (C and D) RIF-2 alone, and (E) RIF-2 + LPEs. (B) Cells in OMV-induced rosettes express Rosetteless and are tightly packed. Anti-tubulin antibodies (white) highlight the cell body, and anti-Rosetteless antibodies (magenta) stain the Rosetteless protein in the center of rosettes (32). (C) Four-celled rosettes induced by RIF-2 are tightly packed, whereas larger rosettes induced by RIF-2 alone (D) appear disorganized with cells spaced farther apart. (E) Rosettes induced by the RIF-2 + LPE mix are large and closely packed and phenocopy rosettes induced by OMVs. All rosettes were fixed 22 h after treatment.
The hypothesis that RIF-2–induced rosettes exhibit less structural integrity than rosettes induced by either OMVs or RIF-2 + LPEs was supported by observations made using high-resolution microscopy (Fig. 5 B–E). Cells in OMV-induced rosettes were tightly packed and properly localized a specific marker of rosette development, the C-type lectin protein Rosetteless (32), to the extracellular matrix-rich center of the rosette (Fig. 5B). Although four-cell rosettes induced by RIF-2 alone showed close cell packing, cells in all larger rosettes induced by RIF-2 (e.g., those with five to seven cells/rosette) were spaced farther apart than those in OMV-induced rosettes of equivalent size (Fig. 5D). Despite a “loose” morphology, RIF-2–induced rosettes secreted Rosetteless protein, demonstrating that they had properly initiated rosette development. Importantly, induction with RIF-2 + the LPE mix restored a robust rosette morphology with the cells tightly packed together, phenocopying OMV-induced rosettes. Thus, although RIFs alone are sufficient to initiate rosette development, LPEs promote structural stability during rosette development and thereby facilitate rosette maturation (Fig. 6).
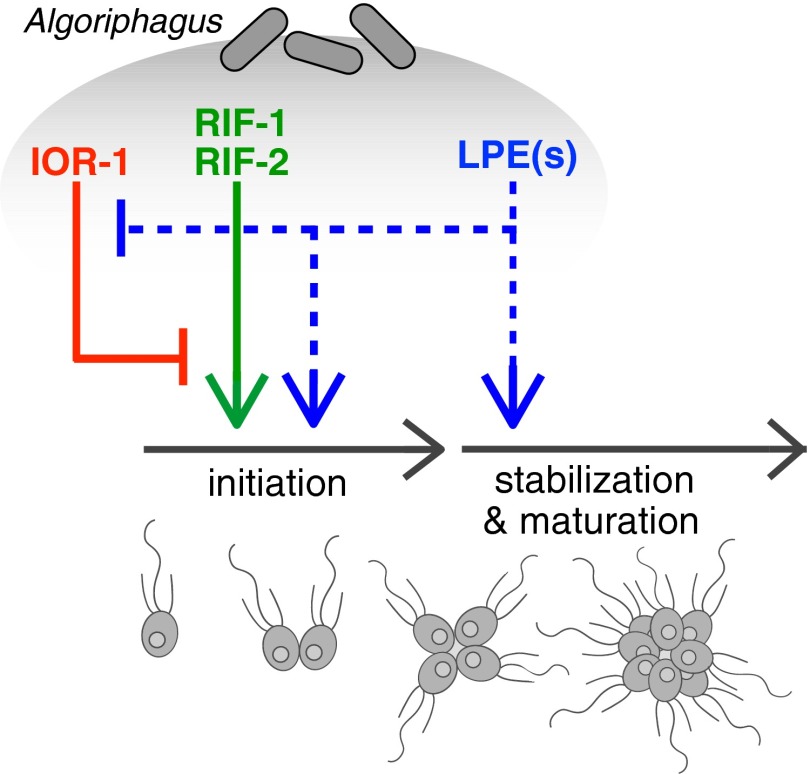
Fig. 6.
Multiple bacterial inputs regulate rosette development in S. rosetta. Algoriphagus produces three chemically distinct classes of lipids—sulfonolipids, LPEs, and a capnine—that interact to alternately induce, enhance, or inhibit rosette development in S. rosetta. The sulfonolipids RIF-1 and RIF-2 are sufficient to initiate rosette development in S. rosetta, although rosettes induced by RIFs alone are restricted in size, potentially because of their sensitivity to shear. Complete rosette maturation requires the synergistic activities of RIFs and LPEs. Although LPEs have no detectable activity on their own, they enhance RIF activity and facilitate the growth of larger and more structurally stable rosettes, perhaps by regulating downstream pathways important for rosette maturation. Although the molecular mechanisms by which LPEs regulate rosette development are unknown (indicated by dashed lines), multiple lines of evidence suggest that LPEs act both to promote the initiation of rosette development and, separately, to promote the subsequent maturation of rosettes. Algoriphagus also produces the inhibitory molecule IOR-1, which inhibits the rosette-inducing activity of RIFs (44). Importantly, when S. rosetta is exposed simultaneously to RIFs and the synergistic LPEs, mature rosettes develop even in the presence of IOR-1.
Discussion
Animals rely on bacteria for everything from proper metabolism to the stimulation of immune system development to the regulation of gut morphogenesis (19, 50). Bacterial cues even direct major life history transitions in animals with many marine invertebrates producing motile larvae that will not settle and undergo morphogenesis until they encounter the appropriate environmental bacteria (51). In one of the most dramatic examples of cross-talk between bacteria and an animal, Vibrio fischeri bacteria are recruited into crypts in the juvenile Hawaiian bobtail squid, where the bacteria then trigger postembryonic morphogenesis of the “light organ” (20). The widespread phylogenetic distribution of bacterially regulated developmental processes in animals suggests that such interactions may have been pivotal during the origin and early evolution of animals (19, 52).
As the number of animal developmental processes influenced by bacteria grows, detailed molecular characterization of the relevant bacterially produced cues promises to reveal the regulatory logic underlying host–microbe interactions. Through the study of rosette development in a close relative of animals, S. rosetta, we have found three classes of structurally distinct lipids produced by Algoriphagus that are interpreted by S. rosetta as activators, synergistic enhancers, and inhibitors of development (Fig. 6). When tested alone, activating RIFs elicit relatively low levels of rosette development and the synergistic LPEs have no detectable activity. However, when used in combination, the activating RIFs + synergizing LPEs induce levels of rosette development in S. rosetta that recapitulate those induced by live Algoriphagus (Figs. 2 and 4). Moreover, although the Algoriphagus capnine IOR-1 is a potent antagonist of the RIFs (44), the synergistic activities of the RIFs and LPEs overcome the inhibitory activities of IOR-1, potentially explaining why endogenous IOR-1 does not prevent robust rosette induction.
We hypothesize that the reliance of S. rosetta on multiple inputs from Algoriphagus prevents the developmental switch to rosette development under suboptimal conditions. The commitment to rosette development requires a trade-off: rosette development is a lengthy process and although rosettes are potentially more efficient than single cells in the capture of planktonic bacteria (36, 37), they are poor swimmers (53) and therefore likely to be less effective at dispersal and escape from certain predators (e.g., amoebae). Moreover, the aquatic world in which choanoflagellates live is patchy (54) with the diversity and density of bacteria dramatically varying between local microenvironments. In animals, the integration of multiple signals is fundamental to the robustness of many developmental decisions, including the establishment of the body axis during early embryogenesis (55–58) and the progressive specification of cell fates (59–61). Likewise, the multi-input regulatory module that controls development in S. rosetta may act to ensure that rosette development is not initiated under the wrong environmental conditions or in response to the wrong bacterial cues.
The integration of multiple bacterial inputs is also essential for proper animal development in two well-studied host–microbe models. In the Hawaiian bobtail squid, two molecules (LPS and TCT) produced by V. fischeri act synergistically to trigger light organ maturation (10), and in mice, several bacterial cell-wall molecules (LPS, PGN, and polysaccharide A) together shape the development of the immune system of the gut (12, 13, 62). The finding that rosette development in S. rosetta requires the integration of a network of bacterial lipids extends this phenomenon to the closest living relatives of animals. Ultimately, as the molecular underpinnings of more host–microbe interactions are fully elucidated, the mechanisms by which bacteria influence their animal hosts may be found to be as intricate and complex as those regulating animal development, with microbial communities providing mixtures of activating, enhancing, and inhibitory cues.
Materials and Methods
Detailed methods are provided in SI Appendix, SI Materials and Methods.
Choanoflagellate Husbandry and Rosette Development Bioassays.
SrEpac was propagated in 5% Sea Water Complete media. For all rosette development bioassays, cultures of SrEpac were diluted to a density of ∼104–105 cells/mL before treatment with bacterial fractions and purified lipids.
Quantifying Rosette Development.
To determine the percentage of cells in rosettes, the relative numbers of single cells and cells within rosettes were scored until a total of 500 cells had been counted per technical replicate. To quantify rosette size, the number of cells in each rosette was counted for each group of four or more cells with organized polarity relative to a central focus (with each cell oriented with the apical flagellum pointing outward) after exposure to shear. Rosette integrity was also characterized by immunofluorescence microscopy.
Fractionation, Isolation, and Characterization of Algoriphagus Lipids.
Ethyl-acetate–extracted Algoriphagus lipids were separated by reversed-phase HPLC and tested for induction of rosette development in SrEpac. Subsequent rounds of activity-guided fractionation led to preparations that were sufficiently pure for characterization by LC-MS/MS and NMR.
Supplementary Material
Acknowledgments
We thank M. Abedin, D. Booth, B. Davidson, and J. Rawls for critical reading of the manuscript; R. Alegado for early discussions; L. Wetzel for help with “violin” plots; and J. Wang at the Harvard Small Molecule Mass Spec Facility for help with high-resolution mass spectrometry. N.K. is a senior fellow in the Integrated Microbial Biodiversity Program of the Canadian Institute for Advanced Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605015113/-/DCSupplemental.
References
- 1.Carroll SB. Chance and necessity: The evolution of morphological complexity and diversity. Nature. 2001;409(6823):1102–1109. doi: 10.1038/35059227. [DOI] [PubMed] [Google Scholar]
- 2.Tyler S. Epithelium: The primary building block for metazoan complexity. Integr Comp Biol. 2003;43(1):55–63. doi: 10.1093/icb/43.1.55. [DOI] [PubMed] [Google Scholar]
- 3.King N. The unicellular ancestry of animal development. Dev Cell. 2004;7(3):313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Stanley SM. An ecological theory for the sudden origin of multicellular life in the late precambrian. Proc Natl Acad Sci USA. 1973;70(5):1486–1489. doi: 10.1073/pnas.70.5.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McFall-Ngai M. The development of cooperative associations between animals and bacteria: Establishing détente among domains. Am Zool. 1998;38(4):592–608. [Google Scholar]
- 6.Narbonne GM. The Ediacara biota: Neoproterozoic origin of animals and their ecosystems. Annu Rev Earth Planet Sci. 2005;33(1):421–442. [Google Scholar]
- 7.Knoll AH. The multiple origins of complex multicellularity. Annu Rev Earth Planet Sci. 2011;39(1):217–239. [Google Scholar]
- 8.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292(5519):1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 9.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99(24):15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koropatnick TA, et al. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306(5699):1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 11.Webster NS, et al. Metamorphosis of a scleractinian coral in response to microbial biofilms. Appl Environ Microbiol. 2004;70(2):1213–1221. doi: 10.1128/AEM.70.2.1213-1221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456(7221):507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 14.Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4570–4577. doi: 10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss BL, Maltz M, Aksoy S. Obligate symbionts activate immune system development in the tsetse fly. J Immunol. 2012;188(7):3395–3403. doi: 10.4049/jimmunol.1103691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch TCG. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu Rev Microbiol. 2013;67(1):499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- 17.An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156(1-2):123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shikuma NJ, et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science. 2014;343(6170):529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254(5037):1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 21.Lang BF, O’Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr Biol. 2002;12(20):1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 22.Carr M, Leadbeater BSC, Hassan R, Nelson M, Baldauf SL. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci USA. 2008;105(43):16641–16646. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451(7180):783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25(4):664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- 25.Leadbeater BSC, Morton C. A microscopical study of a marine species of Codosiga James-Clark (Choanoflagellata) with special reference to the ingestion of bacteria. Biol J Linn Soc Lond. 1974;6(4):337–347. [Google Scholar]
- 26.Nielsen C. Six major steps in animal evolution: Are we derived sponge larvae? Evol Dev. 2008;10(2):241–257. doi: 10.1111/j.1525-142X.2008.00231.x. [DOI] [PubMed] [Google Scholar]
- 27.Richter DJ, King N. The genomic and cellular foundations of animal origins. Annu Rev Genet. 2013;47:509–537. doi: 10.1146/annurev-genet-111212-133456. [DOI] [PubMed] [Google Scholar]
- 28.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8(6):411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 29.Arendt D, Benito-Gutierrez E, Brunet T, Marlow H. Gastric pouches and the mucociliary sole: Setting the stage for nervous system evolution. Philos Trans R Soc Lond, B, Biol Sci. 2015;370(1684):20150286. doi: 10.1098/rstb.2015.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fairclough SR, Dayel MJ, King N. Multicellular development in a choanoflagellate. Curr Biol. 2010;20(20):R875–R876. doi: 10.1016/j.cub.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dayel MJ, et al. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol. 2011;357(1):73–82. doi: 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin TC, Greaney AJ, Wetzel L, King N. The Rosetteless gene controls development in the choanoflagellate S. rosetta. eLife. 2014;3:e04070. doi: 10.7554/eLife.04070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haeckel E. The gastrea-theory, the phylogenetic classification of the animal kingdom and the homology of the germ-lamelle. Q J Microsc Sci. 1874;14:142–165. [Google Scholar]
- 34.Alegado RA, et al. Complete genome sequence of Algoriphagus sp. PR1, bacterial prey of a colony-forming choanoflagellate. J Bacteriol. 2011;193(6):1485–1486. doi: 10.1128/JB.01421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alegado RA, et al. A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. eLife. 2012;1:e00013. doi: 10.7554/eLife.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dayel MJ, King N. Prey capture and phagocytosis in the choanoflagellate Salpingoeca rosetta. PLoS One. 2014;9(5):e95577. doi: 10.1371/journal.pone.0095577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roper M, Dayel MJ, Pepper RE, Koehl MAR. Cooperatively generated stresslet flows supply fresh fluid to multicellular choanoflagellate colonies. Phys Rev Lett. 2013;110(22):228104. doi: 10.1103/PhysRevLett.110.228104. [DOI] [PubMed] [Google Scholar]
- 38.Kreft JM. 2010. Effects of forming multicellular colonies or attaching to surfaces on feeding rates of the choanoflagellate Salpingoeca rosetta, ed Kohl M. MA thesis (University of California, Berkeley)
- 39.Levin TC, King N. Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta. Curr Biol. 2013;23(21):2176–2180. doi: 10.1016/j.cub.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartke N, Hannun YA. Bioactive sphingolipids: Metabolism and function. J Lipid Res. 2009;50(Suppl):S91–S96. doi: 10.1194/jlr.R800080-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbanat DR, Godchaux W, III, Polychroniou G, Leadbetter ER. Biosynthesis of a sulfonolipid in gliding bacteria. Biochem Biophys Res Commun. 1985;130(2):873–878. doi: 10.1016/0006-291x(85)90497-8. [DOI] [PubMed] [Google Scholar]
- 42.Godchaux W, III, Leadbetter ER. Sulfonolipids are localized in the outer membrane of the gliding bacterium Cytophaga johnsonae. Arch Microbiol. 1988;150:42–47. [Google Scholar]
- 43.Beemelmanns C, et al. Synthesis of the rosette-inducing factor RIF-1 and analogs. J Am Chem Soc. 2014;136(29):10210–10213. doi: 10.1021/ja5046692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantley AM, Woznica A, Beemelmanns C, King N, Clardy J. Isolation and synthesis of a bacterially-produced inhibitor of rosette development in choanoflagellates. J Am Chem Soc. 2016;138(13):4326–4329. doi: 10.1021/jacs.6b01190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon Y, Lee S, Oh D-C, Kim S. Simple determination of double-bond positions in long-chain olefins by cross-metathesis. Angew Chem Int Ed Engl. 2011;50(36):8275–8278. doi: 10.1002/anie.201102634. [DOI] [PubMed] [Google Scholar]
- 46.Ishii I, Fukushima N, Ye X, Chun J. Lysophospholipid receptors: Signaling and biology. Annu Rev Biochem. 2004;73:321–354. doi: 10.1146/annurev.biochem.73.011303.073731. [DOI] [PubMed] [Google Scholar]
- 47.Makide K, et al. Novel lysophosphoplipid receptors: Their structure and function. J Lipid Res. 2014;55(10):1986–1995. doi: 10.1194/jlr.R046920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foster JW, Spector MP. Lipids and sterols. In: Moat A, editor. Microbial Physiology. Wiley-Liss; New York: 2003. pp. 450–472. [Google Scholar]
- 49.Grzelczyk A, Gendaszewska-Darmach E. Novel bioactive glycerol-based lysophospholipids: New data—new insight into their function. Biochimie. 2013;95(4):667–679. doi: 10.1016/j.biochi.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 50.Sommer F, Bäckhed F. The gut microbiota: Masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 51.Hadfield MG. Biofilms and marine invertebrate larvae: What bacteria produce that larvae use to choose settlement sites. Annu Rev Mar Sci. 2011;3(1):453–470. doi: 10.1146/annurev-marine-120709-142753. [DOI] [PubMed] [Google Scholar]
- 52.Alegado RA, King N. Bacterial influences on animal origins. Cold Spring Harb Perspect Biol. 2014;6(11):a016162. doi: 10.1101/cshperspect.a016162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirkegaard JB, Marron AO, Goldstein RE. Motility of colonial choanoflagellates and the statistics of aggregate random walkers. Phys Rev Lett. 2016;116(3):038102. doi: 10.1103/PhysRevLett.116.038102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stocker R. Marine microbes see a sea of gradients. Science. 2012;338(6107):628–633. doi: 10.1126/science.1208929. [DOI] [PubMed] [Google Scholar]
- 55.Kimelman D, Kirschner M. Synergistic induction of mesoderm by FGF and TGF-beta and the identification of an mRNA coding for FGF in the early Xenopus embryo. Cell. 1987;51(5):869–877. doi: 10.1016/0092-8674(87)90110-3. [DOI] [PubMed] [Google Scholar]
- 56.Ruiz i Altaba A, Melton DA. Interaction between peptide growth factors and homoeobox genes in the establishment of antero-posterior polarity in frog embryos. Nature. 1989;341(6237):33–38. doi: 10.1038/341033a0. [DOI] [PubMed] [Google Scholar]
- 57.Bachiller D, et al. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403(6770):658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- 58.Niehrs C. Regionally specific induction by the Spemann-Mangold organizer. Nat Rev Genet. 2004;5(6):425–434. doi: 10.1038/nrg1347. [DOI] [PubMed] [Google Scholar]
- 59.Carmena A, Gisselbrecht S, Harrison J, Jiménez F, Michelson AM. Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 1998;12(24):3910–3922. doi: 10.1101/gad.12.24.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson EH, Cameron RA, Ransick A. Specification of cell fate in the sea urchin embryo: Summary and some proposed mechanisms. Development. 1998;125(17):3269–3290. doi: 10.1242/dev.125.17.3269. [DOI] [PubMed] [Google Scholar]
- 61.Halfon MS, et al. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103(1):63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 62.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.