Abstract
Tumors develop in a complex microenvironment alongside numerous cell types that impact their survival. Immune cells make up a large proportion of these accessory cells and many are known to promote tumor progression. Macrophages, in particular, are associated with poor patient prognosis and are therefore potential candidates for therapeutic targeting in cancer. However, to develop successful strategies to target macrophages, it is important to clarify whether these cells are derived from blood-borne precursors or a tissue-resident population. Parabiosis, or the surgical connection of two mice resulting in a shared blood circulation, allows the distinction between these two cellular sources. Here, we describe the use of parabiosis to define cell ontogeny in a mouse model of breast cancer.
Materials and Reagents
Mice: Female age- and weight-matched congenically-marked MMTV-PyMT mice [see Franklin et al. (2014) for specific strain information]
Injectable anesthesia (ketamine/xylazine cocktail: 150 mg/kg/15 mg/kg)
Antiseptic solution (Betadine)
70% ethanol
Ophthalmic ointment (Paralube)
Analgesic (Buprenorphine, 0.1 mg/kg)
Non-wetting sterile water (HydroGel)
Sterile saline solution (0.9% NaCl) 9. 1x PBS
10x PBS
Bovine serum albumin (Sigma-Aldrich, catalog number: A2153)
Sodium azide (Sigma-Aldrich, catalog number: S2002)
Collagenase Type III (Worthington Biochemicals, catalog number: LS004182)
Percoll (Sigma-Aldrich, catalog number: P1644)
HBSS + calcium chloride + magnesium chloride (Life Technologies, Gibco®, catalog number: 14025)
Deoxyribonuclease I from bovine pancreas (Sigma-Aldrich, catalog number: DN25)
CD45.1 APC-eFluor® 780 (eBioscience, catalog number: 47-0453-82)
CD45.2 PerCP-Cyanine5.5 (eBioscience, catalog number: 45-0454-82)
MHC Class II (I-A/I-E) FITC (eBioscience, catalog number: 11-5321-82)
CD11b Pacific Blue™ (Life Technologies, Invitrogen™, catalog number: RM2828)
Ly-6C APC (BD Pharmingen, catalog number: 560595)
Ly-6G PE (BD Pharmingen, catalog number: 551461)
Fc block (2.4G2), (BD Pharmingen, catalog number: 553142)
RPMI 1640 (Life Technologies, Gibco®, catalog number: 11875-093)
Digestion buffer (see Recipes)
100% Percoll (see Recipes)
44% Percoll (see Recipes)
66% Percoll (see Recipes)
FACS buffer (see Recipes)
Equipment
Non-absorbable black nylon suture (4-0, 13mm, 3/8 circle) (Ethicon, catalog number: 1854G)
Wound clip applier, wound clip remover, wound clips (Autoclip, 9 mm) (Braintree Scientific, catalog number: ACS KIT)
Cotton-tipped applicators (3 inch, wood shaft) (Fisherbrand, catalog number: 23-400-105)
Sterile gauze (Fisherbrand, catalog number: 22-028-558)
Needle holder with locking mechanism (Fine Science Tools)
Scissors (Fine Science Tools)
Curved forceps (2 pairs) (Fine Science Tools)
Razor blades
Slides (Corning Incorporated, catalog number: 2948-75X25)
Heating lamp
Heating pads
Electrical shaver
Surgical drapes/sterile pads (Fisherbrand)
6 well non-tissue culture treated plates (Falcon®)
70 micron cell strainers (Falcon®, catalog number: 352350)
50 ml conical tubes (Falcon®)
15 ml conical tubes (Falcon®)
3 ml syringe (BD Bioscience)
U-bottom 96 well plates (Falcon®)
Desktop centrifuge (Sorvall)
37 °C water bath
Flow cytometer (BD LSR II or similar)
Procedure
A. Parabiosis
This surgical procedure will result in the formation of microvasculature at the connection site and thus a shared blood circulation between two animals during the development of oncogene-induced mammary tumors.
Sterilize all surgical tools by autoclaving or soaking in 70% ethanol for at least 30 min.
Lay out all sterile tools and necessary materials on clean surgical drapes/sterile pads.
Warm a clean cage with heating lamp for use during recovery.
Anesthetize mice using ketamine/xylazine cocktail (IP). The animals should be fully sedated (as measured by no response to a toe pinch) before proceeding. Throughout anesthesia, apply ophthalmic ointment to eyes as needed.
After mice are anesthetized, shave the right flank of one partner and the left flank of the other partner. All mice should be kept on heating pads during surgery.
Start procedure on first partner. Lay the mouse on its side on sterile gauze and sterilize the shaved area to prepare for incision, alternating antiseptic solution and 70% ethanol three times, using a fresh cotton-tipped applicator for each application. Make a small incision in the skin over the thigh, about 0.5 cm dorsally from the knee joint. Insert the tip of the scissors into the incision and open and close the nose of the scissors to pull the skin away from the inner fascia covering the leg muscles. This will allow you to expose the muscles and patellar tendon of the hindlimb for suturing in later steps.
Cut in a straight line from this incision up to the shoulder just past the elbow joint. Repeat opening and closing of the scissors the entire length of the incision, being careful to leave the body wall intact.
Holding the ankle joint of the hindlimb between your thumb and forefinger, carefully maneuver the knee out from the skin so that the patella is exposed and turned upwards, facing you. Insert the needle of the non-absorbable suture between the tibia and fibula from the inside of the leg to the outside, about 1/3 of the way below the knee. Avoid piercing major blood vessels. Pull the suture through until 2–3 inches of the free end remain (this free end will be used to tie to the other partner, so make sure there is enough length for you to comfortably perform two throws of an instrument tie square knot). Tie off the suture with the needle holder, using an instrument tie square knot (two throws of the knot is sufficient). There will be two ends of the suture radiating from the knot, the 2–3 inch free end and the end connected to the needle. Cut the needle end as close to the knot as possible, leaving the free end available to tie in subsequent steps. Try to secure the knots on the outside of the thigh as this will make connecting the two partners easier. Detailed instructions on how to perform surgical ties can be found at http://www.bumc.bu.edu/surgery/training/technical-training/instrument-tie/.
Repeat this procedure on the forelimb, using the remaining length of the suture (needle end) from step A8. Insert the suture needle between the radius and ulna, right below the elbow, from inside to outside.
Keeping the 1st partner on the heating pad, laying on its side with the incision facing upwards, make a matching incision on the 2nd partner. Repeat the same procedure as the 1st partner. You will now have 4 loose suture ends with all of the knots facing upwards.
To connect the two mice, lay them both on their backs, with their abdomens facing upwards, and their incisions facing each other. Insert the suture needle under the patellar tendon of the 1st partner from the inside of the knee to the outside and then under the patellar tendon of the 2nd partner from the outside to the inside (one continuous motion). Tie off suture (two throws) and cut both loose ends as close to the knot as possible to minimize irritation from suture ends. Be careful not to accidentally tie the suture ends from step A8 into the knot.
To connect the hindlimb sutures, flip mice over so they are laying on their stomachs. Use the needle holder to perform 2 throws of an instrument tie square knot on the suture ends from step A8 and cut loose ends. Keeping the mice on their stomachs, use the needle holder to tie together the forelimb sutures from step A9 (2 throws of an instrument tie square knot) and cut loose ends.
After these sutures are tied together, the mice will be in their final connected position so take care to line up forelimbs and hindlimbs in a way that will make ambulation as easy as possible. The two partners should now be tightly and stably held together with their body walls in contact with each other.
Close the skin incisions using 9 mm wound clips. This procedure is best performed by two people, one person to hold the skin together, and one to apply the wound clips. Using two curved end forceps, hold the skin flaps from the dorsal sides of both mice up and away from their body walls. At this step it is very important to roll the outer edges of the skin flaps away each other as to avoid clipping any remaining hair into the wound. This also prevents the skin from curling into the wound. Apply the first wound clip where the inside of the skin flaps touch each other, approximately 0.25 cm from the edge of the incision, starting from the tail end and working towards the head, placing each wound clip right next to the previous clip. By pulling the skin up while clipping, you prevent clipping into the body wall or connecting the mice too tightly together. Once clipping on the dorsal side of the wound is complete, flip the mice onto their backs and apply clips to the ventral side of the wound, holding the skin together during clipping as previously described. Extra clips may need to be applied near the forelimbs and hindlimbs to fully close the wound.
After clipping is complete, lay the mice on their stomachs, on sterile gauze, in the warmed cage. The heating lamp must be used until the mice are fully awake and ambulating.
Inject each partner with 500 μl sterile saline (sub-Q) immediately following surgery and 2x daily for 4–7 days post-surgery.
Inject each partner with analgesic (IP) immediately following surgery and 1x daily for 1–2 days post-surgery.
Provide non-wetting sterile water in the bottom of the cage for 1 week following surgery and provide moistened food for the duration of the experiment.
If maintaining the mice for more than 1 month, remove the wound clips at 3–4 weeks using a wound clip remover.
B. Tumor isolation and processing
After mammary tumors become palpable, euthanize the mice and remove the tumors from the unconnected side of each partner. Do not isolate mammary glands/tumors from the connected side. Special care must be taken to avoid disrupting the lymph nodes in order to prevent contamination of the tumor prep with lymph node leukocytes. This is especially important for the mammary gland that is found next to the inguinal lymph node. It is often best to avoid removing this tumor entirely. In addition, some mammary glands will not have palpable tumors, yet the pair of mice will have to be euthanized due to the size of one or more tumors. In this case, all tumors and mammary glands may be pooled (only pool tumors within one individual mouse), or analyzed separately.
Remove tumors and place in 1x PBS in a 6 well plate on ice.
Process tumors by cutting into small pieces with a razor blade on top of a glass slide and incubate tumors in 10–15 ml digestion buffer (see Recipes), for 1¼ h in 37 °C water bath. Vortex tubes every 15–20 min.
After digestion, pour tumors and digestion buffer over a 70 micron cell strainer into a 6 well plate. Mash tumors through cell strainer with the back of a plunger from a 3 ml syringe. Wash filter with 1x PBS and move cell strainer to the next well. Continue mashing, washing, and moving the cell strainer to the next well of the plate until the majority of tumor tissue has been dissociated.
Transfer digested and filtered tumor tissue to a 50 ml conical tube using a serological pipette, wash all wells with 1x PBS to collect any remaining cells, and spin down at 650 x g for 6 min in desktop centrifuge (total volume will be 40–50 ml).
Pour off supernatant and resuspend cell pellet in 10 ml 44% Percoll and transfer to a 15 ml conical tube.
Slowly underlayer with 3 ml 66% Percoll in the bottom of the tube using a 5 ml serological pipette, pulling out the pipette as the Percoll is deposited, taking care not to disturb the interface between the 44% and 66% Percoll.
Spin at 1,500 x g, 30 min, 4 °C, NO BRAKE/lowest brake setting.
Remove debris at the top of the tube, along with 3–5 ml 44% Percoll. Collect cells at interface “buffy coat” using a p1000 micropipette and transfer into a clean 15 ml conical tube. Be generous with the volume collected at this step (4–5 ml total), while avoiding the red blood cell pellet at the bottom of the tube. Spin down at 650 x g for 5 min.
Wash with 10 ml 1x PBS, spin down at 650 x g for 5 min.
C. FACS analysis of immune infiltrates
Resuspend the immune cell pellet in an adequate volume of 1x PBS to transfer into 96 well plate for FACS staining, approximately 100 μl/well, and place on ice.
-
Prepare antibody cocktail in FACS buffer, 50 μl/well. The antibodies listed below are for the identification of myeloid cells, including macrophages, monocytes, and neutrophils.
CD45.1 APC-eFluor780, 1:400
CD45.2 PerCP-Cyanine5.5, 1:300
MHC Class II (I-A/I-E) FITC, 1:1,200
CD11b Pacific Blue, 1:800
Ly-6C APC, 1:500
Ly-6G PE, 1:500
Fc block (2.4G2), 1:400
-
Spin down plate in desktop centrifuge at 500 x g for 3 min and discard supernatant by flicking the plate. Resuspend each cell pellet in 50 μl antibody cocktail. Incubate plate, covered, on ice for 30 min.
Wash 2x in 150 μl of 1x PBS and resuspend in 1x PBS for analysis using a flow cytometer.
Representative data
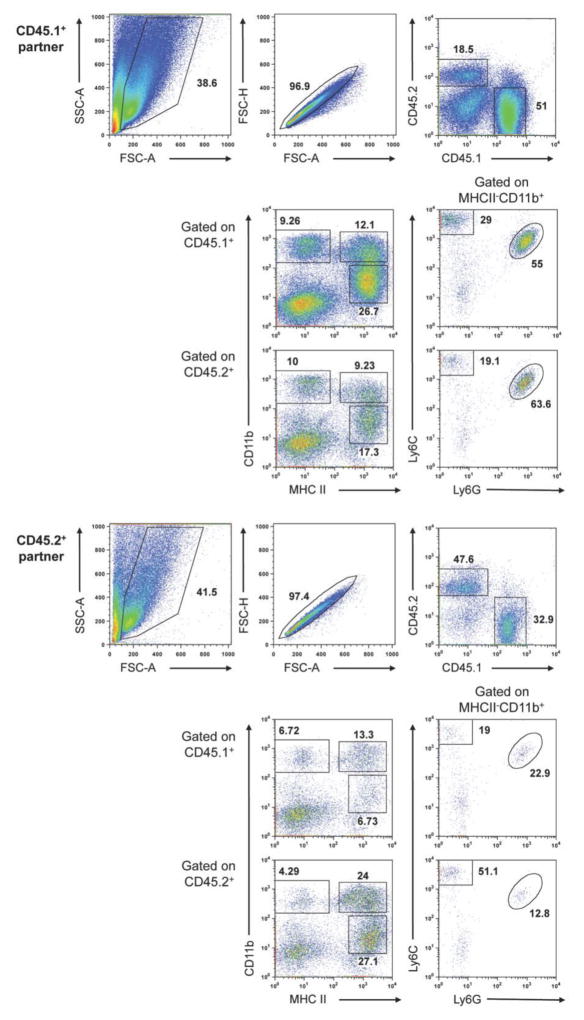
Figure 1. Blood-borne precursors contribute to macrophage populations in developing PyMT tumors.
Representative data expected from a parabiosis experiment. Two weight-matched, 8-week-old, congenically-marked female PyMT mice were surgically connected and maintained for approximately 3 months. The mammary tissue/tumors were removed from the unconnected side of each partner and pooled (tissue/tumors were pooled within each mouse, but not between mice). Tumors were processed as described above. The gated populations are defined as follows: CD45+MHCII+CD11bhi, mammary tissue macrophage (MTMs), CD45+MHCII+CD11blo, tumor-associated macrophages (TAMs), CD45+MHCII−CD11bhiLy6C+Ly6G+, neutrophils, and CD45+MHCII−CD11bhiLy6C+Ly6G−, monocytes.
Notes
This protocol was developed using the MMTV-PyMT tumor model, however it can likely be utilized to investigate other models. Regardless of the model used, all mice must be congenically-marked in order to distinguish cells originating from the individual partners. Additionally, the use of male mice should be avoided if possible due to the likelihood of fighting after surgical connection.
If the MMTV-PyMT model is used, the animals must be connected as early as possible (6–8 weeks) so that the surgery is performed before the tumors have become palpable. This is important from both a practical standpoint, as the surgery is performed near the mammary glands, and an experimental standpoint, as it would be difficult to interpret data from mice connected during later stages of tumor development.
The relative percentage of CD45.1/CD45.2 chimerism in the mammary tissue/tumors depends on both tumor stage of each mouse upon surgery (6–8 weeks is optimal) and upon sacrifice. Invariably, one partner will have more developed tumors than the other and this will be reflected in the degree of chimerism, number of infiltrating cells obtained, and mammary tissue macrophage (MTM): tumor-associated macrophage (TAM) ratio. For example, in the representative data shown above, the CD45.1+ partner had more advanced tumors and therefore had a lower degree of chimerism, contained more infiltrating cells, and had a lower MTM:TAM ratio (TAMs, rather than MTMs, are the dominant macrophage population in advanced PyMT tumors [see Franklin et al. (2014) for additional information].
The CD45.1−CD45.2− population observed after gating on single, live cells (see Representative data) represents tumor cells and is more prominent in advanced or necrotic tumors.
Recipes
-
Digestion buffer
1x HBSS
Collagenase III (~280 U/ml)
DNase I (4 μg/ml)
-
100% Percoll
45 ml Percoll
5 ml 10x PBS
-
44% Percoll
22 ml “100%” Percoll
28 ml 1x PBS
-
66% Percoll
33 ml “100%” Percoll
17 ml RPMI (to provide color, makes interface more clear)
-
FACS buffer
1x PBS
1% BSA
0.02% Sodium azide
Acknowledgments
We thank Kang Liu for her teaching of the parabiosis technique. This work was supported by the Cancer Research Institute Tumor Immunology Predoctoral Fellowship Training Grant (R.A.F), Cancer Research Institute Clinic and Laboratory Integration Program Grant (M.O.L.), and the American Cancer Society Research Scholar Award (M.O.L.).
References
- 1.Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, Pamer EG, Li MO. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]