Abstract
We describe a new experimental approach to investigate touch sensation in the model organism C. elegans using light field deconvolution microscopy. By combining fast volumetric image acquisition with controlled indentation of the organism using a high sensitivity force transducer, we are able to simultaneously measure activity in multiple touch receptor neurons expressing the calcium ion indicator GCaMP6s. By varying the applied mechanical stimulus we show how this method can be used to quantify touch sensitivity in C. elegans. We describe some of the challenges of performing light field calcium imaging in moving samples and demonstrate that they can be overcome by simple data processing.
OCIS codes: (180.2520) Fluorescence microscopy, (170.6900) Three-dimensional microscopy, (170.3880) Medical and biological imaging
1. Introduction
Mechanosensation, the sensory faculty by which an organism is able to detect and respond to mechanical stimuli, is essential for a wide range of behaviours and regulatory processes. Touch sensation, in particular, is critical in enabling an organism to gather information and interact with its external environment [1–3]. However, despite substantial research [4–7] there remain considerable gaps in our understanding of the fundamental mechanisms by which mechanical stresses and strains give rise to neuronal and behavioural responses. Significant advances in our knowledge of the neurobiology of touch sensation have come from experimental studies performed using the nematode Caenorhabditis elegans [8–11]. C. elegans has a relatively simple, well-characterized anatomy and physiology and is optically transparent. Combined with the availability of a wide variety of mutants, which allow study of the function of various genes, many of which have homologs in the human genome, such properties make it well suited to studies of touch sensation.
Fundamental to an understanding of touch sensation is the relationship between the applied stimulus, the biomechanical properties of the organism and the resulting neuronal response. Genetically encoded calcium indicators (GECIs), such as the range of GCaMP fluorescent reporters [12], are powerful tools for monitoring the activity of individual neurons. By modifying an organism to express GECIs, neuronal activity can be inferred from measured variations in brightness of individual touch receptor neurons (TRNs) in time-lapse fluorescence microscopy images. Critically for mechanosensation studies, and in contrast to alternative methods such as patch-clamp electrophysiology [13], this can be achieved without compromising the structural integrity and biomechanical processes of the organism. However the use of GECIs also presents some significant experimental challenges. Principal among these is the need to simultaneously capture images of multiple neurons from across the organism at sufficient speed and spatial resolution, which is typically very difficult using conventional microscopy approaches.
In recent years a number of high speed volumetric microscopic imaging techniques have been developed and successfully used for calcium imaging, one notable example being light sheet microscopy (LSM) [14]. However, despite its speed, LSM is a sequential imaging technique with a 3D volume of the specimen reconstructed from a series of 2D images acquired at different times. Importantly for the present application, LSM systems typically require the use of two or more objective lenses, for formation of the light sheet and imaging of the illuminated volume, limiting scope for physical access to the sample. Multifocal microscopy [15] enables imaging of multiple planes throughout an extended volume using a custom diffraction grating, however is restricted in terms of the number and separation of image planes. Finally, rapid beam scanning techniques [16] have been demonstrated for calcium imaging, but the need to define neuron positions for fluorescence excitation presents problems when using such an approach with moving samples.
Light field microscopy [17] is an alternative volumetric imaging technique in which different perspective views of the sample are captured simultaneously by placing a microlens array in the native image plane of a conventional widefield microscope system. The information in a single light field image can then be used to render images focused to different depths within the specimen [18]. The trade off for this additional information is a significant reduction in lateral spatial resolution, owing to undersampling of the intermediate image by the lenslet array. However, it has recently been shown that exploiting the dense angular sampling of the object using 3D deconvolution can significantly increase spatial resolution [19], allowing reconstruction of a 3D image volume with sufficient spatial resolution to segment individual neurons [20].
In this article we describe an experimental setup for light field calcium imaging under controlled sample micromanipulation using a microforce sensing probe. We consider some of the particular challenges associated with calcium imaging in moving samples and apply the system to investigate the activation of posterior TRNs in C. elegans. We show that the system can be used to simultaneously monitor activity of multiple neurons in response to mechanical stimuli and how this capability can provide new insights into mechanosensation.
2. Materials and methods
2.1 Light field microscope and micromanipulation system
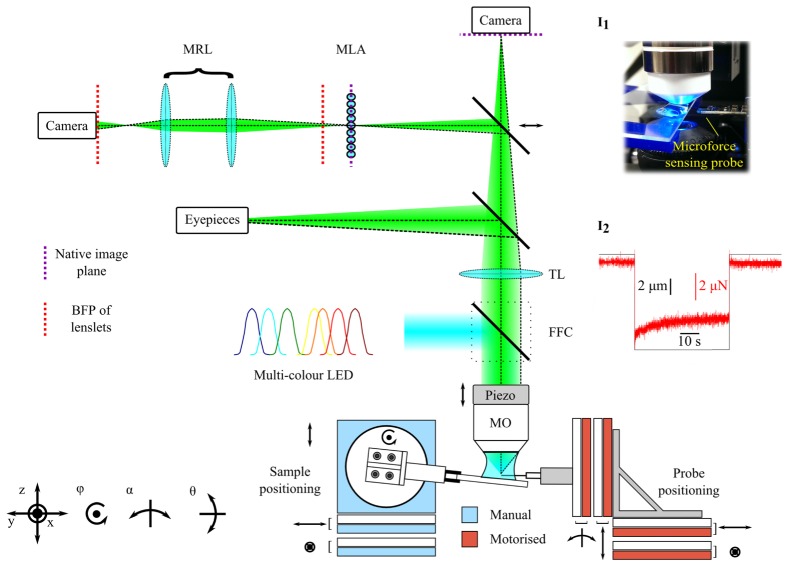
All mechanical stimulation and imaging experiments were performed using an upright epifluorescence microscope (BX51WI, Olympus) modified for light field imaging by inserting a microlens array (MLA-S125-F30, RPC Photonics) at the native image plane (Fig. 1). A macro relay lens with variable zoom and focus (MVL7000, Thorlabs) was used to image the back focal plane of the lenslets onto the image sensor of a scientific CMOS camera (ORCA-Flash 4.0v2, Hamamatsu Photonics). All these auxiallary components were mounted on an optical breadboard, supported on pillars on top of the same optical table as the microscope system. We found that it was necessary to adjust the relay lens for different fluorescence emission wavelengths due to longitudinal chromatic aberration in the system. The microlens array was mounted in a 5-axis positioning stage to facilitate tip-tilt alignment with the optical axis and azimuthal alignment of the microlenses with the camera chip. The latter alignment step, while not essential, simplified the reconstruction of light field images. An adjustable steering mirror allowed the emitted light to be directed to a second camera for capture of comparison widefield images.
Fig. 1.
Schematic digram of light field microscope and micromanipulation system. Acronyms: MRL – macro relay lens; MLA – microlens array; TL – tube lens; FFC – fluorescence filter cube; MO – microscope objective. Inset I1: photograph showing microscope objective lens, sample and microforce sensing probe. Inset I2: typical force (red) and displacement (black) data versus time for a indentation-retraction experiment.
The effective spatial resolution of a light field microscope is determined by the choice of objective lens and microlens array [17]. We choose a 60x/1.0 water immersion objective lens (LUMPLFLN 60XW, Olympus) and an f/30 microlens array (matching the image-side f-number of the objective lens) comprised of square microlenses with a pitch of 125 μm. The irradiance at the native image plane of the microscope is given by integrating the radiance passing through the objective lens over all ray angles [21]. In practice, this means a conventional image of the object can be computed by performing a summation of the pixels under each lenslet subimage. Shearing the 4D light field parallel to the prior to performing this summation allows digital refocusing or reconstruction of images at different depths [21]. However, the effective lateral resolution in these images is limited by the relatively coarse sampling of the intermediate image by the microlenses. In our case, spatial frequencies greater than 0.24 μm−1, which is well below the band limit of the microscope objective lens, are undersampled. In order to further improve spatial resolution we use the 3D deconvolution image reconstruction approach described in [19]. This method makes use of the fact that the angular sampling of the light field by the lenslets means that effective lateral spatial sampling increases for out of focus object planes. Based on the resolution criterion proposed in [19] we anticipate a lateral resolution of approximately 1 μm for objects offset by 4 μm from the native object plane (front focal plane of the objective lens).
Mechanical stimulation of C. elegans specimens was performed using a microforce sensing probe (FT-S100, FemtoTools) consisting of a 3 mm long tapered tungsten filament with a tip, approximately 4 μm in diameter, coupled to a capacitive force sensor. This device offers a nominal force resolution of 5 nN at 10 Hz over a range of ± 100 µN. The probe was mounted on a stack of high precision piezoelectric stages (ECS industrial line, Attocube), which allowed closed loop positioning of the tip with 4 degrees of freedom (xyz and α), with a nominal repeatability of 50 nm / 50 µ°, a positioning resolution of 1 nm / 1 µ° and travel of up to 30 mm / 10°. Samples were positioned in the field of view of the microscope using manual kinematic stages. In order to avoid contact with other objects on the slide during approach to the specimen, and minimize the potential for accidental damage of the force transducer through collision with the substrate, the microscope slide was tilted by approximately 5°-10° with respect to the focal plane ().
2.2 Calibration and image reconstruction
Each raw light field image was processed to reconstruct a focal series of images as described in [20]. Prior to reconstruction, each recorded light field was corrected to remove pincushion/barrel distortion introduced by the macro lens using a radial warping function of the form , where r and are the original and corrected pixel coordinates respectively and k is a distortion parameter. In practice, k was empirically determined by comparing the magnitude of the 2D Fourier transform (FT) of light field images of a uniform fluorescent layer before and after warping. In the absence of distortion, the FT should comprise a uniform array of bright spots with a spacing corresponding to the separatrion of the subimages formed by the microlens array (Fig. 2(a), right panel). Barrel and pincushion distortion result in a smearing out of these points (Fig. 2(a), left panel). The mapping of the pupil subimages onto the camera sensor in the corrected image of the same uniform sample was then found experimentally (Fig. 2(b)). Prior to deconvolution, each corrected light field image was re-sampled to contain 15x15 pixels within each lenslet sub-image. Finally, a z-stack was reconstructed from the corrected, resampled light field via eight iterations of a Richard-Lucy deconvolution algorithm with an initial PSF kernel computed using the Fresnel diffraction integral evaluated using the nominal optical properties of the system.
Fig. 2.

(a) Magnitude of Fast Fourier Transform of a raw light field image of a uniform fluorescent layer captured before (left) and after (right) correction for pincushion / barrel distortion. (b) Corrected raw light field for a uniform fluorescent layer; yellow circles show experimentally determined size and pitch of pupil subimages.
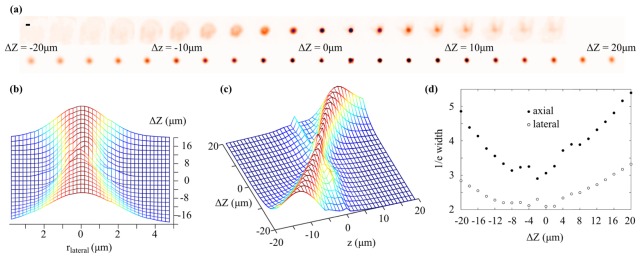
We tested the microscope by imaging a 2.5 μm diameter yellow-green fluorescent microsphere (Fluosphere, Invitrogen) mounted on a glass microscope slide. Although larger than the expected spatial resolution of the microscope, this microsphere is similar in size to the cell body of an individual TRN and, as such, is ideal for testing the performance of the system for our application. Figure 3(a) shows conventional widefield (top row) images of the microsphere captured as the objective lens is displaced axially () up to 20 μm either side of the native object plane. The lower row of images show the microsphere as it appears in the in-focus plane of a focal series reconstruction, illustrating that the additional angular information captured in each raw light field image enables effective imaging of objects away from the native object plane. As well as an increase in the apparent size of the bead as it is displaced from the native object plane there is also a decrease in its brightness, which has important implications for analysing calcium signals during axial displacement of neurons (see section 2.3). Figure 3(b) and Fig. 3(c) show lateral and axial line profiles drawn through the centre of the bead in the reconstructed z-stack as the objective lens is displaced. As expected, a decrease in lateral and axial spatial resolution away from the native object plane results in a broadening of the line profiles. Image reconstruction artefacts result in irregular features in the line profiles when the bead is close to the native object plane (). The axial displacement of the bead between reconstructed z-stacks was somewhat different to that expected from the 2 μm stepping of the objective lens, implying axial compression in the reconstructed images. We attribute this to differences between the nominal and actual properties of the optical components in the system and imaging aberrations. Although these effects are not critical for the present application, we note that they could be corrected through improved characterization and design of the optical system and also through digital correction of aberrations prior to reconstruction [22]. Figure 3(d) shows the 1/e width of these line profiles. Reconstruction artefacts result in a non monotonic variation in the 1/e width close to the native object plane, however more than ~6 μm from the native object plane the 1/e width of both lateral and axial profiles increases approximately linearly, suggesting a corresponding decrease in spatial resolution.
Fig. 3.
(a) Images of a 2.5 μm diameter green fluorescent microsphere acquired as the objective lens is displaced axially. Top row: conventional widefield images. Bottom row: in-focus plane extracted from reconstructed light field focal series. Scale bar in upper left corner is 3 µm. (b) Lateral and (c) axial line profiles taken through the centre of the bead in the focal series reconstruction as the objective lens is axially displaced. (d) Lateral (unfilled circles) and axial (filled circles) 1/e width of the bead image in the focal series reconstruction as the objective lens is displaced axially.
2.3 C. elegans preparation and experimental protocol
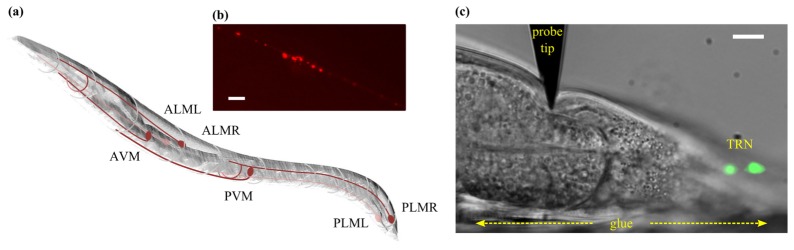
The C. elegans organism contains six TRNs tightly coupled to the worms’s cuticle; each neuron being composed of a cell body and a long neurite which innervates approximately half the length of the animal [13] (Fig. 4(a)). Mechano-electrical transduction in C. elegans is mediated by degenerin/epithelial sodium channels containing the proteins MEC-4 and MEC-10 [23]. Fluorescence microscopy images reveal that these channel proteins form discrete puncta along the length of the TRN neurites (Fig. 4(b)). Once these channels are activated the membrane depolarizes, which in turn results in changes in intracellular calcium concentration that can be detected with a GECI [24].
Fig. 4.
(a) Schematic diagram of an adult stage C.elegans organism showing the locations of the six identified touch receptor neurons. (b) Epifluorescence image of tagRFP labelled MEC-4 protein puncta in a C. elegans TRN. Scale bar is 5 µm. (c) DIC image (grayscale) of a C.elegans organism combined with a maximum intensity projection of a epifluorescence z-stack showing the two posterior gentle touch neurons (green) during a typical indentation with the microforce sensing probe. Scale bar is 12 µm.
We performed a series of mechanosensory stimulation experiments on C. elegans organisms modified to express the calcium indicator GCaMP6s in all six TRNs using the plasmid mec-4::nls-RSET-GCaMP6s:SL2:nls-TagRFP::unc-54utr. The GCaMP6s construct contains a nuclear localizing signal, meaning that the concentration of GCaMP should be highest in the nucleus of each TRN, although in practice we also observed significant fluorescence signal throughout the cell body. Prior to imaging, an agarose pad was placed on top of a microscope slide. Worms were then fixed in position on the pad using Dermabond glue, applied along one side of the body, before being immersed in an imaging solution prepared from 145 mM NaCl, 5 mM KCl, 1 mM CaCl2, 20 mM D-glucose and 10 mM Hepes buffer. All worms were picked at L4 stage and imaged as young adults. We found that GCaMP6s was most clearly visible in the posterior TRNs, PLML and PLMR (see Fig. 4(a)), and for this reason we focused exclusively on these in this study.
In each experiment, the posterior TRNs were first located using differential interference contrast (DIC) and epifluorescence images of the worm. The tip of the force probe was then brought close to the outer cuticle of the animal at the desired location. For a given sample, the positions of the two posterior TRNs depended on the orientation of the animal on the microscope slide; however, we found that they were generally located at different depths within the animal, typically separated by approximately 10 μm along the optical axis. In order to avoid the reconstruction artefacts and degraded spatial resolution close to the native object plane, we shifted the objective lens such that both TRNs were situated on the same side of the native object plane, with the closest neuron defocused by a distance of approximately 4 μm. As well as allowing for the reconstruction of good quality images over a range large enough to encompass both TRNs, we found that this defocus offset was sufficient to accommodate small axial displacements during indentation of the organism.
Epifluorescence light field images were recorded at 10 frames per second during a programmed mechanical stimulation sequence comprising indentation of the animal with the force probe, hold and retraction of the force probe. During imaging, the position of the force probe, read from optical encoders on the probe positioning stages, and the force measured along the probe axis were recorded simultaneously in order to correlate the observed response of the TRN and the mechanical stimulus.
In the absence of any mechanical contact with the sample, there were significant variations in measured force as the probe tip moved through the buffer droplet. In addition to viscous drag forces exerted on the moving tip, we observed a steady state force offset which depended on the position of the tip within the buffer droplet. This was likely due to sensitivity of the force transducer to the transverse buoyancy force exerted on the probe tip, which itself depended on the submerged tip volume. In order to correct for this effect, a reference force trace was subtracted from each measured sample force trace. Such a reference force trace was captured by offsetting the position of the probe tip away from the worm before moving it through the same displacement sequence used to indent the organism. By making the probe position offset just large enough to avoid direct mechanical contact with the worm we were able to measure, and subsequently correct for, underlying variations in measured force due to probe movement through the buffer droplet under conditions similar to those encountered during specimen indentation.
Calcium transients were extracted from the reconstructed z-stacks in the following way. First, an axial projection was computed for each TRN by summing the slices containing the neuron of interest. After normalisation, the projection images were spatially registered to subpixel accuracy using a rigid body template matching algorithm [25]. The raw calcium signal was then taken as the mean pixel value within a region of interest containing the neuron. In order to correct for photobleaching of the GFP, the sequence was detrended using a decaying exponential function fit to the part of the image sequence captured prior to indentation. Finally, intensity offsets in the signal trace due to axial displacement of the TRN during indentation and stimulus withdrawal were corrected based on the mean signal computed over sections of the trace free from calcium transients.
3. Results and discussion
We performed a series of mechanical stimulation and light field calcium imaging experiments on various C. elegans specimens. In this section we present two examples of the results, demonstrating our ability to simultaneously measure the activity of multiple neurons at different depths and investigate limits for touch sensitivity in the organism.
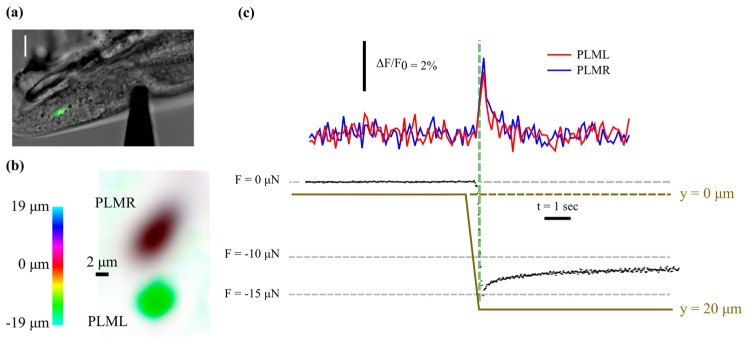
Figure 5(a) shows a specimen indented to a depth of 13 µm along its mid plane at a distance of approximately 38 µm from the centre of the PLMR cell body. Due to the shallow depth of field, only the PLMR is visible in the conventional epifluorescence image (green). Figure 5(b) shows a colour coded depth projection computed from the focal series reconstructed from the light field image, clearly illustrating both TRNs; the centres of which are axially separated by a distance of approximately 9 µm. The corrected calcium signal from the cell bodies of the two posterior TRNs in response to the ramp and hold indentation is shown in the top part of Fig. 5(c), indicating simultaneous activation of both neurons. Analysis of the reconstructed z-stack indicates that the neurons were displaced axially by approximately 2 µm by the indentation, which is significantly larger than the nominal depth of field of the objective lens. The gradual reduction in the magnitude of the compressive force measured by the probe suggests stress relaxation due to viscous energy dissipation and an exponential fit to the force trace gives a steady state force value of −11.7 µN. From brightfield optical images the probe tip has an approximately planar face and a circular cross section with a diameter of 4.5 µm. Based on this simplified geometry, the steady state mechanical contact pressure (force divided by probe contact area) applied to the organism is approximately 0.7 MPa.
Fig. 5.
(a) DIC image showing indentation of C.elegans specimen using microforce sensing probe with conventional epifluorescence image of PLMR overlaid in green. Due to the shallow depth of field the PLML is not visible. Scale bar is 10 µm. (b) Colour coded depth projection created from the reconstructed focal series showing the cell bodies of the two posterior TRNs. (c) Measured force (black data points), probe position (tan solid line) and average brightness of the two TRNs (top) versus time, indicating that both neurons are activated by the stimulus.
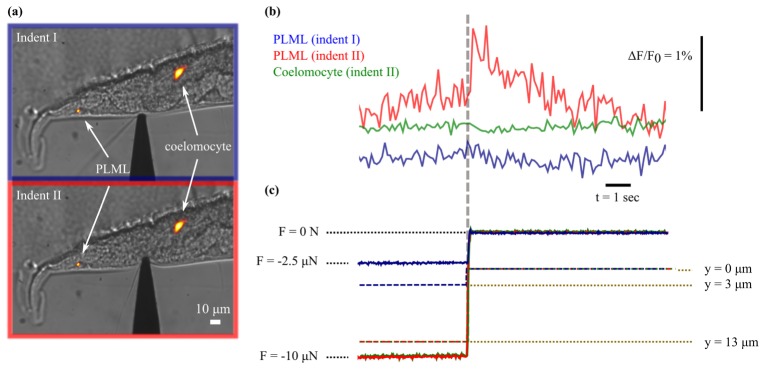
In order to demonstrate how our approach can be applied to investigate touch sensitivity in C. elegans, we indented an organism to different depths in an attempt to establish a contact pressure threshold for TRN activation. Figure 6(a) shows DIC images of the animal with the cuticle indented to depths of 3 μm (indentation I) and 13 μm (indentation II) along the mid plane using the microforce sensing probe. Using the same approximation to the tip geometry as previously, the steady state force measurements of −2.4 μN and −10.2 μN imply an approximate applied contact pressure of 0.2 MPa and 0.6 MPa at the two indentation depths. Due to significant movement of the organism on the slide, we first pinned it against the set glue using the microforce probe and then measured the calcium signal in the PLML in response to retraction of the probe. Previous studies [26] have found that the mechanoreceptor currents generated in C. elegans TRNs are similar in response to both application and removal of a mechanical stimulus. Similarly, experiments using the GECI cameleon [24] found that the TRNs responded to an increase or decrease in the pressure applied to the organism, but not to a constant pressure. Taken together these results suggest that the TRNs respond primarily to the change in magnitude of the mechanical stimulus. Figure 6(b) (top) shows the measured GCaMP response, indicating a small, but significant, increase in brightness following withdrawl of the probe after the 13 μm indentation. In contrast, there is no apparent brightness increase following withdrawl of the probe after the 3 μm indentation, suggesting that the threshold for touch sensation lies between these two limits. A previous study [27], in which a cantilever with a 10 µm diameter spherical tip was used to indent C.elegans specimens from above, reported a behavioural (avoidance) response at an indentation depth of 0.44 µm and a force of 0.49 µN. Direct comparison with our result is difficult due to the significant difference in the size and shape of the indenters, although this result indicates that the organism is somewhat more sensitive than our result would suggest. However, as the posterior TRN neurites run along the ‘left’ and ‘right’ sides of the worm (corresponding to the top and bottom of the organism as it is mounted in our experiments), this difference could be explained by a higher sensitivity to stimulation applied closer to the mechanosensory channels.
Fig. 6.
(a) DIC images showing indentation of a C.elegans specimen to depth of approximately 3 μm (top) and 13 μm (bottom) with a microforce sensing probe approximately 110 μm from the tip of the tail. Maximum intensity projection of reconstructed light field (displayed with a red hot colour map) overlaid to show positions of PLML and coelomocyte. (b) Total brightness of the cell body of the PLML indicates that the TRN is only activated by the withdrawl of the probe following the larger indentation (red line). The signal from a nearby GFP transgenic marker (green line) is constant despite similar displacement following withdrawal of the probe. (c) Corresponding force (solid lines) and probe position (dashed lines) measured during the end of the hold phase of indentation and after probe retraction.
To verify the effectiveness of our detrending and intensity offset correction routine, and provide additional confidence that the observed brightness changes are due to neuronal activation and not an artefact caused by motion of the PLML, we also analysed images of a stable fluorescent transgenic marker (a posterior coelomocyte expressing GFP) located close to the TRNs. The green intensity trace shown in Fig. 6(b) shows the integrated fluorescent intensity in the coelomocyte, during the same probe retraction which resulted in the apparent activation of the PLML, and was computed using the same signal analysis procedure as used to analyse calcium transients within the TRNs. As expected, despite undergoing similar displacement to the PLML, there is no apparent increase in signal from the coelomocyte following withdrawal of the probe. The increased signal-to-noise ratio in the coelomocyte intensity trace reflects the fact that this feature is significantly brighter than the TRNs. We also note that decaying exponentials fitted to the calcium transients in Fig. 5(c) and Fig. 6(b) have half-lives of 0.5 – 1 second, which are consistent with those reported elsewhere for GCaMP6s [12].
The observed variation in brightness of the GCaMP6s reporter in our results is relatively modest (1%-3%), compared to that reported in some other calcium ion imaging experiments [12]. This likely reflects the properties of the reporter and the magnitude of calcium ion concentration changes in the TRNs, rather than any fundamental limitation of the imaging method; although a decrease in the image signal-to-noise ratio owing to the use of 3D deconvolution is expected. We also note that previous work [24] on mechanosensation in C. elegans using the Förster resonance energy transfer-based GECI cameleon, found that the response of the reporter to the type of step indentation-retraction we applied in our study was relatively weak and much lower than the response to a rapid buzz stimulus.
4. Summary and conclusions
By combining light field microscopy and micromanipulation for the first time, we have demonstrated a novel approach for investigating touch sensation in C. elegans. The fact that a single light field exposure can be used to reconstruct a 3D image with micrometre level spatial resolution is particularly important for this application as it allows simultaneous monitoring of calcium ion signals from multiple neurons at different depths and also accommodation of axial movement of individual neurons following mechanical manipulation of the specimen.
As expected, given the spatially varying point spread function (PSF) of a light field microscope, we find that the reconstructed image depends on the axial position of the object with respect to the native object plane. However, intracellular calcium ion signals derived from the brightness of the fluorescent reporter expressed within the neuron can be effectively corrected to remove these effects. Alternatively such effects could, in principle, be compensated by dividing the signal from the GECI by the corresponding signal from a second fluorophore coexpressed in the TRNs. Our attempts to implement this approach were hampered by substantial axial chromatic aberration which made it difficult to focus the camera on the back focal plane of the lenslet array for both GFP (GCaMP) and RFP (control) emission wavelengths. Such a focus offset significantly degraded the reconstructed light field images. Our experimental setup could be modified for ratiometric calcium ion imaging using a two colour image splitter, with adjustable optical paths, mounted in front of the camera. Alternatively the field of view of the system could be preserved using a dichroic mirror to send emission from one of the fluorophores to a second camera synchronized to the first. Further improvements to the system hardware could also include the use of phase mask in front of the camera. Combined with a reliable estimate of the modified PSF, this has been shown to be effective in improving deconvolved light field images close to the native object plane [28].
Building on our proof of principle experiments by performing a wider range of spatio-temporal manipulations will allow further investigation into mechanosensation. In particular, further measurements to establish touch sensitivity in C. elegans combined with a biomechanical model of the organism [27, 29] could provide significant insights into the conversion of mechanical to electrochemical energy by mechanosensory ion channels.
Although the 3D deconvolution method employed here is relatively time consuming, with typical image reconstructions taking several minutes per time point, light field rendering methods can be performed in real time [30] to yield perspective or multifocal views of the specimen at moderate spatial resolution, providing a greater sense of depth perception when performing contact-based interactions at the micrometre scale. Combined with suitable labelling, or the use of label free contrast mechanisms, light field microscopy has significant potential to support a range of biological and biomedical micromanipulation applications including single-cell manipulation and intracellular injection.
Acknowledgments
This work was funded by an Advanced Grant from the European Research Council (ERC-2009-AdG, MicroNanoTeleHaptics) and a Capital for Great Technologies grant (EP/K005030/1) from the UK’s Engineering and Physical Sciences Research Council. We would like to thank Douglas S. Kim, HHMI, Janelia Farm Research Campus, for providing the GCaMP6s construct for C. elegans transgenic production and Martin Chalfie, Columbia University, for providing the mec-4::tagrfp worms.
References and links
- 1.Chalfie M., “Neurosensory mechanotransduction,” Nat. Rev. Mol. Cell Biol. 10(1), 44–52 (2009). 10.1038/nrm2595 [DOI] [PubMed] [Google Scholar]
- 2.Lumpkin E. A., Marshall K. L., Nelson A. M., “The cell biology of touch,” J. Cell Biol. 191(2), 237–248 (2010). 10.1083/jcb.201006074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darian-Smith I., “The Sense of Touch: Performance and Peripheral Neural Processes,” in Comprehensive Physiology (John Wiley & Sons, Inc., 2011). [Google Scholar]
- 4.Lesniak D. R., Marshall K. L., Wellnitz S. A., Jenkins B. A., Baba Y., Rasband M. N., Gerling G. J., Lumpkin E. A., “Computation identifies structural features that govern neuronal firing properties in slowly adapting touch receptors,” eLife 3, e01488 (2014). 10.7554/eLife.01488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas A., Manivannan M., Srinivasan M. A., “Multiscale Layered Biomechanical Model of the Pacinian Corpuscle,” IEEE Trans. Haptics 8(1), 31–42 (2015). 10.1109/TOH.2014.2369416 [DOI] [PubMed] [Google Scholar]
- 6.Biswas A., Manivannan M., Srinivasan M. A., “Vibrotactile Sensitivity Threshold: Nonlinear Stochastic Mechanotransduction Model of the Pacinian Corpuscle,” IEEE Trans. Haptics 8(1), 102–113 (2015). 10.1109/TOH.2014.2369422 [DOI] [PubMed] [Google Scholar]
- 7.Kumar S., Liu G., Schloerb D. W., Srinivasan M. A., “Viscoelastic Characterization of the Primate Finger Pad In Vivo by Microstep Indentation and Three-Dimensional Finite Element Models for Tactile Sensation Studies,” J. Biomech. Eng. 137(6), 061002 (2015). 10.1115/1.4029985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalfie M., Sulston J., “Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans,” Dev. Biol. 82(2), 358–370 (1981). 10.1016/0012-1606(81)90459-0 [DOI] [PubMed] [Google Scholar]
- 9.Driscoll M., Chalfie M., “The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration,” Nature 349(6310), 588–593 (1991). 10.1038/349588a0 [DOI] [PubMed] [Google Scholar]
- 10.Chelur D. S., Ernstrom G. G., Goodman M. B., Yao C. A., Chen L., O’ Hagan R., Chalfie M., “The mechanosensory protein MEC-6 is a subunit of the C. elegans touch-cell degenerin channel,” Nature 420(6916), 669–673 (2002). 10.1038/nature01205 [DOI] [PubMed] [Google Scholar]
- 11.Huang M., Gu G., Ferguson E. L., Chalfie M., “A stomatin-like protein necessary for mechanosensation in C. elegans,” Nature 378(6554), 292–295 (1995). 10.1038/378292a0 [DOI] [PubMed] [Google Scholar]
- 12.Chen T.-W., Wardill T. J., Sun Y., Pulver S. R., Renninger S. L., Baohan A., Schreiter E. R., Kerr R. A., Orger M. B., Jayaraman V., Looger L. L., Svoboda K., Kim D. S., “Ultrasensitive fluorescent proteins for imaging neuronal activity,” Nature 499(7458), 295–300 (2013). 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman M. B., Lindsay T. H., Lockery S. R., Richmond J. E., “Chapter 14 - Electrophysiological Methods for Caenorhabditis elegans Neurobiology,” in Methods in Cell Biology, Joel H. R., Andrew S., eds. (Academic Press, 2012), pp. 409–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahrens M. B., Orger M. B., Robson D. N., Li J. M., Keller P. J., “Whole-brain functional imaging at cellular resolution using light-sheet microscopy,” Nat. Methods 10(5), 413–420 (2013). 10.1038/nmeth.2434 [DOI] [PubMed] [Google Scholar]
- 15.Abrahamsson S., Chen J., Hajj B., Stallinga S., Katsov A. Y., Wisniewski J., Mizuguchi G., Soule P., Mueller F., Dugast Darzacq C., Darzacq X., Wu C., Bargmann C. I., Agard D. A., Dahan M., Gustafsson M. G. L., “Fast multicolor 3D imaging using aberration-corrected multifocus microscopy,” Nat. Methods 10(1), 60–63 (2012). 10.1038/nmeth.2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grewe B. F., Langer D., Kasper H., Kampa B. M., Helmchen F., “High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision,” Nat. Methods 7(5), 399–405 (2010). 10.1038/nmeth.1453 [DOI] [PubMed] [Google Scholar]
- 17.Levoy M., Ng R., Adams A., Footer M., Horowitz M., “Light field microscopy,” ACM Trans. Graph. 25(3), 924–934 (2006). 10.1145/1141911.1141976 [DOI] [Google Scholar]
- 18.Levoy M., Zhang Z., McDowall I., “Recording and controlling the 4D light field in a microscope using microlens arrays,” J. Microsc. 235(2), 144–162 (2009). 10.1111/j.1365-2818.2009.03195.x [DOI] [PubMed] [Google Scholar]
- 19.Broxton M., Grosenick L., Yang S., Cohen N., Andalman A., Deisseroth K., Levoy M., “Wave optics theory and 3-D deconvolution for the light field microscope,” Opt. Express 21(21), 25418–25439 (2013). 10.1364/OE.21.025418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prevedel R., Yoon Y.-G., Hoffmann M., Pak N., Wetzstein G., Kato S., Schrödel T., Raskar R., Zimmer M., Boyden E. S., Vaziri A., “Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy,” Nat. Methods 11(7), 727–730 (2014). 10.1038/nmeth.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng R., “Fourier slice photography,” ACM Trans. Graph. 24(3), 735–744 (2005). 10.1145/1073204.1073256 [DOI] [Google Scholar]
- 22.Hanrahan P., Ng R., “Digital Correction of Lens Aberrations in Light Field Photography,” in International Optical Design, Technical Digest (CD) (Optical Society of America, 2006), WB2. [Google Scholar]
- 23.Chen Y., Bharill S., Isacoff E. Y., Chalfie M., “Subunit composition of a DEG/ENaC mechanosensory channel of Caenorhabditis elegans,” Proc. Natl. Acad. Sci. U.S.A. 112(37), 11690–11695 (2015). 10.1073/pnas.1515968112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki H., Kerr R., Bianchi L., Frøkjaer-Jensen C., Slone D., Xue J., Gerstbrein B., Driscoll M., Schafer W. R., “In Vivo Imaging of C. elegans Mechanosensory Neurons Demonstrates a Specific Role for the MEC-4 Channel in the Process of Gentle Touch Sensation,” Neuron 39(6), 1005–1017 (2003). 10.1016/j.neuron.2003.08.015 [DOI] [PubMed] [Google Scholar]
- 25.Schneider C. A., Rasband W. S., Eliceiri K. W., “NIH Image to ImageJ: 25 years of image analysis,” Nat. Methods 9(7), 671–675 (2012). 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Hagan R., Chalfie M., Goodman M. B., “The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals,” Nat. Neurosci. 8(1), 43–50 (2005). 10.1038/nn1362 [DOI] [PubMed] [Google Scholar]
- 27.Petzold B. C., Park S.-J., Mazzochette E. A., Goodman M. B., Pruitt B. L., “MEMS-based force-clamp analysis of the role of body stiffness in C. elegans touch sensation,” Integr Biol (Camb) 5(6), 853–864 (2013). 10.1039/c3ib20293c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen N., Yang S., Andalman A., Broxton M., Grosenick L., Deisseroth K., Horowitz M., Levoy M., “Enhancing the performance of the light field microscope using wavefront coding,” Opt. Express 22(20), 24817–24839 (2014). 10.1364/OE.22.024817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S.-J., Goodman M. B., Pruitt B. L., “Analysis of nematode mechanics by piezoresistive displacement clamp,” Proc. Natl. Acad. Sci. U.S.A. 104(44), 17376–17381 (2007). 10.1073/pnas.0702138104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levoy M., Hanrahan P., “Light field rendering,” in Proceedings of the 23rd annual conference on Computer graphics and interactive techniques , (ACM, 1996), pp. 31–42. [Google Scholar]